Abstract
Background
Production and health traits are central in cattle breeding. Advances in next-generation sequencing technologies and genotype imputation have increased the resolution of gene mapping based on genome-wide association studies (GWAS). Thus, numerous candidate genes that affect milk yield, milk composition, and mastitis resistance in dairy cattle are reported in the literature. Effect-bearing variants often affect multiple traits. Because the detection of overlapping quantitative trait loci (QTL) regions from single-trait GWAS is too inaccurate and subjective, multi-trait analysis is a better approach to detect pleiotropic effects of variants in candidate genes. However, large sample sizes are required to achieve sufficient power. Multi-trait meta-analysis is one approach to deal with this problem. Thus, we performed two multi-trait meta-analyses, one for three milk production traits (milk yield, protein yield and fat yield), and one for milk yield and mastitis resistance.
Results
For highly correlated traits, the power to detect pleiotropy was increased by multi-trait meta-analysis compared with the subjective assessment of overlapping of single-trait QTL confidence intervals. Pleiotropic effects of lead single nucleotide polymorphisms (SNPs) that were detected from the multi-trait meta-analysis were confirmed by bivariate association analysis. The previously reported pleiotropic effects of variants within the DGAT1 and MGST1 genes on three milk production traits, and pleiotropic effects of variants in GHR on milk yield and fat yield were confirmed. Furthermore, our results suggested that variants in KCTD16, KCNK18 and ENSBTAG00000023629 had pleiotropic effects on milk production traits. For milk yield and mastitis resistance, we identified possible pleiotropic effects of variants in two genes, GC and DGAT1.
Conclusions
Multi-trait meta-analysis improves our ability to detect pleiotropic interactions between milk production traits and identifies variants with pleiotropic effects on milk production traits and mastitis resistance. In particular, this should contribute to better understand the biological mechanisms that underlie the unfavorable genetic correlation between milk yield and mastitis.
Background
Holstein is an important cattle breed in the Danish dairy production and much effort has gone in the genetic improvement of its milk production and functional traits. Intense selection for increased milk yield has negative consequences on the udder health of cows [1]. Unfavorable genetic correlations between milk production and clinical mastitis (from 0.21 to 0.55) have been reported [2]. A genetic correlation between two traits could be due to the pleiotropic action of genetic variants or the correlation (i.e., linkage disequilibrium (LD)) between causal variants. The identification of a quantitative trait locus (QTL) that affects simultaneously milk yield and udder health can help reveal some of the genetic basis of the genetic connection between milk production and mastitis resistance. In combination with specific genetic tests, this information can contribute to reduce the unfavorable correlated response on mastitis due to selection that focused on improving milk production traits by differentially weighting variants based on their favorable or unfavorable effects on the two traits.
One application of genome-wide association studies (GWAS) is to detect pleiotropic effects for the QTL identified from single-trait analysis. If a genomic region is significant for two or more traits, it may be due to causal variants that are in LD and affect individual traits (linkage), or that these traits are affected by the same variant (pleiotropy). The number of segregating variants in a population is large, but finite. The proportion of the segregating variants that are associated with the genetic variation of complex traits is unknown. However, traits often appear to be associated with the same or closely-linked variants in the genome [3, 4], which strongly suggests that, at least some of the underlying causal variants, affect several traits. Therefore, the primary aim of this analysis was to determine whether QTL associated with more than one trait were indeed pleiotropic. We used our previous GWAS results (summary statistics) of milk production traits [3] and mastitis resistance [4] to perform a multi-trait meta-analysis for scanning lead SNPs associated with three milk production traits or with milk yield and mastitis resistance for pleiotropy. In combination with a bivariate analysis, we examined the possible pleiotropic nature of the QTL identified.
Methods
Animals and phenotypes
We used de-regressed proofs (DRP) based on estimated breeding values (EBV) [5, 6] for milk, fat, protein yields and mastitis resistance (udder health index, which is an index for clinical mastitis from first to third lactation) for about 5000 Nordic Holstein (HOL) bulls. Nordic Cattle Genetic Evaluation (https://www.nordicebv.info/) provided the EBV.
Genotype and sequence data
We performed an association analysis using imputed whole-genome sequence (WGS) data. All bulls (~ 5000) were genotyped with the BovineSNP50 BeadChip SNP array (54 k) versions 1 or 2 (Illumina Inc., San Diego, CA). Imputation to WGS variants was described earlier by Iso-Touru et al. [7]. Briefly, 54k genotypes were imputed to WGS variants by a 2-step approach. First, using a multi-breed reference of 3383 animals (1222 Holstein (HOL), 1326 Nordic Red cattle (RDC) and 835 Jersey (JER)) that had been genotyped with the Illumina Bovine HD SNP array (Illumina Inc., San Diego, CA), all the animals were imputed to the high-density (HD) level. Next, the imputed HD genotypes were imputed to the WGS level using a multi-breed reference of 1228 animals: 1148 WGS from Run4 of the 1000 Bull Genomes Project (288 Holstein, 56 Red, and 61 Jersey, as well as 743 individuals from various breeds) [8] and 80 animals from Aarhus University (23 HOL, 30 RDC, and 27 JER). Imputation to HD genotypes was done with the IMPUTE2 v2.3.1 software [9], and imputation to the whole-genome level with the Minimac2 software [10]. The average accuracy (r2-values from Minimac2) was 0.85 for across-breed imputation. Imputation accuracy was previously reported by Wu et al. [11].
Before phasing and imputation, we filtered the 54 k and HD genotypes based on an SNP call rate higher than 85% and an animal call rate higher than 90%. The imputed sequence data included 22,751,039 bi-allelic variants. For each breed, SNPs with a minor allele frequency (MAF) lower than 1% or with a highly significant deviation from Hardy–Weinberg proportions (p < 1.0−6) were excluded. After quality filtering, 16,503,508 SNPs remained for analysis.
Single-SNP association analysis for a trait
The GWAS summary statistics were from two previous association analyses [3, 4] and, here, we provide a brief description of the GWAS method used. The genetic relationship matrix (GRM) was estimated using imputed HD genotypes by the GCTA software [12]. We followed the leave-one-chromosome-out approach [13] to build a kinship matrix that was specific to the analysis of each chromosome by leaving out markers on that chromosome to avoid loss of power due to double-counting (fitting the SNP as a fixed effect for testing associations and as a random effect as part of the GRM) [14].
First, we performed a single-SNP GWAS analysis using GCTA [12] for each chromosome. A Bonferroni multiple-testing correction was applied to control for false-positive associations: a SNP was significant if its test probability p-value, , was less than 0.05/ , where is the number of SNPs. This corresponds to a trait-wise nominal type 1 error-rate of 5%. Here, the significance threshold value was − log10() with million SNPs. We identified the lead SNPs for each independent QTL signal on a chromosome by iteratively fitting the allele dosages of the lead SNPs identified in the previous runs as covariate (for details see [3, 4]).
Genetic variance explained by the identified QTL
We used GCTA [12] to estimate the genetic variance explained by all the identified QTL together for each trait. First, we extracted the genotype for all lead SNPs identified from the GWAS and generated the first GRM. Next, we excluded all SNPs within the 2.5-Mb region around each lead SNP and estimated the second GRM with the remaining SNPs. Finally, we estimated the genetic variance explained by each of these two groups of variants for each trait by fitting two GRM in a linear mixed model.
Defining a QTL region
QTL regions were defined as continuous regions that include a lead SNP with a − log10(p) > 8.50. The start and the end of the QTL region were determined based on the following considerations: (1) the value of 3 was subtracted from the -log10(p) value of the lead SNP; (2) from the remaining SNPs, we identified those that were located furthest to the left and right with a− log10(p) value no less than 3 units below the − log10(p) of the lead SNP of the region; the positions of these SNPs were taken as boundaries of the QTL region, but if they were further than 0.25 Mb (left or right from the lead SNP), then the size of the QTL region was limited by 0.25 Mb.
Estimation of genetic correlations
We used a linkage disequilibrium score regression approach as implemented in the LDSC software [15] to estimate the genetic correlation between traits using GWAS summary statistics. For polygenic traits, the more a SNP is in LD with other genetic variants, the greater is its chance of being correlated with causal variants, and the higher is its expected association test statistic. Exploiting this relationship allows the estimation of SNP-based heritability when using association test statistics for a single trait or the estimation of SNP-based co-heritability when combining association test statistics from two traits. The LD score of a SNP is the sum of the LD (r2) of the SNP with other SNPs and, thus, can be regarded as a measure of the genetic variation that is ‘tagged’ by the SNP. First, we calculated the LD scores for each variant using WGS data of Holstein animals from Run6 in the 1000 Bull Genome Project [8] and of additional Holstein individuals from Aarhus University. Then, GWAS summary statistics from our previous studies [3, 4] were converted to the input format of the LDSC software using the accompanying script munge_sumstats.py (part of LDSC software). The reformatted summary statistics were used to calculate genetic correlations between traits.
Multi-trait meta-analysis
A multi-trait meta-analysis was performed using the approximate multi-trait test statistic described by Bolormaa et al. [16]. Effects of a SNP across all traits were calculated and combined with the genomic correlation matrix between traits to perform a multi-trait test with a number of degrees of freedom equal to the number of traits. The formula to compute the multi-trait statistic for SNP was , where is a vector of signed test statistics for the association of lead SNP with each trait obtained by single trait GWAS, and is the inverse of the genomic correlation matrix for all traits. The same Bonferroni-corrected significance threshold as in the single trait association analyses (i.e., − log10() > 8.5) was applied in the multi-trait analyses.
Single-SNP bivariate association analysis
A single-SNP bivariate association analysis was carried out for each lead SNP from the multi-trait meta-analysis. The bivariate model used for a SNP is as follows
where subscripts and indicate traits 1 and 2 in the analysis, are the vectors of phenotypes for trait , is the general mean for trait , is a vector of genotype doses for the lead SNP, is the allele substitution effect of the lead SNP for trait , is a design matrix relating phenotypic observations to polygenic effects for trait , is a vector of the random polygenic effects with a multivariate normal distribution , where is the additive relationship matrix and is the polygenic covariance matrix, and is a vector of mutually independent residual terms with a multivariate normal distribution , where is the residual variance for trait , and is an identity matrix of appropriate dimensions. The model was fit by AI-REML using DMU [17].
Pleiotropy vs. variants in linkage disequilibrium
A bivariate model might help to distinguish between a variant that affects both traits (via different paths), and a variant that has an effect on one trait that is mediated through another trait. In a bivariate model, the effects of SNPs are expected to be significant for both traits in the first scenario, but only for one of the traits in the second scenario. To distinguish between pleiotropic effects and effects of distinct variants in LD, we conducted bivariate analyses (as described above) for the lead SNPs that were detected in the multi-trait meta-analysis. The lead SNPs that showed genome-wide significance for at least one of the traits in the bivariate analyses and a significance p < 1.18e−4 (=0.05/ , where is equal to number of traits (i.e. 4) times the number of unique lead SNPs (i.e. 106) identified across all traits) for the other trait were considered to have a pleiotropic effect on both traits.
Candidate genes underlying the associated genomic regions
Annotations for the lead SNPs for each QTL region from the single-trait analyses and the meta-analysis along with the genes that harbor the lead SNP were determined via the cow (Bos taurus) genome assembly UMD3.1 [18]. We used the variant effect predictor (VEP) software (ver. 92.0) [19] to predict the functional consequences of the lead SNPs and identify the closest gene.
Results
Single-variant association analysis and genetic correlation
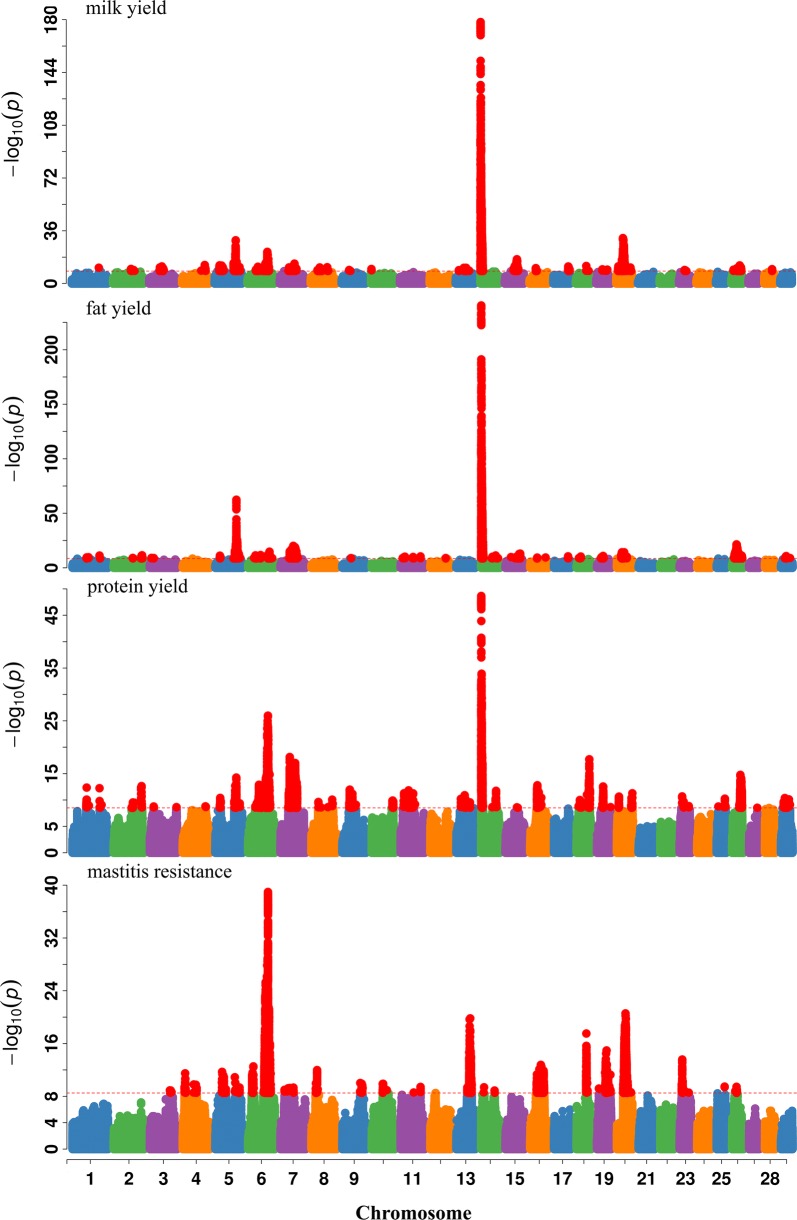
Previously, we published the results of a GWAS for milk production traits and mastitis resistance [3, 4], which are summarized in Fig. 1 and Table 1. We identified 27 independent association signals on 18 chromosomes for fat yield (FY), 34 association signals on 22 chromosomes for protein yield (PY), 26 association signals on 20 chromosomes for milk yield (MY), and 22 association signals on 18 chromosomes for mastitis resistance (MR). Several QTL detected for different traits were located in close proximity. Table 2 lists the genetic correlations between MY, FY, PY and MR as estimated by LDSC. Moderate to strong genetic correlations between MY, FY and PY were observed but unfavorable genetic correlations between each of the three milk production traits and MR were found, as reported previously [2, 20].
Fig. 1.
Manhattan plots for single-trait GWAS for fat yield (FY), protein yield (PY), milk yield (MY) and mastitis resistance (MR). Each color corresponds to an autosome. The horizontal red dotted line shows the genome-wide Bonferroni corrected significance threshold [− log10(p) = 8.5]. Base positions refer to the UMD 3.1.1 [18] bovine genome assembly
Table 1.
Summary of the GWAS results for milk production traits and mastitis resistance
| Traits | Number of QTL | V (lead SNP)/V (DRP)a (%) |
|---|---|---|
| Fat yield | 27 | 28.57 |
| Protein yield | 34 | 16.76 |
| Milk yield | 26 | 21.50 |
| Mastitis resistance | 22 | 14.54 |
aPercentage of genetic variance explained by the identified QTL, V(lead SNP) is the variance explained by the lead SNPs, whereas V(DRP) is the variance of de-regressed breeding values (DRP)
Table 2.
Genetic correlations between milk yield (MY), fat yield (FY), protein yield (PY) and mastitis resistance (MR) estimated from GWAS summary statistics
| Traits | MY | FY | PY | MR |
|---|---|---|---|---|
| MY | 0.40 (0.14) | 0.78 (0.03) | − 0.35 (0.04) | |
| FY | 0.56 (0.17) | − 0.20 (0.05) | ||
| PY | − 0.27 (0.04) |
Numbers in parentheses are standard errors of genetic correlation estimates
Three-trait meta-analysis for fat, protein, and milk yields
We examined the overlap between QTL regions for FY, PY, and MY (Table 3). Some of the overlapping QTL regions did not contain any genes, such as the two regions 20,035,379–20,534,779 bp and 93,703,737–93,762,020 bp on Bos taurus autosome (BTA)5, and 2044,412–2049,435 bp on BTA14 (Table 3). In contrast, the QTL intervals on BTA14 and 19 included several overlapping regions that included many genes.
Table 3.
Overlapping QTL intervals identified based on single-trait GWAS for milk yield (MY), fat yield (FY) and protein yield (PY)
| Chr. | Region | Traits | Genes |
|---|---|---|---|
| 2 | 85393563–86241732 | FY, MY | ENSBTAG00000047452, GTF3C3, CCDC150, PGAP1, ANKRD44 |
| 5 | 20035379–20534779 | FY, PY | – |
| 5 | 93703737–93762020 | FY, PY, MY | – |
| 5 | 93762020–94198670 | FY,PY | MGST1, SLC15A5 |
| 5 | 93698481–93703737 | FY, MY | LMO3 |
| 14 | 1545264–1583427 | FY, PY, MY | bta-mir-2308, ARHGAP39 |
| 14 | 1448510–1545264 | FY, MY | ZNF16, C14H8orf33, ZNF34, ZNF7, COMMD5, RPL8 |
| 14 | 1549272–2044412 | FY, PY, MY | bta-mir-2308, C14H8orf82, LRRC14, RECQL4, GPT, PPP1R16A, KIFC2, VPS28, SLC52A2, TMEM249, SCX, ENSBTAG00000044406, ENSBTAG00000039978, HGH1, MAF1, SHARPIN, CYC1, GPAA1, EXOSC4, ENSBTAG00000015040, GRINA, PARP10, LRRC24, MFSD3, FOXH1, CYHR1, TONSL, SLC39A4, CPSF1, ADCK5, FBXL6, SCRT1, DGAT1, HSF1, MROH1, WDR97, SPATC1, ARHGAP39, BOP1, OPLAH |
| 14 | 2044412–2049435 | FY, MY | – |
| 14 | 67731848–68231920 | FY, MY | KCNS2, STK3, ENSBTAG00000046739 |
| 16 | 31268349–31746789 | FY, MY | ENSBTAG00000044816, H3F3C, ENSBTAG00000021109, ENSBTAG00000042363, SCCPDH, TFB2M, SMYD3, CNST |
| 19 | 27156952–27692965 | FY, PY, MY | PSMB6, GLTPD2, VMO1, CXCL16, MED11, ARRB2, bta-mir-2338, ALOX15, ALOX12E, ENSBTAG00000047925, RNASEK, C19H17orf49, bta-mir-497, bta-mir-195, BCL6B, SLC16A13, CLEC10A, ASGR1, ENSBTAG00000042630, ACADVL, bta-mir-324, PHF23, GABARAP, ELP5, CLDN7, SLC2A4, EIF5A, GPS2, NEURL4, ENSBTAG00000045892, ACAP1, KCTD11, PLD2, TM4SF5, ZMYND15, PELP1, ALOX12, SLC16A11, ASGR2, DLG4, DVL2, CTDNEP1, YBX2, MINK1 |
| 19 | 26625240–27156952 | FY, PY | ENSBTAG00000025126, MIS12, C1QBP, RPAIN, bta-mir-199c, ENSBTAG00000013906, SPAG7, ENO3, ENSBTAG00000004913, SLC25A11, GP1BA, CHRNE, C19H17orf107, NLRP1, DERL2, DHX33, SCIMP, ZFP3, KIF1C, INCA1, CAMTA2, PFN1, NUP88, RABEP1, MINK1 |
| 19 | 27692965–27773922 | FY, MY | PLSCR3, TMEM256, NLGN2, SPEM1, TMEM102, CHRNB1, TNK1, FGF11, ZBTB4 |
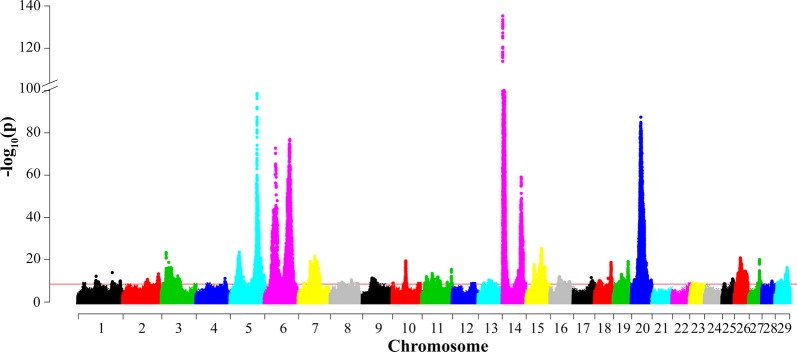
We performed a multi-trait meta-analysis for FY, PY and MY to examine if the lead SNP affected multiple milk production traits (Fig. 2). In total, we identified 59 association signals across 27 chromosomes (Table 4). One peak on BTA5, two peaks on BTA6, two peaks on BTA14 and one peak on BTA20 showed strong association signals in the meta-analysis. The strongest signal was located on BTA14 and resulted from the well-known and previously described SNPs BTA14:1802,265 (rs109234250) and BTA14:1802,266 (rs109326954) in the DGAT1 gene [21, 22]. These two SNPs were also the lead SNPs in the single-trait analyses for FY and MY with a –log10(p) value greater than 240 and 178, respectively. These two SNPs were in complete LD and had identical p values for both traits. The single-trait analysis for PY did not identify these two causal variants as the ‘lead’ SNP. Instead, the strongest associated SNP in this region for PY was SNP BTA14:1835,440 (rs208567981) with –log10(p) = 48.66. This variant was located within the BOP1 gene, but very close to DGAT1 [3], whereas the two causal variants (BTA14:1802,265 and BTA14:1802,266) had –log10(p) = 47.99 in the analysis for PY [3].
Fig. 2.
Manhattan plot of the multi-trait meta-analysis for milk, fat and protein yields. The red horizontal line indicates the genome-wide significance level [− log10(p) = 8.5]. Base positions refer to the UMD 3.1.1 [18] bovine genome assembly
Table 4.
The lead SNP and its nearest genes in the multi-trait meta-analysis of milk yield (MY), fat yield (FY) and protein yield (PY)
| Chr | Lead SNP (bp) | rsid | − log10(p) | Nearest gene (distance) | Annotation | Single trait significanta |
|---|---|---|---|---|---|---|
| 1 | 63177947 | rs42409534 | 12.19 | ENSBTAG00000046854 (7850,66 bp) | Intergenic | FY, PY |
| 1 | 71250238 | rs210007164 | 9.51 | TFRC (9830 bp) | Intergenic | FY |
| 1 | 120470021 | rs109519395 | 13.92 | AGTR1 (67,012 bp) | Intergenic | FY, MY, PY |
| 1 | 148567669 | rs108957710 | 9.97 | ENSBTAG00000046447 | Upstream | Novel |
| 2 | 86095020 | rs110457337 | 10.76 | ANKRD44 | Intron | FY, PY |
| 2 | 124837669 | – | 13.34 | PTPRU | Intron | FY, PY |
| 3 | 15459025 | rs132797166 | 23.45 | GBA | Intron | Novel |
| 4 | 101547644 | – | 11.18 | CHRM2 | Upstream | MY |
| 5 | 31335325 | rs447206924 | 23.61 | LALBA (12,536 bp) | Intergenic | MY |
| 5 | 93945991 | rs208248675 | 126.98 | MGST1 | Intron | FY, MY |
| 6 | 38031954 | rs454966322 | 72.75 | ABCG2 (1371 bp) | Downstream | FY, MY |
| 6 | 88442678 | rs211069111 | 76.92 | SLC4A4 | Intron | PY |
| 7 | 57287990 | – | 21.70 | KCTD16 | Intron | FY, MY, PY |
| 8 | 40627140 | rs381636155 | 9.38 | bta-mir-2471 (64,540 bp) | Intergenic | MY |
| 8 | 73877814 | rs381189656 | 10.50 | ENSBTAG00000010829 (171 bp) | Upstream | MY, PY |
| 8 | 93065787 | rs211543235 | 8.88 | GRIN3A | Intron | PY |
| 9 | 33179984 | rs211241852 | 11.33 | SLC35F1 (83,834 bp) | intergenic | MY, PY |
| 10 | 1989907 | rs109274615 | 9.01 | ENSBTAG00000047622 (109,990 bp) | Intergenic | MY |
| 10 | 46591115 | rs211044009 | 19.41 | USP3 (20,126 bp) | Intergenic | Novel |
| 10 | 93933304 | – | 8.73 | SEL1L | Intron | PY |
| 11 | 15339847 | rs110446044 | 12.01 | TTC27 | Intron | FY, PY |
| 11 | 35270918 | rs109956079 | 13.52 | ENSBTAG00000027786 (241,298 bp) | Intergenic | PY |
| 11 | 55692712 | rs208208268 | 11.86 | CTNNA2 | Intron | FY, PY |
| 11 | 63203084 | rs110286816 | 9.96 | ENSBTAG00000046117 (87,338 bp) | intergenic | Novel |
| 11 | 88771449 | rs109730673 | 9.41 | ENSBTAG00000047976 (150,709 bp) | Intergenic | FY |
| 11 | 103301805 | rs110788821 | 15.43 | PAEP | Intron | Novel |
| 12 | 75309869 | rs43128997 | 8.78 | ENSBTAG00000026070 (148,028 bp) | Intergenic | Novel |
| 13 | 20194953 | rs41602070 | 9.06 | ITGB1 (53,992 bp) | Intergenic | PY |
| 13 | 37208793 | rs385962673 | 10.43 | MKX (10,635 bp) | Intergenic | MY, PY |
| 13 | 46391099 | rs42442665 | 10.03 | ADARB2 | Intron | MY |
| 13 | 60701113 | rs108986421 | 8.61 | RSPO4 (9143 bp) | Intergenic | PY |
| 14 | 1802265 | rs109234250 | 705.26 | DGAT1 | Missense | FY, MY, PY |
| 14 | 1802266 | rs109326954 | 705.26 | DGAT1 | Missense | FY, MY, PY |
| 14 | 66649826 | rs445616049 | 59.08 | MGC148714 (2105 bp) | Downstream | Novel |
| 15 | 28357864 | rs208325660 | 17.79 | RNF214 | Missense | Novel |
| 15 | 53640417 | rs380720492 | 25.33 | ARHGEF17 | Synonymous | MY |
| 15 | 66103726 | rs41775109 | 16.64 | EHF (72,762 bp) | Intergenic | FY |
| 16 | 32262983 | rs384531912 | 12.03 | SMYD3 | Intron | PY |
| 16 | 49798794 | rs380930173 | 9.40 | AJAP1 (33,340 bp) | intergenic | PY |
| 16 | 67736535 | rs385935762 | 9.64 | SWT1 | intron | Novel |
| 17 | 62543160 | rs211016475 | 11.60 | TBX5 | Intron | FY |
| 18 | 15058306 | rs211386971 | 10.04 | VPS35 | Intron | PY |
| 18 | 46583596 | rs110398208 | 11.19 | UPK1A (662 bp) | Upstream | MY, PY |
| 18 | 57064406 | rs433130247 | 18.74 | MYBPC2 | Intron | PY |
| 19 | 27522927 | rs210334173 | 13.09 | ASGR1 (6170 bp) | Intergenic | FY, PY |
| 19 | 51383847 | rs136067046 | 19.23 | FASN (1075 bp) | Upstream | Novel |
| 20 | 9608408 | rs519502268 | 10.57 | CARTPT (183,253 bp) | Intergenic | MY, PY |
| 20 | 31909478 | rs385640152 | 112.79 | GHR | Missense | FY, MY |
| 20 | 69006609 | rs134612291 | 10.41 | IRX1 (466,030 bp) | Intergenic | PY |
| 22 | 60087443 | rs462776871 | 8.51 | EEFSEC | Intron | Novel |
| 23 | 10974968 | rs136158431 | 9.29 | FGD2 (56,619 bp) | Intergenic | PY |
| 23 | 25076739 | rs210864958 | 8.70 | GCM | Downstream | MY |
| 25 | 6984796 | rs110991040 | 8.71 | RBFOX1 (298,008 bp) | Intergenic | Novel |
| 25 | 36299420 | rs210351939 | 10.92 | ACHE (1112 bp) | Upstream | Novel |
| 26 | 20527926 | rs453802222 | 20.87 | ENSBTAG00000023629 (175 bp) | Downstream | FY |
| 26 | 37869471 | rs458256022 | 14.61 | KCNK18 | Synonymous | FY, MY, PY |
| 27 | 36212352 | rs208675276 | 20.10 | GPAT4 | 5_prime_UTR | Novel |
| 28 | 34972377 | rs137526033 | 9.90 | ZMIZ1 (60,078 bp) | Intergenic | MY |
| 29 | 21075705 | rs382642281 | 10.42 | ENSBTAG00000000853 (161,330 bp) | Intergenic | FY, PY |
| 29 | 41825511 | – | 16.32 | STX5 | Intron | Novel |
The distance in base pairs from the nearest gene is in brackets
aNovel hit from multi-trait meta-analysis, not identified by any of the single trait analysis
The multi-trait meta-analysis can help to deal with accuracy of the single-trait analysis. The causal variant known in GHR (F279Y) [23] was the lead SNP on BTA20 from the meta-analysis (Table 4). However, in the single-trait analysis for FY and MY, the causative variant did not emerge as the lead SNP [3]. In addition, on BTA5, we detected the second lead SNP at BTA5:31,335,325 (rs447206924, Table 4). The nearest gene to this lead SNP is LALBA, which encodes α-lactalbumin. The multi-trait meta-analysis helped to pinpoint this known causal gene whereas both the single-trait analysis for MY [3] and the overlapping QTL regions between milk production traits (Table 3) failed to do so.
The lead SNPs detected in the meta-analysis were either lead SNPs from the single-trait analyses (18 lead SNPs) or those the most closely located to the lead SNPs identified by the single-trait analyses (Table 4). Moreover, the meta-analysis identified 16 additional association signals that were not genome-wide significant in the single-trait analyses (Table 4). We searched the mammalian phenotype database [24] to verify the candidate genes that were suggested by the multi-trait meta-analysis. In addition to DGAT1, MGST1, ABCG2 and GHR, we identified one more gene with biological support, GPAT4. The term in the mammalian phenotype database showed that certain alleles of the GPAT4 gene cause “abnormal milk composition” in mouse [25].
Two-trait meta-analysis for milk yield and mastitis resistance
Two overlapping QTL regions for MY and MR were detected in this study on BTA5 and 6 (Table 5). The QTL region on BTA5 harbors several genes and that on BTA6 (88.6 to 89.1 Mb) harbors the GC and NPFFR2 genes, which have been reported to be associated with clinical mastitis in cows [26].
Table 5.
Genes located within the overlapping QTL regions detected in the single-trait GWAS between milk yield and mastitis resistance
| Chr | Region | Genes |
|---|---|---|
| 5 | 30202453–31258920 | bta-mir-2425, DNAJC22, TROAP, PRPH, TUBA1C, TUBA1A, DHH, RHEBL1, bta-mir-2426, PRKAG1, DDN, WNT1, WNT10B, FKBP11, CCDC65, CACNB3, ENSBTAG00000047525, BCDIN3D, NCKAP5L, TMBIM6, FMNL3, PRPF40B, MCRS1, KCNH3, C1QL4, ENSBTAG00000037775, TUBA1B, LMBR1L, RND1, DDX23, ADCY6, CCNT1, SPATS2, KMT2D, FAM186B |
| 6 | 88598011–89097608 | NPFFR2, GC |
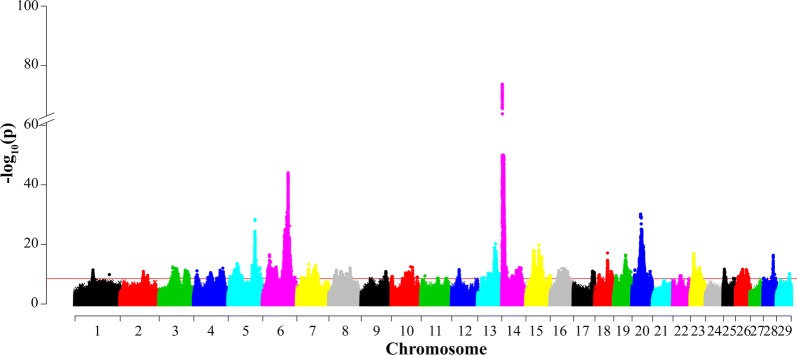
The most significant signal in the meta-analysis was located on BTA14:1793,616 (Table 6 and Fig. 3) and 1735 bp upstream of DGAT1. We believe that this signal was caused by the two known causal mutations in DGAT1. However, this lead SNP was significant only in the single-trait analysis for MY, but not for MR (Table 6). The second strongest association signal was located on BTA6:88,729,872 in the GC gene. The third strongest association signal was on BTA5:93,953,487, close to MGST1 but this lead SNP was significant only in the single-trait analysis for MY, and not in that for MR (Table 6).
Table 6.
The lead SNP and its nearest genes in the multi-trait meta-analysis of milk yield (MY) and mastitis resistance (MR)
| Chr | Lead SNP (bp) | rsid | − log10(p) | Nearest gene (distance) | Annotation | Single-trait significanta |
|---|---|---|---|---|---|---|
| 1 | 62876378 | – | 11.43 | ENSBTAG00000046854 (483,497 bp) | Intergenic | Novel |
| 2 | 81179721 | rs110121625 | 10.92 | TMEFF2 | Intron | MY |
| 2 | 96376739 | – | 8.92 | RF00001 (13,015 bp) | Intergenic | Novel |
| 3 | 47643148 | rs378327566 | 12.42 | PTBP2 (732,492 bp) | Intergenic | MY |
| 3 | 91848036 | – | 11.21 | USP24 | Intron | Novel |
| 4 | 10938389 | rs211526380 | 11.12 | TFPI2 (97,090 bp) | Intergenic | MR |
| 4 | 59012300 | rs136891032 | 10.53 | ENSBTAG00000020620 (287,400 bp) | Intergenic | Novel |
| 4 | 101547644 | – | 12.03 | CHRM2 (4968 bp) | Upstream | MY |
| 5 | 31352419 | – | 13.58 | LALBA (2537 bp) | Downstream | MR, MY |
| 5 | 93953487 | rs210234664 | 28.36 | MGST1 (3325 bp) | Upstream | MY |
| 6 | 23474516 | rs109255104 | 16.50 | MANBA | Intron | MR |
| 6 | 88729872 | rs109803407 | 44.04 | GC | Intron | MR, MY |
| 7 | 18085661 | rs133896398 | 8.98 | FBN3 (9071 bp) | Intergenic | Novel |
| 7 | 41607423 | rs208385619 | 13.33 | ENSBTAG00000039706 (16,819 bp) | Intergenic | MR, MY |
| 7 | 65370850 | rs109644389 | 12.99 | GLRA1 (258,214 bp) | Intergenic | MY |
| 8 | 25684799 | – | 11.38 | ADAMTSL1 | Intron | MR |
| 8 | 41148951 | – | 10.90 | ENSBTAG00000014467 (197,503 bp) | Intergenic | MY |
| 8 | 61253437 | rs43552270 | 10.23 | MELK (22,888 bp) | Intergenic | Novel |
| 8 | 73877814 | rs381189656 | 12.11 | ENSBTAG00000010829 (171 bp) | Upstream | MY |
| 9 | 86108587 | rs209751747 | 10.90 | SAMD5 (87,968 bp) | Intergenic | MR |
| 10 | 2752616 | rs209970861 | 9.26 | YTHDC2 (318,445 bp) | Intergenic | Novel |
| 10 | 39050124 | – | 8.56 | ENSBTAG00000004692 (137,772 bp) | Intergenic | Novel |
| 10 | 49633928 | rs43587750 | 10.44 | RORA | Intron | Novel |
| 10 | 67479321 | rs109694327 | 12.49 | SAMD4A | Intron | MR |
| 10 | 74236917 | rs380306966 | 12.27 | SNAPC1 (58,791 bp) | Intergenic | Novel |
| 10 | 90272296 | rs381454149 | 9.06 | ADCK1 (137,647 bp) | Intergenic | Novel |
| 11 | 13025259 | – | 9.43 | DYSF | Intron | Novel |
| 11 | 58017848 | rs380589113 | 8.64 | LRRTM4 (415,739 bp) | Intergenic | Novel |
| 11 | 88743727 | rs380133715 | 8.53 | ID2 (156,561 bp) | Intergenic | MR |
| 12 | 25763482 | rs210185748 | 11.55 | NBEA (29,504 bp) | Intergenic | Novel |
| 13 | 21477118 | rs135125951 | 8.87 | RF00026 (80,513 bp) | Intergenic | Novel |
| 13 | 36822330 | rs379020207 | 10.39 | MPP7 | Intron | MY |
| 13 | 46391099 | rs42442665 | 10.13 | ADARB2 | intron | MY |
| 13 | 62017506 | rs211080099 | 20.20 | PDRG1 (2545 bp) | Upstream | MR |
| 13 | 76532010 | rs42057265 | 10.31 | ZMYND8 | Intron | Novel |
| 14 | 1793616 | rs384957047 | 183.77 | DGAT1 (1735 bp) | Upstream | MY |
| 14 | 36478894 | rs43757971 | 9.46 | XKR9 (1379 bp) | Upstream | Novel |
| 14 | 61344981 | rs42484846 | 12.22 | ZFPM2 | Intron | MR |
| 15 | 27475189 | rs208648732 | 18.08 | RF00285 (140,736 bp) | Intergenic | Novel |
| 15 | 45384070 | rs43100874 | 19.84 | ENSBTAG00000048176 (4269 bp) | Upstream | MY |
| 15 | 84666672 | rs382250433 | 9.54 | MS4A13 (102 bp) | Upstream | Novel |
| 16 | 30309951 | rs384258494 | 11.26 | STUM | Intron | MR |
| 16 | 47836093 | rs207941573 | 11.92 | ACOT7 | Intron | MR |
| 16 | 59908679 | rs451830006 | 10.94 | BRINP2 (45,408 bp) | Intergenic | MR |
| 17 | 66530413 | rs209525123 | 11.00 | CORO1C | Intron | MY |
| 18 | 13625299 | rs209154036 | 9.53 | ZNF469 (115,323 bp) | Intergenic | Novel |
| 18 | 43909571 | rs464881101 | 17.12 | ENSBTAG00000004994 (17,651 bp) | Intergenic | MR |
| 18 | 57501622 | – | 10.88 | KLK14 (2325 bp) | upstream | Novel |
| 19 | 7941510 | rs209798151 | 9.51 | TRIM25 | Intron | Novel |
| 19 | 27442452 | rs483221509 | 9.52 | bta-mir-497 (689 bp) | Upstream | MY |
| 19 | 41169414 | rs134338592 | 16.40 | WIPF2 | Intron | MR |
| 20 | 10123208 | rs207633790 | 11.45 | GTF2H2 (4035 bp) | Downstream | MY |
| 20 | 29996719 | – | 30.18 | MRPS30 (75,496 bp) | Intergenic | MY |
| 20 | 63369153 | rs133899283 | 10.93 | FAM173B (241,983 bp) | Intergenic | MY |
| 22 | 25185357 | rs110721487 | 9.50 | CNTN6 | Intron | Novel |
| 23 | 11294868 | – | 16.92 | CMTR1 (8638 bp) | Intergenic | MR |
| 23 | 32139475 | rs477621057 | 12.23 | CARMIL1 | Intron | Novel |
| 25 | 3655364 | rs379765871 | 11.68 | CDIP1 | Intron | Novel |
| 25 | 35354412 | rs383829107 | 8.62 | CUX1 | Intron | MR |
| 26 | 24938054 | rs460832137 | 11.68 | SFR1 (7550 bp) | Intergenic | MY |
| 26 | 37716420 | rs381336935 | 11.55 | SHTN1 (26,088 bp) | Intergenic | MY |
| 28 | 1921500 | rs383708617 | 8.69 | RF00001 (26,734 bp) | Intergenic | Novel |
| 28 | 34972377 | rs137526033 | 16.36 | ZMIZ1 (60,078 bp) | Intergenic | MY |
| 29 | 45895253 | rs209161829 | 10.19 | POLD4 (4347 bp) | Downstream | Novel |
The distance in base pairs from the nearest gene is in brackets
aNovel hit from multi-trait meta-analysis, not identified by single trait analysis
Fig. 3.
Manhattan plot for the multi-trait analysis of milk yield and mastitis resistance of Nordic Holstein cattle. The red horizontal line indicates the genome-wide significance level [− log10(p) = 8.5]. Base positions refer to the UMD 3.1.1 [18] bovine genome assembly
Seventeen of the 64 lead SNPs from the meta-analysis were also lead SNPs for either MY or MR. Most of the remaining lead SNPs were close to the lead SNP in the single-trait analysis [3, 4]. In addition to DGAT1 and LALBA, we found one more candidate gene, ZFPM2, with biological support in the mammalian phenotype database [24]. LALBA encodes one of the major milk protein, α-lactalbumin. Both LALBA and ZFPM2 are related to the term “abnormal mammary gland morphology”.
Pleiotropy vs. closely linked variants
To examine if there was evidence for pleiotropic effects of the associated variants, we conducted bivariate analyses for the lead SNPs detected in the multi-trait meta-analysis. The lead SNPs that were genome-wide significant for at least one of the traits from the bivariate analyses are in Table 7. We concluded that a SNP might have pleiotropic effects if it also showed significance (p < 1.18e−4) for the second trait.
Table 7.
Results of the bivariate analyses with genome-wide significance for at least one trait
| Chr | BP | rsid | Trait1 | Trait2 | P1 | P2 | Indication of pleiotropya |
|---|---|---|---|---|---|---|---|
| 1 | 120470021 | rs109519395 | MY | FY | 4.36E−06 | 7.66E−11 | Yes |
| 5 | 93945991 | rs208248675 | MY | FY | 3.58E−21 | 1.33E−38 | Yes |
| 7 | 57287990 | rs380779883 | MY | FY | 4.47E−12 | 1.63E−17 | Yes |
| 14 | 1802265 | rs109234250 | MY | FY | 3.97E−124 | 6.43E−200 | Yes |
| 14 | 1802266 | rs109326954 | MY | FY | 3.88E−124 | 6.69E−200 | Yes |
| 15 | 66103726 | rs41775109 | MY | FY | 0.67E−01 | 1.85E−09 | No |
| 20 | 31909478 | rs385640152 | MY | FY | 2.65E−13 | 4.06E−08 | Yes |
| 26 | 20527926 | rs453802222 | MY | FY | 6.44E−08 | 8.67E−16 | Yes |
| 26 | 37869471 | rs458256022 | MY | FY | 2.33E−09 | 4.55E−08 | Yes |
| 1 | 63177947 | rs42409534 | MY | PY | 5.55E−07 | 4.94E−11 | Yes |
| 1 | 120470021 | rs109519395 | MY | PY | 2.57E−06 | 2.12E−10 | Yes |
| 5 | 93945991 | rs208248675 | MY | PY | 1.90E−21 | 1.82E−06 | Yes |
| 6 | 88442678 | rs211069111 | MY | PY | 2.09E−01 | 1.10E−13 | No |
| 7 | 57287990 | rs380779883 | MY | PY | 2.05E−12 | 9.62E−19 | Yes |
| 14 | 1802265 | rs109234250 | MY | PY | 2.31E−125 | 1.30E−28 | Yes |
| 14 | 1802266 | rs109326954 | MY | PY | 2.26E−125 | 1.28E−28 | Yes |
| 18 | 57064406 | rs433130247 | MY | PY | 5.44E−06 | 1.36E−11 | Yes |
| 20 | 31909478 | rs385640152 | MY | PY | 3.02E−13 | 1.30E−04 | No |
| 26 | 37869471 | rs458256022 | MY | PY | 2.63E−09 | 3.75E−11 | Yes |
| 1 | 63177947 | rs42409534 | FY | PY | 2.94E−08 | 3.06E−11 | Yes |
| 1 | 120470021 | rs109519395 | FY | PY | 7.12E−11 | 2.62E−10 | Yes |
| 5 | 93945991 | rs208248675 | FY | PY | 1.43E−38 | 1.08E−06 | Yes |
| 6 | 88442678 | rs211069111 | FY | PY | 4.24E−01 | 1.37E−13 | No |
| 7 | 57287990 | rs380779883 | FY | PY | 1.52E−17 | 2.56E−18 | Yes |
| 14 | 1802265 | rs109234250 | FY | PY | 4.65E−201 | 3.93E−29 | Yes |
| 14 | 1802266 | rs109326954 | FY | PY | 4.85E−201 | 3.88E−29 | Yes |
| 15 | 66103726 | rs41775109 | FY | PY | 1.59E−09 | 6.91E−01 | No |
| 18 | 57064406 | rs433130247 | FY | PY | 5.93E−01 | 1.10E−11 | No |
| 26 | 20527926 | rs453802222 | FY | PY | 1.49E−15 | 1.85E−06 | Yes |
| 26 | 37869471 | rs458256022 | FY | PY | 5.38E−08 | 3.29E−11 | Yes |
| 5 | 93953487 | rs210234664 | MY | MR | 1.07E−21 | 1.60E−03 | No |
| 6 | 88729872 | rs109803407 | MY | MR | 5.65E−14 | 5.04E−33 | Yes |
| 11 | 88743727 | rs380133715 | MY | MR | 1.33E−01 | 7.37E−10 | No |
| 13 | 62017506 | rs211080099 | MY | MR | 8.25E−01 | 4.38E−15 | No |
| 14 | 1793616 | rs384957047 | MY | MR | 1.61E−123 | 8.06E−06 | Yes |
| 14 | 1802667 | – | MY | MR | 3.41E−123 | 8.16E−06 | Yes |
| 14 | 1804647 | rs109162116 | MY | MR | 1.97E−123 | 8.15E−06 | Yes |
| 14 | 1805963 | rs211282745 | MY | MR | 1.89E−123 | 8.14E−06 | Yes |
| 14 | 1808145 | rs135258919 | MY | MR | 1.97E−123 | 8.33E−06 | Yes |
| 14 | 1817975 | rs135805021 | MY | MR | 1.97E−123 | 9.08E−06 | Yes |
| 14 | 1818125 | rs383356863 | MY | MR | 1.96E−123 | 9.09E−06 | Yes |
| 14 | 1819475 | rs208211113 | MY | MR | 1.89E−123 | 9.23E−06 | Yes |
| 14 | 1825125 | rs208113678 | MY | MR | 2.58E−123 | 1.05E−05 | Yes |
| 16 | 47836093 | rs207941573 | MY | MR | 1.19E−01 | 5.69E−11 | No |
| 16 | 59908679 | rs451830006 | MY | MR | 4.85E−02 | 2.93E−10 | No |
| 18 | 43909571 | rs464881101 | MY | MR | 4.84E−01 | 8.09E−11 | No |
| 20 | 29996719 | rs43116343 | MY | MR | 6.08E−13 | 2.41E−02 | No |
| 26 | 3771642 | – | MY | MR | 1.24E−10 | 4.07E−02 | No |
aSNPs showing genome-wide significance for at least one of the traits and significance (p < 1.18e-4) for the second trait
For MY and FY, as expected, we found that the two consecutive missense mutations in DGAT1 had pleiotropic effects. In addition, we found six other QTL with evidence of pleiotropic effects. On BTA1, we detected the SNP BTA1:120,470,021 with pleiotropic effects on MY and FY. This SNP is located in an intergenic region, close to AGTR1 (they are 67,012 bp apart). The lead SNPs, BTA5:93,945,991 and BTA7:57,287,990, were each located in an intron of MGST1 and KCTD16, respectively (Table 5). On BTA20, we found that the lead SNP BTA20:31,909,478 located in the GHR gene had pleiotropic effects on MY and FY, as previously described [23]. On BTA26, there were two SNPS, BTA26:20,527,926 and BTA26:37,869,471 that were located respectively near ENSBTAG00000023629 and in KCNK18 (synonymous variant).
In addition to DGAT1 and MGST1, variants in KCTD16 and KCNK18 and an intergenic variant BTA1:120,470,021 were associated with MY and PY. Since these SNPs were also associated with MY and FY, this was the indication that the above-mentioned four genes and one SNP have pleiotropic effects on MY, FY and PY. Meanwhile, BTA1:63,177,947 also showed possible pleiotropic effects for MY, FY and PY, located in an intergenic region close to the gene ENSBTAG00000046854.
Apart from the variants in DGAT1, only one SNP had significant effects on both MY and MR, i.e. BTA6:88,729,872 (Table 7), which is located in an intron of the GC gene.
Discussion
Overlapping QTL for three milk production traits
The bivariate analyses showed that the QTL for three milk production traits detected in the single-trait analyses and located on BTA5, 14, 20, and 26 might have pleiotropic effects. The univariate analysis identified overlapping QTL regions for all three milk production traits MY, FY and PY on BTA5, 14, 19, 18, and 26, for MY and FY on BTA2, 5, 14, 16, and 19, and for FY and PY on BTA5 and 19. However, without a joint analysis of two traits, it is not possible to determine whether the causal variants in the overlapping regions are the same ones or not.
BTA14 has been widely explored for genes and QTL related to economically important traits (e.g., [27, 28]), including MY, FY and PY. Recently, Nayeri et al. [29] reported that the region between 1.4 and 2.9 Mb on BTA14 was significantly associated with milk, fat and protein production, and with protein and fat deviation in Canadian Holstein cattle. Our findings support their conclusion that this region on BTA14 is strongly associated with milk production traits.
Segregation of QTL that affect milk production traits on BTA5 has already been reported [30, 31]. Based on an association analysis of a large outbred population, Littlejohn et al. [32] reported that a region on BTA5 at 93.9 Mb had pleiotropic effects on milk protein, fat, and lactose yield, milk volume and milk protein and lactose percentage. A 50-kbp interval that contained 632 variants was centered on the SNP with the most significant p value (g.93945738C > T) in the MGST1 gene. The C allele associated with high milk fat percentage was also associated with increased FY and protein percentage and decreased PY and milk volume. Kemper et al. [33] obtained similar results for the same region with impacts on a subset of the same milk composition traits. These results are consistent with our study that revealed that the QTL on BTA5 at 93.9 Mb had pleiotropic effects on FY, MY and PY (Table 3).
Overlapping QTL between milk yield and mastitis resistance
The univariate analysis identified two overlapping QTL regions (30.2–31.3 Mb on BTA5 and 88.6–89.1 Mb on BTA6) for MY and MR. However, the bivariate analysis showed that only the QTL on BTA6 was significantly associated with both traits. As shown in Table 3, effects for these traits had opposite directions—an unfavorable effect on MY and a favorable effect on MR. An unfavorable genetic correlation between milk production and clinical mastitis has been reported [20, 34] and the 88.6-89.1 Mb region on BTA6 contributes to this correlation. The QTL for mastitis resistance on BTA6 in this region is consistent with previous reports. In Nordic Holstein cattle, the region most significantly associated with mastitis was on BTA6 at 88.97 Mb [26]. The same region was also associated with mastitis in Nordic Red Cattle, but not in Danish Jersey Cattle [26]. This region includes two genes, GC and NPFFR2 that encode the vitamin D-binding protein precursor (88,695,940 to 88,739,180 bp) and the neuropeptide FF receptor 2 (89,052,210 to 89,059,348 bp), respectively, which can be involved in mastitis.
Sodeland et al. [35] identified a QTL for clinical mastitis on BTA6 in Norwegian Red Cattle with the most significant SNP, BTA-119376, being located at 90,670,190 bp. Klungland et al. [36] also reported a QTL for clinical mastitis on BTA6 and Ogorevc et al. [37] showed that BTA6 harbors several QTL for mastitis resistance. Moreover, the results by Nielsen et al. [38] point to a region on BTA6 near 90 Mb (containing the cluster of casein genes that encode around 80% of the proteins in cow milk) that is associated with milk production traits and mastitis in Norwegian Red cattle.
Estimation of genetic correlations using GWAS summary statistics
In this study, we estimated the genetic correlations based on GWAS summary statistics using LDSC regression [15]. There are several advantages for using this approach in cattle: (1) LDSC can estimate a genetic correlation based on GWAS summary statistics, which bypasses the limitation of sharing primary data that are the property of industrial partners; and (2) the genetic parameter estimates obtained by using LDSC in human populations are close to the estimates available from quantitative genetic analyses from previous reports. LDSC regression was first applied on human data [15]. LDSC functions well with the LD structure of the human genome. However, the LD structure in cattle is quite different, i.e. LD is much more extensive in cattle than in humans [39]. Using a linear animal test-day model, Hinrichs et al. estimated genetic correlations of 0.29, 0.30 and 0.34 between mastitis resistance and MY, FY, and PY, respectively [40]. In Norwegian Red cattle, a genetic correlation of 0.25 was reported between clinical mastitis and PY [41], whereas in Danish Holstein cattle, it was equal to 0.33 [42]. In another study on Norwegian cattle, Simianer et al. [43] estimated a genetic correlation of 0.472 between mastitis and MY. In our study, we estimated genetic correlations ranging from moderate to high between the milk production traits. Likewise, Hoekstra et al. [44] reported genetic correlations of 0.39 between MY and FY, 0.86 between MY and PY, and 0.56 between FY and PY in Dutch Black and White cows. Another study from the UK Holstein found genetic correlations of 0.69 between MY and FY, 0.88 between MY and PY, and 0.80 between FY and PY [45]. Based on the comparison of our estimates (MY and FY: 0.40, MY and PY: 0.78, FY and PY: 0.56) with those from these previous studies, we conclude that the LDSC approach with summary statistics from GWAS is reliable for the estimation of genetic parameters in cattle.
The most significant genes (candidate genes)
DGAT1
In our study, the QTL around 1.6 and 2.1 Mb on BTA14 had the strongest association with milk production traits (MY, PY and FY). The previously reported two missense SNPs (rs109326954 at 1802,266 bp and rs109234250 at 1802,265 bp) resulting in an amino acid change (K232A) were among the top associated variants in the QTL interval on BTA14. However, these two causal variants were not the lead SNPs for MY and PY in the single-trait association study. Imperfect imputation was mentioned as one possible reason by Iso-Touru et al. [7], who obtained similar results (the causal variant at 1802,266 bp not being the most significantly associated SNP) in Nordic Red Cattle. Both the multi-trait meta-analysis and the bivariate analysis indicated these two SNPs as the top associated variants (Tables 4 and 7). This was consistent with previously reported results on the contribution of these DGAT1 polymorphisms to variation in milk production traits in cattle [21, 22]. The bivariate analysis confirmed the pleiotropic effect of DGAT1 on FY, PY and MY. In addition, we detected pleiotropic effects of DGAT1 on MR, which was also reported previously [46].
MGST1
Raven et al. [47] identified a highly significant QTL on BTA5 at 85–110 Mb for milk production traits, where one of the lead SNPs was located within 3000 bp from MGST1. Previously, a GWAS in Nordic Red Cattle [7] reported a region associated with FY around 93,945,694 bp on BTA5 and MGST1 was proposed as candidate gene. Another study [48] found a QTL for MY in the same region i.e. between 92.1 and 93 Mb on BTA5. Although MGST1 is known to bind fatty acids directly, this activity appears to be related to its role as a detoxification enzyme [49], thus the mechanism that would explain an association with milk lipid synthesis/secretion on MY remains unknown. In our study, we observed pleiotropic effects of this QTL on FY and MY.
Novel candidate genes
Several genes showed large pleiotropic effects on multiple milk production traits. For a few other genes identified in our study, data in the mammalian phenotype database [24] provided strong support for a possible biological effect on the traits analyzed. For example, a mutation in GPAT4 is responsible for “abnormal milk composition” in mouse. ZFPM2 is related to the term “abnormal mammary gland morphology”. In the bivariate analysis, we found that KCTD16, which is associated with residual feed intake in pigs and meat quality in cattle [48], had pleiotropic effects on FY, PY and MY. Finally, KCNK18 showed pleiotropic effects on PY and MY but no obvious biological mechanism linking KCNK18 to milk production traits was found in the literature.
Conclusions
In this study, we performed a multi-trait meta-analysis and detected several SNPs that affect both milk production traits and mastitis resistance in dairy cattle, which shows the high power of this approach to detect potential pleiotropy effects compared with the subjective assessment of overlapping single-trait QTL regions. Further confirmation of the lead SNPs from the multi-trait meta-analysis shortened the list of those with possible pleiotropic effects. Bivariate analysis can indicate the pleiotropic effect of a variant. We observed that DGAT1 and MGST1 had pleiotropic effects on milk production traits, and GC had pleiotropic effects on MY and MR. In addition, our results suggest that KCTD16 and KCNK18 might have pleiotropic effects on all three milk production traits analyzed. Our findings add to the knowledge about the genetic determination of milk production traits and mastitis resistance in cattle.
Acknowledgements
We are grateful to the Nordic Cattle Genetic Evaluation (NAV, Aarhus, Denmark) for providing the phenotypic data used in this study and Viking Genetics (Randers, Denmark) for providing samples for genotyping. The 1000 Bull Genomes Project is kindly acknowledged for sharing WGS data. Magdalena Dusza acknowledges Erasmus fellowship for Higher Education Learning Traineeship.
Authors’ contributions
GS, ZC, MD, BG, and MSL conceived and designed the study. GS, MD and ZC analyzed the data. MD and ZC wrote the manuscript. MSL and BG contributed materials, analysis tools and participated in the discussion. All authors read and approved the final manuscript.
Funding
This work was partly funded as part of the research project, ‘Genomics in herds’, which is financed by Viking Genetics and Nordic Cattle Genetic Evaluation, and partly by the Center for Genomic Selection in Animals and Plants (GenSAP), which is financed by Innovation Fund Denmark (Grant: 0603-00519B). However, none of these funding institutes had any input into the design, data analyses, and data interpretation of this study.
Availability of data and materials
Genome assembly data were taken from publicly available sources. The assembly UMD_3.1.1 is available for download from NCBI. Part of the whole-genome sequencing data from the 1000 Bull Genomes Project are publically available at NCBI using SRA no. SRP039339 and for the rest, the Board of the 1000 Bull Genome Consortium should be contacted. All annotation information was obtained from a publicly available source (http://www.ensembl.org). Whole-genome sequences from Aarhus University and individual SNP genotype data are available only upon agreement with the breeding organization and should be requested directly from the authors.
Ethics approval and consent to participate
Neither the collection of biological samples nor animal handling were performed for this study. Before conducting this study, consent for use of data was obtained where required.
Consent for publication
Not applicable.
Competing interests
The authors declare that they no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zexi Cai and Magdalena Dusza authors contributed equally to this study
Contributor Information
Zexi Cai, Email: zexi.cai@mbg.au.dk.
Magdalena Dusza, Email: duszamagda@gmail.com.
Bernt Guldbrandtsen, Email: bernt.guldbrandtsen@mbg.au.dk.
Mogens Sandø Lund, Email: mogens.lund@mbg.au.dk.
Goutam Sahana, Email: goutam.sahana@mbg.au.dk.
References
- 1.Oltenacu PA, Broom DM. The impact of genetic selection for increased milk yield on the welfare of dairy cow. Anim Welfare. 2010;19:39–49. [Google Scholar]
- 2.Heringstad B, Chang YM, Gianola D, Klemetsdal G. Genetic association between susceptibility to clinical mastitis and protein yield in norwegian dairy cattle. J Dairy Sci. 2005;88:1509–1514. doi: 10.3168/jds.S0022-0302(05)72819-8. [DOI] [PubMed] [Google Scholar]
- 3.Cai Z, Guldbrandtsen B, Lund MS, Sahana G. Dissecting closely linked association signals in combination with the mammalian phenotype database can identify candidate genes in dairy cattle. BMC Genet. 2019;20:15. doi: 10.1186/s12863-019-0717-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cai Z, Guldbrandtsen B, Lund MS, Sahana G. Prioritizing candidate genes post-GWAS using multiple sources of data for mastitis resistance in dairy cattle. BMC Genomics. 2018;19:656. doi: 10.1186/s12864-018-5050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goddard M. A method of comparing sires evaluated in different countries. Livest Prod Sci. 1985;13:321–331. doi: 10.1016/0301-6226(85)90024-7. [DOI] [Google Scholar]
- 6.Schaeffer LR. Model for international evaluation of dairy sires. Livest Prod Sci. 1985;12:105–115. doi: 10.1016/0301-6226(85)90084-3. [DOI] [Google Scholar]
- 7.Iso-Touru T, Sahana G, Guldbrandtsen B, Lund MS, Vilkki J. Genome-wide association analysis of milk yield traits in Nordic Red Cattle using imputed whole genome sequence variants. BMC Genet. 2016;17:55. doi: 10.1186/s12863-016-0363-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daetwyler HD, Capitan A, Pausch H, Stothard P, van Binsbergen R, Brondum RF, et al. Whole-genome sequencing of 234 bulls facilitates mapping of monogenic and complex traits in cattle. Nat Genet. 2014;46:858–865. doi: 10.1038/ng.3034. [DOI] [PubMed] [Google Scholar]
- 9.Howie B, Marchini J, Stephens M. Genotype imputation with thousands of genomes. G3 (Bethesda) 2011;1:457–470. doi: 10.1534/g3.111.001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchsberger C, Abecasis GR, Hinds DA. Minimac2: faster genotype imputation. Bioinformatics. 2015;31:782–784. doi: 10.1093/bioinformatics/btu704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu X, Guldbrandtsen B, Lund MS, Sahana G. Association analysis for feet and legs disorders with whole-genome sequence variants in 3 dairy cattle breeds. J Dairy Sci. 2016;99:7221–7231. doi: 10.3168/jds.2015-10705. [DOI] [PubMed] [Google Scholar]
- 12.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang J, Zaitlen NA, Goddard ME, Visscher PM, Price AL. Advantages and pitfalls in the application of mixed-model association methods. Nat Genet. 2014;46:100–106. doi: 10.1038/ng.2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Listgarten J, Lippert C, Kadie CM, Davidson RI, Eskin E, Heckerman D. Improved linear mixed models for genome-wide association studies. Nat Methods. 2012;9:525–526. doi: 10.1038/nmeth.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics Consortium et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bolormaa S, Pryce JE, Reverter A, Zhang Y, Barendse W, Kemper K, et al. A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. PLoS Genet. 2014;10:e1004198. doi: 10.1371/journal.pgen.1004198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madsen P, Jensen J, Labouriau R, Christensen OF, Sahana G. DMU—a package for analyzing multivariate mixed models in quantitative genetics and genomics. In Proceedings of the 10th World Congress on Genetics Applied to Livestock Production: 17–22 August 2014; Vancouver.
- 18.Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, et al. A whole-genome assembly of the domestic cow, Bostaurus. Genome Biol. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lund MS, Jensen J, Petersen PH. Estimation of genetic and phenotypic parameters for clinical mastitis somatic cell production deviance, and protein yield in dairy cattle using Gibbs sampling. J Dairy Sci. 1999;82:1045–1051. doi: 10.3168/jds.S0022-0302(99)75325-7. [DOI] [PubMed] [Google Scholar]
- 21.Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, et al. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Res. 2002;12:222–231. doi: 10.1101/gr.224202. [DOI] [PubMed] [Google Scholar]
- 22.Grisart B, Farnir F, Karim L, Cambisano N, Kim JJ, Kvasz A, et al. Genetic and functional confirmation of the causality of the DGAT1 K232A quantitative trait nucleotide in affecting milk yield and composition. Proc Natl Acad Sci USA. 2004;101:2398–2403. doi: 10.1073/pnas.0308518100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rahmatalla SA, Muller U, Strucken EM, Reissmann M, Brockmann GA. The F279Y polymorphism of the GHR gene and its relation to milk production and somatic cell score in German Holstein dairy cattle. J Appl Genet. 2011;52:459–465. doi: 10.1007/s13353-011-0051-3. [DOI] [PubMed] [Google Scholar]
- 24.Bult CJ, Eppig JT, Kadin JA, Richardson JE, JA Blake, Mouse Genome Database G The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:724–728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beigneux AP, Vergnes L, Qiao X, Quatela S, Davis R, Watkins SM, et al. Agpat6—a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium. J Lipid Res. 2006;47:734–744. doi: 10.1194/jlr.M500556-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sahana G, Guldbrandtsen B, Thomsen B, Holm LE, Panitz F, Brondum RF, et al. Genome-wide association study using high-density single nucleotide polymorphism arrays and whole-genome sequences for clinical mastitis traits in dairy cattle. J Dairy Sci. 2014;97:7258–7275. doi: 10.3168/jds.2014-8141. [DOI] [PubMed] [Google Scholar]
- 27.Ashwell MS, Van Tassell CP, Sonstegard TS. A genome scan to identify quantitative trait loci affecting economically important traits in a US Holstein population. J Dairy Sci. 2001;84:2535–2542. doi: 10.3168/jds.S0022-0302(01)74705-4. [DOI] [PubMed] [Google Scholar]
- 28.Wibowo TA, Gaskins CT, Newberry RC, Thorgaard GH, Michal JJ, Jiang Z. Genome assembly anchored QTL map of bovine chromosome 14. Int J Biol Sci. 2008;4:406–414. doi: 10.7150/ijbs.4.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nayeri S, Sargolzaei M, Abo-Ismail MK, May N, Miller SP, Schenkel F, et al. Genome-wide association for milk production and female fertility traits in Canadian dairy Holstein cattle. BMC Genet. 2016;17:75. doi: 10.1186/s12863-016-0386-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bennewitz J, Reinsch N, Grohs C, Leveziel H, Malafosse A, Thomsen H, et al. Combined analysis of data from two granddaughter designs: a simple strategy for QTL confirmation and increasing experimental power in dairy cattle. Genet Sel Evol. 2003;35:319–338. doi: 10.1186/1297-9686-35-3-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khatkar MS, Thomson PC, Tammen I, Raadsma HW. Quantitative trait loci mapping in dairy cattle: review and meta-analysis. Genet Sel Evol. 2004;36:163–190. doi: 10.1186/1297-9686-36-2-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Littlejohn MD, Tiplady K, Fink TA, Lehnert K, Lopdell T, Johnson T, et al. Sequence-based association analysis reveals an MGST1 eQTL with pleiotropic effects on bovine milk composition. Sci Rep. 2016;6:25376. doi: 10.1038/srep25376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kemper KE, Reich CM, Bowman PJ, Vander Jagt CJ, Chamberlain AJ, Mason BA, et al. Improved precision of QTL mapping using a nonlinear Bayesian method in a multi-breed population leads to greater accuracy of across-breed genomic predictions. Genet Sel Evol. 2015;47:29. doi: 10.1186/s12711-014-0074-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heringstad B, Chang YM, Gianola D, Klemetsdal G. Genetic analysis of clinical mastitis, milk fever, ketosis, and retained placenta in three lactations of Norwegian red cows. J Dairy Sci. 2005;88:3273–3281. doi: 10.3168/jds.S0022-0302(05)73010-1. [DOI] [PubMed] [Google Scholar]
- 35.Sodeland M, Kent M, Olsen H, Opsal M, Svendsen M, Sehested E, et al. Quantitative trait loci for clinical mastitis on chromosomes 2, 6, 14 and 20 in Norwegian Red cattle. Anim Genet. 2011;42:457–465. doi: 10.1111/j.1365-2052.2010.02165.x. [DOI] [PubMed] [Google Scholar]
- 36.Klungland H, Sabry A, Heringstad B, Olsen HG, Gomez-Raya L, Vage DI, et al. Quantitative trait loci affecting clinical mastitis and somatic cell count in dairy cattle. Mamm Genome. 2001;12:837–842. doi: 10.1007/s00335001-2081-3. [DOI] [PubMed] [Google Scholar]
- 37.Ogorevc J, Kunej T, Razpet A, Dovc P. Database of cattle candidate genes and genetic markers for milk production and mastitis. Anim Genet. 2009;40:832–851. doi: 10.1111/j.1365-2052.2009.01921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nilsen H, Olsen H, Hayes B, Nome T, Sehested E, Svendsen M, et al. Characterization of a QTL region affecting clinical mastitis and protein yield on BTA6. Anim Genet. 2009;40:701–712. doi: 10.1111/j.1365-2052.2009.01908.x. [DOI] [PubMed] [Google Scholar]
- 39.Bovine HapMap Consortium. Gibbs RA, Taylor JF, Van Tassell CP, Barendse W, Eversole KA, et al. Genome-wide survey of SNP variation uncovers the genetic structure of cattle breeds. Science. 2009;324:528–532. doi: 10.1126/science.1167936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hinrichs D, Stamer E, Junge W, Kalm E. Genetic analyses of mastitis data using animal threshold models and genetic correlation with production traits. J Dairy Sci. 2005;88:2260–2268. doi: 10.3168/jds.S0022-0302(05)72902-7. [DOI] [PubMed] [Google Scholar]
- 41.Heringstad B, Klemetsdal G, Ruane J. Clinical mastitis in Norwegian cattle: frequency, variance components, and genetic correlation with protein yield. J Dairy Sci. 1999;82:1325–1330. doi: 10.3168/jds.S0022-0302(99)75356-7. [DOI] [PubMed] [Google Scholar]
- 42.Hansen M, Lund MS, Sørensen MK, Christensen LG. Genetic parameters of dairy character, protein yield, clinical mastitis, and other diseases in the Danish Holstein cattle. J Dairy Sci. 2002;85:445–452. doi: 10.3168/jds.S0022-0302(02)74093-9. [DOI] [PubMed] [Google Scholar]
- 43.Simianer H, Solbu H, Schaeffer L. Estimated genetic correlations between disease and yield traits in dairy cattle. J Dairy Sci. 1991;74:4358–4365. doi: 10.3168/jds.S0022-0302(91)78632-3. [DOI] [PubMed] [Google Scholar]
- 44.Hoekstra J, van der Lugt AW, van der Werf JHJ, Ouweltjes W. Genetic and phenotypic parameters for milk production and fertility traits in upgraded dairy cattle. Livest Prod Sci. 1994;40:225–232. doi: 10.1016/0301-6226(94)90090-6. [DOI] [Google Scholar]
- 45.Kadarmideen HN, Thompson R, Coffey MP, Kossaibati MA. Genetic parameters and evaluations from single- and multiple-trait analysis of dairy cow fertility and milk production. Livest Prod Sci. 2003;81:183–195. doi: 10.1016/S0301-6226(02)00274-9. [DOI] [Google Scholar]
- 46.Manga I, Říha H. The DGAT1 gene K232A mutation is associated with milk fat content, milk yield and milk somatic cell count in cattle. Arch Anim Breed. 2011;54:257–263. doi: 10.5194/aab-54-257-2011. [DOI] [Google Scholar]
- 47.Raven LA, Cocks BG, Hayes BJ. Multibreed genome wide association can improve precision of mapping causative variants underlying milk production in dairy cattle. BMC Genomics. 2014;15:62. doi: 10.1186/1471-2164-15-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mai M, Sahana G, Christiansen F, Guldbrandtsen B. A genome-wide association study for milk production traits in Danish Jersey cattle using a 50 K single nucleotide polymorphism chip. J Anim Sci. 2010;88:3522–3528. doi: 10.2527/jas.2009-2713. [DOI] [PubMed] [Google Scholar]
- 49.Iida A, Saito S, Sekine A, Harigae S, Osawa S, Mishima C, et al. Catalog of 46 single-nucleotide polymorphisms (SNPs) in the microsomal glutathione S-transferase 1 (MGST1) gene. J Hum Genet. 2001;46:590–594. doi: 10.1007/s100380170026. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genome assembly data were taken from publicly available sources. The assembly UMD_3.1.1 is available for download from NCBI. Part of the whole-genome sequencing data from the 1000 Bull Genomes Project are publically available at NCBI using SRA no. SRP039339 and for the rest, the Board of the 1000 Bull Genome Consortium should be contacted. All annotation information was obtained from a publicly available source (http://www.ensembl.org). Whole-genome sequences from Aarhus University and individual SNP genotype data are available only upon agreement with the breeding organization and should be requested directly from the authors.