Abstract
Background
A number of studies have demonstrated the critical role of long non-coding RNA gastric cancer high expressed transcript 1 (GHET1) in many cancers. This meta-analysis provides an evidence-based evaluation of the prognostic role of GHET1 in cancer.
Materials and methods
Literature searches were conducted in several databases including Medline, Cochrane, EMBASE, CNKI, and Wanfang. The pooled odds ratio (OR) and hazard ratio (HR) with 95% confidence interval (CI) were used to evaluate the role of GHET1 in cancer. The study protocol was registered at PROSPERO (ID: CRD42018111252).
Results
Sixteen studies, containing 1315 patients, were analyzed in this meta-analysis. The pooled results indicated that GHET1 overexpression was significantly associated with poor overall survival (OS) and disease-free survival (DFS) in cancer. Moreover, up-regulation of GHET1 expression predicted larger tumor size, positive lymph node metastasis, positive distant metastasis, and advanced TNM (tumor-node-metastases) stage in human cancers.
Conclusion
There is a significant correlation between up-regulation of GHET1 and both poor prognosis and advanced clinicopathological cancer characteristics. GHET1 may be a potential prognostic predictor for human cancers.
Keywords: GHET1, Meta-analysis, Cancers, Prognosis
Background
Despite great progress in cancer diagnosis and treatment, there were still 18.1 million new cancer cases and 9.6 million cancer deaths worldwide in 2018 [1]. The treatment prognosis of most cancers remains poor; one of the primary reasons for this is the lack of specific biomarkers for the early diagnosis of most cancers [2]. Therefore, it is necessary to identify novel prognostic markers of cancer for potential clinical application [3–5].
Recently, many studies have indicated that long non-coding RNAs (lncRNAs) play a crucial role in the progression of cancer [6–10]. Some lncRNAs are involved in the modulation of cancer proliferation, invasion, and metastasis. In addition, several studies have found that lncRNAs are potential cancer-specific prognostic biomarkers [2, 11–13]. The lncRNA gastric cancer high expressed transcript 1 (GHET1) is located on chromosome 7q36.1 and was originally found to be highly expressed in gastric cancer [14]. In gastric cancer, the up-regulation of GHET1 promotes tumor cell proliferation in vitro and in vivo by physically binding to IGF2BP1, thereby enhancing the interaction between c-Myc mRNA and IGF2BP1; this can enhance the stability of c-Myc mRNA [14].
Clinically, several studies have indicated that up-regulation of GHET1 is associated with poor prognosis and advanced clinical features in several cancers [14–24]. Most existing studies suggest that GHET1 might be a potential biomarker for predicting the prognosis of human cancers. However, due to limitations such as small sample sizes and discrete outcomes, the findings of a single study may not accurately capture the phenomenon under examination [14–24]. Thus, we undertook a systematic review and meta-analysis of all eligible studies to perform an evidence-based evaluation of the prognostic role of GHET1 in cancer.
Materials and methods
This systematic meta-analysis was conducted accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines. It has been registered with PROSPERO (ID: CRD42018111252).
Literature search and selection
Literature searches were conducted in several databases, including Medline, Cochrane, EMBASE, CNKI, and Wanfang, up until April 15th, 2019. The search strategy was as follows: “GHET1” OR “lncRNA GHET1“ OR “gastric cancer high expressed transcript 1“) AND (“cancer“ OR “neoplasm“ OR “tumor“ OR “carcinoma“). The search was limited to English and Chinese studies. The references of relevant studies were also retrieved to avoid missing any potentially eligible studies.
Inclusion and exclusion criteria
The inclusion criteria for this meta-analysis were as follows: (1) detection of GHET1 expression in human cancers by quantitative real-time PCR (qRT-PCR); (2) patients in the study were divided into subgroups based on different GHET1 expression levels; (3) prognosis or clinicopathological feature of GHET1 was reported; (4) hazard ratios (HRs) and 95% confidence intervals (CI) were able to be obtained directly or indirectly from the article. In addition, the exclusion criteria were as follows: (1) reviews, editorials, conference reports, case reports, and meta-analyses; (2) non-human tissue studies; (3) studies only investigating the molecular mechanisms of GHET1; (4) duplicate publications. The titles and abstracts were first evaluated based on the inclusion and exclusion criteria; the full texts of those reports that appeared to meet the criteria were then further evaluated.
Data extraction and quality assessment
Two researchers (Dingding Wang and Xiaolian Fang) independently extracted data from the selected studies according to uniform data extraction standards; any disagreements were settled by consensus with a third investigator (Hong Zhang). Extracted data included: first author’s name, publication year, country or region, sample size, cancer type, method for detection of GHET1, cut-off values, treatment data, disease-free survival (DFS), overall survival (OS), and clinical stage of cancer. If the HRs and 95% CIs for DFS or OS were not available in the paper, the data were indirectly extracted from survival curves, based on the approach described previously [25]. The Newcastle–Ottawa Scale (NOS) was used to assess the methodological quality of each study. A study was considered to be high quality if the NOS score was greater than or equal to 6; otherwise, it was considered to be a low-quality study.
Public data and tools
TCGA data, including RNAseqV2 and clinical data, were extracted from the TCGA Data Portal and UCSC Xena project, according to the publication guidelines (http://cancergenome.nih.gov/publications/publicationguidelines). GEPIA was used to analyze RNAseq data. Differential expression analysis was conducted using one-way ANOVA. Survival analysis was performed using the Kaplan–Meier method and log-rank test.
Statistical analysis
Pooled HRs and 95% CIs were used to assess the relationship between GHET1 expression and prognosis. Odd ratios and 95% CIs were combined to evaluate the relationships between GHET1 expression and clinicopathological factors. If the 95% CI of the combined OR or HR did not overlap 1, the result was considered statistically significant. The I2 and Q tests were used to evaluate the heterogeneity of the meta-analysis. If I2 > 50% or P < 0.05, heterogeneity was considered statistically significant and a random effects model was chosen; otherwise, a fixed effects model was utilized. We also performed a sensitivity analysis to evaluate the stability of the combined results. The Begg’s test was used to assess potential publication bias. All statistical calculations were performed using RevMan 5.3 software and STATA software version 14.2 (StataCorp LLC, College Station, TX, USA). Moreover, Engauge Digitizer 10.0 was utilized to extract HRs and 95% CIs from the Kaplan–Meier survival curves. A P value of less than 0.05 was considered to be statistically significant.
Results
Study characteristics
A preliminary literature search was conducted based on the established search strategy; a total of 107 articles were obtained. After removing 75 duplicates, the titles and abstracts of the remaining 32 articles were evaluated according to the inclusion criteria. Following this, six studies were excluded. The full texts of the remaining 26 articles were reviewed; this resulted in the exclusion of 10 papers due to a lack of eligible prognostic or clinicopathological information. Finally, a total of 16 studies were included in the meta-analysis (Fig. 1).
Fig. 1.
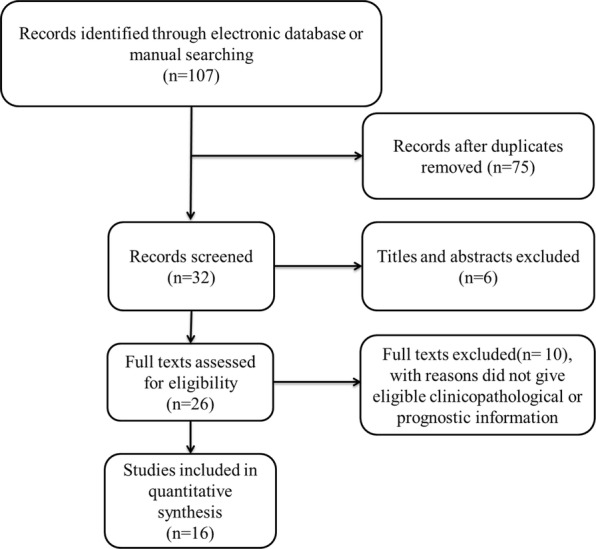
Flaw chart of literature search and selection
The basic characteristics of the patients in these 16 studies were extracted and are summarized in Table 1. A total of 1315 patients were included in the current meta-analysis; all patients were from China. The sample size of each individual study ranged from 42 to 182, and the publication years ranged from 2014 to 2019. Fourteen of the articles were published in English and two were published in Chinese. GHET1 expression in all studies was assessed using qRT-PCR. Moreover, 13 studies provided cut-off definitions for high or low GHET1 expression groups, including the median and median ratio. However, the remaining three studies did not provide explicit cut-off values. The types of cancer evaluated in the evaluated studies included: bladder cancer, breast cancer, cervical cancer, esophageal squamous cell carcinoma, gastric cancer, hepatocellular carcinoma, non-small cell lung cancer, osteosarcoma, pancreatic cancer, and renal cell carcinoma. Patients in 14 cohorts underwent surgical treatments; however, the therapeutic approach adopted in two of the studies was not available. Meanwhile, 15 studies reported the clinical stage of the patients, 12 studies reported OS, and four studies reported DFS. The NOS score ranged from 7 to 9, with an average score of 7.7; this indicates that each of the studies employed high-quality methodology.
Table 1.
Basic characteristics of the included studies
| Author | Year | Country | Sample size (n) | Cancer type | Cut-off value | Treatments | Outcomes | HR statistic | NOS |
|---|---|---|---|---|---|---|---|---|---|
| F. Yang | 2014 | China | 42 | GC | Median | Surgery | CP, OS | Survival curve | 7 |
| H. Liu | 2018 | China | 86 | ESCC | Median | Surgery | CP, OS | Survival curve | 8 |
| H.F. Liu | 2017 | China | 55 | ESCC | Median | Surgery | CP | NA | 8 |
| H.Y. Zhou | 2017 | China | 64 | PADC | NA | Surgery | CP | NA | 8 |
| J. Li | 2017 | China | 179 | HCC | NA | Surgery | CP, DFS, OS | Survival curve | 8 |
| L. Jin | 2017 | China | 68 | HCC | NA | Surgery | CP, OS | Survival curve | 7 |
| L.J. Li | 2014 | China | 80 | BLC | Median | Surgery | OS | Survival curve | 7 |
| Q.C. Zhang | 2019 | China | 94 | cervical cancer | Median | NA | CP, OS | Data in paper | 7 |
| Q.M. Shen | 2018 | China | 105 | NSCLC | Median | NA | CP, DFS, OS | Survival curve | 7 |
| R. Song | 2018 | China | 60 | BC | Median | Surgery | CP, OS | Survival curve | 8 |
| W. Yang | 2018 | China | 60 | osteosarcoma | Median | Surgery | CP, OS | Survival curve | 7 |
| W.J. Xie | 2019 | China | 40 | RCC | Median ratio | Surgery | CP | NA | 8 |
| Y. Xia | 2016 | China | 42 | GC | Median | Surgery | CP | NA | 8 |
| Y.P. Zhang1 | 2018 | China | 106 | HCC | Median | Surgery | CP, DFS, OS | Survival curve | 8 |
| Y.P. Zhang2 | 2018 | China | 182 | HCC | Median | Surgery | CP, DFS, OS | Data in paper | 9 |
| Z.B. Guan | 2018 | China | 52 | NSCLC | Median | Surgery | CP, OS | Data in paper | 8 |
BC breast cancer, BLC bladder cancer, ESCC esophageal squamous cell carcinoma, GC gastric cancer, HCC hepatocellular carcinoma, PADC pancreatic cancer, NSCLC non-small lung cancer, qRT-PCR quantitative reverse transcription polymerase chain reaction, CP clinicopathological parameters, OS overall survival, DFS disease-free survival, NOS Newcastle–Ottawa Scale, NA not available
Association between GHET1 expression and prognosis
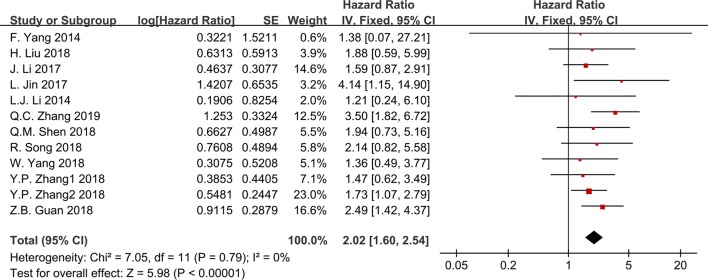
Twelve studies reported OS for eight types of cancer based on GHET1 expression in 1114 patients. As shown in Fig. 2, a fixed-effects model was adopted since there was no statistical heterogeneity (I2 = 0.0%, P = 0.783). The pooled HR for the high GHET1 expression group versus the low group was 2.037 (95% CI 1.626–2.551, P < 0.001). This pooled result indicates a significant association between overexpressed GHET1 and poor OS.
Fig. 2.
Meta-analysis for the association between GHET1 expression and OS
In order to further explore the potential prognostic value of GHET1, a series of subgroup analyses were performed based on cancer type, sample size, cut-off value, treatment, and NOS score. The results indicated that, regardless of the cancer type, sample size, cut-off value, treatment, and NOS score, the up-regulation of GHET1 was significantly correlated with poor OS in all subgroup analyses (Table 2).
Table 2.
Subgroup analysis for the association between GHET1 expression and OS
| Subgroups | No. of studies | No. of patients | Pooled HR (95% CI) | PHet | I2 (%) | P value |
|---|---|---|---|---|---|---|
| Cancer type | ||||||
| Digestive system | 6 | 663 | 1.751 [1.274, 2.408] | 0.844 | 0.0 | 0.001 |
| Respiratory system | 2 | 157 | 2.363 [1.515, 3.685] | 0.656 | 0.0 | < 0.001 |
| Others | 4 | 294 | 2.377 [1.505, 3.754] | 0.357 | 7.2 | < 0.001 |
| Cut-off value | ||||||
| Median | 10 | 867 | 2.067 [1.615, 2.646] | 0.802 | 0.0 | < 0.001 |
| Others | 2 | 247 | 1.894 [1.096, 3.274] | 0.185 | 43.0 | 0.022 |
| Treatments | ||||||
| Surgery | 10 | 915 | 1.890 [1.475, 2.420] | 0.899 | 0.0 | < 0.001 |
| Others | 2 | 199 | 2.918 [1.698, 5.015] | 0.324 | 0.0 | < 0.001 |
| Sample size (n) | ||||||
| ≤ 80 | 5 | 282 | 2.312 [1.574, 3.394] | 0.725 | 0.0 | < 0.001 |
| > 80 | 7 | 832 | 1.906 [1.444, 2.517] | 0.609 | 0.0 | < 0.001 |
| NOS score | ||||||
| ≤ 7 | 6 | 449 | 2.456 [1.602, 3.765] | 0.524 | 0.0 | < 0.001 |
| > 7 | 6 | 665 | 1.895 [1.454, 2.470] | 0.850 | 0.0 | < 0.001 |
HR hazard ratio, CI confidence interval
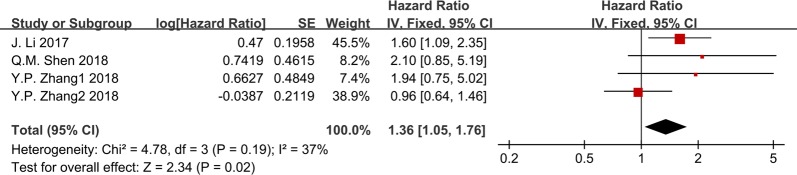
Moreover, DFS was reported in four studies with a total of 572 patients. There was no statistical heterogeneity, and a fixed-effect model was utilized (I2 = 37.1%, P = 0.190). The pooled results indicated that elevated GHET1 expression was significantly correlated with shorter DFS (HR = 1.362, 95% = 1.051–1.765, P = 0.020, Fig. 3). Overall, these results indicate that GHET1 might be an independent factor associated with survival of cancer patients.
Fig. 3.
Meta-analysis for the association between GHET1 expression and DFS
Associations between GHET1 expression and clinicopathological parameters
To explore the associations between GHET1 expression and clinicopathological features, further meta-analysis of seven studies was conducted. The pooled results are shown in Table 3. Compared to low GHET1 expression, high GHET1 expression level was statistically correlated with larger tumor size (P < 0.001, fixed model), positive lymph node metastasis (P < 0.001, fixed model), positive distant metastasis (P < 0.001, fixed model), and advanced clinical stage (P < 0.001, fixed model). However, there were no statistically significant relationships between GHET1 expression level and age (P = 0.452, fixed model), gender (P = 0.925, fixed model), and histological differentiation (P = 0.467, random model). These findings demonstrate statistically significant associations between up-regulation of GHET1 and advanced clinicopathological features of cancer.
Table 3.
Meta-analysis results for the association between GHET1 expression and clinicopathological characteristics
| Variables | Studies (n) | Patient (n) | Pooled OR (95% CI) | PHet | I2 (%) | P | Model |
|---|---|---|---|---|---|---|---|
| Age (old: young) | 15 | 1235 | 0.915 [0.725, 1.154] | 0.608 | 0.0 | 0.452 | Fixed |
| Gender (male: female) | 13 | 1081 | 0.987 [0.750, 1.299] | 0.828 | 0.0 | 0.925 | Fixed |
| Tumor size (large: small) | 13 | 1143 | 2.794 [2.197, 3.554] | 0.101 | 35.2 | < 0.001 | Fixed |
| Differentiation (poor: well) | 12 | 1010 | 1.266 [0.671, 2.388] | 0.000 | 77.4 | 0.467 | Random |
| Lymph node metastasis (yes: no) | 9 | 596 | 4.018 [2.816, 5.734] | 0.151 | 33.4 | < 0.001 | Fixed |
| Distant metastasis (yes: no) | 6 | 342 | 3.994 [2.028, 7.864] | 0.296 | 18.1 | < 0.001 | Fixed |
| TNM stage (III/IV: I/II) | 13 | 1125 | 3.721 [2.889, 4.793] | 0.336 | 10.9 | < 0.001 | Fixed |
Random random-effects model, Fixed fixed-effects model, OR odds ratio, CI confidence interval
Publication bias and sensitivity analysis
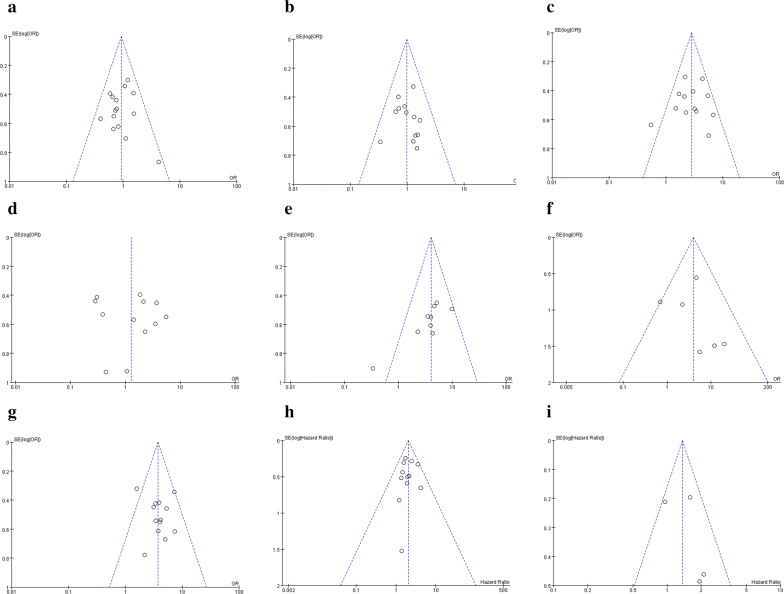
The Begg’s test was conducted to evaluate publication bias among the studies. As shown in Fig. 4a–i, regardless of age (Pr > |z| = 0.692), gender (Pr > |z| = 0.583), tumor size (Pr > |z| = 0.583), lymph node metastasis (Pr > |z| = 0.076), distant metastasis (Pr > |z| = 0.452), TNM stage (Pr > |z| = 0.760), histological differentiation (Pr > |z| = 0.732), DFS (Pr > |z| = 0.734), and OS (Pr > |z| = 1.000), the funnel plots of the Begg’s test showed no obvious asymmetry. This indicates publication bias did not affect the pooled results in the meta-analysis.
Fig. 4.
Funnel plots for the meta-analyses of the association between GHET1 expression and clinicopathological parameters or prognosis; a, age; b, gender; c, tumor size; d, differentiation; e, lymph node metastasis; f, distant metastasis; g, clinical stage; h, OS; i, DFS
In addition, to assess the stability of the combined results, we performed a sensitivity analysis of OS. The sensitivity analysis indicated that no individual study changed the combined results; thus, the OS results can be considered reliable (Fig. 5).
Fig. 5.
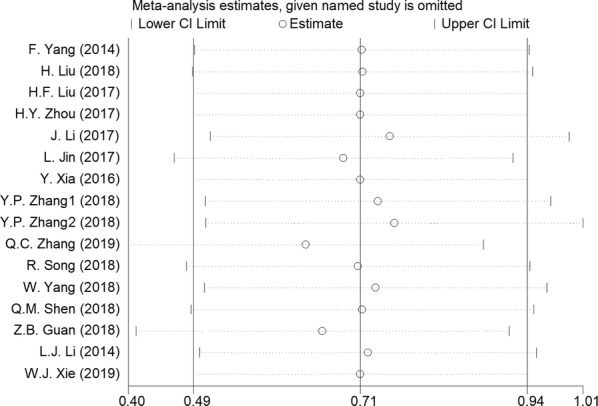
Sensitivity analysis for the meta-analysis of the association between lncRNA DANCR expression and OS
Validation of GHET1 in TCGA dataset
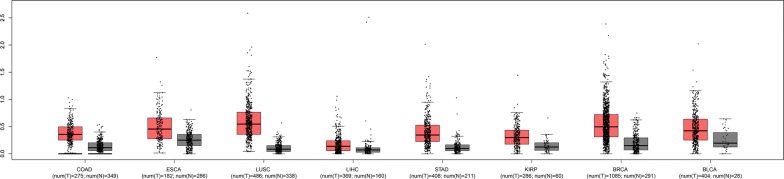
To further verify our results, the expression of GHET1 in eight types of cancer was evaluated using RNAseqV2 and TCGA clinical data. The results indicated that GHET1 was up-regulated in most cancers, including lung squamous cell carcinoma (LUSC), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), stomach adenocarcinoma (STAD), colon adenocarcinoma (COAD), breast invasive carcinoma (BRCA), bladder urothelial carcinoma (BLCA), and esophageal carcinoma (ESCA) (|Log2FC| Cutoff > 1, q-value < 0.01, Fig. 6). Further, the GHET1 high expression indicated poor prognosis in several cancer (Additional file 1: Fig. S1). We merged the expression and prognosis data for cancers of the digestive system, respiratory system, and urinary system, including BLCA, COAD, ESCA, STAD, LIHC, pancreatic adenocarcinoma (PAAD), lung adenocarcinoma (LUAD), LUSC, kidney chromophobe (KICH), and KIRP. According to the median GHET1 expression level, 3036 patients with 10 cancer types were classified into two groups. As demonstrated in Fig. 7, the GHET1 high expression group had shorter OS than the GHET1 low expression group, confirming that over-expression of GHET1 is correlated with poor OS in various human cancers (P < 0.001).
Fig. 6.
The expression levels of GHET1 in eight types of cancer tissues and normal tissues in TCGA cohort
Fig. 7.
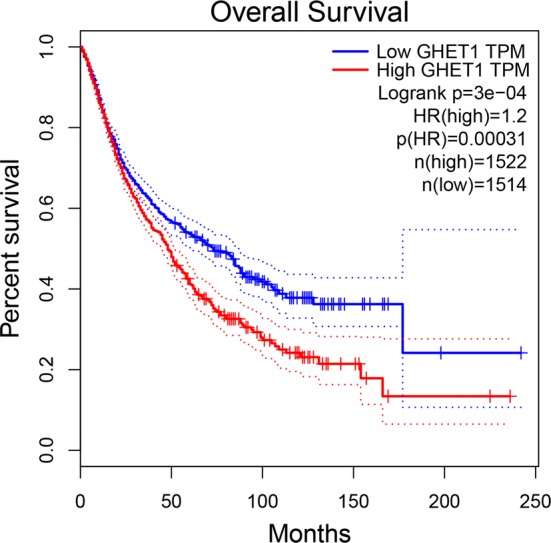
Overall survival plots of GHET1 in TCGA cohort, including BLCA, COAD, ESCA, KICH, KIRP, LIHC, LUAD, LUSC, PAAD and STAD (n = 3077, Log-rank P < 0.001)
Discussion
Accumulating evidence has demonstrated that lncRNAs act as crucial regulators of almost all aspects of physiological and pathological processes [26–30]. Multiple studies have also indicated that lncRNAs contribute to the carcinogenesis and progression of several tumors [6–10]. Recently, several studies have reported that lncRNA GHET1 might be related to prognosis in cancer patients [14–24, 31]. Therefore, we performed this meta-analysis of 16 eligible studies to systematically evaluate the prognostic value of GHET1 in all cancers.
In the present study, we evaluated the prognostic value of GHET1 in cancer. The pooled HR indicated that GHET1 overexpression was significantly associated with poor OS and DFS in cancer. Moreover, further subgroup analyses indicated that elevated GHET1 expression was significantly correlated with OS in each subgroup, regardless of the analysis model, sample size, cut-off value, treatment, cancer type, and NOS score. The pooled data illustrated that GHET1 overexpression was significantly associated with larger tumor size, positive lymph node metastasis, positive distant metastasis, and advanced TNM stage. Unexpectedly, we failed to identify an association between GHET1 expression and histological differentiation. However, these results might be not reliable because there was significant heterogeneity among the included studies. Overall, high GHET1 expression was an unfavorable risk factor for survival outcomes in patients with cancer; thus, GHET1 might be a valuable biomarker for a variety of cancers. To our knowledge, this research is the first meta-analysis focusing on the prognostic value of GHET1 in human cancers.
Many studies have tried to illustrate the correlation between high GHET1 expression and cancer prognosis; however, the molecular mechanism of GHET1 remained unclear [14, 17–19, 23, 24]. Feng et al. found that GHET1 overexpression promotes gastric cancer cell proliferation by binding to IGF2BP1 and enhancing the stability of c-Myc mRNA [14]. Further, GHET1 overexpression could prohibit cellular apoptosis by promoting the expression of Bcl-2 and could contribute to the development of multidrug resistance by promoting the expression of MDR1 and MRP1 in gastric cancer [16]. Xia et al. study revealed that down-regulation of GHET1 could prohibit the G1-S phase transition of the cell cycle in gastric cancer cells by modulating the expression of P21, cyclin, and CDK [21]. Moreover, upregulation of GHET1 could be induced by hypoxia in gastric cancer cells, and the depletion of GHET1 c significantly enhanced the CpG island methylation of EGFR, which plays a crucial role in the metastasis of cancers [32]. In hepatocellular carcinoma, Ding et al. found that high GHET1 expression could be activated by H3K27 acetylation, and could promote the progression and migration of cancer by physically binding to ATF1 [20]. In addition, Jin et al. showed that GHET1 can bind to the enhancer of EZH2 and recruit PRC2 to the promoter region of KLF2; KLF2 acts as a tumor suppressor in hepatocellular carcinoma and is epigenetically repressed [24]. As for lung cancer, Guan et al. revealed that GHET1 depletion inhibited the proliferation, invasion, and epithelial-mesenchymal transition (EMT) of cancer cells by inhibiting the LATS1/YAP pathway [19]. Additionally, several studies have reported that GHET1 can promote EMT in esophageal squamous cell carcinoma, breast cancer, colorectal cancer, osteosarcoma, renal cell carcinoma, and bladder cancer [17, 18, 22, 31, 33, 34]. Other studies have also shown that GHET1 promotes the progression of cancer and might be a therapeutic target of cancer [35, 36].
The present meta-analysis has several limitations. First, this meta-analysis included only 16 studies, and all of these studies were from China. Therefore, the results might only apply to Asian or Chinese patients, which may limit the representativeness of the results. The validation tests using data from TCGA make up this disadvantage in some extent. Second, the HRs and 95% CIs in several studies could not be directly obtained. Thus, we extracted the data from the Kaplan–Meier curve in these studies, which might introduce statistical errors. Finally, the sample sizes of some cancer types in this meta-analysis were limited; this may have contributed to the heterogeneity and may have affected the reliability of the pooled results for some cancer types. Nevertheless, the results of this meta-analysis should be verified by studies evaluating more cancer types with larger sample sizes.
Conclusions
In sum up, the up-regulation of lncRNA GHET1 expression was significantly associated with poor OS, poor DFS, and advanced clinicopathological characteristics in various cancers. GHET1 can be considered to be a promising prognostic predictor for human cancers. However, high-quality studies with larger samples sizes and those encompassing more cancer types are still needed to verify these conclusions.
Supplementary information
Additional file 1: Fig. S1. OS plots for each TCGA cohort.
Acknowledgements
We thank the overall participates in our research for their help.
Abbreviations
- BC
Breast cancer
- BLC
Bladder cancer
- ESCC
Esophageal squamous cell carcinoma
- GC
Gastric cancer
- HCC
Hepatocellular carcinoma
- PADC
Pancreatic cancer
- NSCLC
Non-small lung cancer
- qRT-PCR
Quantitative reverse transcription polymerase chain reaction
- CP
Clinicopathological parameters
- OS
Overall survival
- DFS
Disease-free survival
- HR
Hazard ratios
- OR
Odds ratio
- CI
Confidence intervals
- NOS
Newcastle–Ottawa Scale
Authors’ contributions
Designing the study: HGL Preparing the manuscript: DDW Concept of study: HZ Edit manuscript: DDW and XLF Prepare figures: XZ and HZ Data analysis: DDW and HZ Statistics: XLF. All authors read and approved the final manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 81502493) and Beijing Municipal Administration of Hospitals Clinical Medicine Development of Special Funding (No. ZYLX201814).
Availability of data and materials
The datasets during and/or analysis during the current study available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Dingding Wang and Hong Zhang have contributed equally to this article
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12935-020-01189-9.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Ng L, Poon RT, Pang R. Biomarkers for predicting future metastasis of human gastrointestinal tumors. CMLS. 2013;70(19):3631–3656. doi: 10.1007/s00018-013-1266-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JL, Cho KH, Park EC, Cho WH. A single measure of cancer burden combining incidence with mortality rates for worldwide application. APJCP. 2014;15(1):433–439. doi: 10.7314/apjcp.2014.15.1.433. [DOI] [PubMed] [Google Scholar]
- 4.Qin L, Chen C, Chen L, Xue R, Ou-Yang M, Zhou C, Zhao S, He Z, Xia Y, He J, et al. Worldwide malaria incidence and cancer mortality are inversely associated. Infect Agents Cancer. 2017;12:14. doi: 10.1186/s13027-017-0117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fidler MM, Gupta S, Soerjomataram I, Ferlay J, Steliarova-Foucher E, Bray F. Cancer incidence and mortality among young adults aged 20–39 years worldwide in 2012: a population-based study. Lancet Oncol. 2017;18(12):1579–1589. doi: 10.1016/S1470-2045(17)30677-0. [DOI] [PubMed] [Google Scholar]
- 6.Lin C, Yang L. Long noncoding RNA in cancer: wiring signaling circuitry. Trends Cell Biol. 2017;28(4):287–301. doi: 10.1016/j.tcb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013;45(8):1895–1910. doi: 10.1016/j.biocel.2013.05.030. [DOI] [PubMed] [Google Scholar]
- 8.Yang G, Lu X, Yuan L. LncRNA: a link between RNA and cancer. Biochem Biophys Acta. 2014;1839(11):1097–1109. doi: 10.1016/j.bbagrm.2014.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139(2):269–280. doi: 10.1002/ijc.30039. [DOI] [PubMed] [Google Scholar]
- 10.Bhan A, Mandal SS. LncRNA HOTAIR: a master regulator of chromatin dynamics and cancer. Biochem Biophys Acta. 2015;1856(1):151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou M, Zhao H, Wang Z, Cheng L, Yang L, Shi H, Yang H, Sun J. Identification and validation of potential prognostic lncRNA biomarkers for predicting survival in patients with multiple myeloma. J Exp Clin Cancer Res. 2015;34:102. doi: 10.1186/s13046-015-0219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giulietti M, Righetti A, Principato G, Piva F. LncRNA co-expression network analysis reveals novel biomarkers for pancreatic cancer. Carcinogenesis. 2018;39(8):1016–1025. doi: 10.1093/carcin/bgy069. [DOI] [PubMed] [Google Scholar]
- 13.Ning L, Li Z, Wei D, Chen H, Yang C. LncRNA, NEAT1 is a prognosis biomarker and regulates cancer progression via epithelial-mesenchymal transition in clear cell renal cell carcinoma. Cancer Biomarkers. 2017;19(1):75–83. doi: 10.3233/CBM-160376. [DOI] [PubMed] [Google Scholar]
- 14.Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y, Fang G. Long non-coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c-Myc mRNA stability. FEBS J. 2014;281(3):802–813. doi: 10.1111/febs.12625. [DOI] [PubMed] [Google Scholar]
- 15.Zhou HY, Zhu H, Wu XY, Chen XD, Qiao ZG, Ling X, Yao XM, Tang JH. Expression and clinical significance of long-non-coding RNA GHET1 in pancreatic cancer. Eur Rev Med Pharm Sci. 2017;21(22):5081–5088. doi: 10.26355/eurrev_201711_13822. [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Bo P, Liu L, Zhang X, Li J. Overexpression of long non-coding RNA GHET1 promotes the development of multidrug resistance in gastric cancer cells. Biomed Pharm. 2017;92:580–585. doi: 10.1016/j.biopha.2017.04.111. [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Zhen Q, Fan Y. LncRNA GHET1 promotes esophageal squamous cell carcinoma cells proliferation and invasion via induction of EMT. Int J Biol Markers. 2017;32(4):e403–e408. doi: 10.5301/ijbm.5000304. [DOI] [PubMed] [Google Scholar]
- 18.Li LJ, Zhu JL, Bao WS, Chen DK, Huang WW, Weng ZL. Long noncoding RNA GHET1 promotes the development of bladder cancer. Int J Clin Exp Pathol. 2014;7(10):7196–7205. [PMC free article] [PubMed] [Google Scholar]
- 19.Guan ZB, Cao YS, Li Y, Tong WN, Zhuo AS. Knockdown of lncRNA GHET1 suppresses cell proliferation, invasion and LATS1/YAP pathway in non small cell lung cancer. Cancer Biomarkers. 2018;21(3):557–563. doi: 10.3233/CBM-170431. [DOI] [PubMed] [Google Scholar]
- 20.Ding G, Li W, Liu J, Zeng Y, Mao C, Kang Y, Shang J. LncRNA GHET1 activated by H3K27 acetylation promotes cell tumorigenesis through regulating ATF1 in hepatocellular carcinoma. Biomed Pharm. 2017;94:326–331. doi: 10.1016/j.biopha.2017.07.046. [DOI] [PubMed] [Google Scholar]
- 21.Xia Y, Yan Z, Wan Y, Wei S, Bi Y, Zhao J, Liu J, Liao DJ, Huang H. Knockdown of long noncoding RNA GHET1 inhibits cellcycle progression and invasion of gastric cancer cells. Mol Med Rep. 2018;18(3):3375–3381. doi: 10.3892/mmr.2018.9332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song R, Zhang J, Huang J, Hai T. Long non-coding RNA GHET1 promotes human breast cancer cell proliferation, invasion and migration via affecting epithelial mesenchymal transition. Cancer Biomarkers. 2018;22(3):565–573. doi: 10.3233/CBM-181250. [DOI] [PubMed] [Google Scholar]
- 23.Shen QM, Wang HY, Xu S. LncRNA GHET1 predicts a poor prognosis of the patients with non-small cell lung cancer. Eur Rev Med Pharm Sci. 2018;22(8):2328–2333. doi: 10.26355/eurrev_201804_14823. [DOI] [PubMed] [Google Scholar]
- 24.Jin L, He Y, Tang S, Huang S. LncRNA GHET1 predicts poor prognosis in hepatocellular carcinoma and promotes cell proliferation by silencing KLF2. J Cell Physiol. 2018;233(6):4726–4734. doi: 10.1002/jcp.26257. [DOI] [PubMed] [Google Scholar]
- 25.Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8:16. doi: 10.1186/1745-6215-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013;339(2):159–166. doi: 10.1016/j.canlet.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 27.Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP, et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature. 2017;548(7667):343–346. doi: 10.1038/nature23451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson-Smith AC, Gingeras TR, Haerty W, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iwakiri J, Hamada M, Asai K. Bioinformatics tools for lncRNA research. Biochem Biophys Acta. 2016;1859(1):23–30. doi: 10.1016/j.bbagrm.2015.07.014. [DOI] [PubMed] [Google Scholar]
- 30.Shibayama Y, Fanucchi S, Magagula L, Mhlanga MM. lncRNA and gene looping: what’s the connection? Transcription. 2014;5(3):e28658. doi: 10.4161/trns.28658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou J, Li X, Wu M, Lin C, Guo Y, Tian B. Knockdown of long noncoding RNA GHET1 inhibits cell proliferation and invasion of colorectal cancer. Oncol Res. 2016;23(6):303–309. doi: 10.3727/096504016X14567549091305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Z, Wang R, Zhang T, Dong X. Hypoxia/lncRNA-AK123072/EGFR pathway induced metastasis and invasion in gastric cancer. Int J Clin Exp Med. 2015;8(11):19954–19968. [PMC free article] [PubMed] [Google Scholar]
- 33.Xie W, Chen Q, Liu X, Ma M, Yang X, Gong B, Sun T, Chen J. Silencing of the long non-coding RNA GHET1 inhibits cell proliferation and migration of renal cell carcinoma through epithelial-mesenchymal transition. Oncol Lett. 2019;17(3):3173–3180. doi: 10.3892/ol.2019.9967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang W, Shan Z, Zhou X, Peng L, Zhi C, Chai J, Liu H, Yang J, Zhang Z. Knockdown of lncRNA GHET1 inhibits osteosarcoma cells proliferation, invasion, migration and EMT in vitro and in vivo. Cancer Biomarkers. 2018;23(4):589–601. doi: 10.3233/CBM-181863. [DOI] [PubMed] [Google Scholar]
- 35.Huang H, Liao W, Zhu X, Liu H, Cai L. Knockdown of long noncoding RNA GHET1 inhibits cell activation of gastric cancer. Biomed Pharm. 2017;92:562–568. doi: 10.1016/j.biopha.2017.05.088. [DOI] [PubMed] [Google Scholar]
- 36.Ni W, Luo L, Zuo P, Li RP, Xu XB, Wen F, Hu D. lncRNA GHET1 down-regulation suppresses the cell activities of glioma. Cancer Biomarkers. 2018;23(1):9–22. doi: 10.3233/CBM-171002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. OS plots for each TCGA cohort.
Data Availability Statement
The datasets during and/or analysis during the current study available from the corresponding author on reasonable request.