Abstract
Invasive fungal infections (IFIs) occur predominantly in immunocompromised individuals but can also be seen in previously well persons. The human innate immune system recognizes key components of the fungal cell wall as foreign resulting in a myriad of signaling cascades. This triggers release of antifungal molecules as well as adaptive immune responses, which kill or at least contain the invading fungi. However, these defences may fail in hosts with primary or secondary immunodeficiencies resulting in IFIs. Knowledge of a patient's immune status enables the clinician to predict the fungal infections most likely to occur. Moreover, the occurrence of an opportunistic mycosis in a patient without known immunocompromise usually should prompt a search for an occult immune defect. A rapidly expanding number of primary and secondary immunodeficiencies associated with mycoses has been identified. An investigative approach to determining the nature of these immunodeficiencies is suggested to help guide clinicians encountering patients with IFI. Finally, promising adjunctive immunotherapy measures are currently being investigated in IFI.
Keywords: Fungal immunology, medical mycology, invasive fungal infection, inborn errors of immunity, biologic agents
Introduction
Fungi are eukaryotic microorganisms that are ubiquitous in the environment and include yeasts, moulds, mushrooms, polypores, plant parasitic rusts, and smuts.1 There are an estimated 100,000 described species, but the actual global fungal diversity is estimated at 0.8–5.1 M species.1,2 Fungi play fundamental ecological roles as decomposers, pathogens of plants and animals, and they drive carbon cycling in soils and mediate mineral nutrition of plants.2 We encounter them every day—in plant fruits and roots, in soil and air, on animals, and in fresh and salt water. In a recent study of soil collected from 365 sites globally analyzed by pyrosequencing, 963,458 (94.5%) sequences and 80,486 (85.4%) operational taxonomic units were classified as fungi.2
Despite our multiple encounters with fungi throughout our daily lives, through gardening or composting, food mould on the kitchen bench or refrigerator, or merely by inhaling, serious human fungal infections are quite rare in individuals with healthy immune systems. However, the increase in the numbers of immunosuppressed persons has led to the worldwide emergence of invasive mycoses as major causes of morbidity and mortality. There are an estimated 3 million (M) cases of chronic and 0.25 M cases of invasive pulmonary aspergillosis, 0.22 M cases of cryptococcal meningitis complicating human immunodeficiency virus (HIV)/AIDS, 0.7 M cases of invasive candidiasis, 0.5 M of Pneumocystis jirovecii pneumonia, 0.1 M cases of disseminated histoplasmosis, over 10 M cases of fungal asthma and 1 M cases of fungal keratitis, which occur annually worldwide.3 The most significant opportunistic invasive mycoses include cryptococcosis, candidiasis, pneumocystosis, aspergillosis, and mucormycosis, with a mortality rate in some settings approaching up to 75-95%, and endemic mycoses including histoplasmosis, coccidioidomycosis, penicilliosis (now talaromycosis), paracoccidioidomycosis, and blastomycosis.4
Here we discuss in brief the fungal cell wall, why and how the human host discerns fungal cell from host cell, and the adaptation measures with which fungi have countered. We discuss how modern medicine breaches natural human barriers to fungal infections contributing to invasive candidiasis and other rare IFIs. We summarize primary and secondary acquired immunodeficiencies, including new biologic agents and suggest an investigative approach for clinicians to explore for the presence of immunodeficiency. Finally, we highlight a few promising adjunctive immunotherapy measures currently being studied in IFI.
The clinical challenges of invasive fungal infection
IFI are myriad and their presentations so protean that one often must have a high clinical suspicion to make a diagnosis. Recognizing classic hosts such as those infected with HIV or with a hematological malignancy is relatively easy, but unfortunately, IFIs can surprise by occurring in previously well individuals! The diagnosis of IFIs is most perplexing and often further belated in patients with rare or unknown underlying risk factors.
An accurate diagnosis of an IFI may require advanced medical imaging, microbiological sampling, surrogate fungal blood markers, and a tissue biopsy, as per the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group definitions5—each of which can have logistic challenges. Invasive aspergillosis (IA) remains the most commonly diagnosed invasive mould infection in patients with hematological malignancies and solid organ transplant (SOT) recipients. However, clinicians need to have an awareness of the emergence of mucormycosis, as well as infections due to a myriad of rare moulds such as species of Chrysosporium, Exophiala, Ochroconis, Rhizopus, and Scopulariopsis. Moreover, while Candida and Cryptococcus are the most common yeasts to cause infections, rare yeasts, such as Malassezia, Rhodotorula, Trichosporon, Geotrichum, Kodamaea, and Saccharomyces also occur (reviewed in6,7). Furthermore, in patients from endemic regions, thermally dimorphic mycoses, including histoplasmosis, coccidioidomycosis, blastomycosis, talaromycosis, and emergomycosis must be entertained (reviewed in8).
Determining the infecting fungus beyond the more common Candida, Aspergillus, and Cryptococcus species requires morphological expertise supported in many cases by molecular identification. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) is commonly used in modern laboratories to rapidly identify bacteria and yeasts, but its performance with moulds is limited by the availability and size of reference libraries.9,10 In a recent study comparing three libraries, the highest rate of species identification was seen with the Mass spectrometry identification online library where 72% of 221 moulds were identified. In comparison, the rate was a dismal 19.5% with the National Institutes of Health library and 13.6% with the Bruker mould library.11 More importantly, fungi are sometimes difficult to culture—a prerequisite to the use of MALDI-TOF.
Molecular methods (e.g., DNA sequencing) are currently the gold standard for the identification of fungi to the species level, but these are expensive, require specialized equipment or expertise, and are not available in most clinical laboratories. As it is commonly a send-away test, turnaround time is slow, and accuracy may be limited by available primer sets and extraction techniques. Its potential to identify the presence of fungi directly from patient samples without the need for microbiological culture is attractive, although determining whether the organism found by molecular identification is pathogenic, commensal, or even a contaminant requires clinical nuance. Aspergillus polymerase chain reaction (PCR) is the most advanced fungal molecular tool and is verging on being incorporated in the upcoming revised IFI diagnostic definitions but still suffers from issues of standardization, turnaround time, and clinical interpretation (reviewed in12).
Beyond the difficult therapeutic decisions of selecting the most optimal antifungal therapy agent(s), clinicians are then faced with explaining why this specific patient presented with this specific fungal infection and at this specific time. The degree of immunosuppression in advanced HIV/AIDS is often inferred and largely explained by a low CD4+ T-cell count. The impact of chemotherapeutic agents may be largely due to neutropenia and other drug-specific immunosuppressant effects. However, assessing the immune state of the host beyond these two patient groups remains challenging, particularly outside of research settings. Understanding how the immunocompetent host recognizes and counteracts fungi is key to understanding situations where defences fail and mycoses result.
The fungal cell wall structure – simplified
Animals and fungi are kingdoms in the domain Eukaryota;13 thus, from a cellular point of view, humans and fungi have more in common than is often appreciated. One major exception is nearly all fungi have a cell wall. It follows, then, that when encountering a fungal infection, innate immune systems of animals react predominantly to the foreign fungal cell wall components.
The fungal cell wall is predominantly composed of carbohydrate polymers interspersed with glycoproteins.14 The three major components, found in all medically important fungi studied, are β-glucans (homopolymers of glucose with β-1,3-glucans forming the scaffold and β-1,6 glucans forming the branches), chitin (a homopolymer of N-acetylglucosamine that provides structural stability), and mannans (mannose chains of varying lengths and configurations, often linked to proteins) (reviewed in14,15). Some fungi have other cell wall glycans such as chitosan (deacetylated chitin), α-glucan, and galactomannan. While these components intermingle in the cell wall, structurally, chitin and chitosan tend to be situated in the inner portion near the plasma membrane, glucans in the middle, and mannans on the outer portion.15
Breaching the natural barriers to fungal invasion with modern medicine
The human host is usually prepared to defend against fungal infections from multiple routes of natural exposure, most commonly via sinopulmonary, gastrointestinal, and dermatological routes. Each site has unique anatomical and cellular host defences, such as alveolar macrophages in the lungs and epithelial barriers in the skin and vagina. Breaching these natural anatomical barriers are particularly common means by which IFIs develop. The increasing use of invasive devices, especially intravascular central lines but also cardiac pacemakers, ventricular assist devices, urological stents, and joint prostheses has resulted in a contaminant increase in catheter-related bloodstream infection and other nosocomial fungal infections.
Candida spp. are the most common fungal pathogens causing serious hospital-acquired infections.16 Certain Candida spp., especially Candida albicans, are part of the human microbial flora; hence, most candidal infections are endogenous in origin. In recent data pooled from the national healthcare safety network encompassing more than 17,000 healthcare facilities in the United States, Candida spp. were ranked fourth for all hospital-acquired infections, third for central line-associated bloodstream infections, second for catheter-associated urinary tract infections and tenth for surgical site infections.17
Most nosocomial outbreaks of invasive candidiasis and invasive infections with other yeasts have been associated with intravascular catheters.18 A central venous catheter is present in at least 70% of non-neutropenic patients with candidemia at the time that the diagnostic blood culture is obtained (reviewed in16). In neonates, known risk factors for candidemia include very low birthweight, prematurity, central venous catheter use, necrotizing enterocolitis, total parenteral nutrition, and prior or prolonged broad-spectrum antibacterial drug use (reviewed in19).
Non-albicans, nosocomial Candida outbreaks have included C. parapsilosis20,21 and recently C. krusei in South Africa.19 In a study of 290 neonates or preterm infants and 17 paediatric patients with complicated gastrointestinal surgery based in Southern Italy, six blood stream infections were caused by Malassezia furfur and four by Candida spp. (1.4%; two C. parapsilosis, one C. albicans, and one C. glabrata).22 Rarer nosocomial fungal infection outbreaks include a multistate outbreak of Exserohilum rostratum infections, caused by epidural, paraspinal, and peripheral joint injections of fungus-contaminated vials of methylprednisolone;23,24Sarocladium kiliense bloodstream infections in 67 patients at eight hospitals in Santiago, Chile, found to be associated with intravenous ondansetron,25 and Saprochaete clavata hypothesized to be associated with apheresis platelet concentrates26 (reviewed in27,28). These cases emphasize that fungi that normally have very low virulence can cause serious infections if allowed access to sterile parts of the body by disruption of natural anatomic barriers.
Candida auris, a multiresistant organism first reported in patients with ear infections in 2009,29,30 followed by blood stream infections in 2011,31 has been associated with multiple nosocomial outbreaks globally.32–34 It is often misidentified by standard microbiological techniques. C. auris can be difficult to treat as isolates are usually resistant to fluconazole and have variable susceptibility to other azoles, amphotericin B and echinocandins.34
Recognition by the innate immune system
Our immune system has evolved very sophisticated adaptive and innate systems to counteract and defend against fungi. Innate effector cells, mainly macrophages and neutrophils, are the first line of defense against inhaled fungal spores. Innate fungal recognition is accomplished via sensing of cell wall and intracellular pathogen-associated molecular patterns (PAMPs) by soluble, membrane-bound, and intracellular pattern recognition receptors (PRRs) of myeloid and epithelial cells.35 Innate recognition of fungi leads to signal transduction events, which in turn leads to functional responses such as phagocytosis, cytokine and chemokine release, antimicrobial activity, and involvement and modulation of the adaptive immune system.35
Innate immune system PRRs consist of four major classes: C-type lectin receptors (CLRs), Toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD)-like receptors, and retinoic acid inducible gene I (RIG-I)-like receptors.14 Of these, CLRs appear to be most important for antifungal defenses—they bind glycans using conserved carbohydrate-recognition domains and play major roles in recognizing fungal cell wall components (β-glucans and mannans). CLRs important in fungal recognition include Dectin-1 and -2, which recognize β-1,3 glucan and α-mannans, respectively, and mannose receptors, which recognize exposed mannose residues (reviewed in36,37). A newly described CLR, MelLec, senses melanin on fungal conidia.38 Soluble CLRs may be found circulating in the serum and other body fluids or exist as transmembrane receptors on immune cells such as monocytes, dendritic cells, and neutrophils.36
Receptors recognizing cell wall exposed mannans include two CLR, the mannose receptor (CD206, also known as the macrophage mannose receptor) and Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin (DC-SIGN or CD209).14 Other mannose receptors include langerin and Dectin-2. Receptors on dendritic cells that recognize mannose facilitate uptake of mannosylated antigens by dendritic cells for presentation to T cells.
One of the most studied CLRs is Dectin-1.39 This transmembrane receptor is highly expressed on myeloid cells and has specificity for β-1,3-glucans. Upon receptor engagement, various signalling cascades are triggered, including the Caspase recruitment domain-containing protein (CARD)-9 dependent cascade, resulting in phagocytosis, the respiratory burst, and cytokine/chemokine gene induction (reviewed in14). In addition to Dectin-1, CARD9 serves as a critical adapter protein for Dectin-2 and mincle.40 Hence, patients with CARD9 mutations have a more severe phenotype than those with Dectin-1 mutations. Other receptors that recognize β-glucans include complement receptor (CR) 3, and the scavenger receptors, CD5, CD36, and scavenger receptor class F member 1 (SCARF1) (reviewed in14,41). Recently, two additional β-glucan receptors were described; natural killer cell activating receptor NKp3042 and ephrin type-A receptor 2 (EphA2), an oral epithelial cell PRR.43
Surface components on fungi also stimulate TLR2 and TLR4. TLRs initiate intracellular signaling pathways using adapter proteins, which ultimately activate the transcription factor, nuclear factor kappa-light-chain-enhancer of activated B cells (NF-kB), and interferon-regulatory factors.44 Important TLR-dependent cellular responses that promote antifungal immunity include type I interferons (IFNs) and the proinflammatory cytokines tumor necrosis factor (TNF)-α and interleukin (IL)-12 (reviewed in37). Defects in IL-1 receptor associated kinase-4 (IRAK4) and myeloid differentiation factor 88 (MyD88) lead to susceptibility to infections with bacteria such as pneumococcus, TLR3 signaling defects are specific for susceptibility to herpes simplex virus type 1 encephalitis, while mutations in nuclear factor-B essential modulator (NEMO) and other downstream mediators generally induce broader susceptibility to bacteria, viruses, and fungi (reviewed in44,45). Patients with NEMO mutations have presented with PJP infection.46,47 An increased risk for IA in allogeneic stem cell transplantation was suggested with a specific donor TLR4 haplotype.48 Single nucleotide polymorphisms (SNPs) in Dectin-1 and DC-SIGN were combined with other established clinical risk factors and Aspergillus PCR screening results as a predictive model for IA, in a retrospective cohort of high-risk haematology patients.49 While testing donors for specific polymorphisms predisposing to mycoses has not been adopted in routine clinical care, in the future, this type of immunogenomic study could be used to inform decisions regarding antifungal prophylaxis.
Fungi are potent activators of the complement system. Complement activation fragment C5a, which is formed in response to Candida infection, induces the cellular release of the inflammatory cytokines IL-6 and IL-1β, thereby augmenting the host pro-inflammatory response.50 Further, serum antibodies to fungal cell wall components, particularly mannans and β-glucans, can initiate classical pathway activation upon binding and promote recognition by phagocytic Fc receptors (FcRs). Activation of the lectin pathway occurs when recognition of exposed mannans by mannose-binding lectin (MBL) triggers MBL-associated serine proteases.14 SNPs of the gene coding for MBL are present in nearly 40% of the general population, resulting in reduced cross-linking and secretion of nonfunctional MBL. Heterozygotes and compound heterozygotes for this gene record a serum MBL one third and one-sixth that of normal homozygotes. Patients with IA had significantly lower serum MBL levels compared to febrile immunocompromised controls, and a larger proportion met the definition of MBL deficiency.51
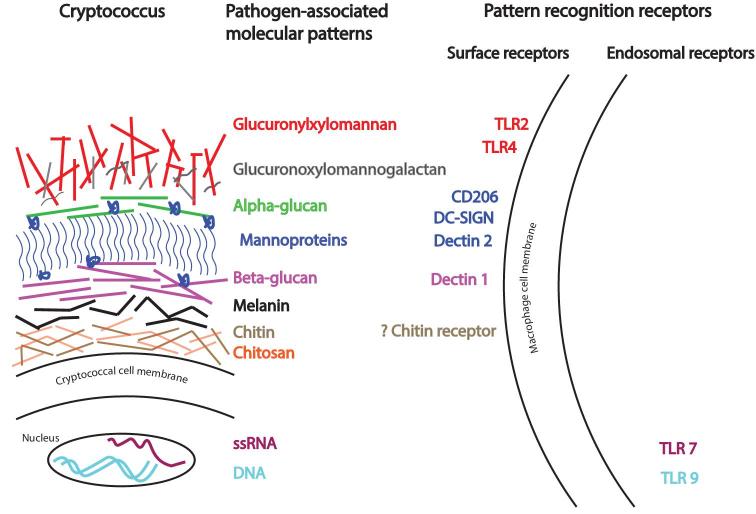
From the above discussion, it is apparent that multiple innate receptors are stimulated by fungi and an immune cell may recognize multiple ligands on a fungus. For example, immune sensing of C. albicans is mediated by multiple recognition systems including Dectin-1-3, Mincle, mannose receptor, DC-SIGN, CR3, FcRs, MBL, and Langerin (reviewed in36). Similarly, several CLRs are involved in the recognition of Aspergillus spp. This includes Dectin-1 and -2, mannose receptor, DC-SIGN, MBL and the lung surfactant proteins (SP) SP-A and SP-D (reviewed in36). Each ligand-receptor system activates specific signaling pathways, with some being proinflammatory, while others are anti-inflammatory. In addition, receptor crosstalk occurs. Thus, the overall host response to fungal infection depends on multiple factors, including the host immune status, site of infection, fungal morphotype (e.g., yeast vs hyphae), cell wall complexity, and fungal virulence traits. The response occurs in a coordinated way involving innate, adaptive and trained immunity.52 A comprehensive review of fungal PRRs and associated fungal PAMPs can be found in Lionakis and Levitz35 and in Goyal et al.36 A simplified illustration of cryptococcal cell wall components acting as PRRs and their respective PAMPs is provided as Figure 1.
Figure 1.
The architecture of the cryptococcal cell wall and capsule, highlighting selected pathogen-associated molecular patterns and their respective pattern recognition receptors. Engagement of receptors leads to activation of signaling pathways that result in cytokine release and other cellular events. CD206 (also known as mannose receptor); DC-SIGN, Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin (also known as CD209); TLR, Toll-like receptor.
Fungal avoidance of innate receptors
While the extent varies depending on the fungal species, mannans sitting on the outer surface shield β-glucans, chitin and chitosan, thereby impairing recognition of these ligands. Some fungi further ‘‘mask’’ ligands to reduce the stimulation of innate immunity. For example, Cryptococcus spp. have a capsule composed of glucuronoxylomannan and galactoxylomannan, which serves as its major virulence factor (reviewed in53). The capsule completely encompasses the cell wall and in the absence of opsonization, encapsulated C. neoformans are poorly phagocytosed.
Importantly, fungal cell walls are dynamic, constantly remodeling structures. By varying the distribution of their components and their cross-linking, they enable morphologic changes to allow for external stress, cellular division, and mating.53 For example, the more virulent Histoplasma chemotype II strain uses α-glucans to mask cell wall β-glucans and secretes Eng1, an endoglucanase which enzymatically prunes exposed β-glucans. Together, this results in reduced Dectin-1 recognition of yeasts and reduced cytokine production by phagocytes.54 The ability to change morphotype by alteration of its surface glycans results in reduced host recognition, reduction in inflammatory response, and increased virulence. Thus, despite the fundamental constructs of fungal cell walls, each fungus presents unique challenges to the host. The host must contend with myriad inter- and intra-species fungal cell wall variations.
Primary immunodeficiencies/ inborn errors of immunity
Primary immunodeficiency diseases are a group of disorders caused by inborn genetic defects in the cells or tissues of the immune system. While severe primary immunodeficiency states usually manifest in childhood, they may occasionally be diagnosed in seemingly immunocompetent adult patients presenting with a severe IFI. Classical primary immunodeficiencies have tended to be of Mendelian inheritance, where single genes confer susceptibility to multitude of infections, in a completely penetrant manner and with a predictable effect on immune cells. Since then, there has been a growing number of monogenic single-gene inborn errors of immunity recognized, with selective susceptibility only to specific infective pathogens.55–57 Routine assessment of these patients with simple investigations looking for immune defects is often unrevealing. More sophisticated analysis of individual patients may identify inborn errors of immunity such as mutations in the genes encoding for CARD9,40,58–64 Dectin-1,65,66 signal transducer and activator of transcription (STAT)-1, 3, or 4,67–70 and IL-12 receptor beta-1.71 As discussed below, other patients have been found to have anti-cytokine antibodies such as auto-antibodies to IFNγ.72–74 GM-CSF,75,76 IL-17,70,77–80 and IL-2278,79 (reviewed in35).
The International Union of Immunological Societies (IUIS) Inborn Errors of Immunity Committee not only publishes and updates a comprehensive classification of the inborn errors of immunity81 but also has a clinician-targeted publication with helpful phenotypic descriptions.82 In its most recent iteration published in 2018, it describes 320 inborn errors, classified into nine categories, with helpful flowcharts, gene names, and phenotypic descriptions. These categories are: combined immunodeficiencies (CID) (cellular and humoral), CID with syndromic features, predominantly antibody deficiencies, diseases with immune dysregulation such as hemophagocytosis, phagocytosis related congenital disorders, defects in intrinsic and innate immunity, autoinflammatory disorders, complement deficiencies, and phenocopies of primary immunodeficiencies. The phenotypic descriptions include some predisposition to bacterial, viral, mycobacterial, and fungal infection.
A detailed knowledge of each signaling cascade is beyond the scope of this review. An update on the immunological and clinical features of inborn errors of immunity that result in mucosal and systemic fungal infection susceptibility has been reviewed by Lionakis and Levitz35. However, illustrative examples of CARD9 deficiency and IFNγ/IL-12 pathway impairment are discussed.
CARD9 loss of function is associated with fungal susceptibility but not viral or bacterial susceptibility. It also may exhibit surprising tissue tropism as evidenced by the particularly severe central nervous system manifestations that CARD9-deficient patients often develop. CARD9 deficiency has been reported in families where multiple individuals have been diagnosed with chronic mucocutaneous candidiasis (CMC), deep dermatophytosis,59Candida encephalitis60,62,64 and the rarely invasive Exophiala spinifera infection.61 While there are many homozygous mutations reported, mostly in consanguineous families, none of the heterozygous individuals was symptomatic, consistent with an autosomal recessive inheritance (reviewed in60). Detailed studies of patients with CARD9 mutations with extrapulmonary aspergillosis and cerebral candidiasis show impaired proinflammatory cytokines and an inability to accumulate neutrophils in the affected tissue.63,83 Administration of granulocyte-macrophage colony stimulating factor (GM-CSF) has been attempted to improve the outcome of a few CARD9 deficient patients with CNS candidiasis58,84,85 but may lead to differential response dependent on the specific affected signaling pathway.
The IL-12/ IFNγ signalling cascade is particularly important in clearing intracellular fungal and bacterial pathogens such as Cryptococcus, Histoplasma, and mycobacteria. Activated macrophages secrete IL-12, which stimulates NK and T cells to produce IFNγ. This cytokine then stimulates macrophage IFNγ receptors, resulting in nuclear translocation of STAT1 and the transcription of IFNγ-related genes. Defects include IFNγ receptor 1 or 2 gene (IFNγR1 or IFNγR2), IL-12 receptor beta-1 gene (IL12RB1), IL12B gene, and STAT1 gene and have variously been associated with CMC, histoplasmosis, cryptococcosis, coccidioidomycosis, and nontuberculous mycobacterial infections (reviewed in35).
IFN-gamma autoantibodies
Apart from genetic defects in the IFNγ/IL-12 axis, acquired autoantibodies against IFNγ have been found in patients presenting with non-tuberculous mycobacteria (NTM) infections86–88 and fungal infections including cryptococcosis,73 talaromycosis,72,74 and histoplasmosis.89 Anti-IFNγ autoantibodies has been associated with HLA-DRB1*16:02 and HLA-DQB1*05:0286 and has a predominance in East Asians.86,88 Rituximab, a chimeric monoclonal antibody against human CD20 primarily found on B cells, has been tried in four patients with high anti-IFNγ antibodies and NTM with promising clinical and immunological responses.90
Idiopathic CD4 lymphopenia
Idiopathic CD4 lymphopenia (ICL) is a rare, heterogeneous syndrome, diagnosed after the exclusion of agents known to depress CD4 T cell counts including HIV.91 It is usually identified when previously well patients present with an opportunistic infection. Current studies use a CD4 cutoff of less than 300 cells/ml or less than 20% of T cells.92–94 In a study of 39 patients with ICL, many had concurrent CD8 lymphopenia, low B cell or low NK cell numbers, and 5 (12.8%) recorded low cell numbers in all four subtypes.94 Cryptococcosis and NTM infections were the most frequent presenting opportunistic infection.94 In a separate study of 25 ICL patients presenting with infection, B cell (CD19+), CD8+, NK cell subsets were also deficient in 19 (76%), 13 (52%), and 7 (28%), respectively.92 Up to 20% of patients with ICL were identified incidentally when lymphopenia is noted on routine blood work.92 An open-label Phase I/IIa dose-escalation study of recombinant human IL-7 in nine patients with ICL found this therapy to be safe.93 There was a resultant increase in peripheral CD4 T-cells at week 3, which peaked at week 5 and was sustained to week 12.93
Secondary immunodeficiencies
Having survived past childhood, adults who develop serious IFIs usually have secondary immunodeficiencies. Secondary immunodeficiency disease states occur when the immune system is compromised due to an environmental factor such as HIV/AIDS or as a consequence of immunosuppressive therapy in association with organ transplantation, cancer, or autoimmune illnesses.
Those with HIV/AIDS with advanced CD4-lymphopenia are classically at risk for cryptococcosis, pneumocystis, oesophageal candidiasis, and the endemic mycoses. Apart from CD4 depletion, HIV leads to B, NK, and dendritic cell (DC) dysfunction, which all contribute to susceptibility to microbial pathogens, including fungi (reviewed in95). Another complication seen in HIV-infected persons coinfected with IFI is immune reconstitution inflammatory syndrome (IRIS) where infective symptoms may paradoxically recur or newly occur after the commencement of antiretroviral therapy. The diagnosis of IRIS is made clinically in the patient who presents with an exaggerated and/or atypical inflammatory response which is temporally associated with the restoration of immune function and in whom an alternative diagnosis has been excluded.96 IRIS has been described as a complication of many IFI, including cryptococcosis, histoplasmosis, pneumocystis, and talaromycosis (reviewed in96).
Hematological stem cell transplant (HSCT) recipients are particularly at risk for pulmonary IA though rarer organisms have been reported. Data from the Transplant-associated infection surveillance network (TRANSNET) database of IFIs in HSCT recipients from 2001 to 2006, showed IA was the most common IFI (43%), followed by invasive candidiasis (28%), zygomycosis (8%), fusariosis (3%), pneumocystosis (2%), and cryptococcosis (0.6%) and a multitude of rare moulds and yeasts.97A. fumigatus caused 44% of the 425 cases of aspergillosis, and concerningly C. glabrata was the most common organism causing candidiasis (33%).97 In a more recent but smaller study of four HSCT centers in the United States, Candida spp. accounted for 34% of the 54 IFIs, Aspergillus spp. for 32%, followed by Mucorales (13%), Pneumocystis (6%), Exophiala (4%), and Alternaria (2%).98 The wide range of fungal pathogens seen in the severely immunosuppressed patients underlines the importance of good mycology diagnostics. This is especially critical as many of these rarer fungi are resistant to commonly used empiric antifungal agents. Dismissing rare fungi as contaminants in the immunosuppressed host is perilous.
Adding to the complexity of treatment-induced immunodeficiency in HSCT and SOT patients, a number of studies highlights the importance of genetic polymorphisms in modulating IFI susceptibility, reviewed in Herrero-Sanchez et al.99 For example, pentraxin-3 is a soluble PRR produced by phagocytes that binds to and opsonizes Aspergillus conidia. Donor and recipient SNPs in the pentraxin-3 gene increase IA susceptibility in allogeneic HSCT100,101 and SOT, respectively.102 This suggests that well-defined immunodeficiency states may be compounded by subtle primary immunological defects.
New biologic agents
There has been a proliferation of new biologic agents in clinical practice many of which have been associated with susceptibility to specific and in some cases unusual fungal infections (reviewed in35,103–106). Keeping pace with these novel agents, particularly their cellular and humoral impact, is challenging. There are two European Society of Clinical Microbiology and Infectious Diseases (ESCMID) publications which summarize the infection risks of these agents; Reinwald et al.105 review drugs impacting intracellular signalling pathways and Redelman-Siti et al.104 review immune checkpoint inhibitors, cell adhesion inhibitors, sphingosine-1-phosphate receptor modulators,and proteasome inhibitors. Examples of these agents and their respective fungal susceptibility are reviewed in Table 1.
Table 1.
| Inhibitor mechanism | Example(s) | Fungal susceptibility |
|---|---|---|
| TNF-α inhibitors | infliximab, etanercept, adalimumab, certolizumab | histoplasmosis, PJP (reviewed in107), cryptococcosis, coccidioidomycosis, and blastomycosis |
| IL-6R | tocilizumab | cryptococcosis108, PJP (reviewed in107) |
| IL-17A | secukinumab ixekizumab brodalumab | candidiasis (reviewed in109) |
| IL-23/IL-17 blockers | ustekinumab | mucosal candidiasis (reviewed in109) |
| Janus kinase inhibitors - JAK1/2 and 1/3 inhibitors | tofacitinib ruxolitinib baricitinib | cryptococcosis110–112, talaromycosis113, candidiasis114 |
| Bruton's tyrosine kinase (BTK) inhibitor | ibrutinib (reviewed in115) | cryptococcosis116,117, PJP118,119, aspergillosis, mucormycosis |
| Vascular endothelial growth factor (VEGF) | bevacizumab | aspergillosis120, fusariosis121 |
| α-4/β-7 integrin | natalizumab vedolizumab | cryptococcosis122,123, candidiasis124 |
| Anti-programmed cell death 1 (PD-1) / PDL-1 | nivolumab pembrolizumab | fusariosis125 |
| Sphingosine-1- phosphate receptor | fingolimod | cryptococcosis126–128, histoplasmosis129 |
| Cytotoxic T-Lymphocyte associated protein 4 (CTLA-4) | ipilimumab | PJP130 |
| Ubiquitin-proteosome | bortezomib | PJP131 |
| Ras/PI3K/Akt/ mTOR | idelalisib, buparlisib, rigosertib, duvelisib | PJP132 |
| BCR-ABL, c-Kyt, other off-target kinases | imatinib, dasatinib, sorafenib | candidiasis133, PJP134, rhodoturulosis135 |
| B cell lymphoma (BCL)-2 | venetoclax | aspergillosis136, candidiasis136, PJP137 |
The utility of giving antifungal prophylaxis when using many of these agents is only starting to be defined. However, anti-pneumocystis prophylaxis is recommended for patients receiving idelalisib throughout the entire course of therapy and for 2 to 6 months after discontinuation and those on Cytotoxic T-Lymphocyte Associated Protein 4 (CTLA-4), programmed cell death 1 (PD-1)/PD-ligand 1 (PDL1) blockade who are expected to receive 20 mg of prednisone (or equivalent) for at least 4 weeks.104,105
A suggested simple clinical algorithm to assess primary and secondary immunodeficiency in the setting of IFI
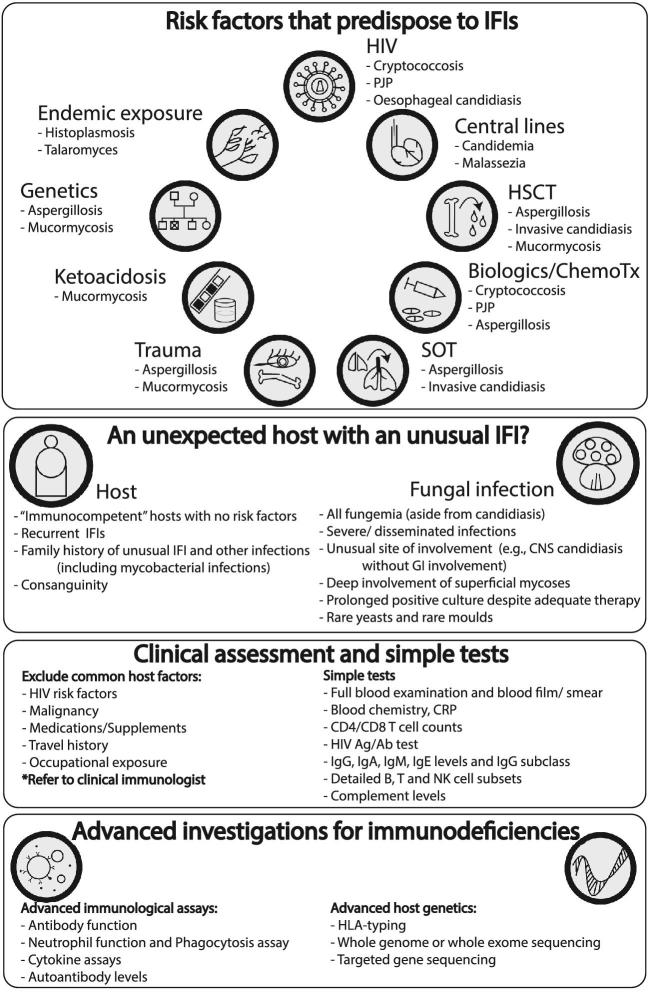
Many individuals presenting with a new IFI will have a known secondary immunodeficiency, such as HIV/AIDS or receipt of immunosuppressive agents. Those with candidemia may have a predisposing central venous catheter. Diagnosing the presence of a secondary immunodeficiency condition such as an undiagnosed haematological malignancy, HIV/AIDS, or solid organ neoplasia is important in all persons presenting with a newly diagnosed IFI. A thorough clinical history and examination is critical in guiding targeted investigation including medical imaging. Simple blood investigations include a complete blood count (CBC) and blood smear, HIV testing, CD4/CD8/ natural killer (NK) cell subsets by flow cytometry, immunoglobulin levels, complement testing, and supportive ancillary tests such as nonspecific inflammatory markers and albumin levels (Fig. 2). This list is not comprehensive and a referral to a clinical immunologist will assist in guiding these investigations.
Figure 2.
A suggested simple clinical algorithm to assess for primary and secondary immunodeficiency in the setting of IFI. ChemoTx, chemotherapeutics; CNS, central nervous system; CRP, C-reactive protein; GI, gastrointestinal; HIV, human immunodeficiency virus; HLA, human leucocyte antigen; HSCT, hematological stem cell transplant; IFI, invasive fungal infections; Ig, immunoglobulin; PJP, Pneumocystis jirovecii pneumonia; SOT, solid organ transplant.
In particular, a thorough clinical assessment of all previous infections should be obtained. For example, multiple episodes of bacterial, viral, and fungal infections in a child or young adult might suggest a combined immunodeficiency. While the types of infections will inform the specific testing, an evaluation of humoral immunity by immunoglobulin, complement and B cell numbers, and adaptive immunity by T and NK cell subset and function are important. Advanced flow cytometry is often required in the diagnostic algorithm. Details of the various diagnostic tools required are reviewed elsewhere.138–141
Patients with unexplained IFI and/ or atypical mycobacterial disease may have an IL-12/ IFNγ pathway defect or IFNγ autoantibodies or other rarer inborn errors of immunity. Patients with IFI without other bacterial or viral or parasitic should be further divided into those limited to chronic mucocutaneous candidiasis (CMC), those with mould infections, or both. Those with just CMC most often have an IL-17 pathway defect. Patients with unusual CNS IFI (e.g., CNS candidiasis in the absence of gastrointestinal involvement) and infections predominantly limited to fungi, should be considered for CARD9 mutations. These inborn errors of immunity are best assessed by whole genome sequencing (WGS), whole exome sequencing (WES), or targeted gene sequencing.140,142–144 A two-tiered investigative approach has been suggested: first focusing on the common, actionable, and severe primary immunodeficiency diseases, followed by a genome-wide approach using either WES or WGS with a large, frequently updated gene candidate list.142 An approach to a patient presenting with an IFI is illustrated in Figure 2.
There are no cost-effectiveness studies to guide or justify these tests, and it is often difficult to know how far to investigate for immunodeficiencies. Access to some of these investigations is also currently limited to research laboratories; thus, a personalized and pragmatic approach is necessary. We advocate that individuals without a known risk factor for IFI, particularly when presenting with a severe and or recurrent IFI, be evaluated for an undiagnosed immunodeficiency, preferably in consultation with an expert clinical immunologist. A diagnosis of primary immunodeficiency may lead to increased awareness of infections, potential prophylaxis strategies and occasionally direct therapeutics. Further, familial linkages may assist early diagnosis in other family members.
Immunotherapeutics for invasive fungal infections
The challenges and promise of immunotherapeutics in IFI have been recently reviewed in Armstrong-James et al.145 In HIV-infected patients with cryptococcal meningitis, adjuvant exogenous IFNγ was shown to be safe and improved clearance rates of cryptococci from the cerebrospinal fluid, although there was no impact on mortality rates compared with standard therapy.146,147 While the use of adjuvant exogenous IFNγ in cryptococcal meningitis has not been adopted as part of routine care, it may have utility in selected difficult to treat patients.
In a prospective open-label study, eight patients with aspergillosis and invasive candidiasis received recombinant IFNγ thrice a week for 2 weeks in addition to standard antifungal therapy. This led to clinical improvement and an increase in HLA-DR positive monocyte levels.148 This strategy was also used in a patient with ICL and progressive cryptococcal meningitis who had a good clinical and immunological response.149 Interestingly an open-label study published more than 10 years ago, explored IFNγ use in 32 HSCT recipients with IFIs and found this to be relatively safe. Only one episode of lymphopenia was reported and this reversed with the cessation of IFNγ.150 No worsening of graft-versus-host-disease (GVHD) occurred, and indeed, about a third of those with acute and chronic GVHD recorded improvement.150 Two successful case reports of adjuvant IFNγ in the treatment of refractory IFI in pediatric patients with acute lymphoblastic leukemia have been recently reported: a 3 year-old with disseminated Candida dublinensis151 and an 8 year-old with disseminated Aspergillus terreus.152 These studies, while promising, must be interpreted cautiously due to the lack of randomized control groups.
An elegant example of translational science is the successful use of topical imiquimod in the treatment of four patients with chromoblastomycosis, a difficult to treat, chronic progressive subcutaneous mycoses most often caused by Fonsecaea pedrosoi.153F. pedrosoi was shown to be recognized by CLRs but not TLRs,154 resulting in defective inflammatory response and chronicity. Imiquimod is a synthetic compound that stimulates both the innate and acquired immune pathways through activation of TLR7.153 Its topical administration led to complete cure in one patient, and partial improvement in three patients, though concerningly these showed morphological transformation from a yeast form to a filamentous form.153
Immune checkpoint inhibitors restore T-cell responses and thus have been recently proposed to be of potential utility in refractory fungal infections, which are controlled by T-cell effector function. Recently, IFNγ dosed thrice weekly and a single dose of 250 mg nivolumab, an anti-PD1 inhibitor, was used to treat severe mucormycosis. This resulted in a rise in absolute lymphocyte count, monocyte HLA-DR expression, and CD8 T-cells, a decrease in T-cell PD-1 expression and clinical recovery.155
The need to keep pace with new therapeutic agents which exploit and expose the host immune system is difficult but important. The gap between research settings and the reality of clinical practice remain wide, particularly in the realm of immunogenomics. The data and literature are growing exponentially. Discoveries are occurring so rapidly that clinicians need a basic framework of fungal immunology assisted by some helpful resources as reference. An openness to embrace scientific collaborators should further enhance clinical care.
Acknowledgements
C.C.C. is supported by the Australian National Health and Medical Research Council (NHMRC) Early Career Fellowship APP1092160. S.M.L. is supported by NIH grants RO1 AI025780, RO1 AI139615, and RO1 AI125045.
Declaration of interest
The authors report no conflict of interest. The authors alone are responsible for the content and the writing of the paper.
References
- 1. Blackwell M. The fungi: 1, 2, 3 … 5.1 million species? Am J Bot. 2011; 98: 426–438. [DOI] [PubMed] [Google Scholar]
- 2. Tedersoo L, Bahram M, Polme S et al.. Fungal biogeography: global diversity and geography of soil fungi. Science. 2014; 346: 1256688. [DOI] [PubMed] [Google Scholar]
- 3. Bongomin F, Gago S, Oladele RO, Denning DW. Global and multi-national prevalence of fungal diseases-estimate precision. J Fungi (Basel). 2017; 3: E57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012; 4: 165rv113. [DOI] [PubMed] [Google Scholar]
- 5. De Pauw B, Walsh TJ, Donnelly JP et al.. Revised definitions of invasive fungal disease from the European Organization for Research and Treatment of Cancer/Invasive Fungal Infections Cooperative Group and the National Institute of Allergy and Infectious Diseases Mycoses Study Group (EORTC/MSG) Consensus Group. Clin Infect Dis. 2008; 46: 1813–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Husain S, Silveira FP, Azie N, Franks B, Horn D. Epidemiological features of invasive mold infections among solid organ transplant recipients: PATH Alliance(R) registry analysis. Med Mycol. 2017; 55: 269–277. [DOI] [PubMed] [Google Scholar]
- 7. Arendrup MC, Boekhout T, Akova M et al.. ESCMID and ECMM joint clinical guidelines for the diagnosis and management of rare invasive yeast infections. Clin Microbiol Infect. 2014; 20: 76–98. [DOI] [PubMed] [Google Scholar]
- 8. Sil A, Andrianopoulos A. Thermally dimorphic human fungal pathogens: polyphyletic pathogens with a convergent pathogenicity trait. Cold Spring Harb Perspect Med. 2014; 5: a019794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wattal C, Oberoi JK, Goel N, Raveendran R, Khanna S. Matrix-assisted laser desorption ionization time of flight mass spectrometry (MALDI-TOF MS) for rapid identification of micro-organisms in the routine clinical microbiology laboratory. Eur J Clin Microbiol Infect Dis. 2017; 36: 807–812. [DOI] [PubMed] [Google Scholar]
- 10. Sanguinetti M, Posteraro B. Identification of molds by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2017; 55: 369–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stein M, Tran V, Nichol KA et al.. Evaluation of three MALDI-TOF mass spectrometry libraries for the identification of filamentous fungi in three clinical microbiology laboratories in Manitoba, Canada. Mycoses. 2018; 61: 743–753. [DOI] [PubMed] [Google Scholar]
- 12. Barnes RA, White PL, Morton CO et al.. Diagnosis of aspergillosis by PCR: Clinical considerations and technical tips. Med Mycol. 2018; 56: 60–72. [DOI] [PubMed] [Google Scholar]
- 13. Burki F. The eukaryotic tree of life from a global phylogenomic perspective. Cold Spring Harb Perspect Biol. 2014; 6: a016147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Levitz SM. Innate recognition of fungal cell walls. PLoS Pathog. 2010; 6: e1000758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Levitz SM, Huang H, Ostroff GR, Specht CA. Exploiting fungal cell wall components in vaccines. Semin Immunopathol. 2015; 37: 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pappas PG, Kauffman CA, Andes DR et al.. Clinical practice guideline for the management of candidiasis: 2016 Update by the Infectious Diseases Society of America. Clin Infect Dis. 2016; 62: e1–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Weiner LM, Webb AK, Limbago B et al.. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016; 37: 1288–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suleyman G, Alangaden GJ. Nosocomial fungal infections: epidemiology, infection control, and prevention. Infect Dis Clin North Am. 2016; 30: 1023–1052. [DOI] [PubMed] [Google Scholar]
- 19. van Schalkwyk E, Iyaloo S, Naicker SD et al.. Large outbreaks of fungal and bacterial bloodstream infections in a neonatal unit, South Africa, 2012–2016. Emerg Infect Dis. 2018; 24: 1204–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H, Zhang L, Kudinha T et al.. Investigation of an unrecognized large-scale outbreak of Candida parapsilosis sensu stricto fungaemia in a tertiary-care hospital in China. Sci Rep. 2016; 6: 27099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Almirante B, Rodriguez D, Cuenca-Estrella M et al.. Epidemiology, risk factors, and prognosis of Candida parapsilosis bloodstream infections: case-control population-based surveillance study of patients in Barcelona, Spain, from 2002 to 2003. J Clin Microbiol. 2006; 44: 1681–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iatta R, Cafarchia C, Cuna T et al.. Bloodstream infections by Malassezia and Candida species in critical care patients. Med Mycol. 2014; 52: 264–269. [DOI] [PubMed] [Google Scholar]
- 23. Kainer MA, Reagan DR, Nguyen DB et al.. Fungal infections associated with contaminated methylprednisolone in Tennessee. N Engl J Med. 2012; 367: 2194–2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Smith RM, Schaefer MK, Kainer MA et al.. Fungal infections associated with contaminated methylprednisolone injections. N Engl J Med. 2013; 369: 1598–1609. [DOI] [PubMed] [Google Scholar]
- 25. Etienne KA, Roe CC, Smith RM et al.. Whole-genome sequencing to determine origin of multinational outbreak of Sarocladium kiliense bloodstream infections. Emerg Infect Dis. 2016; 22: 476–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vaux S, Criscuolo A, Desnos-Ollivier M et al.. Multicenter outbreak of infections by Saprochaete clavata, an unrecognized opportunistic fungal pathogen. MBio. 2014; 5: e02309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bougnoux ME, Brun S, Zahar JR. Healthcare-associated fungal outbreaks: new and uncommon species, new molecular tools for investigation and prevention. Antimicrob Resist Infect Control. 2018; 7: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duran Graeff L, Seidel D, Vehreschild MJ et al.. Invasive infections due to Saprochaete and Geotrichum species: report of 23 cases from the FungiScope Registry. Mycoses. 2017; 60: 273–279. [DOI] [PubMed] [Google Scholar]
- 29. Satoh K, Makimura K, Hasumi Y, Nishiyama Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009; 53: 41–44. [DOI] [PubMed] [Google Scholar]
- 30. Kim MN, Shin JH, Sung H et al.. Candida haemulonii and closely related species at 5 university hospitals in Korea: identification, antifungal susceptibility, and clinical features. Clin Infect Dis. 2009; 48: e57–61. [DOI] [PubMed] [Google Scholar]
- 31. Lee WG, Shin JH, Uh Y et al.. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 2011; 49: 3139–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams E, Quinn M, Tsay S et al.. Candida auris in healthcare facilities, New York, USA, 2013–2017. Emerg Infect Dis. 2018; 24: 1816–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eyre DW, Sheppard AE, Madder H et al.. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018; 379: 1322–1331. [DOI] [PubMed] [Google Scholar]
- 34. Lockhart SR, Etienne KA, Vallabhaneni S et al.. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017; 64: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lionakis MS, Levitz SM. Host Control of Fungal Infections: lessons from basic studies and human cohorts. Annu Rev Immunol. 2018; 36: 157–191. [DOI] [PubMed] [Google Scholar]
- 36. Goyal S, Castrillon-Betancur JC, Klaile E, Slevogt H. The Interaction of human pathogenic fungi with C-type lectin receptors. Front Immunol. 2018; 9: 1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Drummond RA, Gaffen SL, Hise AG, Brown GD. Innate defense against fungal pathogens. Cold Spring Harb Perspect Med. 2014; 5: a019620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Stappers MHT, Clark AE, Aimanianda V et al.. Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus. Nature. 2018; 555: 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brown GD, Gordon S. Immune recognition. A new receptor for beta-glucans. Nature. 2001; 413: 36–37. [DOI] [PubMed] [Google Scholar]
- 40. Lionakis MS, Holland SM. CARD9: at the intersection of mucosal and systemic antifungal immunity. Blood. 2013; 121: 2377–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Canton J, Neculai D, Grinstein S. Scavenger receptors in homeostasis and immunity. Nat Rev Immunol. 2013; 13: 621–634. [DOI] [PubMed] [Google Scholar]
- 42. Li SS, Ogbomo H, Mansour MK et al.. Identification of the fungal ligand triggering cytotoxic PRR-mediated NK cell killing of Cryptococcus and Candida. Nat Commun. 2018; 9: 751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swidergall M, Solis NV, Lionakis MS, Filler SG. EphA2 is an epithelial cell pattern recognition receptor for fungal beta-glucans. Nat Microbiol. 2018; 3: 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Picard C, Casanova JL, Puel A. Infectious diseases in patients with IRAK-4, MyD88, NEMO, or IkappaBalpha deficiency. Clin Microbiol Rev. 2011; 24: 490–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maglione PJ, Simchoni N, Cunningham-Rundles C. Toll-like receptor signaling in primary immune deficiencies. Ann N Y Acad Sci. 2015; 1356: 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Salt BH, Niemela JE, Pandey R et al.. IKBKG (nuclear factor-kappa B essential modulator) mutation can be associated with opportunistic infection without impairing Toll-like receptor function. J Allergy Clin Immunol. 2008; 121: 976–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hanson EP, Monaco-Shawver L, Solt LA et al.. Hypomorphic nuclear factor-kappaB essential modulator mutation database and reconstitution system identifies phenotypic and immunologic diversity. J Allergy Clin Immunol. 2008; 122: 1169–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bochud PY, Chien JW, Marr KA et al.. Toll-like receptor 4 polymorphisms and aspergillosis in stem-cell transplantation. N Engl J Med. 2008; 359: 1766–1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. White PL, Parr C, Barnes RA. Predicting invasive aspergillosis in hematology patients by combining clinical and genetic risk factors with early diagnostic biomarkers. J Clin Microbiol. 2018; 56: e01122–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Cheng SC, Sprong T, Joosten LA et al.. Complement plays a central role in Candida albicans-induced cytokine production by human PBMCs. Eur J Immunol. 2012; 42: 993–1004. [DOI] [PubMed] [Google Scholar]
- 51. Lambourne J, Agranoff D, Herbrecht R et al.. Association of mannose-binding lectin deficiency with acute invasive aspergillosis in immunocompromised patients. Clin Infect Dis. 2009; 49: 1486–1491. [DOI] [PubMed] [Google Scholar]
- 52. Netea MG, van der Meer JW. Trained immunity: an ancient way of remembering. Cell Host Microbe. 2017; 21: 297–300. [DOI] [PubMed] [Google Scholar]
- 53. Doering TL. How sweet it is! Cell wall biogenesis and polysaccharide capsule formation in Cryptococcus neoformans. Annu Rev Microbiol. 2009; 63: 223–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ray SC, Rappleye CA. Flying under the radar: Histoplasma capsulatum avoidance of innate immune recognition. Semin Cell Dev Biol. 2018; 89: 91–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Bucciol G, Moens L, Bosch B et al.. Lessons learned from the study of human inborn errors of innate immunity. J Allergy Clin Immunol. 2018; 143: 507–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Casanova JL. Human genetic basis of interindividual variability in the course of infection. Proc Natl Acad Sci U S A. 2015; 112: E7118–7127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Casanova JL. Severe infectious diseases of childhood as monogenic inborn errors of immunity. Proc Natl Acad Sci U S A. 2015; 112: E7128–7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gavino C, Cotter A, Lichtenstein D et al.. CARD9 deficiency and spontaneous central nervous system candidiasis: complete clinical remission with GM-CSF therapy. Clin Infect Dis. 2014; 59: 81–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lanternier F, Pathan S, Vincent QB et al.. Deep dermatophytosis and inherited CARD9 deficiency. N Engl J Med. 2013; 369: 1704–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lanternier F, Mahdaviani SA, Barbati E et al.. Inherited CARD9 deficiency in otherwise healthy children and adults with Candida species-induced meningoencephalitis, colitis, or both. J Allergy Clin Immunol. 2015; 135: 1558–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lanternier F, Barbati E, Meinzer U et al.. Inherited CARD9 deficiency in 2 unrelated patients with invasive Exophiala infection. J Infect Dis. 2015; 211: 1241–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Glocker EO, Hennigs A, Nabavi M et al.. A homozygous CARD9 mutation in a family with susceptibility to fungal infections. N Engl J Med. 2009; 361: 1727–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Rieber N, Gazendam RP, Freeman AF et al.. Extrapulmonary Aspergillus infection in patients with CARD9 deficiency. JCI Insight. 2016; 1: e89890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Alves de Medeiros AK, Lodewick E, Bogaert DJ et al.. Chronic and invasive fungal infections in a family with CARD9 deficiency. J Clin Immunol. 2016; 36: 204–209. [DOI] [PubMed] [Google Scholar]
- 65. Overton NL, Simpson A, Bowyer P, Denning DW. Genetic susceptibility to severe asthma with fungal sensitization. Int J Immunogenet. 2017; 44: 93–106. [DOI] [PubMed] [Google Scholar]
- 66. Cunha C, Di Ianni M, Bozza S et al.. Dectin-1 Y238X polymorphism associates with susceptibility to invasive aspergillosis in hematopoietic transplantation through impairment of both recipient- and donor-dependent mechanisms of antifungal immunity. Blood. 2010; 116: 5394–5402. [DOI] [PubMed] [Google Scholar]
- 67. Chandesris MO, Melki I, Natividad A et al.. Autosomal dominant STAT3 deficiency and hyper-IgE syndrome: molecular, cellular, and clinical features from a French national survey. Medicine (Baltimore). 2012; 91: e1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Toubiana J, Okada S, Hiller J et al.. Heterozygous STAT1 gain-of-function mutations underlie an unexpectedly broad clinical phenotype. Blood. 2016; 127: 3154–3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. van de Veerdonk FL, Plantinga TS, Hoischen A et al.. STAT1 mutations in autosomal dominant chronic mucocutaneous candidiasis. N Engl J Med. 2011; 365: 54–61. [DOI] [PubMed] [Google Scholar]
- 70. Puel A, Cypowyj S, Marodi L, Abel L, Picard C, Casanova JL. Inborn errors of human IL-17 immunity underlie chronic mucocutaneous candidiasis. Curr Opin Allergy Clin Immunol. 2012; 12: 616–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ouederni M, Sanal O, Ikinciogullari A et al.. Clinical features of candidiasis in patients with inherited interleukin 12 receptor beta1 deficiency. Clin Infect Dis. 2014; 58: 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Tang BS, Chan JF, Chen M et al.. Disseminated penicilliosis, recurrent bacteremic nontyphoidal salmonellosis, and burkholderiosis associated with acquired immunodeficiency due to autoantibody against gamma interferon. Clin Vaccine Immunol. 2010; 17: 1132–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chetchotisakd P, Anunnatsiri S, Nithichanon A, Lertmemongkolchai G. Cryptococcosis in anti-interferon-gamma autoantibody-positive patients: a different clinical manifestation from HIV-infected patients. Jpn J Infect Dis. 2017; 70: 69–74. [DOI] [PubMed] [Google Scholar]
- 74. Pruetpongpun N, Khawcharoenporn T, Damronglerd P et al.. Disseminated Talaromyces marneffei and Mycobacterium abscessus in a patient with anti-interferon-gamma autoantibodies. Open Forum Infect Dis. 2016; 3: ofw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kuo CY, Wang SY, Shih HP et al.. Disseminated cryptococcosis due to anti-granulocyte-macrophage colony-stimulating factor autoantibodies in the absence of pulmonary alveolar proteinosis. J Clin Immunol. 2017; 37: 143–152. [DOI] [PubMed] [Google Scholar]
- 76. Arai T, Inoue Y, Akira M, Nakata K, Kitaichi M. Autoimmune pulmonary alveolar proteinosis following pulmonary aspergillosis. Intern Med. 2015; 54: 3177–3180. [DOI] [PubMed] [Google Scholar]
- 77. Yamazaki Y, Yamada M, Kawai T et al.. Two novel gain-of-function mutations of STAT1 responsible for chronic mucocutaneous candidiasis disease: impaired production of IL-17A and IL-22, and the presence of anti-IL-17F autoantibody. J Immunol. 2014; 193: 4880–4887. [DOI] [PubMed] [Google Scholar]
- 78. Puel A, Doffinger R, Natividad A et al.. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010; 207: 291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Kisand K, Boe Wolff AS, Podkrajsek KT et al.. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010; 207: 299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sarkadi AK, Tasko S, Csorba G, Toth B, Erdos M, Marodi L. Autoantibodies to IL-17A may be correlated with the severity of mucocutaneous candidiasis in APECED patients. J Clin Immunol. 2014; 34: 181–193. [DOI] [PubMed] [Google Scholar]
- 81. Picard C, Bobby Gaspar H, Al-Herz W et al.. International union of immunological societies: 2017 primary immunodeficiency diseases committee report on inborn errors of immunity. J Clin Immunol. 2018; 38: 96–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Bousfiha A, Jeddane L, Picard C et al.. The 2017 IUIS phenotypic classification for primary immunodeficiencies. J Clin Immunol. 2018; 38: 129–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Drummond RA, Collar AL, Swamydas M et al.. CARD9-dependent neutrophil recruitment protects against fungal invasion of the central nervous system. PLoS Pathog. 2015; 11: e1005293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Gavino C, Hamel N, Zeng JB et al.. Impaired RASGRF1/ERK-mediated GM-CSF response characterizes CARD9 deficiency in French-Canadians. J Allergy Clin Immunol. 2016; 137: 1178–1188. [DOI] [PubMed] [Google Scholar]
- 85. Drummond RA, Zahra FT, Natarajan M et al.. GM-CSF therapy in human CARD9 deficiency. J Allergy Clin Immunol. 2018; 142: 1334–1338. [DOI] [PubMed] [Google Scholar]
- 86. Chi CY, Lin CH, Ho MW et al.. Clinical manifestations, course, and outcome of patients with neutralizing anti-interferon-gamma autoantibodies and disseminated nontuberculous mycobacterial infections. Medicine (Baltimore). 2016; 95: e3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Valour F, Perpoint T, Senechal A et al.. Interferon-gamma autoantibodies as predisposing factor for nontuberculous mycobacterial infection. Emerg Infect Dis. 2016; 22: 1124–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Patel SY, Ding L, Brown MR et al.. Anti-IFN-gamma autoantibodies in disseminated nontuberculous mycobacterial infections. J Immunol. 2005; 175: 4769–4776. [DOI] [PubMed] [Google Scholar]
- 89. van de Vosse E, van Wengen A, van der Meide WF, Visser LG, van Dissel JT. A 38-year-old woman with necrotising cervical lymphadenitis due to Histoplasma capsulatum. Infection. 2017; 45: 917–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Browne SK, Zaman R, Sampaio EP et al.. Anti-CD20 (rituximab) therapy for anti-IFN-gamma autoantibody-associated nontuberculous mycobacterial infection. Blood. 2012; 119: 3933–3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Laurence J. T-cell subsets in health, infectious disease, and idiopathic CD4+ T lymphocytopenia. Ann Intern Med. 1993; 119: 55–62. [DOI] [PubMed] [Google Scholar]
- 92. Regent A, Autran B, Carcelain G et al.. Idiopathic CD4 lymphocytopenia: clinical and immunologic characteristics and follow-up of 40 patients. Medicine (Baltimore). 2014; 93: 61–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sheikh V, Porter BO, DerSimonian R et al.. Administration of interleukin-7 increases CD4 T cells in idiopathic CD4 lymphocytopenia. Blood. 2016; 127: 977–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Zonios DI, Falloon J, Bennett JE et al.. Idiopathic CD4+ lymphocytopenia: natural history and prognostic factors. Blood. 2008; 112: 287–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chang CC, Crane M, Zhou J et al.. HIV and co-infections. Immunol Rev. 2013; 254: 114–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chang CC, French MA. Immune reconstitution inflammatory syndrome in invasive fungal infections: what we know and what we need to know? Curr Clin Micro Rpt. 2016; 3: 63–70. [Google Scholar]
- 97. Kontoyiannis DP, Marr KA, Park BJ et al.. Prospective surveillance for invasive fungal infections in hematopoietic stem cell transplant recipients, 2001–2006: overview of the Transplant-Associated Infection Surveillance Network (TRANSNET) Database. Clin Infect Dis. 2010; 50: 1091–1100. [DOI] [PubMed] [Google Scholar]
- 98. Schuster MG, Cleveland AA, Dubberke ER et al.. Infections in hematopoietic cell transplant recipients: results from the organ transplant infection project, a multicenter, prospective, cohort study. Open Forum Infect Dis. 2017; 4: ofx050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Herrero-Sanchez MC, Angomas EB, de Ramon C et al.. Polymorphisms in receptors involved in opsonic and non-opsonic phagocytosis and the risk of infection in oncohematological patients. Infect Immun. 2018. doi:10.1128/IAI.00709-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Cunha C, Aversa F, Lacerda JF et al.. Genetic PTX3 deficiency and aspergillosis in stem-cell transplantation. N Engl J Med. 2014; 370: 421–432. [DOI] [PubMed] [Google Scholar]
- 101. Fisher CE, Hohl TM, Fan W et al.. Validation of single nucleotide polymorphisms in invasive aspergillosis following hematopoietic cell transplantation. Blood. 2017; 129: 2693–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Wojtowicz A, Lecompte TD, Bibert S et al.. PTX3 Polymorphisms and Invasive mold infections after solid organ transplant. Clin Infect Dis. 2015; 61: 619–622. [DOI] [PubMed] [Google Scholar]
- 103. Vallabhaneni S, Chiller TM. Fungal infections and new biologic therapies. Curr Rheumatol Rep. 2016; 18: 29. [DOI] [PubMed] [Google Scholar]
- 104. Redelman-Sidi G, Michielin O, Cervera C et al.. ESCMID Study Group for Infections in Compromised Hosts (ESGICH) Consensus Document on the safety of targeted and biological therapies: an infectious diseases perspective (immune checkpoint inhibitors, cell adhesion inhibitors, sphingosine-1-phosphate receptor modulators and proteasome inhibitors). Clin Microbiol Infect. 2018; 24: S95–S107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Reinwald M, Silva JT, Mueller NJ et al.. ESCMID Study Group for Infections in Compromised Hosts (ESGICH) Consensus document on the safety of targeted and biological therapies: an infectious diseases perspective (intracellular signaling pathways: tyrosine kinase and mTOR inhibitors). Clin Microbiol Infect. 2018; 24: S53–S70. [DOI] [PubMed] [Google Scholar]
- 106. Kourbeti IS, Ziakas PD, Mylonakis E. Biologic therapies in rheumatoid arthritis and the risk of opportunistic infections: a meta-analysis. Clin Infect Dis. 2014; 58: 1649–1657. [DOI] [PubMed] [Google Scholar]
- 107. Takeuchi T, Kameda H. The Japanese experience with biologic therapies for rheumatoid arthritis. Nat Rev Rheumatol. 2010; 6: 644–652. [DOI] [PubMed] [Google Scholar]
- 108. Nishioka H, Takegawa H, Kamei H. Disseminated cryptococcosis in a patient taking tocilizumab for Castleman's disease. J Infect Chemother. 2018; 24: 138–141. [DOI] [PubMed] [Google Scholar]
- 109. Saunte DM, Mrowietz U, Puig L, Zachariae C. Candida infections in patients with psoriasis and psoriatic arthritis treated with interleukin-17 inhibitors and their practical management. Br J Dermatol. 2017; 177: 47–62. [DOI] [PubMed] [Google Scholar]
- 110. Seminario-Vidal L, Cantrell W, Elewski BE. Pulmonary cryptococcosis in the setting of tofacitinib therapy for psoriasis. J Drugs Dermatol. 2015; 14: 901–902. [PubMed] [Google Scholar]
- 111. Liu J, Mouhayar E, Tarrand JJ, Kontoyiannis DP. Fulminant Cryptococcus neoformans infection with fatal pericardial tamponade in a patient with chronic myelomonocytic leukaemia who was treated with ruxolitinib: case report and review of fungal pericarditis. Mycoses. 2018; 61: 245–255. [DOI] [PubMed] [Google Scholar]
- 112. Wysham NG, Sullivan DR, Allada G. An opportunistic infection associated with ruxolitinib, a novel janus kinase 1,2 inhibitor. Chest. 2013; 143: 1478–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Chan JF, Chan TS, Gill H et al.. Disseminated infections with Talaromyces marneffei in non-AIDS patients given monoclonal antibodies against CD20 and kinase inhibitors. Emerg Infect Dis. 2015; 21: 1101–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Chen Y, Gong FY, Li ZJ et al.. A study on the risk of fungal infection with tofacitinib (CP-690550), a novel oral agent for rheumatoid arthritis. Sci Rep. 2017; 7: 6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Ghez D, Calleja A, Protin C et al.. Early-onset invasive aspergillosis and other fungal infections in patients treated with ibrutinib. Blood. 2018; 131: 1955–1959. [DOI] [PubMed] [Google Scholar]
- 116. Swan CD, Gottlieb T. Cryptococcus neoformans empyema in a patient receiving ibrutinib for diffuse large B-cell lymphoma and a review of the literature. BMJ Case Rep. 2018. doi:10.1136/bcr-2018-224786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Messina JA, Maziarz EK, Spec A, Kontoyiannis DP, Perfect JR. Disseminated cryptococcosis with brain involvement in patients with chronic lymphoid malignancies on ibrutinib. Open Forum Infect Dis. 2017; 4: ofw261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Lee R, Nayernama A, Jones SC, Wroblewski T, Waldron PE. Ibrutinib-associated Pneumocystis jirovecii pneumonia. Am J Hematol. 2017; 92: E646—E648. [DOI] [PubMed] [Google Scholar]
- 119. Ahn IE, Jerussi T, Farooqui M, Tian X, Wiestner A, Gea-Banacloche J. Atypical Pneumocystis jirovecii pneumonia in previously untreated patients with CLL on single-agent ibrutinib. Blood. 2016; 128: 1940–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Williams J, Lim R, Tambyah P. Invasive aspergillosis associated with bevacizumab, a vascular endothelial growth factor inhibitor. Int J Infect Dis. 2007; 11: 549–550. [DOI] [PubMed] [Google Scholar]
- 121. Ruiz N, Fernandez-Martos C, Romero I et al.. Invasive fungal infection and nasal septum perforation with bevacizumab-based therapy in advanced colon cancer. J Clin Oncol. 2007; 25: 3376–3377. [DOI] [PubMed] [Google Scholar]
- 122. Valenzuela RM, Pula JH, Garwacki D, Cotter J, Kattah JC. Cryptococcal meningitis in a multiple sclerosis patient taking natalizumab. J Neurol Sci. 2014; 340: 109–111. [DOI] [PubMed] [Google Scholar]
- 123. Gundacker ND, Jordan SJ, Jones BA, Drwiega JC, Pappas PG. Acute cryptococcal immune reconstitution inflammatory syndrome in a patient on natalizumab. Open Forum Infect Dis. 2016; 3: ofw038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Gutwinski S, Erbe S, Munch C, Janke O, Muller U, Haas J. Severe cutaneous Candida infection during natalizumab therapy in multiple sclerosis. Neurology. 2010; 74: 521–523. [DOI] [PubMed] [Google Scholar]
- 125. Tokumo K, Masuda T, Miyama T et al.. Nivolumab-induced severe pancytopenia in a patient with lung adenocarcinoma. Lung Cancer. 2018; 119: 21–24. [DOI] [PubMed] [Google Scholar]
- 126. Seto H, Nishimura M, Minamiji K et al.. Disseminated cryptococcosis in a 63-year-old patient with multiple sclerosis treated with fingolimod. Intern Med. 2016; 55: 3383–3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Forrestel AK, Modi BG, Longworth S, Wilck MB, Micheletti RG. Primary cutaneous cryptococcus in a patient with multiple sclerosis treated with fingolimod. JAMA Neurol. 2016; 73: 355–356. [DOI] [PubMed] [Google Scholar]
- 128. Achtnichts L, Obreja O, Conen A, Fux CA, Nedeltchev K. Cryptococcal meningoencephalitis in a patient With multiple sclerosis treated with fingolimod. JAMA Neurol. 2015; 72: 1203–1205. [DOI] [PubMed] [Google Scholar]
- 129. Veillet-Lemay GM, Sawchuk MA, Kanigsberg ND. Primary cutaneous histoplasma capsulatum Infection in a patient treated with fingolimod: a case report. J Cutan Med Surg. 2017; 21: 553–555. [DOI] [PubMed] [Google Scholar]
- 130. Arriola E, Wheater M, Krishnan R, Smart J, Foria V, Ottensmeier C. Immunosuppression for ipilimumab-related toxicity can cause pneumocystis pneumonia but spare antitumor immune control. Oncoimmunology. 2015; 4: e1040218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Wondergem MJ, Grunberg K, Wittgen BP, Sonneveld P, Zweegman S. Interstitial pneumonitis caused by Pneumocystis jirovecii pneumonia (PCP) during bortezomib treatment. Histopathology. 2009; 54: 631–633. [DOI] [PubMed] [Google Scholar]
- 132. Jones JA, Robak T, Brown JR et al.. Efficacy and safety of idelalisib in combination with ofatumumab for previously treated chronic lymphocytic leukaemia: an open-label, randomised phase 3 trial. Lancet Haematol. 2017; 4: e114–e126. [DOI] [PubMed] [Google Scholar]
- 133. Speletas M, Vyzantiadis TA, Kalala F et al.. Pneumonia caused by Candida krusei and Candida glabrata in a patient with chronic myeloid leukemia receiving imatinib mesylate treatment. Med Mycol. 2008; 46: 259–263. [DOI] [PubMed] [Google Scholar]
- 134. Chang H, Hung YS, Chou WC. Pneumocystis jiroveci pneumonia in patients receiving dasatinib treatment. Int J Infect Dis. 2014; 25: 165–167. [DOI] [PubMed] [Google Scholar]
- 135. Coppola R, Zanframundo S, Rinati MV et al.. Rhodotorula mucilaginosa skin infection in a patient treated with sorafenib. J Eur Acad Dermatol Venereol. 2015; 29: 1028–1029. [DOI] [PubMed] [Google Scholar]
- 136. DiNardo CD, Pratz KW, Letai A et al.. Safety and preliminary efficacy of venetoclax with decitabine or azacitidine in elderly patients with previously untreated acute myeloid leukaemia: a non-randomised, open-label, phase 1b study. Lancet Oncol. 2018; 19: 216–228. [DOI] [PubMed] [Google Scholar]
- 137. Stilgenbauer S, Eichhorst B, Schetelig J et al.. Venetoclax in relapsed or refractory chronic lymphocytic leukaemia with 17p deletion: a multicentre, open-label, phase 2 study. Lancet Oncol. 2016; 17: 768–778. [DOI] [PubMed] [Google Scholar]
- 138. Boldt A, Bitar M, Sack U. Flow cytometric evaluation of primary immunodeficiencies. Clin Lab Med. 2017; 37: 895–913. [DOI] [PubMed] [Google Scholar]
- 139. Seleman M, Hoyos-Bachiloglu R, Geha RS, Chou J. Uses of next-generation sequencing technologies for the diagnosis of primary immunodeficiencies. Front Immunol. 2017; 8: 847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Richardson AM, Moyer AM, Hasadsri L, Abraham RS. Diagnostic tools for inborn errors of human immunity (primary immunodeficiencies and immune dysregulatory diseases). Curr Allergy Asthma Rep. 2018; 18: 19. [DOI] [PubMed] [Google Scholar]
- 141. Lehman H, Hernandez-Trujillo V, Ballow M. Diagnosing primary immunodeficiency: a practical approach for the non-immunologist. Curr Med Res Opin. 2015; 31: 697–706. [DOI] [PubMed] [Google Scholar]
- 142. Stray-Pedersen A, Sorte HS, Samarakoon P et al.. Primary immunodeficiency diseases: Genomic approaches delineate heterogeneous Mendelian disorders. J Allergy Clin Immunol. 2017; 139: 232–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rae W, Ward D, Mattocks C et al.. Clinical efficacy of a next-generation sequencing gene panel for primary immunodeficiency diagnostics. Clin Genet. 2018; 93: 647–655. [DOI] [PubMed] [Google Scholar]
- 144. Belkadi A, Bolze A, Itan Y et al.. Whole-genome sequencing is more powerful than whole-exome sequencing for detecting exome variants. Proc Natl Acad Sci U S A. 2015; 112: 5473–5478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Armstrong-James D, Brown GD, Netea MG et al.. Immunotherapeutic approaches to treatment of fungal diseases. Lancet Infect Dis. 2017; 17: e393–e402. [DOI] [PubMed] [Google Scholar]
- 146. Jarvis JN, Meintjes G, Rebe K et al.. Adjunctive interferon-gamma immunotherapy for the treatment of HIV-associated cryptococcal meningitis: a randomized controlled trial. AIDS. 2012; 26: 1105–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Pappas PG, Bustamante B, Ticona E et al.. Recombinant interferon- gamma 1b as adjunctive therapy for AIDS-related acute cryptococcal meningitis. J Infect Dis. 2004; 189: 2185–2191. [DOI] [PubMed] [Google Scholar]
- 148. Delsing CE, Gresnigt MS, Leentjens J et al.. Interferon-gamma as adjunctive immunotherapy for invasive fungal infections: a case series. BMC Infect Dis. 2014; 14: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Netea MG, Brouwer AE, Hoogendoorn EH et al.. Two patients with cryptococcal meningitis and idiopathic CD4 lymphopenia: defective cytokine production and reversal by recombinant interferon- gamma therapy. Clin Infect Dis. 2004; 39: e83–87. [DOI] [PubMed] [Google Scholar]
- 150. Safdar A, Rodriguez G, Ohmagari N et al.. The safety of interferon-gamma-1b therapy for invasive fungal infections after hematopoietic stem cell transplantation. Cancer. 2005; 103: 731–739. [DOI] [PubMed] [Google Scholar]
- 151. Buddingh EP, Leentjens J, van der Lugt J et al.. Interferon-gamma immunotherapy in a patient with refractory disseminated candidiasis. Pediatr Infect Dis J. 2015; 34: 1391–1394. [DOI] [PubMed] [Google Scholar]
- 152. Assendorp EL, Gresnigt MS, Sprenkeler EGG et al.. Adjunctive interferon-gamma immunotherapy in a pediatric case of Aspergillus terreus infection. Eur J Clin Microbiol Infect Dis. 2018; 37: 1915–1922. [DOI] [PubMed] [Google Scholar]
- 153. de Sousa Mda G, Belda W Jr., Spina R et al.. Topical application of imiquimod as a treatment for chromoblastomycosis. Clin Infect Dis. 2014; 58: 1734–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Sousa Mda G, Reid DM, Schweighoffer E et al.. Restoration of pattern recognition receptor costimulation to treat chromoblastomycosis, a chronic fungal infection of the skin. Cell Host Microbe. 2011; 9: 436–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Grimaldi D, Pradier O, Hotchkiss RS, Vincent JL. Nivolumab plus interferon-gamma in the treatment of intractable mucormycosis. Lancet Infect Dis. 2017; 17: 18. [DOI] [PubMed] [Google Scholar]