Abstract
DNA methylation, catalyzed by DNA methyltransferases (DNMTs), is a heritable epigenetic mark, participating in numerous physiological processes. DNMT3A is of particular relevance to hematopoietic differentiation, because DNMT3A mutations are strongly related to hematopoietic malignancies. Additionally, DNMT3A deficiency has been reported to increase the hematopoietic stem cell pool by limiting their differentiation. Our previous study demonstrated that complete loss of DNMT3A resulted in anemia, while DNMT3A haploinsufficiency caused an elevated population of erythrocytes in the content of oncogenic KRAS. Since erythropoiesis is tightly regulated via the erythropoietin (EPO)-mediated RAS-RAF-MEK-ERK1/2 pathway, the question arises whether DNMT3A cooperates with RAS signaling to modulate erythropoiesis. Human leukemia cell lines were used, with differentiation capabilities towards megakaryocyte and erythroid lineages. Overexpression of DNMT3A was found to enhance erythrocytic differentiation of K562 cells, while DNMT3A knockdown suppressed differentiation. Furthermore, higher DNMT3A expression was detected in late-stage mouse erythroblasts along with the DNMT3A translocation to the nucleus. Further studies demonstrated that both ERK1/2-DNMT3A interaction and serine-255 phosphorylation in DNMT3A led to DNMT3A translocation into the nucleus, and modulated erythrocytic differentiation. Our results not only explore the critical role of DNMT3A in erythropoiesis, but also provide a new insight into ERK1/2-DNMT3A interaction in the hematopoietic system.
Keywords: Erythropoiesis, DNA methyltransferase, DNMT3A, ERK1/2
Introduction
DNA methylation is an important epigenetic mechanism that regulates numerous physiological and pathological processes. It is catalyzed by DNA methyltransferases (DNMTs), which covalently add a methyl group at the 5-carbon of the cytosine ring, to form 5-methylcytosine (5-mC). DNMT1 primarily maintains pre-existing DNA methylation patterns, whereas DNMT3A and DNMT3B carry out de novo DNA methylation. In contrast, DNMT2 has the potential to methylate RNA instead of DNA. DNMT3-like protein (DNMT3L) lacks the catalytic domain and functions as an accessory protein of DNMT3s [1,2]. Although current studies mainly focus on DNA methylation at a repressed promoter and its consequences, there is much evidence that DNA methylation can be modulated by extracellular stimuli [1]. Thus, we are interested in the crosstalk between signaling pathways and DNMTs in physiological processes.
Hematopoiesis in vertebrates is sustained throughout life, because a few hematopoietic stem cells (HSCs) continuously regenerate blood cells [3]. Previous studies have demonstrated that DNA methylation patterns are associated with cellular plasticity and the lineage specification of both hematopoietic stem cells (HSCs) and progenitor cells [4-7]. Erythropoiesis is a process by which erythrocytes (red blood cells or RBCs) are produced from HSCs [8,9]. Previous studies showed that DNA methylation plays an important role in modulating the expression of globin genes. Maturation of RBCs is associated with increased expression of α- and β-globin genes. The β-globin locus consists of five genes: ε, Gγ, Aγ, δ and β [10]. In non-erythroid cells, these genes are hypermethylated, which results in transcriptional silence. During erythropoiesis, individual β-globin genes, corresponding to embryonic (ε), fetal (GγAγ) and adult (δ, β) stages, are expressed in a sequential fashion, e.g. the embryonic and fetal genes are ultimately silenced under hypermethylation and adult genes are activated [11]. Accordingly, erythropoietin (EPO) is the key regulator of erythropoiesis. The binding of EPO to its receptor triggers the activation of STAT5, PI3K/Akt, and RAS-RAF-MEK-ERK pathways [12,13]. Notably, injection of EPO induces the expression of DNMT3A and DNMT3B in the hippocampus [14]. These studies suggest the potential for crosstalk between DNMTs and EPO-mediated signaling pathways during erythropoiesis.
It has long been recognized that abnormal DNA methylation is strongly associated with tumorigenesis. The first high-penetrance mutation in the DNMT family (DNMT3A) was identified in patients with acute myeloid leukemia (AML) [15,16]. Moreover, twelve major cancer genome sequencing projects, from the Cancer Genome Atlas (TCGA), further reveal the importance of DNMT3A mutations to hematopoietic malignancies [17]. Loss of Dnmt3a, in the murine hematopoietic system, progressively impairs the differentiation ability of HSCs, while simultaneously expanding HSC numbers [18]. After a prolonged latency, Dnmt3a deficiency leads to multiple hematopoietic diseases with lethal results [19,20]. Studies of hematopoietic defects showed that loss of Dnmt3a induces extramedullary/stress erythropoiesis [19], and progressively leads to macrocytic anemia [20] in these affected mice. However, Socolovsky’s group (2011) showed that downregulation of Dnmt3a and Dnmt3b are coincident with the progressive loss of DNA methylation in the differentiating erythroblasts, isolated from fetal liver cells [21]. This controversy elicited significant questions about whether DNMT3A affects erythrocytic differentiation. In addition, our previous results indicated that loss of Dnmt3a promotes myeloid malignancies, while Dnmt3a haploinsufficiency induces T-cell acute lymphoblastic leukemia (T-ALL) in the cell context with oncogenic Kras expression. However, a discrepancy about DNMT3A in RBC production arose in leukemic mice. Complete loss of Dnmt3a cooperates with oncogenic Ras to promote severe anemia [19,22,23], while deletion of one-allele of Dnmt3a in KrasG12D hematopoietic cells elevates the counts of RBCs, hemoglobin and hematocrit [22]. This discrepancy raised the issue of whether EPO downstream RAS pathway interacts with DNMT3A to regulate erythropoiesis.
Recent reports indicate the importance of protein phosphorylation to DNMT3A activity and function [24,25]. CK2 can phosphorylate DNMT3A to modulate its localization to heterochromatin [24]. Another report indicated that fibroblast growth factor (FGF) modulates chondrogenesis through extracellular-signal-regulated kinase 1/2 (ERK1/2)-mediated DNMT3A phosphorylation on serine-255 residue (S255). ERK1/2 interacts with either leucine-373 (L373) and/or L637 residues in DNMT3A proteins. This interaction is critical for efficient S255 phosphorylation of DNMT3A [25]. Due to the importance of ERK1/2 to the RAS signaling in EPO-regulated erythropoiesis, we focused on whether ERK1/2 interplays with DNMT3A to modulate erythrocytic differentiation. In order to test our hypothesis, human K562 and HEL cell lines were used. These two cell lines are of the erythroleukemia type, with the capability to differentiate toward erythroid or megakaryocyte lineages [26-30], so they exhibit comparable differentiation potential to human CD34+ hematopoietic stem/progenitor cells [27,29]. In this study, knockdown of DNMT3A interrupted erythrocytic differentiation. We further demonstrated that DNMT3A directly interacted with ERK1/2, and that S255 phosphorylation of DNMT3A was critical for both DNMT3A translocation and erythrocytic differentiation. Our results not only resolved previous controversy about the role of DNMT3A in erythropoiesis but also provided a new insight into the interaction between signaling molecules and DNMTs in hematopoiesis.
Materials and methods
Antibodies
The antibodies against DNMT3A (Cat. 3598), ERK1/2 (Cat. 9102), phospho-ERK1/2 (Cat. 9101), phospho-MAPK/CDK substrates (Cat. 2325) and normal rabbit IgG (Cat. 2729) were purchased from Cell Signaling Technology (MA, U.S.). Peroxidase-conjugated anti-mouse IgG (Cat. 115-035-003), anti-rabbit IgG (Cat. 111-035-003) and rhodamine red-conjugated anti-rabbit IgG (Cat. 111-295-003) were obtained from Jackson Immunoresearch Laboratories (PA, U.S.). The anti-GAPDH antibody (Cat. NB300-221) was bought from Novus Biologicals (CO, U.S.), and the anti-HA antibodies (Cat. SC-805, and Cat. SC-7392) were obtained from Santa Cruz Biotechnology (TX, U.S.).
Plasmids and construction
The pCMV6-DNMT3A-Myc-DDK plasmid (#RG213064) was purchased from OriGene Technologies Inc. (MD, U.S.). To stably express N-terminally HA-tagged DNMT3A protein, DNMT3A cDNA was amplified using the polymerase chain reaction (PCR), and then inserted into a pcDNA3-HA2 vector, which was a gift from Dr. Wey-Jinq Lin (Institute of Biopharmaceutical Sciences, National Yang-Ming University, Taiwan) [27,29], to obtain the pcDNA3-HA2-DNMT3A plasmid. The overlap-extension PCR method for site-directed mutagenesis was used to generate various DNMT3A mutants. The scramble shRNA and human DNMT3A shRNAs, including A2 (TRCN0000035754), D2 (TRCN0000035756), and E2 (TRCN0000035757), were obtained from the National RNAi Core Facility (Academia Sinica, Taipei City, Taiwan). All plasmids were verified by DNA sequencing, which was carried out by Genomics (New Taipei City, Taiwan).
Cell culture, clone selection and differentiation
Human chronic myelogenous leukemia (CML) K562 and erythroleukemia HEL 92.1.7 (HEL) cells were obtained from the Bioresource Collection and Research Center (BCRC) (Hsinchu City, Taiwan). K562 cells (4×104 cells/ml) were cultivated in Iscove’s Modified Dulbecco’s Medium (IMDM), supplemented with 10% fetal bovine serum (FBS), 100 IU/ml penicillin and 100 IU/ml streptomycin, and were passaged every two days. HEL cells (1×105 cells/ml) were cultivated in Roswell Park Memorial Institute (RPMI) 1640 medium, supplemented with 10% FBS, 1 mM sodium pyruvate, 100 IU/ml penicillin and 100 IU/ml streptomycin, and were passaged every 2 to 3 days. For erythrocytic differentiation, K562 and HEL cells were treated with 2 or 0.1 mM sodium butyrate (NaB), respectively. Hemoglobin expression was examined using benzidine/hydrogen peroxide solution [29]. Transfection was performed using LipofectamineTM 2000 reagent (Thermo Fisher Scientific Inc., MA, U.S.) according to the manufacturer’s instruction. The HA-DNMT3A cell clones, HA-3A-E1 and HA-3A-K1, were selected with 0.5 mg/ml of G418.
Flow cytometry and cell sorting
To analyze erythroblasts in murine bone marrow (BM), the pure C57BL/6 mice (8-10 weeks) were purchased from the Laboratory Animal Center of National Yang-Ming University. All animal experiments were conducted in accordance with the Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee (IACUC) of National Yang-Ming University (IACUC number: 1041226). BM cells were carefully isolated from tibia and femur. The specific antibodies against CD71 (Clone C2) and TER119 (Clone Ter119) were purchased from BD PharmingenTM. The subgroups of erythroblasts were analyzed using flow cytometry, which were performed as previously described [19,21,31]. The CD71highTER119high (early-stage erythroblast, subgroup II) and CD71lowTER119high (late-stage erythroblast, subgroup IV) were isolated for RNA extraction and immunostaining. The stained cells were analyzed and sorted out on a BD FACSMelody (BD Biosciences, U.S.).
Immunoprecipitation
Cell lysates were incubated with antibodies, overnight, gently shaking at 4°C. Then, samples were incubated with protein G agarose for 3 hours, gently shaking at 4°C. Beads were centrifuged (800×g) for 5 min at 4°C, then washed with cold PBS buffer containing 0.05% tween-20 once, then with cold PBS twice. Finally, 4X SDS-PAGE sample buffer was added, and heated to 95°C. These products were either immediately analyzed using western blot analysis or stored at -80°C.
Immunostaining
Cells, including K562 cells and erythroblasts, were fixed with 4% paraformaldehyde. After blocking, the fixed cells were incubated with anti-HA (1:100) or anti-DNMT3A (1:50) antibodies overnight. Subsequently, they were visualized with AlexaFluor 488-conjugated anti-mouse IgG antibodies (1:200) to detect HA-DNMT3A proteins in K562 cells, or rhodamine red-conjugated anti-rabbit IgG antibodies (1:100) to detect endogenous DNMT3A proteins in erythroblasts. These cells were counterstained with DAPI (50 μg/ml) for nuclear DNA. Coverslips were mounted on slides with Fluoromount G (Southern Biotechnology Associates, Inc., AL, U.S.). The cells were examined using a confocal laser scanning microscope (Zeiss LSM 880) at a magnification of 100X.
Statistical analysis
Statistical analyses were performed with GraphPad Prism 6.0 (GraphPad Software, Inc., San Diego, CA, U.S.). Data are presented as means ± s.d. All experiments were performed at least three times. We compared the results, of the treatment groups and controls, using Student’s t-test, the two-way analysis of variance (ANOVA) followed by the Tukey’s multiple-comparison posttest, or the χ2 (Chi-Squared) test. Differences between groups were considered to be significant at a P value of <0.05.
Results
The expression level of DNMT3A modulates erythrocytic differentiation
A previous report showed that NaB can induce erythrocytic differentiation by K562 cells [29]. In our study, treatment by NaB, for 24 hours, significantly increased DNMT3A protein expression (Figure 1A). In confirmation, the expression of Dnmt3a messages was detected, in different stages of erythroblast. Compared to early-stage erythroblasts (CD71highTER119high, Subgroup II), the Dnmt3a expression was significantly increased in the more mature erythroblasts (CD71lowTER119high, Subgroup IV) (Figure 1B). To examine the role of DNMT3A in erythrocytic differentiation, DNMT3A cDNA was inserted into the pcDNA3-HA2 vector [27] to express the hemagglutinin (HA)-tagged DNMT3A proteins. Two K562 cell clones with higher HA-DNMT3A expressions, HA-3A-E1 and HA-3A-K1, were selected (Figure 1C). Stable expression of HA-DNMT3A in K562 cells further promoted NaB-induced hemoglobin production (Figure 1D, lower panel), which was detected using a benzidine/hydrogen peroxide solution (Figure 1D, upper panel). In addition, three DNMT3A shRNAs were transiently expressed in K562 cells. Two DNMT3A shRNAs, sh3A-D2 and sh3A-E2, exhibited a higher knockdown efficiency, compared to sh3A-A2 (Figure 1E, upper panel). Downregulation of DNMT3A significantly decreased NaB-induced erythrocytic differentiation in a dose-dependent manner (Figure 1E, lower panel). Taken together, our results not only demonstrate the critical role of DNMT3A in erythrocytic differentiation, but also indicate that human K562 cells can be a reliable cellular model for studying the regulation of DNMT3A, during erythropoiesis.
Figure 1.
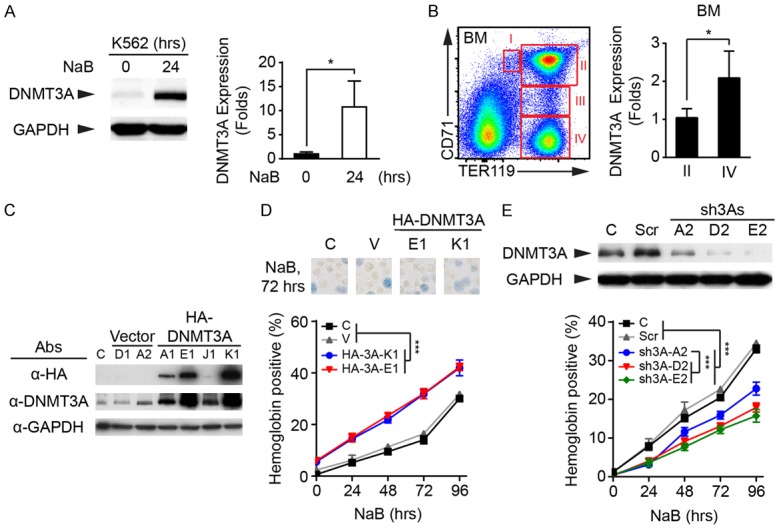
The increasing of DNMT3A expression promotes erythrocytic differentiation. (A) Human K562 cells were treated with NaB for 24 hours. Cells were harvested, then subjected to western blot analysis. The expression of DNMT3A was detected using anti-DNMT3A antibodies. The level of GAPDH proteins was used as a loading control (Left panel). The DNMT3A expression was quantified, then normalized to GAPDH (Right panel). (B) Different stages of erythroblasts (subgroup I-IV) were analyzed, using flow cytometry, and subgroup II (CD71highTER119high) and IV (CD71lowTER119high) were isolated for RNA extraction. The expression of Dnmt3a message was measured using the quantitative polymerase chain reaction (qPCR). (C) The empty vector and pcDNA3-HA2-DNMT3A plasmid were stably expressed in K562 cells. The ectopic DNMT3A proteins were detected using specific antibodies against HA-tag, and all DNMT3A proteins were detected using anti-DNMT3A antibodies. The level of GAPDH proteins was used as a loading control. (C) wild-type (WT) K562 cells. (D) The control (WT) and various K562 stable clones were treated with NaB for 72 hours. The hemoglobin expression were examined using benzidine/hydrogen peroxide solution (Upper panel). Overexpression of DNMT3A significantly increased erythrocytic differentiation (Lower panel). (E) Scrambled shRNA (Scr) and three DNMT3A shRNAs were transiently expressed in K562 cells. The DNMT3A level was detected using western blot analysis (Upper panel). The control (WT) and transfected cells were treated with NaB, and the expression of hemoglobin was measured (Lower panel). All experiments were repeated at least three times, and one representative set of western data is shown. The quantified data were presented as means ± s.d. (A and B) The Student’s t-test and (D and E) two-way ANOVA followed by the Tukey’s multiple-comparison posttest were used for statistical analysis. *P<0.05; ***P<0.001.
Induction of erythrocytic differentiation influences intracellular localization of DNMT3A
Although most reports showed that DNMT3A methylates genomic DNA in the nucleus, some suggested that DNMT3A can translocate into the cytoplasm or mitochondria [32,33]. Thus, we hypothesized that the intracellular localization of DNMT3A proteins might be changed during erythrocytic differentiation. K562 cells were transiently transfected with pcDNA3-HA2-DNMT3A plasmid, and then treated with NaB. Cells were harvested at controlled intervals, cytospun on a coverslip, and subjected to immunostaining with anti-HA antibodies. From confocal microscopic analysis, around 31% and 67% of untreated cells revealed cytoplasmic and diffuse (localized in both cytoplasm and nucleus) DNMT3A patterns, respectively. Six hours after NaB treatment, ~45% of cells exhibited nuclear HA-DNMT3A staining, and ~48% of cells showed diffuse HA-DNMT3A distributions (Figure 2 and Table 1). The intracellular distribution of DNMT3A, in early- and late-stage erythroblasts, was analyzed. Most early-stage erythroblasts (CD71highTER119high, Subgroup II) exhibited cytoplasmic and diffuse DNMT3A patterns, while around 50% of late-stage erythroblasts (CD71lowTER119high, Subgroup IV) showed nuclear DNMT3A staining (Figure 3). These results indicated that induction of erythrocytic differentiation would promote the translocation of DNMT3A into the nucleus.
Figure 2.
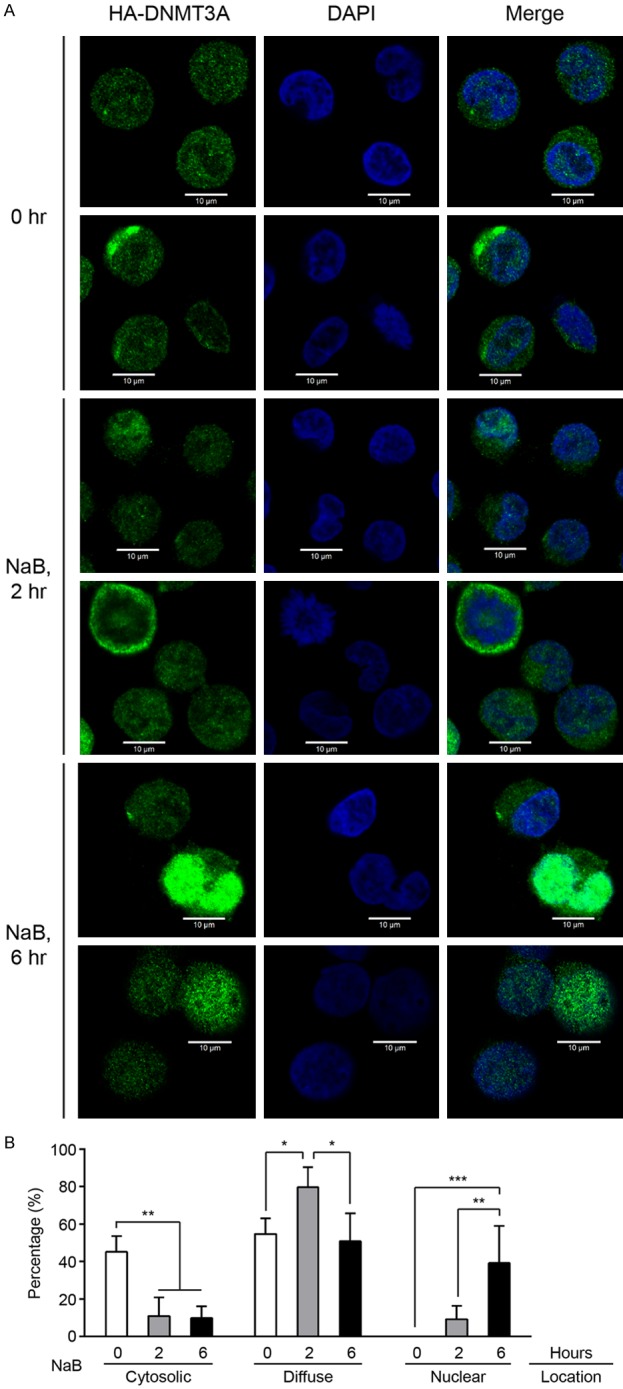
Induction of erythrocytic differentiation promotes the translocation of DNMT3A into the nucleus in K562 cells. A. The HA-DNMT3A-expressed cells were treated with NaB, then harvested at the indicated times for cytospin preparation. After fixation and blocking, the HA-DNMT3A proteins were detected using anti-HA antibodies, and then examined using confocal laser scanning microscopy. DAPI was used to label nuclear DNA. B. The quantified data were presented as means ± s.d. The two-way ANOVA followed by the Tukey’s multiple-comparison posttest was used for statistical analysis. *P<0.05; **P<0.01; ***P<0.001.
Table 1.
The intracellular distribution of various HA-DNMT3A proteins
| NaB | Loc. | WT | S255A | S255D | S255E | L373A | ||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
||||||
| Num. (%) | Num. (%) | χ2 Test | Num. (%) | χ2 Test | Num. (%) | χ2 Test | Num. (%) | χ2 Test | ||
| 0 hr | Cyto. | 51/166 (30.7%) | 14/36 (38.9%) | Compare to: | 3/33 (9.1%) | Compare to: | 5/37 (13.5%) | Compare to: | 34/38 (89.5%) | Compare to: |
| Diff. | 111/166 (66.9%) | 22/36 (61.1%) | WT: ns | 22/33 (66.7%) | WT: *** | 21/37 (56.8%) | WT: *** | 4/38 (10.5%) | WT: *** | |
| Nuc. | 4/166 (2.4%) | 0/36 (0%) | 8/33 (24.2%) | S255A: *** | 11/37 (29.7%) | S255A: *** | 0/38 (0%) | S255A: *** | ||
| S255D: ns | S255D: *** | |||||||||
| S255E: *** | ||||||||||
| 2 hr | Cyto. | 12/105 (11.4%) | 5/33 (31.7%) | Compare to: | 5/33 (15.2%) | Compare to: | 3/31 (9.7%) | Compare to: | 19/36 (52.8%) | Compare to: |
| Diff. | 84/105 (66.9%) | 28/41 (68.3%) | WT: *** | 18/33 (54.5%) | WT: ns | 21/31 (67.7%) | WT: ns | 17/36 (47.2%) | WT: *** | |
| Nuc. | 9/105 (8.6%) | 0/41 (0%) | 10/33 (30.3%) | S255A: *** | 7/31 (22.6%) | S255A: *** | 0/36 (0%) | S255A: ns | ||
| S255D: ns | S255D: *** | |||||||||
| S255E: *** | ||||||||||
| 6 hr | Cyto. | 8/110 (7.3%) | 22/37 (59.5%) | Compare to: | 3/33 (9.1%) | Compare to: | 5/31 (16.2%) | Compare to: | 27/42 (64.3%) | Compare to: |
| Diff. | 53/110 (48.2%) | 15/37 (40.5%) | WT: *** | 23/33 (69.7%) | WT: * | 20/31 (64.5%) | WT: ** | 15/42 (35.7%) | WT: *** | |
| Nuc. | 49/110 (44.5%) | 0/37 (0%) | 7/33 (21.2%) | S255A: *** | 6/31 (19.4%) | S255A: *** | 0/42 (0%) | S255A: ns | ||
| S255D: ns | S255D: *** | |||||||||
| S255E: *** | ||||||||||
Abbreviation: Loc.: Location; Cyto.: Cytoplasm; Diff.: Diffuse; Nuc.: Nucleus; Num.: Number. The χ2 (Chi-Squared) test were used for statistical analysis. *P<0.05; **P<0.01; ***P<0.001; ns: no significance.
Figure 3.
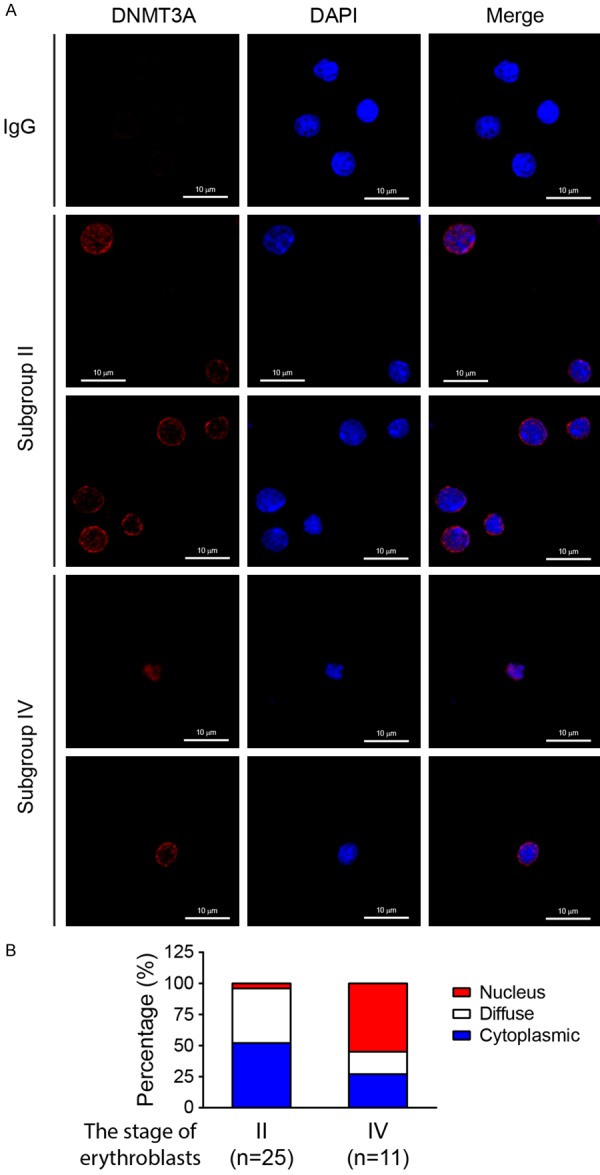
The nuclear accumulation of DNMT3A proteins is found in the late-stage of erythroblasts. A. The erythroblasts in subgroup II (CD71highTER119high) and IV (CD71lowTER119high) were isolated, and cytospun on a coverslip. After fixation and blocking, the endogenous DNMT3A proteins were detected, using anti-DNMT3A antibodies, and then examined using confocal laser scanning microscopy. DAPI was used to label nuclear DNA. B. The quantified data were presented. Subgroup II: n=25; Subgroup IV: n=11.
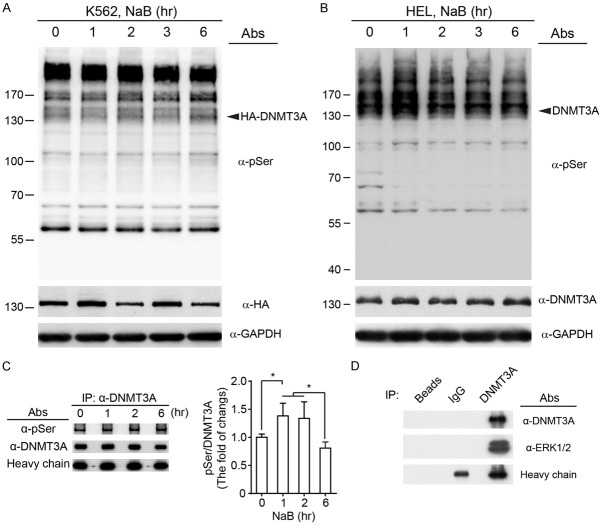
DNMT3A proteins interact with ERK1/2
The RAS-RAF-MEK-ERK1/2 cascade is one of the major downstream pathways of EPO signaling [12,13]. The serine/threonine kinase, ERK1/2, is the member of mitogen-activated protein kinase (MAPK) family, and participate in hematopoiesis and leukemogenesis [34]. A previous study has demonstrated that ERK1/2 can phosphorylate DNMT3A on residue S255 to participate in FGF-modulated chondrogenesis [25]. To test the association between DNMT3A and ERK1/2 in erythrocytic differentiation, we firstly applied the specific antibody against phosphorylated serine residues on MAPK/CDK substrates. Induction of erythrocytic differentiation seemed to dynamically promote the serine phosphorylation for ectopic HA-DNMT3A proteins in K562 cells (Figure 4A), as well as for endogenous DNMT3A proteins in HEL cells (Figure 4B). Next, the endogenous DNMT3A proteins were immunoprecipitated from HEL cells at intervals. The level of serine phosphorylation on DNMT3A proteins was increased after NaB treatment, peaking at one to two hours after treatment (Figure 4C). To examine whether DNMT3A was directly modulated by ERK1/2, DNMT3A proteins were immunoprecipitated from HEL cell lysates. Notably, ERK1/2 proteins were co-immunoprecipitated with DNMT3A proteins. In contrast, the protein G agarose beads and non-specific IgG interacted with neither DNMT3A nor ERK1/2 (Figure 4D). Taken together, these results suggested that DNMT3A proteins might be posttranslationally regulated by ERK1/2-mediated serine phosphorylation during erythropoiesis.
Figure 4.
DNMT3A protein is phosphorylated on serine residues, and interacts with ERK1/2. (A) The transfected K562 cells and (B) HEL cells were treated with NaB, and cells were harvested at the indicated times. The phosphorylated serine residues, on MAPK/CDK substrates, were detected (Upper panel). The ectopic and endogenous DNMT3A proteins were immunoprecipitated, using (A) anti-HA and (B) anti-DNMT3A antibodies, respectively. GAPDH was used as a loading control. (C) Endogenous DNMT3A proteins were immunoprecipitated from HEL cells. The serine phosphorylation of DNMT3A proteins was detected, quantified, and normalized to heavy chains. (D) The ERK1/2 proteins were co-immunoprecipitated with DNMT3A from HEL cell lysates. The protein G agarose beads and normal rabbit IgG were used as immunoprecipitation controls. All experiments were repeated at least three times, and one representative set of western data is shown. The two-way ANOVA followed by the Tukey’s multiple-comparison posttest was used for statistical analysis. The quantified results were presented as means ± s.d. *P<0.05; **P<0.01.
Either interaction or phosphorylation by ERK1/2 enhances erythrocytic differentiation by affecting intracellular localization of DNMT3A
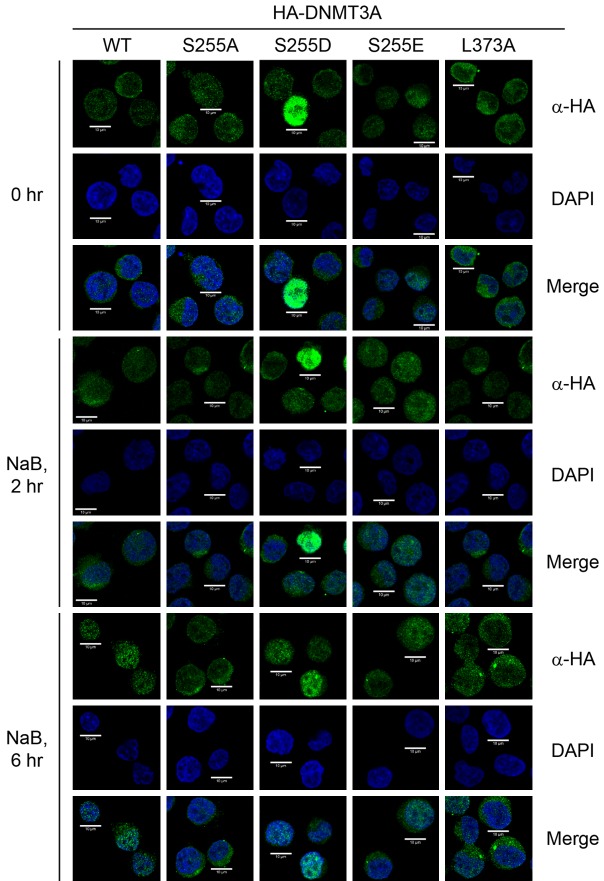
Following a previous report, which demonstrated that CK2 phosphorylates DNMT3A to modulate its localization [24], we hypothesized that the intracellular localization of DNMT3A proteins might be influenced by ERK1/2-mediated serine phosphorylation during erythrocytic differentiation. Kumar & Lassar (2014) refer to the generation of various DNMT3A mutations, including the DNMT3A S255A mutation (a dephosphomimetic mutant), S255D and S255E mutations (phosphomimetic mutants), as well as the DNMT3A L373A mutation (one of the ERK1/2 docking sites is mutated to decrease the interaction between DNMT3A and ERK1/2) [25]. Ectopic expression of DNMT3A phosphomimetic mutants (S255D and S255E) was found to enhance erythrocytic differentiation. However, overexpression of dephosphomimetic mutant (S255A) repressed differentiation (Figure 5A and 5B). In addition, partial disruption of ERK1/2-DNMT3A interaction (L373A) also decreased erythrocytic differentiation (Figure 5C and 5D).
Figure 5.
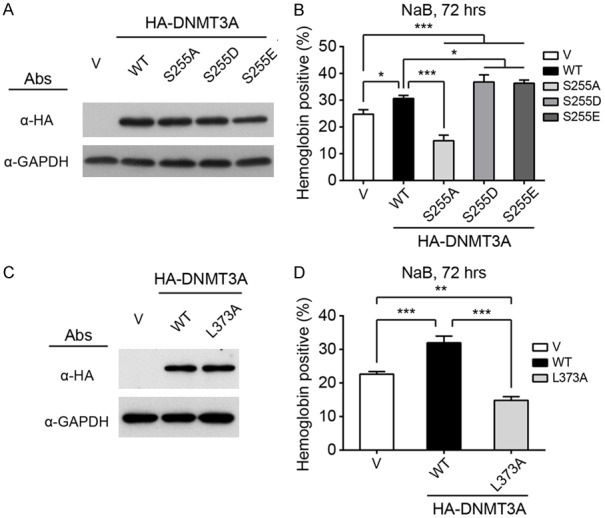
The interaction with and phosphorylation of DNMT3A by ERK1/2 are important for erythrocytic differentiation. A and C. HA-tagged WT and mutant DNMT3As were transiently expressed in K562 cells. The expression of WT and mutant DNMT3A proteins were detected using western blot analysis. GAPDH was used as a loading control. B and D. Hemoglobin expression was detected after 72-hour NaB treatment. All experiments were repeated at least three times, and one representative set of western data is shown. Differentiation data were presented as means ± s.d. The two-way ANOVA followed by the Tukey’s multiple-comparison posttest was used for statistical analysis. *P<0.05; **P<0.01; ***P<0.001.
Next, various DNMT3A mutants were used to investigate whether ERK1/2 modulates intracellular localization of DNMT3A. Notably, partial disruption of ERK1/2 docking (L373A) or the impairment of ERK1/2-mediated phosphorylation (S255A) significantly suppressed the translocation of DNMT3A into the nucleus. In contrast, phosphomimetic mutations (S255D and S255E) promoted nuclear accumulation of DNMT3A during differentiation (Figure 6 and Table 1). Taken together, our results demonstrated that DNMT3A modulates erythrocytic differentiation via ERK1/2-dependent protein-protein interaction and phosphorylation, which governs the intracellular localization of DNMT3A.
Figure 6.
Either interaction with or phosphorylation of DNMT3A by ERK1/2 modulates intracellular localization of DNMT3A. The HA-DNMT3A-expressed cells were treated with NaB, then harvested at the indicated times for cytospin preparation. After fixation and blocking, the HA-DNMT3A proteins were detected using anti-HA antibodies, and then examined using confocal laser scanning microscopy. DAPI was used to label nuclear DNA. One represented figure for each treatment is shown.
Discussion
Erythropoiesis is a complex and tightly-regulated process, which generates RBCs to maintain homeostasis. Using human cell lines and murine erythroblasts from BM cells, combined with overexpression and knockdown approaches, we demonstrated that upregulation of DNMT3A would promote erythrocytic differentiation (Figure 1). However, Socolovsky’s group (2011) found downregulation of Dnmt3a and Dnmt3b in differentiating erythroblasts isolated from fetal liver cells [21]. The mismatch in findings between the Socolovsky group and ours might be resulted from different sources of erythroblasts, because previous study indicated the developmental differences between neonatal and adult human erythropoiesis [35]. RAS-RAF-MEK-ERK1/2 signaling is one of the EPO-mediated signaling pathways involved in erythropoiesis [12,13]. Here, our results demonstrated that ERK1/2 interacts with DNMT3A, and this interaction, plus S255 phosphorylation, act to control nuclear DNMT3A accumulation, and thus to regulate erythropoiesis (Figure 7). In addition to a previous report showing that inhibition of ERK1/2 reduces DNMT3A expression in rat amygdala [36], our results provide another mechanism by which ERK1/2 modulates the intracellular distribution of DNMT3A. Our findings also explain discrepancies in results obtained by previous study of Dnmt3a+/-; KrasG12D and Dnmt3a-/-; KrasG12D mice [22].
Figure 7.
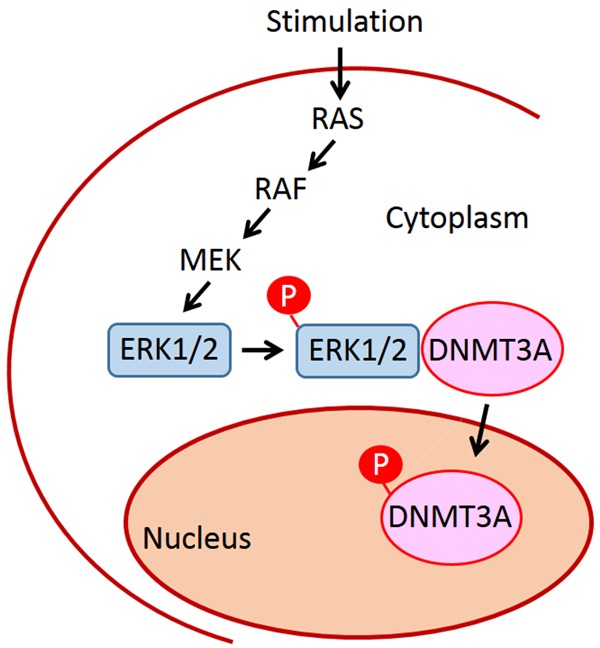
Schematic illustration summarizes that EPO’s downstream signaling molecule, ERK1/2, interacts with DNMT3A to promote the serine phosphorylation of DNMT3A, followed by nuclear accumulation of DNMT3A, to affect erythropoiesis.
Our data might provide a pathophysiological mechanism for the ERK1/2-DNMT3A axis in progression of polycythemia vera (PV). PV is a subcategory of myeloproliferative neoplasm (MPN), and the pathophysiological mechanism is not fully understood. PV is characterized by erythrocytosis, associated with the presence of the activating JAK2V617F mutation [37]. Constitutive JAK2 signaling usually activates numerous signaling molecules, including ERK1/2 [38]. In addition to PV, the JAK2V617F mutation is highly prevalent in other subgroups of MPN, including primary myelofibrosis (PMF) and essential thrombocythemia (ET) [37]. Similar to the progression of PV in patients, conditional expression of Jak2V617F from the endogenous promoter, in murine HSCs, is sufficient to develop MPN resembling human PV, and some mice further transform into a PMF-like phenotype [39,40]. Besides JAK2 mutations, recent studies identify DNMT3A mutations in a cohort of PV patients [41-43]. Loss of Dnmt3a cooperates with Jak2V617F to accelerate the transformation of PV into PMF in recipient mice [44]. In addition, abnormal activation of RAS and PI3K pathways are associated with increased proliferation and resistance to apoptosis of erythroid precursor cells, isolated from PV patients with JAK2V617F mutation [45]. Notably, combined JAK/MEK inhibition suppressed the activation of the MEK-ERK1/2 pathway in Jak2V617F mutant mice, with increased efficacy, and caused reversal of myelofibrosis to an extent not seen with JAK inhibitors [46]. These results raise the possibility of manipulation of the ERK1/2-DNMT3A axis to control PV progression. In addition, our previous report demonstrated that loss of Dnmt3a promotes myeloid malignancies, while Dnmt3a haploinsufficiency induces T-ALL in the context of oncogenic Kras [22]. Our finding, in this new study, might provide a novel insight into how DNMT3A cooperates with oncogenic signaling pathways to transform HSCs into different types of hematopoietic malignancies.
During chondrogenesis, Wnt signaling induces DNA methylation in the Sox9 promoter to repress differentiation. FGF signaling blocks the recruitment of DNMT3A to the Sox9 promoter by inducing the interaction between ERK1/2 and DNMT3A, and promoting phosphorylation of DNMT3A [25]. In contrast, our data show that ERK1/2-mediated DNMT3A S255 phosphorylation promotes the translocation of DNMT3A into the nucleus to increase differentiation (Figure 7). This suggests that diverse upstream signaling pathways differentially influence the consequence of ERK1/2-mediated DNMT3A phosphorylation. Indeed, ERK1/2 have been demonstrated to be involved in different cell functions, such as cell proliferation and apoptosis. Different upstream signaling pathways might affect cytosolic retention of activated ERK1/2 and ERK1/2 translocation to the nucleus, resulting in different cell fates [47]. In addition, CK2 phosphorylates DNMT3A to modulate its localization to heterochromatin [24]. Various cyclin-dependent kinases (CDKs) can phosphorylate DNMT1 to modulate the enzyme activity and protein stability of DNMT1 [48]. Taken together, multiple signaling pathways can directly regulate diverse aspects of DNMTs, such as intracellular localization, activity and stability, through protein phosphorylation. Still, further study is needed for a deeper understanding of the intricate interaction of signaling pathways that govern DNMTs to comprehensively elucidate the complex role of DNA methylation in pathophysiological processes.
Acknowledgements
We are grateful to Dr. Wey-Jinq Lin (National Yang-Ming University, Taiwan) for providing pcDNA3-HA2 vector. We thank the Instrumentation Research Center of National Yang-Ming University (Taiwan) for use of the shared services to complete this research. We also thank the National RNAi Core Facility at Academia Sinica in Taiwan for providing shRNA reagents and related services. This research was supported by funding from Ministry of Science and Technology (MOST 105-2628-B-010-010-MY3 and MOST 108-2320-B-010-043-MY3) (Y.I.C.), a grant from Yen Tjing Ling Medical Foundation (CI-106-11), the grant from Ministry of Education, Aim for the Top University Plan (105AC-P624 and 106AC-P624) (Y.I.C.), and “Yin Yen-Liang Foundation Development and Construction Plan” of the School of Medicine, National Yang-Ming University (Y.I.C.).
Disclosure of conflict of interest
None.
References
- 1.Smith ZD, Meissner A. DNA methylation: roles in mammalian development. Nat Rev Genet. 2013;14:204–220. doi: 10.1038/nrg3354. [DOI] [PubMed] [Google Scholar]
- 2.Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi: 10.3389/fonc.2014.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iwasaki H, Akashi K. Hematopoietic developmental pathways: on cellular basis. Oncogene. 2007;26:6687–6696. doi: 10.1038/sj.onc.1210754. [DOI] [PubMed] [Google Scholar]
- 4.Hogart A, Lichtenberg J, Ajay SS, Anderson S NIH Intramural Sequencing Center. Margulies EH, Bodine DM. Genome-wide DNA methylation profiles in hematopoietic stem and progenitor cells reveal overrepresentation of ETS transcription factor binding sites. Genome Res. 2012;22:1407–1418. doi: 10.1101/gr.132878.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hodges E, Molaro A, Dos Santos CO, Thekkat P, Song Q, Uren PJ, Park J, Butler J, Rafii S, McCombie WR, Smith AD, Hannon GJ. Directional DNA methylation changes and complex intermediate states accompany lineage specificity in the adult hematopoietic compartment. Mol Cell. 2011;44:17–28. doi: 10.1016/j.molcel.2011.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deaton AM, Webb S, Kerr AR, Illingworth RS, Guy J, Andrews R, Bird A. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21:1074–1086. doi: 10.1101/gr.118703.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bock C, Beerman I, Lien WH, Smith ZD, Gu H, Boyle P, Gnirke A, Fuchs E, Rossi DJ, Meissner A. DNA methylation dynamics during in vivo differentiation of blood and skin stem cells. Mol Cell. 2012;47:633–647. doi: 10.1016/j.molcel.2012.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dzierzak E, Philipsen S. Erythropoiesis: development and differentiation. Cold Spring Harb Perspect Med. 2013;3:a011601. doi: 10.1101/cshperspect.a011601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hattangadi SM, Wong P, Zhang L, Flygare J, Lodish HF. From stem cell to red cell: regulation of erythropoiesis at multiple levels by multiple proteins, RNAs, and chromatin modifications. Blood. 2011;118:6258–6268. doi: 10.1182/blood-2011-07-356006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levings PP, Bungert J. The human beta-globin locus control region. Eur J Biochem. 2002;269:1589–1599. doi: 10.1046/j.1432-1327.2002.02797.x. [DOI] [PubMed] [Google Scholar]
- 11.Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26:6697–6714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- 12.Fisher JW. Erythropoietin: physiology and pharmacology update. Exp Biol Med. 2003;228:1–14. doi: 10.1177/153537020322800101. [DOI] [PubMed] [Google Scholar]
- 13.Wojchowski DM, Menon MP, Sathyanarayana P, Fang J, Karur V, Houde E, Kapelle W, Bogachev O. Erythropoietin-dependent erythropoiesis: new insights and questions. Blood Cells Mol Dis. 2006;36:232–238. doi: 10.1016/j.bcmd.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Yu N, Liu J, Yi G, Ye F, Xiao J, Guo F. DNA methylation is necessary for erythropoietin to improve spatial learning and memory in SAMP8 mice. Exp Gerontol. 2015;69:111–115. doi: 10.1016/j.exger.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hundal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yan XJ, Xu J, Gu ZH, Pan CM, Lu G, Shen Y, Shi JY, Zhu YM, Tang L, Zhang XW, Liang WX, Mi JQ, Song HD, Li KQ, Chen Z, Chen SJ. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nature Genet. 2011;43:309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 17.Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, Leiserson MD, Miller CA, Welch JS, Walter MJ, Wendl MC, Ley TJ, Wilson RK, Raphael BJ, Ding L. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502:333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa JP, Godley LA, Li W, Goodell MA. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2011;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang YI, You X, Kong G, Ranheim EA, Wang J, Du J, Liu Y, Zhou Y, Ryu MJ, Zhang J. Loss of Dnmt3a and endogenous Kras cooperate to regulate hematopoietic stem and progenitor cell functions in leukemogenesis. Leukemia. 2015;29:1847–1856. doi: 10.1038/leu.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guryanova OA, Lieu YK, Garrett-Bakelman FE, Spitzer B, Glass JL, Shank K, Martinez AB, Rivera SA, Durham BH, Rapaport F, Keller MD, Pandey S, Bastian L, Tovbin D, Weinstein AR, Teruya-Feldstein J, Abdel-Wahab O, Santini V, Mason CE, Melnick AM, Mukherjee S, Levine RL. Dnmt3a regulates myeloproliferation and liver-specific expansion of hematopoietic stem and progenitor cells. Leukemia. 2016;30:1133–1142. doi: 10.1038/leu.2015.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shearstone JR, Pop R, Bock C, Boyle P, Meissner A, Socolovsky M. Global DNA demethylation during mouse erythropoiesis in vivo. Science. 2011;334:799–802. doi: 10.1126/science.1207306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang YI, Kong G, Ranheim EA, Tu PS, Yu YS, Zhang J. Dnmt3a haploinsufficiency cooperates with oncogenic Kras to promote an early-onset T-cell acute lymphoblastic leukemia. Am J Transl Res. 2017;9:1326–1334. [PMC free article] [PubMed] [Google Scholar]
- 23.Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, Challen GA, Li W, Wheeler D, Rebel VI, Goodell MA. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deplus R, Blanchon L, Rajavelu A, Boukaba A, Defrance M, Luciani J, Rothe F, Dedeurwaerder S, Denis H, Brinkman AB, Simmer F, Muller F, Bertin B, Berdasco M, Putmans P, Calonne E, Litchfield DW, de Launoit Y, Jurkowski TP, Stunnenberg HG, Bock C, Sotiriou C, Fraga MF, Esteller M, Jeltsch A, Fuks F. Regulation of DNA methylation patterns by CK2-mediated phosphorylation of Dnmt3a. Cell Rep. 2014;8:743–753. doi: 10.1016/j.celrep.2014.06.048. [DOI] [PubMed] [Google Scholar]
- 25.Kumar D, Lassar AB. Fibroblast growth factor maintains chondrogenic potential of limb bud mesenchymal cells by modulating DNMT3A recruitment. Cell Rep. 2014;8:1419–1431. doi: 10.1016/j.celrep.2014.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Andersson LC, Nilsson K, Gahmberg CG. K562--a human erythroleukemic cell line. Int J Cancer. 1979;23:143–147. doi: 10.1002/ijc.2910230202. [DOI] [PubMed] [Google Scholar]
- 27.Chang YI, Hua WK, Yao CL, Hwang SM, Hung YC, Kuan CJ, Leou JS, Lin WJ. Protein-arginine methyltransferase 1 suppresses megakaryocytic differentiation via modulation of the p38 MAPK pathway in K562 cells. J Biol Chem. 2010;285:20595–20606. doi: 10.1074/jbc.M109.092411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conde I, Pabon D, Jayo A, Lastres P, Gonzalez-Manchon C. Involvement of ERK1/2, p38 and PI3K in megakaryocytic differentiation of K562 cells. Eur J Haematol. 2010;84:430–440. doi: 10.1111/j.1600-0609.2010.01416.x. [DOI] [PubMed] [Google Scholar]
- 29.Hua WK, Chang YI, Yao CL, Hwang SM, Chang CY, Lin WJ. Protein arginine methyltransferase 1 interacts with and activates p38alpha to facilitate erythroid differentiation. PLoS One. 2013;8:e56715. doi: 10.1371/journal.pone.0056715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin P, Papayannopoulou T. HEL cells: a new human erythroleukemia cell line with spontaneous and induced globin expression. Science. 1982;216:1233–1235. doi: 10.1126/science.6177045. [DOI] [PubMed] [Google Scholar]
- 31.Anderson SA, Nizzi CP, Chang YI, Deck KM, Schmidt PJ, Galy B, Damnernsawad A, Broman AT, Kendziorski C, Hentze MW, Fleming MD, Zhang J, Eisenstein RS. The IRP1-HIF-2alpha axis coordinates iron and oxygen sensing with erythropoiesis and iron absorption. Cell Metab. 2013;17:282–290. doi: 10.1016/j.cmet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Li A, Zhang W, Huang Z, Wang J, Yi B. High glucose-induced cytoplasmic translocation of Dnmt3a contributes to CTGF hypo-methylation in mesangial cells. Biosci Rep. 2016;36 doi: 10.1042/BSR20160141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wong M, Gertz B, Chestnut BA, Martin LJ. Mitochondrial DNMT3A and DNA methylation in skeletal muscle and CNS of transgenic mouse models of ALS. Front Cell Neurosci. 2013;7:279. doi: 10.3389/fncel.2013.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung E, Kondo M. Role of Ras/Raf/MEK/ERK signaling in physiological hematopoiesis and leukemia development. Immunol Res. 2011;49:248–268. doi: 10.1007/s12026-010-8187-5. [DOI] [PubMed] [Google Scholar]
- 35.Yan H, Hale J, Jaffray J, Li J, Wang Y, Huang Y, An X, Hillyer C, Wang N, Kinet S, Taylor N, Mohandas N, Narla A, Blanc L. Developmental differences between neonatal and adult human erythropoiesis. Am J Hematol. 2018;93:494–503. doi: 10.1002/ajh.25015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Monsey MS, Ota KT, Akingbade IF, Hong ES, Schafe GE. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One. 2011;6:e19958. doi: 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine RL, Wadleigh M, Cools J, Ebert BL, Wernig G, Huntly BJ, Boggon TJ, Wlodarska I, Clark JJ, Moore S, Adelsperger J, Koo S, Lee JC, Gabriel S, Mercher T, D’Andrea A, Frohling S, Dohner K, Marynen P, Vandenberghe P, Mesa RA, Tefferi A, Griffin JD, Eck MJ, Sellers WR, Meyerson M, Golub TR, Lee SJ, Gilliland DG. Activating mutation in the tyrosine kinase JAK2 in polycythemia vera, essential thrombocythemia, and myeloid metaplasia with myelofibrosis. Cancer Cell. 2005;7:387–397. doi: 10.1016/j.ccr.2005.03.023. [DOI] [PubMed] [Google Scholar]
- 38.Meyer SC, Levine RL. Molecular pathways: molecular basis for sensitivity and resistance to JAK kinase inhibitors. Clin Cancer Res. 2014;20:2051–2059. doi: 10.1158/1078-0432.CCR-13-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mullally A, Lane SW, Ball B, Megerdichian C, Okabe R, Al-Shahrour F, Paktinat M, Haydu JE, Housman E, Lord AM, Wernig G, Kharas MG, Mercher T, Kutok JL, Gilliland DG, Ebert BL. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell. 2010;17:584–596. doi: 10.1016/j.ccr.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mullally A, Poveromo L, Schneider RK, Al-Shahrour F, Lane SW, Ebert BL. Distinct roles for long-term hematopoietic stem cells and erythroid precursor cells in a murine model of Jak2V617F-mediated polycythemia vera. Blood. 2012;120:166–172. doi: 10.1182/blood-2012-01-402396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jain P, Verstovsek S, Wang W, Loghavi S, Torres HA, Estrov Z, Patel KP, Pemmaraju N. DNMT3A, TET2, and JAK2 mutations in polycythemia vera following long-term remission of secondary acute myeloid leukemia. Leuk Lymphoma. 2016;57:1969–1973. doi: 10.3109/10428194.2015.1122785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lundberg P, Karow A, Nienhold R, Looser R, Hao-Shen H, Nissen I, Girsberger S, Lehmann T, Passweg J, Stern M, Beisel C, Kralovics R, Skoda RC. Clonal evolution and clinical correlates of somatic mutations in myeloproliferative neoplasms. Blood. 2014;123:2220–2228. doi: 10.1182/blood-2013-11-537167. [DOI] [PubMed] [Google Scholar]
- 43.Quintas-Cardama A, Abdel-Wahab O, Manshouri T, Kilpivaara O, Cortes J, Roupie AL, Zhang SJ, Harris D, Estrov Z, Kantarjian H, Levine RL, Verstovsek S. Molecular analysis of patients with polycythemia vera or essential thrombocythemia receiving pegylated interferon alpha-2a. Blood. 2013;122:893–901. doi: 10.1182/blood-2012-07-442012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacquelin S, Straube J, Cooper L, Vu T, Song A, Bywater M, Baxter E, Heidecker M, Wackrow B, Porter A, Ling V, Green J, Austin R, Kazakoff S, Waddell N, Hesson LB, Pimanda JE, Stegelmann F, Bullinger L, Dohner K, Rampal RK, Heckl D, Hill GR, Lane SW. Jak2V617F and Dnmt3a loss cooperate to induce myelofibrosis through activated enhancer-driven inflammation. Blood. 2018;132:2707–2721. doi: 10.1182/blood-2018-04-846220. [DOI] [PubMed] [Google Scholar]
- 45.Laubach JP, Fu P, Jiang X, Salter KH, Potti A, Arcasoy MO. Polycythemia vera erythroid precursors exhibit increased proliferation and apoptosis resistance associated with abnormal RAS and PI3K pathway activation. Exp Hematol. 2009;37:1411–1422. doi: 10.1016/j.exphem.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stivala S, Codilupi T, Brkic S, Baerenwaldt A, Ghosh N, Hao-Shen H, Dirnhofer S, Dettmer MS, Simillion C, Kaufmann BA, Chiu S, Keller M, Kleppe M, Hilpert M, Buser AS, Passweg JR, Radimerski T, Skoda RC, Levine RL, Meyer SC. Targeting compensatory MEK/ERK activation increases JAK inhibitor efficacy in myeloproliferative neoplasms. J Clin Invest. 2019;130:1596–1611. doi: 10.1172/JCI98785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mebratu Y, Tesfaigzi Y. How ERK1/2 activation controls cell proliferation and cell death: is subcellular localization the answer? Cell Cycle. 2009;8:1168–1175. doi: 10.4161/cc.8.8.8147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lavoie G, St-Pierre Y. Phosphorylation of human DNMT1: implication of cyclin-dependent kinases. Biochem Biophys Res Commun. 2011;409:187–192. doi: 10.1016/j.bbrc.2011.04.115. [DOI] [PubMed] [Google Scholar]