Abstract
BACKGROUND
Checkpoint inhibition has demonstrated clinical efficacy in a variety of solid tumors. Reports of programmed death ligand 1 (PD-L1) expression in glioblastoma are highly variable (ranging from 6% to 88%) and its role as a prognostic marker has yielded conflicting results.
OBJECTIVE
To validate the prevalence and prognostic role of PD-L1 expression in a large cohort of diffuse gliomas according to the 2016 revised WHO classification.
METHODS
Using tissue microarrays, we compared 5 PD-L1 monoclonal antibodies (n = 56) and validated expression (n = 183) using quantitative immunohistochemistry (IHC) and RNA in situ hybridization (RISH). Expression data from The Cancer Genome Atlas (TCGA) and published studies were compared with clinical outcome. Multiplexed immunophenotyping was used to identify PD-L1+ cell populations in post-treatment glioblastoma.
RESULTS
Using a 5% cut-off, PD-L1 expression was significantly associated with a poor prognosis in both histologically defined (n = 125, log-rank P < .001) and recurrent isocitrate dehydrogenase (IDH)-wildtype glioblastoma (n = 60, log-rank P = .015). PD-L1 remained a significant negative prognosticator in Cox regression analysis (hazard ratio: 1.96, P = .021). Analysis of TCGA data confirmed decreased overall survival in recurrent non–glioma CpG island methylator phenotype (G-CIMP) glioblastoma (n = 12, log-rank P = .023), but not in glioblastoma as a group (n = 444, log-rank P = .135). PD-L1 RISH showed a significant correlation with IHC (P < .0001). PD-L1 was observed in the proliferating perivascular stem cell and immune niche of post-treatment glioblastoma.
CONCLUSION
A 5% PD-L1 expression cut-off identified a subset of glioblastoma that is associated with a worse clinical outcome. This association remained significant within the newly defined IDH-wildtype classification. These findings could have implications for patient stratification in future clinical trials of PD-1/PD-L1 blockade.
Keywords: PD-L1, PD-1, Immunohistochemistry, RNA, In situ hybridization, Glioblastoma, Immunotherapy
ABBREVIATIONS
- CI
confidence interval
- G-CIMP
glioma CpG island methylator phenotype
- HR
hazard ratio
- IHC
immunohistochemistry
- IDH
isocitrate dehydrogenase
- OS
overall survival
- PD-L1
programmed death ligand 1
- RISH
RNA in situ hybridization
- TCGA
The Cancer Genome Atlas
- TMA
tissue microarray
- WHO
World Health Organization
The glioblastoma microenvironment is particularly immunosuppressive, including many secreted and cell-based immune suppressive mechanisms.1 Programmed death ligand-1 (PD-L1) is a labile, inducible transmembrane receptor ligand that facilitates immune system evasion through co-ligation of its receptor, PD-1, on activated T cells.2 Upregulation of PD-L1 on tumor cells has been proposed as a mechanism of immune escape in gliomas,3 and its detection at the protein level has been previously demonstrated.4-6 However, study characteristics (grading, sample size) and technical considerations (assays, cut-offs) have resulted in highly variable rates of expression—ranging from 6.1% to 88% in larger studies.4,7 Furthermore, the role of PD-L1 as a prognostic marker in gliomas, independent of predicting treatment response, remains contentious. Initial evidence of an association with poor survival8 has been followed by mixed results in larger studies.4,5 Recent evidence in gliomas, however, suggests an inverse association of PD-L1 expression with mutations in the isocitrate dehydrogenase (IDH) gene,9 which could potentially confound the observed differences in overall survival (OS).
A specific cut-off for PD-L1 expression has not been established in glioblastoma, and it remains to be seen what role PD-L1 expression has in patient selection in future clinical trials which include PD-1/PD-L1 blockade—as either a predictive or prognostic marker. Here, we sought to address the prognostic role of PD-L1 in recurrent glioblastoma in a large tumor cohort according to the updated 2016 World Health Organization (WHO) classification of diffuse gliomas. Additionally, we sought to localize cellular expression of PD-L1 in the tumor microenvironment using multiplex immunofluorescence in post-treatment glioblastoma. These results confirm recent findings of a PD-L1/IDH-wildtype association and validate the poor prognosis associated with PD-L1 within the IDH-wildtype molecular subtype in recurrent glioblastoma.
METHODS
NIH Cohort and Tumor Classification
Formalin-fixed paraffin-embedded tumor samples were obtained from NIH retrospectively from 2000 to 2016. The cohort consisted of 183 individual patient tissues, after excluding repeat tissues in 16 paired patient samples. Tissue procurement, tissue microarray (TMA) construction (large core 4.0 mm), and tumor classification was carried out as previously described.10 Briefly, using an integrated approach, tumors were re-classified from the original diagnosis according to the updated 2016 revised fourth edition of the WHO Classification of Central Nervous System Tumors.11 Classification was largely based on immunohistochemistry (IHC), with fluorescent in situ hybridization and polymerase chain reaction (PCR) incorporated where available. A probabilistic approach combining age, clinical history, and tumor grade was used to assign IDH-wildtype status in tumors negative for staining with an IDH R132H mutant-specific antibody.12 See Pratt et al10 for methodology, antibodies (IDH1, ATRX, p53, H3K27M), and molecular markers (IDH, 1p/19q co-deletion) used in the current study. Results of O-6-methylguanine DNA methyltransferase (MGMT) promoter methylation analysis had not been routinely assessed in this retrospective cohort and was not included as a comparison. Clinical characteristics and pathological diagnoses, including updated tumor classification, are listed in Table 1.
TABLE 1.
Clinical and Pathological Characteristics of the NIH Cohort and Distribution of PD-L1 Expression
| PD-L1 IHC (SP263) n(%) | ||||
|---|---|---|---|---|
| Total n = 183a | ≥1% n = 54 (29.5) | ≥5% n = 43 (23.4) | ≥25% n = 28 (15.3) | |
| Age (yr) | ||||
| Median (range) | 48 (4-75) | 52 (4-74) | 53 (18-74) | 53 (23-74) |
| Sex | ||||
| Female | 57 (31) | 16 (30) | 14 (33) | 6 (21) |
| Male | 124 (69) | 38 (70) | 29 (67) | 22 (79) |
| Presentation | ||||
| Primaryb | 46 (25) | 13 (24) | 11 (26) | 9 (3) |
| Recurrent/post-therapy | 137 (75) | 41 (76) | 32 (74) | 19 (68) |
| Diagnosis (WHO 2016) | ||||
| LGG | 6 (3.3) | 0 (0) | 0 (0) | 0 (0) |
| AA, IDHmut | 21 (11.6) | 1 (1.8) | 0 (0) | 0 (0) |
| AO, IDH-mut/1p19q codeleted | 5 (2.7) | 2 (3.7) | 1 (2.3) | 1 (3.6) |
| Glioblastoma, IDHwt | 81 (44.2) | 37 (68.5) | 30 (69.7) | 20 (71.4) |
| Glioblastoma, IDHmut | 13 (7.1) | 2 (3.7) | 1 (2.3) | 0 (0) |
| Glioblastoma, NOS | 31 (16.9) | 12 (22.2) | 11 (25.6) | 7 (25) |
| DMG, H3K27Mmut | 16 (8.8) | 0 (0) | 0 (0) | 0 (0) |
LGG, low-grade (diffuse) glioma (WHO grade II); AA, anaplastic astrocytoma; AO, anaplastic oligodendroglioma; DMG, diffuse midline glioma.
an = 5 cases did not have available clinical information.
bPrior to chemotherapy or radiotherapy.
Glioblastoma, NOS: WHO grade IV diffuse gliomas with negative IDH R132H staining and an alternative IDH1 or IDH2 mutation probability between 11 and 89%.
Not shown: AANOS (not otherwise specified, n = 2); AAIDHwt (n = 1); AOANOS (anaplastic oligoastrocytoma, n = 1); AONOS (n = 1); DMG non-H3K27M (n = 5).
Immunohistochemistry
Staining was performed on a Ventana Benchmark Ultra (Ventana Medical Systems, Tucson, Arizona; IDH1 R132H, ATRX, p53, H3K27M, CD163, PD-1) and Leica Bond-Max (Leica Biosystems, Bannockburn, Illinois; PD-L1: SP142, SP263, E1L3N, 28-8, CAL10) automated immunostainer according to the manufacturers’ instructions. See Table, Supplemental Digital Content 1 for antibody names, dilutions, and source. Detailed staining protocol is outlined in Supplemental Methods and Results, Supplemental Digital Content 2.
PD-L1 Western Blot Methods
Western blots were performed to validate the specificity of SP142 (1:500, Spring Bioscience M4420, Pleasanton, California), E1L3N (1:1000, Cell Signaling Technology 13684S, Danvers, Massachusetts), and 28-8 (1:3000, Abcam ab205921, Cambridge, United Kingdom), all rabbit anti-human PD-L1 antibodies (see Supplemental Methods and Results, Supplemental Digital Content 2 for details).
RNA In Situ Hybridization
RNAscope® (Advanced Cell Diagnostics, Newark, California) allows for visualization and analysis of expressed RNA in deparaffinized tissue with targeted probes. RNA tagging was performed per manufacturer protocol. Details of staining, probes used, and hybridization are outlined in the Supplemental Methods and Results, Supplemental Digital Content 2.
Digital Automated Analysis
Following slide review, PD-L1 positive cases with IHC and ISH were subjected to automated analysis (Supplemental Methods and Results, Supplemental Digital Content 2). Membranous immunoreactivity with IHC was analyzed using the Aperio Membrane algorithm (v9) and results expressed as the percentage of positive cells over total number of cells within the region of interest (ROI) (Figure, Supplemental Digital Content 3). For RNA ISH (RISH), cores with <1 dot per 10 tumor cells were considered negative (per manufacturer-recommended scoring system) and were not subjected to automated quantification.
Multiplex Fluorescence Immunohistochemistry
Multiplex fluorescence IHC was performed on 5-μm-thick paraffin sections using iterative antibody staining, stripping, and re-staining steps to accumulate 14-plex biomarker imaging data (Supplemental Methods and Results, Supplemental Digital Content 2 and Table, Supplemental Digital Content 4).
TCGA Microarray Datasets
Data from The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov/) were acquired and visualized through the GlioVis portal (http://gliovis.bioinfo.cnio.es).13 Agilent 4502A array data from TCGA glioblastoma multiforme samples (n = 444) were used for survival analyses. A small subset of these samples that were recurrent and had a non-glioma CpG island methylator phenotype (non G-CIMP; verified to be wildtype for IDH) were further analyzed (n = 12). In addition to TCGA data, published datasets with PD-L1 gene expression data (n = 456) acquired using the Affymetrix 2.0 platform14-18 were aggregated and combined for analysis. Median RNA values were used for stratification in all gene expression analyses.
Statistical Analyses
The Kaplan–Meier method was used to estimate the survival curves, median survival time, and 95% confidence intervals, and the log-rank test was used to test for differences in survival functions between dichotomized PD-L1 groups (high vs low expression using 5% and the median as a cut-off for the protein and gene expression data, respectively). The association between IDH status (IDH-wildtype vs IDH-mutant) and PD-L1 was evaluated using Fisher's exact test for dichotomized PD-L1 and Wilcoxon 2-sample test for continuous PD-L1. The Cox proportional hazards regression model was used for both continuous and dichotomized PD-L1 (Supplemental Methods and Results, Supplemental Digital Content 2). A P-value < .05 was considered statistically significant.
RESULTS
Cohort Demographics and Tumor Classification
Baseline patient characteristics, including diagnoses and detailed clinical features, have been previously described.10 A total of 213 tumor specimens were initially included in the TMAs, including surgical and postmortem archival specimens. Six cases proved inadequate due to tissue loss, insufficient tumor content, or a diagnosis that is outside of the scope of this study (eg circumscribed/localized gliomas). ATRX, a nuclear marker that is particularly sensitive to fixation and storage duration, was used as a surrogate for antigen viability and to reduce false-negative cases. Cases that showed absence of ATRX expression in non-neoplastic cells (ie, endothelia, neurons, glia; n = 9) were excluded from further analyses. Of the initial cases, a total of 183 unique tumor samples remained for final evaluation (see Table 1 for demographics and WHO diagnoses of samples; n = 5 cases did not have available clinical information). The median age of the NIH cohort was 48 (range: 4-75 yr of age). The majority of cases were postchemotherapy and/or postradiotherapy (n = 137 recurrent vs n = 46 primary). IDH-wildtype diffuse gliomas (n = 102) constituted the majority of the integrated diagnoses in the cohort: glioblastoma (n = 81), diffuse midline glioma, H3K27M-mutant (n = 16) and wildtype (n = 5), and anaplastic astrocytoma (n = 1). Glioblastoma samples negative for IDH R132H immunostaining and where IDH status could not be confidently predicted (glioblastoma, NOS) constituted 24.8% (n = 31) of glioblastoma samples.
PD-L1 Expression Shows Clone-Dependent Variability and is Associated with IDH Mutation Status
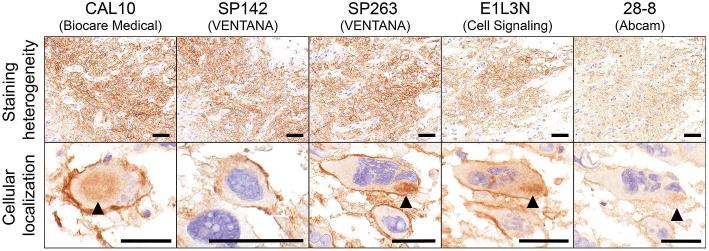
An initial comparison of 5 PD-L1 monoclonal antibodies was performed in n = 56 TMA cases. Antibodies showed some variability in staining intensity and distribution (Figure 1). However, all clones demonstrated some degree of cell surface (membranous) staining with variable amounts of cytoplasmic staining (most notable with CAL10, SP263, and E1L3N). Clones SP263, SP142, and CAL10 showed the highest and most robust signal-to-noise ratio, while E1L3N and 28-8 were either less intense or showed higher background staining. The SP263 clone was chosen for further analysis due to its strong and robust signal in the tissues tested.
FIGURE 1.
Comparison of 5 monoclonal antibodies to PD-L1 in glioblastoma. Both distribution of staining (staining heterogeneity) and cellular localization were compared. CAL10, SP142, and SP263 showed concordant intensity and extent of staining. All clones showed cell surface (membranous) staining. Cytoplasmic immunoreactivity (arrowhead) was more evident with CAL10, SP263, and E1L3N. Clone 28-8 was comparatively weaker in intensity. Scale bar = 50 μm.
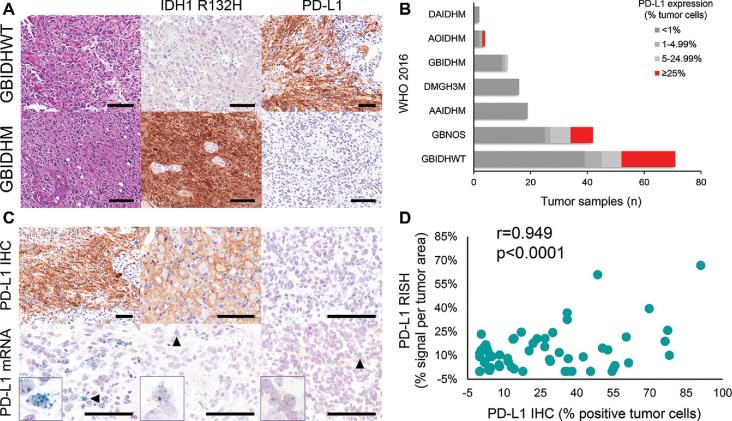
In our cohort, PD-L1 was expressed in 23.5% (43/183) of diffuse gliomas using a 5% cut-off (Table 1). The median percentage of expression in positive tumors was 26% (range 2-91%), with 29.5% of tumors expressing PD-L1 in >1% of cells, 23.5% in >5%, and 15.3% in >25%. Notably, of those expressing PD-L1 in >25% of cells, 96.4% were glioblastoma and 95.5% were wildtype for IDH in cases where IDH status (IDH-wildtype vs IDH-mutant tumors) was established (Figures 2A and 2B). Across all diagnoses, PD-L1 expression was significantly associated with IDH status (for dichotomized PD-L1, PD-L1 ≥ 5% was more frequent in IDH-wildtype (90.1%) than in IDH-mutant tumors (9.1%), Fisher's exact P = .001; for continuous PD-L1, Wilcoxon 2-sample test P = .004). Among glioblastomas with any level of PD-L1 expression, the vast majority were wildtype for IDH (n = 37, 94.8%) compared to IDH-mutant (n = 2, 5.1%).
FIGURE 2.
PD-L1 IHC across 2016 WHO histomolecular subtypes and RISH correlation. A, Representative images of IDH-wildtype (GBIDHWT) and IDH-mutant (GBIDHM) glioblastoma stained with anti-PD-L1 (SP263). B, Stacked bar chart showing PD-L1 IHC distributed across WHO 2016 diagnoses in the NIH cohort (n = 183). C, Overall, PD-L1 staining intensity (not quantified in the current study) was associated with an increase in PD-L1 RISH signals. D, Across the entire cohort (n = 183), quantitative PD-L1 IHC and mRNA showed a significant positive correlation. Scale bar = 100 μm.
PD-L1 IHC and RNA ISH
PD-L1 mRNA detection by RISH showed a range of amplicon frequencies and distribution. Both individual “dots” and, less frequently, large signal aggregates representing coalescence of signals were observed (Figure 2C). PD-L1 (SP263) IHC expression showed a significant correlation with mRNA ISH (n = 183, r = 0.949, P < .0001; Figure 2D). Of the PD-L1 positive cases by IHC with evaluable tissue for RNA (n = 54), 50 had detectable ISH expression (protein+/RNA–, n = 4; Figure, Supplemental Digital Content 5); 2 cases were not evaluable by ISH due to tissue loss. In ISH positive cases (n = 52), only 2 cases showed an absence of staining with PD-L1 IHC (protein–/RNA+, n = 2). Results of PD-L1 western blot evaluation are illustrated in Figure, Supplemental Digital Content 6.
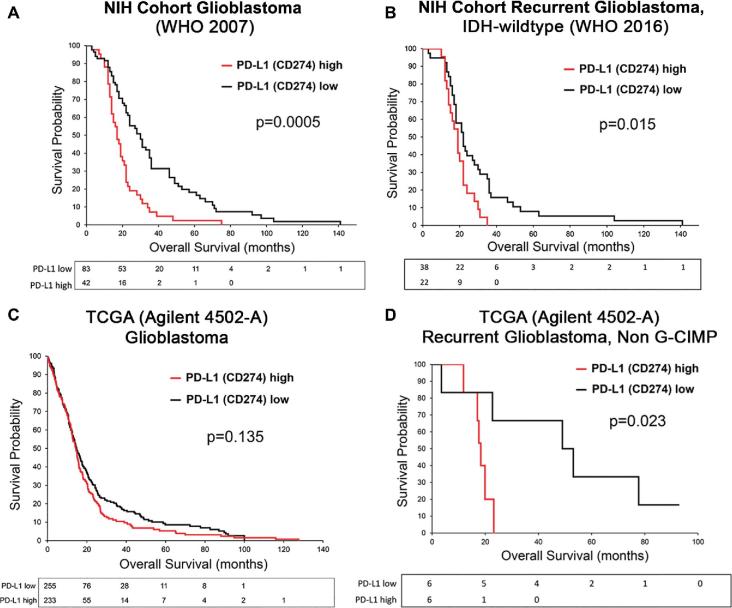
PD-L1 is Associated With a Poor Clinical Outcome in Recurrent Glioblastoma, Independent of IDH Mutation Status
In all histologically defined glioblastomas (WHO 2007), membranous PD-L1 expression in ≥5% of cells was significantly associated with shorter OS (log-rank P < .001; Figure 3A). There is evidence to suggest PD-L1 can be induced by genotoxic stress (eg, radiotherapy).19 Additionally, detection of increased macrophages has been associated with decreased survival in glioblastoma.20 To create a more homogeneous sample for analysis, tumors were matched for grade (IV), treatment (post radio or chemotherapy), and IDH mutation status (glioblastoma, IDH-wildtype, WHO 2016). In subgroup analysis, PD-L1 remained significantly associated with poor outcome in survival analysis of recurrent IDH-wildtype glioblastoma (n = 60, log-rank P = .015, Figure 3B). After exclusion of 2 subjects identified as outliers, the log-rank test P-value was slightly reduced (P = .006, n = 58). PD-L1 protein expression showed a weak, but significant association with PD-1 expression (Spearman r = 0.320, P = .013) and a borderline association with macrophage infiltration (CD163; Spearman r = 0.247, P = .059). Thus, Cox regression model was applied to further assess PD-L1 as a poor prognosticator using age, sex, CD163, and PD-1 as covariates, which were dropped from the model based on significance level of 0.15 (Table 2). PD-L1 continued to be associated with poor clinical outcome: the hazard of death for those with high PD-L1 expression was almost 2 times compared to those with low (<5%) PD-L1 expression (hazard ratio [HR]: 1.96; 95% confidence interval [CI]: 1.11-3.45; P = .021; Table 2). When evaluated as a continuous variable, for each 5% increase in PD-L1 expression, the hazard of death increased by an estimated 11.3% (HR: 1.11; 95% CI: 1.03-1.20; P = .006).
FIGURE 3.
Kaplan–Meier survival estimates comparing high and low PD-L1 expression in glioblastoma. A, Differences in survival curves from tumors meeting the 2007 WHO histologic criteria for glioblastoma (5% PD-L1 cut-off). B, Differences in survival curves from recurrent IDH-wildtype glioblastoma reclassified according to the 2016 WHO nomenclature (5% PD-L1 cut-off). C, TCGA survival curves, classified only as glioblastoma, stratified by median PD-L1 mRNA expression. D, Survival curves from TCGA patients after filtering for recurrent, non–G-CIMP (IDH-wildtype) tumors (median PD-L1 mRNA cut-off).
TABLE 2.
Cox Regression Analysis of OS in Recurrent IDH-wildtype Glioblastoma (NIH cohort, n = 60)a,b
| HR | 95% CI | P-value | |
|---|---|---|---|
| Binary or dichotomized variables | |||
| Sex (F vs M) | 1.010 | 0.580-1.760 | .9706 |
| Surgery (biopsy vs resection) | 1.398 | 0.555-3.521 | .4773 |
| PD-L1 IHC (≥5% vs. <5%) | 1.957 | 1.11-3.45 | .0208 |
| PD-L1 mRNA (0 vs >0) | 0.838 | 0.498-1.41 | .5047 |
| KPS n = 57 (≥80 vs <80) | 1.140 | 0.587-2.23 | .6920 |
| EOR n = 56 (GTR vs non-GTR) | 1.368 | 0.787-2.380 | .2666 |
| Continuous variables | |||
| Age (unit = 1) | 1.008 | 0.987-1.03 | .4494 |
| PD-L1 IHC (unit = 5%) | 1.113 | 1.03-1.20 | .0063 |
| PD-L1 mRNA (unit = 0.01) | 1.015 | 0.988-1.04 | .2775 |
| CD163 (unit = 0.1) | 1.167 | 0.9-1.51 | .2432 |
| PD-1 (unit = 0.1) | 0.923 | 0.81-1.05 | .2297 |
Note: (a) Graphical and statistical tests did not suggest any deviation from the proportionality assumption for any variables.
(b) Age, sex, CD163, and PD-1 were considered as covariates, but they were dropped from the regression model based on α = 0.15.
Microarray Datasets
Previous studies using data from the TCGA have shown conflicting results in the association of PD-L1 gene expression with survival. Agilent-4502A platform expression data were analyzed in a manner replicating our own TMA study. In all glioblastomas (WHO 2007, n = 444), high PD-L1 mRNA (median cut-off: 0.520) was not associated with decreased OS (log-rank P = .135; Figure 3C). This was further confirmed after aggregating published glioblastoma gene expression datasets (n = 456, median cut-off: 6.897; log-rank P = .162). However, after filtering samples for treatment and IDH status in the TCGA dataset, we found that high PD-L1 mRNA (median cut-off = 0.242) was associated with poor OS in a small cohort of recurrent, non–G-CIMP glioblastoma (n = 12; high PD-L1 median OS: 17.95 mo; low PD-L1 median OS: 51.05 mo; log-rank P = .023; Figure 3D), supporting our IHC findings in the NIH cohort.
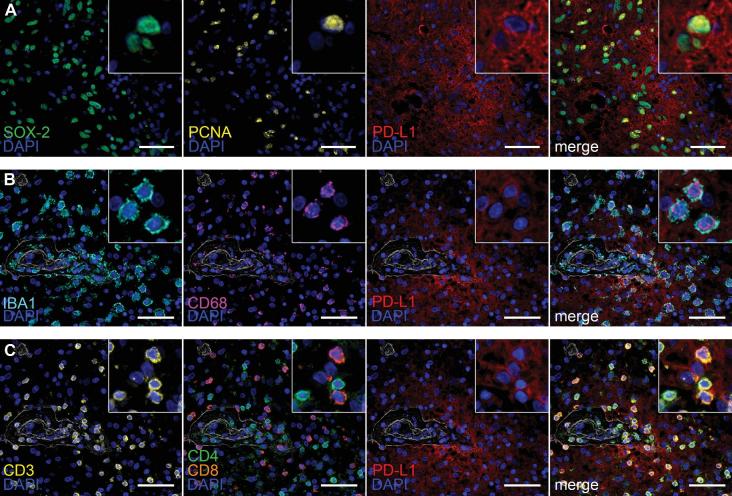
Multiplex Immunoprofiling Reveals PD-L1 Expression in Proliferating GSCs and Variable Expression in the Immune Niche
PD-L1 is a ubiquitous marker where expression has been found in various reactive, inflammatory, and neoplastic states. Deep multiplex immunoprofiling in 4 glioblastomas treated with checkpoint inhibition (nivolumab, n = 2; ipilimumab, n = 2) revealed heterogeneous expression of PD-L1 (SP142) within tumor cells. PD-L1 showed prominent perivascular co-localization with the stem cell antigens SOX-2 (Figure 4A) and nestin (not pictured). This population also co-localized with the proliferation marker PCNA. Tumor areas expressing mature glial antigens S100β and GFAP showed comparatively weaker co-expression with PD-L1 (Figure, Supplemental Digital Content 7). Admixed IBA-1/CD68+ macrophages showing variable expression of PD-L1 were also observed, predominantly within the perivascular compartment (Figure 4B). Within the immune microenvironment, we also show that PD-L1 is expressed in both effector (CD3/CD8+) and helper (CD3/CD4+) T cells (Figure 4C).
FIGURE 4.
Multiplex immunofluorescence of the tumor microenvironment in post-treatment glioblastoma with inflammatory response to checkpoint inhibition. A, Immunolabeling with the nuclear stem cell marker SOX-2 (green), proliferation marker PCNA (yellow), and PD-L1 (red; SP142). PD-L1/SOX-2/PCNA+ cells are depicted in the merged image. B, Representative images of perivascular immune cells co-expression of IBA1 (aqua), CD68 (purple), and PD-L1 (red). C, Representative images of T-cells (CD3, yellow) showing co-expression of both CD4 helper T-cells (green) and CD8 cytotoxic T cells (orange) with PD-L1 (red). Nuclei are counterstained with 4',6-diamidino-2-phenylindole (DAPI; blue). Scale bar = 50 μm.
DISCUSSION
Despite the initial promise of anti-PD-1 checkpoint inhibition in preclinical glioma models, it is becoming apparent that such therapies are only likely to be successful in a subset of glioblastoma patients. This may be due to several factors. Studies have suggested that tumors with high somatic mutational loads are more susceptible to checkpoint inhibition.21 However, the mutational load in glioblastoma is, on average, much lower than that in the cancers that have responded to checkpoint inhibition. Only 3.5% of glioblastoma samples had a high mutational burden in a recent report.22 Mismatch repair deficiencies that also increase susceptibility to checkpoint inhibition are similarly rare in glioblastoma patients. Even in the successful trials of these checkpoint inhibitors in other cancers, these therapies seem to only be effective in a small subset of patients.23 Clinical trials for glioblastoma are in need of accurate ways to appropriately stratify patients according to prognostic factors and, if possible, predict which patients will respond to checkpoint inhibition.
In this study, after stringent antibody validation steps, we compared 6 commercially available antibodies and, generally, found concordance with positivity or negativity for membranous staining in glioblastoma tissue, similar to findings by Gaule et al24 in non-small cell lung cancer (NSCLC) tissue. We also, for the first time in glioblastoma, assessed PD-L1 mRNA expression by ISH. In doing so, we found a strong correlation between PD-L1 protein and in situ RNA expression. While we did observe a small number of PD-L1+ cases with an absence of RNA staining, this may be due to a degradation of global mRNA expression, due to the age of these samples. Alternatively, such mRNA–protein dissociation could be a result of membrane stabilization of PD-L1 protein due to interactions of this protein with the CMTM gene family, as it has been found that this regulation is temporally uncoupled from PD-L1 mRNA expression.25 Our study suggests that most commercially available antibodies seem suitable for PD-L1 evaluation, though we suggest that the use of SP142 or SP263 may increase the ease of interpretation of results.
We found that 23.4% of patient samples expressed PD-L1 at a biologically relevant ≥5% cutoff, which is less than the 38% found by Nduom et al5 and the 32% found by Lee et al.26 Archival clinical information is no longer available for the TMA referenced in Nduom et al,5 unfortunately, so we are unable to assess whether the differences in PD-L1 expression might be accounted for by differences in treatment status or IDH status of their samples. Lee et al26 exclusively evaluated newly diagnosed glioblastoma patients, while our patient population was enriched for recurrent glioblastoma patients. Berghoff et al4,9 reported much higher PD-L1 positivity in their original4 and follow-up manuscript,9 though these results are outliers as compared to the other recently published reports, referenced above. Despite this discrepancy in PD-L1-expression percentages, Berghoff et al9 did also demonstrate a PD-L1 and IDH-wildtype association.9
Nivolumab failed to show clinical efficacy in recurrent glioblastoma patients in a recent phase III randomized trial (CheckMate-143).27 Notably, PD-L1 expression (≥1%) was reported in both treatment cohorts (26.1% and 18.9% in the nivolumab and bevacizumab treatment arms, respectively).28 As PD-L1 expression assessment by IHC had not been standardized prior to that study, it is difficult to assess whether PD-L1 expression was predictive of response in that trial. It is also possible that in recurrent, IDH-wildtype glioblastoma patients, PD-L1 positivity is not predictive of response but remains a prognostic factor, meaning that Checkmate 143 underestimated the therapeutic benefit of this treatment in a less favorable patient cohort. Our findings suggest that, for future trials, a 5% PD-L1 staining threshold may result in more meaningful patient stratification and subset analysis.
Using a cut-off of 5%, we found PD-L1 expression is associated with decreased OS in histologically defined glioblastoma, consistent with results from Nduom et al.5 Furthermore, we show that this association remained significant in subgroup analysis of recurrent IDH-wildtype glioblastoma, a previously unreported finding, as prior reports have focused on the survival of newly diagnosed glioblastoma patients.4,26 Our results thus support the use of a 5% staining threshold for delineating prognosis in glioblastoma in the post-treatment setting.
Establishing PD-L1 expression by IHC as a prognostic biomarker in gliomas has been contentious. Analysis of RNA and transcriptome datasets have also yielded variable results, and may partly be due to the use of different TCGA source material (ie, array platform – Affymetrix, Agilent, RNAseq) and methods of patient separation. Additional analyses will need to be performed in fully clinically annotated datasets in order to further validate these findings.
The tumor microenvironment plays an integral role in glioma pathogenesis and behavior through interaction with regulatory, effector, and helper T cells, microglia, macrophages, and myeloid-derived suppresor cells (MDSCs).29 Immune cells derived from the monocyte lineage are known to make up a significant portion of the tumor mass in glioblastoma.30 In a multicancer study, Taube et al31 demonstrated a significant correlation between immune cell infiltrates and tumor PD-L1 expression. The authors also demonstrated clinical “benefit” (objective response or stable disease for ≥6 mo) with nivolumab in tumors with PD-L1+ immune cells. In a limited sample of post-nivolumab glioblastoma, we found PD-L1 showed co-expression with stem cell antigens in tumor cells and, less frequently, on macrophages and T-cells in the perivascular compartment. Cross-talk between glioma stem cells (GSCs) and tumor associated macrophages has been implicated in tumor progression and treatment resistance.32 Our findings suggest preferential PD-L1 expression within the proliferating GSC population, which may underlie tumor aggressiveness in a subset of post-treatment glioblastoma. However, our restricted sample size requires this to be confirmed in larger studies.
CONCLUSION
Clinical trials that include targeting of PD-1/PD-L1 are ongoing in glioblastoma patients (NCT02658981, NCT03014804, NCT02798406). Interim findings from a phase 2 trial investigating single-agent durvalumab (MEDI4736, NCT02336165) showed promising activity in recurrent glioblastoma. Recently, a phase 2 trial investigating efficacy of a novel IgG1 monoclonal antibody to PD-L1 (avelumab) in recurrent glioblastoma commenced patient recruitment. There are also large randomized trials of checkpoint inhibition targeting PD-1 alongside standard therapy in newly diagnosed glioblastoma patients.33,34 We hope that our work will help to standardize PD-L1 evaluation across future clinical trials so that the impact of PD-L1 expression can be prospectively assessed for predictive or prognostic relevance.
Disclosures
This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government. The authors have no personal, financial, or institutional interest in any of the drugs, materials, or devices described in this article.
Supplementary Material
Supplemental Digital Content 1. Table. Antibody names, source and dilutions used in the study for standard IHC.
Supplemental Digital Content 2. Supplemental Methods and Results. Supplemental Methods for: Digital Automated Analysis; Immunohistochemistry; RNA In Situ Hybridization; Multiplex Fluorescence Immunohistochemistry; Multiplex Fluorescence Immunohistochemistry Image Acquisition; Western Blot Methods; and Statistical Analyses. Also contains Supplemental Results for Western Blot assay.
Supplemental Digital Content 3. Figure. Representative images of PD-L1 expression and automated quantification. (A) PD-L1 IHC (SP263) and the corresponding mark-up image using the Aperio Membrane algorithm (v9). (B) PD-L1 mRNA (RNAscope) showing glioma cells with multiple amplicons (blue-green chromogen) and the corresponding mark-up image using the Aperio Color Deconvolution algorithm (v9).
Supplemental Digital Content 4. Table. Primary antibodies for multiplex IHC-P staining of human glioblastoma slides.
Supplemental Digital Content 5. Figure. Representative images of PD-L1 protein-RISH discrepant cases (A, B).
Supplemental Digital Content 6. Figure. Western blot validation of anti-PD-L1 antibodies SP142, E1L3N, and 28-8 using PD-L1 protein isolated from HEK93 cells transfected with differing concentrations of lipofectamine. Zero lipofectamine group and actin detection used as negative and positive controls.
Supplemental Digital Content 7. Figure. Low-power images of glioblastoma stained with anti-PD-L1 (SP142), GFAP (A), and S-100 (B).
COMMENTS
Immune checkpoint inhibitors targeting PD-1 and PD-L1 have proven effective in multiple types of cancer, especially in melanoma where progress, measured by overall survival, is extraordinary, in fact “head-spinning”.1 Given the complex, immunosuppressive microenvironment of glioblastoma,2,3,4 the use of checkpoint inhibitors for this disease is challenging; as a single agent, these drugs are unlikely to increase overall survival, except perhaps in a small subset of patients.5,6,7,8
The current report indicates that expression of PD-L1, the tumor ligand for PD-1, is a negative prognostic marker in recurrent IDH-wildtype glioblastoma. After validating the immunohistochemical (IHC) detection of PD-L1 with RNA-ISH methodology, the authors suggest a 5% PD-L1 cutoff value by IHC as an independent prognostic index. The authors note that other factors that increase susceptibility to checkpoint inhibition, eg, high somatic mutational load and mismatch repair deficiencies, are uncommon in glioblastomas. Nonetheless, in the CheckMate 143 trial, a small subset of patients (8%) did respond to the PD-1 inhibitor, nivolumab, and showed a superior, durable response (11.1 months) compared to the VEGF inhibitor, bevacizumab (5.3 months).5 In this and other trials, it remains unknown whether the use of a 5% PD-L1 staining threshold could serve as a predictive biomarker and thereby lessen treatment failure. Future studies are needed. However, the current study lays the foundation by validating a clinically available PD-L1 assay in glioblastoma and demonstrating its prognostic significance. Ultimately, because of the severely immunosuppressive microenvironment of glioblastoma, additional immunomodulatory strategies, eg, T-cell CAR,3 IL-6 targeted therapy,4 and others, hold promise in combination with immune checkpoint inhibitors.
Steven Brem
Stephen Bagley
Arati Desai
Gerald Linette
MacLean Nasrallah
Philadelphia, Pennsylvania
References
- 1. Schuchter LM. Adjuvant melanoma therapy – head-spinning progress. N Engl J Med. 2017;377:1888– 1890. [DOI] [PubMed] [Google Scholar]
- 2. Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B. Immune microenvironment of gliomas. Lab Invest. 2017;97:498–518. [DOI] [PubMed] [Google Scholar]
- 3. O’Rourke DM, Nasrallah MP, Desai A, et al.. A single dose of peripherally-infused T cells redirected to EGFRvIII with a chimeric antigen receptor mediates antigen loss and induces adaptive resistance in patients with recurrent glioblastoma, 2017. Sci Transl Med. 2017;9(399):pii: eaaa0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Q, He Z, Huang M, Liu T, et al.. Vascular niche IL-6 macrophage M2 polarization in glioblastoma through HIF-2α. 2018. Nat Commun. 2018;9(1):559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Filey AC, Henriquez M, Dey M. Recurrent glioma clinical trial, CheckMate-143: the game is not over yet. Oncotarget. 2017;8:91 779-91 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Berghoff AS, Kiesel B, Widhalm G, et al.. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol. 2017;19:1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nduom EK, Wei J, Yaghi NK, et al.. PD-L1 expression and prognostic impact in glioblastoma. Neuro-Oncol. 2016;18:195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chamberlain MC, Kim BT. Nivolumab for patients with recurrent glioblastoma progressing on bevacizumab: a retrospective case series. J Neurooncol. 2017;133:561-569. [DOI] [PubMed] [Google Scholar]
As FDA approvals for immunotherapy treatments across many cancer types continue to accrue, the search continues for immune-based therapies that will improve the lives of patients with glioblastoma (GBM). Regarding anti-PD-1/PD-L1 checkpoint blockade approaches, these agents have been such “game-changers” in non-central nervous system cancers that it is critically important to explore the biology of PD-L1 and the effects of treatment in GBM patients. In this study, the authors expand previous work on the incidence of PD-L1 expression in GBM1,2 by describing its expression in a large cohort of recurrent, rather than newly diagnosed, patients. Interestingly, patients with a threshold of PD-L1 expression of at least 5% exhibited poorer overall survival.
In the long term, it is likely that combination treatments rather than anti-PD-1/PD-L1 monotherapy will be needed to improve clinical outcomes in GBM. However, in the near term, how can we improve the likelihood that anti-PD-1/PD-L1 therapy may be more efficacious? As with any trial consideration, it is imperative to be judicious with regard to patient selection. First, although the 5% threshold was correlated with natural history, what is the threshold in the recurrent setting that may presage improved response to checkpoint blockade therapy? As observed with clinical trials of anti-PD-1 in metastatic NSCLC, patients with tumors with at least 50% PD-L1 expression showed an improved overall survival to anti-PD-1 therapy,3 whereas those with at least 5% PD-L1 expression in a separate study did not.4
Secondly, the authors point out that higher mutational burdens have been correlated with improved response to checkpoint blockade but that the incidence of the hypermutated genotype is low in GBM. On the contrary, it is important to note that somatic hypermutation is seen in nearly 20% of patients with recurrent GBM, an observation made by a number of independent groups over the last 11 years.5,6,7,8 Moreover, several groups have described objective responses in adult9 and pediatric10 GBM patients with hypermutated genotypes treated with anti-PD-1 immunotherapy. Thus, it is crucial to identify these patients at recurrence, and clinical trial efforts are under development to test the hypothesis that these patients are more susceptible to checkpoint blockade.
Gavin P. Dunn
St. Louis, Missouri
REFERENCES
- 1. Nduom EK, Weller M, Heimberger AB. Immunosuppressive mechanisms in glioblastoma. Neuro Oncol. 2015;17(suppl 7):vii9-vii14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dong H, Strome SE, Salomao DR, et al.. Erratum: Tumor-associated B7-H1 promotes T-cell apoptosis: a potential mechanism of immune evasion. Nat Med. 2002;8(8):793-800. [DOI] [PubMed] [Google Scholar]
- 3. Wintterle S, Schreiner B, Mitsdoerffer M, et al.. Expression of the B7-related molecule B7-H1 by glioma cells: a potential mechanism of immune paralysis. Cancer Res. 2003;63(21):7462-7467. [PubMed] [Google Scholar]
- 4. Berghoff AS, Kiesel B, Widhalm G, et al.. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nduom EK, Wei J, Yaghi NK, et al.. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016;18(2):195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Parsa AT, Waldron JS, Panner A, et al.. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13(1):84-88. [DOI] [PubMed] [Google Scholar]
- 7. Garber ST, Hashimoto Y, Weathers S-P, et al.. Immune checkpoint blockade as a potential therapeutic target: surveying CNS malignancies. Neuro Oncol. 2016;18(10):1357-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu Y, Carlsson R, Ambjorn M, et al.. PD-L1 expression by neurons nearby tumors indicates better prognosis in glioblastoma patients. J Neurosci. 2013;33(35):14231-14245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Berghoff AS, Kiesel B, Widhalm G, et al.. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol. 2017;19(11):1460-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pratt D, Pittaluga S, Palisoc M, et al.. Expression of CD70 (CD27L) is associated with epithelioid and sarcomatous features in IDH-Wild-Type glioblastoma. J Neuropathol Exp Neurol. 2017;76(8):697-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Louis DN, Perry A, Reifenberger G, et al.. The 2016 world health organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803-820. [DOI] [PubMed] [Google Scholar]
- 12. Chen L, Voronovich Z, Clark K, et al.. Predicting the likelihood of an isocitrate dehydrogenase 1 or 2 mutation in diagnoses of infiltrative glioma. Neuro Oncol. 2014;16(11):1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bowman RL, Wang Q, Carro A, Verhaak RG, Squatrito M. GlioVis data portal for visualization and analysis of brain tumor expression datasets. Neuro Oncol. 2017;19(1):139-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Madhavan S, Zenklusen JC, Kotliarov Y, Sahni H, Fine HA, Buetow K. Rembrandt: helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gravendeel LA, Kouwenhoven MC, Gevaert O, et al.. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065-9072. [DOI] [PubMed] [Google Scholar]
- 16. Murat A, Migliavacca E, Gorlia T, et al.. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015-3024. [DOI] [PubMed] [Google Scholar]
- 17. Ducray F, de Reynies A, Chinot O, et al.. An ANOCEF genomic and transcriptomic microarray study of the response to radiotherapy or to alkylating first-line chemotherapy in glioblastoma patients. Mol Cancer. 2010;9 (1):234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vital AL, Tabernero MD, Castrillo A, et al.. Gene expression profiles of human glioblastomas are associated with both tumor cytogenetics and histopathology. Neuro Oncol. 2010;12(9):991-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adams DL, Adams DK, He J, et al.. Sequential tracking of PD-L1 expression and RAD50 induction in circulating tumor and stromal cells of lung cancer patients undergoing radiotherapy. Clin Cancer Res. 2017;23(19):5948-5958. [DOI] [PubMed] [Google Scholar]
- 20. Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 2008;216(1):15-24. [DOI] [PubMed] [Google Scholar]
- 21. Le DT, Uram JN, Wang H, et al.. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hodges TR, Ott M, Xiu J, et al.. Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro Oncol. 2017;19(8):1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wolchok JD, Kluger H, Callahan MK, et al.. Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med. 2013;369(2):122-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gaule P, Smithy JW, Toki M, et al.. A Quantitative comparison of antibodies to programmed cell death 1 ligand 1. JAMA Oncol. 2016;3(2):256-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mezzadra R, Sun C, Jae LT, et al.. Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature. 2017;549(7670):106-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee KS, Lee K, Yun S, et al.. Prognostic relevance of programmed cell death ligand 1 expression in glioblastoma. J Neurooncol. 2018;136(3):453-461. [DOI] [PubMed] [Google Scholar]
- 27. Reardon DA, Omuro A, Brandes AA, et al.. OS10.3 randomized phase 3 study evaluating the efficacy and safety of nivolumab vs bevacizumab in patients with recurrent glioblastoma: CheckMate 143. Neuro Oncol. 2017;19(suppl_3):iii21-iii21. [Google Scholar]
- 28. Reardon DA, Omuro A, Brandes AA, et al.. Unpublished work. Information presented during presentation “randomized phase 3 study evaluating the efficacy and safety of nivolumab vs bevacizumab in patients with recurrent glioblastoma: CheckMate 143”. 5th Quadrennial Meeting of the World Federation of Neuro-Oncology Societies. Zurich, Switzerland: 2017. [Google Scholar]
- 29. Gieryng A, Pszczolkowska D, Walentynowicz KA, Rajan WD, Kaminska B. Immune microenvironment of gliomas. Lab Invest. 2017;97(5):498-518. [DOI] [PubMed] [Google Scholar]
- 30. Charles NA, Holland EC, Gilbertson R, Glass R, Kettenmann H. The brain tumor microenvironment. Glia. 2011;59(8):1169-1180. [DOI] [PubMed] [Google Scholar]
- 31. Taube JM, Klein A, Brahmer JR, et al.. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti-PD-1 therapy. Clin Cancer Res. 2014;20(19):5064-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wu A, Wei J, Kong LY, et al.. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro Oncol. 2010;12(11):1113-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Weller M, Vlahovic G, Khasraw M, et al.. A randomized phase 2, single-blind study of temozolomide (TMZ) and radiotherapy (RT) combined with nivolumab or placebo (PBO) in newly diagnosed adult patients (pts) with tumor O6-methylguanine DNA methyltransferase (MGMT)-methylated glioblastoma (GBM)—CheckMate-548. Ann Oncol. 2016;27(suppl_6):356TiP-356TiP. [Google Scholar]
- 34. Sampson JH, Omuro AMP, Preusser M, et al.. A randomized, phase 3, open-label study of nivolumab versus temozolomide (TMZ) in combination with radiotherapy (RT) in adult patients (pts) with newly diagnosed, O-6-methylguanine DNA methyltransferase (MGMT)-unmethylated glioblastoma (GBM): CheckMate-498. J Clin Oncol. 2016;34(15_suppl):TPS2079-TPS2079. [Google Scholar]
References
- 1. Nduom EK, Wei J, Yaghi NK, et al.. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016;18(2):195-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Berghoff AS, Kiesel B, Widhalm G, et al.. Programmed death ligand 1 expression and tumor-infiltrating lymphocytes in glioblastoma. Neuro Oncol. 2015;17(8):1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Reck M, Rodriguez-Abreu D, Robinson AG, et al.. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med. 2016;375(19):1823-1833. [DOI] [PubMed] [Google Scholar]
- 4. Carbone DP, Reck M, Paz-Ares L, et al.. First-Line Nivolumab in Stage IV or Recurrent Non-Small-Cell Lung Cancer. N Engl J Med. 2017;376(25):2415-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cahill DP, Levine KK, Betensky RA, et al.. Loss of the mismatch repair protein MSH6 in human glioblastomas is associated with tumor progression during temozolomide treatment. Clin Cancer Res. 2007;13(7):2038-2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Johnson BE, Mazor T, Hong C, et al.. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim H, Zheng S, Amini SS, et al.. Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res. 2015;25(3):316-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J, Cazzato E, Ladewig E, et al.. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Johanns TM, Miller CA, Dorward IG, et al.. Immunogenomics of Hypermutated Glioblastoma: A Patient with Germline POLE Deficiency Treated with Checkpoint Blockade Immunotherapy. Cancer Discov. 2016;6(11):1230-1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bouffet E, Larouche V, Campbell BB, et al.. Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol. 2016;34(19):2206-2211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.