Abstract
This study aimed to identify candidate biomarkers for predicting outcomes in nonalcoholic fatty liver disease (NAFLD) and hepatocellular carcinoma (HCC). Using Gene Expression Omnibus and The Cancer Genome Atlas (TCGA) databases, we identified common upregulated differential expressed genes (DEGs) in patients with NAFLD/nonalcoholic steatohepatitis (NASH) and HCC and conducted survival analysis of these upregulated DEGs with HCC outcomes. Two common upregulated DEGs including squalene epoxidase (SQLE) and EPPK1 messenger RNA (mRNA) were significantly upregulated in NAFLD, NASH, and HCC tissues, both in GSE45436 (P < .001) and TCGA profile (P < .001). Both SQLE and EPPK1 mRNA were upregulated in 15.56% and 8.06% patients with HCC in TCGA profile. Overexpression of SQLE in tumors was significantly associated with worse overall survival (OS) and disease-free survival (DFS) in patients with HCC (log-rank P = .027 and log-rank P = .048, respectively), while no statistical significances of OS and DFS were found in EPPK1 groups (both log-rank P > .05). For validation, SQLE upregulation contributed to significantly worse OS in patients wih HCC using Kaplan-Meier plotter analysis (hazard ratio = 1.43, 95% confidence interval: 1.01-2.02, log-rank P = .043). In addition, high level of SQLE significantly associated with advanced neoplasm histologic grade, advanced AJCC stage, and α-fetoprotein elevation (P = .036, .045, and .029, respectively). Squalene epoxidase is associated with OS and DFS and serves as a novel prognostic biomarker for patients with HCC.
Keywords: squalene epoxidase, hepatocellular carcinoma, α-fetoprotein, survival, AJCC stage
Introduction
Liver cancer, comprising 75% to 85% cases of hepatocellular carcinoma (HCC), is predicted to be the sixth most commonly diagnosed cancer and the fourth leading cancer-related deaths worldwide in 2018, with about 841 000 new cases and 782 000 deaths annually.1-4 In addition, the incidence of HCC will continue to rise until 2030 based on a the Surveillance, Epidemiology, and End Results (SEER) program registry projects study.5 Precise estimation of prognosis plays a critical role in treatment decision in patients with HCC. Thus, identifying novel biomarkers for predicting HCC prognosis and revealing target for treatment are urgently required.6,7
The rising prevalence of obesity is considered a contributory factor to the observed increasing incidence of HCC.8 Emerging evidences revealed that paralleling the epidemic of metabolic syndrome and obesity worldwide,9 nonalcoholic fatty liver disease (NAFLD) and nonalcoholic steatohepatitis (NASH) served as independent risk factors for HCC, even in the absence of cirrhosis.10-13 The pathogenesis of HCC arising in the context of NAFLD as underlying liver disease has not been well elucidated.14 The molecular classifications of HCCs did not provide a signature pathognomonic for HCC in NAFLD/NASH.15 In this study, we identified common upregulated differential expressed genes (DEGs) in patients with NAFLD/NASH and HCC and conducted survival analysis of these upregulated DEGs with HCC outcomes, in hope of providing screening suggestions of NAFLD/NASH and useful insights into the pathogenesis and progression of HCC.
Materials and Methods
Source of Data
GSE59045 and GSE45436 profiles were obtained from Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/) database. Patients with NAFLD/NASH in GSE59045 were grouped according to liver tissue histology: 6 patients in group I (<5% steatosis), 4 in group II (NAFLD, 30% to 50% steatosis), and 5 in group III (NASH).16 GSE45436 were composed with GSE45267, GSE45434 (training sets), and GSE45435 (validation set). Eighteen frozen tumorous and adjacent nontumorous liver tissues were used for gene expression profiling study in GSE60502 and 14 pairs of HCC tissues and corresponding noncancerous tissues were isolated and purified in GSE84402. Microarray analyses were performed to identify genes expressed differentially among groups. Affymetrix Human Gene Expression Array platform was used for microarray analysis in GSE59045, Affymetrix Human Genome U133A Array was used in GSE60502, and Affymetrix Human Genome U133 Plus 2.0 Array platforms were used in GSE45436 and GSE84402.
Screening Commonly Upregulated DEGs
The gene expression data were processed using the Robust Multi-array Average algorithm. To investigate DEGs in transcriptome between groups including normal, NAFLD, and NASH in morbidly obese patients, and tumor and adjacent normal tissues in patients with HCC, Affy, AffyPLM, and Limma packages were used for quality assessment and identifying DEGs in each GEO profile based on the microarray platform. The criteria for selection of DEGs were set as ∣log2FC∣> 1 and adjusted P value <.05. To identify upregulated DEGs, log2FC > 1 and adjusted P value <.05 were set. To identify commonly upregulated DEGs among GSE59045 and GSE45436, E Chart online service (http://www.ehbio.com/ImageGP/index.php/Home/Index/index.html) for Venn diagram was used.
Survival Analysis
Liver Hepatocellular Carcinoma (The Cancer Genome Atlas [TCGA], Provisional) database in cBioPortal for cancer genomics web service was used for identifying potential candidate biomarkers for predicting the overall survival (OS) and disease-free survival (DFS) of patients with HCC.17,18 Messenger RNA (mRNA) expression levels calculated by log2 calculation were compared based on clinical attribute in patients with HCC. To evaluate associations between candidate biomarkers and survival and clinicopathological features in patients with HCC, gene data with Z scores and clinical data of patients with HCC in Liver Hepatocellular Carcinoma (TCGA, Provisional) database were downloaded from cBioPortal and matched using VLOOKUP index in Excel, Microsoft Office 2016. After excluding 10 patients with liver histology of hepatocholangiocarcinoma (n = 7) and fibrolamellar carcinoma (n = 3) and 6 patients without gene expression levels, 361 patients with HCC were included in the analysis. Additionally, the Kaplan-Meier plotter online service (http://kmplot.com/analysis/)19 was used for validation of candidates with auto select best cutoff and OS in patients with HCC.
Statistical Analysis
Differences of gene expression between the individual groups were analyzed using Mann-Whitney U test, χ2 test, and Ridit analysis based on variables types. PASW Statistics software version 23.0 from SPSS Inc (Chicago, Illinois) was used. A 2-tailed P < .05 was considered significant for all tests.
Results
Identification of Commonly Upregulated DEGs in NAFLD, NASH, and HCC Tumors
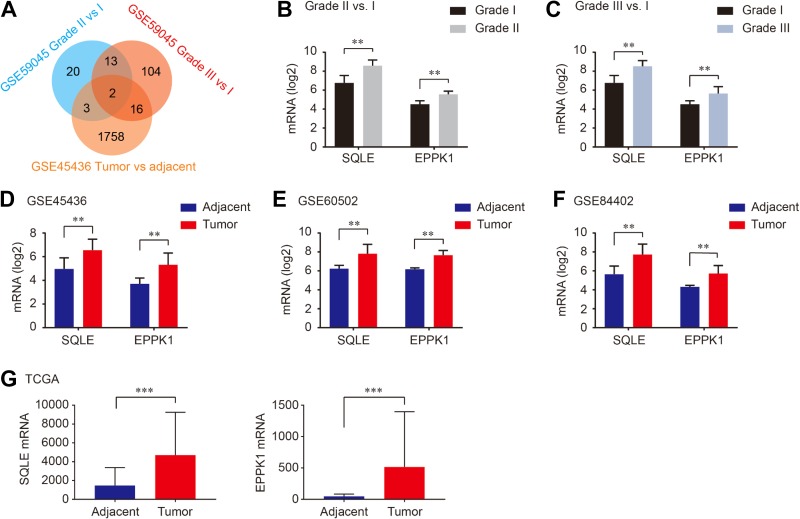
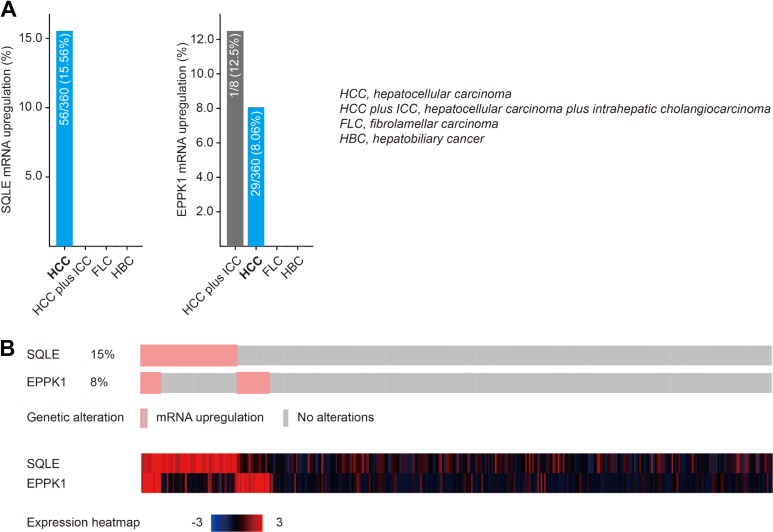
Gene expression in liver of morbidly obese patients was conducted in GSE59045. We compared upregulated DEGs between patients with NAFLD/NASH and obese patients with liver histology <5% steatosis. Then we identified upregulated DEGs in tumor and nontumor tissues from patients with HCC using GSE45436 profile. As shown in Figure 1, 2 common upregulated DEGs including squalene epoxidase (SQLE) and EPPK1 were identified in NAFLD, NASH, and HCC tumors (Figure 1A). As shown in Figure 1B and C, SQLE and EPPK1 mRNA were significantly overexpressed in patients with NAFLD and NASH compared to that in obese cases <5% steatosis (all P < .01; Figure 1B and C). As we expect, SQLE and EPPK1 mRNA were significantly upregulated in tumor tissues in patients with HCC in GSE45436, GSE60502, and GSE84402 (all P <.01; Figure 1D-F), which was validated in TCGA (both P < .001; Figure 1G). In addition, SQLE and EPPK1 mRNA were upregulated in 15.56% and 8.06% patients with HCC in TCGA profile using cBioPortal online analysis (Figure 2A). The genetic alteration and heatmap of SQLE and EPPK1 are described in Figure 2B.
Figure 1.
Two commonly upregulated DEGs including SQLE and EPPK1 were identified between GSE59045 and GSE45436 (A), SQLE mRNA and EPPK1 mRNA were overexpressed in steatosis grade II and grade III compared with grade I (B and C), SQLE mRNA and EPPK1 mRNA were upregulated in tumor tissues than those in adjacent tissues in patients with HCC (D-F), which was validated in TCGA database (G). DEGs indicates differential expressed genes; HCC, hepatocellular carcinoma; mRNA, messenger RNA; SQLE, squalene epoxidase; TCGA, The Cancer Genome Atlas.
Figure 2.
Upregulation frequency of SQLE and EPPK1 mRNA in different liver cancer types using cBioPortal online analysis. mRNA indicates messenger RNA; SQLE, squalene epoxidase.
Associations Between SQLE and EPPK1 and HCC Survivals
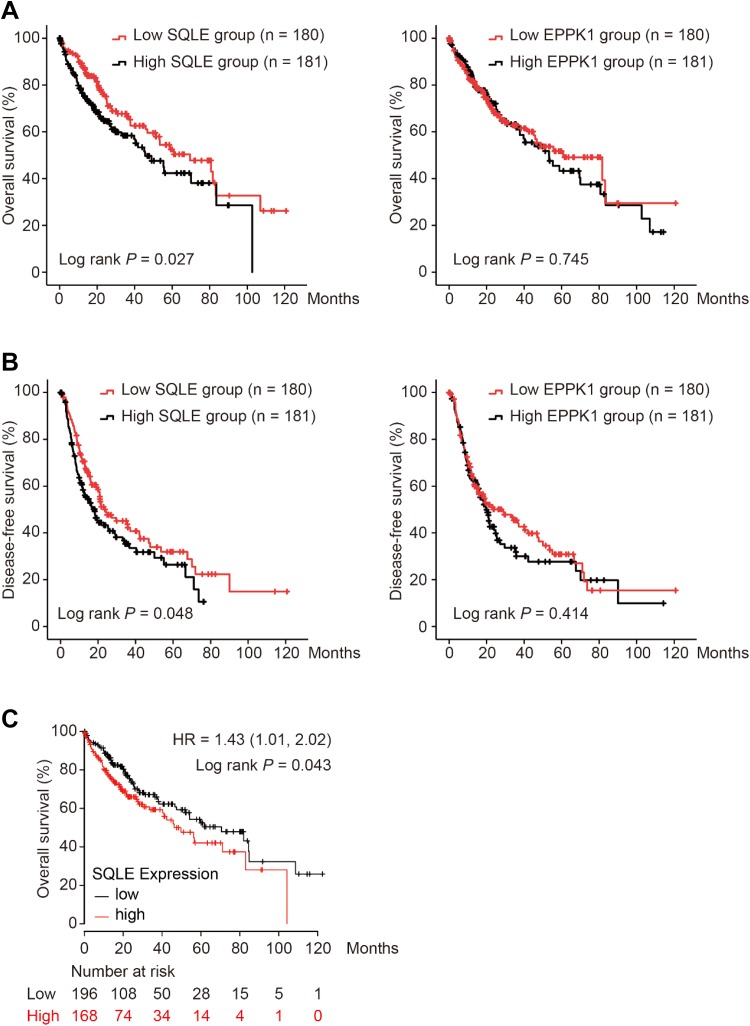
Using Liver Hepatocellular Carcinoma (TCGA, Provisional) database in cBioPortal for cancer genomics, we grouped patients with HCC with median cutoffs of SQLE and EPPK1. As shown in Figure 3, overexpression of SQLE in tumors was significantly associated with worse OS in patients with HCC (log-rank P = .027; Figure 3A), while no difference in OS was found in EPPK1 groups (log-rank P = .745; Figure 3A). Moreover, high-level SQLE in tumor tissues was significantly associated with poor DFS in patients with HCC (log-rank P = .048; Figure 3B), and no statistical significance was observed in DFS comparison in EPPK1 groups (log-rank P = .414; Figure 3B). For validation, we performed OS analysis using Kaplan-Meier plotter. As shown in Figure 3C, SQLE upregulation contributed to significantly worse OS in patients with HCC (hazard ratio = 1.43, 95% confidence interval = 1.01-2.02, log-rank P = .043; Figure 3C).
Figure 3.
Comparison of overall survival (A) and disease-free survival (B) in patients with HCC grouped by SQLE and EPPK1 median cutoffs in TCGA database, and overall survival analysis for validation of SQLE was performed using Kaplan-Meier plotter (C). HCC indicates hepatocellular carcinoma; SQLE, squalene epoxidase; TCGA, The Cancer Genome Atlas.
Associations Between SQLE and Clinicopathological Characteristics in Patients With HCC
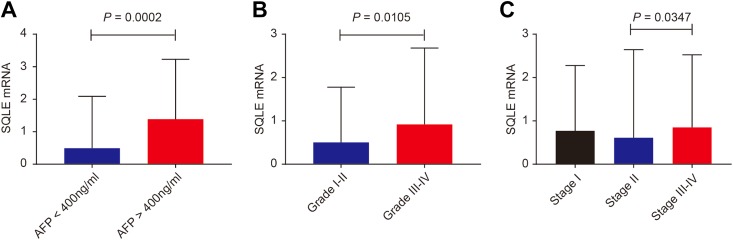
Clinicopathological characteristics comparison grouped by SQLE median are summarized in Table 1. High level of SQLE significantly contributed to advanced neoplasm histologic grade, advanced AJCC stage, and α-fetoprotein (AFP) elevation (P = .036, .045, and .029, respectively; Table 1). In addition, we compared SQLE mRNA expression levels based on neoplasm histologic grade, AJCC stage, and AFP level. As shown in Figure 4, SQLE mRNA was significantly overexpressed in patients with AFP >400 ng/mL and neoplasm histologic grade III-IV compared to those with AFP <400 ng/mL and neoplasm grade I-II (P = .0002 and P = .0105, respectively; Figure 4A and B). Compared to patients with HCC with AJCC stage II, those with AJCC stage III-IV had significantly higher SQLE mRNA levels (P = .0347; Figure 4C).
Table 1.
Characteristics of Patients With HCC Between SQLE High and SQLE Low Groups.
| Variables | SQLE Expression Level | P Value | |
|---|---|---|---|
| Low (n = 180) | High (n = 181) | ||
| Gender, male (%) | 117 (65.0) | 127 (70.2) | .294 |
| Age, median (IQR), years | 61 (18) | 60 (19) | .849 |
| BMI, kg/m2, n (%) | .491 | ||
| <18.5 | 13 (7.2) | 8 (4.4) | |
| 18.5-24.99 | 75 (41.7) | 77 (42.5) | |
| 25-29.99 | 42 (23.3) | 46 (25.4) | |
| >30 | 38 (21.1) | 29 (16.0) | |
| Race, n (%) | .14 | ||
| Asian | 70 (38.9) | 86 (47.5) | |
| White | 96 (53.3) | 80 (44.2) | |
| Black or African American | 7 (3.9) | 11 (6.1) | |
| Tumor status, n (%) | .415 | ||
| With tumor | 51 (28.3) | 58 (32.0) | |
| Tumor free | 117 (65.0) | 110 (60.8) | |
| Family history of cancer, n (%) | 56 (31.1) | 53 (29.3) | .705 |
| Hepatocarcinoma risk factors, n (%) | .255 | ||
| Hepatitis virus infection | 50 (27.8) | 61 (33.7) | |
| Alcohol consumption | 36 (20.0) | 34 (18.8) | |
| Hepatitis virus plus alcohol consumption | 25 (13.9) | 14 (7.8) | |
| Nonalcoholic fatty liver disease | 8 (4.4) | 7 (3.9) | |
| No risk factors | 38 (21.1) | 48 (26.5) | |
| Neoplasm histologic grade, n (%) | .036 | ||
| G1 | 28 (15.6) | 25 (13.8) | |
| G2 | 87 (48.3) | 84 (46.4) | |
| G3 | 60 (33.3) | 61 (33.7) | |
| G4 | 1 (0.6) | 11 (6.1) | |
| AJCC stage, n (%) | .045 | ||
| I | 78 (43.3) | 89 (49.2) | |
| II | 49 (27.2) | 33 (18.2) | |
| III | 33 (18.3) | 51 (28.2) | |
| IV | 1 (0.6) | 3 (1.7) | |
| Vascular invasion, n (%) | .362 | ||
| Macro | 9 (5.0) | 7 (3.9) | |
| Micro | 49 (27.2) | 40 (22.1) | |
| None | 98 (54.4) | 102 (56.4) | |
| Child-Pugh classification, n (%) | .784 | ||
| A | 105 (58.3) | 107 (59.1) | |
| B | 10 (5.6) | 11 (6.1) | |
| C | 1 (0.6) | 0 (0) | |
| NA | 64 (35.6) | 63 (34.8) | |
| AFP > 400 ng/mL, n (%) | 24 (13.3) | 40 (22.1) | .029 |
| Platelet, ×103/mm3, n (%) | .98 | ||
| <100 | 9 (5.0) | 7 (3.9) | |
| 100-199 | 62 (34.4) | 51 (28.2) | |
| 200-299 | 49 (27.2) | 46 (25.4) | |
| 300-399 | 14 (7.8) | 14 (7.8) | |
| >400 | 24 (13.3) | 20 (11.0) | |
| New tumor event after initial treatment, n (%) | 46 (25.6) | 47 (26.0) | .929 |
| Ishak fibrosis status, n (%) | .676 | ||
| No fibrosis | 32 (17.8) | 40 (22.1) | |
| Portal fibrosis | 17 (9.4) | 14 (7.8) | |
| Fibrous septa | 13 (7.2) | 15 (8.3) | |
| Nodular formation/incomplete cirrhosis | 5 (2.8) | 4 (2.2) | |
| Cirrhosis | 39 (21.7) | 31 (17.1) | |
| NA | 74 (41.1) | 77 (42.5) | |
| Hepatic inflammation, n (%) | .9 | ||
| None | 60 (33.3) | 54 (29.8) | |
| Mild | 54 (30.0) | 43 (23.8) | |
| Severe | 10 (5.6) | 8 (4.4) | |
| NA | 56 (31.1) | 76 (42.0) | |
| Follow up, median (IQR), years | 0.41 (1.8) | 0.27 (1.4) | .116 |
Abbreviations: AJCC, American Joint Committee on Cancer; AFP, α-fetoprotein; BMI, body mass index; HCC, hepatocellular carcinoma; IQR, interquartile range; NA, not available; SQLE, squalene epoxidase.
Figure 4.
Squalene epoxidase mRNA levels grouped by α-fetoprotein (A), neoplasm histologic grade (B) and AJCC stage (C). mRNA indicates messenger RNA
Diseases, Genes, and Functions Associated With SQLE
We searched diseases associated with SQLE in DisGeNET database. As shown in Table 2, liver neoplasm, liver carcinoma, and hypercholesterolemia were all included in the top 10 scored diseases associated with SQLE (Table 2). However, the scores were low and PMIDs were few. Therefore, more studies evaluating associations between SQLE and liver cancer and cholesterol synthesis are needed in future.
Table 2.
Top 10 Scored Disease Associations for SQLE in DisGeNET Database.
| Disease | Entry Name | Score | PMIDs |
|---|---|---|---|
| C0023904 | Liver Neoplasms, Experimental | 0.2 | 1 |
| C1458155 | Mammary Neoplasms | 0.003 | 1 |
| C0006142 | Malignant Neoplasm of Breast | <0.001 | 2 |
| C0678222 | Breast Carcinoma | <0.001 | 2 |
| C0600139 | Prostate Carcinoma | <0.001 | 2 |
| C0376358 | Malignant Neoplasm of Prostate | <0.001 | 2 |
| C2239176 | Liver Carcinoma | <0.001 | 2 |
| C0020443 | Hypercholesterolemia | <0.001 | 1 |
| C1335302 | Pancreatic Ductal Adenocarcinoma | <0.001 | 1 |
| C1301034 | Pancreatic Intraepithelial Neoplasia | <0.001 | 1 |
Abbreviation: SQLE, squalene epoxidase.
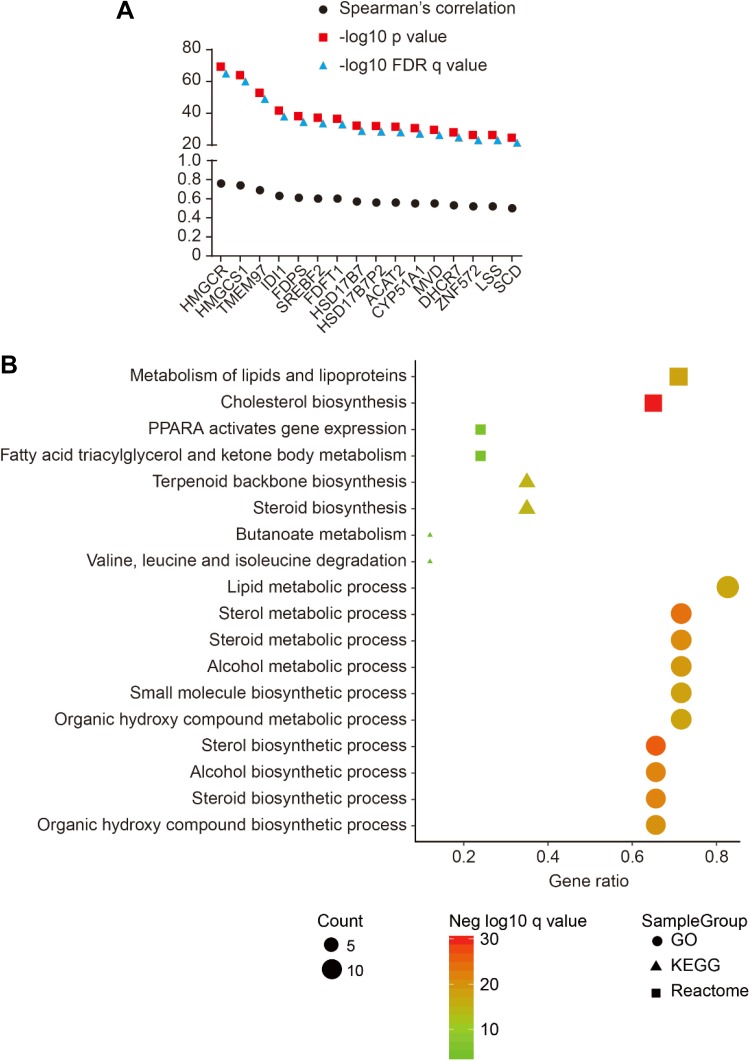
In Liver Hepatocellular Carcinoma (TCGA, Provisional) database, we searched coexpressed genes of SQLE with Spearman correlations >0.5; totally 16 genes were identified (Figure 5A). In Gene Set Enrichment Analysis database, most SQLE-coexpressed genes were enriched in Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, GO biological process, and Reactome associated with cholesterol, sterol/steroid biosynthesis processes (Figure 5B).
Figure 5.
Coexpressed genes of SQLE with Spearman correlation >0.5 (A) and KEGG, GO, and Reactome enrichment of SQLE-coexpressed genes in GSEA (B). GSEA, Gene Set Enrichment Analysis; SQLE, squalene epoxidase.
Discussion
Hepatocellular carcinoma can arise in the context of noncirrhotic liver in patients with NAFLD/NASH, suggesting a specific carcinogenic pathway. Pathology studies have also described steatohepatitic HCC as a specific histological variant.20-22 Patients are often diagnosed with HCC in the advanced NAFLD stage because of the absence of efficient surveillance policies in this population. Management of patients with HCC with NAFLD is also complicated by comorbidities, mainly cardiac disease and diabetes, which negatively affect eligibility for radical treatments.23 Hence, finding commonly upregulated DEGs in NAFLD/NASH and HCC tumors might provide novel insights into the pathogenesis, progression, and therapeutic strategy in patients with HCC.
In our analysis, we found that SQLE and EPPK1 were commonly upregulated DEGs during chronic liver disease process including NAFLD, NASH, and HCC. Unfortunately, further analysis demonstrated that only SQLE was significantly associated with HCC survivals and clinicopathological features including AFP elevation and advanced tumor stages. Located in the endoplasmic reticulum, SQLE was found to be the one of the key rate-limiting enzymes in the cholesterol biosynthesis.24-26 Squalene epoxidase catalyzes the first oxygenation step of the cholesterol biosynthetic pathway, the conversion of squalene to 2,3-oxidosqualene.24,27,28 Recent studies have shown that SQLE is involved in the development and metastasis of the tumorigenesis. A report by Sui et al indicated that the expression of SQLE was upregulated in the HCC tissues. And, overexpression of SQLE in HCC cells promoted cell proliferation and migration, while downregulation of SQLE inhibited the tumorigenicity of HCC cells in vitro and in vivo. Mechanistically, SQLE positively regulated the extracellular signal-regulated kinase signaling.29 Squalene epoxidase exerts its oncogenic effect via its metabolites, cholesteryl ester and nicotinamide adenine dinucleotide phosphate. Squalene epoxidase is expressed at very low levels in most of the noncholesterolemic tissues and is found in greatest abundance in liver. Increased SQLE expression promotes the biosynthesis of cholesteryl ester, which induces NAFLD-HCC cell growth.30 Suppression of tumor growth by blockade of SQLE function is associated with decreased cholesteryl ester concentrations, restoration of PTEN expression, and inhibition of AKT-mTOR.30 Considering previous study, we assumed that associated with NAFLD progression, SQLE might contribute to the disease aggressiveness of patients with HCC, especially in NAFLD/NASH population. We also suggested further basic study focusing mechanisms of SQLE in the pathogenesis and progression of NAFLD-related patients with HCC.
In breast cancer, frequent MYC gene substantial coamplification and aberrant methylation of SQLE promoter were observed and reduced patient survival.31,32 Overexpression of SQLE was more prevalent in aggressive breast cancer and was an independent prognostic factor of unfavorable outcome. Inhibition of SQLE resulted in a copy-dosage correlated decrease in cell viability.33,34 Squalene epoxidase and other genes involved in cholesterol biosynthesis were consistently associated with radioresistance in the pancreatic cancer.35,36 In addition, low expression level of SQLE was associated with a better prognosis in patients with colorectal cancer.37 Moreover, differential expression of SQLE was confirmed in tumor tissue in human primary lung squamous cell carcinoma and prostate acinar cancer.38,39 The expression of SQLE mRNA was closely correlated with poor differentiation, clinical stages, lymphatic metastasis, and OS in squamous cell lung carcinoma.40 Also, SQLE could induce epithelial-to-mesenchymal transition by regulating of miR-133b in esophageal squamous cell carcinoma.41
Existing literatures mainly focused on the potential mechanisms of SQLE in the promotion of NAFLD and HCC. The correlations between SQLE and HCC have not yet been illustrated. Our research was of great importance in investigating the oncogenic effects of SQLE in patients with HCC. Additionally, our findings should be considered in the context of its limitations. First, SQLE was examined in transcription levels, not in protein levels. Second, no mechanisms of these genes were conducted, such as gene silencing approaches. Based on our results and previous publications, we proposed that SQLE is an oncogene in many human malignances including NAFLD-HCC and repurposing SQLE inhibitors targeting SQLE-induced cholesterol synthesis pathway may be a promising approach for the prevention and treatment of NAFLD-HCC.42-44
Supplemental Material
Supplemental Material, upregulated_DEGs_in_GSE59045_and_GSE45436 for High Squalene Epoxidase in Tumors Predicts Worse Survival in Patients With Hepatocellular Carcinoma: Integrated Bioinformatic Analysis on NAFLD and HCC by Tingting Shen, Yunfei Lu and Qin Zhang in Cancer Control
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article
ORCID iD: Yunfei Lu, MD, PhD  https://orcid.org/0000-0002-9292-2517
https://orcid.org/0000-0002-9292-2517
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. [DOI] [PubMed] [Google Scholar]
- 2. Heimbach JK, Kulik LM, Finn RS, et al. AASLD guidelines for the treatment of hepatocellular carcinoma. Hepatology. 2018;67(1):358–380. [DOI] [PubMed] [Google Scholar]
- 3. Omata M, Cheng AL, Kokudo N, et al. Asia-Pacific clinical practice guidelines on the management of hepatocellular carcinoma: a 2017 update. Hepatol Int. 2017;11(4):317–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. [DOI] [PubMed] [Google Scholar]
- 5. Petrick JL, Kelly SP, Altekruse SF, McGlynn KA, Rosenberg PS. Future of hepatocellular carcinoma incidence in the United States forecast through 2030. J Clin Oncol. 2016;34(15):1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guo W, Tan HY, Wang N, Wang X, Feng Y. Deciphering hepatocellular carcinoma through metabolomics: from biomarker discovery to therapy evaluation. Cancer Manag Res. 2018;10(4):715–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang B, Finn RS. Personalized clinical trials in hepatocellular carcinoma based on biomarker selection. Liver Cancer. 2016;5(3):221–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marengo A, Rosso C, Bugianesi E. Liver cancer: connections with obesity, fatty liver, and cirrhosis. Annu Rev Med. 2016;67:103–117. [DOI] [PubMed] [Google Scholar]
- 9. Said A, Ghufran A. Epidemic of non-alcoholic fatty liver disease and hepatocellular carcinoma. World J Clin Oncol. 2017;8(6):429–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baffy G, Brunt EM, Caldwell SH. Hepatocellular carcinoma in non-alcoholic fatty liver disease: an emerging menace. J Hepatol. 2012;56(6):1384–1391. [DOI] [PubMed] [Google Scholar]
- 11. Marrero JA, Fontana RJ, Su GL, Conjeevaram HS, Emick DM, Lok AS. NAFLD may be a common underlying liver disease in patients with hepatocellular carcinoma in the United States. Hepatology. 2002;36(6):1349–1354. [DOI] [PubMed] [Google Scholar]
- 12. Starley BQ, Calcagno CJ, Harrison SA. Nonalcoholic fatty liver disease and hepatocellular carcinoma: a weighty connection. Hepatology. 2010;51(5):1820–1832. [DOI] [PubMed] [Google Scholar]
- 13. Klein S, Dufour JF. Nonalcoholic fatty liver disease and hepatocellular carcinoma. Hepat Oncol. 2017;4(3):83–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zoller H, Tilg H. Nonalcoholic fatty liver disease and hepatocellular carcinoma. Metabolism. 2016;65(8):1151–1160. [DOI] [PubMed] [Google Scholar]
- 15. Schulze K, Imbeaud S, Letouze E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47(5):505–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. du Plessis J, van Pelt J, Korf H, et al. Association of adipose tissue inflammation with histologic severity of nonalcoholic fatty liver disease. Gastroenterology. 2015;149(3):635–648. e614. [DOI] [PubMed] [Google Scholar]
- 17. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Szasz AM, Lanczky A, Nagy A, et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7(31):49322–49333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salomao M, Remotti H, Vaughan R, Siegel AB, Lefkowitch JH, Moreira RK. The steatohepatitic variant of hepatocellular carcinoma and its association with underlying steatohepatitis. Hum Pathol. 2012;43(5):737–746. [DOI] [PubMed] [Google Scholar]
- 21. Shibahara J, Ando S, Sakamoto Y, Kokudo N, Fukayama M. Hepatocellular carcinoma with steatohepatitic features: a clinicopathological study of Japanese patients. Histopathology. 2014;64(7):951–962. [DOI] [PubMed] [Google Scholar]
- 22. Yeh MM, Liu Y, Torbenson M. Steatohepatitic variant of hepatocellular carcinoma in the absence of metabolic syndrome or background steatosis: a clinical, pathological, and genetic study. Hum Pathol. 2015;46(11):1769–1775. [DOI] [PubMed] [Google Scholar]
- 23. Degasperi E, Colombo M. Distinctive features of hepatocellular carcinoma in non-alcoholic fatty liver disease. Lancet Gastroenterol Hepatol. 2016;1(2):156–164. [DOI] [PubMed] [Google Scholar]
- 24. Nagai M, Sakakibara J, Wakui K, et al. Localization of the squalene epoxidase gene (SQLE) to human chromosome region 8q24.1. Genomics. 1997;44(1):141–143. [DOI] [PubMed] [Google Scholar]
- 25. Nusbaum C, Mikkelsen TS, Zody MC, et al. DNA sequence and analysis of human chromosome 8. Nature. 2006;439(7074):331–335. [DOI] [PubMed] [Google Scholar]
- 26. Chugh A, Ray A, Gupta JB. Squalene epoxidase as hypocholesterolemic drug target revisited. Prog Lipid Res. 2003;42(1):37–50. [DOI] [PubMed] [Google Scholar]
- 27. Ha J, Kwon S, Hwang JH, et al. Squalene epoxidase plays a critical role in determining pig meat quality by regulating adipogenesis, myogenesis, and ROS scavengers. Sci Rep. 2017;7(1):16740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gill S, Stevenson J, Kristiana I, Brown AJ. Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 2011;13(3):260–273. [DOI] [PubMed] [Google Scholar]
- 29. Sui Z, Zhou J, Cheng Z, Lu P. Squalene epoxidase (SQLE) promotes the growth and migration of the hepatocellular carcinoma cells. Tumour Biol. 2015;36(8):6173–6179. [DOI] [PubMed] [Google Scholar]
- 30. Liu D, Wong CC, Fu L, et al. Squalene epoxidase drives NAFLD-induced hepatocellular carcinoma and is a pharmaceutical target. Sci Transl Med. 2018;10(437):9. [DOI] [PubMed] [Google Scholar]
- 31. Parris TZ, Kovacs A, Hajizadeh S, et al. Frequent MYC coamplification and DNA hypomethylation of multiple genes on 8q in 8p11-p12-amplified breast carcinomas. Oncogenesis. 2014;3:e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Helms MW, Kemming D, Pospisil H, et al. Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease. Br J Cancer. 2008;99(5):774–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown DN, Caffa I, Cirmena G, et al. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci Rep. 2016;6(1):19435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Polycarpou-Schwarz M, Gross M, Mestdagh P, et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene. 2018;37(34):4750–4768. [DOI] [PubMed] [Google Scholar]
- 35. Souchek JJ, Baine MJ, Lin C, et al. Unbiased analysis of pancreatic cancer radiation resistance reveals cholesterol biosynthesis as a novel target for radiosensitisation. Br J Cancer. 2014;111(6):1139–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harada T, Chelala C, Crnogorac-Jurcevic T, Lemoine NR. Genome-wide analysis of pancreatic cancer using microarray-based techniques. Pancreatology. 2009;9(1-2):13–24. [DOI] [PubMed] [Google Scholar]
- 37. Yuen HF, McCrudden CM, Huang YH, et al. TAZ expression as a prognostic indicator in colorectal cancer. PLoS One. 2013;8(1):e54211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Y, Sun W, Zhang K, et al. Identification of genes differentially expressed in human primary lung squamous cell carcinoma. Lung Cancer. 2007;56(3):307–317. [DOI] [PubMed] [Google Scholar]
- 39. Jardel P, Debiais C, Godet J, Irani J, Fromont G. Ductal carcinoma of the prostate shows a different immunophenotype from high grade acinar cancer. Histopathology. 2013;63(1):57–63. [DOI] [PubMed] [Google Scholar]
- 40. Zhang HY, Li HM, Yu Z, Yu XY, Guo K. Expression and significance of squalene epoxidase in squamous lung cancerous tissues and pericarcinoma tissues. Thorac Cancer. 2014;5(4):275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Qin Y, Zhang Y, Tang Q, Jin L, Chen Y. SQLE induces epithelial-to-mesenchymal transition by regulating of miR-133b in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai). 2017;49(2):138–148. [DOI] [PubMed] [Google Scholar]
- 42. Cirmena G, Franceschelli P, Isnaldi E, et al. Squalene epoxidase as a promising metabolic target in cancer treatment. Cancer Lett. 2018;425:13–20. [DOI] [PubMed] [Google Scholar]
- 43. Gotteland JP, Loubat C, Planty B, Junquero D, Delhon A, Halazy S. Sulfonamide derivatives of benzylamine block cholesterol biosynthesis in HepG2 cells: a new type of potent squalene epoxidase inhibitors. Bioorg Med Chem Lett. 1998;8(11):1337–1342. [DOI] [PubMed] [Google Scholar]
- 44. Sawada M, Matsuo M, Hagihara H, et al. Effect of FR194738, a potent inhibitor of squalene epoxidase, on cholesterol metabolism in HepG2 cells. Eur J Pharmacol. 2001;431(1):11–16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, upregulated_DEGs_in_GSE59045_and_GSE45436 for High Squalene Epoxidase in Tumors Predicts Worse Survival in Patients With Hepatocellular Carcinoma: Integrated Bioinformatic Analysis on NAFLD and HCC by Tingting Shen, Yunfei Lu and Qin Zhang in Cancer Control