SUMMARY
Tafazzin (TAZ) is a mitochondrial transacylase that remodels the mitochondrial cardiolipin into its mature form. Through a CRISPR screen, we identified TAZ as necessary for the growth and viability of acute myeloid leukemia (AML) cells. Genetic inhibition of TAZ reduced stemness and increased differentiation of AML cells both in vitro and in vivo. In contrast, knockdown of TAZ did not impair normal hematopoiesis under basal conditions. Mechanistically, inhibition of TAZ decreased levels of cardiolipin but also altered global levels of intracellular phospholipids, including phosphatidylserine, which controlled AML stemness and differentiation by modulating toll-like receptor (TLR) signaling.
Graphical Abstract
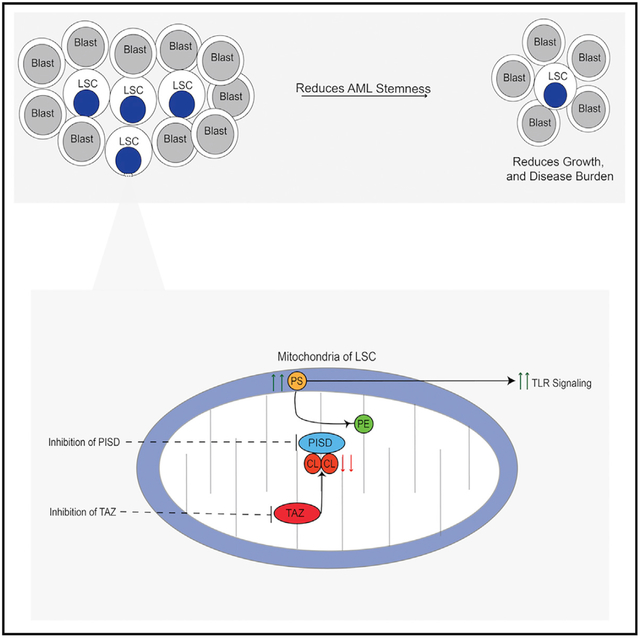
In Brief
Seneviratne et al. performed a CRISPR screen and identified tafazzin (TAZ) as important for the growth of leukemia cells. The inhibition of TAZ specifically reduced the stemness of leukemia cells by increasing phosphatidylserine levels and activating toll-like receptor signaling.
INTRODUCTION
Hematopoiesis is a hierarchical process in which hematopoietic stem cells give rise to differentiated precursors and mature blood cells. Like hematopoiesis, acute myeloid leukemia (AML) is also hierarchical with leukemic stem and progenitor cells giving rise to mature and differentiated leukemic blasts (Corces-Zimmerman et al., 2014; Jan et al., 2012; McCracken et al., 2016).
AML cells and stem cells have unique mitochondrial and metabolic features with greater reliance on oxidative phosphorylation (Cole et al., 2015; Kuntz et al., 2017; Lagadinou et al., 2013; Skrtić et al., 2011). Inhibiting mitochondrial processes such as mitochondrial protein translation impairs oxidative phosphorylation and is selectively cytotoxic to AML cells and stem cells (Skrtić et al., 2011). Thus, we conducted a genome-wide CRISPR screen for mitochondrial pathways whose inhibition targets AML cells and identified TAZ (Figure 1A).
Figure 1. CRISPR Screens Identify TAZ as an Essential Gene for the Growth and Viability of AML Cells.
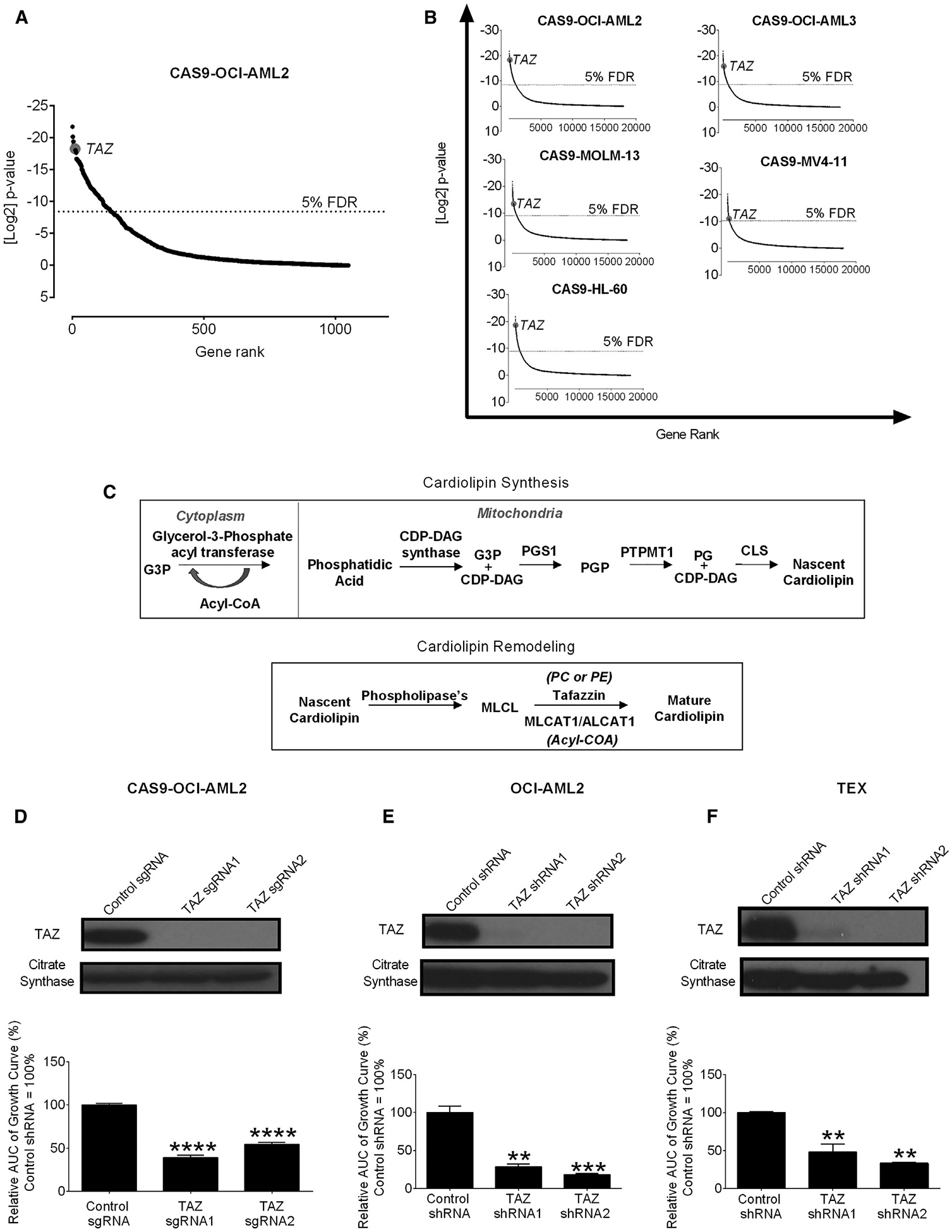
(A) Results of a dropout screen in Cas9-OCI-AML2 cells. Positive-hits were identified at a false discovery rate (FDR) <5%.
(B) The rank of TAZ in screens of OCI-AML2, OCI-AML3, MOLM-13, MV4–11, and HL-60 cells from the published CRISPR dropout screens by Tzelepis et al. (2016).
(C) A model of the enzymatic steps involved in cardiolipin synthesis and remodeling, where TAZ utilizes phosphatidylcholine (PC) or phosphatidylethanolamine (PE) as acyl chain donors to reacylate monolysocardiolipin (MLCL).
(D) Proliferation of CAS-9-OCI-AML2 cells after CRISPR-mediated knockout of TAZ. The relative area under the curve (AUC) of viable cell counts over 12 days are shown. Control sgRNA = 100%. Data are mean ± SEM (N = 3). ****p ≤ 0.0001 by one-way ANOVA and Dunnett’s post hoc test.
(E and F) Proliferation and TAZ protein expression of OCI-AML2 (E) and TEX cells (F) after shRNA-mediated TAZ knockdown. The relative AUC of viable cell counts over 12 days are shown (control shRNA = 100%). Data are mean ± SD (n = 2) of a representative experiment from 3 independent experiments. **p ≤ 0.01, ***p ≤ 0.001 by one-way ANOVA and Dunnett’s post hoc test.
Tafazzin (TAZ) is an X-linked gene that encodes a mitochondrial transacylase that is required for the production of the mitochondrial phospholipid cardiolipin (Lu and Claypool, 2015; Lu et al., 2016; Paradies et al., 2014). Cardiolipin is synthesized by a cascade of enzymes located in the cytoplasm and inner mitochondrial membrane (Figure 1C). Once synthesized, nascent cardiolipin is remodeled into a mature cardiolipin molecule (Claypool and Koehler, 2012; Lu and Claypool, 2015; Malhotra et al., 2009). TAZ is the enzyme responsible for the majority of cardiolipin remodeling under physiological conditions. Mutations or loss of TAZ leads to reductions in mature cardiolipin and increases the remodeling intermediate monolysocardiolipin (MLCL) (Claypool and Koehler, 2012; Lu and Claypool, 2015; Malhotra et al., 2009). Our objective was to characterize the role of TAZ in AML and hematopoiesis.
RESULTS
A CRISPR Screen Identifies TAZ as Essential for Growth and Viability of AML Cells
CAS9-expressing human OCI-AML2 leukemia cells were transduced with a library of 91,320 small guide RNAs (sgRNAs) in barcoded lentiviral vectors targeting 17,232 nuclear-encoded genes. Cells were harvested, genomic DNA was isolated, and the relative abundance of sgRNAs was determined by sequencing barcodes 17 days after transduction. sgRNAs able to reduce the viability or growth of OCI-AML2 cells were inferred to be those not represented in the final cell population. In analyzing the data, we focused on the sgRNAs targeting the 1,049 nuclear-encoded mitochondrial proteins. Top hits included sgRNAs targeting dihydroorotate dehydrogenase (DHODH), B cell lymphoma 2 (BCL2), and components of the mitochondrial ribosome, all of which have been previously shown to be necessary for the growth and viability of AML cells and stem cells (Table S1) (Skrtić et al., 2011; Sykes et al., 2016). We also identified the cardiolipin remodeling enzyme TAZ among the top 1% of mitochondrial hits (Figure 1A; Table S1). Of note, TAZ was also in the top 0.5% of hits in the whole genome. We also analyzed data from previously published CRISPR screens (Tzelepis et al., 2016) and identified TAZ as essential for the growth and viability of the leukemic cell lines OCI-AML2, OCI-AML3, MOLM-13, MV4–11, and HL-60 (Figure 1B).
TAZ is involved in the production of the mitochondrial-specific phospholipid cardiolipin (Claypool and Koehler, 2012; Lu and Claypool, 2015; Lu et al., 2016; Paradies et al., 2014) (Figure 1C) and we focused our investigation on this target. Using individual sgRNA, we confirmed that knockout of TAZ reduced the growth of CAS9-OCI-AML2 cells, thus validating the findings from our screen (Figure 1D).
Genetic Deficiency of TAZ Decreases Stemness and Increases Differentiation in AML
To understand how TAZ affects the growth of leukemia, OCI-AML2, TEX, U-937, and K562 leukemia cells were transduced with two independent short hairpin RNA (shRNA) targeting TAZ or control sequences in lentiviral vectors. Similar to knockout of TAZ by CRISPR, TAZ knockdown inhibited the growth of the tested leukemia cells (Figures 1E, 1F, S1A, and S1B). TAZ knockdown induced cell-cycle arrest but not apoptosis as measured by Annexin V staining (Figures S1C–S1F).
Leukemia stem and progenitor cells initiate and drive the propagation of AML, so we assessed the effects of TAZ knockdown on AML stem and progenitors and differentiation. First, we analyzed global changes in gene expression by RNA sequencing after TAZ knockdown in OCI-AML2 cells. The gene expression profile of TAZ knockdown AML cells was compared with genes that characterize primary AML stem cells (LSC+) and bulk cells (LSC−) (Ng et al., 2016). TAZ knockdown increased expression of genes associated with LSC− cells and decreased expression of genes associated with LSC+ cells (Figures 2A and 2B). As shown in Shlush et al. (2017), unsupervised clustering can be used to divide the TCGA AML cohort into two distinct clusters: one TCGA cluster is similar to stem- and progenitor-like cells, and the other TCGA cluster is similar to more mature myeloid cells (Figure S2A). Per the LSC gene set enrichment analysis, TAZ knockdown increased the expression of genes associated with the TCGA myeloid-like cluster and decreased the expression of genes in the TCGA stem- and progenitor-like cluster (Figure S2B). Besides, the gene expression after TAZ knockdown was compared to 20 different sub-fractions of normal hematopoietic cells from 38 human samples (Novershtern et al., 2011). Genes associated with more differentiated myeloid populations such as granulocyte-neutrophils were increased, after TAZ knockdown (Figures S2C and S2D).
Figure 2. TAZ Knockdown Reduces AML Stemness.
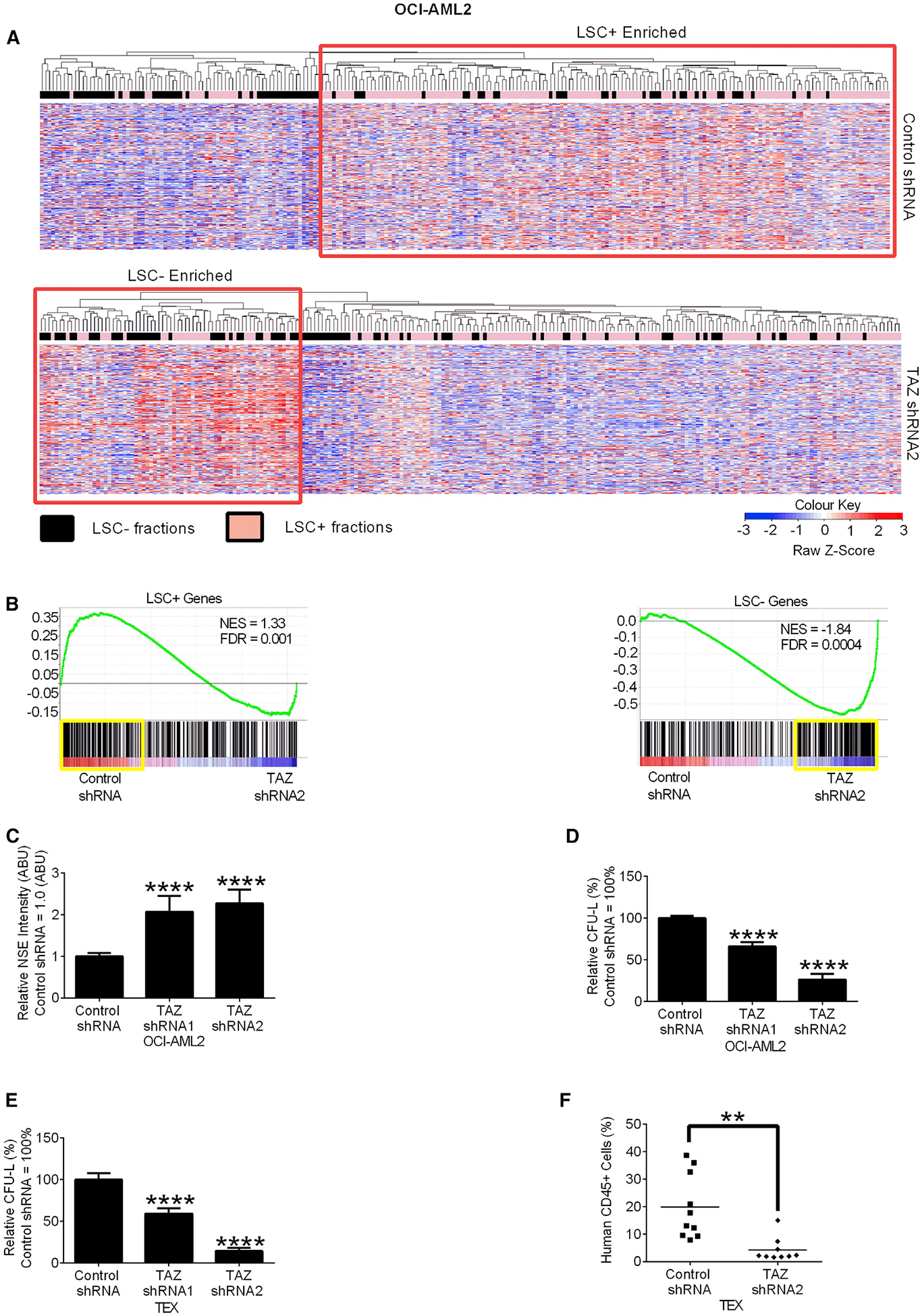
(A) Heatmap of standardized Z score expression of the 500 most highly upregulated genes in OCI-AML2 cells after TAZ shRNA knockdown. Rows represent genes and columns represent LSC+ (pink bars) or LSC− (black bars) fractions.
(B) Gene set enrichment analysis (GSEA) of OCI-AML2 cells from (A). The normalized enrichment scores (NES), and false discovery rates (FDRs) are indicated in each GSEA plot.
(C) Non-specific esterase (NSE) staining of OCI-AML2 cells after TAZ shRNA knockdown. Data are relative mean ± SEM (N = 4, control shRNA = 1.0 ABU). ****p ≤ 0.0001 by one-way ANOVA and Dunnett’s post hoc test.
(D and E) Clonogenic growth of OCI-AML2 (D) or TEX (E) after TAZ shRNA knockdown. Data are relative mean ± SEM (N = 3, control shRNA = 100%). ****p ≤ 0.0001 by one-way ANOVA and Dunnett’s post hoc test.
(F) CD45+TEXcell engraftment after TAZ shRNA knockdown. Bar represents mean (n = 10 mice control shRNA group, n = 9 mice TAZ shRNA2 group). **p ≤ 0.01 by Student’s t test.
See also Figure S2.
Given the changes in gene expression, we characterized the cellular phenotype of OCI-AML2 cells after TAZ knockdown. Non-specific esterase (Figure 2C), a marker of monocytic differentiation in vitro, was increased, although no changes in macrophage markers CD11b and CD14 were seen (Figures S2E and S2F). Increased differentiation was also observed when TAZ knockdown cells were grown in vivo, with increased expression of the myeloid granule protein, lysozyme (Sykes et al., 2016) (Figure S2G).
We then evaluated the effects of TAZ knockdown on stem and progenitor cell functions both in vitro and in vivo. In methylcellulose assays, TAZ knockdown reduced the clonogenic growth of OCI-AML2 and TEX cells. The reduction in clonogenic growth persisted upon serial re-plating (Figures 2D, 2E, and S2H). In xenotransplantation assays, TAZ knockdown in TEX cells reduced bone marrow engraftment in NOD-SCID-GF mice, suggesting that TAZ knockdown reduces AML propagation by reducing AML stemness both in vitro and in vivo (Figure 2F).
TAZ Knockdown Mice Have Normal Hematopoiesis
To assess the effects of TAZ inhibition on normal hematopoiesis in vivo, we analyzed doxycycline-inducible TAZ knockdown (iDOX-Taz-KD) mice. (Acehan et al., 2011; Soustek et al., 2011). 7.6- to 14.3-week-old adult iDOX-Taz-KD were fed DOX to induce Taz knockdown. After 12–19.4 weeks of DOX induction, we measured levels of TAZ and phospholipid levels. We observed reduced TAZ protein levels and an expected increase of the MLCL:cardiolipin ratio in bone marrow mononuclear cells (Figure 3A).
Figure 3. TAZ Knockdown Mice Are Viable with Normal Hematopoiesis but Have Reduced Numbers of Hematopoietic Stem Cells after Stress.
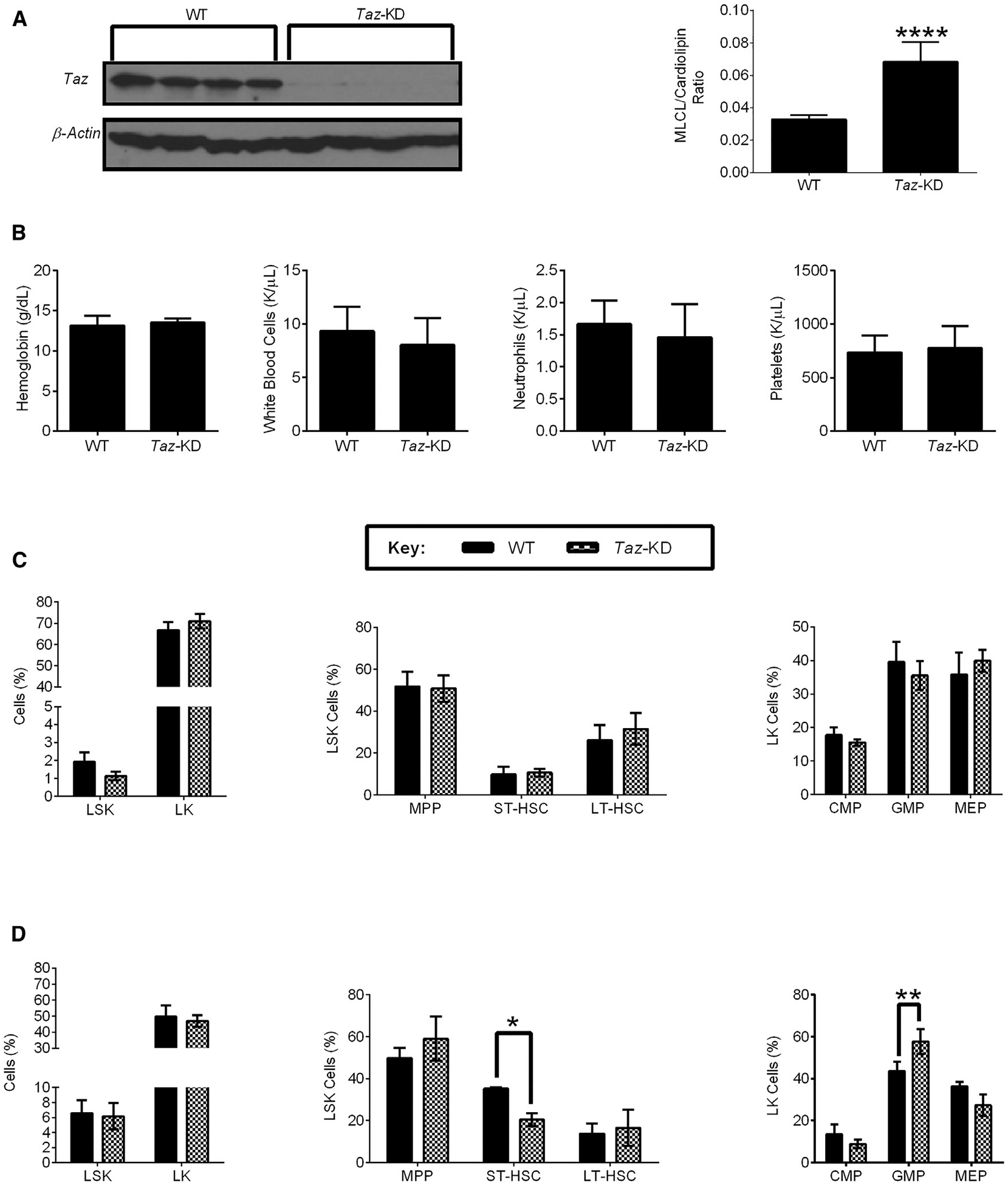
(A) TAZ expression and the MLCL:Cardiolipin ratio of 7.6- to 14.3-week-old wild-type (WT) and iDox-Taz-shRNA transgenic mice treated with doxycycline for 12.9–19.4 weeks to induce TAZ knockdown. Data are mean ± SEM (N = 3 mouse groups, n = 7 WT mice, and n = 6 Taz-KD mice). ****p ≤ 0.0001 by Student’s t test.
(B) Hemoglobin, white blood cells, neutrophils, and platelets of WT and Taz-KD mice treated with doxycycline as in (A). Data are mean ± SEM (N = 6 mouse groups, n = 13 WT mice, and n = 14 Taz-KD mice).
(C) The frequency of Lin−cKit+Sca1+ (LSK), Lin−cKit+ (LK), multipotent progenitor cells (MPP, CD48+,CD150−), short-term hematopoietic stem cells (ST-HSC, CD48+,CD150+), long-term hematopoietic stem cells (LT-HSC, CD48−,CD150+), common myeloid progenitor cells (CMP, CD34+, FcγR−), granulocyte-macrophage progenitor cells (GMP, CD34+, FcγR+), megakaryocyte-erythrocyte progenitors (MEP, CD34−, FcγR−) of WT and Taz-KD mice treated with doxycycline as in (A). Data are mean ± SEM (N = 4 mouse groups, n = 10 WT mice, and n = 8 Taz-KD mice).
(D) The frequency of LSK, LK, MPP, ST-HSC, LT-HSC, CMP, GMP, MEP in WT and Taz-KD 15 days after being injected with 5-FU. Both WT and Taz-KD mice were treated with doxycycline as in (A). Data are mean ± SEM (N = 2 mouse groups, n = 5 WT mice, and n = 6 Taz-KD mice). *p ≤ 0.05, **p ≤ 0.01 by two-way ANOVA and Bonferroni post hoc test.
See also Figure S3.
We then analyzed the hematopoietic system in these mice. Taz knockdown mice had normal levels of hemoglobin, white blood cells, neutrophils, and platelets (Figure 3B). We also enumerated the frequency of hematopoietic stem and progenitor cells by flow cytometry. Taz knockdown mice had normal levels of long-term hematopoietic stem cells (LT-HSC), short-term hematopoietic stem cells (ST-HSC), multipotent progenitor cells (MPP), mega-karyocyte-erythroid progenitor cells (MEP), common myeloid progenitor cells (CMP), and granulocyte/monocyte progenitor cells (GMP) (Figure 3C; Table S2).
To determine the role of TAZ in hematopoietic stress, wild-type (WT) or Taz knockdown mice were treated with 200 mg/kg of 5-fluorouracil (5-FU), and changes in white blood cells, neutrophils, lymphocytes, erythrocytes, and platelets were monitored (Figures S3A–S3F). WT and Taz-KD mice demonstrated reversible cytopenias after 5-FU treatment. Compared to controls, Taz knockdown mice showed decreased numbers of ST-HSC and increased numbers of GMP after 5-FU treatment (Figure 3D; Table S2). However, we observed little or no change in clonogenic growth in primary and serial re-plating experiments (Figure S3G). Thus, the lack of effect on stem cell function contrasts with the effects on AML cells.
TAZ Knockdown Alters Levels of Cellular Phospholipids
To understand the effects of TAZ knockdown on cardiolipin levels and composition in leukemia, we knocked down TAZ in OCI-AML2 cells and measured levels of MLCL and cardiolipin. As expected, TAZ knockdown increased MLCL:cardiolipin ratio (Figure 4A) in AML cells. TAZ also determined the final acyl composition of cardiolipin (Lu and Claypool, 2015; Lu et al., 2016; Paradies et al., 2014). Interestingly, TAZ knockdown AML cells had more cardiolipin acyl species with >5 double bonds (Figure 4B), as well as longer MLCL and cardiolipin acyl chain lengths (Figures S4A and S4B). Also, the predominant cardiolipin species in AML switched from 68:4 to 70:5 (Figures S4C–S4H). Cardiolipin binds to and activates caspase-8, which cleaves pro-apoptotic BID into t-bid, and facilitates extrinsic apoptosis (Gonzalvez et al., 2008; 2013). Mutation or deletion of TAZ renders cells resistant to extrinsic apoptosis. Similarly, TAZ knockdown rendered OCI-AML2 cells resistant to extrinsic apoptosis (Figure S4I). Thus, taken together, TAZ knockdown produced functionally significant reductions in levels of the phospholipid cardiolipin.
Figure 4. TAZ Knockdown Reduces TAZ Activity and Increases Phosphatidylserine, which Are Functionally Important in the Reduction of AML Stemness.
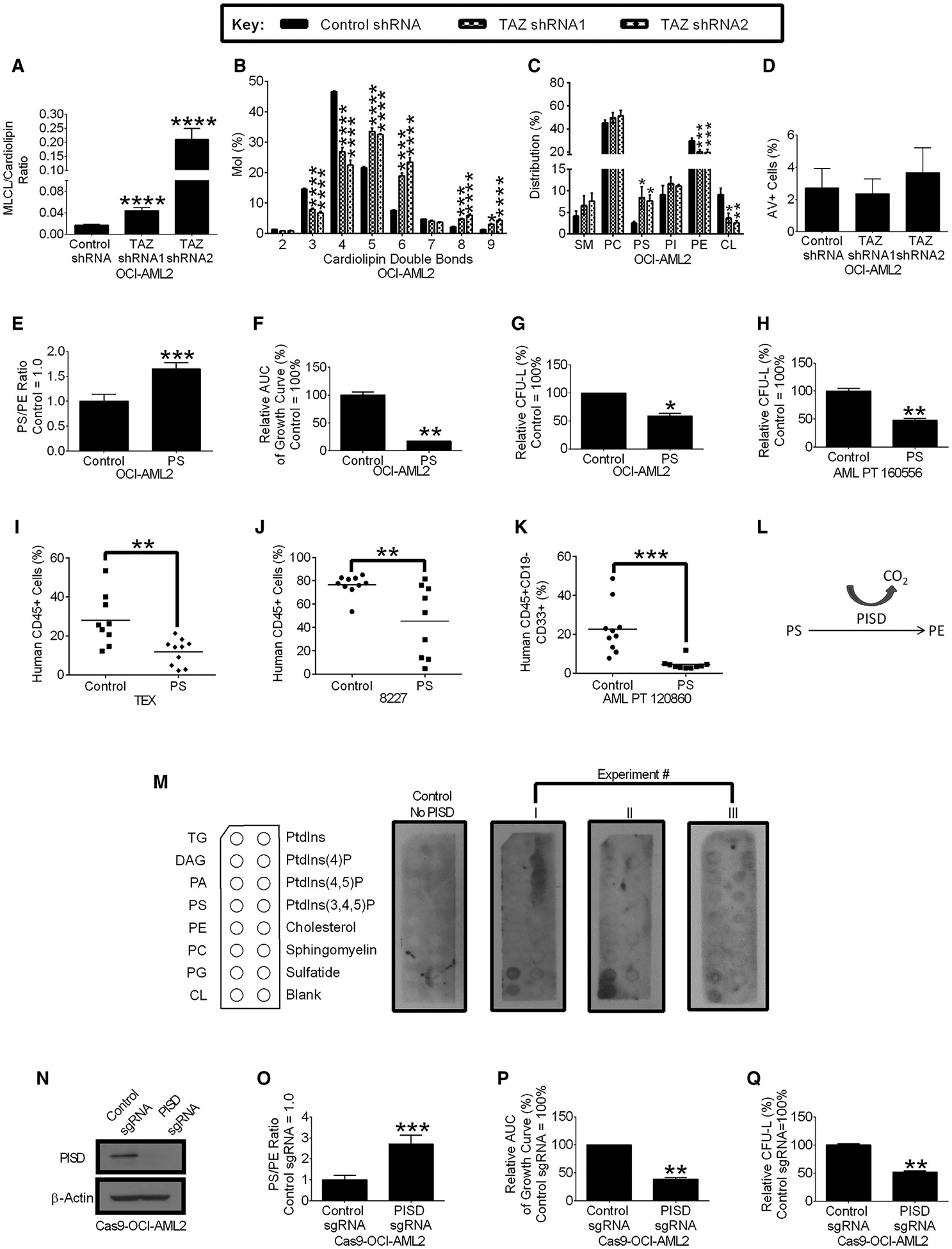
(A and B) The relative MLCL:CL ratio (A) and cardiolipin double bonds (B) in OCI-AML2 cells after TAZ knockdown. Data are mean ± SEM (N = 3). *p ≤ 0.05, ***p ≤ 0.001, ****p ≤ 0.0001 by Student’s t test (ML:CL ratio) or two-way ANOVA and post hoc Dunnett’s test (cardiolipin double bonds).
(C) Composition of sphingomyelin (SM), phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylinositol (PI), phosphatidylethanolamine (PE), and cardiolipin (CL) in OCI-AML2 cells after TAZ knockdown. Data are mean ± SEM (N = 3). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001 by two-way ANOVA, and Dunnett’s post hoc test.
(D) Extracellular PS levels in OCI-AML2 cells following TAZ knockdown. Data are mean ± SEM of 3 independent experiments.
(E) PS:PE ratio in OCI-AML2 cells supplemented with 25 μM PS or vehicle control. (Vehicle control = 1.0.) Data are mean ± SD (n = 4). ***p ≤ 0.001 by Student’s t test.
(F) Cell proliferation of OCI-AML2 cells supplemented with 25 μM PS or vehicle control. The relative AUC of viable cell counts over 14 days are shown. Data are relative mean ± SD of a representative experiment from 3 independent experiments (Vehicle control = 100%.) **p ≤ 0.01 by Student’s t test.
(G) Clonogenic growth of OCI-AML2 cells pre-treated with 25 μM PS or vehicle control before being seeded in methylcellulose medium without PS. Data are relative mean ± SD of a representative experiment from 3 independent experiments (Vehicle control = 100%.) *p ≤ 0.05 by Student’s t test.
(H) Clonogenic growth of primary AML cells pre-treated with 25 μM PS or vehicle control before seeded in methylcellulose medium without PS. Data are relative mean ± SD (n = 2, 0 μM = 100%). **p ≤ 0.01 by Student’s t test.
(I and J) Engraftment of TEX cells (I) or 8227 cells (J) treated with PS (25 μM) in NOD-SCID-GF mice. Bar represents mean. (n = 9–10 mice vehicle control group, n = 9–10 mice). **p ≤ 0.01 by Student’s t test.
(K) Engraftment of primary AML cells treated with PS (75 μM) or vehicle controls in NOD-SCID-GF mice. Bar represents mean. (n = 10 mice vehicle control group, n = 10 mice PS group.) ***p ≤ 0.001 by Student’s t test.
(L) The role of PS decarboxylase (PISD), where PISD decarboxylates PS to produce PE.
(M) Immunoblots measuring recombinant PISD protein bound to lipids. Data from 3 independent experiments are shown.
(N–Q) PISD protein expression (N) and PS:PE ratio (O) of Cas9-OCI-AML2 cells after CRISPR-mediated PISD knockout. Data are mean ± SD (n = 4). ***p ≤ 0.001 by Student’s t test.
(P) Proliferation of Cas9-OCI-AML2 cells after CRISPR-mediated PISD knockout. The relative AUC of viable cell counts over 15 days are shown. Data are mean ± SD of a representative experiment from three independent experiments (control sgRNA = 100%). **p ≤ 0.001 by Student’s t test.
(Q) Clonogenic growth of Cas9-OCI-AML2 cells after CRISPR-mediated PISD knockout. Data are relative mean ± SD of a representative experiment from three independent experiments (control sgRNA = 100%). **p ≤ 0.01 by Student’s t test.
See also Figures S4 and S5.
Cardiolipin is required for the stability, proper localization, and efficient function of respiratory chain enzymes (Paradies et al., 2014). However, we observed no decrease in basal oxygen consumption (Figure S4J), electron transport chain reserve capacity (Figure S4K), cellular ROS production (Figure S4L), mitochondrial mass (Figure S4M), mitochondrial structure by electron microscopy (Figure S4N), or mitochondrial length by confocal microscopy (Figure S4O). Thus, the change in cardiolipin levels after TAZ knockdown was not sufficient to impact mitochondrial structure or oxidative phosphorylation. It is noteworthy that, we did observe a decrease in glycolysis, which is consistent with differentiation (Figure S4P) (Gu et al., 2016).
Phospholipids are organized in a highly interconnected network, and perturbations in one species affect levels of other phospholipids (Köberlin et al., 2015). Therefore, we measured changes in cellular phospholipids by high-performance thin-layer chromatography (HPLC) after TAZ knockdown in OCI-AML2 and observed decreased levels of cardiolipin, as well as phosphatidylethanolamine (PE), increased levels of phosphatidylserine (PS), but no change in phosphatidylcholine (PC), phosphatidylinositol (PI), and sphingomyelin (SM) (Figure 4C). Of note, the increase in PS was intracellular, and no increase in PS was detected on the cell surface (Figure 4D).
We also evaluated changes in PS after TAZ knockdown in primary AML cells and the primary AML culture system 8227 (Lechman et al., 2016). TAZ knockdown increased intracellular PS in primary AML and 8227 cells (Figures S5A–S5C).
Increasing Intracellular PS Decreases AML Stemness and Increases Differentiation in AML
We then determined whether the changes in PE and PS were functionally essential to explain the effects of TAZ knockdown on AML. First, we tested whether the supplementation of PE or the PE substrate lysophosphatidylethanolamine (LPE) could protect cells from TAZ knockdown. Despite increasing PE levels, we saw no protection from TAZ knockdown by supplementing AML cells with PE or LPE (Figures S5D and S5F). Next, we tested whether supplementing cells with PS could increase differentiation and decrease stemness in AML. Supplementing AML cells with PS increased intracellular levels of PS and the PS:PE ratio (Figure 4E) and decreased AML cell growth (Figure 4F). Furthermore, pre-treatment of leukemia cells with PS reduced AML clonogenic growth (Figures 4G and 4H) and engraftment into mice (Figures 4I–K). PS decarboxylase (PISD) converts PS to PE in the inner mitochondrial membrane (Figure 4L), and we discovered that recombinant PISD binds preferentially to cardiolipin and the cardiolipin moiety phosphatidylglycerol (Figure 4M). Knockdown of TAZ decreased levels of PISD protein but not mRNA (Figure S5G) and did not change the levels of the PS synthesis enzymes PSS1 and PSS2 (Figure S5H). Therefore, to further characterize the effects of increasing PS on AML stemness and differentiation we evaluated the effects of PISD inhibition in AML. First, we knocked out PISD in CAS9-OCI-AML2 cells using sgRNA sequences in lentiviral vectors. Knockout of PISD was confirmed by immunoblotting (Figure 4N). The knockout of PISD increased the PS:PE ratio (Figure 4O) and reduced the growth (Figure 4P) and clonogenic growth (Figures 4Q and S5I) of CAS9-OCI-AML2 cells, without decreasing basal oxygen consumption or electron-transport chain reserve capacity in OCI-AML2 cells (Figures S5J and S5K). Furthermore, the overexpression of PISD rescued the differentiation phenotype of TAZ knockdown AML cells (Figures S5L and S5M).
As a chemical approach to inhibit PISD and increase intracellular levels of PS, we tested the reported inhibitor of Plasmodium falciparum PISD, MMV007285 (Figure 5A) (Choi et al., 2016; Lucantoni et al., 2013). Consistent with its effects as a PISD inhibitor, MMV007285 increased the PS:PE ratio in OCI-AML2 cells (Figure 5B). Concentrations of MMV007285 that increased PS also decreased growth (Figure 5C) and increased differentiation as evidenced by increase in non-specific esterase staining and CD11b expression (Figures 5D and 5E).
Figure 5. The PISD Inhibitor MMV007285 Reduces AML Growth and Stemness.
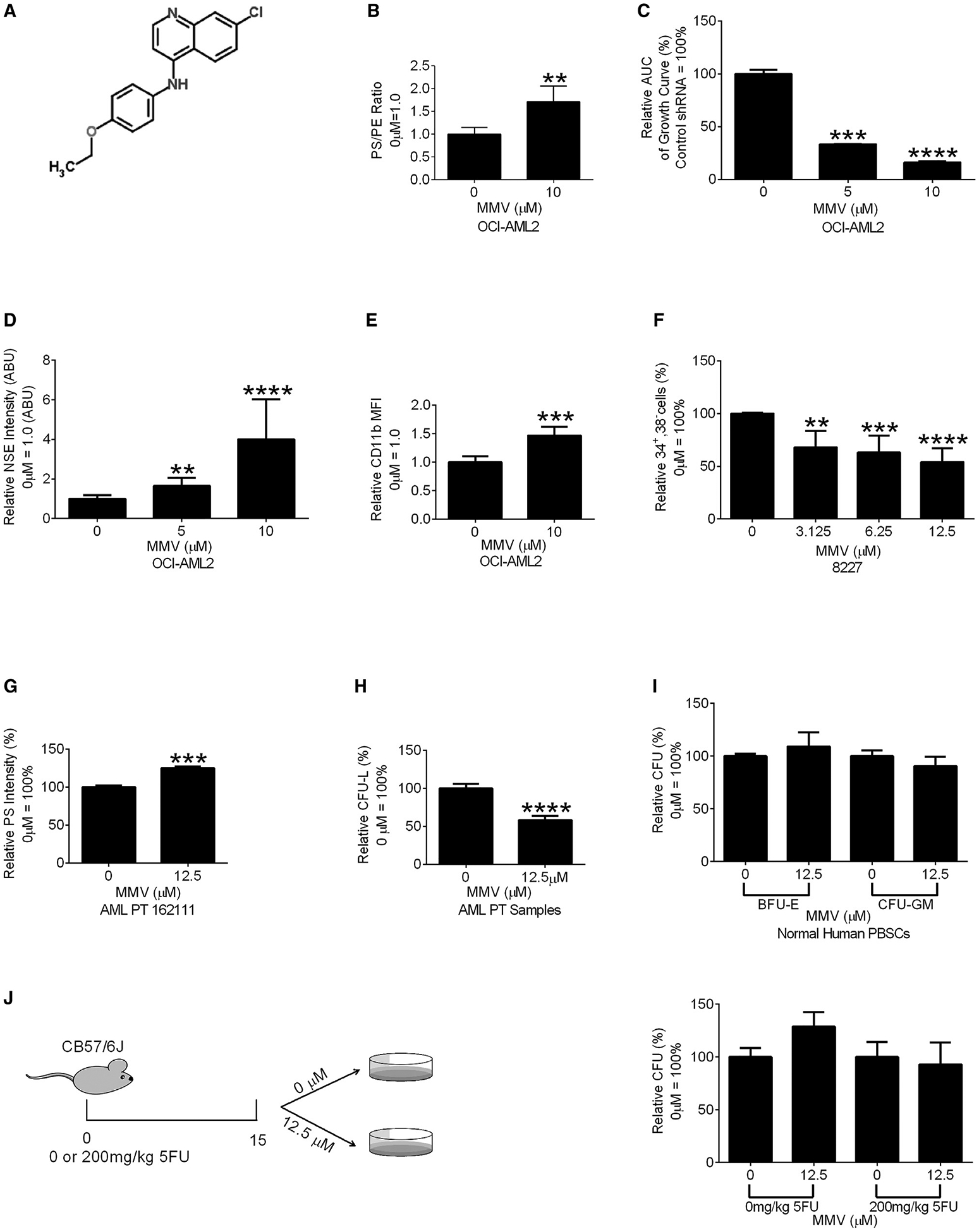
(A) Chemical structure of MMV007285.
(B) PS:PE ratio in OCI-AML2 cells treated with MMV007285 (10 μM) or vehicle controls. Data are relative mean ± SEM of three independent experiments (0 μM = 1.00). **p ≤ 0.01 by Student’s t test.
(C) Cell proliferation of OCI-AML2 cells treated with increasing concentrations of MMV007285 or vehicle control. Relative AUC of viable cell counts over 14 days are shown. Data are the relative mean ± SD (0 μM = 100%) of one of three independent experiments. ***p ≤ 0.001, ****p ≤ 0.0001 by one-way ANOVA and Dunnett’s post hoc test.
(D) NSE staining of OCI-AML2 cells incubated with MMV007285 or the vehicle control. Data represent relative mean ± SEM of three independent experiments (0 μM = 1 ABU). **p ≤ 0.01, ****p ≤ 0.0001 by Student’s t test.
(E) CD11b expression of OCI-AML2 cells incubated with MMV007285 or vehicle control. Data represent relative mean fluorescent intensity (MFI) ± SEM of two independent experiments (0 μM = 1.0). ***p ≤ 0.001 by Student’s t test.
(F) Frequency of viable CD34+,38− in MMV007285 and vehicle-treated samples. Data represent relative mean ± SEM of three independent experiments (0 μM = 100%). **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001 by one-way ANOVA and Dunnett’s post hoc test.
(G) Intracellular PS levels of primary AML cells treated with MMV007285 or the vehicle control. Data represent relative MFI ± SD (n = 3, control = 100%). ***p ≤ 0.001 by Student’s t test.
(H) Clonogenic growth of primary AML cells treated with MMV007285 or vehicle control for 48 h. Data represent mean ± SEM of three independent experiments (0 μM = 100%). ****p ≤ 0.0001 by a Student’s t test.
(I) Burst forming unit-erythroid (BFU-E) and colony forming unit-granulocyte/monocyte (CFU-GM) of normal hematopoietic cells treated with MMV007285 or vehicle control. Data represent mean ± SEM of three independent experiments (0 μM = 100%).
(J) Clonogenic growth of bone marrow cells from 21- to 29-week C567BL/6J mice injected with 200 mg/kg of 5-FU and treated with MMV007285 or vehicle control. Data represent mean ± SEM of two independent experiments (0 μM = 100%).
To understand the effects of inhibiting PISD and increasing PS on stemness in primary AML cells, we tested MMV007285 in the 8227 primary AML culture system. 8227 cells are arranged in a hierarchy of bulk and stem cells with the stem cells enriched in the CD34+38− fraction. Treatment with MMV007285 decreased the abundance of the CD34+38− stem cells (Figure 5F).
We also examined the effects of MMV007285 on the clonogenic growth of primary AML (N = 3) and normal hematopoietic cell (N = 3) samples. MMV007285 increased PS and inhibited the clonogenic growth of primary AML cells (Figures 5G and 5H) but did not affect the clonogenic growth of normal hematopoietic cells (Figure 5I). MMV007285 also did not alter the clonogenic growth of normal murine hematopoietic cells under conditions of hematopoietic stress (Figure 5J).
Increasing Intracellular PS Increases Toll-like Receptor Activity
To investigate how TAZ knockdown and increases in PS reduce AML stemness and increase differentiation, we characterized the genes that changed after the knockdown of TAZ in OCI-AML2 cells. Among the genes upregulated after TAZ knockdown, we observed a strong enrichment in toll-like receptor (TLR) and other immune activation pathways (false discovery rate [FDR] ≤0.01, Figure 6A). TAZ knockdown increased TLR4 and TLR8 expression as well as expression of their downstream cytokine mediators IL6 and INFβ (Figure 6B). Similar changes in TLR4 and TLR8 signaling were observed after inhibiting PISD at time points before differentiation was observed (Figure 6C).
Figure 6. TAZ-KD and PISD Inhibitor MMV007285 Activates TLR Signaling.
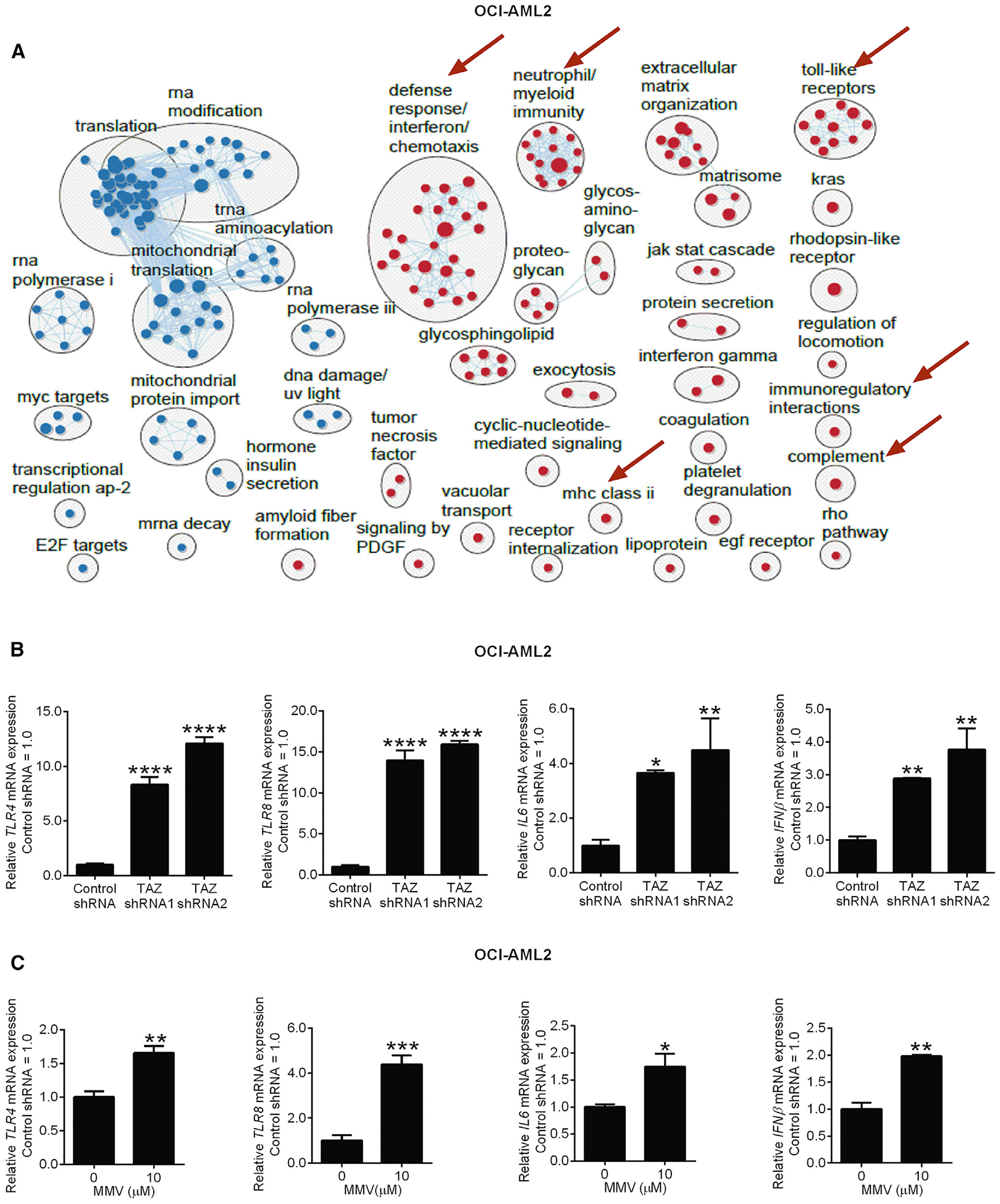
(A) Pathway enrichment analysis using GSEA, and visualization using Cytoscape EnrichmentMap in OCI-AML2 cells after TAZ knockdown. Circles (nodes) represent pathways, clusters represent biological processes, and lines connect pathways with common genes. Red nodes represent pathways that are upregulated, and blue nodes represent pathways that are downregulated in TAZ shRNA samples compared to control (FDR ≤0.01). Red arrows indicate upregulated immune pathways.
(B) qRT-PCR of toll-like receptor 4 (TLR4), toll-like receptor 8 (TLR8), interleukin-6 (IL6), interferon-beta (IFNβ) in OCI-AML2 cells after TAZ knockdown. Data are relative mean ± SD (n = 2–3, control shRNA = 1.0). *p ≤ 0.05, **p ≤ 0.01, ****p ≤ 0.0001 by one-way ANOVA and Dunnett’s post hoc test.
(C) qRT-PCR of toll-like receptor 4 (TLR4) and toll-like receptor 8 (TLR8), IL6, and IFNβ in OCI-AML2 cells treated with MMV007285 or vehicle control. Data are mean ± SD (n = 2–3, 0 μM = 1.0). *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, by Student’s t test.
See also Figure S6.
Treatment of AML cells with the TLR8 agonist, CL075 increased differentiation and decreased growth of AML cells similar to TAZ knockdown (Figures S6A–S6C). Moreover, the combination of the TLR8 agonist CL075 with the PISD inhibitor MMV007285 increased expression of TLR8 more than either drug alone and synergistically decreased growth of AML cells (Figures S6D and S6E). Collectively, these data suggest that the observed TLR activation is functionally important in promoting AML differentiation.
Increasing Intracellular PS Decreases AML Growth and Stemness In Vivo
Next, we investigated whether altering PS affects AML growth in vivo. We knocked down TAZ in 8227 cells as well as primary AML samples (N = 4) and injected control and knockdown cells into the femurs of immunodeficient mice. TAZ knockdown reduced the engraftment potential (Pei et al., 2018) of these primary cells into the mouse marrow (Figures 7A–7E and S7A–S7E).
Figure 7. Increased PS Reduced Leukemia Burden in Xenograft Models of Human Leukemia.
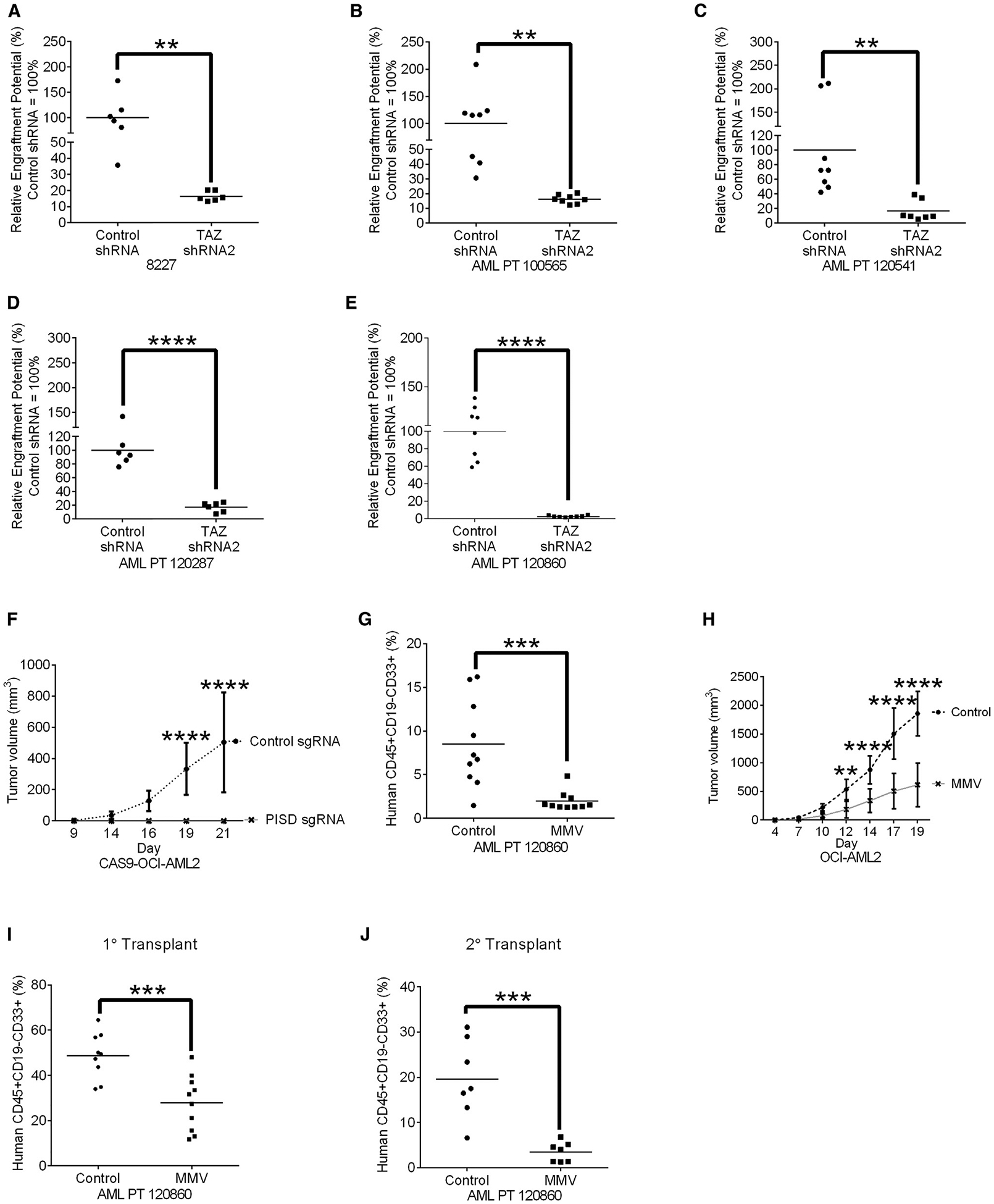
(A) Engraftment of 8227 cells in NOD-SCID-GF mice after TAZ knockdown. Bar represents mean engraftment potential (n = 6 mice/group, control shRNA = 100%), **p ≤ 0.01 by Student’s t test.
(B–E) Engraftment of primary AML cells from patients 100565 (B), 120541 (C), 120287 (D), and 120860 (E) in NOD-SCID mice after TAZ knockdown. Bar represents mean engraftment potential (n = 6–8 mice/group, control shRNA = 100%), **p ≤ 0.01, ****p ≤ 0.0001 by Student’s t test.
(F) Tumor growth of Cas9-OCI-AML2 cells after PISD knockout. Data represent mean ± SD (n = 8 mice/group). ****p ≤ 0.0001 by two-way ANOVA and Bonferroni’s post hoc test.
(G) Primary AML cell engraftment in NOD-SCID after pre-treatment in vitro of cells for 48 h with 12.5 μM of MMV00728. Data represent mean ± SD (n = 10 mice/group). ***p ≤ 0.01 by Student’s t test.
(H) Tumor growth of OCI-AML2 cells xenografts into SCID mice treated with 300 mg/kg of MMV007285 or vehicle control twice daily for 5 days/week orally (n = 10 mice/group). Data represent mean ± SD, **p ≤ 0.01. ****p ≤ 0.0001 by two-way ANOVA and Bonferroni’s post hoc test.
(I) Engraftment of primary AML cells in NOD-SCID mice treated once daily with MMV007285 (150 mg/kg orally) for 5 days/week Bar represents mean (n = 9 control group, n = 10 MMV007285 group). ***p ≤ 0.001, as determined by Student’s t test
(J) Secondary engraftment of primary AML cells from (I). Bar represents mean (n = 7 mice/group). ***p ≤ 0.001, as determined by Student’s t test.
See also Figure S7.
We also knocked out PISD in CAS9-OCI-AML2 cells and monitored their growth, to determine the role of PISD in leukemia propagation in vivo. CAS9-OCI-AML2 cells lacking PISD failed to form tumors in SCID mice (Figure 7F).
When we treated primary AML cells with the PISD inhibitor MMV007285 or vehicle control for 48 h and injected an equal number of viable cells into sub-lethally irradiated NOD-SCID mice. Primary AML cells pre-treated with MMV007285 had significantly lower levels of leukemic engraftment compared to primary AML cells pre-treated with the vehicle control (Figure 7G), suggesting that pre-treatment with MMV007285 was sufficient to reduce the leukemia-initiating cell frequency in AML.
Although MMV007285 has a short half-life (Figure S7F), we evaluated the anti-leukemic efficacy and toxicity of systemic treatment with MMV007285 in mouse models of leukemia. SCID mice xenografted with OCI-AML2 cells were treated with MMV007285 or vehicle control. Treatment with MMV007285 delayed the growth of OCI-AML2 cells in mice (Figure 7H) without a more than 10% reduction in body weight (Figure S7G) or alterations in serum chemistry measuring liver, muscle, or renal toxicity (Figure S7G).
Last, we treated mice engrafted with primary AML cells with MMV007285. AML cells were injected intrafemorally into NOD-SCID mice. Eleven days after injection, mice were treated with MMV007285 or vehicle control for 4 weeks. After treatment, levels of AML engraftment in the marrow of the mice was determined by flow cytometry. Treatment of mice with MMV007285 reduced levels of leukemic engraftment compared to controls with no evidence of toxicity (Figures 7I and S7H). We then harvested leukemia cells from the marrow of primary mice and injected these cells into secondary mice. The leukemia grafts from MMV00285-treated mice demonstrated an 80% reduction in engraftment compared to vehicle-treated controls, indicating that MMV007285 inhibits both leukemia propagation and targets leukemia stem cells (Figure 7J).
DISCUSSION
TAZ is a transacylase responsible for the generation of mature cardiolipin (Lu and Claypool, 2015). Through a CRISPR screen to identify genes necessary for the growth and viability of AML cells, we identified TAZ as a top hit. Cardiolipin is a mitochondrial phospholipid that undergoes a remodeling process, whereby enzymes such as TAZ alter the acyl chain composition of nascent cardiolipin to produce mature cardiolipin (Lu and Claypool, 2015). Interestingly, both nascent and mature cardiolipin can support oxidative phosphorylation (Baile et al., 2014). The remodeling intermediate MLCL is a byproduct of this reaction, and an optimal MLCL cardiolipin ratio, which is normally very low, is essential for the stability of respiratory chain supercomplexes, which are necessary for the optimal function of the respiratory chain and oxidative phosphorylation (Baile et al., 2014; Cheneval et al., 1983; Claypool and Koehler, 2012; Claypool et al., 2006, 2008; Eble et al., 1990; Lu and Claypool, 2015; Lu et al., 2016; Paradies et al., 2014; Shinzawa-Itoh et al., 2007). Knockdown of TAZ in AML cells increased MLCL levels, cardiolipin acyl chain length, cardiolipin acyl chain double bonds, and decreased total cardiolipin levels, consistent with reductions in TAZ function. However, despite reductions in levels of cardiolipin and acyl chain composition, mitochondrial structure and oxidative phosphorylation were preserved. Therefore, the increase in MLCL and reductions in cardiolipin after inhibiting TAZ were not sufficient to impair mitochondrial function, in keeping with the ability of nascent cardiolipin to support respiratory chain function. Alternatively, MLCL acyltransferase 1 (MLCLAT1) and acyl-CoA:lysocardiolipin acyltransferase-1 (ALCAT1) also contribute to cardiolipin remodeling, so the redundancy in the cardiolipin acyl chain remodeling pathway potentially maintained sufficient levels of cardiolipin to preserve mitochondrial function (Claypool and Koehler, 2012; Taylor and Hatch, 2003, 2009; Taylor et al., 2012; Xu et al., 2006). However, we did observe an increase in oxidative phosphorylation and a decrease in glycolysis after TAZ knockdown. Thus, while AML cells have an increased reliance on mitochondrial metabolism (Cole et al., 2015; Skrtić et al., 2011; Sriskanthadevan et al., 2015), the reductions in AML growth after TAZ inhibition could not be explained by the changes in glycolysis and oxidative phosphorylation observed, but instead these metabolic changes seem to highlight the effects of differentiation.
Barth syndrome is an inherited disorder due to inactivating mutations in the X-linked Taz gene. Individuals with Barth syndrome are viable but have mitochondrial defects that lead to cardiac dysfunction and skeletal muscle weakness (Acehan et al., 2011; Saric et al., 2016). Patients with Barth syndrome also experience transient episodes of neutropenia, but the mechanism for this hematologic toxicity has not been explained to date. We showed that knockdown of TAZ did not affect normal hematopoiesis under basal conditions in mice. Interestingly, when subjected to hematopoietic stress, TAZ knockdown mice had a 40% decrease in ST-HSC and 10% increase in GMP cells, although no change in clonogenic growth was observed. Thus, our results provide a potential mechanism for the transient episodes of neutropenia seen in Barth syndrome. As reported for other conditions that lead to chronic or cyclic neutropenia, repeated episodes of hematopoietic stress can reduce the ST-HSC reserve and produce neutropenia (Bartels et al., 2016; Colijn and Mackey, 2005; Haurie et al., 1998; Mitroulis et al., 2017).
Lipid metabolites are organized in a highly interconnected network, and alterations in levels of one lipid species affect the levels of other intracellular lipids (Köberlin et al., 2015). For example, genetic perturbations in the sphingomyelin synthesis pathway revealed that the synthesis of the glycerophospholipids PS and PE are co-regulated, while PC synthesis is co-regulated with phosphatidylglycerol (Köberlin et al., 2015). Furthermore, ceramide and sphingomyelin synthesis are co-regulated with glycerophospholipids (Köberlin et al., 2015). In this study, we demonstrated that the levels of PS are influenced by cardiolipin metabolism.
Altering cellular lipid composition modulates the activity of some cell signaling pathways including TLR pathways (Huang et al., 2011; Kay and Grinstein, 2013; Köberlin et al., 2015). We have shown that increasing PS levels activates TLR4 and TLR8 signaling. Interestingly, TLR signaling is a known regulator of hematopoietic and AML differentiation (Hayashi et al., 2010; Ignatz-Hoover et al., 2015; Köberlin et al., 2015; Megías et al., 2012; Nagai et al., 2006). The stimulation of TLR4 drives myeloid differentiation during emergency hematopoiesis in response to infection (Hayashi et al., 2010; Megías et al., 2012; Nagai et al., 2006). Also, the TLR8 agonist R848 promotes AML differentiation and reduces tumor burden both in vivo and in vitro (Ignatz-Hoover et al., 2015).
Increasing intracellular levels of PS by supplementing cells with this phospholipid decreased AML growth and stemness, and thus phenocopied the effects of TAZ inhibition. These results are consistent with a prior study where PS supplementation promoted the osteogenic differentiation of mesenchymal stromal cells (Xu et al., 2013). We also increased intracellular PS and reduced AML stemness by genetically or chemically inhibiting PISD. As a chemical approach, we used the PISD inhibitor MMV007285 (Choi et al., 2016; Lucantoni et al., 2013). MMV007285 was initially identified through a screen for inhibitors of malarial PISD, but our results show that MMV007285 also targets the mammalian form of PISD. Reducing levels of PISD in breast cancer cells induced their differentiation (Keckesova et al., 2017). Thus, similar to breast cancer inhibiting PISD in AML reduced stemness and promoted differentiation and could be a therapeutic strategy for AML. Although a useful tool to study PISD in vitro, MMV007285 has a short half-life when administered systemically to mice. Therefore, developing more potent and stable PISD inhibitors based on other scaffolds is a priority to fully assess the efficacy and toxicity of PISD inhibition in vivo and evaluate PISD as a potential therapeutic target for AML.
In summary, our study suggests that phospholipid composition is critical for leukemia, stemness, initiation, and propagation. Although AML is genetically diverse, phospholipid metabolism may be amendable for therapeutic exploitation.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and request for resources and reagents should be directed and will be fulfilled by the Lead Contact, Dr. Aaron Schimmer (aaron.schimmer@uhn.ca).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human Cell Lines
OCI-AML2, and K562 cells were maintained in IMDM (Iscove’s modified Dulbecco’s medium), supplemented with 10% FBS (fetal bovine serum), 100 units/mL penicillin and 100 μg/mL of streptomycin. TEX cells obtained from John Dick’s lab (Warner et al., 2005) were cultured in IMDM, with 20% FBS, 2 mM L-glutamine, 100 units/mL penicillin and 100 μg/mL of streptomycin, 20 ng/mL stem cell factor (SCF), and 2 ng/mL interleukin-3 (IL-3). U-937 cell were maintained in RPMI 1640 medium, with 10% FBS, 100 units/mL penicillin and 100 μg/mL of streptomycin. K562 and U-937 cells were authenticated via STR profiling. 8227 cells were also provided by John Dick’s lab (Princess Margaret Cancer Centre, University Health Network, Toronto, Canada) (Lechman et al., 2016) and were cultured in X-VIVO-10, with 20% bovine serum albumin-insulin-transferrin (BIT), Fms-related tyrosine kinase 3 ligand (Flt3-L, 50 ng/ml), interleukin-6 (IL-6,10 ng/ml), SCF (50 ng/ml), thrombopoietin (TPO, 25 ng/ml), IL-3 (10 ng/ml), granulocyte colony-stimulating factor (G-CSF, 10 ng/ml). Lentiviral packing cells (293T) were cultured in DMEM with 10% FCS for seeding, and DMEM with 10% FCS, 100 units/mL penicillin and 100 μg/mL of streptomycin, as well as 1% BSA for harvesting of virus. All cell lines were maintained at 37°C, supplemented with 5% CO2. The sex and age of the patients from whom the cell lines were generated are indicated in Table S3.
Animals
Six-twelve week Male or female immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NOD-SCID-GF) mice used to transplant TEX and 8227 cells were obtained from Dr. Connie J. Eaves and bred in our facility (Nicolini et al., 2004). Eight-twelve week female immunodeficient NOD.CB17-Prkdcscid/J (NOD-SCID) and 6–8 week immunodeficient male Prkdcscid (SCID) mice, used for the transplantation of primary AML and OCI-AML2 cells respectively were obtained from the Ontario Cancer Institute. Mice were randomly assigned to each experimental group.
Immunocompetent B6.Cg-Gt(ROSA)26Sortm37(H1/tetO-RNAi:Taz)Art/ZkhuJ doxycycline-Inducible-TAZ-Knockdown (iDOX-Taz-KD) transgenic mice were acquired from Dr. Zaza Khuchua, at the University of Cincinnati. In collaboration with TaconicArtemis GmbH Dr. Khuchua generated the iDOX-Taz-KD mice, in a C57BL/6J genetic background as previously described (Acehan et al., 2011). iDOX-Taz-KD transgenic mice transferred to our facility, were backcrossed with female immunocompetent C57BL/6J mice for 7–10 generations. To maintain iDOX-Taz-KD transgenic mice on a C57BL/6J genetic background, iDOX-Taz-KD male mice were bred with either wild-type littermates or C57BL/6J female mice. iDOX-Taz-KD mice were weaned onto a standard chow diet and genotyped for the iDOX-Taz-shRNA transgene by PCR of the tail genomic DNA (Table S4). Note that, iDOX-Taz-KD transgenic mice were born viable at the expected Mendelian ratio and displayed normal fertility. Litter mates were randomly assigned to experimental groups. Twenty-one to twenty-nine week C57BL/6J mice were used to asses the effect MMV007285 has on hematopoietic cells subjected to stress.
During all experiments, the weights of the mice were approximately 18–30 g with no animals loosing greater than 10% body weight. All animals were housed in microisolator cages with temperature-controlled conditions under a 12-hour light/dark cycle with free access to drinking water, and food. Only one experimental procedure was performed in each mouse, and all mice used were drug naive prior to the experiment. Furthermore, all animal studies were performed in accordance with the Ontario Cancer Institute Animal Use Protocol (AUP): # 1251.33 (NOD-SCID-GF, NOD-SCID, and SCID) and AUP # 2244.12 (iDOX-Taz-KD transgenic mice).
TAZ-KD Induction in iDOX-Taz-KD Mice
TAZ-knockdown was induced by feeding 7.6–14.3 week old iDOX-Taz-KD transgenic mice with DOX-containing chow (625 mg/kg chow), formulated by Purina Mills, for 12.9–19.4 weeks. Non-transgenic littermates were fed DOX-containing chow and used as wild-type (WT) controls.
Primary AML and Normal Hematopoietic Cells
Primary human AML samples were obtained from peripheral blood or the bone marrow of consenting male or female AML patients, with a malignant cell frequency of 80% among mononuclear cells. Differential density centrifugation was used to isolate AML cells. Peripheral blood stem cells (PBSCs) were obtained from healthy consenting male or female volunteers, donating PBSCs for allogenic stem cell transplantation. PBSCs were isolated by G-CSF stimulation, and leukopheresis. Both primary AML cells, and PBSCs were frozen in alpha MEM + 5% FBS or 90% FBS +15U/mL of heparin + 10% DMSO. The University Health Network institutional review boards approved the collection and use of human tissue for this study (Research Ethics Board protocol #15–9324). As per regulation, all specimens were de-identified. Each experiment was performed using a single aliquot from a donor. Information about the patients who were the source of the cells are indicated in Table S5.
Primary AML Cell Cultures for Transduction
Cells were thawed in a 37°C water bath, washed once in media composed of IMDM 10% FBS, 100 units/mL penicillin, and 100 μg/mL of streptomycin. Primary AML cells were resuspended in X-VIVO 10 supplemented with 20% BIT, 50 ng/ml Flt3-L, 10 ng/mL IL-6, 50 ng/mL SCF, 25 ng/mL TPO, 10 ng/mL IL-3,10 ng/mL G-CSF at a concentration of 5.0 × 105 cells/mL before being transduced.
METHODS DETAILS
Plasmids
The 90K gRNA library developed in Hart et al., 2015 was obtained from the Moffat lab. Whereas, the TAZ, PISD, and control guide RNA (sgRNA) oligonucleotides were synthesized and cloned into the pLCKO lentiviral vector using the BspAI/BfuAI sites as described previously (Hart et al., 2015). The coding sequence of sgRNA’s targeting TAZ (gene id: 6901), PISD (gene id: 23761), and the LacZ genes (Control) are listed in Table S6.
For the transduction of AML cell lines, the TAZ shRNA constructs in the hairpin-pLKO.1 vector were purchased from Sigma-Aldrich® as glycerol bacterial stocks. The coding sequence of shRNAs targeting TAZ (accession number NM_000116), and the control shRNA targeting the GFP sequence (GFP, accession number clonetechGfp_587s1c1) are listed in Table S6.
To transduce 8227 cell and primary AML patient samples, TAZ shRNA sequences were first modified in order to be cloned into the hairpin-pRS19 vector using the restriction enzyme BsbI. The coding sequence of shRNAs targeting TAZ (accession number NM_000116), and the control shRNA non-targeting sequence are listed in Table S6.
The PISD gene (Accession number NM_014338) from the Myc-DDK-tagged PISD ORF (OriGene #RC200269) was cloned into the pLenti-EF1a-C-Myc-DDK-IRES-Bsd plasmid (OriGene, #PS100085), using restriction enzymes AsisI and MluI.
All sgRNA, shRNA, and overexpression plasmids were validated by Sanger sequencing before its use in downstream experiments.
Lentiviral Packing
For library virus production lentivirus was made in a 175 cm2 flask format, by transfecting 11 × 106 packaging cells (293T) with a three-plasmid system (lentiviral pLCKO vector containing the library, packing plasmid with: gag, pol, and rev genes, and envelope plasmid) (Hart et al., 2015; Moffat et al., 2006).
sgRNA and shRNA plasmids used in AML cell lines were isolated using the E.N.Z.A® Plasmid Midi Kit system (Omega bio-tek, GA, USA) from glycerol bacteria stocks, and then quantified by the NanoDrop (ThermoScientific, MA, USA) spectrophotometer. Lentivirus was made in a 25 cm2 flask format, by 293T cells with a three-plasmid system (hairpin-pLKO.1 vector/guide-pLCKO vector, packing plasmid with: gag, pol, and rev genes, and envelope plasmid) (Cole et al., 2015; Simpson et al., 2012).
Whereas, cloned shRNA plasmids used in primary AML patient samples were isolated using the E.N.Z.A® Plasmid Midi Kit system (Omega bio-tek, GA, USA), and then quantified by the NanoDrop (ThermoScientific, MA, USA) spectrophotometer. Lentivirus was made in a 175 cm2 flask format, by transfecting 293T cells with a three-plasmid system (hairpin-pRS19 vector, packing plasmid with: gag, pol, and rev genes, and envelope plasmid) (Cole et al., 2015; Simpson et al., 2012). The virus was concentrated using the Lenti-X Concentrator as per manufacturer’s instructions.
CAS9-OCI-AML2 Cell Line Generation
CAS9-OCI-AML2 cells were generated by adapting a method previously described (Hart et al., 2015). We first infected OCI-AML2 cells with virus containing a CAS9–2A-Basticidin expressing cassette (Addgene plasmid#73310) and then selected with blasticidin (10 μg/mL) for six days. After selection period CAS9-OCI-AML2 clones were sorted by manual seeding at a concentration of 0.4 cells/well in 96-well plates. Independent clones were isolated, and the CAS9 mRNA levels for each clone was quantified by RT-qPCR. The clone with the highest level of CAS9 mRNA (designated clone 7), was further analyzed by immunoblotting for CAS9 protein levels. Subsequently, the Cas9-expressing clonal population was subject to another round of single cell cloning procedures described above. The clone (designated clone 7.2) from the second round of selection was then characterized by immunoblotting for Cas9 protein expression and selected for screening as well as the TAZ, and PS decarboxylase (PISD) knockout studies as described below.
CRISPR Screen
To identify gRNA that reduced the growth and viability of AML cells, we performed a pooled lentiviral gRNA screen cells as described previously (Hart et al., 2015). CAS9-OCI-AML2 cells were transduced with a pooled lentiviral library consisting of 91,320 sgRNAs in barcoded lentiviral vectors targeting 17,232 nuclear-encoded genes. The day after transduction, cells were treated with puromycin (2 μg/mL) to select transduced cells. Resistant clones were passaged at regular intervals (3–4 days). Cells were harvested at 3 and 17 days post-transduction. Genomic DNA was extracted from cell pellets obtained on day 3 as well as day 17 and sequenced on an Illumina NextSeq500 (Illumina, CA, USA). shRNA depletion was characterized by MAGeCk analysis as described below.
MAGeCk Analysis
sgRNA sequences were first extracted from FASTQ files by trimming and then aligned to a reference library via Bowtie. Mapped sgRNA counts for samples were analyzed using MAGeCK 0.5.5, using default parameters (Li et al., 2014). Essential genes were identified at a FDR < 5%, by comparing of D17 sgRNA to D0 sgRNA.
Viral Infection
CRISPR-sgRNA Knockout
1.0 × 106 CAS9-OCI-AML2 cells were centrifuged and resuspended in 3 mL of medium containing 5 μg/mL of protamine sulfate. 1 mL of virus was added to cells, followed by a 24-hour incubation (37°C, 5% CO2). On the following day, cells were centrifuged and washed, then resuspended in fresh medium with puromycin (1.5 μg/mL). After at least 3 days of selection, equal numbers of cells were plated for growth assay, and counted by trypan blue exclusion staining for a period of 12–15 days post-transduction. To confirm knockout, 5–20 × 106 cells were collected at day 8 post-transduction for immunoblot analysis, as described in the mitochondria protein lysate, and immunoblotting sections below.
shRNA knockdown of AML Cell Lines
Lentiviral infections with shRNA constructs were performed as previously described in Cole et al., 2015. 5 × 106 cells were centrifuged and resuspended in 5 mL of medium containing 5 μg/mL of protamine sulfate. 2 mL of virus was added to cells, followed by a 24-hour incubation (37°C, 5% CO2). On the following day, cells were centrifuged washed, and resuspended in fresh medium with puromycin (1.5 μg/mL for OCI-AML2, 2 μg/mL for TEX, K562, and U-937 cells). After 3 days of selection, equal numbers of cells were plated for growth assay, and counted by trypan blue exclusion staining for a period of 12 days post-transduction. To confirm target knockdown, 5–20 × 106 cells were collected at day 4 post-transduction for immunoblot analysis, as described in mitochondria protein lysate and immunoblotting sections below.
shRNA knockdown of Primary AML Samples
Lentiviral infections with shRNA constructs were performed as previously described (Chan et al., 2015). Wells in a 24-well non-tissue-culture-treated plate were coated with 500 μl of RetroNectin® (20 μg/mL in PBS) per well for 2 hours at room temperature. The wells were blocked with 1 mL of 2% BSA (W/V) for 30 minutes at room temperature. The BSA solution was then aspirated, and concentrated lentiviral particles in HBBS with 25 mM HEPS were added to each well at a volume of 0.5 mL. The plate was then centrifuged at 3,000 rpm for 5 hours at room temperature to promote the attachment of lentiviral particles to RetroNectin®. After aspirating 0.4 mL of the viral supernatant, 5 × 105 cells were added to each well in 1 mL of X-VIVO 10 (20% BIT, 50 ng/ml Flt3-L, 10 ng/mL IL6, 50 ng/mL SCF, 25 ng/mL TPO, 10 ng/mL IL3, 10 ng/mL G-CSF). The plate was then centrifuged again at 1,300 rpm for 10 minutes at room temperature to promote the interaction between the cells and lentiviral particles, and then transferred to a 37°C incubator to initiate lentiviral infection. Twenty-four hours afterward, the cells were resuspended in fresh media at a concentration of 1 × 106 cells/mL and seeded in a 24-well plate at 1 mL per well. TAZ Knockdown was confirmed by quantitative reverse transcriptase-real time polymerase chain reaction (qRT-PCR) 5–7 days after transduction, as described in the qRT-PCR section.
PISD Overexpression
OCI-AML2 cells were transduced with lentiviral particles containing PISD or control vector sequences, and then selected with 10 μg/ml blasticidin for 8 days. After selection, PISD or empty vector overexpressing cells were transduced with TAZ or control shRNA constructs as described in shRNA knockdown of AML cell lines section. Eleven days after transduction with the PISD vector sequence PISD overexpression was confirmed by qRT-PCR, and cell phenotype was characterized by non-specific esterase staining, as described in the qRT-PCR and non-specific esterase sections below.
Mitochondrial Protein Lysates
Mitochondrial lysates were made to quantify TAZ, using the cytochrome c releasing apoptosis kit (Abcam, Cambridge, UK), as per manufacturer’s instructions, with minor modifications. We harvested confluent cells and froze these cells on dry ice. Cell pellets (10–20 × 106 cells) were then thawed on ice and lysed in distilled water containing protease inhibitors, by hypotonic shock for 15 minutes at 4°C. The Cytosolic Extraction Buffer Mix containing DTT and protease inhibitors were added to the cell lysate, and samples were then centrifuged at 800 g at 4°C for 20 minutes. The supernatant containing mitochondria was collected and centrifuged at 10,000 g for 30 minutes at 4°C. Next, the supernatant was discarded, and the mitochondrial pellet was resuspended in ice-cold Mitochondrial Extraction Buffer, with DTT and protease inhibitors. Finally, mitochondria were lysed by adding 2X Laemmli buffer. The protein concentration in mitochondrial lysates was quantified using the DC protein determination kit (Bio-Rad, CA, USA) before immunoblotting.
Whole Cell Protein Lysates
For all other immunoblots whole cell lysates were used. To make whole cell lysates, cells (5 × 106) were washed with PBS followed by cell lysis in RIPA buffer. Protein concentration was measured by Bradford method (Protein assay dye, Bio-Rad, CA, USA).
Immunoblotting
Equal amounts of protein were loaded and fractionated on 10%–12% SDS-polyacrylamide gels. Proteins were separated on SDS-PAGE and transferred to nitrocellulose membranes or PVDF prior to antibody treatment. Blots were blocked with 5% milk TBST then incubated overnight with an appropriate primary antibody. Membranes were then blocked with 5% milk TBST, and further probed using an appropriate secondary antibody conjugated to HRP, and then developed using Enhanced chemiluminescence (ECL) detection (ThermorFisher Scientific, MA, USA). The antibodies used for immunoblotting can be found in the key resource table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-TAZ (F-7) | Santa Cruz Biotechnology | Cat# Sc-365810 RRID: AB_10842049 |
| Rabbit polyclonal anti-PISD | Proteintech Group | Cat# 16401-1-AP, RRID:AB_2186673 |
| Rabbit monoclonal PTDSS1 antibody | Abcam | Cat# ab157222 |
| Rabbit monoclonal PSS2 antibody | Abcam | Cat# ab183504 |
| Rabbit monoclonal anti-citrate synthase | Abcam | Cat# ab129095 |
| Mouse monoclonal anti-beta-Actin (AC-15) | Santa Cruz Biotechnology | Cat# sc-69879, RRID:AB_1119529 |
| Rabbit polyclonal anti-beta Tubulin (H-235) | Santa Cruz Biotechnology | Cat# sc-9104, RRID:AB_2241191 |
| Sheep Anti-Mouse IgG, Whole Ab ECL Antibody, HRP Conjugated | GE Healthcare | Cat# NA931, RRID:AB_772210 |
| Donkey Anti-Rabbit IgG, Whole Ab ECL Antibody, HRP Conjugated | GE Healthcare | Cat# NA934, RRID:AB_772206 |
| BB515 Rat Anti-Mouse Ly-6A/E | BD Biosciences | Cat# 565397 |
| Pe/Cy7 Rat anti-mouse CD117 (c-Kit) | BioLegend | Cat# 105814 RRID: AB_2131136 |
| PE Rat anti-mouse CD150 (SLAM) | BioLegend | Cat# 115904 RRID: AB_313683 |
| APC Hamster anti-mouse CD48 | ThermorFisher Scientific | Cat# 17-0481-82 RRID: AB_469408 |
| APC/Cy7 Rat anti-mouse CD16/32 | BioLegend | Cat# 101328 RRID: AB_2104158 |
| eFluor 450 Rat anti-mouse CD34 | ThermoreFisher Scientific | Cat# 48-0341-82 RRID: AB_2043837 |
| APC Mouse anti-human CD11b | BD Biosciences | Cat# 340937 |
| APC Mouse anti-human CD14 | BD Biosciences | Cat# 555399 |
| CD34 | BD Biosciences | Cat# 348053, RRID:AB_2228982 |
| CD38 | BD Biosciences | Cat# 342371, RRID:AB_400453 |
| CD122 | Lab Stock | N/A |
| Biological Samples | ||
| Human AML Patient Samples | Leukemia Biobank, Princess Margaret Hospital | N/A |
| Normal Peripheral Blood Stem Cells (PBSC’s) | Leukemia Biobank, Princess Margaret Hospital | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Iscove’s Modification Dulbecco’s Modification Eagle’s Medium (IMDMEM) | Wisent | Cat# 319-701-CL |
| RPMI 1640 | Wisent | Cat# 350-700-CL |
| Alpha MEM | Lab Stock | N/A |
| MyeloCult H5100 | STEMCELL Technologies | Cat# 05150 |
| X-VIVO 10 Chemically Defined, Serum-free Hematopoietic Cell Medium | Lonza | Cat# 04-380Q |
| Dulbecco’s Modification Eagle’s Medium (DMEM) | Wisent | Cat# 319-005-CL |
| Fetal Bovine Serum | Sigma-Aldrich | Cat# F1051 |
| Recombinant Human SCF Protein (TEX) | R&D Systems | 255-sc/cf., R&D, |
| Recombinant Human IL-3 Protein (TEX) | R&D Systems | 203-IL |
| L-Glutamine 200mM (TEX) | ThermoFisher Scientific | Cat# 25030081 |
| Recombinant Human IL-3 (8227, and Primary Sample Transductions) | PeproTech | Cat# 200-03 |
| Recombinant Human SCF (78227, and Primary Sample Transductions) | PeproTech | Cat# 300-07 |
| Recombinant Human Flt3-Ligand (8227, and Primary Sample Transductions) | PeproTech | Cat# 300-19 |
| Recombinant Human TPO (8227, and Primary Sample Transductions) | PeproTech | Cat# 300-18 |
| Recombinant Human IL-6 (8227, and Primary Sample Transductions) | PeproTech | Cat# 200-06 |
| BIT 9500 Serum Substitute (8227, and Primary Sample Transductions) | STEMCELL Technologies | Cat# 09500 |
| Human GM-CSF, premium grade (primary samples) | Miltenyi Biotec | Cat# 130-093-866 |
| Human IL-6 (primary samples) | Miltenyi Biotec | Cat# 130-095-365 |
| Human Flt3-Ligand (primary samples) | Miltenyi Biotec | Cat# 130-096-479 |
| Human IL-7 (primary samples) | Miltenyi Biotec | Cat# 130-095-363 |
| Granulocyte colony stimulating factor (G-CSF), [Filgrastim] | Amgen Canada | CAS#121181-53-1 |
| Puromycin HCL | Sigma-Aldrich | Cat# P8833 |
| Protamine Sulfate | MP Biomedicals | Cat#194729 |
| Laemmli buffer | Lab Stock | N/A |
| RIPA buffer | Lab Stock | N/A |
| TBS (10X) | Wisent | Cat# 311-030-LL |
| Thermo Scientific Pierce ECL western blotting substrate | ThermorFisher Scientific | Cat# PI32106 |
| Annexin V-FITC Reagent | CEDARLANE, BioVision | Cat# 1001-1000 |
| Propidium Iodide | CEDARLANE, BioVision | Cat# 1056-1 |
| Annexin V Binding Buffer | Lab Stock | N/A |
| Sodium Fluoride | Sigma-Aldrich | Cat# 919 |
| Rodent Chow with 625mg doxycycline/kg chow | Ren’s Pets Depot | Cat# 58MY-1811541 |
| Red Blood Cell Lysing Buffer Hybri-Max | Sigma-Aldrich | Cat# R7757 |
| Lineage Cell Depletion Kit mouse | MACS Miltenyi Biotec | Cat# 130-090-858 |
| autoMACs Rinsing Solution | MACS Miltenyi Biotec | Cat# 130-091-222 |
| MACs BSA Stock Solution | MACS Miltenyi Biotec | Cat# 130-091-376 |
| 7AAD | BD Biosciences | Cat# 559925 |
| UltraComp eBeads | affymetrix eBioscience | Cat# 01-2222-42 |
| mSCF | Miltenyi Biotec | Cat# 130-101-697 |
| mIL-6 | Miltenyi Biotec | Cat# 130-096-683 |
| mIL-3 Conditioned Media | Karasuyama and Melchers, 1998 | N/A |
| 5-Fluorouracil | Sigma-Aldrich | Cat# F6627-10G |
| Chloroform (UHPLC/MS) | Fisher Scientific | Cat# C6074 |
| Methanol CHROMASOLV, Riedel-de Haen (UHPLC/MS) | Fisher Scientific | Cat# 60046513 |
| Sodium-Phosphate Monobasic Anhydrous (UHPLC/MS) | Fisher Scientific | Cat# BP329-500 |
| Alfa Aesar Acetonitrile (UHPLC/MS) | Fisher Scientific | Cat# AA47138K7 |
| Isopopanol, Optima (UHPLC/MS) | Fisher Scientific | Cat# A4614 |
| Formic Acid, Optima (UHPLC/MS) | Fisher Scientific | Cat# A11750 |
| Amonium Formate (UHPLC/MS) | Sigma-Aldrich | Cat# 70221-100G-F |
| Standard (UHPLC/MS): 14:0 Cardiolipin | Avanti, Polar Lipids | Cat# 710332C |
| Human Recombinant KillerTRAIL | Alexis Biochemicals | Cat# 201-123-C500 |
| Cyclohexamide | Sigma-Aldrich | Cat# C7698 |
| Oligomycin | Sigma-Aldrich | Cat# 75351 |
| FCCP | Cayman Chemical | Cat# 15218 |
| Corning Cell-Tak | FisherScientific | Cat# C354240 |
| Seahorse XF assay medium | Agilent | Cat# 102365-100 |
| Paraformaldehyde | Electron Microscopy Sciences | Cat# RT19200 |
| 70% Glutaraldehyde | Electron Microscopy Sciences | Cat# 16360 |
| Sodium Phosphate Monobasic monohydrate (NaH2P04. H2O Buffer) | Sigma-Aldrich | Cat# S9638 |
| Sodium Phosphate Dibasic (Na2H2P04, Buffer) | Lab Stock | Cas# 7558-79-4 |
| 1N Sodium Hydroxide [NaOH] | Lab stock | Cas# 1310-73-2 |
| Osmium tetraoxide [OsO4] | Electron Microscopy Sciences | Cat# RT19100 |
| Embed-812 kit | Electron Microscopy Sciences | Cat# 14120 |
| 300 Mesh Copper grids | Electron Microscopy Sciences | Cat# G300H-Cu |
| Uranyl Acetate | Electron Microscopy Sciences | Cat# 22400 |
| Lead Nitrate [Pb(NO3)2] for staining for electron microscopy | Electron Microscopy Sciences | Cat# RT17900 |
| Sodium Citrate [Na3C6H5O7·2H2O] for staining | Electron Microscopy Sciences | Cat# RT21140 |
| MitoTracker Red CMXRos | ThermoFisher Scientific | Cat# M7512 |
| Poly-L-lysine | Sigma-Aldrich | Cat#P4707 |
| Chloroform (CHCL3, Spot Densitometry) | Sigma-Aldrich | Cat# 34854 |
| Methanol (CH3OH, Spot Densitometry) | Sigma-Aldrich | Cat# 34860 |
| Acetic Acid (C3H6O, Spot Densitometry) | Canadawide Scientific | Cat# CWA0302-4 |
| Standard (Spot Densitometry): Sphingomyelin | Sigma-Aldrich | Cat# S0756 |
| Standard (Spot Densitometry): L-α-Phosphatidylcholine | Sigma-Aldrich | Cat# P3556 |
| Standard (Spot Densitometry): L-α-Phosphatidyl-L-serine | Sigma-Aldrich | Cat# P0474 |
| Standard (Spot Densitometry): L-α-Phosphatidylinositol | Sigma-Aldrich | Cat# P6636 |
| Standard (Spot Densitometry): L-α-Phosphatidylethanolamine | Sigma-Aldrich | Cat# P7943 |
| Standard (Spot Densitometry): Cardiolipin Sodium Salt from Bovine Heart | Sigma-Aldrich | Cat# C0563 |
| Copper (II) sulfate pentahydrate (Spot Densitometry) | Sigma-Aldrich | Cat# C-6283 |
| O-Phosphoric Acid (Spot Densitometry) | Fisher Scientific | Cat# A242-212 |
| PBS (Spot Densitometry) | Sigma-Aldrich | Cat# P3813 |
| H20 (Spot Densitometry) | Fisher Scientific | Cat# 2745 |
| HPTLC plates (Spot Densitometry) | Sigma-Aldrich | Cat# 1.05633.0001 |
| Chloroform | Lab Stock | Cas# 67-66-3 |
| HPLC methanol | Sigma-Aldrich | Cat# 646377 |
| Dimethyl sulfoxide (DMSO) | Sigma-Aldrich | Cat# D8418 |
| Tween 80 | Sigma-Aldrich | Cat# P4780 |
| Propylene Glycol (PEG) | Sigma-Aldrich | Cat# P4347 |
| Methanol | BioShop Canada | Cat# MET302 |
| Soy PS | Avanti Polar Lipids | Cat# 870336P |
| Soy PE | Avanti Polar Lipids | Cat# 840024P |
| Egg Lyso PE | Avanti Polar Lipids | Cat# 860081P |
| Saponin | Sigma-Aldrich | 47036-50G-F |
| Bovine Serum Albumin | Sigma-Aldrich | Cat# A9647 |
| 16% paraformaldehyde (CAT#15710) | Electron Microscopy Sciences | Cat# 15710 |
| AnnexinV Alexa-647 | ThermoFisher Scientific | Cat# A23204 |
| ProLong Gold Antifade Mountant | ThermoFisher Scientific | Cat# P36934 |
| SuperScript IV Reverse Transcriptase | ThermoFisher Scientific | Cat# 18090050 |
| 7-chloro-N-(4-ethoxyphenyl)-4-quinolinamine (MMV007285) | This paper | Jae-Yeon Choi et. al, 2016 |
| CL075 | TOCRIS | Cat# 6142 |
| Formic acid (Pharmacokinetic Studies) | Sigma-Aldrich | Cat# 33015 |
| CellTiter 96 Aqueous MTS Reagent Powder | Promega | Cat# G1111 |
| RetroNectin Recombinant Human Fibronectin Fragment | Takara | Cat# T100A |
| Acetonitrile, Optima, Fisher Chemical (Pharmacokinetic Studies) | ThermoFisher Scientific | Cat# A996-4 |
| Critical Commercial Assays | ||
| E.N.Z.A Plasmid Midi Kit | Omega BIO-TEK | Cat# D6904-04 |
| Cytochrome c Releasing Apoptosis Assay Kit | abcam | Cat# ab65311 |
| α-Naphthyl Acetate (Non-Specific Esterase) | Sigma-Aldrich® | Cat# 91A-1KT |
| Protein Assay Dye | Bio-Rad | Cat# 5000006 |
| DC Protein Assay kit II | Bio-Rad | Cat#5000112 |
| MethoCult H4100 | STEMCELL Technologies | Cat# 04100 |
| MethoCult H4434 | STEMCELL Technologies | Cat# 04434 |
| MethoCult GF M3534 | STEMCELL Technologies | Cat# 03534 |
| Bioanalyzer RNA Analysis Kit | Agilent Technologies | Cat# 5067-1511 |
| KAPA Library Complete Quantification Kit for ABI Prism genetic analyzer and Illumina | KAPABiosystems | Cat# KK4835 |
| Agilent High Sensitivity DNA Kit | Agilent Technologies | Cat# 5067-4626 |
| NEBNext Ultra Directional RNA Library Prep Kit for Illumia | New England BioLabs | Cat# E7420S |
| HiSeq SBS kit V4 250 cycle kit | Illumina | Cat# FC-401-4003 |
| Carboxy-H2DCFDA | ThermoFisher Scientific | Cat# C400 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| Power SYBR Green PCR Master Mix | ThermoFisher Scientific | Cat# 4367659 |
| Lenti-X Concentrator | Takara | Cat# 631231 |
| Lipid strips | Echelon Biosciences | Cat# P-6002 |
| Fatty acid-free Bovine Serum Albumin | Sigma-Aldrich® | Cat# A7030 |
| PISD recombinant protein | OriGene | Cat# TP761730 |
| Deposited Data | ||
| AML2 gene expression after TAZ-KD | This paper | GEO: GSE107045 |
| Experimental Models: Cell Lines | ||
| Human: OCI-AML2 | Wang C et al., 1989 | N/A |
| Human: TEX | Warner et al., 2005 | N/A |
| Human: K562 | ATCC | Cat# CCL-243 |
| Human: U937 | ATCC | Cat# CRL-1593 |
| 8227 cells | Lechman et al., 2016 | N/A |
| 293T/17 | ATCC | Cat# CRL-11268 |
| Experimental Models: Organisms/Strains | ||
| Mouse: NOD.Cg-Prkdcscid II2rgtm1WjI Tg(CMV-IL3,CSF2,KITLG)1Eav/MloySzJ (NOD-SCID-GF) | Nicolini et al., 2004 | N/A |
| Mouse: B6.Cg-Gt(ROSA)26Sortm37(H1/tetO-RNAi:Taz)Arte/ZkhuJ doxycycline-Inducible-TAZ-Knockdown (iDOX-Taz-KD) | Acehan et al., 2011 | N/A |
| Mouse: C57BL/6J | Jackson Laboratory | Cat# 000664; RRID: IMSR_JAX:000664 |
| Mouse: NOD.CB17-Prkdcscid/J (NOD-SCID) | Jackson Laboratory | Cat# 001303 |
| Mouse: Prkdcscid (SCID) | In house | RRID: IMSR_ARC: SCID |
| Oligonucleotides | ||
| Primers for TAZ, PISD, 18srRNA, LYZ, iDOX-Taz-shRNA, ND1, HGB, TLR4, TLR8, IL6, and IFNβ | See Table | Table S4 |
| sgRNAs targeting TAZ, PISD, and control sequences | See Table | Table S6 |
| shRNAs targeting TAZ, and control sequences | See Table | Table S6 |
| Recombinant DNA | ||
| pCMV-R8.74psPAX2 Packaging Plasmid | Moffat et al., 2006 | N/A |
| VSV-G/pMD2.G Envelope Plasmid | Moffat et al., 2006 | N/A |
| pLKO.1-Puro-shRNA lentiviral plasmid | Moffat et al., 2006 | N/A |
| Cas9-2A-Blast lentivrial plasmid | Hart et al., 2015 | N/A |
| pLCKO-Puro-sgRNA lentiviral plasmid | Hart et al., 2015 | Addgene #73311 |
| pRS19-Puro-GFP-shRNA lentiviral plasmid | Chan et al., 2015 | N/A |
| TKO CRISPR library V1 | Hart et al., 2015 | N/A |
| pLenti-EF1a-C-Myc-DDK-IRES-Bsd plasmid | OriGene | Cat# PS100085 |
| Software and Algorithms | ||
| MAGeCk | Lietal., 2014,_S1_Reference34 | https://sourceforge.net/projects/mageck/ |
| ImageJ | NIH | https://imagej.nih.gov/ij/ |
| GraphPad Prism | GraphPad Software version 6.03 for Windows, GraphPad Software, La Jolla California USA | https://www.graphpad.com/ |
| FLOWJO | FlowJo v 7.7.1, TreeStar, Ashland Oregon USA | https://www.flowjo.com/ |
| Xcalibur v.2.1 | Thermo-Fisher Scientific | Part# XCALI-97213 |
| Chromeleon Xpress v.7.2 | Thermo-Fisher Scientific | Cat# CHROMELEON7 |
| NIST MS Search v.2.0 | NIST | https://chemdata.nist.gov/mass-spc/ms-search/ |
| FastQC v.0.11.2 | Babraham Institute | http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Trim Galore v. 0.4.0. | Babraham Institute | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| FastQ-Screen v.0.4.3 | Babraham Institute | http://www.bioinformatics.babraham.ac.uk/projects/fastq_screen/ |
| RSeQC v. 2.3.7 | http://rseqc.sourceforge.net/ | |
| Tophat v. 2.0.11 | John’s Hopkins University | https://ccb.jhu.edu/software/tophat/index.shtml |
| htseq-v.0.6.1p2 | Simon Anders | https://ccb.jhu.edu/software/tophat/index.shtml |
| DESeq v.1.18.0 | Bioconductor | http://bioconductor.org/packages/release/bioc/html/DESeq.html |
| edgeR, R package, v.3.16.5 (Differential Gene Expression Analysis, Control shRNA Vs. TAZ-KD) | Bioconductor | http://www.bioconductor.org/packages/release/bioc/html/edgeR.html |
| edgeR, R package, v.3.28.21 (Differential Gene Expression Analysis, LSC-versus LSC+, TCGA clusters) | Bioconductor | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| Gene Set Enrichment Analysis Software | Broad Institute | http://software.broadinstitute.org/gsea/msigdb/ |
| Cytoscape 3.6.1 | The Cytoscape Consortium | http://www.cytoscape.org/?gclid=CjwKCAjw_tTXBRBsEiwArqXyMp1y9jXNftKoksBpO7Q2BCKzWsvEGPteq7VRCpoVHH22tEsWgCnb7BoCnL4QAvD_BwE |
| EnrichmentMap 3.1 | The Cytoscape Consortium | https://enrichmentmap.readthedocs.io/en/docs-3.1/ |
| AutoAnnotate 1.2. | The Cytoscape Consortium | http://apps.cytoscape.org/apps/autoannotate |
| affy, R package, affy_1.38.1 | Bioconductor | https://www.bioconductor.org/packages/release/bioc/html/affy.html |
| sva, R package, sva_3.6.0 | Bioconductor | http://bioconductor.org/packages/release/bioc/html/sva.html |
| Other | ||
| LSC+ and LSC− Geneset | Ng et al., 2016 | GEO: GSE76008 |
| TCGA Repository | National Cancer Institute | https://portal.gdc.cancer.gov/ |
| DMAP | Novershtern et al., 2011 | GEO: GSE24759 |
| Bader Lab Database | This paper | http://baderlab.org/GeneSets, version from September 2017 |
Basal Apoptosis
OCI-AML2 cells were transduced with shRNA in lentiviral vectors targeting TAZ or control sequences. Seven days after transduction, cells were harvested, and cell death was measured by flow cytometry with annexin V fluorescein isothiocyanate (FITC) and propidium iodide (PI) R-Phycoerythrin (PE, Biovision Research Products, CA, USA) staining according to the manufacturer’s instructions. Flow cytometry data were acquired using a LSRFORTESSA X20 (BD Biosciences, FL, USA) flow cytometer and frequency of annexin V, PI negative cells were quantified with the FlowJo software (TreeStar, OR, USA)
Cell Cycle Analysis
OCI-AML2 cells were transduced with shRNA in lentiviral vectors targeting TAZ or control sequences. Seven days after transduction, cells were harvested, washed with cold PBS, and fixed in absolute ethanol and PBS (80% ethanol, 20% PBS) for a minimum of one hour. Afterward, cells were washed in cold PBS, and treated with 5 μg/mL of DNase-free RNase A (Invitrogen, CA, USA) at 37°C for 30 minutes, then incubated in PBS containing 5 μg/mL of propidium iodide (PI) for 5–10 minutes at room temperature. DNA content was measured using flow cytometry (FACs CANTO, BD, FL, USA) and analyzed with FlowJo software (TreeStar, OR, USA).
Colony Formation Assays
Leukemia Cell Lines
Five (TAZ knockdown OCI-AML2, and TEX cells) or nine (PISD knockout CAS9-OCI-AML2 cells) days after transduction cells were plated at equal concentrations (CAS9-OCI-AML2 and OCI-AML2 = 750 cells; TEX cells = 2,000) in duplicate 35mm dishes. (Nunclon, Rochester, USA) to a final volume of 1 mL/dish in MethoCult H4100 media (StemCell Technologies, BC, Canada) supplemented with 30% FCS (CAS9-OCI-AML2 and OCI-AML2) or MethoCult H4100 media (StemCell Technologies, BC, Canada) supplemented with 30% FCS, 20 ng/mLSCF, and 2 ng/mL IL-3 (TEX cells). After incubating the dishes for5 (CAS9-OCI-AML2), 7 (OCI-AML2), or 10 (TEX) days at 37°C, 5% CO2 with 95% humidity, the number of colonies containing 10 or more cells were counted on an inverted microscope. The mean of the duplicate plates for each condition are presented. During serial re-plating colonies were removed from MethoCult by vortexing and washing in PBS, cells were counted and then re-plated in MethoCult every 5 (PISD knockout CAS9-OCI-AML2 cells) or 7 (TAZ knockdown TEX cells) days. The mean of the duplicate plates for each condition are presented.
To assess clonogenic growth after PS treatment, OCI-AML2 cell were pre-treated with 25 μM of PS or the vehicle control for 9 days. Afterward, treated OCI-AML2 cells were platted at equal volumes in duplicate 35mm dishes. (Nunclon, NY, USA) to a final volume of mL/dish in MethoCult H4100 media (StemCell Technologies, BC, Canada) supplemented with 30% FCS without PS. After 5 days of incubation at 37° C, 5% CO2 with 95% humidity, the number of colonies containing 10 or more cells were counted on an inverted microscope. The mean of the duplicate plates for each condition are presented.
Primary Samples
Fresh AML mononuclear cells and normal peripheral blood stem cells (PBSCs) (4 × 105 AML cells, 2 × 105 PBSCs) were incubated with MMV007285, PS or vehicle control for 48 hours in MyelocultH5100, supplemented with 100 ng/mL SCF, 10 ng/mL Flt3-L, 20 ng/mL IL-7, 10 ng/mL IL-3, 20 ng/mL IL-6, 20 ng/mL G-CSF, 20 ng/mL GM-CSF. Treated PBSC’s or AML patient samples were platted in MethoCult H4434 medium (StemCell Technologies, BC, Canada). After incubating the dishes for 7 days (AML) or 2 weeks (normal hematopoietic cells) at 37°C with 5% CO2 and 95% humidity, AML colonies containing 10 or more cells and PBSC colonies containing more than 50 cells were counted. The mean of duplicate plates for each condition are presented.
To assess the clonogenic growth of mouse hematopoietic progenitor cells after TAZ knockdown and hematopoietic stress, 40000 bone marrow cells cells/ml/dish from WT or TAZ-KD mice treated with 0 or 200 mg/Kg of 5-FU were platted on MethoCult GF M3534, and incubated at 37°C, 5% CO2 with 95% humidity, 15 days after 5-FU treatment. Six days after, colonies containing 50 or more cells were enumerated. During serial re-plating, colonies were removed from MethoCult GF M3534 by vortexing and washing in PBS. Cells were counted and 40000 cells/ml/dish replated on MethoCult GF M3534. After 6 days in culture, colonies containing 50 cells or more cells were enumerated. The mean of the duplicate plates for each condition are presented.
The effect MMV007285 has on hematopoietic cells subjected to stress was characterized by incubating 160000 bone marrow cells isolated from C57BL/6J mice 15 days after treatment with 0 or 200 mg/Kg in StemSpan SFEM media (50 ng/mL mSCF, 50 ng/mL mIL6,1:320 mIL3 conditioned media) containing 0 or 12.5 μM MMV007285 for 72 hours. After pre-treatment the cells were washed and plated on MethoCult GF M3534 with 0 or 12.5 μM of MMV007285, then incubated at 37°C, 5% CO2 with 95% humidity for 6 days. Finally, colonies containing 50 or more cell were enumerated. The mean of the duplicate plates for each condition are presented.
RNA-Sequencing
Total RNA was isolated from OCI-AML2 cells using the RNeasy Plus Mini Kit (QIAGEN, Hilden, Germany) 7 days after transduction with shRNA targeting TAZ or control sequences. The experimental design consisted of 2 biological replicates of control cells and 2 biological replicates of TAZ knockdown samples. Quality of total RNA samples were checked on an Agilent Bioanalyzer 2100 RNA Nano chip (CA, USA) following Agilent Technologies’ recommendation. Concentration was measured by Qubit RNA HS Assay on a Qubit fluorometer (ThermoFisher Scientific, MA, USA). RNA library preparation was performed following the NEBNext Ultra Directional Library (MA, USA) Preparation Protocol. Briefly, 800 ng of total RNA was used as the input material and enriched for poly-A mRNA, fragmented into the 200–300-bases range for 4 minutes at 94°C and converted to double stranded cDNA, end-repaired and adenylated at the 3′ to create an overhang A to allow for ligation of Illumina adapters with an overhang T; library fragments were amplified under the following conditions: initial denaturation at 98°C for 10 s, followed by 13 cycles of 98°C for 10 s, 60°C for 30 s and 72°C for 30 s, and finally an extension step for 5 minutes at 72°C; at the amplification step, each sample were amplified with a different barcoded adapters to allow for multiplex sequencing. One ml of the final RNA libraries was loaded on a Bioanalyzer 2100 DNA High Sensitivity chip (Agilent Technologies, CA, USA) to check for size; RNA libraries were quantified by qPCR using the KAPA Library Quantification Illumina/ABI Prism Kit protocol (MA, USA). Libraries were pooled in equimolar quantities and paired-end sequenced on 1 lane of a High Throughput Run Mode flowcell with the V4 sequencing chemistry on an Illumina HiSeq 2500 platform (CA, USA) following Illumina’s recommended protocol to generate paired-end reads of 126-bases in length. RNA-sequencing data was used to perform differential gene expression analysis as described below.
Differential Gene Expression Analysis
Prior to analysis, read adapters and low quality ends were removed using Trim Galore v. 0.4.0. Reads were aligned against hg19 using Tophat v. 2.0.11. Read counts per gene were obtained through htseq-count v.0.6.1p2 in the mode “intersection_nonempty.” After removing genes whose cpm (counts per million reads) were less than 0.5 in at least one sample, edgeR R package v.3.16.5 was used to normalize the data using the TMM (trimmed mean of M values) method and to estimate differential expression by applying the generalized linear model likelihood ratio test between the control OCI-AML2 samples and the TAZ-KD OCI-AML2 samples. A score that ranks genes in TAZ knockdown samples from the most upregulated to the most downregulated compared to control shRNA samples was calculated using the formula ‘−log10(pvalue) * sign(logFC)’. Pathway, LSC+/LSC− signature, DMAP signature, and PERT deconvolution analyses was performed using the differential expression data generated.
Pathway Analysis
The rank list genes generated for TAZ knockdown and control shRNA samples was used for Gene set enrichment analysis (GSEA) with a setting of 1000 permutations, and default parameters. The pathway database from the Bader lab (http://baderlab.org/GeneSets, version of September 2017) which contains the Gene Ontology, Biological Process, as well as Msigdb-c2 and −c3, IOB, NetPAth, HumanCyc, Reactome and Panther was used. Results were visualized in Cytoscape 3.6.1 using EnrichmentMap 3.1, clustered and annotated using AutoAnnotate 1.2. Cluster labels were manually edited for clarity. Pathway analysis was validated by measuring mRNA levels of TLR4, TLR8, IL6, and IFNβ in TAZ knockdown or MMV treated samples, by qRT-PCR as described in the qRT-PCR section.
TCGA Unsupervised Clustering
The TCGA AML RNaseq data were retrieved from the GDC portal (https://portal.gdc.cancer.gov/). Read counts were normalized using the TMM method in the edgeR R package. The TCGA samples were hierarchically clustered using the Ward linkage method and Euclidean distance on the basis of the scaled log2(CPM) of the 5,334 genes with standard deviation > 1. AU (approximately unbiased) cluster probability, computed by multiscale bootstrap resampling, was calculated using the R package pvclust.
LSC+/LSC− Signature Analysis
Illumina beadchip transcriptomics data containing LSC+ and LSC− sorted AML fractions were obtained from the Gene Expression Omnibus data portal (GSE76008) (Ng et al., 2016) and differential expression between LSC+ and LSC− fractions was calculated using a moderated t test available in the limma R package 3.28.21, incorporating array batch effects into the linear model. First, the 500 most highly upregulated genes in TAZ knockdown and control shRNA samples were compared to normalized gene expression of LSC+/LSC− fractions, then subsequently visualized using the heatmap.2 function in R by setting row normalization as true. In addition, the top upregulated in the LSC+ fractions to top downregulated in comparison to LSC− fractions was scored using the formula ‘−log10(pvalue) *sign(logFC)’. The 400 highest scoring in LSC+ and the 400 highest scoring LSC− genes were used as signature gene set for LSC+ and LSC− samples respectively. Then gene set enrichment analysis (GSEA) (Broad Institute, MA, USA) with 1000 permutations and default parameters was used to measure the enrichment of genes upregulated in TAZ knockdown and control samples in the LSC+ or LSC− gene signatures.
DMAP Signature Analysis
Changes in gene expression after TAZ knockdown were mapped to the Gene Expression Omnibus dataset GSE24759 (DMAP) (Novershtern et al., 2011), containing Affymetrix GeneChip HT-HG_U133A Early Access Array gene expression data of 20 distinct hematopoietic cell states. GSE24759 data were background corrected using Robust Multi-Array Average (RMA), quantile normalized using the expresso function of the affy Bioconductor package (affy_1.38.1, R 3.0.1), array batch corrected using the ComBat function of the sva package (sva_3.6.0), and then standardized using Z score for each gene across samples. Bar graphs were created by selecting genes that were up- (top 500) TAZ knockdown cells as well as control shRNA cells, and summing the number of standardized data points that were above (> 0) or below (< 0) the mean for each DMAP cell population, corrected by the number of samples per population and results obtained from 1,000 random selections of DMAP genes.
PERT deconvolution analysis
The PERT deconvolution was run on the TMM-normalized CPM data from TAZ knockdown and control samples (Qiao et al., 2012). The batch-corrected linear RMA-normalized data from the GSE24759 (DMAP) data were used as the reference profile. The vector theta from the PERT output was used to estimate the percentage of reference populations within each TAZ knockdown and control samples. The theta percentage of each biological replicate of the same populations was summed. A high theta value for one population means a high estimated presence of the profile in the TAZ knock down or control sample.
Non-Specific Esterase Staining
Cytospin slides were prepared by centrifuging OCI-AML2 cells (1 × 105) 7–11 days after transduction (TAZ shRNA and PISD overexpression plasmids respectively) onto glass slides, which were then subsequently stained for nonspecific esterase (NSE) activity, using the staining kit from Sigma-Aldrich, as per manufacturer’s instructions. The monocyte non-specific esterase inhibitor sodium fluoride was used to confirm the specificity of the reaction. Finally, glass slides were dried at room temperature, mounted in Clear Mount and scanned using the Aperio ScanScope AT2 (Leica, Wetzlar, Germany), and then analyzed by ImageJ as described below.
NSE Staining Analysis Using ImageJ
Aperio ImageScope (Leica, Wetzlar, Germany) was used to select 5 random sections from the scanned AML cell images. ImageJ was used to quantify the intensity of NSE staining. The average of the 5 sections for each condition, from three independent experiments is presented.
Cell Surface Phenotype of OCI-AML2
OCI-AML2 cells were co-immunostained with the viability dye 7AAD and anti-human CD11b or 7AAD and anti-human CD14. Flow cytometry data were acquired using a LSRFORTESSA X20 (BD Biosciences, FL, USA) flow cytometer and analyzed with the FlowJo software (TreeStar, OR, USA).
RNA Isolation and Quantitative Reverse Transcriptase-Real Time Polymerase Chain Reaction
Total RNA was isolated from leukemia cells using the RNeasy Plus Mini Kit (QIAGEN), and cDNA was prepared using SuperScript IV Reverse Transcriptase (ThermoreFisher, MA, USA). Equal amounts of cDNA for each sample were added to a prepared master mix (Power SYBR Green PCR Master mix; Applied Biosystems, CA, USA). Quantitative real time polymerase chain (qRT-PCR) reactions were performed on an ABI Prism 7900 sequence detection system (Applied Biosystems, CA, USA). The relative abundance of a transcript was represented by the threshold cycle of amplification (CT), which is inversely correlated to the amount of target RNA/first-strand cDNA being amplified. To normalize for equal amounts of cDNA we assayed the transcript levels of 18srRNA gene. The comparative CT method was calculated as per manufacturer’s instructions. Primers that were used are listed in Table S4.
Lipid Extraction Protocols
Ultra-High Performance Liquid Chromatography/Mass Spectrometry (UHPLC/MS)
Frozen leukemia cells (day 13 post-transduction), or mouse bone marrow samples (12.9–19.4 weeks after doxycycline treatement) were resuspended in PBS, and then added to 2:1 chloroform:methanol (v/v) on ice. Samples were vortexed, and 400 μL of 0.2 M sodium-phosphate buffer was added to induce layer separation. After inversion, samples were centrifuged for 5 minutes at 1734g. The organic layer, which contains the lipids, was collected. An additional 2mL of chloroform was added to the aqueous layer. Samples were re-vortexed, re-centrifuged, and the organic layer was collected and combined with the organic layer of the first extraction. The methanol/water layer was discarded. Samples were stored in chloroform at 4°C until further analysis.
Densitometry Analysis
Lipids from TAZ knockdown (day 8 post-transduction), PISD knockout (15 days post-transduction), PS supplemented (day 7 post supplementation), PE/LPE supplemented (day 4 post supplementation) or MMV treated (day 7 post treatment) leukemia cell lines were extracted for densitometric analysis as described previously (Folch et al., 1957). Briefly, Samples were resuspended in PBS, and mixed with 1:2 chloroform:methanol (v/v). H20 (HPLC grade) was added to increase separation. Afterward, samples were mixed then centrifuged for 6 minutes at 1700 g. The organic layer, which contains all the lipids was collected for the characterization of phospholipid distribution, using densitometric analysis.
UHPLC/MS Characterization of MLCL and CL
UHPLC/MS analysis was performed as previously described in (Bradley et al., 2017), with modification to detect MLCL and CL. Samples were dried under N2 gas and resuspended in 100 μL 65:35:5 acetonitrile:isopropanol:water (v/v/v) with 0.1% formic acid. A Dionex UltiMate 3000 UHPLC System was used (Dionex Corporation, Bannockburn, IL, USA), and was coupled to a Thermo Q-Extractive Quadrupole-Orbitrap mass spectrometer (ThermoFisher Scientific, Waltham, MA, USA). A reversed-phase, binary multistep, ultrahigh-performance liquid chromatography (UHPLC) protocol was used with a C18 Ascentis Express column with dimensions of 15cm × 2.1mm × 2.0 μm (Sigma-Aldrich®, MO, USA). The mobile phase consisted of A: 60:40 acetonnitrile:water (v/v), 10mM ammonium formate, and 0.1% formic acid, and B: 90:10 isopopanol:acetonitrile (v/v), 10mM ammonium formate and 0.1% formic acid. The gradient protocol used was as follows: from 0 – 1.5 minutes it was 32% B, from 1.5 – 4 minutes 45% B, from 4 – 8 minutes 50% B, from 8 – 18 minutes55% B, from 18 – 20 minutes 60% B, from 20–35 minutes 70% B, from 35 – 40 minutes 95% B, from 40 – 45 minutes 95% B, from 45 – 47 minutes B was decreased to 32%, and allowed to equilibrate until the 48 minute mark. The flow was set to 260 μL/minute, column temperature at 45°C, and tray temperature at 4°C. The injection volume was 5 μL. The mass spectrometer was operated in negative electrospray ionization mode, 35,000 resolution, with a scan range of m/z 200 to 2,000, spray voltage of −3.0 kV, sheath gas flow rate of 35 units, and capillary temperature of 300°C. MS/MS experiments were done under data-dependent conditions with top 5 ions and 17,500 resolution, and the normalized collision energy was 17.5. Thermo Xcalibur QualBrowser software (version 2.1; ThermoFisher Scientific, MA, USA) was used for extracting ion chromatograms, integrating peak areas, and exporting MS-MS spectra. Chromeleon Xpress (version 7.2; ThermoFisher Scientific, MA, USA) was used to monitor and control the Dionex UHPLC settings. MS/MS spectra were analyzed using the NIST MS search program (version 2.0; National Institute of Standards and Technology, MD, USA) and LipidBlast library. Extracted ion profiles were integrated for 20 cardiolipin species and 8 mono-lyso cardiolipin species using their deprotonated molecular ion m/z ratios ([M-H]−) within a 0.2Da window (Table S7).
Densitomertic Analysis of Phospholipids
Lipids extracted as described above, were spotted onto high-performance thin layer chromatography plates (HPTLC; 5633–5, EMD Chemicals, Darmstadt, Germany) and individual phospholipids were separated using a chloroform:methanol:acetic acid:water (100:75:7:4) solvent system (Stefanyk et al., 2010). A reference standard containing a mixture of sphigomyelin, phosphatidylcholine, PS, phosphatidylinositol, phosphatidylethanolamine, and cardiolipin was spotted alongside each plate to correctly identify the phospholipid species. After allowing the solvent to run up each plate for 45 minutes, the plates were then charred at 180°C with a 10% (w/v) copper (II) sulfate in 8% phosphoric acid solution for 15 minutes (Churchward et al., 2008). Images of the HPTLC plates were captured using a CCD camera on a Fluorchem 5500 imaging station (Alpha Innotech, CA, USA) under reflective white light, and then quntified as described in densitometric phospholipid analysis section below.
Densitometric Phospholipid Analysis
Densitometry analyses were then performed using Image StudioTM (LI-COR, NB, USA) and the percent distribution of each phospholipid specie was calculated by dividing the densitometric value for the individual phospholipid specie by the sum of the denistometric values for all phospholipid species combined.
Quantification of Extracellular PS
OCI-AML2 cells were transduced with shRNA in lentiviral vectors targeting TAZ or control sequences. Seven days after transduction, cells were harvested, and cell death was measured by flow cytometry with annexin V fluorescein isothiocyanate (FITC) and propidium iodide (PI) (Biovision Research Products, CA, USA) staining according to the manufacturer’s instructions. Flow cytometry data were acquired using a LSRFORTESSA X20 (BD Biosciences, FL, USA) flow cytometer and frequency of annexin V positive cells were quantified by with the FlowJo software (TreeStar, OR, USA)
PS Quanification by Confocal Microscopy
Cytospin slides were prepared by centrifuging 10,000 TAZ knockdown 8227 (day 11 post-transduction) or primary AML (day 7 post-transduction) cells onto glass slides, which were then fixed in 4% paraformaldehyde for 5 minutes, permeabalized (0.2% saponin) for 20 minutes, and then stained with annexin V Alexa-647 (1:15) for 1 hour, and DAPI (1:500) for 2 minutes, and then mounted with anti-fade solution. Finally, images were acquired using the Olympus FV1000 (Tokoyo, Japan) confocal microscope, and the amount of PS was quantified as described below.
PS Quantification Using ImageJ
Olympus FV1000 was used to select 10–13 random sections from acquired AML cell images. ImageJ was used to quantify the number of DAPI+ cells, and intensity of PS staining. Then, the average PS intensity per cell of each image aquired was quantified by ImageJ using the formula:
The average of the 10–13 sections for each condition is presented.
Sensitivity to Extrinsic Apoptosis
OCI-AML2 cells transduced with shRNA in lentivrial vectors targeting TAZ or control sequences were then co-treated with 2.5 μg/mL of cyclohexamide and increasing concentrations of TRAIL for 16 hours, 13 days after transduction. Cells were stained with annexin V and PI staining (BD Biosciences, FL, USA) according to the manufacturer’s instructions. Flow cytometry data were acquired using a LSRFORTESSA X20 (BD Biosciences, FL, USA) flow cytometer and frequency of annexin V, PI negative cells were quantified with the FlowJo software (TreeStar, OR, USA)
Seahorse
Oxygen consumption rate (OCR), and extracellular acidification rate (ECAR) were measured in AML cells after TAZ or PISD knockdown using the Seahorse XF-96 analyzer (Seahorse Bioscience, MA, USA). OCR was measured as a marker of electron transport chain activity. Six days (TAZ knockdown) or 8 days (PISD knockout) after transduction cells were resuspended in unbuffered medium and seeded at 1 × 105 cells/well in Cell-Tak coated (0.15 mg/well) XF96 plates. Cells were equilibrated in the unbuffered α-MEM medium (supplemented with 2% FCS) or XF Assay medium (supplemented with 4.5 g/l (25mM) glucose + 1mM pyruvate) for 45 minutes at 37°C in a CO2-free incubator before being transferred to the XF96 analyzer. Spare reserve capacity of the mitochondrial respiratory chain was measured by treating cells with oligomycin, and FCCP (carbonyl cyanide p-trifluoromethoxyphenylhydrazone) in succession.
Cellular Reactive Oxygen Species
Four days after transduction with TAZ or control shRNA sequences cells were stained with Carbocy-H2DCFDA in PBS buffer at 37°C for 30 minutes, and resuspended in PBS with the viability dye PI to assess the reactive oxygen species produced by viable cells. Data was acquired by LSRFortessaX20 (BD Biosciences, FL, USA), and MFI of Carbocy-H2DCFDA in PI negative cells were analyzed using FloJo (TreeStar, OR, USA).
Mitochondrial Mass
Eight days after transduction with TAZ or control shRNA sequences mitochondrial mass was characterized by quantifying the mtDNA to nuclear DNA ratio. mtDNA and nuclear DNA was extracted using the DNeasy Blood and Tissue kit (QIAGEN, MD, USA) from AML cell lines transduced with lentiviral vectors with shRNA targeting TAZ, or control sequences. The relative amount of mtDNA to nuclear DNA was measured using qRT-PCR, using primer pairs for mitochondrial ND1, and nuclear HGB outlined in Table S4.
Live Imaging
The mitochondrial morphology of cells transduced with shRNA in lentiviral vectors targeting TAZ or control sequences, was assessed in live cells by labeling the mitochondria with 1 μM Mito Tracker and Red CMXRos (ThermoFisher Scientific, MA, USA) for 30 minutes at 37°C, 7 days after transduction. Cells were washed twice with complete media and transferred to a poly-L-lysine (Sigma-Aldrich®, MO, USA) coated imaging plate. Images were acquired on spinning disc confocal microscope VisiScope CSU-W1 Olympus IX83 (Tokyo, Japan). Mitochondria shape was characterized by quantifying mitochondrial aspect ratio as described below.
Aspect Ratio
For mitochondrial shape assessment an aspect ratio of each mitochondrion was measured, confocal z-stacks were collected and projected as a z-project in ImageJ (NIH, Bethesda, MD). The population median, quartiles, minimum, and maximum of fields (40–60 cells) are expressed as a boxplot.
Electron Microscopy
The mitochondrial morphology of cells transduced with shRNA in lentiviral vectors targeting TAZ or control sequences, was assessed by transmission electron microscopy (TEM). To perform TEM, cells were harvested at day 8 post-transduction, and fixed with a modified Graham-Karnovsky’s fixative-4% paraformaldehyde plus 1% glutaraldehyde in 0.1 M phosphate buffer pH 7.2 (PB)-for 30 minutes at room temperature, and 4°C overnight. Cells were then washed with phosphate buffer (PB, NaH2P04. H2O + Na2H2PO4) 3 times for 15 minutes. Next, cells were post-fixed with 1% osmium tetroxide buffered with PB for 1 hour and washed again using PB twice for 20 minutes, dehydrated with an ethanol series then infiltrated with propylene oxide. The samples were then resin embedded with Epon, which was polymerized at 40°C for 48 hours. Solid epoxy blocks were sectioned on a Reichert Ultracut E microtome to 90 nm thickness, collected on 300 mesh copper grids and counterstained with uranyl acetate and lead citrate. A Hitachi H7000 (Hitachi, Tokyo, Japan) transmission electron microscope with a beam current of 25 mm, was used to view the sections. Images of the sections were acquired by the Advanced Microscopy Techniques (AMT)-CCD camera (AMT, MA, USA). A composite of representative images acquired are shown in Figure S4.
Lipid-Protein Overlay Assay
Binding of PISD to lipid strips (Echelon Biosciences) was performed as per manufacturer’s instructions. Briefly, strips were blocked in 3% fatty acid-free BSA in TBS (25 mM Tris, 150 mM NaCl pH7.2) followed by incubation with 1 μg/mL of PISD recombinant protein in 1% fatty acid-free BSA in TBS for 16 hours at 4°C. Strips were washed extensively in TBST (0.05% Tween-20 in TBS) buffer to remove unbound proteins, and incubated with PISD antibody for 16 hours at 4°C. PISD protein bound to lipid strips were detected by immunoblotting.
Lipid Supplementation and Growth Analysis
All lipids were purchased from Avanti Polar lipids, lysophosphatidylethanolamine (LPE), phosphatidylethanolamine (PE), PS. A stock solution of LPE (18mM) and PE (25mM) were made by dissolving lipid powder in 65 chloroform: 35 methanol: 8 water (v:v:v), whereas a 25mM stock solution of PS was made by dissolving PS powder in 70% methanol: 20% TWIN-80: 10% Propylene Glycol (PEG) (v:v:v). After lipid supplementation, equal numbers of cells were plated for growth assay, and counted by trypan blue exclusion staining for a period of 8–14 days.
MMV Treatment and Growth Analysis
Cell lines or primary samples were seeded at 1 × 105 cells/mL in 20 mL of medium in T75 flasks. Cells were then treated with increasing concentrations of MMV007285 or DMSO control. Fresh MMV or vehicle control was added every 3 days during the incubation period. After MMV treatment, equal numbers of cells were plated for growth assay, and counted by trypan blue exclusion staining for a period of 14 days
8227 Flow Cytometry
8227 cells were co-immunostained with the viability dye Aqua Dead (ThermoFisher Science, MA, USA), anti-human antibodies recognizing CD34, CD38, Flow cytometry data were acquired using a LSRFORTESSA X20 (BD Biosciences, FL, USA) flow cytometer and frequency of viable 34+,38− cells were analyzed with the FlowJo software (TreeStar, OR, USA).
PS Quanification by Flow Cytometry
Five days after MMV treatment primary AML cells were fixed in 4% paraformaldehyde for 5 minutes, permeabalized (0.2% saponin) for 20 minutes, and then stained with Annexin V FITC (1:25) for 1 hour. Flow cytometry data were acquired using a LSRFORTESSA X20 (BD Biosciences, FL, USA) flow cytometer and annexin V MFI was analyzed with the FlowJo software (TreeStar, OR, USA).
MMV and CL075 Synergism Studies
OCI-AML2 leukemia cells were seeded in 96 well plates, and then treated with increasing concentrations of CL075 with and without increasing concentrations of MMV007285. Cells were incubated at 37°C with 5% CO2 for 72 hours. Cell growth and viability was determined using the MTS assay (Promega), as per manufacturer’s instructions. For each combination excess over Bliss additivism (EOBA) values were calculated as previously described (Borisy et al., 2003).
Animal Studies
Hematopoiesis in WT and Taz-KD mice
In order to understand the role of TAZ in normal hematopoiesis under basal conditions, adult (7.6–14.3) week IDOX-TAZ-KD mice were treated with doxycycline for 12.9–19.4 weeks, and then sacrificed.
To understand the role of TAZ in hematopoietic stress, doxycycline treated IDOX-TAZ-KD mice were injected intraperitoneally (IP) with 200mg/kg of 5-FU (Sigma-Aldrich®, MO, USA), dissolved in phosphate-buffered saline (GIBCO, MA, USA). Peripheral blood was collected at 0, 3, 7, 10, 15, and 23 days after 5-FU injection from the tail vein into EDTA-containing tubes. White blood cell, neutrophil, lymphocyte, red blood cell, and platelet counts were enumerated using the HEMAVET 950FS (Drew Scientific Inc, FL, USA). All mice were sacrificed 15 or 23 days after 5-FU treatment.
At the time of sacrifice, blood was isolated by cardiac puncture for complete blood counts, and hematopoietic cells were flushed from the femur and tibiae of WT and iDOX-TAZ-KD mice, using DMEM + 2% FCS. Red cell were lysed, and the remaining cells were passed through a 100μM strainer. Bone marrow samples were depleted of mature myeloid and lymphoid lineages (Lin−) using magnetic bead separation with biotin-conjugated antibodies CD5, CD45R (B220), CD11b, Anti-Gr-1 (Ly-6G/C), 7–4 and Ter-119 (Miltenyl Biotec, mouse lineage depletion kit).
Lin− cells were co-immunostained with anti-mouse antibodies recognizing Sca1, CKIT, CD48, CD150, CD34, and FCyR, and the viability dye 7AAD. Flow cytometry data were acquired using a FACSCanto II (BD Biosciences) flow cytometer and analyzed with the FlowJo software (TreeStar, OR, USA).
TEX and 8227 Engraftment
Equal numbers of TEX or 8227 cells (1–2 × 106) transduced with shRNA sequences in lentiviral vectors targeting TAZ or control sequences or treated with vehicle control or PS, were injected into the right femur of sublethally irradiated NOD-SCID-GF mice with human IL-3, GM-CSF, and Steel factor (SF) (Nicolini et al., 2004). Five weeks after injection, mice were sacrificed, and the percentage of human CD45+ was enumerated in TEX and PS supplemented 8227 cells by flow cytometry. Whereas in 8227 cells transduced with TAZ shRNA, engraftment of cells into the bone marrow was assessed by measuring GFP+ CD45+cells by flow cytometry, and then calculating relative human cell engraftment as described in the calculation of engraftment potential section.
Stability and Pharmacokinetics of MMV007285
The concentration of intact MMV007285 in blood plasma was measured using Waters Xevo Quadruple Time-of-flight (Q-Tof) hybrid mass spectrometer (MS) system coupled with ACQUITY ultra-performance liquid chromatography (UPLC) (Waters Limited, Mississauga, Canada). Chromatographic separations were carried out on an ACQUITY UPLC BEH C18 (2.1 × 50 mm, 1.7 μm) column (Waters Limited, ON, Canada). The mobile phase was 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). Compound concentrations were measured relative to a freshly prepared calibration curve.
Subcutaneous AML Xenografts
To understand the role of TAZ in AML differentiation and PISD in AML growth in vivo, OCI-AML2 or CAS9-OCI-AML2 cells were first transduced with shRNA targeting TAZ/control sequences, or sgRNA targeting PISD/control sequences respectively. 1 × 106 transduced cells were then in injected subcutaneously into the flanks of male SCID mice (Ontario Cancer Institute). Differentiation after TAZ knock down was evaluated by measuring differentiation markers in sub-cutaneous tumors 8 days after injection, whereas role of PISD in tumor growth was characterized by measuring tumor volume every 2–3 days.
The in vivo anti-tumor activity and toxicity of MMV007285 was also assessed by injecting OCI-AML2 human leukemia cells (1 × 106) subcutaneously into the flanks of male SCID mice (Ontario Cancer Institute). When the tumors were palpable, mice were treated with MMV007285 or vehicle control (5% DMSO, 47.5% PEG400, 10% Tween80, 37.5% H20) by oral gavage (300 mg/kg twice daily for 5 of 7 days) for 10 days (n = 10/group). Tumor volume was measured every 2–3 days. Nineteen after injection mice body weight was measured, afterward mice were sacrificed, peripheral blood was collected then alkaline phosphatase (ALP), aspartate transaminase (AST), bilirubin (Bili), creatine kinase (CK), creatinine (Cr) levels were measured by Idexx Laboratories (Ontario, Canada).
Primary AML Engraftment Models
The role of TAZ in leukemia stem cells in vivo was characterized by transducing primary AML patient samples with TAZ shRNA or control shRNA containing lenti-viral vectors. NOD-SCID mice were conditioned with 208 rad of irradiation from a 137 rad source, and a 200 mg of anti-mouse CD122 48 hours before transplantation. Forty-eight hours after transduction 3×105–1×106 cells were injected into the right femurs of conditioned NOD-SCID mice, 6–8 weeks after, mice were sacrificed, cells were flushed from the femora, and then stained with CD33. The percentage of human GFP+, CD33+ cells in the bone marrow was determined by flow cytometry. The engraftment of transduced human AML cells into the bone marrow was assessed by enumerating relative human cell engraftment as described in the calculation of engraftment potential section.
To asses effect of MMV on leukemia initiating cells in vivo, a frozen aliquot of Primary AML patient samples were thawed, and pre-treated with MMV007285 or DMSO for 48 hours, in IMDM supplemented with 20% FCS, 100 units/mL penicillin and 100 μg/mL of streptomycin before transplantation. The day before transplantation, NOD-SCID mice conditioned as described above. Then, 2 × 106 viable treated cells were injected into the right femurs of 10 week-old female NOD-SCID mice. Six weeks after, mice were sacrificed, cells were flushed from the femora. Engraftment of human AML cells into the bone marrow of was assessed by enumerating the percentage of human hematopoietic myeloid cells with the cell surface signature of CD45+CD33+CD19− by flow cytometry using the BD Biosciences FACSCanto. Data were analyzed with FlowJo version 7.7.1 (TreeStar, OR, USA).
Whereas, to characterize the of MMV007285 on AML patient stem cell propagation properties in vivo, a frozen aliquot of Primary AML patient samples were thawed, counted and re-suspended in PBS, and 2 × 106 viable trypan blue negative cells were injected into the right femurs of NOD-SCID mice, conditioned as described above. Eleven days after transplantation mice were treated with MMV007285 (150mg/kg) by oral gavage or vehicle control (n = 9 mice vehicle control group, n = 10 mice MMV007285 group) once daily for 5 of 7 days for 5 weeks. Mice body weight was measured at the end of the experiment (41 days after AML patient sample transplantation). Afterward mice were sacrificed, and AML cell engraftment bone marrow was measured as described above. Also, at sacrifice, peripheral blood was collected and then alkaline phosphatase, asparate transaminase, bilirubin, creatine kinase, creatinine levels were measured by Idexx Laboratories (Ontario, Canada).
The effects of MMV007285 on leukemia stem cell disease initiating properties were further characterized by injecting equal numbers of human leukemia cells isolated from the bone marrow of MMV007285 or DMSO treated mice, into secondary NOD-SCID mice that were conditioned as described above, but not treated with MMV007285. Six week later, human leukemia cell engraftment in the bone marrow was measured by flow cytometric analysis as described above.
Calculation of Engraftment Potential
Relative engraftment was measured as described in Pei et al. (2018). Using flow cytometry, we first measured % of GFP+ cells within the 7AAD− gate to determine the % of shRNA-expressing cells injected (%GFP-injected). After engraftment, we also measured % of viable GFP+, human CD33+ (primary AML samples) or % of GFP+, human CD45+ (8227) cells to determine the % of shRNA-expressing cells engrafted (%GFP-engrafted). For each experimental group, we first calculated engraftment value TAZ shRNA or control shRNA using the following formula: TAZ shRNA or control shRNA = %GFP-engrafted ÷ %GFP-injected for each mouse. This calculation yielded an array of engraftment values for both control shRNA and TAZ shRNA groups. To calculate the final relative engraftment potential score, each engraftment value was normalized to the mean engraftment of the control shRNA group. These scores were plotted to compare the relative engraftment potential between control and TAZ shRNA.
QUANTIFICATION AND STATISTICAL ANALYSIS
In all figures, n represents technical replicates and N represents independent biological replicates. Blinded analysis was implemented into the experimental design when possible. Data are mean ± standard error of the mean (SEM) or mean ± standard deviation (SD). Prism Graph Pad 6.0 was used to perform statistical analysis and data plotting unless otherwise specified in figure legends. A Two-Way ANOVA followed by a post hoc Dunnett’s test was used to compare two or more variables between three groups, and a Two-Way ANOVA followed by a post hoc Bonferroni test was used to compare two or more variables between two groups. A One-Way ANOVA followed by a post hoc Dunnett’s test was used to compare one variable between three groups unless otherwise stated. Finally, an unpaired Student’s t test was used to compare the mean between two groups. The level of significance is indicated as follows: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, ****p ≤ 0.0001. A detailed description of the experimental setup and statistical analysis is in the figure legends.
DATA AVAILABILITY
AML gene expression after TAZ knock down has been deposited in the Gene Expression Omnibus database under the accession code GEO: GSE107045.
Supplementary Material
Highlights.
Inhibiting TAZ leads to increased levels of PS in AML cells
TAZ and PS regulate AML stemness
Reducing TAZ or increasing PS decreases AML stemness and activates TLR signaling
Increasing PS is a potential therapeutic strategy for AML
ACKNOWLEDGMENTS
We thank Jill Flewelling (Princess Margaret Cancer Center) for administrative assistance. We also thank The Center of Applied Genomics at Sick Kids for performing RNA sequencing and analysis, as well as the Microscopy Imaging Laboratory for performing electron microscopy. This work was supported by the Canadian Institutes of Health Research, the Ontario Institute of Cancer Research with funding provided by the Ontario Ministry of Research and Innovation, the Princess Margaret Cancer Centre Foundation, the Ministry of Long Term Health and Planning in the Province of Ontario, and the NIH (R01HL108882 and R01GM111548 to S.M.C.). A.D.S. holds the Barbara Baker Chair Leukemia and Related Diseases. A.K.S. holds the Fredrick Banting and Charles Best Canada Graduate Scholarships-Doctoral Award from the Canadian Institute of Health as well as the McLaughlin Award from the University of Toronto. M.X. holds the Princess Margaret Hospital Postdoctoral Fellowship and Lady Tata Foundation Postdoctoral Fellowship.
Footnotes
DECLARATION OF INTERESTS
A.D.S. has received honorariums from Novartis, Jazz, and Otsuka Pharmaceuticals and research support from Medivir AB and Takeda. A.D.S. owns stock in Abbvie Pharmaceuticals.
SUPPLEMENTAL INFORMATION
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.stem.2019.02.020.
REFERENCES
- Acehan D, Vaz F, Houtkooper RH, James J, Moore V, Tokunaga C, Kulik W, Wansapura J, Toth MJ, Strauss A, and Khuchua Z (2011). Cardiac and skeletal muscle defects in a mouse model of human Barth syndrome. J. Biol. Chem 286, 899–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baile MG, Sathappa M, Lu YW, Pryce E, Whited K, McCaffery JM, Han X, Alder NN, and Claypool SM (2014). Unremodeled and remodeled cardiolipin are functionally indistinguishable in yeast. J. Biol. Chem 289, 1768–1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels M, Murphy K, Rieter E, and Bruin M (2016). Understanding chronic neutropenia: life is short. Br. J. Haematol 172, 157–169. [DOI] [PubMed] [Google Scholar]
- Borisy AA, Elliott PJ, Hurst NW, Lee MS, Lehar J, Price ER, Serbedzija G, Zimmermann GR, Foley MA, Stockwell BR, and Keith CT (2003). Systematic discovery of multicomponent therapeutics. Proc. Natl. Acad. Sci. USA 100, 7977–7982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley RM, Mardian EB, Bloemberg D, Aristizabal Henao JJ, Mitchell AS, Marvyn PM, Moes KA, Stark KD, Quadrilatero J, and Duncan RE (2017). Mice Deficient in lysophosphatidic acid acyltransferase delta (Lpaatδ)/acylglycerophosphate acyltransferase 4 (Agpat4) Have Impaired Learning and Memory. Mol. Cell. Biol Published online October 27, 2017. 10.1128/MCB.00245-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan SM, Thomas D, Corces-Zimmerman MR, Xavy S, Rastogi S, Hong WJ, Zhao F, Medeiros BC, Tyvoll DA, and Majeti R (2015). Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat. Med 21, 178–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheneval D, Müller M, and Carafoli E (1983). The mitochondrial phosphate carrier reconstituted in liposomes is inhibited by doxorubicin. FEBS Lett. 159, 123–126. [DOI] [PubMed] [Google Scholar]
- Choi JY, Kumar V, Pachikara N, Garg A, Lawres L, Toh JY, Voelker DR, and Ben Mamoun C (2016). Characterization of Plasmodium phosphatidylserine decarboxylase expressed in yeast and application for inhibitor screening. Mol. Microbiol 99, 999–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward MA, Brandman DM, Rogasevskaia T, and Coorssen JR (2008). Copper (II) sulfate charring for high sensitivity on-plate fluorescent detection of lipids and sterols: quantitative analyses of the composition of functional secretory vesicles. J. Chem. Biol 1, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, and Koehler CM (2012). The complexity of cardiolipin in health and disease. Trends Biochem. Sci 37, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, McCaffery JM, and Koehler CM (2006). Mitochondrial mis-localization and altered assembly of a cluster of Barth syndrome mutant tafaz-zins. J. Cell Biol 174, 379–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Oktay Y, Boontheung P, Loo JA, and Koehler CM (2008). Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. J. Cell Biol 182, 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole A, Wang Z, Coyaud E, Voisin V, Gronda M, Jitkova Y, Mattson R, Hurren R, Babovic S, Maclean N, et al. (2015). Inhibition of the Mitochondrial Protease ClpP as a Therapeutic Strategy for Human Acute Myeloid Leukemia. Cancer Cell 27, 864–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colijn C, and Mackey MC (2005). A mathematical model of hematopoiesis: II. Cyclical neutropenia. J. Theor. Biol 237, 133–146. [DOI] [PubMed] [Google Scholar]
- Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, and Majeti R (2014). Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc. Natl. Acad. Sci. USA 111, 2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eble KS, Coleman WB, Hantgan RR, and Cunningham CC (1990). Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J. Biol. Chem 265, 19434–19440. [PubMed] [Google Scholar]
- Folch J, Lees M, and Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem 226, 497–509. [PubMed] [Google Scholar]
- Gonzalvez F, Schug ZT, Houtkooper RH, MacKenzie ED, Brooks DG, Wanders RJ, Petit PX, Vaz FM, and Gottlieb E (2008). Cardiolipin provides an essential activating platform for caspase-8 on mitochondria. J. Cell Biol 183, 681–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F, D’Aurelio M, Boutant M, Moustapha A, Puech JP, Landes T, Arnauné-Pelloquin L, Vial G, Taleux N, Slomianny C, et al. (2013). Barth syndrome: cellular compensation of mitochondrial dysfunction and apoptosis inhibition due to changes in cardiolipin remodeling linked to tafazzin (TAZ) gene mutation. Biochim. Biophys. Acta 1832, 1194–1206. [DOI] [PubMed] [Google Scholar]
- Gu W, Gaeta X, Sahakyan A, Chan AB, Hong CS, Kim R, Braas D, Plath K, Lowry WE, and Christofk HR (2016). Glycolytic Metabolism Plays a Functional Role in Regulating Human Pluripotent Stem Cell State. Cell Stem Cell 19, 476–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. (2015). High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 163, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Haurie C, Dale DC, and Mackey MC (1998). Cyclical neutropenia and other periodic hematological disorders: a review of mechanisms and mathematical models. Blood 92, 2629–2640. [PubMed] [Google Scholar]
- Hayashi EA, Granato A, Paiva LS, Bertho AL, Bellio M, and Nobrega A (2010). TLR4 promotes B cell maturation: independence and cooperation with B lymphocyte-activating factor. J. Immunol 184, 4662–4672. [DOI] [PubMed] [Google Scholar]
- Huang BX, Akbar M, Kevala K, and Kim HY (2011). Phosphatidylserine is a critical modulator for Akt activation. J. Cell Biol 192, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatz-Hoover JJ, Wang H, Moreton SA, Chakrabarti A, Agarwal MK, Sun K, Gupta K, and Wald DN (2015). The role of TLR8 signaling in acute myeloid leukemia differentiation. Leukemia 29, 918–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan M, Snyder TM, Corces-Zimmerman MR, Vyas P, Weissman IL, Quake SR, and Majeti R (2012). Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci. Transl. Med 4, 149ra118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasuyama H, and Melchers F (1998). Establishment of mouse cell lines which constitutively secrete large quantities of interleukin 2, 3, 4 or 5, using modified cDNA expression vectors. Eur. J. Immunol 18, 97–104. [DOI] [PubMed] [Google Scholar]
- Kay JG, and Grinstein S (2013). Phosphatidylserine-mediated cellular signaling. Adv. Exp. Med. Biol 991, 177–193. [DOI] [PubMed] [Google Scholar]
- Keckesova Z, Donaher JL, De Cock J, Freinkman E, Lingrell S, Bachovchin DA, Bierie B, Tischler V, Noske A, Okondo MC, et al. (2017). LACTB is a tumour suppressor that modulates lipid metabolism and cell state. Nature 543, 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köberlin MS, Snijder B, Heinz LX, Baumann CL, Fauster A, Vladimer GI, Gavin AC, and Superti-Furga G (2015).A Conserved Circular Network of Coregulated Lipids Modulates Innate Immune Responses. Cell 162, 170–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz EM, Baquero P, Michie AM, Dunn K, Tardito S, Holyoake TL, Helgason GV, and Gottlieb E (2017). Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med 23, 1234–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagadinou ED, Sach A, Callahan K, Rossi RM, Neering SJ, Minhajuddin M, Ashton JM, Pei S, Grose V, O’Dwyer KM, et al. (2013). BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell 12, 329–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechman ER, Gentner B, Ng SW, Schoof EM, van Galen P, Kennedy JA, Nucera S, Ciceri F, Kaufmann KB, Takayama N, et al. (2016). miR-126 Regulates Distinct Self-Renewal Outcomes in Normal and Malignant Hematopoietic Stem Cells. Cancer Cell 29, 214–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Xu H, Xiao T, Cong L, Love MI, Zhang F, Irizarry RA, Liu JS, Brown M, and Liu XS (2014). MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YW, and Claypool SM (2015). Disorders of phospholipid metabolism: an emerging class of mitochondrial disease due to defects in nuclear genes. Front. Genet 6, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YW, Galbraith L, Herndon JD, Lu YL, Pras-Raves M, Vervaart M, Van Kampen A, Luyf A, Koehler CM, McCaffery JM, et al. (2016). Defining functional classes of Barth syndrome mutation in humans. Hum. Mol. Genet 25, 1754–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucantoni L, Duffy S, Adjalley SH, Fidock DA, and Avery VM (2013). Identification of MMV malaria box inhibitors of plasmodium falciparum early-stage gametocytes using a luciferase-based high-throughput assay. Antimicrob. Agents Chemother 57, 6050–6062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra A, Edelman-Novemsky I, Xu Y, Plesken H, Ma J, Schlame M, and Ren M (2009). Role of calcium-independent phospholipase A2 in the pathogenesis of Barth syndrome. Proc. Natl. Acad. Sci. USA 106, 2337–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken MN, George BM, Kao KS, Marjon KD, Raveh T, and Weissman IL (2016). Normal and Neoplastic Stem Cells. Cold Spring Harb. Symp. Quant. Biol 81, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megías J, Yáñez A, Moriano S, O’Connor JE, Gozalbo D, and Gil ML (2012). Direct Toll-like receptor-mediated stimulation of hematopoietic stem and progenitor cells occurs in vivo and promotes differentiation toward macrophages. Stem Cells 30, 1486–1495. [DOI] [PubMed] [Google Scholar]
- Mitroulis I, Chen LS, Singh RP, Kourtzelis I, Economopoulou M, Kajikawa T, Troullinaki M, Ziogas A, Ruppova K, Hosur K, et al. (2017). Secreted protein Del-1 regulates myelopoiesis in the hematopoietic stem cell niche. J. Clin. Invest 127, 3624–3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. (2006). A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell 124, 1283–1298. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Garrett KP, Ohta S, Bahrun U, Kouro T, Akira S, Takatsu K, and Kincade PW (2006). Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity 24, 801–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimmer AD, et al. (2016). A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature 540, 433–437. [DOI] [PubMed] [Google Scholar]
- Nicolini FE, Cashman JD, Hogge DE, Humphries RK, and Eaves CJ (2004). NOD/SCID mice engineered to express human IL-3, GM-CSF and Steel factor constitutively mobilize engrafted human progenitors and compromise human stem cell regeneration. Leukemia 18, 341–347. [DOI] [PubMed] [Google Scholar]
- Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, Habib N, Yosef N, Chang CY, Shay T, et al. (2011). Densely interconnected transcriptional circuits control cell states in human hematopoiesis. Cell 144, 296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies G, Paradies V, Ruggiero FM, and Petrosillo G (2014). Cardiolipin and mitochondrial function in health and disease. Antioxid. Redox. Signal 20, 1925–1953. [DOI] [PubMed] [Google Scholar]
- Pei S, Minhajuddin M, Adane B, Khan N, Stevens BM, Mack SC, Lai S, Rich JN, Inguva A, Shannon KM, et al. (2018). AMPK/FIS1-Mediated Mitophagy Is Required for Self-Renewal of Human AML Stem Cells. Cell Stem Cell. 23, 86–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao W, Quon G, Csaszar E, Yu M, Morris Q, and Zandstra PW (2012). PERT: a method for expression deconvolution of human blood samples from varied microenvironmental and developmental conditions. PLoS Comput. Biol 8,e1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saric A, Andreau K, Armand AS, Møller IM, and Petit PX (2016). Barth Syndrome: From Mitochondrial Dysfunctions Associated with Aberrant Production of Reactive Oxygen Species to Pluripotent Stem Cell Studies. Front. Genet 6, 359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, et al. (2007). Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 26, 1713–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Mitchell A, Heisler L, Abelson S, Ng SWK, Trotman-Grant A, Medeiros JJF, Rao-Bhatia A, Jaciw-Zurakowsky I, Marke R, et al. (2017). Tracing the origins of relapse in acute myeloid leukemia to stem cells. Nature 547, 104–108. [DOI] [PubMed] [Google Scholar]
- Simpson CD, Hurren R, Kasimer D, MacLean N, Eberhard Y, Ketela T, Moffat J, and Schimmer AD (2012). A genome wide shRNA screen identifies α/β hydrolase domain containing 4 (ABHD4) as a novel regulator of anoikis resistance. Apoptosis 17, 666–678. [DOI] [PubMed] [Google Scholar]
- Skrtić M, Sriskanthadevan S, Jhas B, Gebbia M, Wang X, Wang Z, Hurren R, Jitkova Y, Gronda M, Maclean N, et al. (2011). Inhibition of mitochondrial translation as a therapeutic strategy for human acute myeloid leukemia. Cancer Cell 20, 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soustek MS, Falk DJ, Mah CS, Toth MJ, Schlame M, Lewin AS, and Byrne BJ (2011). Characterization of a transgenic short hairpin RNA-induced murine model of Tafazzin deficiency. Hum. Gene Ther 22, 865–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriskanthadevan S, Jeyaraju DV, Chung TE, Prabha S, Xu W, Skrtic M, Jhas B, Hurren R, Gronda M, Wang X, et al. (2015). AML cells have low spare reserve capacity in their respiratory chain that renders them susceptible to oxidative metabolic stress. Blood 125, 2120–2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanyk LE, Coverdale N, Roy BD, Peters SJ, and LeBlanc PJ (2010). Skeletal muscle type comparison of subsarcolemmal mitochondrial membrane phospholipid fatty acid composition in rat. J. Membr. Biol 234,207–215. [DOI] [PubMed] [Google Scholar]
- Sykes DB, Kfoury YS, Mercier FE, Wawer MJ, Law JM, Haynes MK, Lewis TA, Schajnovitz A, Jain E, Lee D, et al. (2016). Inhibition of Dihydroorotate Dehydrogenase Overcomes Differentiation Blockade in Acute Myeloid Leukemia. Cell 167, 171–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WA, and Hatch GM (2003). Purification and characterization of monolysocardiolipin acyltransferase from pig liver mitochondria. J. Biol. Chem 278, 12716–12721. [DOI] [PubMed] [Google Scholar]
- Taylor WA, and Hatch GM (2009). Identification of the human mitochondrial linoleoyl-coenzyme A monolysocardiolipin acyltransferase (MLCL AT-1). J. Biol. Chem 284, 30360–30371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor WA, Mejia EM, Mitchell RW, Choy PC, Sparagna GC, and Hatch GM(2012). Human trifunctional proteinalphalinks cardiolipin remodeling to beta-oxidation. PLoS ONE 7, e48628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, et al. (2016). A CRISPR Dropout Screen Identifies Genetic Vulnerabilities and Therapeutic Targets in Acute Myeloid Leukemia. Cell Rep. 17, 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JK, Wang JC, Takenaka K, Doulatov S, McKenzie JL, Harrington L, and Dick JE (2005). Direct evidence for cooperating genetic events in the leukemic transformation of normal human hematopoietic cells. Leukemia 19, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Xu Y, Malhotra A, Ren M, and Schlame M (2006). The enzymatic function of tafazzin. J. Biol. Chem 281, 39217–39224. [DOI] [PubMed] [Google Scholar]
- Xu C, Zheng Z, Fang L, Zhao N, Lin Z, Liang T, Zhai Z, and Zhou J (2013). Phosphatidylserine enhances osteogenic differentiation in human mesenchymal stem cells via ERK signal pathways. Mater. Sci. Eng C 33, 1783–1788. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
AML gene expression after TAZ knock down has been deposited in the Gene Expression Omnibus database under the accession code GEO: GSE107045.


