Abstract
The essential roles of long noncoding RNA (lncRNA) have been identified by emerging literature in the non-small cell lung cancer (NSCLC). However, the role of lncRNA hyaluronan synthase 2 antisense 1 (HAS2-AS1) in the NSCLC tumorigenesis is not clear. Here, we investigate the role and mechanism of HAS2-AS1 in the NSCLC tumorigenesis. In the NSCLC tissue and cells, HAS2-AS1 was found to be up-regulated, which, in turn, indicated the poor prognosis of NSCLC patients. Functional experiments illustrated that HAS2-AS1 promoted the proliferation, invasion and gefitinib chemotherapy resistance of NSCLC cells. In vivo, HAS2-AS1 knockdown suppressed the tumor growth. Mechanically, HAS2-AS1 recruited the lysine-specific demethylase 1 (LSD1) to the EphB3 promoter region to inhibit its transcription. In conclusion, this finding elucidates the essential roles of HAS2-AS1 in the NSCLC tumorigenesis, providing a possible treatment strategy for the NSCLC.
Keywords: Non-small cell lung cancer, HAS2-AS1, EphB3, LSD1
Introduction
Lung cancer, especially non-small-cell lung carcinoma (NSCLC) that accounting for 85% of all cases, remains the leading cause of tumor-related death worldwide [1,2]. The histological classification contains adenocarcinoma (LUAD) and squamous cell carcinoma (LUSC) [3]. Although multiple therapeutic methods are proposed to overcome the tenacious foe, there are still lots of problems for the treatment, such as chemotherapy resistance, recurrence and metastasis [4]. As well known, the 5-year overall survival of NSCLC is as low as 15%, calling for accurate agents targeting the authentic headstream of NSCLC.
Emerging literature indicates the critical roles of the long noncoding RNAs (lncRNAs) in the lung cancer [5,6]. The biological roles of lncRNAs are involved in the proliferation, cycle modification, drug resistance and epithelial mesenchymal transformation. For example, lncRNA FEZF1-AS1 is up-regulated in NSCLC tissues and cells and FEZF1-AS1 epigenetically repress E-cadherin via binding with LSD1 and EZH2 [7]. LncRNA LINC00460 is upregulated in the gefitinib-resistant NSCLC tissue and cells and LINC00460 facilitates the multidrug-resistant-related proteins (P-gp, BCRP, and MRP1) and the 50% inhibitive concentration of gefitinib [8]. Therefore, these finding could indicate that lncRNAs regulate the NSCLC tumorigenesis by epigenetically modifying the tumor behavior.
LncRNA HAS2-AS1 has been identified to act as the oncogene in the human cancer [9-11]. For example, in the epithelial ovarian cancer, HAS2-AS1, which is activated by the transcription factor CREB1, regulates the proliferation, invasion and tumor growth of EOC cells in vitro and in vivo [12]. In this research, we found that lncRNA HAS2-AS1 is up-regulated in the NSCLC tissue and cells. HAS2-AS1 exerts an oncogenic role via repressing the EphB3 in the NSCLC.
Materials and methods
Cancer tissue collection
We collected total thirty pairs of NSCLC and their adjacent normal tissues from individuals who were diagnosed to be NSCLC and then underwent the surgical excision at The Second Affiliated Hospital of Dalian Medical University. Clinicopathological characteristics of these individuals were shown in the Table 1. All collected tissue samples were immediately extracted and then snap-frozen in liquid nitrogen and stored at -80°C. This study obtained all the patients’ written informed consent and was approved by the Ethics Committee of The Second Affiliated Hospital of Dalian Medical University.
Table 1.
Correlation within HAS2-AS1 level and clinicopathological feature of NSCLC patients
| No | HAS2-AS1 | p | ||
|---|---|---|---|---|
|
| ||||
| Low No=14 | High No=16 | |||
| Gender | 0.531 | |||
| Male | 17 | 8 | 9 | |
| Female | 13 | 6 | 7 | |
| Age (years) | 0.403 | |||
| ≥ 65 | 16 | 6 | 10 | |
| < 65 | 14 | 8 | 6 | |
| Primary location | 0.712 | |||
| Left | 16 | 7 | 9 | |
| Right | 14 | 7 | 7 | |
| Tumor size | 0.075 | |||
| ≥ 4 cm | 16 | 5 | 11 | |
| < 4 cm | 14 | 9 | 5 | |
| Differentiation | 0.001* | |||
| Well, moderate | 11 | 8 | 3 | |
| Poor | 19 | 6 | 13 | |
| TNM | 0.012* | |||
| I-II | 12 | 8 | 4 | |
| III/IV | 18 | 6 | 12 | |
| Lymphonode metastasis | 0.568 | |||
| No | 15 | 8 | 7 | |
| Yes | 15 | 6 | 9 | |
P < 0.05 represents statistical difference.
Cells and culture
NSCLC cell lines (A549, H1299, SK-MES-1, H460), as well as the normal bronchial epithelial cells (NHBE) were obtained from American Type Culture Collection (ATCC, USA). For the cellular culture, cells were cultured in DMEM (GIBCO-BRL, Thermo Fisher Scientific, Shanghai, China) medium supplemented with 10% fetal bovine serum, 100 U/ml penicillin and 100 mg/ml streptomycin (Invitrogen, Carlsbad, CA, USA) at 37°C and 5% CO2.
Transfection
Small interfering RNA (siRNA), short hairpin RNA (shRNA) and the overexpression plasmids (pCDNA3.1 vector) targeting the HAS2-AS1 or EphB3 were synthesized by the GENEWIZ Inc (Guangzhou, China). The siRNA and shRNA sequences were shown in Table S1. Cells were seeded in the six-well plates and then transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions.
Quantitative real-time polymerase chain reaction (qRT-PCR)
After the transfection into NSCLC cells, the total RNAs were extracted using Trizol reagent (Invitrogen, Carlsbad, USA). DNA Reverse Transcriptase Kit (TAKARA, Beijing, China) was utilized for the cDNA synthesis. The PCR was performed using the SYBR Green PCR Master One-Mix kit (Trans-Gen, Beijing, China) on Real-time PCR 7300 System (Applied Biosystems, Foster City, USA) Relative quantification of RNA expression was calculated using the 2-ΔΔCT method. The primers of HAS2-AS1 and EphB3 were shown in the Table S1. Relative quantification of RNA expression levels, including HAS2-AS1 and EphB3, were calculated using the 2-ΔΔCT method.
Colony formation and CCK-8 activity analysis
For the colony formation assay, A549 and H460 cells were cultured in the six-well plate and medium containing 10% FBS. After 2 weeks, clones were fixed with methanol and stained with 0.1% crystal violet. For the cellular activity analysis, the CCK-8 assay was performed as previous described to measure the 50% maximal inhibitory concentration (IC50) value of A549 and H460 cells for gefitinib [13].
Transwell assay
After 48 hours of the transfection of A549 and H460, cells (1 × 104 per well) were digested and resuspended in 200 µl serum-free medium in the upper chamber coated with Matrigel (BD Biosciences, San Jose, CA). The 600 µl 10% fetal bovine serum medium (Gibco) was added to the lower chamber. After culture for 48 hours, cells were fixed with 4% paraformaldehyde and then stained with crystal violet for 15 mins. Cell invasion was washed with PBS and the quantification was determined under high-power microscope (Olympus Corporation, Tokyo, Japan).
Western blot analysis
The cells were transfected and culture for 48 hours, then the total protein was extracted using RIPA lysate and determined BCA kit (Beyotime, Shanghai, China). Protein (100 μg) was sent for the electrophoresis on PVDF membrane at room temperature for 1 hour. The membrane was incubated with the primary antibodies (anti-EphB3, ab133742, 1:1000) overnight at low temperature. Horseradish peroxidation incubated with the member for 1 hour. The bands were visualized using chromogenic solution on Immuno Star LD (Wako Pure Chemical, Osaka, Japan).
Subcellular location analysis
PARIS Kit (Life Technologies, CA, USA) was used to extract the RNA fractions in nuclear and cytosolic according to the manufacturer’s instructions. The quantification of these fractions were measured by the RT-qPCR. GAPDH or U1 RNA acted the control in cytoplasm or nuclear fraction.
RNA binding protein immunoprecipitation (RIP)
Cell lysate were centrifugated and incubated with RIP buffer. The A/G magnetic beads were coated with anti-Ago2 or negative control anti-IgG antibody. Following incubation overnight at 4°C, the isolation of RNA was detected by qRT-PCR.
Chromatin immunoprecipitation (ChIP)
ChIP was performed as previously described [14]. Cell were treated with formaldehyde and incubated with LSD1 and H3K4me2-specific antibody (Millipore) or IgG as control for 10 min to generate DNA-protein cross-links overnight at 4°C. The beads were eluted and then DNA was purified using a spin column and quantified using qRT-PCR.
Mice in vivo xenograft assay
Five week old female athymic BALB/c nude mice were purchased from Shanghai SIPPR-BK Laboratory Animal Co. Ltd. (Shanghai, China). A549 cells were stably transfected with sh-HAS2-AS1 or empty vectors. Then, total of 5 × 106 cells were subcutaneously injected into the posterior flank of mice. Every three days, the length and width of tumor were examined, and the volumes was calculated using the equation: 0.5 × length × width2. After 27 days, the subcutaneous growth of tumor was detected. The protocol was approved by the Committee on the Ethics of Animal Experiments of The Second Affiliated Hospital of Dalian Medical University.
Statistical analysis
All experimental data were analyzed by SPSS version 19.0 and graphed by GraphPad Prism version 7.0. The results were presented as mean ± SD (standard deviation, SD). Comparison within two groups was analyzed by Student’s t-test and the one-way ANOVA for multiple comparison. P < 0.05 was considered as the significance.
Results
HAS2-AS1 indicated the poor prognosis of NSCLC patients
The level of HAS2-AS1 was calculated by the RT-PCR in the NSCLC tissue, revealing the significant overexpression of HAS2-AS1 (Figure 1A; Table 1). In the advanced pathological grading of NSCLC, the level of HAS2-AS1 was remarkedly up-regulated compared with the benignant pathological grading (Figure 1B). Receiver operating characteristic (ROC) curve revealed the high diagnostic efficiency (AUC=0.83) of HAS2-AS1 for NSCLC patients (Figure 1C). Survival analysis was analyzed via the Kaplan-Meier method, suggesting that the high level of HAS2-AS1 indicated the bad outcome and lower survival rate (Figure 1D). These finding illustrated that HAS2-AS1 indicated the poor prognosis of NSCLC patients.
Figure 1.
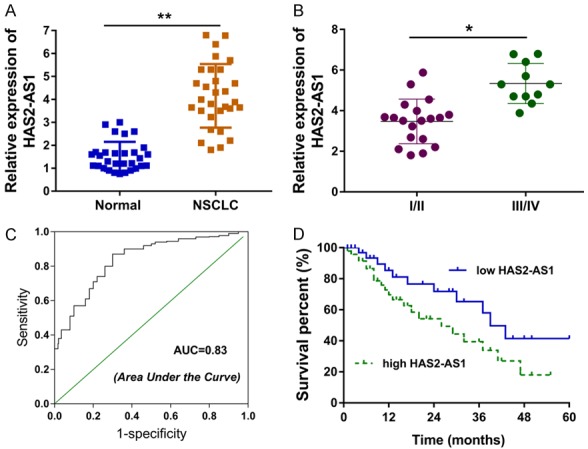
HAS2-AS1 indicated the poor prognosis of NSCLC patients. A. RT-PCR revealed the level of HAS2-AS1 in the NSCLC tissue and adjacent normal tissue. B. The level of HAS2-AS1 in the advanced or benignant pathological grading of NSCLC. C. Receiver operating characteristic (ROC) curve revealed the high diagnostic efficiency (AUC=0.83) of HAS2-AS1 for NSCLC patients. D. Survival analysis was analyzed via the Kaplan-Meier method for NSCLC patients. Data are presented as means ± SD of three independent experiments. **P < 0.01.
HAS2-AS1 promotes the NSCLC tumorigenesis and gefitinib resistance of NSCLC
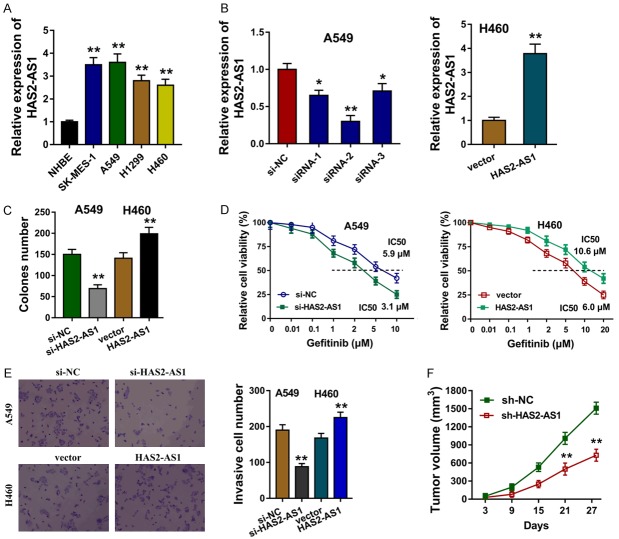
We here investigated the tumorous roles of HAS2-AS1 on NSCLC cells in vivo and vitro. The expression levels of HAS2-AS1 in the NSCLC cells were explored in the first step, presenting the overexpression of HAS2-AS1 (Figure 2A). The small interfering RNAs (siRNAs) targeting HAS2-AS1 were synthesized to silence the HAS2-AS1 expression, and the plasmids were transfected to up-regulate the HAS2-AS1 expression (Figure 2B). Colony formation assay elucidated that the HAS2-AS1 silencing inhibited the clone number, while the HAS2-AS1 over-expression enhanced the clone number (Figure 2C). Gefitinib chemotherapy resistance of NSCLC cells was performed CCK-8, revealing that HAS2-AS1 silencing decreased the 50% maximal inhibitory concentration (IC50) value for gefitinib in A549 cells, and HAS2-AS1 over-expression enhanced the IC50 in H460 cells (Figure 2D). Transwell invasion assay showed that HAS2-AS1 silencing repressed the invaded cells and HAS2-AS1 over-expression increased the quantity (Figure 2E). Xenograft in vivo mice assay showed that the stably HAS2-AS1 silencing by shRNA could remarkedly repress the tumor growth (Figure 2F). These finding could conclude that HAS2-AS1 promotes the NSCLC tumorigenesis and gefitinib resistance of NSCLC.
Figure 2.
HAS2-AS1 promotes the NSCLC tumorigenesis and chemotherapy resistance of NSCLC. A. RT-PCR showed the expression levels of HAS2-AS1 in the NSCLC cells (SK-MES-1, A549, H1299, H460). B. The small interfering RNAs (siRNAs) and plasmids specially targeting HAS2-AS1 were synthesized to silence or enhance the HAS2-AS1 expression. C. Colony formation assay elucidated the clone number. D. Gefitinib chemotherapy resistance and the 50% maximal inhibitory concentration (IC50) of NSCLC cells was performed CCK-8 in A549 cells and H460 cells. E. Transwell invasion assay showed the invaded cells. F. Xenograft in vivo mice assay showed the tumor growth with stable HAS2-AS1 silencing by shRNA. Data are presented as means ± SD of three independent experiments. **P < 0.01.
HAS2-AS1 repressed the EphB3 via recruiting LSD1
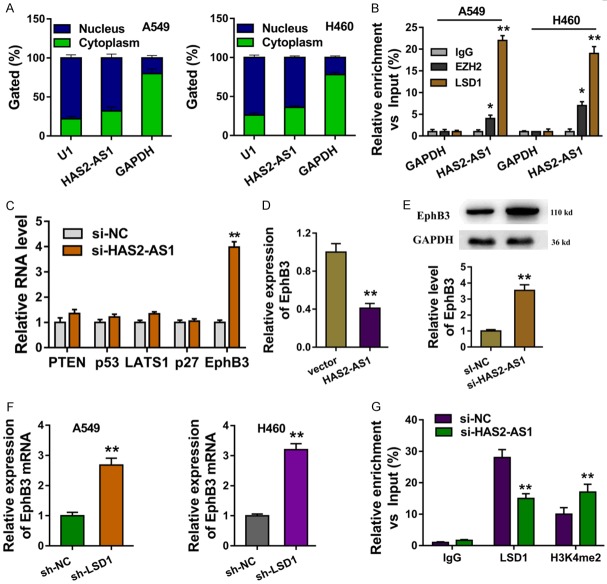
The subcellular location of HAS2-AS1 was analyzed, elucidating that HAS2-AS1 was mainly located in the nucleus more than in the cytoplasm (Figure 3A). RNA binding protein immunoprecipitation (RIP) presented that LSD1 and EZH2 could bind with the HAS2-AS1, however the LSD1 much more remark (Figure 3B). We selected several potential downstream targets of HAS2-AS1 and then measured the expression level of them after the HAS2-AS1 silencing (Figure 3C). The HAS2-AS1 overexpression repressed the EphB3 mRNA (Figure 3D). Western blot showed that HAS2-AS1 knockdown up-regulated the EphB3 protein (Figure 3E). RT-PCR elucidated that the LSD1 shRNA could up-regulated the EphB3 mRNA (Figure 3F). Chromatin immunoprecipitation (ChIP) showed that LSD1 and H3K4me2 occupied the promoter regions of EphB3, while HAS2-AS1 knockdown decreased the occupancy of LSD1 and H3K4me2 (Figure 3F). Therefore, these results illustrated that HAS2-AS1 repressed the EphB3 via recruiting LSD1.
Figure 3.
HAS2-AS1 repressed the EphB3 via recruiting LSD1. A. The subcellular location of HAS2-AS1 was analyzed for the nucleus or cytoplasm fraction. B. RNA binding protein immunoprecipitation (RIP) presented the binding of LSD1 and EZH2 with the HAS2-AS1. C. The several selected potential downstream targets of HAS2-AS1 were measured RT-PCR. D. The EphB3 mRNA was measured by HAS2-AS1 overexpression. E. EphB3 protein was measured by western blot with HAS2-AS1 knockdown or not. F. RT-PCR elucidated the EphB3 mRNA with LSD1 shRNA transfection. G. Chromatin immunoprecipitation (ChIP) showed the occupancy of LSD1 and H3K4me2 of the promoter regions of EphB3. Data are presented as means ± SD of three independent experiments. **P < 0.01.
EphB3 acted the target of HAS2-AS1 in the NSCLC tumorigenesis
Previous research found that the HAS2-AS1 could target the EphB3 via recruiting LSD1. In the subsequent investigation, we co-transfected the EphB3 silencing plasmids (sh-EphB3) into the A549 cells to elucidate the roles of HAS2-AS1 and EphB3. In the NSCLC tissue sample, we found that EphB3 level was down-regulated compared with the controls (Figure 4A). The interaction analyzed by the Spearman’s rank analysis showed that the EphB3 was negatively correlated with HAS2-AS1 (Figure 4B). Western blot showed that the EphB3 silencing plasmids (sh-EphB3) decreased its protein expression (Figure 4C). Colony formation assay illustrated that the sh-EphB3 transfection recovered the clone number of A549 cells (Figure 4D). Gefitinib chemotherapy resistance revealed that sh-EphB3 transfection rescued the 50% maximal inhibitory concentration (IC50) value for gefitinib (Figure 4E). Transwell invasion assay indicted that sh-EphB3 transfection rescued the invasive ability induced by the HAS2-AS1 knockdown (Figure 4F). Overall, EphB3 acted the target of HAS2-AS1 in the NSCLC tumorigenesis.
Figure 4.
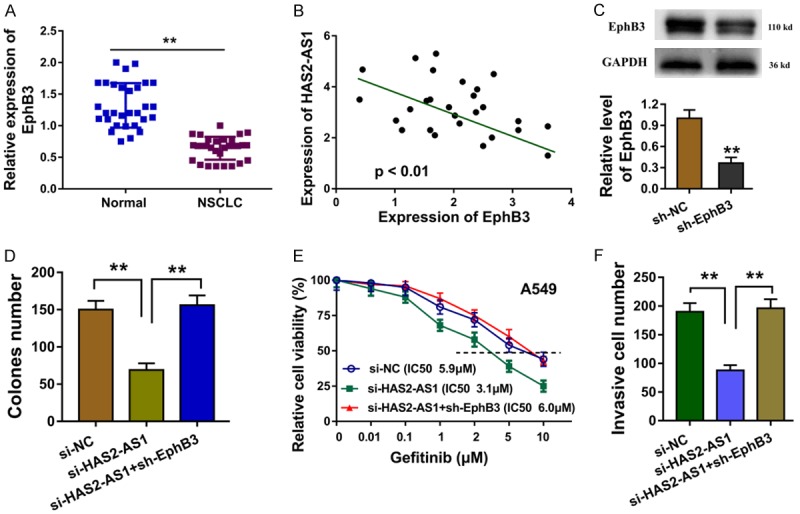
EphB3 acted the target of HAS2-AS1 in the NSCLC tumorigenesis. A. EphB3 level was down-regulated in the NSCLC tissue sample compared with the controls. B. Spearman’s rank analysis showed the negative interaction within EphB3 and HAS2-AS1. C. Western blot showed the EphB3 protein expression by the silencing plasmids (sh-EphB3). D. Colony formation assay illustrated the clone number of A549 cells. E. Gefitinib chemotherapy resistance revealed the 50% maximal inhibitory concentration (IC50) value for gefitinib of A549. F. Transwell invasion assay indicted the invasive ability induced by the HAS2-AS1 knockdown and sh-EphB3 transfection. Data are presented as means ± SD of three independent experiments. **P < 0.01.
Discussion
The roles of ncRNAs in the lung cancer imply the critical functions involving the tumor proliferation, metastasis and chemotherapy resistance [15,16]. LncRNAs are group of transcripts with the longer than 200 nucleotides and non-protein coding potential [17,18]. Given the increasing regulation of lncRNA, more and more published papers report the regulation and function of lncRNA.
In present research, we found that the level of lncRNA HAS2-AS1 was up-regulated in the NSCLC tissue and cells. The diagnostic efficiency of HAS2-AS1 was also effective and astonishing, suggesting the high possibility for HAS2-AS1 to identify the suffering of NSCLC. Moreover, the ectopic high-expression of HAS2-AS1 indicates the bad clinical outcome of NSCLC patients. The gain and loss of functional experiments illustrated that HAS2-AS1 promotes the proliferation, invasion and gefitinib chemotherapy resistance of NSCLC cells. Being combined with the existed reports, we confirmed the oncogenic impact of HAS2-AS1 in the NSCLC tumorigenesis.
Gefitinib is a novel targeted drug for the treatment of locally advanced or metastatic NSCLC patients, especially for patients with exon 19 and 21 mutations in the EGFR gene [19,20]. In this research, we found that HAS2-AS1 could regulate the gefitinib resistance of NSCLC cells. However, the underlying mechanism is still elusive. Chemotherapy resistance is a major and nonnegligible difficulty in the NSCLC treatment [21]. One of the traits of the NSCLC is stubborn and recuperative, causing by the survivability of cancer cells in the long-term drug induction. For example, TP-binding cassette (ABC) transporters are a family of membrane proteins that that significantly regulate the bioavailability anti-drugs and the multidrug resistance (MDR) for chemotherapy [22].
There are much more evidence supporting that the lncRNA could modulate the transcriptional regulation and post-transcriptional regulation in NSCLC. For example, transcription factor Twist1 binds with the promoter of INC01296 and activate its transcriptional level, constructing the INC01296/miR-598/Twist1 positive feedback loop to promote the tumorigenesis of NSCLC [23].
Epigenetic writer lysine-specific demethylase 1 (LSD1) is found to be aberrantly upregulated in series of cancer types. The overexpression of LSD1 is closely correlated to the poor survival [24]. For instance, lncRNA AGAP2-AS1 bind with EZH2 and LSD1 to recruit them to the promoter region of KLF2 and LATS2, thereby repressing their transcriptional activity [14]. LSD1 could directly bind with FBXW7 to destabilize its demethylase activity, and LSD1 acts as the pseudosubstrate to trigger LSD1 ubiquitylation [25]. EphB3 is found to be significantly downregulated in NSCLC tissue and suppresses its metastasis via PP2A/RACK1/Akt signalling complex [26].
LncRNA hyaluronan synthase 2 antisense 1 (HAS2-AS1) is found to be up-regulated in the several human cancer, such as oral squamous cell carcinoma, epithelial ovarian cancer and glioma cells [9,27]. In present research, we found that the oncogenic HAS2-AS1 could recruit the LSD1 to promoter of EphB3. This HAS2-AS1/LSD1/EphB3 axis might be a potential pathogenesis for NSCLC. Taken together, our findings indicate that HAS2-AS1 might act as an oncogene and provide a novel insight and a valuable therapeutic strategy.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Chacon MR, Enrico DH, Burton J, Waisberg FD, Videla VM. Incidence of placebo adverse events in randomized clinical trials of targeted and immunotherapy cancer drugs in the adjuvant setting: a systematic review and meta-analysis. JAMA Netw Open. 2018;1:e185617. doi: 10.1001/jamanetworkopen.2018.5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He T, Mehta AC. Linear endobronchial ultrasound: what’s new? Semin Respir Crit Care Med. 2018;39:649–660. doi: 10.1055/s-0038-1676646. [DOI] [PubMed] [Google Scholar]
- 3.Ma K, Lu Y, Jiang S, Tang J, Li X, Zhang Y. The relative risk and incidence of immune checkpoint inhibitors related pneumonitis in patients with advanced cancer: a meta-analysis. Front Pharmacol. 2018;9:1430. doi: 10.3389/fphar.2018.01430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang D, Hu Y. Long non-coding rna pvt1 competitively binds microRNA-424-5p to regulate CARM1 in radiosensitivity of non-small-cell lung cancer. Mol Ther Nucleic Acids. 2019;16:130–140. doi: 10.1016/j.omtn.2018.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Lu X, He X, Su J, Wang J, Liu X, Xu K, De W, Zhang E, Guo R, Shi YE. EZH2-mediated epigenetic suppression of GDF15 predicts a poor prognosis and regulates cell proliferation in non-small-cell lung cancer. Mol Ther Nucleic Acids. 2018;12:309–318. doi: 10.1016/j.omtn.2018.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Russo V, Paciocco A, Affinito A, Roscigno G, Fiore D, Palma F, Galasso M, Volinia S, Fiorelli A, Esposito CL, Nuzzo S, Inghirami G, de Franciscis V, Condorelli G. Aptamer-miR-34c conjugate affects cell proliferation of non-small-cell lung cancer cells. Mol Ther Nucleic Acids. 2018;13:334–346. doi: 10.1016/j.omtn.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC) Biomed Pharmacother. 2017;95:331–338. doi: 10.1016/j.biopha.2017.08.057. [DOI] [PubMed] [Google Scholar]
- 8.Ma G, Zhu J, Liu F, Yang Y. Long noncoding RNA LINC00460 promotes the gefitinib resistance of nonsmall cell lung cancer through epidermal growth factor receptor by sponging miR-769-5p. DNA Cell Biol. 2019;38:176–183. doi: 10.1089/dna.2018.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao Z, Liang T, Feng S. Silencing of HAS2-AS1 mediates PI3K/AKT signaling pathway to inhibit cell proliferation, migration, and invasion in glioma. J Cell Biochem. 2019 doi: 10.1002/jcb.28430. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 10.Yung Y, Ophir L, Yerushalmi GM, Baum M, Hourvitz A, Maman E. HAS2-AS1 is a novel LH/hCG target gene regulating HAS2 expression and enhancing cumulus cells migration. J Ovarian Res. 2019;12:21. doi: 10.1186/s13048-019-0495-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao F, Chen J, Shi M, Song Y, Chen Z. LncRNA HAND2-AS1 inhibits non-small cell lung cancer migration, invasion and maintains cell stemness through the interactions with TGF-β1. Biosci Rep. 2019;39 doi: 10.1042/BSR20181525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tong L, Wang Y, Ao Y, Sun X. CREB1 induced lncRNA HAS2-AS1 promotes epithelial ovarian cancer proliferation and invasion via the miR-466/RUNX2 axis. Biomed Pharmacother. 2019;115:108891. doi: 10.1016/j.biopha.2019.108891. [DOI] [PubMed] [Google Scholar]
- 13.Liu X, Lu X, Zhen F, Jin S, Yu T, Zhu Q, Wang W, Xu K, Yao J, Guo R. LINC00665 induces acquired resistance to gefitinib through recruiting EZH2 and activating PI3K/AKT pathway in NSCLC. Mol Ther Nucleic Acids. 2019;16:155–161. doi: 10.1016/j.omtn.2019.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li W, Sun M, Zang C, Ma P, He J, Zhang M, Huang Z, Ding Y, Shu Y. Upregulated long non-coding RNA AGAP2-AS1 represses LATS2 and KLF2 expression through interacting with EZH2 and LSD1 in non-small-cell lung cancer cells. Cell Death Dis. 2016;7:e2225. doi: 10.1038/cddis.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu J, Qi X, Liu L, Hu X, Liu J, Yang J, Yang J, Lu L, Zhang Z, Ma S, Li H, Yun X, Sun T, Wang Y, Wang Z, Liu Z, Zhao W. Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids. 2019;16:589–596. doi: 10.1016/j.omtn.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu XH, Wang HF, Wu JB, Wang SS, Tang YJ, Tang YL, Liang XH. Non-coding RNAs derailed: the many influences on the fatty acid reprogramming of cancer. Life Sci. 2019;231:116509. doi: 10.1016/j.lfs.2019.05.065. [DOI] [PubMed] [Google Scholar]
- 17.Do H, Kim W. Roles of oncogenic long non-coding RNAs in cancer development. Genomics Inform. 2018;16:e18. doi: 10.5808/GI.2018.16.4.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Wang H, Zhang M, Jiang S, Zhou C, Fang B, Chen P. Abnormally expressed long non-coding RNAs in prognosis of osteosarcoma: a systematic review and meta-analysis. J Bone Oncol. 2018;13:76–90. doi: 10.1016/j.jbo.2018.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Zhang R, Zhou Y, Song J, Luo W, Tian P, Li W. Combined use of crizotinib and gefitinib in advanced lung adenocarcinoma with leptomeningeal metastases harboring MET amplification after the development of gefitinib resistance: a case report and literature review. Clin Lung Cancer. 2019;20:e251–e255. doi: 10.1016/j.cllc.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 20.Kucharczuk CR, Ganetsky A, Vozniak JM. Drug-drug interactions, safety, and pharmacokinetics of EGFR tyrosine kinase inhibitors for the treatment of non-small cell lung cancer. J Adv Pract Oncol. 2018;9:189–200. [PMC free article] [PubMed] [Google Scholar]
- 21.Roca E, Pozzari M, Vermi W, Tovazzi V, Baggi A, Amoroso V, Nonnis D, Intagliata S, Berruti A. Outcome of EGFR-mutated adenocarcinoma NSCLC patients with changed phenotype to squamous cell carcinoma after tyrosine kinase inhibitors: a pooled analysis with an additional case. Lung Cancer. 2019;127:12–18. doi: 10.1016/j.lungcan.2018.11.016. [DOI] [PubMed] [Google Scholar]
- 22.Zhao H, Huang Y, Shi J, Dai Y, Wu L, Zhou H. ABCC10 plays a significant role in the transport of gefitinib and contributes to acquired resistance to gefitinib in NSCLC. Front Pharmacol. 2018;9:1312. doi: 10.3389/fphar.2018.01312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu L, Wei B, Hui H, Sun Y, Liu Y, Yu X, Dai J. Positive feedback loop of lncRNA LINC01296/miR-598/Twist1 promotes non-small cell lung cancer tumorigenesis. J Cell Physiol. 2019;234:4563–4571. doi: 10.1002/jcp.27235. [DOI] [PubMed] [Google Scholar]
- 24.Lim SY, Macheleidt I, Dalvi P, Schafer SC, Kerick M, Ozretic L, Ortiz-Cuaran S, George J, Merkelbach-Bruse S, Wolf J, Timmermann B, Thomas RK, Schweiger MR, Buettner R, Odenthal M. LSD1 modulates the non-canonical integrin beta3 signaling pathway in non-small cell lung carcinoma cells. Sci Rep. 2017;7:10292. doi: 10.1038/s41598-017-09554-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lan H, Tan M, Zhang Q, Yang F, Wang S, Li H, Xiong X, Sun Y. LSD1 destabilizes FBXW7 and abrogates FBXW7 functions independent of its demethylase activity. Proc Natl Acad Sci U S A. 2019;116:12311–12320. doi: 10.1073/pnas.1902012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li G, Ji XD, Gao H, Zhao JS, Xu JF, Sun ZJ, Deng YZ, Shi S, Feng YX, Zhu YQ, Wang T, Li JJ, Xie D. EphB3 suppresses non-small-cell lung cancer metastasis via a PP2A/RACK1/Akt signalling complex. Nat Commun. 2012;3:667. doi: 10.1038/ncomms1675. [DOI] [PubMed] [Google Scholar]
- 27.Zhu G, Wang S, Chen J, Wang Z, Liang X, Wang X, Jiang J, Lang J, Li L. Long noncoding RNA HAS2-AS1 mediates hypoxia-induced invasiveness of oral squamous cell carcinoma. Mol Carcinog. 2017;56:2210–2222. doi: 10.1002/mc.22674. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.