Summary
Background
An outbreak of acute encephalitis of unknown origin with high case fatality (183 of 329 cases) was reported in children from Andhra Pradesh state in southern India during 2003. We investigated the causative agent.
Methods
Cell lines and peripheral blood lymphocyte co-cultures were used to isolate the causative agent from clinical samples. Identity of the agent was established by electron microscopy and serological and molecular assays.
Findings
Clinical samples tested negative for IgM antibodies to Japanese encephalitis, West Nile, dengue, and measles viruses, and for RNA of coronavirus, paramyxovirus, enterovirus, and influenza viruses. Virus was isolated from six patients with encephalitis and was identified as Chandipura virus by electron microscopy, complement fixation, and neutralisation tests. Chandipura virus RNA was detected in clinical samples from nine patients. Sequencing of five of these RNA samples showed 96·7–97·5% identity with the reference strain of 1965. Chandipura viral antigen and RNA were detected in brain tissue of a deceased child by immunofluorescent antibody test and PCR. Neutralising, IgG, and IgM antibodies to Chandipura virus were present in some patients' serum samples. Serum samples obtained after 4 days of illness were more frequently positive for IgM to Chandipura virus than were those obtained earlier (p<0·001). A similar trend was noted for neutralising antibodies.
Interpretation
Our findings suggest that this outbreak of acute encephalitis in Andhra Pradesh was associated with Chandipura virus, adding to the evidence suggesting that this virus should be considered as an important emerging pathogen.
Introduction
Viral encephalitis is an important global public-health problem. In India, although many encephalitis outbreaks have been associated with Japanese encephalitis virus,1 several outbreaks have remained undiagnosed. One such outbreak was documented in Jamshedpur as early as 1954.2 A group of patients was characterised by sudden onset of high-grade fever (101–106°F), occasional vomiting, rigors, and drowsiness leading to unconsciousness, followed by death in 6–48 h. The age of the affected children ranged from 2·5 months to 15 years. The case fatality rate was 52·3%, and CSF findings were within normal limits. The cause was thought to be viral, but laboratory findings were inconclusive. Subsequently, outbreaks of a similar nature were described from Nagpur, Raipur, Bilaspur, and nearby areas of central India in the years 1958, 1965, 1968, 1978, and 1983. Similar outbreaks were reported from Warangal in Andhra Pradesh in 1997 and 2002. In the absence of a defined cause, these outbreaks were tentatively attributed to Reye's syndrome, dengue, chikungunya, Japanese encephalitis, measles, and so on.3, 4, 5
An outbreak of encephalitis was reported between June and September, 2003, in Andhra Pradesh and adjoining areas in the Maharashtra state of India. We report our investigation into the cause of this outbreak.
Methods
Affected area and patients
Andhra Pradesh is a state in southern India situated between 77–84° east and 13–19° north. It is divided into three main regions: Telangana (ten districts), Rayalseema (four districts), and a coastal region of nine districts. Most of the encephalitis cases were reported from the Telangana region. The state's general population density is 200–250 people per km2. There are three distinct seasons: summer (March to July with temperature range of 36–49°C), monsoon (July to December having an average rainfall of 56·00 mm), and winter from December to February (temperature range 20–40°C).
The state health authorities of Andhra Pradesh undertook surveillance to detect cases of encephalitis, with a broad case definition of acute fever with CNS involvement and negative for other known causes of illness. Clinical samples were collected from three groups: an encephalitis group, based on the case definition used by the state government; a fever group—ie, fever without CNS involvement; and a family-contact group—ie, no fever and no CNS involvement. Samples obtained were: 54 blood samples, 22 throat swabs, ten CSF samples, and one brain aspirate from 55 patients with encephalitis; five blood samples and nine throat swabs from 13 fever cases; and ten blood samples and one throat swab from ten family contacts (including specimens from the brother and mother of a patient who died from encephalitis). The confirmed Chandipura virus encephalitis group consisted of individuals from whose samples we isolated the virus, viral RNA, or reactive IgM antibodies.
Laboratory investigations
The state government did laboratory tests to rule out bacteria and malaria, and to study CSF profiles. We tested for Japanese encephalitis virus, West Nile virus, measles virus, dengue virus, paramyxoviruses, rabies virus, enteroviruses, influenza virus, coronaviruses, and mycoplasma, by use of serological tests, PCR, or both.6, 7, 8, 9 All handling of material and tests were done with appropriate biosafety practices.
Isolation of virus was done by inoculation of clinical specimens (31 throat swabs, five CSF, and one brain aspirate from a patient who died) into Vero, Madin-Darby canine kidney (MDCK), and rhabdomyosarcoma (RD) cell lines, with standard procedures. White blood cells were obtained from ten patients' blood clots by lysing red blood cells, and were co-cultured with phytohaemagglutinin-stimulated peripheral-blood mononuclear cells from normal donors. All cultures inoculated with the clinical material were frequently checked for cytopathic effects. Five CSF samples were also inoculated into the brains of 1-day-old Swiss-albino mice to grow virus.
Tissue culture fluids from cultures showing cytopathic effects were negatively stained with 1% sodium phosphotungstic acid pH 6·0 and examined with the 100 KV mode of a transmission electron microscope (Tecnai 12 Biotwin, FEI, Eindhoven, Netherlands).
Complement fixation tests were done with hyperimmune Chandipura virus antiserum raised against a prototype strain of the virus (653514).10 In-vitro virus neutralisation tests were done in Vero cells as described previously11 with 100 TCID50 (50% tissue culture infective dose) of Chandipura virus. The titre of virus-neutralising antibody was assigned as the reciprocal of the antibody dilution capable of neutralising the virus. Hyperimmune serum raised in mice against a standard strain of Chandipura virus served as a positive control and was used for identification of virus isolate.
An immunofluorescence assay was used to test for virus in brain-tissue suspension, which was spread on glass slides, air dried, acetone fixed, and immunolabelled with mouse anti-Chandipura-virus hyperimmune serum and anti-mouse fluorescein-isothiocynate-conjugate, by use of standard procedures, along with appropriate controls.
RT-PCR for Chandipura virus RNA was done on the basis of the G gene sequence for an Indian Chandipura virus isolate (CHPI-653514, Genebank accession number J04350) The primers shown in the panel were synthesised and used in hemi-nested format for the detection of Chandipura viral RNA in clinical specimens.
Panel. Primers.
CHPG-F2 5′ GTC TTG TGG TTA TGC TTC TGT 3′
CHPG-F3 5′ TGT GTC CGA CCG GGA TCA GAG GT 3′
CHPG-R2 5′ TGA GCA TGA GGT AGC TGT GGAT 3′
PCR products were purified by use of Wizard PCR preps DNA purification Kit (Promega, Madison, USA) according to the manufacturer's instructions, and sequenced with Big Dye Terminator cycle sequencing Ready Reaction Kit (Applied Biosystems, Foster City, USA) and an automatic Sequencer (ABI PRISM 310 Genetic Analyser, Applied Biosystems). Both strands were sequenced.
We did phylogenetic analysis of the partial G gene sequence (395 nt) with MEGA software.12 The Jukes-Cantor algorithm was used with the neighbour-joining method. To assess the reliability of the groupings obtained, we used bootstrap analysis (1000 bootstraps) available in the software (SEQBOOT: 1000 boot strap replications). Greater than 70% bootstrap support was judged to be reliable phylogenetic grouping.
IgM and IgG antibodies against Chandipura virus were detected by locally developed capture ELISA (patent pending). IgM or IgG from patient's serum were captured on wells coated with anti-human IgM or IgG, respectively. Chandipura virus extracted from mouse brain by sucrose-acetone was the source of antigen. Captured antigen was detected with the IgG fraction of polyclonal anti-Chandipura-virus mouse serum conjugated with biotin13 (Sigma Chemicals, St Louis, USA) followed by avidin-horseradish peroxidase. o-phenyline-diamine-hydrogen peroxide was added for colour development. Negative controls included: age-matched serum from apparently healthy children from an area not affected by the outbreak, and serum and CSF from children with flavivirus encephalitis. The cutoff value was determined as mean optical density for negative controls plus 3 SD.
Statistical analysis
For comparison of proportions, Fisher's exact test was used. The Mann-Whitney test was used to compare neutralising antibody titres.
Role of the funding source
The sponsors of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
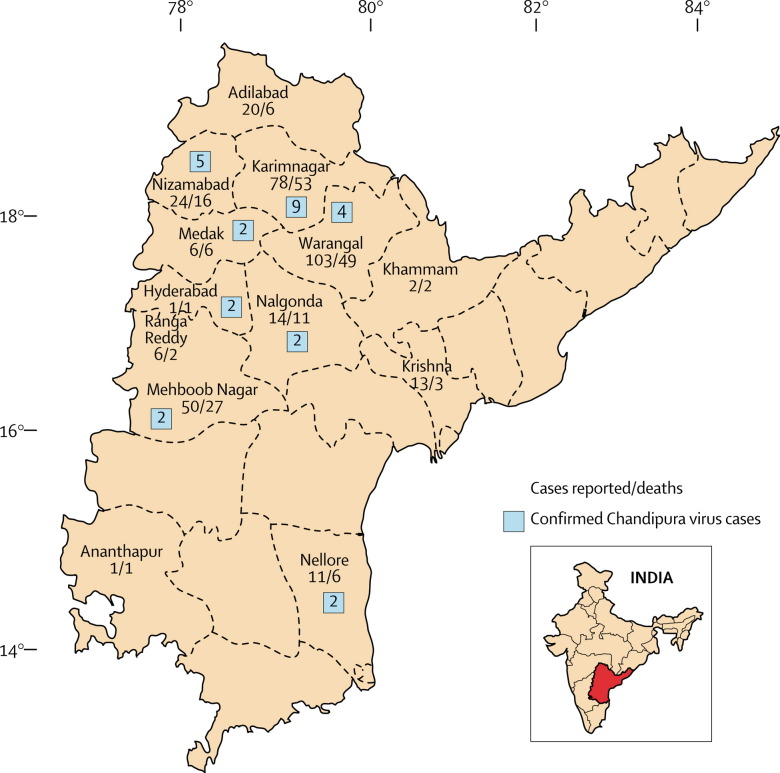
On the basis of the broad case-definition used by the Andhra Pradesh state government, 329 cases of encephalitis were reported between June and September, 2003, with 183 deaths (case fatality rate 55·6%). Most deaths occurred within 48 h of admission to hospital. The distribution of cases was mainly rural, spread over 13 districts of the state, and was spotty without clustering (figure 1 ). Age of patients ranged from 9 months to 14 years. The male to female ratio was 1 to 0·77. Limited data suggested that CSF was usually under pressure; pleocytosis was absent. Neurological sequelae were rare in the children who recovered.
Figure 1.
Map of Andhra Pradesh, India, showing districts affected by encephalitis
The typical clinical manifestations in the group with confirmed Chandipura virus encephalitis (n=28) included rapid onset of fever (28 patients, 100%), followed by vomiting (15, 54%), altered sensorium (25, 89%), convulsions (23, 82%), diarrhoea (five, 18%), neurological deficit (four, 14%), and meningeal irritation (two, 7%).
Brain tissue was obtained after death from only one patient: a 9-year-old, previously healthy girl developed fever, vomiting, and generalised tonic-clonic convulsions, became unconscious, and had severe dehydration. On admission she developed tachycardia, dyspnoea, irregular breathing, bilateral papilloedema, decerebrate posture, and no response to painful stimuli. Blood pressure at admission was 90/60 mm Hg and subsequently dropped to 60 mm Hg/not recordable. The girl was put on a mechanical ventilator. She continued with fever (101°F) and developed bradycardia and pupillary dilatation. Potassium rose substantially in her serum (to 8 mmol/L). The patient was treated with intravenous fluids, antibiotics, phenytoin, diazepam, and mannitol, and died within 24 h of admission.
Routine laboratory investigations screening for Japanese encephalitis virus, West Nile virus, paramyxovirus, coronavirus, measles virus (serum and CSF), dengue virus, rabies virus, influenza viruses, and mycoplasma (throat swab and CSF) yielded negative results.
In the encephalitis group, all ten CSF samples were negative for virus isolations. Virus was isolated from three of 22 throat swabs (one in MDCK; one in RD, Vero, and MDCK; one in Vero and RD), one brain aspirate in RD and Vero cell cultures, and two of ten blood clots in peripheral-blood mononuclear cell co-culture. Five of the six samples from which virus was isolated had been obtained within 4 days of onset of illness. In the fever group, one of eight throat swabs yielded virus in the RD cell line.
Assessment of the culture isolates with transmission electron microscopy showed the presence of bullet-shaped particles that were 150–165 nm long and 50–60 nm wide, with distinct surface projections 9–11 nm in length and a stain-filled canal at the base of the particle—all features suggestive of rhabdoviruses (figure 2 ). Isolates were identified as Chandipura virus by complement fixation and neutralisation tests. Chandipura viral antigen and RNA were detected in brain tissue by immunofluorescence assay (figure 3 ) and PCR, respectively. Four of 21 throat swabs, one of seven CSF samples, five of 25 serum samples, and one brain aspirate were positive for Chandipura virus RNA. One of the five positive serum samples was from the patient who died, described earlier. All nine samples that were positive for RNA had been obtained within 4 days of illness onset (table 1 ). All isolates were also positive for Chandipura viral RNA.
Figure 2.
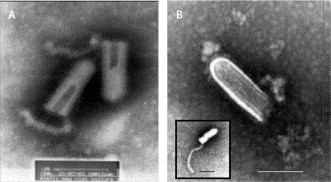
Transmission electron micrographs of primary Chandipura virus isolates from culture
Bar=100 nm in both micrographs. (A) Two negative stained virus particles showing the stain filled canals and basal attachments. (B) Negative stained Chandipura virus particle showing typical vesiculovirus morphology, including the internal ribonucleoprotein coil. Inset shows a virus particle with a released helical ribonucleoprotein coil .
Figure 3.
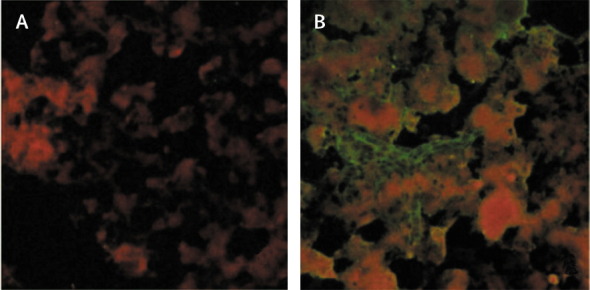
Immunofluorescence of brain smear stained with anti-Chandipura-virus antibody
(A) Fluorescent antibody with normal mouse serum. (B) Fluorescent antibody with anti-Chandipura-virus mouse serum.
Table 1.
Details of patients with confirmed diagnosis of recent Chandipura virus infection
| Age in years (sex) | POD |
CHP-reactive antibodies |
CHP isolation |
CHP PCR |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| IgM | IgG | NAb | TS | Blood clot | TS | Serum | CSF | |||
| Encephalitis | ||||||||||
| 1 | 4(f) | 0 | NEG | NEG | <10 | ·· | POS | ·· | ·· | ·· |
| 2 | 8(m) | 0 | POS | POS | <10 | ·· | ·· | ·· | ·· | ·· |
| 3 | 8 (f) | 0 | NEG | NEG | <10 | NEG (V,R) | ·· | ·· | POS | ·· |
| 4 | 5 (m) | 1 | NEG | NEG | <10 | POS (V,R) | ·· | POS | ·· | ·· |
| 5 | 6 (f) | 1 | NEG | NEG | <10 | NEG (V,R) | NEG | ·· | POS | NEG |
| 6 | 11 (f) | 1 | POS | NEG | <10 | ·· | ·· | ·· | ·· | ·· |
| 7 | 9 (m) | 1 | NEG | NEG | <10 | POS (M) | ·· | POS | ·· | ·· |
| 8 | 5(f) | 1 | NEG | NEG | <10 | ·· | NEG | NEG | POS | ·· |
| 9* | 9 (f) | 2 | NEG | NEG | <10 | ·· | ·· | NEG | POS | POS10 |
| 10 | 4 (f) | 2 | NEG | NEG | <10 | POS(V,R,M) | NEG | POS | NEG | NEG |
| 11 | 2(f) | 3 | POS | NEG | <10 | ·· | ·· | ·· | ·· | ·· |
| 12 | 8 (f) | 3 | NEG | NEG | <10 | ·· | ·· | ·· | POS | ·· |
| 13 | 5 (m) | 4 | POS | NEG | <10 | ·· | ·· | POS | ·· | ·· |
| 14 | 3(f) | 5 | POS | POS | 270 | NEG (V,R,M) | ·· | NEG | ·· | NEG |
| 15 | 4 (f) | 6 | POS | NEG | 30 | ·· | NEG | ·· | ·· | ·· |
| 16 | 5(f) | 6 | POS | NEG | 10 | ·· | ·· | ·· | ·· | ·· |
| 17 | 0.75 (m) | 6 | POS | POS | 540 | ·· | ·· | ·· | ·· | ·· |
| 18 | 4 (m) | 7 | POS | POS | 540 | ·· | ·· | ·· | ·· | ·· |
| 19 | 9(m) | 7 | POS | NEG | 90 | ·· | ·· | ·· | ·· | ··20 |
| 20 | 4(m) | 7 | POS | POS | 90 | NEG (V,R,M) | NEG | NEG | NEG | ·· |
| 21 | 2.5 (f) | 8 | NEG | NEG | <10 | NEG(V,R,M) | POS | NEG | NEG | NEG |
| 22 | 9 (f) | 8 | POS | NEG | 270 | ·· | ·· | ·· | ·· | ·· |
| 23 | 4 (m) | 10 | POS | POS | 270 | ·· | ·· | ·· | ·· | ·· |
| 24 | 11 (f) | 12 | POS | POS | 540 | ·· | ·· | ·· | ·· | ·· |
| 25 | 4 (f) | 14 | POS | POS | 540 | ·· | ·· | ·· | ·· | ·· |
| 26 | 4 (m) | NA | POS | POS | NA | NEG (V,R) | ·· | NEG | ·· | ·· |
| 27 | 3(f) | NA | POS | POS | 540 | ·· | ·· | ·· | ·· | ·· |
| 28 | 4(f) | NA | POS | POS | >270 | ·· | ·· | ·· | ·· | ·· |
| Fever | ||||||||||
| 29 | 9(f) | NA | POS | POS | 270 | ·· | ·· | ·· | ·· | ·· |
| 30 | 11(m) | NA | NA | NA | NA | POS(R) | ·· | ·· | ·· | ·· |
| 31 | 8(m) | 3 | POS | NEG | <10 | ·· | ·· | ·· | ·· | ·· |
| Contacts | ||||||||||
| 32 | 17(m) | ·· | POS | POS | 270 | ·· | ·· | ·· | ·· | ·· |
| 33 | 32(f) | ·· | POS | NEG | 90 | ·· | ·· | ·· | ·· | ·· |
NA=not available
Isolate from brain in Vero and RD. M=male. f=female. Cultures used for isolation: V=Vero, R=RD, M=MDCK, CHP=Chandipura virus. NEG=negative. POS=positive. POD=Post-onset day of illness when specimens were obtained. TS=throat swab. Absence of data in cells denotes that clinical specimens were not available.
Partial G gene sequences of 395 nt (accession number AY5544072–411) representing clinical specimens from five patients showed 96–99·8% identity (mean 97·5%, 95% CI 95·1–99·9; figure 4 ). These sequences were 96·7–97·5% (97·03, 95% CI 96·4–97·6) identical with the 1965 reference sequence. All Chandipura virus sequences clustered together with 100% bootstrap support and were closer to Piry virus than to vesicular stomatitis virus (figure 5 ).
Figure 4.

Nucleotide alignment of reference (CHPI-653514) and epidemic (bold) strains of Chandipura virus partial G gene sequences
Figure 5.
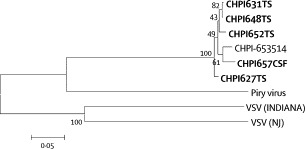
Phylogenetic analysis of partial G gene sequences (395 nt) of viruses from vesiculovirus genus
Viruses: vesicular stomatitis Indiana virus (VSV-Indiana, J02428), vesicular stomatitis New Jersey virus (VSV-NJ, JJ02433), Piry virus (Z15093) and reference Chandipura virus (CHPI-653514, J04350). Five sequences obtained during the present study are shown in bold. Percent bootstrap support is indicated by the values at each node.
Table 1 shows IgM, IgG, and neutralising antibody status for 28 patients with confirmed Chandipura virus encephalitis, three patients with fever, and two contacts. Table 2 shows antibody status in 48 patients with encephalitis, including 25 of the patients with confirmed Chandipura virus infection mentioned in table 1 (data were incomplete for the other three patients). Since no paired samples of serum from the acute and convalescent phases of illness could be obtained from the patients during this outbreak, the results from testing of single-bleed serum samples were assessed in relation to the time after disease onset at which samples were obtained. A smaller proportion (four of 30 [13%]) of serum samples obtained up to 4 days after onset of illness (early samples) were positive for IgM reactive with Chandipura virus, compared with samples obtained later than 4 days after onset of illness (11 of 16 [69%]; p<0·001). A similar trend was also noted for Chandipura virus-reactive IgG antibodies (3% of early samples positive vs 50% of late samples) and neutralising antibodies (7% vs 83%). The geometric mean titres in early versus late samples were significantly different (6·1 vs 87·4, p<0·001). Chandipura virus-reactive IgM antibodies were also found in two of five patients with fever but no CNS involvement, and in two of ten family contacts. Unfortunately, because of the nature of the outbreak response and local resources, we were unable to obtain serum samples from a larger control group.
Table 2.
Antibody status and relation to time of sample collection
|
Encephalitis |
Fever without encephalitis | Contacts without disease | ||
|---|---|---|---|---|
| POD 0–4 | POD >4 | |||
| IgM+,IgG− | 3/30 | 4/16 | 1/5 | 1/10 |
| IgM+, IgG+ | 1/30 | 7/16 | 1/5 | 1/10 |
| IgM−, IgG+ | 0/30 | 1/16 | 0/5 | 2/10 |
| IgM−, IgG− | 26/30 | 4/16 | 3/5 | 6/10 |
| NAb | 2/29 | 15/18 | 2/5 | 7/10 |
| NAb(GMT) | 6·1 | 87·4 | 28·31 | 45·22 |
| NAb (median [range]) | <10 (<10–90) | 135 (<10–540) | (<10–270)* | 60(<10–540) |
Data are number of samples positive/total tested, unless otherwise stated. POD=Post-onset day of illness when specimens were obtained. NAb=Neutralising antibody. GMT=Geometric mean titre. POD data were not available for three patients with encephalitis included in table 1 but not in table 2
Only two values for this set ofdata.
Discussion
The viruses isolated in different cell lines from clinical samples from patients with encephalitis were confirmed as Chandipura virus with various techniques including complement fixation, neutralisation test, and immunofluorescence assay. Analysis of partial G gene sequences showed that the genome of the Chandipura virus strain associated with this outbreak was distinct from that of the reference strain obtained in 1965. Importantly, in one fatal encephalitis case from this outbreak, both Chandipura viral antigen and RNA were detected in situ in the post-mortem brain tissue. The clinical presentation of this patient was representative of the symptoms seen in most of the other patients reported during the present outbreak. Chandipura virus was also subsequently isolated from this tissue. Therefore, the presence of the virus in the brain was probably the cause of CNS pathology leading to encephalitis in this patient. Moreover, the presence of Chandipura virus RNA in nine patients with encephalitis, all from samples obtained before day 4 after onset of illness, suggests an early viraemic phase of the infection process. That five of six virus isolates were obtained from such early samples further strengthens this observation.
Further evidence that Chandipura virus was the primary causal agent for this outbreak is the pattern of immune response to the virus. Substantially higher proportions of positive test results for Chandipura virus-reactive antibodies were noted among samples obtained more than 4 days after onset of illness, compared with samples obtained earlier, suggesting a seroconversion window period and primary exposure to the virus in the people from whom samples were taken early.
Of the 55 cases of encephalitis investigated, evidence of recent infection with Chandipura virus could be established conclusively in 28 cases (51%) based on either the presence of virus or viral RNA, IgM antibodies, or both. The absence of evidence of such an infection in some samples could be because samples were obtained very soon after disease onset. A small proportion of the remaining cases might represent encephalitis due to other causes. The multiple lines of evidence together implicate Chandipura virus as the causative agent associated with the present outbreak of acute encephalitis in the absence of any other identifiable causes.
The presence of IgM reactive with Chandipura virus in two patients with fever and in two family contacts of a patient with encephalitis suggests that infection with the virus may remain asymptomatic or lead to fever without encephalitis; the disease spectrum needs careful study. Evidence of IgG against Chandipura virus detected in adults probably indicates environmental exposure to the virus. Several earlier outbreaks of encephalitis recorded in central India from 1954 onwards that showed similarities to the present outbreak (clinical features, seasonality, and undetermined aetiology) might perhaps have been due to Chandipura virus, which could be endemic in these regions.
Most of the vesiculoviruses are transmitted by sandflies.14 Interestingly, Chandipura virus was detected by PCR in a sandfly pool obtained from the house of an affected patient in the present outbreak (unpublished report). This finding and earlier isolation of the virus from sandflies both in India15 and Africa16 strongly suggest that sandflies might be the vector of this disease. In India, these insects are more prevalent in the early monsoon period, a season that coincided with the present outbreak. The isolation of Chandipura virus from sandflies,15, 16 human beings,17 and vertebrates;18 the presence of reactive antibodies in human beings and other vertebrate hosts;19 and the ability of sandflies and mosquitoes to transmit the virus to susceptible hosts;20, 21, 22, 23 represents a broad eco-cycle of the virus in nature. The previous isolation of the virus from human beings with febrile and encephalopathy4, 17 and association of this virus with the present large encephalitis outbreak, implicates Chandipura virus as an emerging pathogen of substantial public-health importance.
Acknowledgments
Acknowledgments
We express our sincere gratitude to Stuart Nichol (Chief, Molecular Biology Laboratory, Special Pathogens Branch, Division of Viral and Rickettsial Diseases, Centers for Disease Control and Prevention, Atlanta, GA, USA) who gave us a unique opportunity to discuss the data in person and extensive constructive review and suggestions that went into the present manuscript. We also gratefully acknowledge the help and co-operation of the Directorate of Health services, Government of Andhra Pradesh, India. We would also like to sincerely thank S Tikute, S D Pawar, G N Sapkal, Mr Hanumaiah, S V Gangodkar, V M Ayachit, N J Shaikh, S P Shrotri, and B Kundu for their technical support; A Walimbe for statistical assistance, Mr Murlikrishna for photographic help, and M V Joshi for providing standard immune serum samples. Funds were provided by the Indian Council of Medical Research, Ministry of Health and Family Welfare, Government of India.
Contributors
B L Rao isolated and characterised virus, collected and analysed laboratory data, and had substantial intellectual input into manuscript writing. A Basu did electron microscopy and laboratory studies related to primary virus identification, analysed data, and had substantial intellectual input into manuscript writing. N S Wairagkar did clinico-epidemiological studies of the outbreak, examined patients, obtained and analysed clinical material, compiled data, and had substantial intellectual input into manuscript writing. M M Gore did virus isolation studies, planned execution analysis of virus neutralisation data, and had substantial intellectual input into manuscript writing. V A Arankalle designed and did molecular virology experiments and had substantial intellectual input into manuscript writing. J P Thakare did serology tests, including development of ELISAs for Chandipura virus, IgM, and IgG, and took part in manuscript preparation. R Jadi participated in virus isolation studies and manuscript preparation. A K Rao contributed clinical data and took part in patient management, including data analysis. A C Mishra was the group leader, substantially contributed to design, interpretation, and data analysis, and had significant intellectual contribution in developing the manuscript.
References
- 1.Rodrigues FM. Epidemiology of Japanese encephalitis in India: national conference on Japanese encephalitis. Indian J Med Res. 1984;(suppl):1–9. [Google Scholar]
- 2.Khan N. Jamshedpur fever. Indian J Med Sci. 1954;8:597–608. [Google Scholar]
- 3.Rodrigues FM, Patankar MR, Banerjee K. Etiology of the 1965 epidemic of febrile illness in Nagpur city, Maharashtra state, India. Bull World Health Organ. 1972;46:173–179. [PMC free article] [PubMed] [Google Scholar]
- 4.Rodrigues JR, Singh PB, Dave DS. Isolation of Chandipura virus from the blood in acute encephalopathy syndrome. Indian J Med Res. 1983;77:303–307. [PubMed] [Google Scholar]
- 5.John TJ. Outbreak of killer brain disease in children: mystery or missed diagnosis? Indian Pediatr. 2003;40:863–869. [PubMed] [Google Scholar]
- 6.Read SJ, Kurtz KB. Laboratory diagnosis of common viral infections of the central nervous system by using a single multiplex PCR screening assay. J Clin Microbiol. 1999;37:1352–1355. doi: 10.1128/jcm.37.5.1352-1355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephensen CB, Casebolt DB, Gangopadhya NN. Phylogenetic analysis of a highly conserved region of the polymerase gene from 11 coronaviruses and development of a consensus polymerase chain reaction assay. Virus Res. 1999;60:181–189. doi: 10.1016/S0168-1702(99)00017-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chua KB, Bellini WJ, Rota P. Nipah virus: a recently emergent deadly paramyxovirus. Science. 2000;28:1432–1435. doi: 10.1126/science.288.5470.1432. [DOI] [PubMed] [Google Scholar]
- 9.Poddar SK, Espina R, Schnurr DP. Evaluation of a single step multiplex RT-PCR for influenza virus type and subtype detection in respiratory samples. J Clin Lab Anal. 2002;16:163–166. doi: 10.1002/jcla.10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sever JL. Application of microtechniques to viral serological investigation. J Immunol. 1962;88:320–329. [PubMed] [Google Scholar]
- 11.Gore MM, Thakare JP, Deuskar NJ. Ethanethiol sensitive and resistant antibodies to Enterovirus 70 in post conjunctivitis neurological syndromes. Indian J Med Res. 1985;81:343–348. [PubMed] [Google Scholar]
- 12.Kumar S, Tamura K, Jakobson IB. MEGA2·1: molecular evolutionary genetics analysis software. Arizona State University; Tempe, Arizona, USA: 2001. [Google Scholar]
- 13.Nerukar LS, Namba N, Brashears G. Rapid detection of herpes simplex virus in clinical specimen by use of a capture biotin-streptavidin enzyme linked immunosorbent assay. J Clin Microbiol. 1984;20:109–114. doi: 10.1128/jcm.20.1.109-114.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mattos de CA, Mattos de CC, Ruppercht CE. Rhabdoviruses. In: Knipe MD, Howley P, editors. Fields Virology. 4th edn. Lippincott Williams & Wilkins; Pennsylvania, USA: 2001. pp. 1245–1277. [Google Scholar]
- 15.Dhanda V, Rodrigues FM, Ghosh SN. Isolation of Chandipura virus from sandflies in Aurangabad. Indan J Med Res. 1970;58:179–180. [PubMed] [Google Scholar]
- 16.Traore-Lamizana M, Fontenille D, Diallo M. Arbovirus surveillance from 1990 to 1995 in the Barkedji area (Ferlo) of Senegal, a possible natural focus of Rift Valley fever virus. J Med Entomol. 2001;38:480–492. doi: 10.1603/0022-2585-38.4.480. [DOI] [PubMed] [Google Scholar]
- 17.Bhatt PN, Rodrigues FM. Chandipura virus: a new arbovirus isolated in India from patients with febrile illness. Indian J Med Res. 1967;55:1295–1305. [PubMed] [Google Scholar]
- 18.Kemp GE. Viruses other than arenaviruses from West African wild mammals: factors affecting transmission to man and domestic animals. Bull World Health Organ. 1975;52:615–620. [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee K. Emerging Arboviruses of zoonotic and human importance in India. In: Misra, Polasa, editors. Virus Ecology. SE Asian publishers; New Delhi: 1984. pp. 109–121. [Google Scholar]
- 20.Tesh RB, Modi GB. Growth and transovarial transmission of Chandipura virus (Rhabdoviridae:Vesiculovirus) in Phlebotomus papatasi. Am J Trop Med Hyg. 1993;32:621–623. doi: 10.4269/ajtmh.1983.32.621. [DOI] [PubMed] [Google Scholar]
- 21.Rao TR, Singh KRP, Dhanda V. Experimental transmission of Chandipura virus by mosquitoes. Indian J Med Res. 1967;55:1306–1310. [PubMed] [Google Scholar]
- 22.Ilkal MA, Goverdhan MK, Shetty PS. Susceptibility of four species of mosquitoes to Chandipura virus and its detection by immunofluorescence. Acta Virol. 1991;35:27–32. [PubMed] [Google Scholar]
- 23.Kelkar SD. Antibody response to Chandipura virus in experimental animals. Indian J Med Res. 1976;64:814–823. [PubMed] [Google Scholar]