Abstract
MicroRNAs (miRNAs) are small regulatory non-coding RNAs that have been reported to play an important role in the tumorigenesis of many cancers. In addition, miRNAs might serve as new promising biomarkers for diagnosis and prognosis and as effective therapeutic targets for patients with such malignancies. Accordingly, the dysregulation of miR-212-3p has been reported in a variety of human cancers. However, its biological functions and molecular mechanisms high-grade serous ovarian cancer (HGSOG) remain unknown. In this study, we demonstrated that miR-212-3p interacts with MAP3K3 based on bioinformatics-based predictions. Further, MAP3K3 was identified as a direct target gene of miR-212-3p in HGSOC. In addition, overexpression of miR-212-3p in HGSOC inhibited cell proliferation, colony formation, invasion, and migration. In contrast MAP3K3 mitigated the suppressive effects of miR-212-3p on HGSOC cell proliferation, invasion, and migration. Furthermore, miR-212-3p was significantly downregulated in HGSOC tissues compared to expression in normal fallopian tube tissues and was inversely associated with MAP3K3 levels. Accordingly, low miR-212-3p expression was also correlated with poor prognosis for HGSOC patients. In conclusion, miR-212-3p might act as a suppressor of HGSOC carcinogenesis by directly targeting MAP3K3. Therefore, this miRNA could be a novel and effective target for the treatment of patients with HGSOC.
Keywords: miR-212-3p, MAP3K3, high-grade serous ovarian cancer (HGSOC), tumor suppressor
Introduction
Ovarian carcinoma (OC) is one of the most common malignancies of the female genital system and ranks eighth among female cancers worldwide [1,2]. High-grade serous ovarian cancer (HGSOC) is the most common histological type of OC. Because of its undefined pathogenesis, high heterogeneity, few associated therapeutic targets, and chemotherapy resistance, patients with HGSOC typically have poor prognosis and overall survival rates.
Our previous study showed that MAP3K3 is overexpressed and an independent risk factor for prognosis in HGSOC and could continuously activate NF-κB signaling in this disease [3,4]. MAP3K3 promotes OC cell proliferation, chemotherapeutic resistance, the evasion of apoptosis, migration, and invasion in vitro and in vivo. Consistent with our results, MAP3K3 plays a similar oncogenic role in various solid tumors such as breast [5], lung [6], cervical [7], and esophageal cancers [8]. However, the mechanism underlying the overexpression of MAP3K3 in tumors has not been confirmed. Further, due to its important role in the development of mammalian embryos, it cannot be directly knocked out. Therefore, our group further explored small non-coding RNA targets that can regulate MAP3K3 expression.
MicroRNAs (miRNAs) comprise a class of non-coding RNAs of 19-25 nt in length in eukaryotes that are involved in a variety of gene regulatory processes such as the pre-transcriptional control of genes that affect histone modification and mRNA stability [9,10]. MiR-212-3p is located on chromosome 17p13.3 and an increasing number of reports indicate that it can participate in complex regulatory mechanisms that affect the malignant evolution of solid tumors [11,12]. However, with respect to HGSOG, the biological functions and underlying molecular mechanisms of this miRNA remain unknown. Therefore, this study evaluated the expression level and clinical significance of miR-212-3p in HGSOC and investigated the roles of this marker and associated regulatory interactions between miR-212-3p and MAP3K3 in this disease.
Materials and methods
Bioinformatic websites
Four bioinformatic websites were used to analyze and predict miRNAs that interact with MAP3K3, including Targetscan (http://www.targetscan.org/vert_71/), miRDB (http://www.mirdb.org/cgi-bin/search.cgi), StarBase (http://starbase.sysu.edu.cn/), and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php).
Patients
Sixty-three HGSOC tissues and 26 normal fallopian tube tissues were obtained from the First Affiliated Hospital of Shihezi University School of Medicine between 2010 and 2015. Clinical, demographic, and pathologic data were obtained from the medical records of the patients. All tissues were fixed in 10% neutral formalin and paraffin-embedded. Diagnoses were confirmed through hematoxylin-eosin and immunohistochemistry staining by two experienced pathologists, according to the World Health Organization Pathology and Genetics Tumors of the Breast and Female Genital Organs (eighth edition). All subjects provided informed consent, and the study was approved and supervised by the Research Ethics Committee of the First Affiliated Hospital of Shihezi University School of Medicine.
Cell lines and antibodies
The human serous epithelial OC cell lines SKOV3, HeyA8, OVCA433, C13*, OV2008, and A2780 and the human embryonic kidney cell line 293T were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) in a humidified tissue culture incubator containing 5% CO2 at 37°C. SKOV3, HeyA8, and 293T cells were purchased from the Chinese Academy of Sciences Type Culture Collection (Shanghai, China). OVCA433 and A2780 cells were a generous gift from Dr Gang Cheng (Department of Gynecology and Obstetrics, Tongji Hospital of Huazhong University of Science and Technology). OV2008 and C13 cells were from Prof Benjamin K. Tsang (Ottawa Health Research Institute, Ottawa, Canada).
Introduction of miRNA mimics and inhibitors and MAP3K3 plasmid construction
All RNA oligonucleotides used in this study, including the miR-212-3p mimic, inhibitor, and their cognate control RNAs, were purchased from GenePharma (Shanghai, China). The RNA sequences mentioned are listed as follows: miR-212-3p mimic: sense 5’-UAACAGUCUCCAGUCACGGCC-3’ and antisense 5’-CCGUGACUGGAGACUGUUAUU-3’, mimic control: sense 5’-UUCUCCGAACGUG UCACGUTT-3’ and antisense 5’-ACGUGACACGUUCGGAGAATT-3’; miR-212-3p inhibitor: 5’-GGCCGUGACUGGAGACUGUUA-3’; inhibitor control: 5’-CAGUACUUUU-GUGUAGUACAA-3’. The transfection of ovarian cancer cells with Lipofectamin2000 (Life technologies, USA) was performed according to the manufacturer’s instructions. The human MAP3K3 expression vector pBabe-MAP3K3-WT-V5-His (MAP3K3), short hairpin RNA (shRNA) expression vectors specific to MAP3K3, p-super-shRNA-MAP3K3-13 (sh-MAP3K3-13) and p-super-shRNA-MAP3K3-15 (sh-MAP3K3-15), and the random sequence interference expression vector p-super-shRNA-Scramble (sh-scramble) were generous gifts from Prof Jianhua Yang (Baylor College of Medicine, TX, Houston, USA). Construction of the human MAP3K3 expression vector was performed as previously described [4].
RNA extraction and quantitative real-time PCR
Total RNA was extracted from OC cell lines using the miRNeasy Mini Kit (Qiagen, Valencia, CA) and from HGSOC tissues using the miRNeasy FFPE Kit (Qiagen, Valencia, CA) in accordance with the manufacturer’s instructions. cDNA was synthesized with the QuantiTect Reverse Transcription Kit (Qiagen). Quantitative real-time polymerase chain reaction (qRT-PCR) was performed on a 7500 Fast Real-Time PCR System (Life Technologies, Shanghai, China) using a QuantiFast SYBR Green PCR kit (Qiagen) in accordance with the manufacturer’s instructions. The related primers used in this study were prepared by Shanghai Sangon Biotechnology company. The primer sequences are shown in Table 1.
Table 1.
qRT-PCR related primer sequences
| Name | Forward primer (5’-3’) | Reverse primer (5’-3’) |
|---|---|---|
| GADPH | GAGTCAACGGATTTGGTCGT | TTGATTTTGGAGGGATCTCG |
| MAP3K3 | CGAAAGTACACGCGGCAGAT | CAGCAGAGTCTCGGAGGATGTT |
| miR-212-3p | TAACAGTCTCCAGTCACGGC | - |
| U6 | CTCGCTTCGGCAGCACA | AACGCTTCACGAATTTGCGT |
Note: miR-212-3p: hsa-miR-212-3p.
Western blot analysis
Western blotting was performed following standard protocols. The primary antibodies used in this study included rabbit anti-human monoclonal MAP3K3 antibody (1:1000 dilution; Cell signaling technology, CA, USA) and mouse anti-human monoclonal β-actin antibody (1:1000 dilution; Zhongshan Biotechnology, Beijing, China). Goat anti-rabbit/mouse IgG-HRP, used as the secondary antibody, was purchased from Zhongshan Biotechnology (Beijing, China). The protein signals were detected with an enhanced chemiluminescence kit (Thermo, Waltham, MA, USA). β-actin was used as an internal control.
Dual-luciferase reporter gene assay
The miR-212-3p-binding motif in the MAP3K3 3’-UTR was identified by the TargetScan algorithm (http://TargetScan.org). The different fragment sequences were synthesized and then inserted into the pmirGLO-basic vector (OMEGA Engineering Inc.) and co-transfected with wild-type and mutant MAP3K3 plasmids into SKOV3 cells. The 3’-UTR of MAP3K3 was cloned into the luciferase vector and transfected into SKOV3 cells together with miR-212-3p mimics, the miR-212-3p inhibitor, the MAP3K3 plasmid, or the negative control. All vectors were verified by sequencing, and luciferase activities were assessed using a Dual Luciferase Assay Kit (OMEGA Engineering Inc.) in accordance with the manufacturer’s instructions.
RNA immunoprecipitation
An EZMagna RNA immunoprecipitation (RIP) Kit (Millipore, Bedford, MA, USA) was used following the manufacturer’s protocol. SKOV3 cells were lysed in complete RIP lysis buffer (containing proteinase inhibitor and phosphatase inhibitor), and the cell extract was incubated with magnetic beads conjugated with specific antibodies or control IgG for 6 h at 4°C. Beads were washed and incubated with proteinase K to remove proteins. Finally, purified RNA was subjected to qRT-PCR analysis.
Proliferation assays
SKOV3, A2780, and OV2008 cells were incubated and transfected with the different treatments as described previously herein for 48 h, and the Cell Counting Kit-8 (CCK-8) assay was performed to construct a cell growth curve. The optical density (OD) value was determined by measuring the absorbance at 450 nm with a microplate reader at the same time every day until the sixth day from the beginning of cell attachment. The daily OD values of each group of cells were used to produce the cell growth curve.
Migration and invasion assays
Transwell assays were used to test cell migration capacity. The cells were seeded in six-well plates and cultured until full confluence was reached. Uniform scratches were then made in the center of the wells. The plates were washed and cultured in serum-free RPMI1640 medium without a penicillin-streptomycin mixture. Scratches were observed every 0, 12, 24, and 48 h. Statistical analysis of the areas before and after healing was conducted using IPP statistical software.
For invasion assays, 1 × 105 cells in serum-free medium were placed into the upper chamber of an insert coated with Matrigel. Medium containing 10% FBS was added to the lower chamber. After incubation for 24 h, the cells remaining on the upper membrane were removed with cotton wool. Cells that had migrated or invaded through the membrane were fixed with methanol, stained with 0.1% crystal violet, imaged, and counted using an inverted microscope (Olympus, Tokyo, Japan).
Statistical analysis
Differences between groups were assessed by a paired, two tailed Student’s t-test. The Chi-square test was used to analyze the clinicopathological features associated with miR-212-3p expression in HGSOC. The survival curves were drawn using Kaplan-Meier survival plots and tested using log-rank tests. Univariate and multivariate Cox proportional hazards modeling was used to determine the effects of variables on survival. All statistical analyses were performed using SPSS 22.0 software (IBM SPSS, Chicago, IL, USA).
Results
MiR-212-3p directly targets the MAP3K3 3’-UTR
Our previous study demonstrated that MAP3K3 is overexpressed in HGSOC and that it promotes HGSOC proliferation and migration by activating the NF-κB signaling pathway. Nevertheless, the mechanism driving dysregulated MAP3K3 expression and the upstream oncogenic network in HGSOC remained unclear. We thus identified miR-212-3p, which could regulate MAP3K3 expression, based on bioinformatic analysis (Figure 1A, 1B). To investigate whether miR-212-3p directly binds the 3’-UTR of MAP3K3, we cloned the MAP3K3 3’-UTR and its mutants into luciferase reporter plasmids, and co-transfected SKOV3 cell lines with the constructed plasmids and miR-212-3p mimics. As shown in Figure 1C, luciferase activity was decreased in cells co-transfected with miR-212-3p, as compared to that in cells expressing miR-NC, and this was statistically significant (P < 0.01). In addition, mutations in the putative binding site abolished the repression of luciferase activity mediated by miR-212-3p. These findings suggest that miR-212-3p regulates MAP3K3 expression through direct interactions with its 3’-UTR. To further verify whether MAP3K3 is the direct target of miR-212-3p, we performed RNA immunoprecipitation. The result demonstrated that endogenous MAP3K3 pull-down by AGO2 was significantly enriched in miR-212-3p-overexpressing cells (Figure 1D; P < 0.05).
Figure 1.
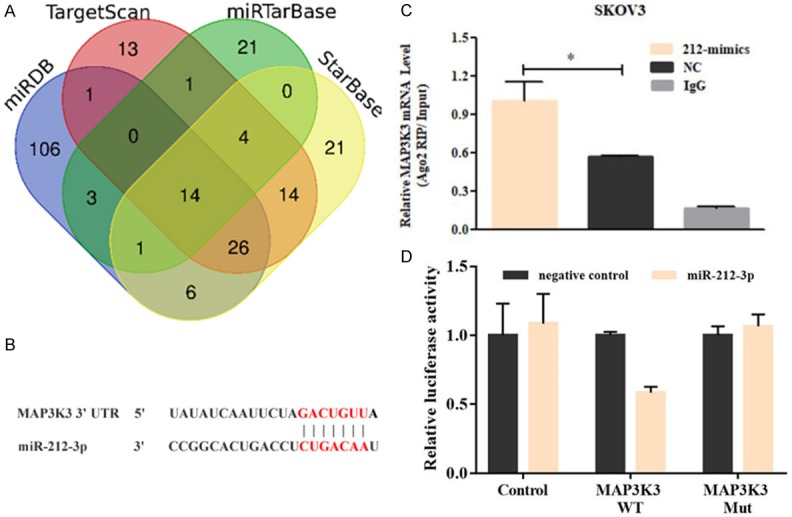
The MAP3K3 gene is a direct target of miR-212-3p in high-grade serous ovarian cancer (HGSOC) cells. A. The target genes of miR-212-3p were predicted using bioinformatics tools (TargetScan, miRTarBase, miRDB, and StarBase databases). B. The binding site involved in the interaction between the MAP3K3 3’-UTR and miR-212-3p was predicted using these databases. C. Dual-luciferase reporter gene assays showed that miR-212-3p binds the 3’-UTR of MAP3K3 and inhibits its expression. D. RNA immunoprecipitation assays demonstrated that endogenous MAP3K3 pull-down by AGO2 was significantly enriched in miR-212-3p-overexpressing cells; IgG was used as the control.
MiR-212-3p suppresses MAP3K3 expression
To confirm the regulatory effect of miR-212-3p on MAP3K3 expression, we first determined the expression levels of MAP3K3 and miR-212-3p in various HGSOC cell lines (Figure 2A). The result showed that MAP3K3 expression was relatively high in A2780 and SKOV3 cell lines, whereas it was relatively low in OV2008 cell lines. Therefore, qRT-PCR and western blot assays were performed to test MAP3K3 expression in A2780 and SKOV3 cell lines after transfecting them with miR-212-3p mimics and in OV2008 cell lines after transfecting them with miR-212-3p inhibitors. The results revealed that MAP3K3 expression was remarkably decreased after transfection with miR-212-3p mimics compared to that in negative controls in A2780 (Figure 2B) and SKOV3 (Figure 2C) cell lines. However, MAP3K3 expression was significantly increased after transfection with a miR-212-3p inhibitor compared to levels with the negative control inhibitor in OV2008 cell lines (Figure 2D). These results indicated that miR-212-3p deregulates the expression level of MAP3K3.
Figure 2.
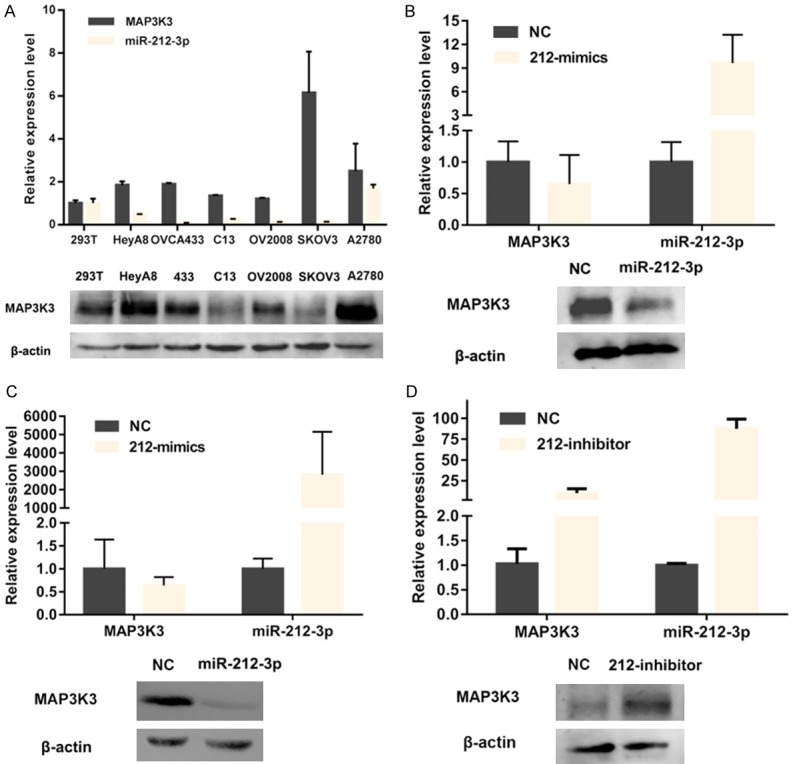
miR-212-3p can downregulate MAP3K3 expression in high-grade serous ovarian cancer (HGSOC) cells. (A) mRNA and protein expression of miR-212-3p and MAP3K3 in six ovarian cancer cell lines and 293T cells. MAP3K3 expression was remarkably decreased after transfection with miR-212-3p mimics in A2780 (B) and SKOV3 (C) cells at both the mRNA and protein levels. MAP3K3 expression was significantly increased after transfection with a miR-212-3p inhibitor in OV2008 (D) cell lines.
MiR-212-3p suppresses HGSOC cell proliferation, migration and invasion
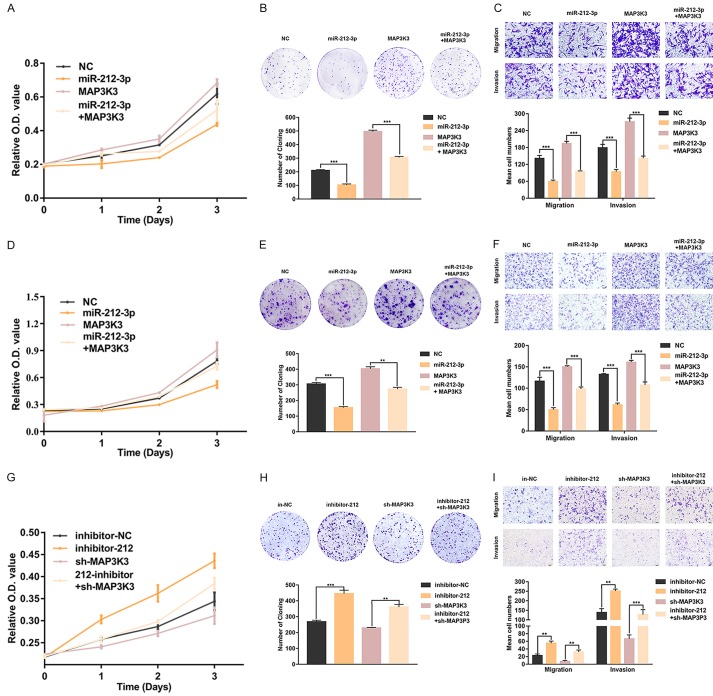
To investigate the function of miR-212-3p with respect to HGSOC biological behaviors, we performed cell proliferation, invasion and migration assays using cell lines transfected with miR-212-3p mimics or inhibitors; specifically, A2780 and SKOV3 cell lines with high endogenous expression of MAP3K3 were transfected with miR-212-3p mimics, whereas OV2008 cells with low endogenous MAP3K3 expression were transfected with a miR-212-3p inhibitor. Based on CCK8 assays, we observed that miR-212-3p markedly reduced cell proliferation at 24, 48, and 72 h, compared to that in negative controls (Figure 3A, 3B). In contrast, the proliferation of cells transfected with the miR-212-3p inhibitor was significantly increased (Figure 3C). Colony forming assays demonstrated that miR-212-3p mimics could promote the formation of A2780 (Figure 3D) and SKOV3 (Figure 3E) cell clones; however clone formation in OV2008 cells was diminished upon inhibiting miR-212-3p (Figure 3F). Furthermore, we also examined the effect of miR-212-3p on cell invasion and migration by performing transwell assays. With this treatment, the invasive and migratory activities of A2780 (Figure 3G) and SKOV3 (Figure 3H) cells were enhanced compared to those in negative control cells, whereas these phenotypes were suppressed in OV2008 cells (Figure 3I). These results indicated that miR-212-3p is involved in the regulation of cell proliferation, invasion and migration.
Figure 3.
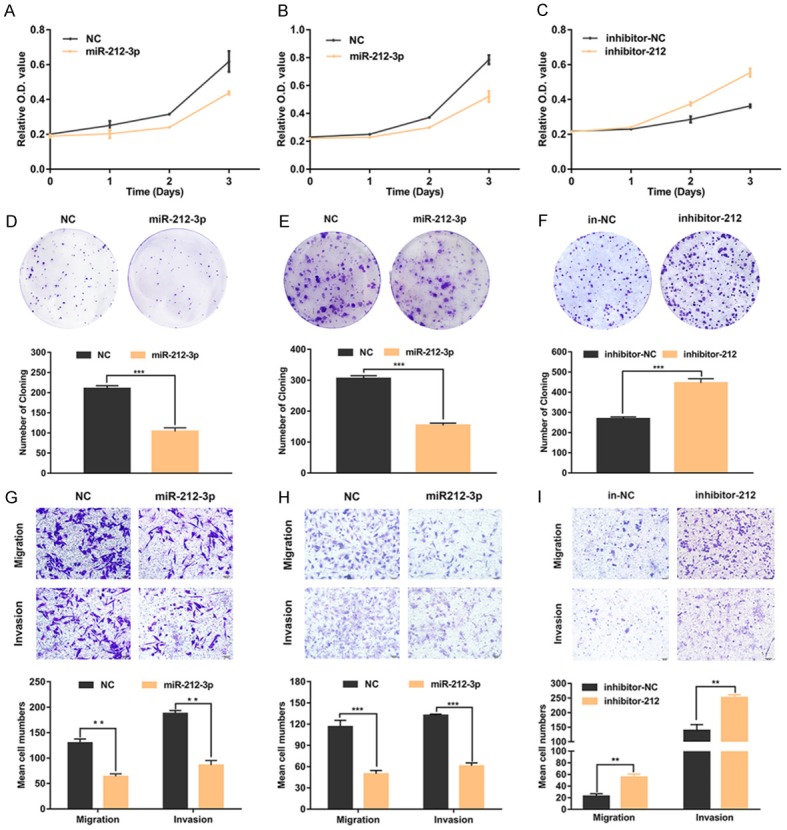
miR-212-3p inhibits high-grade serous ovarian cancer (HGSOC) cell proliferation, invasion, and migration. A2780 (A) and SKOV3 (B) cell proliferation was inhibited as evaluated by the CCK-8 assays after transfecting them with miR-212-3p mimics. OV2008 (C) cell proliferation was enhanced after transfecting cells miR-212-3p inhibitors. MiR-212-3p impaired the colony formation of A2780 (D) and SKOV3 (E) cells. The inhibition of miR-212-3p enhanced the colony formation of OV2008 (F) cells. Migration and invasion abilities of A2780 (G) and SKOV3 (H) cells were decreased with the transfection of miR-212-3p mimics. The migration and invasion abilities of OV2008 cells (I) were increased upon transfection with miR-212-3p inhibitors. Data are presented as the mean ± SD, based on the Student’s t-test for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
MAP3K3 reverses the inhibitory effect of miR-212-3p on HGSOC cell proliferation, migration, and invasion
To determine whether the effect of miR-212-3p on cell proliferation and migration was mediated by the repression of MAP3K3, we respectively transfected cells with negative control (NC), miR-212-3p mimics (212m), and MAP3K3 vectors, and co-transfected MAP3K3 and miR-212-3p mimics into A2780 and SKOV3 cells. The results showed that compared to that in the negative control group, cell proliferation in the miR-212-3p mimics group was decreased, whereas the proliferation of cells transfected with MAP3K3 was enhanced. Cell proliferation in the co-transfected groups was between that in the miR-212-3p mimics group and that in the MAP3K3 groups (Figure 4A, 4D). The same phenomenon was observed with colony forming assays (Figure 4B, 4E) and transwell assays (Figure 4C, 4F). Furthermore, we respectively used the negative control inhibitor, miR-212-3p inhibitors, and sh-MAP3K3 for transfection, and co-transfected sh-MAP3K3 and miR-212-3p inhibitors into OV2008 cells. As shown in Figure 4G, MAP3K3 inhibition significantly attenuated the enhancing effect of miR-212-3p on cell proliferation in OV2008 cells. Similar results were also obtained for colony formation assay (Figure 4H) and transwell assay (Figure 4I). These observations indicated that miR-212-3p can reduce cell proliferation and migration by down-regulating the expression of MAP3K3.
Figure 4.
MAP3K3 reverses the inhibitory effect of miR-212-3p on the proliferation, invasion, and migration abilities of high-grade serous ovarian cancer (HGSOC) cells. In both A2780 and SKOV3 cells, proliferation (A, B), colony formation (D, E), migration, and invasion (G, H) were enhanced after co-transfecting cells with miR-212-3p mimics and MAP3K3 compared to those with only miR-212-3p mimics. OV2008 cell proliferation (C), colony formation (F), migration, and invasion (I) abilities were inhibited by co-transfecting cells with sh-MAP3K3 and miR-212-3p inhibitors compared to those with only miR-212-3p inhibitors. Data are presented as the mean ± SD, based on the Student’s t-test for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
MiR-212-3p is expressed at low levels in HGSOC tissues and is negatively associated with both MAP3K3 expression and poor survival
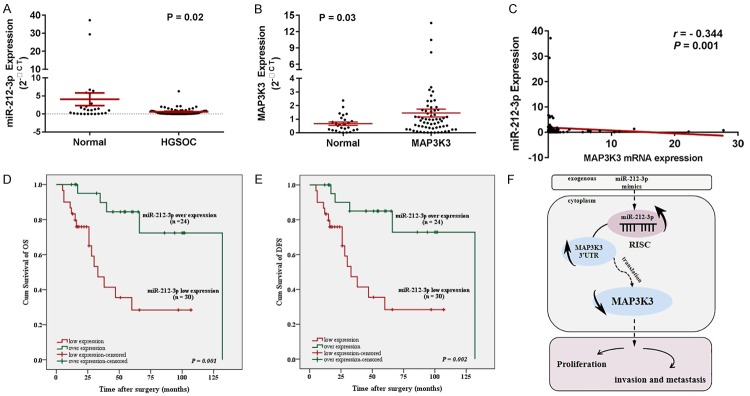
To confirm the role of miR-212-3p in ovarian malignancy, we detected its expression level in 63 HGSOC tissues and 26 normal fallopian tube tissues by qRT-PCR. The results demonstrated that the expression of miR-212-3p was significantly decreased in HGSOC compared to that in normal fallopian tube tissues (Figure 5A; P < 0.05), whereas the expression of MAP3K3 was significantly increased in HGSOC tissues (Figure 5B; P < 0.05). Meanwhile, it was negatively associated with MAP3K3 expression in HGSOC (Figure 5C; r = -0.344, P < 0.001). These results indicated that miR-212-3p might be an upstream factor that induces dysregulated MAP3K3 expression.
Figure 5.
MiR-212-3p is downregulated and negatively associated with poor prognosis and MAP3K3 expression in HGSOC patients. (A) MiR-212-3p was found to be downregulated in HGSOC tissues. (B) MAP3K3 was overexpressed in HGSOC tissues. (C) miR-212-3p expression was negatively associated with MAP3K3 (r = -0.344, P < 0.001). Correlation between miR-212-3p expression and overall survival (OS) (D) or disease-free survival (DFS) (E) in 63 HGSOC patients. The median value of mi-212-3p expression was chosen as the point to classify tumors as low- or high-level expression groups. (F) A proposed model illustrating the function and mechanism of mi-212-3p in modulating HGSOC tumorigenesis. Data are presented as the mean ± SD, based on the Student’s t-test for statistical analysis. *P < 0.05, **P < 0.01, ***P < 0.001.
Further, by investigating the clinicopathological and prognostic significance of miR-212-3p levels in HGSOC patients, results revealed that the downregulation miR-212-3p was significantly correlated with FIGO stage and sensitivity to chemotherapy (Table 2). Furthermore, Kaplan-Meier analysis revealed that low miR-212-3p levels were associated with poor overall survival (OS) (Figure 5D; P = 0.001) and disease-free survival (Figure 5E; P = 0.002) in HGSOC patients. Based on univariate analysis using the Cox regression model, low miR-212-3p expression and FIGO stage were considered strong prognostic indicators of poor OS (Table 3; P = 0.004, P = 0.002, respectively).
Table 2.
The association between miR-212-3p expression and clinicopathological features of HGSOC patients
| Characteristics | n | miR-212-3p expression | χ2 | P | |
|---|---|---|---|---|---|
|
|
|||||
| 63 | Over expression (%) | Low expression (%) | |||
| Age | |||||
| < 53 | 28 | 4 | 24 | 0.352 | 0.553 |
| ≥ 53 | 35 | 7 | 28 | ||
| FIGO stage | |||||
| I-II | 15 | 6 | 9 | 6.94 | 0.008** |
| III-IV | 48 | 5 | 43 | ||
| Chemotherapy response | |||||
| Sensitive | 30 | 9 | 21 | 3.871 | 0.049* |
| Partial | 10 | 0 | 10 | ||
| Unknown | 23 | ||||
| Ascites | |||||
| No | 13 | 1 | 12 | 1.13 | 0.288 |
| Yes | 44 | 9 | 35 | ||
| Unknown | 6 | ||||
p < 0.05;
p < 0.01.
Table 3.
MiR-212-3p is an prognostic biomarker for patients with HGSOC
| Univariate | Multivariate | |||
|---|---|---|---|---|
|
|
|
|||
| HR (95% CI) | P | HR (95% CI) | P | |
| miR-212-3p (low vs overexpression) | 0.206 (0.070-0.611) | 0.004* | 0.151 (0.019-1.214) | 0.075 |
| Age (< 53 vs ≥ 53) | 1.952 (0.931-4.090) | 0.076 | ||
| FIGO stage (I-II vs III-IV) | 7.393 (2.136-25.584) | 0.002** | 5.612 (1.133-27.797) | 0.035* |
| Chemotherapy response (sensitive vs partial) | 1.393 (0.373-4.976) | 0.64 | ||
| Ascites (no vs yes) | 1.567 (0.713-3.443) | 0.264 | ||
p < 0.05;
p < 0.01.
Discussion
OC is one of the most common malignant tumors of female genital organs. In 2015, there were approximately 52,100 new cases of ovarian carcinoma in China and mortality ranked first among all gynecologic malignancies [1,2]. HGSOC is the most common histological type of OC and originates from the epithelial cells of the fallopian tube; tumor cells attach to the surface of the ovary and grow, which facilitates extensive abdominal metastasis. HGSOC also possesses characteristics of genetic instability such as high TP53 and BRCA1/2 mutation rates [13-17]. With the development of treatment in recent years, targeted therapy and immunotherapy have resulted in promising effects on HGSOC. However, because of its unclear pathogenesis, high heterogeneity, few therapeutic targets, and relapse resistance after chemotherapy, clinical treatment effects and OS rates are unsatisfactory for HGSOC patients. Therefore, it is crucial to find new targets for disease prediction and treatment and to elucidate the molecular mechanisms involved in the malignant transformation of HGSOC.
Our previous study demonstrated that MAP3K3 is overexpressed in HGSOC and promotes the proliferation and migration of HGSOC by activating the NF-κB signaling pathway [4]. MAP3K3 plays a similar oncogenic role in HGSOC and various other tumors such as breast, lung, cervical, and esophageal cancers [5-8]. However, the mechanism underlying the effects of MAP3K3 overexpression in tumors has not been confirmed. MiRNAs are involved in a variety of gene-regulatory processes such as the pre-transcriptional control of genes affecting histone modification and mRNA stability. This is achieved via the formation of an RNA-induced silencing complex (RISC) with the 3’-UTR region of targeting mRNA, reducing target mRNA stability and translation or promoting degradation [18,19]. Therefore, we further explored small non-coding RNA targets that can regulate MAP3K3 expression.
In this study, we screened 14 miRNAs that interact with MAP3K3 based on four bioinformatics websites for prediction and cross-analysis. miR-212-3p was chosen and was proven to directly bind MAP3K3 by dual-luciferase reporter gene assays and RNA immunoprecipitation. Furthermore, we found that miR-212-3p overexpression can inhibit the expression of MAP3K3 protein and suppress cell proliferation, invasion, and migration in HGSOC. Accordingly, restored MAP3K3 expression rescued the tumor-suppressive effects of miR-212-3p overexpression in HGSOC. These results suggest that miR-212-3p serves as a tumor suppressor gene in HGSOC carcinogenesis by directly targeting MAP3K3. Several studies demonstrated that miR-212-3p is commonly aberrantly expressed and inhibits cancer cell proliferation and migration by targeting downstream genes in multiple types of human malignancies including renal clear cell carcinoma, intrahepatic cholangiocarcinoma, and osteosarcoma [20-23]. Studies have also shown that miR-212-3p is downregulated in ovarian cancer SKOV3 cells and exerts a tumor suppressor effect by targeting HBEGF [24]. This report is consistent with our findings. miR-212-3p is thus dysregulated in a variety of malignancies including HGSOC and is significantly decreased in tumor tissues, suggesting that it might be involved in the evolution of cancer as a tumor suppressor gene.
In our study, we demonstrated that miR-212-3p is significantly downregulated in HGSOC tissues, which was found to be correlated with poor prognosis in HGSOC patients. Meanwhile, its reduced expression was determined to be correlated with high FIGO stage and chemotherapy resistance. Univariate analysis of COX risk regression showed that low expression of miR-212-3p and high FIGO stage were risk factors for HGSOC. These results suggest that miR-212-3p might represent a novel molecular marker of HGSOC prognosis and could be a new potential therapeutic target for this disease. Many studies have found that miRNAs can be used as molecular markers for the early diagnosis and prognosis of various tumors [25-28]. In NSCLC, miR-505 was also found to be expressed at low levels and was negatively correlated with tumor size, TNM stage, and distant metastasis, indicating that it could become a new target for disease treatment and clinical applications [29]. Some potential clinically valuable miRNAs have also been found for ovarian cancer [30]. For example, miR-181a promotes TGF-β-mediated epithelial-mesenchymal transition by inhibiting its functional target Smad7 and was found to be associated with ovarian cancer recurrence and poor prognosis. In addition, the overexpression of miR-181a promotes OC cell proliferation, migration, invasion, drug resistance, tumor formation in vivo, and metastasis [31]. Based on these studies, a variety of miRNAs including miR-212-3p might become molecular markers for the early diagnosis and prognosis of solid tumors and provide a theoretical basis for potential new clinical therapeutic targets.
In conclusion, miR-212-3p is downregulated in HGSOC tissues and cell lines, and its low-expression is correlated with FIGO stage and chemotherapy resistance. MiR-212-3p also plays a tumor suppressive role in HGSOC by directly targeting MAP3K3. These results suggest that miR-212-3p could be a new therapeutic target for HGSOC patients.
Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. 81660431 and No. 81960465), the high-level talent project of Shihezi University (No. RCZK2018C17), the Youth Science and Technology Innovation Leading Talents Project of Corps (No. 2017CB004) and Xinjiang Production, Construction Corps Key Areas Innovation Team Project (No. 2018CB002). No funding or other benefits related to the subject of this article were received from any commercial entity.
Disclosure of conflict of interest
None.
References
- 1.Ferlay JS, Ervik M, Dikshit R, editors. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC CancerBase No. 11 [J] Lyon, France: International Agency for Research on Cancer; 2013. Available from http://globocan.iarc.fr. [Google Scholar]
- 2.Howlader N, Noone AM, Krapcho M. National Cancer Institute SEER Cancer Statistics Review, 1975-2012, Bethesda, MD [J] . 2015 [Google Scholar]
- 3.Jia W, Dong Y, Tao L, Pang L, Ren Y, Liang W, Jiang J, Cheng G, Zhang WJ, Yuan X, Li F. MAP3K3 overexpression is associated with poor survival in ovarian carcinoma. Hum Pathol. 2016;50:162–169. doi: 10.1016/j.humpath.2015.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Y, Wang SS, Tao L, Pang LJ, Zou H, Liang WH, Liu Z, Guo SL, Jiang JF, Zhang WJ, Jia W, Li F. Overexpression of MAP3K3 promotes tumour growth through activation of the NF-kappaB signalling pathway in ovarian carcinoma. Sci Rep. 2019;9:8401. doi: 10.1038/s41598-019-44835-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan Y, Ge N, Wang X, Sun W, Mao R, Bu W, Creighton CJ, Zheng P, Vasudevan S, An L, Yang J, Zhao YJ, Zhang H, Li XN, Rao PH, Leung E, Lu YJ, Gray JW, Schiff R, Hilsenbeck SG, Osborne CK, Yang J, Zhang H. Amplification and over-expression of MAP3K3 gene in human breast cancer promotes formation and survival of breast cancer cells. J Pathol. 2014;232:75–86. doi: 10.1002/path.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y, Wang L, Liu W, Zhong J, Bai S, Wang Z, Thomas DG, Lin J, Reddy RM, Ramnath N, Carrott PW, Lynch WR, Orringer MB, Chang AC, Beer DG, Chen G. MAP3K3 expression in tumor cells and tumor-infiltrating lymphocytes is correlated with favorable patient survival in lung cancer. Sci Rep. 2015;5:11471. doi: 10.1038/srep11471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao XQ, Lu HS, Zhang L, Chen LL, Gan MF. MEKK3 and survivin expression in cervical cancer: association with clinicopathological factors and prognosis. Asian Pac J Cancer Prev. 2014;15:5271–5276. doi: 10.7314/apjcp.2014.15.13.5271. [DOI] [PubMed] [Google Scholar]
- 8.Hasan R, Sharma R, Saraya A, Chattopadhyay TK, DattaGupta S, Walfish PG, Chauhan SS, Ralhan R. Mitogen activated protein kinase kinase kinase 3 (MAP3K3/MEKK3) overexpression is an early event in esophageal tumorigenesis and is a predictor of poor disease prognosis. BMC Cancer. 2014;14:2. doi: 10.1186/1471-2407-14-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019;20:21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Csak T, Bala S, Lippai D, Satishchandran A, Catalano D, Kodys K, Szabo G. microRNA-122 regulates hypoxia-inducible factor-1 and vimentin in hepatocytes and correlates with fibrosis in diet-induced steatohepatitis. Liver Int. 2015;35:532–541. doi: 10.1111/liv.12633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ju C, Zhou R, Sun J, Zhang F, Tang X, Chen KK, Zhao J, Lan X, Lin S, Zhang Z, Lv XB. LncRNA SNHG5 promotes the progression of osteosarcoma by sponging the miR-212-3p/SGK3 axis. Cancer Cell Int. 2018;18:141. doi: 10.1186/s12935-018-0641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurman RJ, Shih Ie M. The dualistic model of ovarian carcinogenesis: revisited, revised, and expanded. Am J Pathol. 2016;186:733–747. doi: 10.1016/j.ajpath.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas Research Network. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lawrenson K, Song F, Hazelett DJ, Kar SP, Tyrer J, Phelan CM, Corona RI, Rodríguez-Malavé NI, Seo JH, Adler E, Coetzee SG, Segato F, Fonseca MAS, Amos CI, Carney ME, Chenevix-Trench G, Choi J, Doherty JA, Jia W, Jin GJ, Kim BG, Le ND, Lee J, Li L, Lim BK, Adenan NA, Mizuno M, Park B, Pearce CL, Shan K, Shi Y, Shu XO, Sieh W Australian Ovarian Cancer Study Group. Thompson PJ, Wilkens LR, Wei Q, Woo YL, Yan L, Karlan BY, Freedman ML, Noushmehr H, Goode EL, Berchuck A, Sellers TA, Teo SH, Zheng W, Matsuo K, Park S, Chen K, Pharoah PDP, Gayther SA, Goodman MT. Genome-wide association studies identify susceptibility loci for epithelial ovarian cancer in east Asian women. Gynecol Oncol. 2019;153:343–355. doi: 10.1016/j.ygyno.2019.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie EL, Pattnaik S, Beach J, Copeland A, Rashoo N, Fereday S, Hendley J, Alsop K, Brady SL, Lamb G, Pandey A, deFazio A, Thorne H, Bild A, Bowtell DDL. Multiple ABCB1 transcriptional fusions in drug resistant high-grade serous ovarian and breast cancer. Nat Commun. 2019;10:1295. doi: 10.1038/s41467-019-09312-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ballabio S, Craparotta I, Paracchini L, Mannarino L, Corso S, Pezzotta MG, Vescio M, Fruscio R, Romualdi C, Dainese E, Ceppi L, Calura E, Pileggi S, Siravegna G, Pattini L, Martini P, Delle Marchette M, Mangioni C, Ardizzoia A, Pellegrino A, Landoni F, D’Incalci M, Beltrame L, Marchini S. Multisite analysis of high-grade serous epithelial ovarian cancers identifies genomic regions of focal and recurrent copy number alteration in 3q26.2 and 8q24.3. Int J Cancer. 2019;145:2670–2681. doi: 10.1002/ijc.32288. [DOI] [PubMed] [Google Scholar]
- 18.Ghildiyal M, Xu J, Seitz H, Weng Z, Zamore PD. Sorting of drosophila small silencing RNAs partitions microRNA* strands into the RNA interference pathway. RNA. 2010;16:43–56. doi: 10.1261/rna.1972910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kobayashi H, Tomari Y. RISC assembly: coordination between small RNAs and Argonaute proteins. Biochim Biophys Acta. 2016;1859:71–81. doi: 10.1016/j.bbagrm.2015.08.007. [DOI] [PubMed] [Google Scholar]
- 20.Wang D, Zheng J, Liu X, Xue Y, Liu L, Ma J, He Q, Li Z, Cai H, Liu Y. Knockdown of USF1 inhibits the vasculogenic mimicry of glioma cells via stimulating SNHG16/miR-212-3p and linc00667/miR-429 axis. Mol Ther Nucleic Acids. 2019;14:465–482. doi: 10.1016/j.omtn.2018.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Ju C, Zhou R, Sun J, Zhang F, Tang X, Chen KK, Zhao J, Lan X, Lin S, Zhang Z, Lv XB. LncRNA SNHG5 promotes the progression of osteosarcoma by sponging the miR-212-3p/SGK3 axis. Cancer Cell Int. 2018;18:141. doi: 10.1186/s12935-018-0641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou P, Kang Y, Luo J. Hypoxia-mediated miR-212-3p downregulation enhances progression of intrahepatic cholangiocarcinoma through upregulation of Rab1a. Cancer Biol Ther. 2018;19:984–993. doi: 10.1080/15384047.2018.1456608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie C, Chen B, Wu B, Guo J, Cao Y. LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed Pharmacother. 2018;97:1645–1653. doi: 10.1016/j.biopha.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 24.Wei LQ, Liang HT, Qin DC, Jin HF, Zhao Y, She MC. MiR-212 exerts suppressive effect on SKOV3 ovarian cancer cells through targeting HBEGF. Tumour Biol. 2014;35:12427–12434. doi: 10.1007/s13277-014-2560-2. [DOI] [PubMed] [Google Scholar]
- 25.Zheng Y, Liu H, Kong Y. miR-188 promotes senescence of lineage-negative bone marrow cells by targeting MAP3K3 expression. FEBS Lett. 2017;591:2290–2298. doi: 10.1002/1873-3468.12720. [DOI] [PubMed] [Google Scholar]
- 26.Zhao L, Ni X, Zhao L, Zhang Y, Jin D, Yin W, Wang D, Zhang W. MiroRNA-188 acts as tumor suppressor in non-small-cell lung cancer by targeting MAP3K3. Mol Pharm. 2018;15:1682–1689. doi: 10.1021/acs.molpharmaceut.8b00071. [DOI] [PubMed] [Google Scholar]
- 27.Tian T, Lv X, Pan G, Lu Y, Chen W, He W, Lei X, Zhang H, Liu M, Sun S, Ou Z, Lin X, Cai L, He L, Tu Z, Wang X, Tannous BA, Ferrone S, Li J, Fan S. Long noncoding RNA MPRL promotes mitochondrial fission and cisplatin chemosensitivity via disruption of Pre-miRNA processing. Clin Cancer Res. 2019;25:3673–3688. doi: 10.1158/1078-0432.CCR-18-2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lefort K, Brooks Y, Ostano P, Cario-Andre M, Calpini V, Guinea-Viniegra J, Albinger-Hegyi A, Hoetzenecker W, Kolfschoten I, Wagner EF, Werner S, Dotto GP. A miR-34a-SIRT6 axis in the squamous cell differentiation network. EMBO J. 2013;32:2248–2263. doi: 10.1038/emboj.2013.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang H, Lv W, Sun W, Bi Q, Hao Y. miR505 inhibits cell growth and EMT by targeting MAP3K3 through the AKTNFkappaB pathway in NSCLC cells. Int J Mol Med. 2019;43:1203–1216. doi: 10.3892/ijmm.2018.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaluvally-Raghavan P, Zhang F, Pradeep S, Hamilton MP, Zhao X, Rupaimoole R, Moss T, Lu Y, Yu S, Pecot CV, Aure MR, Peuget S, Rodriguez-Aguayo C, Han HD, Zhang D, Venkatanarayan A, Krohn M, Kristensen VN, Gagea M, Ram P, Liu W, Lopez-Berestein G, Lorenzi PL, Borresen-Dale AL, Chin K, Gray J, Dusetti NJ, McGuire SE, Flores ER, Sood AK, Mills GB. Copy number gain of hsa-miR-569 at 3q26.2 leads to loss of TP53INP1 and aggressiveness of epithelial cancers. Cancer Cell. 2014;26:863–879. doi: 10.1016/j.ccell.2014.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parikh A, Lee C, Joseph P, Marchini S, Baccarini A, Kolev V, Romualdi C, Fruscio R, Shah H, Wang F, Mullokandov G, Fishman D, D’Incalci M, Rahaman J, Kalir T, Redline RW, Brown BD, Narla G, DiFeo A. microRNA-181a has a critical role in ovarian cancer progression through the regulation of the epithelial-mesenchymal transition. Nat Commun. 2014;5:2977. doi: 10.1038/ncomms3977. [DOI] [PMC free article] [PubMed] [Google Scholar]