Abstract
Annexins are multifunctional proteins that bind to phospholipid membranes in a calcium-dependent manner. Annexins play a myriad of critical and well-characterized roles in mammals, ranging from membrane repair to vesicular secretion. The role of annexins in the kingdoms of bacteria, protozoa and fungi have been largely overlooked. The fact that there is no known homologue of annexins in the yeast model organism Saccharomyces cerevisiae may contribute to this gap in knowledge. However, annexins are found in most medically important fungal pathogens, with the notable exception of Candida albicans. In this study we evaluated the function of the one annexin gene in Cryptococcus neoformans, a causative agent of cryptococcosis. This gene CNAG_02415, is annotated in the C. neoformans genome as a target of calcineurin through its transcription factor Crz1, and we propose to update its name to cryptococcal annexin, AnnexinC1. C. neoformans strains deleted for AnnexinC1 revealed no difference in survival after exposure to various chemical stressors relative to wild-type strain, as well as no major alteration in virulence or mating. The only alteration observed in strains deleted for AnnexinC1 was a small increase in the titan cells' formation in vitro. The preservation of annexins in many different fungal species suggests an important function, and therefore the lack of a strong phenotype for annexin-deficient C. neoformans indicates either the presence of redundant genes that can compensate for the absence of AnnexinC1 function or novel functions not revealed by standard assays of cell function and pathogenicity.
Keywords: Cryptococcus neoformans, C. neoformans, annexin, AnxC1, CNAG_02415
Introduction
Annexins are a multifunctional family of proteins that bind to phospholipid membranes in a calcium-dependent manner. The bridging of membranes by annexins is calcium dependent and is mediated through a structural domain consisting of about 70 amino acids in consecutive alpha helical conformations known as the ‘annexin fold’ [1].
Annexins have been reported in all domains of life, except Archaebacteria [2, 3]. In plants, annexins are important for immunity and resistance to nutrient stress [4]. In mammals, annexins play a critical role in membrane signalling, encompassing plasma membrane repair, gene regulation, organelle trafficking, endosomal fusion, endocytosis and exocytosis. Therefore, annexins are indispensable for tissue functions such as clotting, immunity, among others [1, 5–7]. In mammals, annexins are found in the nucleus, the cytosol and the cell surface, and are also secreted into the extracellular milieu.
A recent review revealed a paucity of studies about microbial annexins, but the available information suggests that these proteins are critical for the virulence of certain pathogens [8, 9]. Giardin from Giardia [10] is an immunodominant annexin protein [11], suggesting vaccine or antigenic potential for this protein. Administration of giardin to mice induced protective immunity against Giardia and is under investigation for its vaccine potential [8]. The Burkholderia JOSHI_001 protein possesses both a colicinD toxin domain and an annexin domain [9], which reinforces the notion that microbial annexins contribute to microbial virulence.
Very little information is available on fungal annexins. In Neurospora crassa, no phenotype was found during phenotype mutation efforts [12]. The absence of an annexin gene in Saccharomyces cerevisiae and Candida spp. may have contributed to the lack of information on fungal annexins, given that these organisms are the most extensively studied fungi at the cellular level. However, annexin genes can be found in all basidiomycota and the majority of medically important ascomycota. Thus far, annexin deletion strains were characterized in Aspergillus [13–16], a genus with 3 recognizable annexins, and Thermomyces lanuginosus [17]. Deletion of anxc1 in A. niger or anxc3 in A. fumigatus did not result in growth or protein secretion defects [14]. Deletion of anxc3 (previously anxc4) of A. fumigatus was associated with very subtle changes in the protein secretion profile [14]. In T. lanuginosus, annexinC7 is involved in conidia formation and resistance to oxidative stress [17]. In amoeba Dictyostelium discoideum, one annexin gene is found and its deletion led to a delay in growth in low Ca2+ conditions [18].
Cryptococcus neoformans is the causative agent of cryptococcosis, a life-threatening infection that often results in high mortality and morbidity [19]. The cryptococcal genome contains one predicted annexin gene, CNAG_02415, annotated as AnnexinXIV. A literature search for references to AnnexinXIV, fungal annexin or CNAG_02415 found no reports on the function of this gene. Exploration of the available full datasets from high-throughput experiments gave us some clues into the possible roles of AnnexinC1. A genetic screen showed that CNAG_02415 deletion led to altered susceptibility to a range of small molecule compounds [20]. Further, CNAG_02415 (AnnexinC1) is dependent on Crz1, a transcription factor of the calcineurin pathway, which is essential for virulence in C. neoformans [20–22]. Chip-sequencing data identified AnnexinC1 as a direct target of Crz1 [23]. This finding posits a role for AnnexinC1 as a component of the Crz1-calcineurin response, involved in high-temperature growth, cell-wall stability and heavy-metal susceptibility [22, 24]. Available gene and protein expression datasets in C. neoformans detected no alterations in expression of AnxC1 expression upon infection [25], nor was the protein detected as secreted into the extracellular milieu [26, 27]. In a transcriptomic dataset, absence of Rim101 led to an increase in expression of ANXC1 when placed for 3 h in Dulbecco’s Modified Eagle Medium [28, 29], which suggests ANXC1 is additionally regulated by Rim101 [28], an important cell morphology regulator. Based on its regulation by two important fungal signalling pathways, we decided to characterize the function of this gene. We posited that AnxC1 of C. neoformans could play a role in fungal pathogenesis and its life cycle. We found that annexin deletion affected titan cell production but had no contribution to virulence in a mouse model. As with the other fungi cited above, the molecular function of the cryptococcal annexin remains cryptic, but we provide a first step in characterizing its functions in the fungal world.
Methods
Phylogenetic and protein modelling analysis
Conserved domains were detected with NCBI Conserved Domains [29]. Fasta files were downloaded and aligned using muscle [30] with Mega7 software. Phylogenetic analysis was constructed with a maximum likelihood tree, neighbour-join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model. A model of AnnexinC1 was built using Swiss-Model workspace (https://swissmodel.expasy.org/interactive#structure) [31, 32], using AnnexinA5 from Homo sapiens as a template. FungiDB (http://fungidb.org/fungidb/) was used to look for gene expression patterns of CNAG_02415 [33].
Cn strains and deletion strain preparation
The H99 wild-type strain was obtained from Jennifer K. Lodge laboratory (available at the Fungal Genetic Stock Center) and is also identified as JLCN69, originating from an H99E lineage (Jennifer K. Lodge, personal communication) [34]. To confirm some of the phenotypes observed, we used deletion strains and respective wild-types (one in KN99 and the other in H99 background) from freely available libraries, prepared by the laboratories of Suzanne E. Noble and Hiten D. Madhani [35]. Cells were kept in 10 % glycerol frozen stocks. C. neoformans deletion strains were prepared by biolistic transformation [36], using double-joint nourseothricin-split (NAT) markers [37]. We joined the NAT markers with sequences flanking 1000–500 bp of the CNAG_02415 coding region, which were deposited into gold carrier particles and shot into a lawn of C. neoformans. Single colonies were picked from 100 µg ml−1 NAT plates. We selected several clones and further characterized them via PCR to verify insertion in the correct genomic location, as well as Southern blot using DIG-High Prime DNA labelling and Detection starter kit II [Roche to verify only single insertion of the NAT cassette into each of the strains (577 bp)]. PCR 1 amplifies the entire NAT cassette (forward primer: 5′-GCCTGATTGATTTGCTGTGG and reverse primer: GCTTCTCGTTTACAAACAGCGC); PCR 2 is from 994 bp upstream of Annexin gene and 1217 bp internal to ANXC1 (GCCTGATTGATTTGCTGTGG and GCTTCTCGTTTACAAACAGCGC); PCR 3 is from 1140 upstream of insertion site and 290 bp internal to NAT (forward primer: GTGAGGGAAAGAATTCGTCG and reverse primer: TGTGGATGCTGGCGGAGGATA); PCR 4 is from 847 bp internal to NAT and 65 bp downstream of insertion site (forward primer: CTCTTGACGACACGGCTTACCGG and reverse primer: GGTGGACTAAATGGGGTTCAAAGG). A Southern blot probe was constructed with primers CTCTTGACGACACGGCTTACCGG and GCCTTCACGAATTCTTAGGGGC recognizing the NAT cassette. Two independent rounds of biolistic transformation, clone screening by PCR and Southern blot were performed and we selected two clones from each round of biolistic transformation, for a total of four. Deletion strains are always compared to their parental isolate (from the same round of biolistic transformation).
Spotting assays
Yeasts were grown overnight at 30 °C in yeast peptone dextrose (YPD) broth with shaking. Early stationary phase cultures were diluted to 1×106 cells ml−1 and then serially diluted tenfold. Dilutions were spotted (3 µl) into each plate and incubated at 30, 37 or 39 ˚C until visible colonies developed. YPD was purchased from BD Difco. Minimal media (MM) consisted of 15 mM glucose, 10 mM MgSO4, 29.4 mM KH2PO4, 13 mM glycine and 3 µM of thiamine-HCl, with final pH=5.5. All chemicals were purchased from Sigma and used in the concentrations indicated in the figure legend. Assays were performed by incorporating the indicated amount of chemicals in minimal media agar, with the exception of cell-wall stress tests, which were performed in YPD agar. Alkaline stress was induced in MM supplemented 150 mM HEPES and adjusted to indicated pH. Urease secretion was performed in Christensen’s agar and melanin secretion was measured by adding 1 mM L-DOPA to MM agar. Dikaryon mating was induced by mixing equal suspensions of alpha and a strains in V8 juice agar [38].
Antifungal drug resistance
The susceptibility of yeast strains to fluconazole and amphotericin B was assessed by broth microdilution according to CLSI M27-A3 [39]. Briefly, strains were grown at 35 °C for 2 days on Sabouraud dextrose agar, then used to inoculate drug plates to final concentration of 1.5×103 c.f.u. ml−1 (determined by hemocytometer, rather than spectrophotometer). Plates were incubated for 3 days at 35 °C prior to reading and each well imaged (Cellular Technology). Minimal inhibitory concentration (MIC) was determined visually, with fluconazole MIC being the lowest concentration at which growth was at least 50 % inhibited, and amphotericin B MIC being the lowest concentration at which no growth was observed. Each strain was tested in duplicate in three independent experiments.
Macrophage growth assays and capsule measurements
Two macrophages cell lines were used for most experiments: the macrophage-like murine cell line J774.16 [40] and the microglia-like cell line BV2. The BV2 cell line was a kind gift from Herbert W Virgin. J774.16 were kept in DMEM complete media consisting of DMEM (CellGro), 10 % NCTC-109 Gibco medium (LifeTechnologies), 10 % heat-inactivated FBS (Atlanta Biologicals), and 1 % non-essential amino acids (CellGro). Macrophages were plated at a density of 5×104 cells ml−1 the day before infection. C. neoformans, opsonized with 10 µg ml−1 of mAb 18B7 [41] were added at a m.o.i. (target to effector ratio) indicated in the figure legend in a final volume of 250 µl and infection proceeded for 24 h. Macrophages were lysed by resuspending all cells with 10× the volume of distilled water and the number of surviving yeasts was measured via a ‘tadpole’ assay [42]. Briefly, the aqueous suspension of yeast cells and lysed macrophages is serially tenfold diluted in YPD broth and allowed to grow overnight at 30 °C. Colonies are visible in the wells and the number of surviving yeasts is calculated by multiplying the number of colonies by the dilution of each well. For capsule growth, C. neoformans cells were incubated in macrophage media in 37 °C, 5 % CO2 for 24 h, conditions known to induce capsule growth.
Amoeba growth assays
Amoeba infections were performed as described previously [43]. Acanthamoeba castellanii strain 30 234 was obtained from the American Type Culture Collection (ATCC). Cultures were maintained in peptone yeast glucose broth (ATCC medium 712) at 25 °C. C. neoformans was grown in Sabouraud dextrose broth with 120 r.p.m. shaking at 30 °C overnight (16 h) prior to infection of amoeba. A. castellanii cells were washed twice with Dulbecco’s PBS (Corning, Corning, NY, USA), supplemented with Ca2+ and Mg2 + and 1×104 cells seeded in DPBS into each well of 96-well plates. After 1 h adhesion, 1×104 cells of C. neoformans in the same DPBS were added to wells containing amoebae or control wells containing DPBS alone, and the plates were incubated at 25 °C. At 0, 24 and 48 h, the amoebae were lysed by repeated shearing through 27-gauge needles. The lysates were serially diluted, plated on Sabouraud agar, and incubated at room temperature for 48 h for c.f.u. determination. For solid agar matrix infection experiments (pseudohyphae formation), 200 yeast cells were spread on Sabouraud agar, and incubated at 30 °C overnight. Acanthamoeba castellanii at a density of 5×103 cells per plate were randomly spotted on the agar. The plates were incubated at 25 °C for 2–3 weeks until filamentation was observed macroscopically. Filamentation sites were observed for fungal morphology under brightfield illumination microscopy.
Titan cell generation
Titan cells were generated according to the protocol generated by Hommel et al. [44]. Briefly cells were pre-cultured in YPD overnight at 30 °C with slow shaking, washed in MM and diluted to 1×106 cells ml−1 in MM. Cell suspension was placed in closed 1.5 ml microcentrifuge tubes and shaken for 2 days in Eppendorf Thermomixer (Hamburg, Germany) at 30 °C with 800 r.p.m. shaking. Cells were imaged and cell size (based on cell wall) was determined using ICY software [44]. Cells with body size >10 µm were considered titan cells. Results are expressed as median cell size (interquartile range, IQR) or as median frequency % of titan cells for each strain.
Galleria virulence assays
Galleria mellonella larvae were picked based on weight (0.2±0.02 g) and appearance (creamy white in colour). Larvae were starved overnight at room temperature. Yeast strains were inoculated overnight on Sabouraud broth, 30 °C, with shaking. The following day yeasts were washed three times with PBS and adjusted to 1.2×107 cells ml−1. Yeasts were injected on the seventh front paw of the larva with 27G tuberculin needles. Infected larvae were incubated at 30 °C for 15 days and observed daily for lack of movement (death). Control groups of larvae were inoculated with 10 µl of sterile PBS or left untouched. Experiments were repeated three times with experimental groups of 10–12 larvae at a time [45–48].
Animal infections
C57BL/6J mice, aged 8–10 weeks, were obtained from Jackson Laboratories and infected intratracheally as previously described with the indicated c.f.u. per animal in a final volume of 50 µl of sterile PBS [49]. Intranasal experiments were performed by dropping 40 µl of yeast suspension into the mouse nares while under isoflurane anesthesia [44]. Intravenous injections were performed by 40 µl injection in the retroorbital sinus of the animal under isoflurane anesthesia. Mice were monitored daily for signs of stress and deterioration of health throughout the experiment. Animals were euthanized if unable to feed.
Results
Genetic information and previous data
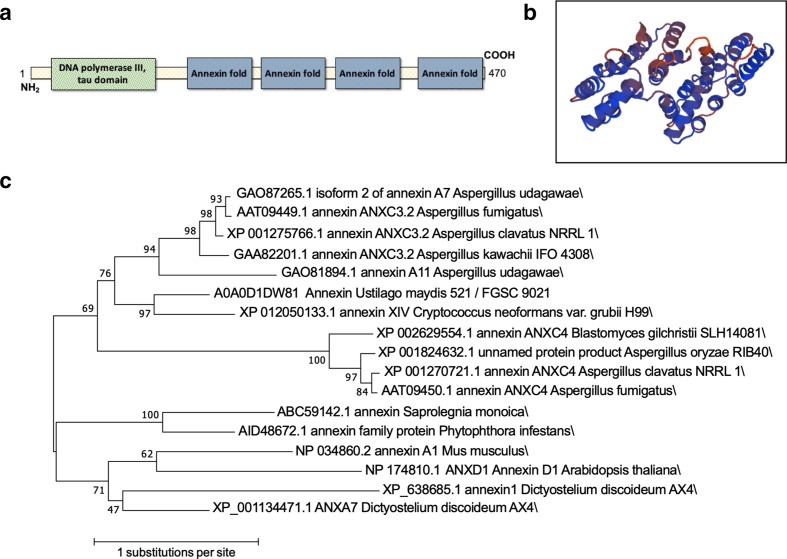
Annexins share a common structural feature known as the annexin domain, a fourfold repeat of alpha helixes made up of about 70 amino acids. This domain binds Ca2+, which in turn mediates phospholipid binding. Despite this common structural feature of the annexin fold, annexins show significant diversity between kingdoms [9, 50]. Phylogenetic analysis shows that Annexins can be grouped into five clades (A through E). Animal annexins (clade A) manifest highly conserved motifs whereas annexins in plants (clade D) show motif diversity [2]. In the kingdom Protista there is even greater diversity among annexins and these organisms generally express more annexin genes than in animal genomes [10]. Bacterial annexin sequences are so diverse that the existence of non-classical annexins was postulated [9]. Mammals and Protista have multiple different annexins in their genomes, but only one annexin has been identified in the cryptococcal genome, the gene product of CNAG_02415 [13]. We performed homology gene searches using DNA and protein sequences of mammalian (Mus musculus), plant (Arabidopsis thaliana), fungal (Aspergillus spp.) and bacterial ‘non-classical’ annexins [9] against the cryptococcal genome to identify other annexin-like genes, but found no other homologue. Thus, we concluded that CNAG_02415 is the only classical annexin in C. neoformans. Classically, fungal annexins are grouped in clade C, clustering with annexins from amoeba such as Dictyostelium discoideum [1]. We extracted the predicted protein sequences of annexins from the fungal kingdom and built a phylogenetic tree (Fig. 1), with annexin representatives of human, mouse, A. thaliana, D. discoideum, and water molds Saprolegnia monoica and Phytophthora infestans for comparison. This tree shows grouping of fungal annexins to each other and separate from annexins from all other groups. The sequence of fungal annexins differs significantly from that of their mammalian and plant counterparts [13], with the third and fourth folds of fungal annexins harbouring unusual sequences [51] while still retaining clear and recognizable annexin folds. CNAG_02415 encodes a protein with four of the characteristic annexin domains, one predicted calcium-binding site (Fig. 1) on the C-terminus, and a NH2 terminal head [1]. The annotation of the tau domain of DNA polymerase III at the N-terminus is cryptic but may be related to nucleotide-binding properties, which have been reported for annexins [52]. This cryptococcal gene is annotated as AnnexinXIV, likely due to homology with the first described fungal annexin in Neurospora crassa [18]. Following nomenclature conventions for Cryptococcus species complex [50], the gene name should be ANX1, but nomenclature conventions of annexins [50] indicate the gene name of ANXC1. In face of this conflict, we tentatively name the gene ANXC1 while the gene product is named as annexinC1 (AnxC1).
Fig. 1.
Predicted structure and phylogeny of cryptococcal annexin. (a) Diagram of location of conserved domains (as predicted by NCBI conserved domains); (b) Prediction of annexin C1 structure (J9VS56) as described in Methods. (c) The evolutionary history was inferred using the maximum likelihood method based on the Le_Gascuel_2008 model [53]. The tree with the highest log likelihood (−7162.32) is shown. The percentage of trees in which the associated taxa clustered together is shown next to the branches. Initial tree(s) for the heuristic search were obtained automatically by applying neighbour-join and BioNJ algorithms to a matrix of pairwise distances estimated using a JTT model, and then selecting the topology with superior log likelihood value. A discrete Gamma distribution was used to model evolutionary rate differences among sites [five categories (+G, parameter=4.6855)]. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The analysis involved 17 amino acid sequences. All positions with less than 95 % site coverage were eliminated. That is, fewer than 5 % alignment gaps, missing data and ambiguous bases were allowed at any position. There were a total of 250 positions in the final dataset. Evolutionary analyses were conducted in mega7 [54].
Generation of a new deletion strain
We constructed AnxC1-deficient strains in two independent rounds of biolistic transformation using a H99 parental strain (lineage E, as indicated in Methods). After individual colony screening we generated ten isolates indistinguishable by PCR and Southern blot (Fig. S1). We randomly selected four of these clones AnxC1-deleted (anxC1∆) and their corresponding parental strain (wild-type H99) for subsequent studies.
Functions of cryptococcal AnnexinC1
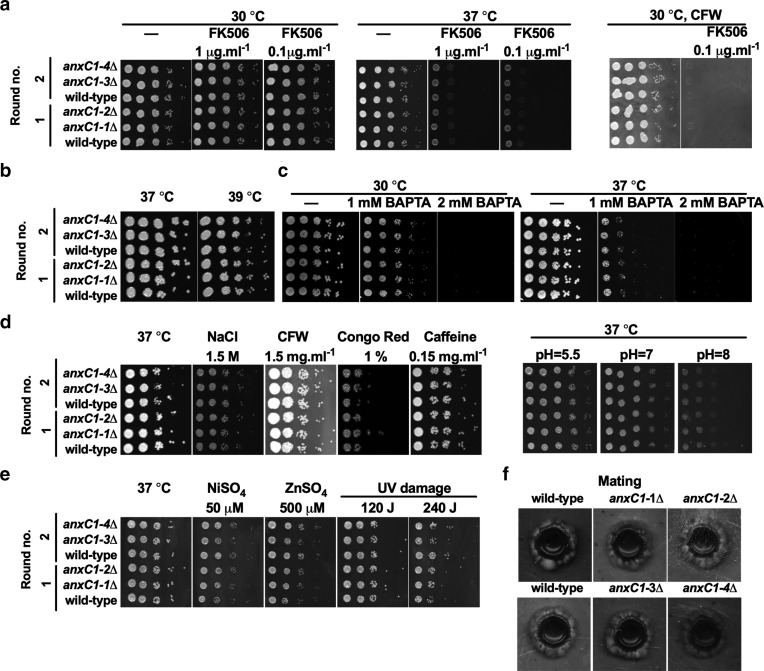
AnnexinC1 is reported to be a target of Crz1, a major component of the calcineurin pathway [22]. Although we did not confirm expression of this protein in yeast cells, key virulence transcription factors regulate its expression, which encouraged us to investigate its functions. To ascertain possible defects in responses dependent on the calcineurin pathway, we performed several phenotypic tests in functions strongly associated with the calcineurin pathway (Fig. 2). We found no alteration in growth when anxC1∆ strains were exposed to the calcineurin inhibitor FK506, high temperature or low calcium conditions due to addition of calcium chelator [1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid (BAPTA)]. The calcineurin pathway is also involved in the defense against cell-wall stress, caused by hyperosmotic stress, calcofluor white (CFW), congo red or caffeine. We found no evidence for a defect in cell-wall integrity for anxC1∆ compared to wild-type when exposed to either of these compounds. We tested the resistance to high pH, given our preliminary analysis had involved Rim101 in AnxC1 regulation, but found no alteration in growth at neutral to basic pH. Additional chemical and physical stresses, such as heavy metals and UV damage, were tested and no alteration in susceptibility to these stresses was detected [22, 55]. In previous work, anxC1∆ deletion strains in an H99 background showed a small increase in susceptibility to cell-wall stress and other chemical stressors [20, 22]. We had access to some of these strains as they originate from an available deletion library in C. neoformans, in an H99 background [35]. We also had access to a bigger deletion library in a KN99 background (from Hiten Madhani laboratory). To confirm the absence of cell-wall stress changes in the deletion strains in a different backgrounds of C. neoformans we tested the deletion strains from the available libraries for FK506 susceptibility, high temperature and the cell-wall stressor CFW. As previously reported we found that the anxC1∆ in the H99 background had a small increase in susceptibility to FK506 and high temperature (39 ˚C) [22]. These defects were not observed in the anxC1∆ from the KN99 background (Fig. S2). To test the contribution of anxC1∆ to a wider range of calcineurin-dependent conditions we measured sexual mating and filamentation efficiency. We detected a similar pattern and timing in mating when we crossed our anxC1∆ strains (α mating type) with an isolate from a mating type. Finally, we tested susceptibility to fluconazole and amphotericin B and determined an MIC was 2 µg ml−1 and 0.5–1 µg ml−1 for all three experiments, respectively, and no difference in MIC between wild-type and deletion strains (Fig. S3).
Fig. 2.
Deletion of AnnexinC1 does not affect phenotypes associated with calcineurin. AnnexinC1 deletion strains are not susceptible to (a) calcineurin inhibitors; (b) high temperature; (c) low calcium concentrations due to BAPTA-chelation of calcium; (d) cell-wall stress or alkaline pH stress; (e) nickel stress or UV damage; (f) dikaryon mating in V8 agar. Two independent transformations were performed and indicated by round no. Shown is one representative experiment.
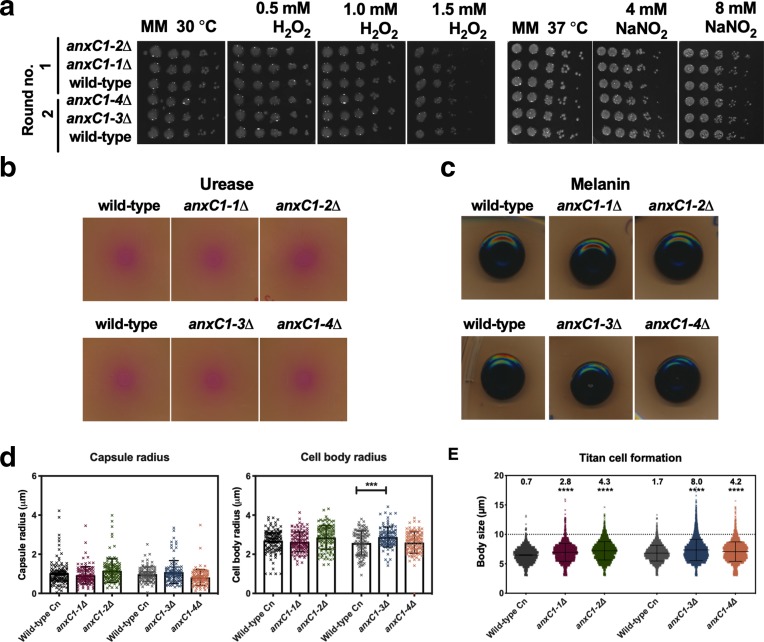
Fungal annexins are poorly characterized and the literature provides no additional clues to their functions. We performed a broad panel of assays to pinpoint the functions of this protein in fungi (Fig. 3). Annexins are involved in oxidative stress in T. lanuginosus [17]. However, we detected no alteration in growth when anxC1∆ strains after exposure to oxidative and nitro-oxidative stress. Alternatively, annexins can be involved in membrane trafficking, i.e. exocytosis and plasma membrane dynamics. We reasoned that this could affect secretion of virulence factors and cellular morphology in stress conditions. We found no defect in urease or melanin secretion or alteration in capsule size in our experimental conditions. These results provide evidence against a defect in secretion of virulence factors and therefore a role of cryptococcal annexin in secretion of these virulence factors. However, we found that one of our deletion strains had an increase in cell body size compared to its parental strain. We detected a significant increase in the number of titan cells produced by anxC1Δ strains in vitro [44] when compared to the parental strain H99, in all experiments performed. These data implicate AnxC1 as a negative regulator of titan cell formation. We note the magnitude of this effect is small and therefore its biological relevance is difficult to ascertain. One possible strategy to confirm influence in titan cell formation would involve engineering a complemented strain. Complemented strains in C. neoformans frequently do not fully reconstitute the wild-type phenotypes, and can display aberrant morphology [56], negating the utility of generating a complemented strain in this instance. Overall, we observed an increase in titan cell formation in AnxC1 deletion strains, but do not decipher the biological relevance of this phenomenon.
Fig. 3.
AnnexinC1 deletion does not affect virulence factors but increases production of titan cells. (a) resistance to oxidative and nitrosative stress in MM agar plates. Two independent transformations were performed and indicated by round no.; (b) urease secretion in Christensen agar plates; (c) melanin secretion in MM with addition of 1 mM L-DOPA; experiments were performed at least three times and one representative experiment is shown. (d) Capsule size or cell size, after 24 h incubation in conditions of mammalian cell culture. ***P<0.001 by one-way ANOVA, with Sidak correction. Experiments were repeated three times with triplicates and shown is the mean and sd of all experiments; (e) titan cell formation (numbers on top represent % of titan cells, defined as cells with cell body size >10 µm). Black bars represent average and sd are shown of one representative experiment, analysing over 1000 individual cells per strain. ****P<0.0001 for unpaired t-test for all strains compared with parental wild-type strains.
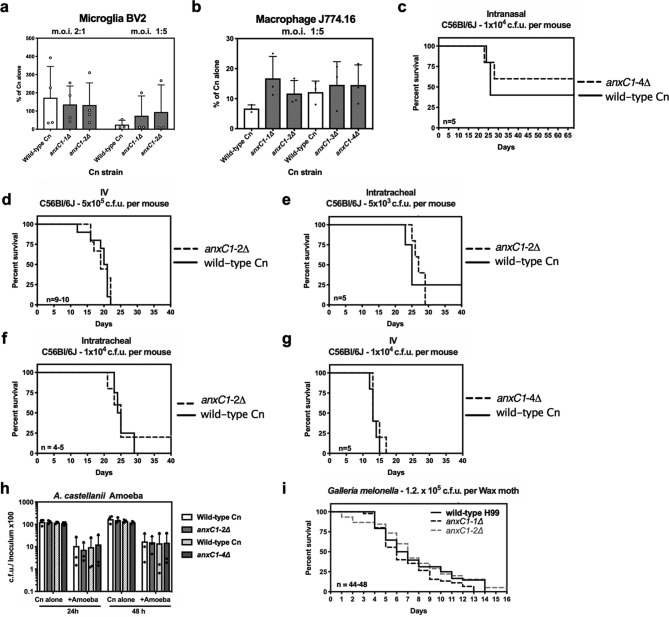
Given the lack of clues to cellular functions of AnxC1 we tested anxc1Δ strains for alterations in virulence in several models of virulence that are routinely performed in our laboratory (Fig. 4). We found a similar survival rate when we exposed both deletion strains and parental strains to murine macrophages. Because disease is a complex interaction between host and pathogen we decided to measure animal survival when challenged with anxC1∆ strains, and we used different routes of infection to cover the broadest range of host–pathogen interactions. Infection of mice via intranasal (IN), intravenous (IV) and intratracheal (IT) routes demonstrated indistinguishable death rate and time to death between anxC1Δ strains and their parental wild-type. C. neoformans when in environmental niches, soil and trees, is exposed to predation by amoeba. We found that anxC1∆ strains had similar survival rates, as compared to wild-type, when ingested by the amoeba Acanthamoeba castellanii. When C. neoformans is exposed to amoeba in a solid agar matrix, it can filament and form pseudohyphae structures [57] (Fu, manuscript in preparation), but we observed no gross differences between the wild-type and the deletion strains in filamentation morphology (data not shown). Finally, in the Galleria mellonella invertebrate infection model, both deletion or wild-type strains showed similar survival kinetics. Based on this data, we conclude that AnxC1 is not required for virulence in the tested models. Our investigations of the roles of cryptococcal annexin showed no clues on the function of annexin, with the exception of a small increase in titan cell formation.
Fig. 4.
AnnexinC1 deletion does not affect virulence of C. neoformans. (a) Survival of C. neoformans (Cn), as measured by tadpole assay, after 24 h infection of BV2, a murine microglia cell line, or (b) J774.16, a murine ascites-derived macrophage cell line; (c) intranasal infection; (d) IV infection; (e–g) intratracheal infections; (h) c.f.u. after interactions with A. castellani. (i) Survival of Galleria melonella challenged with Cn strains. Experiments (a, b and h) were repeated three times with triplicates and shown is mean and sd of all experiments. For survival experiments (c–g, I) n represents number of infected individuals per Cn strain and shown is pooled data from all experiments. No differences were found when testing statistical significance by one-way ANOVA (a, b and h), and log-rank or Gehan–Breslow–Wilcoxon test (c–g,i).
Discussion
The importance of annexins to mammalian and plant biology led us to wonder about the role of fungal annexins, specifically in C. neoformans. The scarcity of knowledge regarding fungal annexins and our preliminary searches implicating annexin in responses to two major regulators of fungal virulence, the calcineurin and the Rim101 pathways, encouraged us to pursue a characterization of the cryptococcal annexin. Based on the most recent annexin nomenclature classifications, we propose that the name annexinXIV should be replaced by AnnexinC1.
Our initial search through available datasets suggested that in C. neoformans, annexin could be involved in the calcineurin pathway, given that it was previously identified as a target of Crz1 and upregulated in high temperature [22]. When testing for phenotypes indicative of deficiencies in calcineurin/Crz1 pathway, AnxC1 is not an effector of the known functions of calcineurin/Crz1. If annexinC1 functions are dependent on calcium it is possible that the high affinity calcium channel Cch1-Mid1 or calcium from ER stores provide sufficient calcium from annexin function [58], a possibility we did not test. Another master regulator of cryptococcal biology and virulence that affects ANXC1 expression levels is Rim101, which is critical for cell-wall integrity and pH response [28], as well as titan cell induction. Titan cells are enlarged C. neoformans cells occurring in mouse lungs during infection and generated in vitro under multiple conditions [44, 59, 60]. The characteristic cell enlargement is associated with increased virulence and has been shown to be negatively regulated by Pkr1 [44] and positively regulated by Rim101 [28]. We observed no alteration in capacity to grow at neutral to basic pH in anxC1Δ strain , but the observed increase in titan cell production in ANXC1-deletion strains posits a negative regulation circuit between Rim101 and AnxC1.
We observe no differences in virulence between anxC1Δ and wild-type strains in both mammalian or invertebrate infection models. Overall, the lack of phenotypes associated with AnnexinC1 was puzzling, particularly given that its transcription is altered by important virulence regulators such as Crz1 and Rim101. Given that the several deletion AnxC1-deletion clones yielded no discernible phenotype on all but one of the phenotypes tested and its effects on titan cell formation were minor, we saw no need to complement the gene since there was no strong phenotype to restore. It may be that annexinC1 functions are much subtler than our assays could detect. For example, in the slime mold D. discoideum, deletion of annexin delays, but does not prevent, repair of the plasma membrane upon laser wounding [61]. An alternative hypothesis is that an additional annexin is present in C. neoformans and that both annexins play redundant roles. However, sequence analysis revealed no other annexin in C. neoformans and therefore if another molecular player is responsible for redundancy in annexin functions, it could not be readily identified by sequence homology. Our data is similar to what has been previously shown for Aspergillus spp., where no obvious phenotypes were observed upon deletion of annexin genes [13, 14, 16]. Likewise, in Neurospora crassa an annexin deleted strain had no gross growth or filamentation defects [62].
In contrast, in the thermophilic fungi Thermomyces lanuginosus a deletion strain of AnxC7 displayed normal growth in standard media, but had increased resistance to cell-wall stress and oxidative stress. This fungal annexin C7 is predicted to localize to fungal mitochondria [17]. These authors also observed that conidia formation was precocious. This observation may be somehow related to the observation that in C. neoformans expression of AnxC1 peaked at the time of bud formation [63], approximately 50 min after separation of cells by centrifugal elutriation, insinuating that cryptococcal annexin, and maybe annexins from most fungal species, are involved in membrane dynamics during cellular replication. This framework would fit within our observation that AnxC1 deletion leads to an increase in the formation of titan cells, which display very atypical size and replication patterns, requiring considerable coordination with cellular membrane dynamics. AnxC1 was not differentially regulated during titan cell formation using Trevijano-Contador et al.'s dataset (the only RNAseq dataset available to date for titan cell formation), generated after 7 and 18 h of titan cell induction as compared to normal sized cells [60]. A role in membrane dynamics would also fit within the generalized function of annexins as a bridge between phospholipid structures, given the dynamic membrane remodelling needed during cellular replication. Future studies should define the cellular localization and regulation of annexinC1 protein to complement the transcriptomic data already available and better define these hypotheses. Given the scarcity of studies focusing on fungal annexins and shortage of (dramatic) phenotypic insights from deletion strains, the role of cryptococcal annexins remains cryptic.
In summary, C. neoformans possesses one single annexin gene as evident from sequence homology analysis. We were not able to identify a molecular function for AnxC1 because comparisons of annexin-deficient to wild-type strains revealed no discernible phenotype with the notable exception of an increase in titan cell formation. Although it is conceivable that a critical function lurks beneath the absence of vivid phenotypes, we were unable to find a strong effect in major phenotypic features of C. neoformans and its virulence in mammalian, invertebrate and protozoal models of infection. In this regard, our experience is similar to that of investigators who have explored the role of annexin in Neurospora crassa and Aspergillus spp. [12, 14, 16]. who have found only weak phenotypes upon deletion of this gene. The explanation for these negative results could lie in the existence of redundant mechanisms to compensate for the absence of annexin or new functions that were not considered in experimental design. In this regard, it is possible that annexins in fungi function very differently than annexins in the other kingdoms of life. Although negative results are disappointing, we take the optimistic view that a gene that is found as a single copy in C. neoformans with homologues throughout the fungal kingdom must be important somewhere in the biology or life cycle of these organisms. Perhaps as new assays are developed to study C. neoformans physiology and mechanisms of virulence, a role for its single annexin will become apparent. Accurate three-dimensional structures of fungal annexin proteins, coupled with in-depth cellular localization and transcriptional studies, would allow in silico structure homology search, which could help resolve the conundrum of specific biological function of cryptococcal annexins.
Supplementary Data
Funding information
A. C. was supported by National Institutes of Health (NIH) awards 5R01HL059842, 5R01AI033774, 5R37AI033142, and 5R01AI052733. E. C. was supported by a Postdoctoral Fellowship from the Johns Hopkins Malaria Research Institute. E. E. F. was supported by RISE grant number: 1R25GM113748-01. The fungal deletion libraries were funded by NIH R01AI100272 to Hiten D. Madhani and deposited in the FGSC (http://www.fgsc.net/).
Author contributions
C. C. and M. M. generated deletion strains, performed the experiments and analysis. C. C. and A. C. conceptualized the experiments and supervised the project. E. C., D. S. G., M. S. F., E. E. F., A. A. performed the phenotypic assays and performed formal analysis. C. C. wrote the original draft. All authors edited, read and approved the final version of the manuscript. A. C. acquired the financial support for the project.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All animal experiments were approved by Johns Hopkins University IACUC under protocol number MO18H152.
Footnotes
Abbreviations: BAPTA, 1,2-bis(o-aminophenoxy)ethane-N,N,N′,N′-tetraacetic acid; c.f.u., colony-forming units; CFW, calcofluor white; L-DOPA, L-3,4-dihydroxyphenylalanine; MIC, Minimal inhibitory concentration; MM, Minimal Media; m.o.i., multiplicity of infection; NAT, nourseothricin; YPD, Yeast Peptone Dextrose;
Three supplementary figures are available with the online version of this article.
Edited by: V. J. Cid and C. Kistler
References
- 1.Gerke V, Moss SE. Annexins: from structure to function. Physiol Rev. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- 2.Clark GB, Morgan RO, Fernandez MP, Roux SJ. Evolutionary adaptation of plant annexins has diversified their molecular structures, interactions and functional roles. New Phytol. 2012;196:695–712. doi: 10.1111/j.1469-8137.2012.04308.x. [DOI] [PubMed] [Google Scholar]
- 3.Davies JM. Annexin-mediated calcium signalling in plants. Plants. 2014;3:128–140. doi: 10.3390/plants3010128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang F, Li S, Yang S, Wang L, Guo W. Overexpression of a cotton annexin gene, GhAnn1, enhances drought and salt stress tolerance in transgenic cotton. Plant Mol Biol. 2015;87:47–67. doi: 10.1007/s11103-014-0260-3. [DOI] [PubMed] [Google Scholar]
- 5.Li R, Tan S, Yu M, Jundt MC, Zhang S, et al. Annexin A2 regulates autophagy in Pseudomonas aeruginosa infection through the Akt1-mTOR-ULK1/2 signaling pathway. J Immunol. 2015;195:3901–3911. doi: 10.4049/jimmunol.1500967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stukes S, Coelho C, Rivera J, Jedlicka AE, Hajjar KA, et al. The membrane phospholipid binding protein annexin A2 promotes phagocytosis and nonlytic exocytosis of Cryptococcus neoformans and impacts survival in fungal infection. J Immunol. 2016;197:1252–1261. doi: 10.4049/jimmunol.1501855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanessa KHQ, Julia MG, Wenwei L, Michelle ALT, Zarina ZRS, et al. Absence of annexin A1 impairs host adaptive immunity against Mycobacterium tuberculosis in vivo. Immunobiology. 2015;220:614–623. doi: 10.1016/j.imbio.2014.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Jenikova G, Hruz P, Andersson MK, Tejman-Yarden N, Ferreira PCD, et al. A1-giardin based live heterologous vaccine protects against Giardia lamblia infection in a murine model. Vaccine. 2011;29:9529–9537. doi: 10.1016/j.vaccine.2011.09.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kodavali PK, Dudkiewicz M, Pikuła S, Pawłowski K. Bioinformatics analysis of bacterial annexins-putative ancestral relatives of eukaryotic annexins. PLoS One. 2014;9:e85428. doi: 10.1371/journal.pone.0085428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantacessi C, Seddon JM, Miller TL, Leow CY, Thomas L, et al. A genome-wide analysis of annexins from parasitic organisms and their vectors. Sci Rep. 2013;3:2893. doi: 10.1038/srep02893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weeratunga SK, Osman A, Hu N-J, Wang CK, Mason L, et al. Alpha-1 giardin is an annexin with highly unusual calcium-regulated mechanisms. J Mol Biol. 2012;423:169–181. doi: 10.1016/j.jmb.2012.06.041. [DOI] [PubMed] [Google Scholar]
- 12.Dunlap JC, Borkovich KA, Henn MR, Turner GE, Sachs MS, et al. Enabling a community to dissect an organism: overview of the Neurospora functional genomics project. Adv Genet. 2007;57:49–96. doi: 10.1016/S0065-2660(06)57002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khalaj K, Aminollahi E, Bordbar A, Khalaj V. Fungal annexins: a mini review. Springerplus. 2015;4:721. doi: 10.1186/s40064-015-1519-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalaj V, Azarian B, Enayati S, Vaziri B. Annexin C4 in A. fumigatus: a proteomics approach to understand the function. J Proteomics. 2011;74:1950–1958. doi: 10.1016/j.jprot.2011.05.018. [DOI] [PubMed] [Google Scholar]
- 15.Khalaj V, Smith L, Brookman J, Tuckwell D. Identification of a novel class of annexin genes. FEBS Lett. 2004b;562:79–86. doi: 10.1016/S0014-5793(04)00186-3. [DOI] [PubMed] [Google Scholar]
- 16.Khalaj V, Hey P, Smith L, Robson GD, Brookman J. The Aspergillus niger annexin, anxC3.1 is constitutively expressed and is not essential for protein secretion. FEMS Microbiol Lett. 2004a;239:163–169. doi: 10.1016/j.femsle.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 17.Xie XL, Yang H, Chen LN, Wei Y, Zhang SH. ANXC7 is a Mitochondrion-Localized annexin involved in controlling Conidium development and oxidative resistance in the thermophilic fungus Thermomyces lanuginosus . Front Microbiol. 2018;9:864. doi: 10.3389/fmicb.2018.01770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Braun EL, Kang S, Nelson MA, Natvig DO. Identification of the first fungal annexin: analysis of annexin gene duplications and implications for eukaryotic evolution. J Mol Evol. 1998;47:531–543. doi: 10.1007/PL00006409. [DOI] [PubMed] [Google Scholar]
- 19.Rajasingham R, Smith RM, Park BJ, Jarvis JN, Govender NP, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown JCS, Nelson J, VanderSluis B, Deshpande R, Butts A, et al. Unraveling the biology of a fungal meningitis pathogen using chemical genetics. Cell. 2014;159:1168–1187. doi: 10.1016/j.cell.2014.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lev S, Desmarini D, Chayakulkeeree M, Sorrell TC, Djordjevic JT. The Crz1/Sp1 transcription factor of Cryptococcus neoformans is activated by calcineurin and regulates cell wall integrity. PLoS One. 2012;7:e51403. doi: 10.1371/journal.pone.0051403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park H-S, Chow EWL, Fu C, Soderblom EJ, Moseley MA, et al. Calcineurin targets involved in stress survival and fungal virulence. PLoS Pathog. 2016;12:e1005873. doi: 10.1371/journal.ppat.1005873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chow EWL, Clancey SA, Billmyre RB, Averette AF, Granek JA, et al. Elucidation of the calcineurin-Crz1 stress response transcriptional network in the human fungal pathogen Cryptococcus neoformans . PLoS Genet. 2017;13:e1006667. doi: 10.1371/journal.pgen.1006667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Steinbach WJ, Reedy JL, Cramer RA, Perfect JR, Heitman J. Harnessing calcineurin as a novel anti-infective agent against invasive fungal infections. Nat Rev Micro. 2007;5:418–430. doi: 10.1038/nrmicro1680. [DOI] [PubMed] [Google Scholar]
- 25.Derengowski LdaS, Paes HC, Albuquerque P, Tavares AH, Fernandes L, et al. The transcriptional response of Cryptococcus neoformans to ingestion by Acanthamoeba castellanii and macrophages provides insights into the evolutionary adaptation to the mammalian host. Eukaryot Cell. 2013;12:761–774. doi: 10.1128/EC.00073-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geddes JMH, Croll D, Caza M, Stoynov N, Foster LJ, et al. Secretome profiling of Cryptococcus neoformans reveals regulation of a subset of virulence-associated proteins and potential biomarkers by protein kinase A. BMC Microbiol. 2015;15:206. doi: 10.1186/s12866-015-0532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodrigues ML, Nakayasu ES, Oliveira DL, Nimrichter L, Nosanchuk JD, et al. Extracellular vesicles produced by Cryptococcus neoformans contain protein components associated with virulence. Eukaryot Cell. 2008;7:58–67. doi: 10.1128/EC.00370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio. 2013;4:e00522-12. doi: 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, et al. CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017;45:D200–D203. doi: 10.1093/nar/gkw1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004b;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benkert P, Biasini M, Schwede T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics. 2011;27:343–350. doi: 10.1093/bioinformatics/btq662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Biasini M, Bienert S, Waterhouse A, Arnold K, Studer G, et al. SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42:W252–W258. doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Basenko E, Pulman J, Shanmugasundram A, Harb O, Crouch K, et al. FungiDB: an integrated bioinformatic resource for fungi and Oomycetes. J Fungi. 2018;4:39. doi: 10.3390/jof4010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janbon G, Ormerod KL, Paulet D, Byrnes EJ, Yadav V, et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014;10:e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, et al. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans . Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McClelland EE, Ramagopal UA, Rivera J, Cox J, Nakouzi A, et al. A small protein associated with fungal energy metabolism affects the virulence of Cryptococcus neoformans in mammals. PLoS Pathog. 2016;12:e1005849. doi: 10.1371/journal.ppat.1005849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim MS, Kim SY, Jung KW, Bahn YS. Targeted gene disruption in Cryptococcus neoformans using double-joint PCR with split dominant selectable markers. Methods Mol Biol. 2012;845:67-84. doi: 10.1007/978-1-61779-539-8_5. [DOI] [PubMed] [Google Scholar]
- 38.Kent CR, Ortiz-Bermúdez P, Giles SS, Hull CM. Formulation of a defined V8 medium for induction of sexual development of Cryptococcus neoformans . Appl Environ Microbiol. 2008;74:6248–6253. doi: 10.1128/AEM.00970-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexander BD. Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeasts. 4 edn. Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 40.Damiani G, Kiyotaki C, Soeller W, Sasada M, Peisach J, et al. Macrophage variants in oxygen metabolism. J Exp Med. 1980;152:808–822. doi: 10.1084/jem.152.4.808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Casadevall A, Cleare W, Feldmesser M, Glatman-Freedman A, Goldman DL, et al. Characterization of a murine monoclonal antibody to Cryptococcus neoformans polysaccharide that is a candidate for human therapeutic studies. Antimicrob Agents Chemother. 1998;42:1437–1446. doi: 10.1128/AAC.42.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch AZ, Koshland DE. A simple colony-formation assay in liquid medium, termed 'tadpoling', provides a sensitive measure of Saccharomyces cerevisiae culture viability. Yeast. 2013;30:501–509. doi: 10.1002/yea.2989. [DOI] [PubMed] [Google Scholar]
- 43.Fu MS, Casadevall A. Divalent metal cations potentiate the predatory capacity of amoeba for Cryptococcus neoformans . Appl Environ Microbiol. 2018;84 doi: 10.1128/AEM.01717-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hommel B, Mukaremera L, Cordero RJB, Coelho C, Desjardins CA, et al. Titan cells formation in Cryptococcus neoformans is finely tuned by environmental conditions and modulated by positive and negative genetic regulators. PLoS Pathog. 2018;14:e1006982. doi: 10.1371/journal.ppat.1006982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bouklas T, Diago-Navarro E, Wang X, Fenster M, Fries BC. Characterization of the virulence of Cryptococcus neoformans strains in an insect model. Virulence. 2015;6:809–813. doi: 10.1080/21505594.2015.1086868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eisenman HC, Duong R, Chan H, Tsue R, McClelland EE. Reduced virulence of melanized Cryptococcus neoformans in Galleria mellonella . Virulence. 2014;5:611–618. doi: 10.4161/viru.29234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mylonakis E, Moreno R, El Khoury JB, Idnurm A, Heitman J, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infect Immun. 2005;73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trevijano-Contador N, Herrero-Fernández I, García-Barbazán I, Scorzoni L, Rueda C, et al. Cryptococcus neoformans induces antimicrobial responses and behaves as a facultative intracellular pathogen in the non mammalian model Galleria mellonella . Virulence. 2015;6:66–74. doi: 10.4161/21505594.2014.986412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivera J, Mukherjee J, Weiss LM, Casadevall A. Antibody efficacy in murine pulmonary Cryptococcus neoformans infection: a role for nitric oxide. J Immunol. 2002;168:3419–3427. doi: 10.4049/jimmunol.168.7.3419. [DOI] [PubMed] [Google Scholar]
- 50.Inglis DO, Skrzypek MS, Liaw E, Moktali V, Sherlock G, et al. Literature-based gene curation and proposed genetic nomenclature for Cryptococcus . Eukaryot Cell. 2014;13:878–883. doi: 10.1128/EC.00083-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Morgan RO, Martin-Almedina S, Iglesias JM, Gonzalez-Florez MI, Fernandez MP. Evolutionary perspective on annexin calcium-binding domains. Biochim Biophys Acta. 2004;1742:133–140. doi: 10.1016/j.bbamcr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 52.Mortimer JC, Laohavisit A, Macpherson N, Webb A, Brownlee C, et al. Annexins: multifunctional components of growth and adaptation. J Exp Bot. 2008;59:533–544. doi: 10.1093/jxb/erm344. [DOI] [PubMed] [Google Scholar]
- 53.Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008;25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 54.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen YL, Lehman VN, Lewit Y, Averette AF, Heitman J. Calcineurin governs thermotolerance and virulence of Cryptococcus gattii . G3. 2013;3:527–539. doi: 10.1534/g3.112.004242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Arras SDM, Chitty JL, Blake KL, Schulz BL, Fraser JA. A genomic safe haven for mutant complementation in Cryptococcus neoformans . PLoS One. 2015;10:e0122916. doi: 10.1371/journal.pone.0122916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Magditch DA, Liu TB, Xue C, Idnurm A. DNA mutations mediate microevolution between host-adapted forms of the pathogenic fungus Cryptococcus neoformans . PLoS Pathog. 2012;8:e1002936. doi: 10.1371/journal.ppat.1002936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vu K, Bautos JM, Gelli A. The Cch1-Mid1 high-affinity calcium channel contributes to the virulence of Cryptococcus neoformans by mitigating oxidative stress. Eukaryot Cell. 2015;14:1135–1143. doi: 10.1128/EC.00100-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dambuza IM, Drake T, Chapuis A, Zhou X, Correia J, et al. The Cryptococcus neoformans titan cell is an inducible and regulated morphotype underlying pathogenesis. PLoS Pathog. 2018;14:e1006978. doi: 10.1371/journal.ppat.1006978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trevijano-Contador N, de Oliveira HC, García-Rodas R, Rossi SA, Llorente I, et al. Cryptococcus neoformans can form titan-like cells in vitro in response to multiple signals. PLoS Pathog. 2018;14:e1007007. doi: 10.1371/journal.ppat.1007007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pervin MS, Itoh G, Talukder MSU, Fujimoto K, Morimoto YV, et al. A study of wound repair in Dictyostelium cells by using novel laserporation. Sci Rep. 2018;8:7969. doi: 10.1038/s41598-018-26337-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Turner GE. Phenotypic analysis of Neurospora crassa gene deletion strains. Methods Mol Biol. 2011;722:191–198. doi: 10.1007/978-1-61779-040-9_14. [DOI] [PubMed] [Google Scholar]
- 63.Kelliher CM, Leman AR, Sierra CS, Haase SB. Investigating conservation of the cell-cycle-regulated transcriptional program in the fungal pathogen, Cryptococcus neoformans . PLoS Genet. 2016;12:e1006453. doi: 10.1371/journal.pgen.1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.