Abstract
The microRNA-29 family, which contains mir-29a, mir-29b, and mir-29c, can promote or resist the development of several types of tumors. However, its role in rhabdomyosarcoma (RMS) has not been determined. In this work, we detected the expression of mir-29a/b/c in RMS. Results showed that the tissues and cell lines in RMS were significantly lower than those in muscle and human skeletal muscle cells, and that these cell lines could also inhibit the proliferation, migration, and invasion and induce apoptosis of RMS cells. Dual-luciferase reporter assay and RNA immunoprecipitation verified the direct binding site between mir-29a/b/c and GEFT. Under the combined actions of mir-29a/b/c and GEFT, the former weakened the promoting effect of GEFT on RMS cells. Finally, mir-29a inhibited the tumorigenesis of subcutaneous xenografts in nude mice and inhibited the mRNA and protein expression levels of GEFT in transplanted tumors. These findings proved that mir-29 inhibits the occurrence of RMS and may be a potential molecular target.
Keywords: Rhabdomyosarcoma, mir-29a/b/c, GEFT, proliferation, migration, invasion, apoptosis
Introduction
Rhabdomyosarcoma (RMS) is a soft-tissue sarcoma from skeletal muscle-lineage cells common among children [1]. The World Health Organization (WHO) guidelines in 2012 categorized RMS into four subtypes according to its genetic, histological, and clinical features: embryonal RMS (ERMS), alveolar RMS (ARMS), pleomorphic RMS, and spindle cell/sclerosing RMS [2]. Although the neoplasm easily develops in several positions limitedly and few patients show metastasis, the prognosis of high-risk groups remains poor, and treatment effects are disheartening. The 5-year progression-free survival for RMS is less than 30% [3,4]. Thus, understanding the molecular mechanisms of the disease is necessary to provide a theoretical basis for molecular targeted therapy.
MicroRNAs (miRNAs) are small noncoding RNA molecules that were first discovered in Candida elegans by Lee; miRNAs are a kind of single-stranded non-coding RNA molecule with a length of about 22 nucleotides that regulate gene expression at the post-transcriptional level [5,6]. MiRNAs can regulate the expression of more than 30% of coding protein genes in the body [7,8]. MiRNAs may affect their target genes as the oncogenes or antioncogenes perform many functions in cell proliferation, migration, and apoptosis [9-11]. These molecules have regulators at upstream or downstream pathways that allow them to play important roles in tumor occurrence and development [9]. For example, the overall survival of a cohort of patients with RMS is correlated with miR-206 level [12]. Mir-206 blocks tumor growth by promoting myogenic differentiation in mice xenotransplanted with RMS [13]. The oncogene N-ras is a target gene of mir-214, which in turn could inhibit N-ras expression in xenograft tumor models [14]. TGF-β1 is negatively correlated with mir-411-5p, and SPRY4 is a target gene of mir-411-5p. These findings were confirmed by other studies [15-26] on RMS (Table 1). Mir-29a/b/c belongs to the mir-29 family, which is the target of the PcG transcription factor Yin Yang 1 (YY1). There have study showed that abnormal mir-29 expression is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma [27]. In addition, studies of mir-29 in different types of tumors have been involved [15,28-63] (Table 2). An in-depth study of this phenomenon may further our understanding of the mechanism of RMS.
Table 1.
MicroRNA targeting genes and their roles in rhabdomyosarcoma
| MicroRNA | Target Gene | Biological Behavior |
|---|---|---|
| Mir-1/206 | c-Met, CCND2, PAX3 | myogenic differentiation, proliferation [15] |
| Mir-133b | PAX3, PTBP1 | proliferation, autophagy [16] |
| Mir-29 | CCND2, YY1, EZH2, E2F7 | proliferation, myogenesis [15,17-19] |
| Mir-26a | EZH2 | myogenesis [20] |
| Mir-27 | PAX3 | proliferation [21] |
| Mir-214 | N-ras | proliferation [14] |
| Mir-450 | ENOX2, PAX9 | pyogenic [22] |
| Mir-411 | SPRY4 | myogenic [23] |
| Mir-203 | P63, LIFR | myogenic, proliferation, invasion, migration [24] |
| Mir-378 | IGF1R | myogenic, apoptosis migration [25] |
| Mir-183 | EGR1, PTEN | migration [26] |
Table 2.
Mir-29 expression in various types of tumors
| Tumor Types | Author | Level | Target Gene |
|---|---|---|---|
| Lung cancer | Volinia S [28] | Down | - |
| Fabbri M [29] | Down | DNMT3A/3B | |
| Morita S [30] | Down | TET1, TDG | |
| Barkley LR [31] | Down | Cdc7, Dbf4 | |
| Qu H [32] | Down | PMP22 | |
| Mizuno K [33] | Down | LOXL2 | |
| Breast cancer | Gebeshuber CA [34] | Up | TTP |
| Parpart S [35] | Down | Bcl-2, Mcl-1 | |
| Down | ADAM12-L | ||
| Zolkiewska A [36] | Down | ADAM12-L | |
| Duhachek-Muggy S [37] | Down | KLF4 | |
| Cittelly DM [38] | Down | NMI | |
| Zhang [39] | Down | CDC42 | |
| Liver cancer | Fang JH [40] | Down | MMP-2 |
| Lin [41] | Down | TET1 | |
| Xing TJ [42] | Down | - | |
| Stomach cancer | Gong J [43] | Down | CCND2, MMP-2 |
| Wang D [44] | Down | - | |
| Glioblastoma | Cortez MA [45] | Down | PDPN |
| Xu H [46] | Down | B7-H3 | |
| Prostate cancer | Peng Ru [47] | Down | - |
| Nishikawa [48] | Down | LAMC1 | |
| Mantle cell lymphoma | Dalton WS [49] | Down | CDK6 |
| Chronic myelogenous leukemia | Hassel [50] | Down | RNase-L |
| Nasopharyngeal carcinoma | Zeng X [51] | Down | - |
| Esophageal squamous cell cancer | Wang JW [52] | Down | cyclin E |
| Osteosarcoma | Qian JX [53] | Down | Bcl-2, Mcl-1, E2F1 |
| Cholangiocarcinoma | Mott JL [54] | Down | Mcl-1 |
| Bladder cancer | Xu XD [55] | Down | Bcl-2, Mcl-1 |
| Clear cell renal cell carcinoma | Atala A [56] | Down | LOXL2 |
| Colorectal cancer | Fu J [57] | Up | KLF4 |
| Inoue A [58] | Down | - | |
| Acute myeloid leukemia | Han YC [59] | Up | - |
| Garzon R [60] | Up | - | |
| Melanoma | Schmitt MJ [61] | Up | - |
| Diffuse large B cell lymphoma | Fang C [62] | Up | - |
| Rhabdomyosarcoma | Li L [15] | Down | CCND2, E2F7 |
| Balkhi MY [63] | Down | HuR, TNFAIP3 |
-; Not to mention.
Materials and methods
Patients and tissue samples
Formalin-fixed and paraffin-embedded tissue samples were retrieved from the database of the Department of Pathology of the First Affiliated Hospital of Shihezi University School of Medicine. The specimens included 16 RMS samples and 9 normal striated muscle samples. The size of the samples met the requirements of the basic experiment with certain representativeness. These samples were obtained from patients who were previously diagnosed through optical microscopy by at least two experienced pathologists in accordance with the WHO classification of tumors. We used the blind method, in which both pathologists were unaware of the original diagnosis of the pathology. Except for the above process, the samples were not included in our inclusion criteria. Informed consent was provided by all of the recruited patients. This study was approved by the Institutional Ethics Committee of the First Affiliated Hospital of Shihezi University School of Medicine and was conducted in accordance with the ethical guidelines of the Declaration of Helsinki.
Cell lines and transfection
We used five cell lines: three RMS cell lines, namely, RD (ERMS), RH30 (ARMS), and PLA802 (ARMS); human embryonal kidney (HEK) 293 cells; and human skeletal muscle satellite cells (HSkMSC). The cells were obtained from Biological Technology Co., Ltd. (Fu Xiang, Shanghai, China). RD, RH30, PLA802, HSkMSC, and HEK 293 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplied with 10% fetal bovine serum (FBS) (Gibco), 100 U/mL penicillin (Sigma), and 100 mg/mL streptomycin (Sigma). All cells were pollution-free and cultured in a humidified 5% CO2 atmosphere at 37°C. Mir-29a/b/c and control miRNA (miR-control) plasmids, 3’-untranslated regions (3’UTR) of wild-type human GEFT [3’UTR-GEFT-WT, GenBank-ID: NM-182947-3utr (mir-29a/b/c)], mutant-type GEFT [3’UTR-GEFT-MUT, GenBank-ID: NM-182947-3utr (mir-29a/b/c)-mut] plasmids, and pRL-TK Renilla plasmid were designed and synthesized by Genechem (Shanghai, China). An endo-free plasmid mini kit (Omega, USA, No. D6950-01) was used to extract the plasmids. Mir-29a/b/c mimic oligoribonucleotides were purchased from Genepharma (Shanghai, P.R. China). The RMS cells were transiently transfected with Lipofectamine 2000 (Invitrogen) for 48 h in accordance with the manufacturer’s protocol.
Quantitative real-time polymerase chain reaction (qRT-PCR) detection
Total RNA was isolated from the paraffin-embedded tissues and cell lines by using the miRNeasy FFPE kit and miRNeasy mini kit (Qiagen, Hilden, Germany, No. 217504 and 217004) according to the manufacturer’s protocol. The complementary DNA (cDNA) of miRNA or mRNA was generated with the miScript II RT kit (Qiagen, Hilden, Germany, No. 218161). qRT-PCR was performed using the miScript SYBR Green PCR kit (Qiagen, Hilden, Germany, No. 218075) or SYBR Green PCR Master Mix (Qiagen, Hilden, Germany) on a 7500 fast real-time PCR system (Applied Biosystems, South San Francisco, CA, USA). The parameters were set in accordance with the kit’s manual. U6 small RNA (U6) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA were used as internal controls. The primer sequences (5’-3’) were as follows: GAPDH-F, ACCCAGAAGACTGTGGATGG; and GAPDH-R, TCTAGACGGCAGGTCAGGTC; GEFT-F, AAACTGAGGCAGACAGTGGTC, and GEFT-R, TTCAGGTGGCTGCGTCTTAT. mir-29a/b/c and U6 primers were obtained from QuantiTect Primer Assays (Qiagen). All reactions were performed in triplicate.
Western blot analysis
Approximately 48 h after transfection in RMS cells, we used radio-immunoprecipitation assay lysis buffer (Solarbio, China) for cell lysis and protein extraction. The proteins were separated on sodium dodecyl sulfate-polyacrylamide gels, electrotransferred onto polyvinylidene fluoride membranes (Immobilon 0.22 μm, Millipore, USA), and immersed in blocking solution containing 5% nonfat milk and 0.1% Tween-20 for 1.5 h. After blocking, the membranes were incubated with GEFT (1:1000, Abcam, MA, USA, No. Ab127690) or β-actin (mouse-derived antibody, Zhongshan JinQiao, Beijing, China, No. IE9A3) at 4°C overnight. We then added the corresponding secondary antibodies (Zhongshan JinQiao, Beijing, China) and incubated the membranes for another 2 h at room temperature. Detection was performed by routine Western blot assay with an enhanced chemiluminescence kit (Thermo Fisher Scientific). All reactions were performed in triplicate.
Cell proliferation assays
We used the CCK-8-kit (Dojindo, Japan) to evaluate cell proliferation in accordance with the manufacturer’s protocol. Approximately 0, 24, 48, and 72 h after transfection, we recorded the absorbance of the solutions at 450 nm. All reactions were performed in triplicate.
Plate clone formation assay
After the RMS cells were transfected successfully at approximately 48 h, we counted 1 × 103 cells and cultivated them in six-well plates for 2 w. The cells were fixed with 4% paraformaldehyde for 20 min and stained with 0.1% crystal violet for 20 min. The number of colonies was counted after drying the plate. All reactions were performed in triplicate.
Cell migration and invasion assays
A 24-well plate containing 8 μm pore size chamber inserts (Costar, Corning, USA) was used to evaluate the migration and invasion of tumor cells in accordance with the manufacturer’s instructions. For the migration assay, 5 × 104 cells transfected for 48 h were seeded in the upper chamber. For the invasion assay, the membrane was coated with Matrigel to form a matrix barrier, and 2 × 105 cells transfected for 48 h were placed in the upper chamber. The cells in 100 μL of serum-free DMEM medium were carefully loaded onto each filter insert (upper chamber), whereas 600 μL of DMEM medium with 10% FBS was added in each lower chamber. The cells were incubated at 37°C for 24 and 48 h for the migration and invasion assays, respectively. The filter inserts were then removed from the chambers. The migrated and invaded cells were fixed and stained with 0.1% crystal violet for 20 min, counted, and photographed using an inverted microscope (Olympus, Japan). All reactions were performed in triplicate.
Apoptosis assay
The apoptosis of mir-29a/b/c was investigated using two methods, namely, terminal deoxy-nucleotidyl transferase dUTP nick end labeling (TUNEL, One Step TUNEL Apoptosis Assay Kit, #C1089; Beyotime, Jiangsu, China) and flow cytometry analysis (eBioscience, USA). We evaluated the apoptosis in RMS cells 48 h after transfection. For the TUNEL assay, the cells were treated with 4% paraform phosphate buffer saline, rinsed with phosphate-buffered saline (PBS), permeabilized by 0.1% Triton X-100 for 5 min, and incubated at 37°C for 1 h following the manufacturer’s protocol. Fluorescence microscopy with 488 nm excitation and 530 nm emission was used to detect positive cells. Red fluorescence was defined as positive apoptotic cells. For flow cytometry analysis, the cells were rinsed with prechilled PBS, digested, and counted. Approximately 1 × 106 cells were placed in a tube and added with 5 μL of annexin V-APC and 10 μL of 7-AAD according to the manufacturer’s instructions. Flow cytometry analysis was conducted to detect apoptosis. All reactions were performed in triplicate.
Wound-healing assay
About 48 h after transfection in six-well plates, we drew a trace on the cell surface with a sterile pipette. We then obtained photographs of the traces by using a microscope (Olympus, Japan) 0, 12, and 24 h after scratching. All reactions were performed in triplicate.
Luciferase reporter assay
We used HEK 293 cells for the luciferase reporter assay. The cells were cotransfected with 0.52 μg of plasmids containing 0.4 μg of mir-29a/b/c, 0.1 μg of 3’UTR-GEFT-MUT/WT of GEFT, and 0.02 μg of pRL-TK in Lipofectamine® 2000 Reagent (Invitrogen). Approximately 48 h after transfection in the 24-well plate, the dual-luciferase reporter assay system (Promega, USA, No. E1910) was used to measure luciferase activity with Renilla luciferase activity as internal reference. All experiments were performed in triplicate.
RNA immunoprecipitation (RIP)
The cells that were transfected successfully were added with 5 μg of Ago2 antibody, and RIP was conducted using the Magna RIP kit (Millipore) according to the manufacturer’s instructions. GEFT mRNA was analyzed by qRT-PCR to detect the interaction with mir-29a/b/c. All reactions were performed in triplicate.
Mouse xenografts
The mir-29a overexpressed RD cells were inoculated under the skin of the right side of 12 nude mice in the two groups (BALB/c nude mice, female, aged 4 weeks, 13-17 g in weight). The size of the samples met the requirements of the basic experiment and had a certain representativeness. The experimental and control groups were randomly assigned. Tumor formation was observed on the 9th day. The diameter of the tumor was measured every 2 days. The tumor volume was calculated, and the tumor growth curve was plotted. The body weight of nude mice was recorded. The tumor was removed and weighed on the 16th day of inoculation. The size of the nude mice after tumor formation was measured by persons unrelated to this experiment in accordance with the blind method principle.
Statistical analysis
SPSS 22.0 software was used for all statistical analyses. Student’s t-test was employed to determine significant differences between the two groups. All results were presented as mean ± standard deviation and met the assumptions of the tests. No estimate of variation was calculated within each group of data. p values < 0.05 were considered statistically significant. All figures were drawn using GraphPad Prism 7.0 (GraphPad Software, CA, USA).
Results
Mir-29 is downregulated in RMS tissues and cell lines
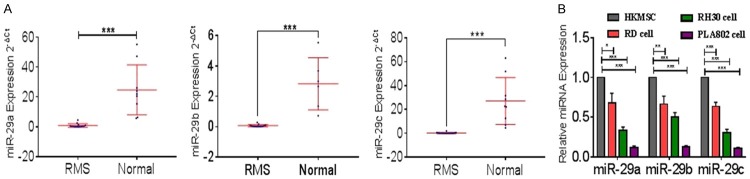
We reviewed the literature [15,63] and found that mir-29 was underexpressed in RMS. We measured mir-29 (mir-29a, mir-29b, and mir-29c) expression in paraffin-embedded tissues and RMS cell lines. Mir-29a, mir-29b, and mir-29c were detected in 15, 14, and 16 tumors and nine, six, and nine normal striated muscle tissues and 4 cell lines, respectively (Figure 1A and 1B).
Figure 1.
Mir-29 is downregulated in RMS tissues and cell lines. A. The expression levels of mir-29 in RMS and normal tissues were measured by qRT-PCR. B. The relative expression of miR-29 in RD, RH30, PLA802, and human skeletal muscle satellite cells were measured by qRT-PCR. ***P < 0.001 according to Student’s t-test; qRT-PCR results were normalized to those of U6.
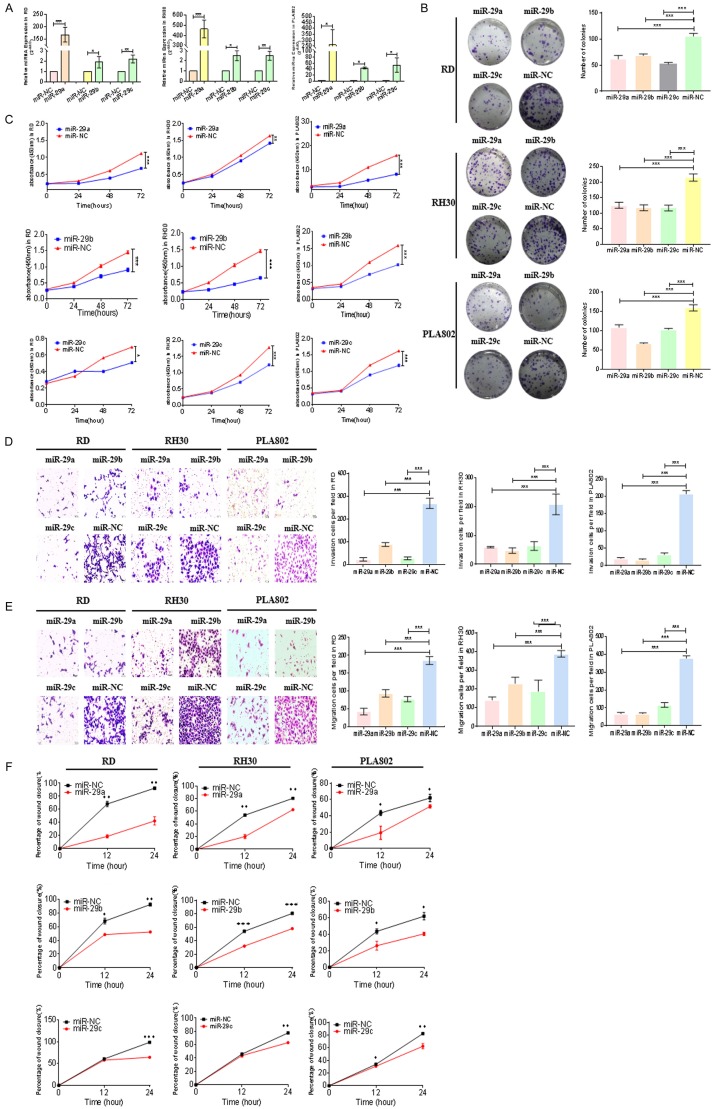
Mir-29 inhibits the proliferation, invasion, and migration in RMS cells
Although mir-29 (mir-29a, mir-29b, and mir-29c) was underexpressed in the samples we detected for RMS, its corresponding biological role is unknown. To clarify the function of mir-29 (mir-29a, mir-29b, and mir-29c), we transfected their mimics into RMS cells (RD, RH30, and PLA802) and detected miRNA expression via qRT-PCR. All miRNAs were overexpressed in the tested cell lines (Figure 2A). Experiments were performed to analyze the proliferative effects of mir-29 (mir-29a, mir-29b, and mir-29c) on RMS cells (RD, RH30, and PLA802). Plate clone formation assay revealed that mir-29 overexpression inhibited the proliferation of these three lines (Figure 2B). CCK-8 assay obtained the same results (Figure 2C). Therefore, mir-29 could inhibit the proliferation of RMS cells. We used Transwell models to analyze the invasion and migration abilities of RMS cells transfected with mir-29 (mir-29a, mir-29b, and mir-29c). Decreased numbers of invasive and migratory cells were found after transfection (Figure 2D and 2E). Wound-healing assay also indicated that mir-29 (mir-29a, mir-29b, and mir-29c) reduces the percentage of wound closure (Figure 2F and Supplementary Figures 1, 2, 3).
Figure 2.
Mir-29 inhibits the proliferation, invasion, and migration of RMS cells. (A) The MiRNA content was measured by qRT-PCR to verify the transfection of mir-29 mimics. (B) The number of colonies in RMS cells (RD, RH30, and PLA802) after treatment with mir-29 mimics were determined through cell cloning experiment. (C) The proliferative abilities of RMS cells (RD, RH30, and PLA802) transfected with mir-29 mimics were determined through CCK8 assay. Transwell systems were used to detect the effect of (D) invasion and (E) migration in RMS cells (RD, RH30, and PLA802) after mir-29 mimic treatment. (F) Effect of mir-29 mimic treatment on the healing ability of RMS cells (RD, RH30, and PLA802). ***P < 0.001 according to Student’s t-test.
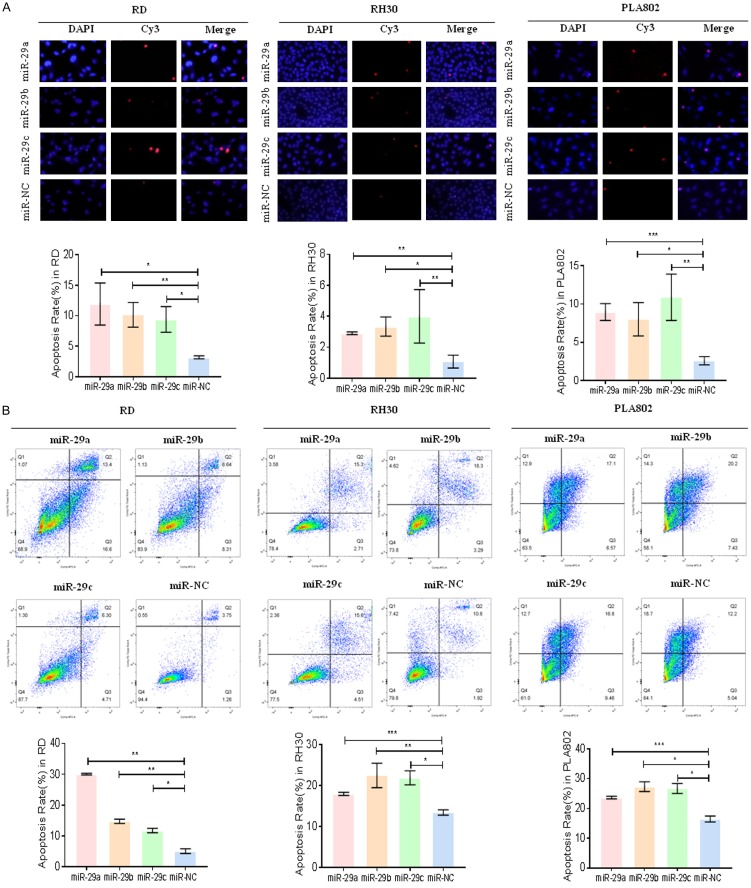
Mir-29 promotes apoptosis in RMS cells
After transfecting the mimics at approximately 48 h, we performed TUNEL and flow cytometry analyses to evaluate apoptosis. As shown in Figure 3A, the number of positive apoptotic cells overexpressing mir-29 (mir-29a, mir-29b, and mir-29c) increased compared with that in the control. Flow cytometry analysis demonstrated the same result (Figure 3B).
Figure 3.
Mir-29 promotes RMS cell apoptosis. A. The situations of apoptosis of RD, RH30, and PLA802 cells after treatment with mir-29 mimics were monitored by TUNEL analysis. B. The results of apoptosis of mir-29 mimic treatment on the flow cytometry of RMS cells (RD, RH30, and PLA802). *P < 0.05 and **P < 0.01 according to Student’s t-test.
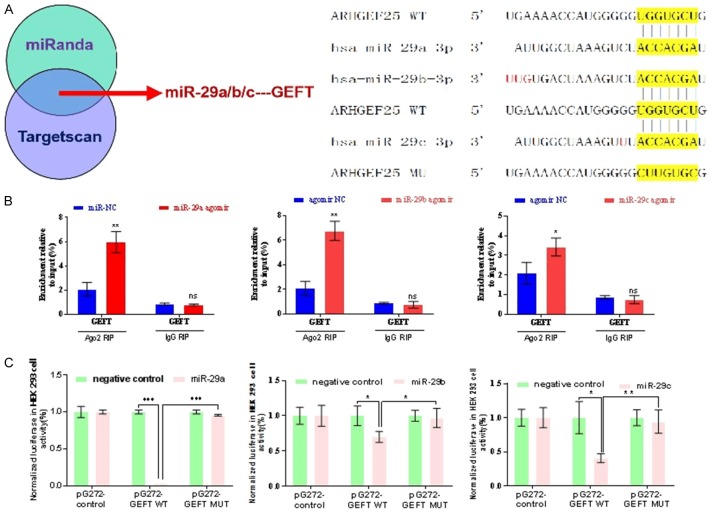
GEFT is targeted by mir-29
We used Targetscan (http://www.targetscan.org/vert_71) and miRanda (http://miranda.org.uk/) to predict the target gene of mir-29 (mir-29a, mir-29b, and mir-29c), and the identified gene is GEFT (Figure 4A). To validate the bioinformatics approach, we used RIP assay and showed that mir-29a, mir-29b, and mir-29c directly repress GEFT (Figure 4B). Luciferase reporter assay results also showed that mir-29 (mir-29a, mir-29b, and mir-29c) likely suppress the luciferase activity of 3’UTR-GEFT-WT (P = 0.044) (Figure 4C).
Figure 4.
GEFT is targeted by mir-29. A. Targetscan and miRanda were used for target gene prediction and schematic of the binding site between mir-29 and 3’UTR of GEFT. B. RIP assay revealed the direct association between mir-29 and GEFT in RH30 cells. C. The binding site between mir-29 and 3’UTR of GEFT was verified by luciferase reporter assay. *P < 0.05 and **P < 0.01 according to Student’s t-test.
Mir-29 overexpression inhibits GEFT levels at mRNA and protein levels
We verified the expression level of GEFT in RMS tissues. Samples from clinical trials were used in the experiment to measure the mRNA expression of GEFT, and high levels of expression were found (Figure 5A). A negative correlation was found between mir-29 (mir-29a, mir-29b, and mir-29c) level and GEFT mRNA expression in RMS tissues (Figure 5B). We also detected the mRNA and protein expression of GEFT when mir-29 (mir-29a, mir-29b and mir-29c) was exogenously upregulated and found that mir-29 (mir-29a, mir-29b, and mir-29c) overexpression could suppress the expression of GEFT mRNA (Figure 5C) and protein (Figure 5D) in cell lines.
Figure 5.
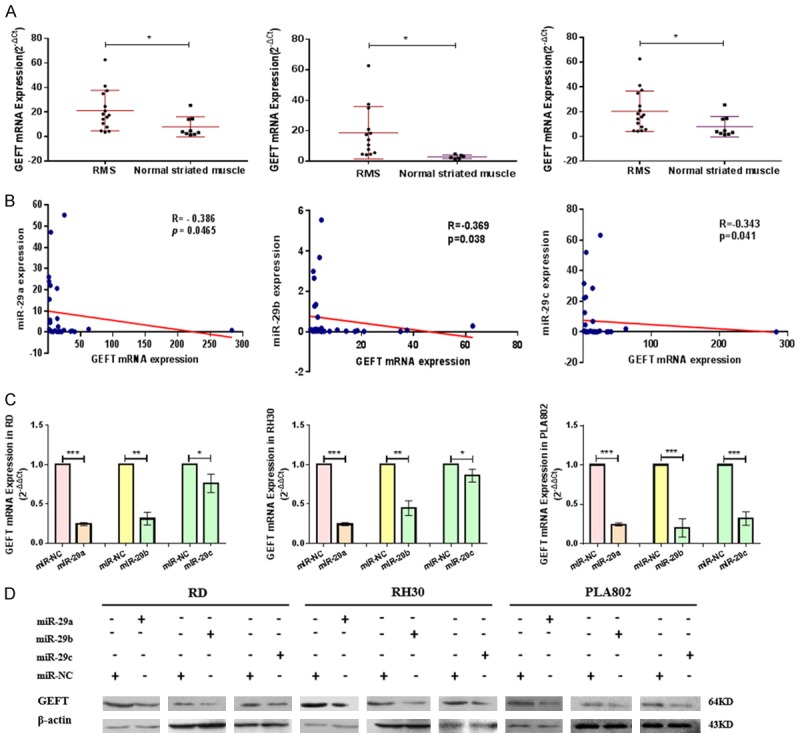
Mir-29 overexpression inhibits GEFT levels at mRNA and protein levels. A. The relative expression of GEFT in RMS tissues was measured by qRT-PCR. B. The correlation analysis of mir-29 and GEFT mRNA in RMS tissues was performed via Spearman’s correlation analysis. C. The expression levels of GEFT mRNA in RD, RH30, and PLA802 after treatment with mir-29 mimics were measured by qRT-PCR. D. The expression levels of GEFT protein in RD, RH30, and PLA802 after treatment with mir-29 mimics were measured by Western blot. *P < 0.05 and **P < 0.01 according to Student’s t-test.
Mir-29 mediates GEFT to restrain the biological functions in RMS cell
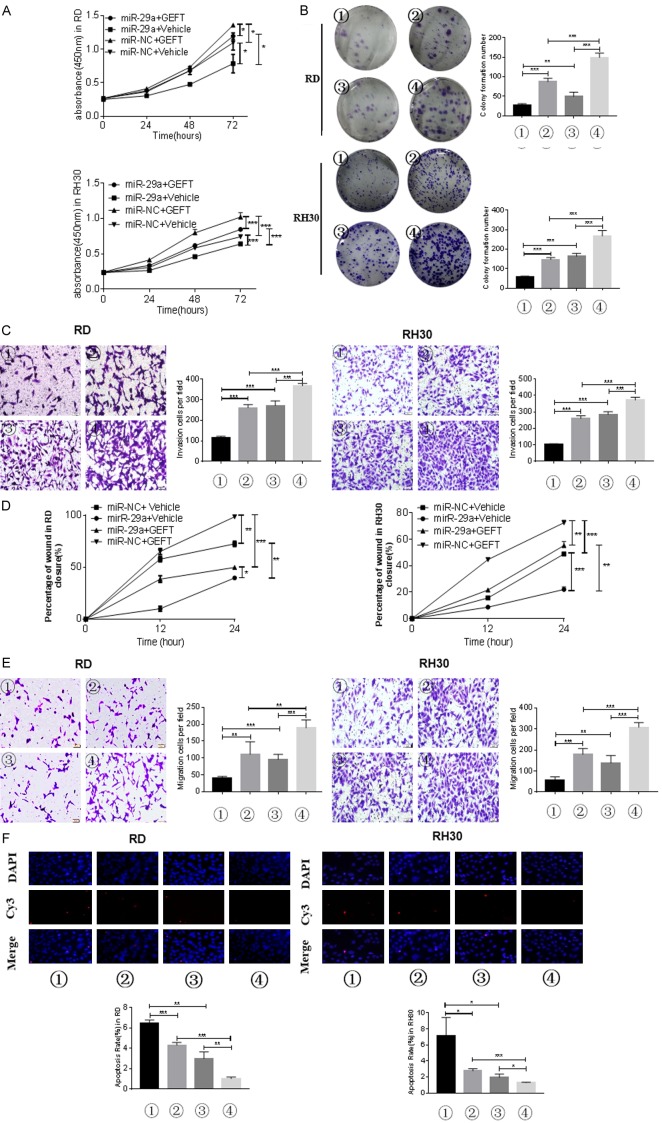
To verify whether the biological function of mir-29 on RMS cell tumorigenesis is mediated by GEFT, we employed cells stably overexpressing GEFT. Western blot was performed to verify the success of this approach (Supplementary Figure 4A). After transfecting the mir-29 (mir-29a, mir-29b, and mir-29c) mimics into the two cell lines (RD and RH30), GEFT protein was detected by Western blot. Mir-29 (mir-29a, mir-29b, and mir-29c) overexpression reversed the expression of GEFT protein in GEFT/vehicle-RD and GEFT/vehicle-RH30 cells (Supplementary Figure 4B). We applied CCK-8 assay and found that the optical density in the group treated with both mir-29a and GEFT at the same time was higher than that in the group treated with mir-29a alone but lower than that in the group treated with GEFT alone (Figure 6A). Plate clone formation assay revealed similar results (Figure 6B). We thus believe that mir-29a could reduce the effect of GEFT on the proliferation of RMS cells. Transwell system assay was used to assess cell invasion and migration. The numbers of invasive cells in the group treated with both mir-29a and GEFT were higher than those in the group treated with mir-29a alone but lower than those in the group treated with GEFT alone (Figure 6C). Moreover, the number of migrated cells increased in the group treated with GEFT alone and decreased in the group treated with mir-29a alone compared with that in the group treated with both mir-29a and GEFT (Figure 6E). Wound-healing assay revealed similar results (Figure 6D and Supplementary Figure 5). Finally, TUNEL staining and flow cytometry analyses showed that the apoptosis rate increased in the group treated with GEFT alone and decreased in the group treated with mir-29a alone compared with that in the group treated with both mir-29a and GEFT (Figure 6F). Similar results were observed with mir-29b and mir-29c (Supplementary Figures 6, 7, 8, 9, 10, 11). Thus, mir-29 (mir-29a, mir-29b, and mir-29c) could attenuate the effect of GEFT on the migration, invasion, and apoptosis of RMS cells.
Figure 6.
Mir-29a mediates GEFT to restrain the biological functions in RMS cell. A. The proliferative abilities of RMS cells (RD and RH30) transfected with mir-29a and GEFT were determined through CCK8 assay. B. The number of colonies of RMS cells (RD and RH30) transfected with mir-29a and GEFT were determined through cell cloning experiment. C. Transwell systems were used to detect the effect of invasion in RMS cells (RD and RH30) after being transfected with mir-29a and GEFT. D. The healing ability was detected to determine the effect of migration in RMS cells (RD and RH30) after being transfected with mir-29a and GEFT. E. Transwell systems were used to detect the effect of migration in RMS cells (RD and RH30) after being transfected with mir-29a and GEFT. F. The results of RD and RH30 cell apoptosis after treatment with mir-29a and GEFT by TUNEL analysis. *P < 0.05, **P < 0.01, and ***P < 0.001 according to Student’s t-test. ① mir-29a+Vehicle, ② mir-NC+Vehicle, ③ mir-29a+GEFT, and ④ mir-NC+GEFT.
Mir-29a inhibits the tumorigenesis of subcutaneous xenograft in nude mice
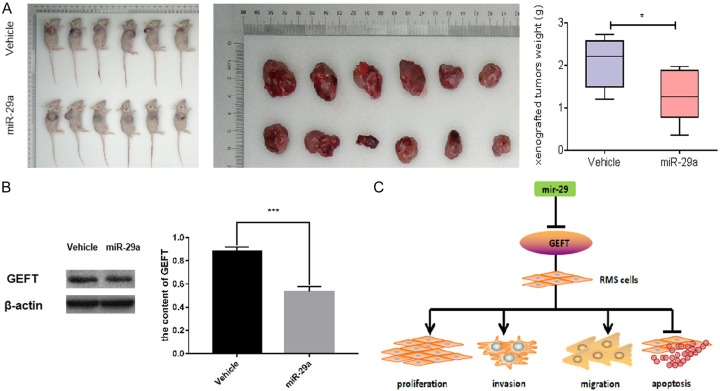
Our studies revealed that the mir-29a expression level in RMS cells and tissues and its effect on some biological behaviors in cells are more obvious than the mir-29b/c expression level. Therefore, we constructed a tumor formation model in nude mice to explore the role of mir-29a further. We inoculated the constructed mir-29a-overexpressing RD cells into the right armpit of nude mice to observe the effect of this molecule on the tumorigenic ability of the xenografts and then detected GEFT expression in the transplanted tumor tissues. The weights of solid tumors in the inoculated group were significantly smaller than those in the control group (Figure 7A). Western blot revealed that the expression level of GEFT protein in the xenograft tumors of the mir-29a-overexpressing RD cell group was lower than that in the control group (Figure 7B). The role of mir-29a in xenograft tumors in nude mice provides further evidence that the mir-29 family includes tumor suppressor genes targeting GEFT.
Figure 7.
Mir-29a inhibits the tumorigenesis of subcutaneous xenograft in nude mice. A. Comparison of tumor volume and weight in mice treated with RD cells overexpressing mir-29a in the control groups. B. Western blot results revealed that the GEFT protein expression with subcutaneous xenograft in nude mice treated with RD cells overexpressing mir-29a and the control group. C. The mechanism of mir-29 targeting GEFT to affect proliferation, invasion, migration, and apoptosis of rhabdomyosarcoma. *P < 0.05 according to Student’s t-test.
Discussion
In this study, we mainly explored the functional significance of the mir-29 family, which is composed of mir-29a, mir-29b, and mir-29c. Mir-29b includes two subfamily members, namely, mir-29b-1 and mir-29b-2, in which mir-29b-1/mir-29a and mir-29b-2/mir-29c are located at chromosomes 1q32.2 and 7q32.3, respectively. Mir-29 is involved in the regulation of various normal physiological activities of the body, such as insulin secretion and cell aging [64,65]. A recent study reported the presence of mir-29 in RMS [15] and its promotion of skeletal muscle cell differentiation [17]. Mir-29 is induced in a nuclear factor kappa B (NF-κB)-dependent manner in the absence of a myogene and promotes skeletal muscle differentiation [66]. NF-κB activation in RMS leads to the overexpression of YY1, which interacts with EZH2, thereby downregulating mir-29b/mir-29c and suppressing myogenesis [66]. YY1 recruits EZH2 to a specific site of the mir-29b/mir-29c promoter that is different from that used during the expansion of normal myoblasts. The mir-29 pathways in RMS may depend on the effects of miRNAs on the expression of Pax3 and proteins regulating the cell cycle, such as CCND2 [15]. Mir-29 also targets the cell cycle regulator E2F7 and regulates cell proliferation [67]. Therefore, mir-29 does affect the occurrence and development of RMS.
To date, the term “epi-miRNAs” is proposed to represent a class of specific tumor suppressor miRNAs that can reduce the expression of epigenetic enzymes and affect the expression of tumor suppressor genes [68]. Mir-29b is an epi-miRNA that can target DNA methyltransferase (DNMTs) and/or regulate members of the DNA demethylation pathway, leading to overall downregulation of DNA methylation in malignant tumor cells [68]. Mir-29b overexpression in acute leukemia cells inhibits the mRNA and protein expression levels of DNMT1, DNMT3B, and DNMT3A. Mir-29b can also target Sp1 and inhibit DNMT1, leading to downregulation of DNA methylation in malignant tumor cells and deletion of the expression of the tumor suppressor gene p15 (INK4b) and ESR1 [69]. The abnormal expression of mir-29 regulates the cell cycle and inhibits apoptosis and DNA methylation in Burkitt’s lymphoma. Mir-29 methylation-mediated epigenetic silencing may also occur in Burkitt’s lymphoma [70]. Further research must seek to determine whether mir-29 is also methylated in RMS.
GEFT is the target gene of mir-29 that we predicted and verified. It is also called ARHGEF25 or p63RhoGEF and is located on chromosome 12q13.3 and belongs to the Rho guanine nucleotide exchange factor (GEF) family [71,72]. GEFT can catalyze Rho GTPases and activate the bound guanosine diphosphates for guanosine triphosphates on Rho proteins, thereby activating these proteins and their downstream targets [73,74]. In our previous study on RMS, we conducted high-resolution array comparative genomic hybridization and found that GEFT overexpression is related to the degree of tumor differentiation. GEFT overexpression in RMS tissues is correlated with poor prognosis [75,76]. The gene may function as an oncogene [75].
In summary, the present work successfully demonstrated that mir-29 targets GEFT to inhibit the proliferation, migration, invasion, and apoptosis of RMS cells (Figure 7C). The results provide a preliminary basis for studies on RMS at the miRNA level and subsequent related research. This study is the first to report that mir-29 targets GEFT expression and is necessary to inhibit the proliferation, migration, invasion, and promote apoptosis of RMS cells. Mir-29 overexpression can effectively destroy the biological behavior of RMS cells. Our research also provides a basis for further research on the diagnosis and treatment of RMS using mir-29 and GEFT as targets. Further studies must be conducted to determine the mechanism of maladjustment of mir-29 expression in RMS. A larger sample size must be employed to determine the effect of mir-29 on RMS. Whether Mir-29a/b/c elicits the observed biological behavior by directly acting on GEFT and its effect on the pathogenesis of RMS must be explored deeply.
Conclusions
We elucidated a new mechanism of RMS pathogenesis. Our data demonstrated the targeting effect of mir-29a/b/c on the GEFT-regulated proliferation, migration, invasion, and apoptosis of RMS cells. Mir-29a/b/c, as a tumor suppressor gene, may provide a new basis for the targeted treatment and evaluation of the prognosis of RMS.
Acknowledgements
The present study was supported by grants from the National Natural Science Foundation of China (grant nos. 81660441 and 81460404).
Disclosure of conflict of interest
None.
Abbreviations
- RMS
rhabdomyosarcoma
- GEFT
Guanine nucleotide exchange factor T
- mir-29
microRNA-29
- WHO
World Health Organization
- ERMS
embryonal rhabdomyosarcoma
- ARMS
alveolar rhabdomyosarcoma
- PRMS
pleomorphic rhabdomyosarcoma
- microRNAs
miRNAs
- YY1
Yin Yang 1
- NF-κB
nuclear factor kappa B
- qRT-PCR
quantitative real-time polymerase chain reaction
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HSkMSC
human skeletal muscle satellite cells
- cDNA
complementary DNA
- PBS
phosphate-buffered saline
- RIP
RNA immunoprecipitation
Supporting Information
References
- 1.Wexler LH, Meyer W, Helman L. Rhabdomyosarcoma. In: Pizzo P, Poplack D, editors. Principles and Practice of Pediatric Oncology. 2011. pp. 923–953. [Google Scholar]
- 2.Fletcher C, Bridge JA, Hogendoorn P, Mertens F. World Health Organization classification of tumours of soft tissue and bone. 4th edition. Lyon: IARC Press; 2013. [Google Scholar]
- 3.Oberlin O, Rey A, Sanchez de Toledo J, Martelli H, Jenney ME, Scopinaro M, Bergeron C, Merks JH, Bouvet N, Ellershaw C, Kelsey A, Spooner D, Stevens MC. Randomized comparison of intensified six-drug versus standard three-drug chemotherapy for high-risk nonmetastatic rhabdomyosarcoma and other chemotherapy-sensitive childhood soft tissue sarcomas: long-term results from the International Society of pediatric oncology MMT95 study. J. Clin. Oncol. 2012;30:2457–2465. doi: 10.1200/JCO.2011.40.3287. [DOI] [PubMed] [Google Scholar]
- 4.Carli M, Colombatti R, Oberlin O, Bisogno G, Treuner J, Koscielniak E, Tridello G, Garaventa A, Pinkerton R, Stevens M. European intergroup studies (MMT4-89 and MMT4-91) on childhood metastatic rhabdomyosarcoma: final results and analysis of prognostic factors. J. Clin. Oncol. 2004;22:4787–4794. doi: 10.1200/JCO.2004.04.083. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.Kinoshita T, Hanazawa T, Nohata N, Okamoto Y, Seki N. The functional significance of microRNA-375 in human squamous cell carcinoma: aberrant expression and effects on cancer pathways. J Hum Genet. 2012;57:556–563. doi: 10.1038/jhg.2012.75. [DOI] [PubMed] [Google Scholar]
- 8.Pillai RS, Bhattacharyya SN, Filipowicz W. Repression of protein synthesis by miRNAs: how many mechanisms? Trends Cell Biol. 2007;17:118–126. doi: 10.1016/j.tcb.2006.12.007. [DOI] [PubMed] [Google Scholar]
- 9.Nana-Sinkam SP, Croce CM. MicroRNA regulation of tumorigenesis, cancer progression and interpatient heterogeneity: towards clinical use. Genome Biol. 2014;15:445. doi: 10.1186/s13059-014-0445-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes J, Peruzzi PP, Lawler S. MicroRNAs in cancer: biomarkers, functions and therapy. Trends Mol Med. 2014;20:460–469. doi: 10.1016/j.molmed.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 11.Auyeung VC, Ulitsky I, McGeary SE, Bartel DP. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Missiaglia E, Shepherd CJ, Patel S, Thway K, Pierron G, Pritchard-Jones K, Renard M, Sciot R, Rao P, Oberlin O, Delattre O, Shipley J. MicroRNA-206 expression levels correlate with clinical behaviour of rhabdomyosarcomas. Br J Cancer. 2010;102:1769–1777. doi: 10.1038/sj.bjc.6605684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taulli R, Bersani F, Foglizzo V, Linari A, Vigna E, Ladanyi M, Tuschl T, Ponzetto C. The muscle-specific microRNA miR-206 blocks human rhabdomyosarcoma growth in xenotransplanted mice by promoting myogenic differentiation. J Clin Invest. 2009;119:2366–2378. doi: 10.1172/JCI38075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang HJ, Liu J, Hua H, Li SE, Zhao J, Yue S, Yu TT, Jin YC, Cheng SY. MiR-214 and N-ras regulatory loop suppresses rhabdomyosarcoma cell growth and xenograft tumorigenesis. Oncotarget. 2014;5:2161–2175. doi: 10.18632/oncotarget.1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Sarver AL, Alamgir S, Subramanian S. Downregulation of microRNAs miR-1, -206 and -29 stabilizes PAX3 and CCND2 expression in rhabdomyosarcoma. Lab Invest. 2012;92:571–583. doi: 10.1038/labinvest.2012.10. [DOI] [PubMed] [Google Scholar]
- 16.Sugiyama T, Taniguchi K, Matsuhashi N, Tajirika T, Futamura M, Takai T, Akao Y, Yoshida K. MiR-133b inhibits growth of human gastric cancer cells by silencing pyruvate kinase muscle-splicer polypyrimidine tract-binding protein 1. Cancer Sci. 2016;107:1767–1775. doi: 10.1111/cas.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winbanks CE, Wang B, Beyer C, Koh P, White L, Kantharidis P, Gregorevic P. TGF-beta regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J Biol Chem. 2011;286:13805–13814. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou L, Wang L, Lu L, Jiang P, Sun H, Wang H. A novel target of microRNA-29, Ring1 and YY1-binding protein (Rybp), negatively regulates skeletal myogenesis. J Biol Chem. 2012;287:25255–25265. doi: 10.1074/jbc.M112.357053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugito N, Taniguchi K, Kuranaga Y, Ohishi M, Soga T, Ito Y, Miyachi M, Kikuchi K, Hosoi H, Akao Y. Cancer-specific energy metabolism in rhabdomyosarcoma cells is regulated by microRNA. Nucleic Acid Ther. 2017;27:365–377. doi: 10.1089/nat.2017.0673. [DOI] [PubMed] [Google Scholar]
- 20.Wong CF, Tellam RL. MicroRNA-26a targets the histone methyltransferase enhancer of zeste homolog 2 during myogenesis. J Biol Chem. 2008;283:9836–9843. doi: 10.1074/jbc.M709614200. [DOI] [PubMed] [Google Scholar]
- 21.Hirai H, Verma M, Watanabe S, Tastad C, Asakura Y, Asakura A. MyoD regulates apoptosis of myoblasts through microRNA-mediated down-regulation of Pax3. J Cell Biol. 2010;191:347–365. doi: 10.1083/jcb.201006025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun MM, Li JF, Guo LL, Xiao HT, Dong L, Wang F, Huang FB, Cao D, Qin T, Yin XH, Li JM, Wang SL. TGF-beta1 suppression of microRNA-450b-5p expression: a novel mechanism for blocking myogenic differentiation of rhabdomyosarcoma. Oncogene. 2014;33:2075–2086. doi: 10.1038/onc.2013.165. [DOI] [PubMed] [Google Scholar]
- 23.Sun M, Huang F, Yu D, Zhang Y, Xu H, Zhang L, Li L, Dong L, Guo L, Wang S. Autoregulatory loop between TGF-beta1/miR-411-5p/SPRY4 and MAPK pathway in rhabdomyosarcoma modulates proliferation and differentiation. Cell Death Dis. 2015;6:e1859. doi: 10.1038/cddis.2015.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Diao Y, Guo X, Jiang L, Wang G, Zhang C, Wan J, Jin Y, Wu Z. miR-203, a tumor suppressor frequently down-regulated by promoter hypermethylation in rhabdomyosarcoma. J Biol Chem. 2014;289:529–539. doi: 10.1074/jbc.M113.494716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Megiorni F, Cialfi S, McDowell HP, Felsani A, Camero S, Guffanti A, Pizer B, Clerico A, De Grazia A, Pizzuti A, Moles A, Dominici C. Deep sequencing the microRNA profile in rh-abdomyosarcoma reveals down-regulation of miR-378 family members. BMC Cancer. 2014;14:880. doi: 10.1186/1471-2407-14-880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarver AL, Li L, Subramanian S. MicroRNA miR-183 functions as an oncogene by targeting the transcription factor EGR1 and promoting tumor cell migration. Cancer Res. 2010;70:9570–9580. doi: 10.1158/0008-5472.CAN-10-2074. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen T, Kuo C, Nicholl MB, Sim MS, Turner RR, Morton DL, Hoon DS. Downregulation of microRNA-29c is associated with hypermethylation of tumor-related genes and disease outcome in cutaneous melanoma. Epigenetics. 2011;6:388–394. doi: 10.4161/epi.6.3.14056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, Volinia S, Guler G, Morrison CD, Chan KK, Marcucci G, Calin GA, Huebner K, Croce CM. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morita S, Horii T, Kimura M, Ochiya T, Tajima S, Hatada I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int J Mol Sci. 2013;14:14647–14658. doi: 10.3390/ijms140714647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barkley LR, Santocanale C. MicroRNA-29a regulates the benzo[a]pyrene dihydrodiol epoxide-induced DNA damage response through Cdc7 kinase in lung cancer cells. Oncogenesis. 2013;2:e57. doi: 10.1038/oncsis.2013.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qu H, Zhu M, Tao Y, Zhao Y. Suppression of peripheral myelin protein 22 (PMP22) expression by miR29 inhibits the progression of lung cancer. Neoplasma. 2015;62:881–886. doi: 10.4149/neo_2015_107. [DOI] [PubMed] [Google Scholar]
- 33.Mizuno K, Seki N, Mataki H, Matsushita R, Kamikawaji K, Kumamoto T, Takagi K, Goto Y, Nishikawa R, Kato M, Enokida H, Nakagawa M, Inoue H. Tumor-suppressive microRNA-29 family inhibits cancer cell migration and invasion directly targeting LOXL2 in lung squamous cell carcinoma. Int J Oncol. 2016;48:450–460. doi: 10.3892/ijo.2015.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gebeshuber CA, Zatloukal K, Martinez J. miR-29a suppresses tristetraprolin, which is a regulator of epithelial polarity and metastasis. EMBO Rep. 2009;10:400–405. doi: 10.1038/embor.2009.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parpart S, Roessler S, Dong F, Rao V, Takai A, Ji J, Qin LX, Ye QH, Jia HL, Tang ZY, Wang XW. Modulation of miR-29 expression by alpha-fetoprotein is linked to the hepatocellular carcinoma epigenome. Hepatology. 2014;60:872–883. doi: 10.1002/hep.27200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Solomon E, Duhachek Muggy S, Sun D, Zolkiewska A. Metalloprotease-disintegrin ADAM12 expression is regulated by notch signaling via microRNA-29. J Biol Chem. 2011;286:21500–21510. doi: 10.1074/jbc.M110.207951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Duhachek-Muggy S, Zolkiewska A. ADA-M12-L is a direct target of the miR-29 and miR-200 families in breast cancer. BMC Cancer. 2015;15:93. doi: 10.1186/s12885-015-1108-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cittelly DM, Finlay-Schultz J, Howe EN, Spoelstra NS, Axlund SD, Hendricks P, Jacobsen BM, Sartorius CA, Richer JK. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene. 2013;32:2555–2564. doi: 10.1038/onc.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang M, Guo W, Qian J, Wang B. Negative regulation of CDC42 expression and cell cycle progression by miR-29a in breast cancer. Open Med (Wars) 2016;11:78–82. doi: 10.1515/med-2016-0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang JH, Zhou HC, Zeng C, Yang J, Liu Y, Huang X, Zhang JP, Guan XY, Zhuang SM. MicroRNA-29b suppresses tumor angio-genesis, invasion, and metastasis by regulating matrix metalloproteinase 2 expression. Hepatology. 2011;54:1729–1740. doi: 10.1002/hep.24577. [DOI] [PubMed] [Google Scholar]
- 41.Lin LL, Wang W, Hu Z, Wang LW, Chang J, Qian H. Negative feedback of miR-29 family TET1 involves in hepatocellular cancer. Med Oncol. 2014;31:291. doi: 10.1007/s12032-014-0291-2. [DOI] [PubMed] [Google Scholar]
- 42.Xing TJ, Jiang DF, Huang JX, Xu ZL. Expression and clinical significance of miR-122 and miR-29 in hepatitis B virus-related liver disease. Genet Mol Res. 2014;13:7912–7918. doi: 10.4238/2014.September.29.4. [DOI] [PubMed] [Google Scholar]
- 43.Gong J, Li J, Wang Y, Liu C, Jia H, Jiang C, Wang Y, Luo M, Zhao H, Dong L, Song W, Wang F, Wang W, Zhang J, Yu J. Characterization of microRNA-29 family expression and investigation of their mechanistic roles in gastric cancer. Carcinogenesis. 2014;35:497–506. doi: 10.1093/carcin/bgt337. [DOI] [PubMed] [Google Scholar]
- 44.Wang D, Fan Z, Liu F, Zuo J. Hsa-miR-21 and Hsa-miR-29 in tissue as potential diagnostic and prognostic biomarkers for gastric cancer. Cell Physiol Biochem. 2015;37:1454–1462. doi: 10.1159/000438514. [DOI] [PubMed] [Google Scholar]
- 45.Cortez MA, Nicoloso MS, Shimizu M, Rossi S, Gopisetty G, Molina JR, Carlotti C Jr, Tirapelli D, Neder L, Brassesco MS, Scrideli CA, Tone LG, Georgescu MM, Zhang W, Puduvalli V, Calin GA. miR-29b and miR-125a regulate podoplanin and suppress invasion in glioblastoma. Genes Chromosomes Cancer. 2010;49:981–990. doi: 10.1002/gcc.20808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu H, Cheung IY, Guo HF, Cheung NK. MicroRNA miR-29 modulates expression of immunoinhibitory molecule B7-H3: potential implications for immune based therapy of human solid tumors. Cancer Res. 2009;69:6275–6281. doi: 10.1158/0008-5472.CAN-08-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ru P, Newhall P, Ray RB, Phillips NJ, Toth K, Steele R. miRNA-29b suppresses prostate cancer metastasis by regulating epithelial-mesenchymal transition signaling. Mol Cancer Ther. 2012;11:1166–1173. doi: 10.1158/1535-7163.MCT-12-0100. [DOI] [PubMed] [Google Scholar]
- 48.Nishikawa R, Goto Y, Kojima S, Enokida H, Chiyomaru T, Kinoshita T, Sakamoto S, Fuse M, Nakagawa M, Naya Y, Ichikawa T, Seki N. Tumor-suppressive microRNA-29s inhibit cancer cell migration and invasion via targeting LAMC1 in prostate cancer. Int J Oncol. 2014;45:401–410. doi: 10.3892/ijo.2014.2437. [DOI] [PubMed] [Google Scholar]
- 49.Zhao JJ, Lin J, Lwin T, Yang H, Guo J, Kong W, Dessureault S, Moscinski LC, Rezania D, Dalton WS, Sotomayor E, Tao J, Cheng JQ. microRNA expression profile and identification of miR-29 as a prognostic marker and pathogenetic factor by targeting CDK6 in mantle cell lymphoma. Blood. 2010;115:2630–2639. doi: 10.1182/blood-2009-09-243147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee TY, Ezelle HJ, Venkataraman T, Lapidus RG, Scheibner KA, Hassel BA. Regulation of human RNase-L by the miR-29 family reveals a novel oncogenic role in chronic myelogenous leukemia. J Interferon Cytokine Res. 2013;33:34–42. doi: 10.1089/jir.2012.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zeng X, Xiang J, Wu M, Xiong W, Tang H, Deng M, Li X, Liao Q, Su B, Luo Z, Zhou Y, Zhou M, Zeng Z, Li X, Shen S, Shuai C, Li G, Fang J, Peng S. Circulating miR-17, miR-20a, miR-29c, and miR-223 combined as non-invasive biomarkers in nasopharyngeal carcinoma. PLoS One. 2012;7:e46367. doi: 10.1371/journal.pone.0046367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ding DP, Chen ZL, Zhao XH, Wang JW, Sun J, Wang Z, Tan FW, Tan XG, Li BZ, Zhou F, Shao K, Li N, Qiu B, He J. miR-29c induces cell cycle arrest in esophageal squamous cell carcinoma by modulating cyclin E expression. Carcinogenesis. 2011;32:1025–1032. doi: 10.1093/carcin/bgr078. [DOI] [PubMed] [Google Scholar]
- 53.Zhang W, Qian JX, Yi HL, Yang ZD, Wang CF, Chen JY, Wei XZ, Fu Q, Ma H. The microRNA-29 plays a central role in osteosarcoma pathogenesis and progression. Mol Biol (Mosk) 2012;46:622–7. [PubMed] [Google Scholar]
- 54.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu XD, Wu XH, Fan YR, Tan B, Quan Z, Luo CL. Exosome-derived microRNA-29c induces apoptosis of BIU-87 cells by down regulating BCL-2 and MCL-1. Asian Pac J Cancer Prev. 2014;15:3471–3476. doi: 10.7314/apjcp.2014.15.8.3471. [DOI] [PubMed] [Google Scholar]
- 56.Atala A. Re: tumour-suppressive microRNA-29s directly regulate LOXL2 expression and inhibit cancer cell migration and invasion in renal cell carcinoma. J Urol. 2016;195:1622. doi: 10.1016/j.juro.2016.02.003. [DOI] [PubMed] [Google Scholar]
- 57.Fu J, Tang W, Du P, Wang G, Chen W, Li J, Zhu Y, Gao J, Cui L. Identifying microRNA-mRNA regulatory network in colorectal cancer by a combination of expression profile and bioinformatics analysis. BMC Syst Biol. 2012;6:68. doi: 10.1186/1752-0509-6-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Inoue A, Yamamoto H, Uemura M, Nishimura J, Hata T, Takemasa I, Ikenaga M, Ikeda M, Murata K, Mizushima T, Doki Y, Mori M. MicroRNA-29b is a novel prognostic marker in colorectal cancer. Ann Surg Oncol. 2015;22(Suppl 3):S1410–1418. doi: 10.1245/s10434-014-4255-8. [DOI] [PubMed] [Google Scholar]
- 59.Han YC, Park CY, Bhagat G, Zhang J, Wang Y, Fan JB, Liu M, Zou Y, Weissman IL, Gu H. microRNA-29a induces aberrant self-renewal capacity in hematopoietic progenitors, biased myeloid development, and acute myeloid leukemia. J Exp Med. 2010;207:475–489. doi: 10.1084/jem.20090831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garzon R, Garofalo M, Martelli MP, Briesewitz R, Wang L, Fernandez-Cymering C, Volinia S, Liu CG, Schnittger S, Haferlach T, Liso A, Diverio D, Mancini M, Meloni G, Foa R, Martelli MF, Mecucci C, Croce CM, Falini B. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci U S A. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmitt MJ, Philippidou D, Reinsbach SE, Margue C, Wienecke-Baldacchino A, Nashan D, Behrmann I, Kreis S. Interferon-gamma-induced activation of Signal Transducer and Activator of Transcription 1 (STAT1) up-regulates the tumor suppressing microRNA-29 family in melanoma cells. Cell Commun Signal. 2012;10:41. doi: 10.1186/1478-811X-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fang C, Zhu DX, Dong HJ, Zhou ZJ, Wang YH, Liu L, Fan L, Miao KR, Liu P, Xu W, Li JY. Serum microRNAs are promising novel biomarkers for diffuse large B cell lymphoma. Ann Hematol. 2012;91:553–559. doi: 10.1007/s00277-011-1350-9. [DOI] [PubMed] [Google Scholar]
- 63.Balkhi MY, Iwenofu OH, Bakkar N, Ladner KJ, Chandler DS, Houghton PJ, London CA, Kraybill W, Perrotti D, Croce CM, Keller C, Guttridge DC. miR-29 acts as a decoy in sarcomas to protect the tumor suppressor A20 mRNA from degradation by HuR. Sci Signal. 2013;6:ra63. doi: 10.1126/scisignal.2004177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pandey AK, Verma G, Vig S, Srivastava S, Srivastava AK, Datta M. miR-29a levels are elevated in the db/db mice liver and its overexpression leads to attenuation of insulin action on PEPCK gene expression in HepG2 cells. Mol Cell Endocrinol. 2011;332:125–133. doi: 10.1016/j.mce.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 65.Ugalde AP, Ramsay AJ, de la Rosa J, Varela I, Marino G, Cadinanos J, Lu J, Freije JM, Lopez-Otin C. Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J. 2011;30:2219–2232. doi: 10.1038/emboj.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang H, Garzon R, Sun H, Ladner KJ, Singh R, Dahlman J, Cheng A, Hall BM, Qualman SJ, Chandler DS, Croce CM, Guttridge DC. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lu L, Zhou L, Chen EZ, Sun K, Jiang P, Wang L, Su X, Sun H, Wang H. A Novel YY1-miR-1 regulatory circuit in skeletal myogenesis revealed by genome-wide prediction of YY1-miRNA network. PLoS One. 2012;7:e27596. doi: 10.1371/journal.pone.0027596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amodio N, Rossi M, Raimondi L, Pitari MR, Botta C, Tagliaferri P, Tassone P. miR-29s: a family of epi-miRNAs with therapeutic implications in hematologic malignancies. Oncotarget. 2015;6:12837–12861. doi: 10.18632/oncotarget.3805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mazzoccoli L, Robaina MC, Apa AG, Bonamino M, Pinto LW, Queiroga E, Bacchi CE, Klumb CE. MiR-29 silencing modulates the expression of target genes related to proliferation, apoptosis and methylation in Burkitt lymphoma cells. J Cancer Res Clin Oncol. 2018;144:483–497. doi: 10.1007/s00432-017-2575-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guo X, Stafford LJ, Bryan B, Xia C, Ma W, Wu X, Liu D, Songyang Z, Liu M. A Rac/Cdc42-specific exchange factor, GEFT, induces cell proliferation, transformation, and migration. J Biol Chem. 2003;278:13207–13215. doi: 10.1074/jbc.M208896200. [DOI] [PubMed] [Google Scholar]
- 72.Bryan BA, Mitchell DC, Zhao L, Ma W, Stafford LJ, Teng BB, Liu M. Modulation of muscle regeneration, myogenesis, and adipogenesis by the Rho family guanine nucleotide exchange factor GEFT. Mol Cell Biol. 2005;25:11089–11101. doi: 10.1128/MCB.25.24.11089-11101.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Momotani K, Somlyo AV. p63RhoGEF: a new switch for G(q)-mediated activation of smooth muscle. Trends Cardiovasc Med. 2012;22:122–127. doi: 10.1016/j.tcm.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith TK, Hager HA, Francis R, Kilkenny DM, Lo CW, Bader DM. Bves directly interacts with GEFT, and controls cell shape and movement through regulation of Rac1/Cdc42 activity. Proc Natl Acad Sci U S A. 2008;105:8298–8303. doi: 10.1073/pnas.0802345105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sun C, Liu C, Li S, Li H, Wang Y, Xie Y, Li B, Cui X, Chen Y, Zhang W, Li F. Overexpression of GEFT, a Rho family guanine nucleotide exchange factor, predicts poor prognosis in patients with rhabdomyosarcoma. Int J Clin Exp Pathol. 2014;7:1606–1615. [PMC free article] [PubMed] [Google Scholar]
- 76.Liu C, Li D, Jiang J, Hu J, Zhang W, Chen Y, Cui X, Qi Y, Zou H, Zhang W. Analysis of molecular cytogenetic alteration in rhabdomyosarcoma by array comparative genomic hybridization. PLoS One. 2014;9:e94924. doi: 10.1371/journal.pone.0094924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.