Abstract
CD34+ myeloid lineage progenitor cells are an important reservoir of latent human cytomegalovirus (HCMV), and differentiation to macrophages or dendritic cells (DCs) is known to cause reactivation of latent virus. Due to its species-specificity, murine models have been used to study mouse CMV (MCMV) latency and reactivation in vivo. While previous studies have shown that MCMV genomic DNA can be detected in the bone marrow (BM) of latently infected mice, the identity of these cells has not been defined. Therefore, we sought to identify and enrich for cellular sites of MCMV latency in the BM haematopoietic system, and to explore the potential for establishing an in vitro model for reactivation of latent MCMV. We studied the kinetics and cellular characteristics of acute infection and establishment of latency in the BM of mice. We found that while MCMV can infect a broad range of haematopoietic BM cells (BMCs), latent virus is only detectable in haematopoietic stem cells (HSCs), myeloid progenitor cells, monocytes and DC-enriched cell subsets. Using three separate approaches, MCMV reactivation was detected in association with differentiation into DC-enriched BMCs cultured in the presence of granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin 4 (IL-4) followed by lipopolysaccharide (LPS) treatment. In summary, we have defined the kinetics and cellular profile of MCMV infection followed by the natural establishment of latency in vivo in the mouse BM haematopoietic system, including the haematopoietic phenotypes of cells that are permissive to acute infection, establish and harbour detectable latent virus, and can be stimulated to reactivate following DC enrichment and differentiation, followed by treatment with LPS.
Keywords: MCMV, hematopoietic cells, latency, reactivation
Introduction
Cytomegalovirus (CMV) remains an important pathogen in immunocompromised patients [1]. In addition to well-established cytopathic effects (CPE) in target organs, CMV infection can induce myelosuppression, thrombocytopenia and haemolytic anemia [2]. Moreover, human CMV (HCMV) infection has been associated with delayed engraftment and graft failure in stem-cell transplant recipients. The suppressive effects on haematopoiesis may reflect a direct inhibition of haematopoietic stem cells and progenitor cells (HSPCs), or alternatively damage to bone marrow (BM) stromal cells that play a supportive role in haematopoiesis [2–4]. CD34+ HSPCs constitute a critical reservoir of latent HCMV in seropositive individuals, and latency is maintained when these HSPCs differentiate along the myeloid lineage into monocytes [5–9]. Reactivation of latent viral genomes occur when infected HSPCs and monocytes differentiate into mature dendritic cells or macrophages, and is characterized by the expression of lytic genes and production of infectious viral progeny [10–14]. Due to the species-specificity of HCMV infection, and the limitations of studying molecular mechanisms responsible for reactivation in vivo in human subjects, we and others have utilized murine models to study CMV reactivation in vivo, while HCMV studies remain restricted to cell and tissue culture in vitro models.
Murine cytomegalovirus (MCMV) is similar to HCMV in many aspects, including the ability to establish latent infection and reactivate from latency, the organization and function of immediate early (IE) genes, and the presence of transcription factor binding sites in major immediate enhancer and promoter (MIEP) regions that respond to inflammatory signalling pathways [15–18]. Therefore, as a result of our ability to infect and manipulate latently infected mice, we and others have utilized in vivo models to study numerous aspects of MCMV infection such as pathogenesis, immunity, latency, reactivation and superinfection [19–25]. Similar to HCMV, MCMV can infect the BM acutely causing myelosuppression, characterized by a reduction in the number of lineage marker negative and c-Kit/CD117 positive (Lin- CD117+) and Lin- CD34+ cells. In addition, MCMV causes decreases in c.f.u-spleen (c.f.u.-S), in c.f.u-granulocyte/macrophage (c.f.u.-GM) in BM cells (BMCs), and in haematocrit and platelet counts in the peripheral blood [26–28]. While MCMV DNA can be detected in the BM of latently infected mice, the cell types that harbour viral latency have remained elusive [29, 30].
In this study, we sought to define and characterize the cellular sites of MCMV latency in the BM haematopoietic system, and to study the potential for establishing an in vitro model of MCMV reactivation from latency through DC enrichment, differentiation followed by treatment with LPS. An in vitro model that allows reactivation of naturally occurring latency has the potential to contribute significantly to our current understanding of the molecular events operative in CMV reactivation.
Methods
Mice and viruses
Three-week-old female specific-pathogen-free BALB/c mice and pregnant BALB/c mice with 15- to 17-day-old embryos were purchased from Jackson Laboratory. Mice were maintained in isolation cages and fed and watered ad libitum. MCMV (Smith strain, ATCC 194-VR) was purchased from America Type Culture Collection (ATCC). Recombinant MCMV RM4503 derived from K181 strain that can express EGFP, driven by a chimeric promoter of HCMV and MCMV, obtained from Dr Edward Mocarski was also used [31]. Viral stocks were generated by routine propagation in mouse salivary gland as previously described [32]. Viral stocks were titrated on confluent monolayers of murine embryonic fibroblasts (MEFs). To establish latency, 3–4-week-old female BALB/c mice were infected by intraperitoneal (IP) injection with 5×105 p.f.u. of Smith strain or 1×107 p.f.u. of RM4503 strain, and housed ≥3 months in the Northwestern University Center for Comparative Medicine (CCM).
Some of our virus stocks of RM4503 have been propagated repeatedly in vitro either in MEF or 3T3 cells for many times, resulting in loss of virulence compared to other MCMV viruses. As a result, when we noticed loss in virulence, we increased the inoculum needed to generate latent mice in order to make more robust comparisons between viruses and experiments. Our benchmark in BALB/c mice using wild-type Smith strain MCMV to create acutely or latently infected mice has traditionally been to use a 100 ul IP injection of 5×106 p.f.u. ml−1 (5×105 p.f.u. inoculum); this is what we used for experiments that utilized the Smith strain. As a result of our titre and virulence data for the stock of RM4503 used in these experiments, we infected mice IP with 100 ul of RM4503 with a titre of 1×108 p.f.u. ml−1 diluted in Dulbecco’s Modified Eagle’s Medium (DMEM).
BMC isolation and separation
Mice were anaesthetized with isoflurane and sacrificed by cervical dislocation, then femurs and tibiae were excised and cleaned of muscle tissue with scalpels. The intact bones were soaked in 70 % ethanol for 3 min for disinfection, and washed with 1× PBS. Then epiphyses were removed with scissors so that the BM was exposed. The BM was flushed out with PBS using a 23-gauge needle attached to a 3 ml syringe. Aggregates were dislodged by passing through a 16-gauge needle, and filtered through a 70 μm nylon strainer. RBCs in the filtrate were lysed with 1× RBC lysis buffer (Biolegend). This solution was filtered through a 70 μm cell strainer to remove aggregates, and washed twice with cold 1× PBS.
Anti-mouse CD19 (6D5), anti-mouse CD3e (145–2 C11), anti-mouse CD49b (DX5), EasySep Mouse FITC-positive Selection Kit, and EasySep Mouse CD11b-positive Selection Kit II used for BMC separation were purchased from StemCell Technologies. Mouse Lineage Cell Depletion Kit, Mouse CD117 Microbeads, Mouse Monocyte Isolation Kit, CD11c Microbeads, CD45 Microbeads, MS Columns, and LS Columns were purchased from Miltanyi Biotec. BMC separation was performed following instructions from the manufacturers.
Cell culture
BALB/c and C57BL/6 mouse embryo fibroblasts (MEFs) were isolated from 15 to 17 day mouse embryos by trypsinization in 0.25 % trypsin with 0.125 % EDTA. The cells were passaged in DMEM with 4.5 g l−1 glucose, 10 % heat inactivated FBS, and 100 IU ml−1 penicillin and streptomycin at 37 °C in a humidified chamber with 5 % CO2.
StemSpan (Stem Cell Technologies) serum-free expansion medium (SFEM) was supplemented with 10 μg ml−1 heparin, 10 ng ml−1 mouse stem-cell factor (SCF), 20 ng ml−1 mouse thrombopoietin (TPO), 10 ng ml−1 human fibroblast growth factor 1 (FGF1), 100 ng ml−1 insulin-like growth factor-binding protein 2 (IGFBP2), and 500 ng ml−1 angiopoietin-like protein 3 (Angptl3), together referred to as STFIA medium, was used to expand the HSPCs[33]. SFEM supplemented with 10 ng ml−1 mouse SCF, 20 ng ml−1 mouse TPO, 10 ng ml−1 human FGF1, and 20 ng ml−1 mouse insulin-like growth factor2 (IGF2) is referred to as STF medium, and was used for short-term BMC maintenance culture [33]. Then, 106 cells ml−1 in STFIA or STF were seeded in a well of six-well ultra-low attachment surface plates (Corning Costar), cultured in a humidified incubator at 37 °C and 5 % CO2, and equal volumes of fresh medium were added in at day 4 and day 7, respectively. Heparin, mouse SCF and mouse TPO were obtained from Stem Cell Technologies. Human FGF1 was obtained from ThermoFisher. IGFBP2, IGF2 and Angptl3 were procured from R and D Systems.
In vitro differentiation of BMCs to cultured bone-marrow-derived macrophages (cBMMs) in the presence macrophage colony-stimulating factor (M-CSF) was performed using the previously described protocol with slight modification [34]. Briefly, BMCs were cultured in bacteriological grade petri dishes in DMEM (1.5 g l−1 glucose) supplemented with 10 % FBS and 20 ng ml−1 mouse M-CSF (Miltenyi Biotec), referred to as M-CSF containing medium. At day 7, the BMMs adhered to the bottom of petri dishes, and were ready for maturation and other experiments.
Cultured bone-marrow-derived DC-enriched (cBMDCe) cells were generated from BMCs using published protocols [35]. Briefly, BMCs were cultured in RPMI1640 supplemented with 10 % FBS, 100 IU ml−1 penicillin/streptomycin, 2 mM l-glutamine or Glutamax, 50 μM 2-mercaptoethanol, 1 % non-essential amino acid, 1 mM sodium pyruvate, 20 ng ml−1 mouse granulocyte-macrophage colony-stimulating factor (GM-CSF), and 5 ng ml−1 interleukin4 (IL-4) (Miltenyi Biotec), which is hereafter referred to as GM-CSF containing medium, for 8 days. The non-adherent cells were collected via gentle pipetting, pelleted through centrifugation, and re-suspended in fresh medium and seeded into a tissue culture petri dish. On day 9, cBMDCe cells were ready for maturation and time-course studies. Both cBMMs and cBMDCe cells can be matured through treatment with 100 ng ml−1 LPS (Sigma Aldrich) for 24 h.
One day before the co-culture experiments, 2×105 MEFs were seeded into each well of six-well plates, up to 5×106 undifferentiated or differentiated BMCs were laid on the top of the MEF monolayer respectively based on the experiment, and nourished with MEF growth medium. Medium change for non-adherent BMCs was carried out through pelleting the floating cells by centrifugation at 250 g for 10 min, re-suspending these cells' pellet in fresh medium, and putting them back into their respective cultures.
Plaque assay
Tissue/cell lysate was made by homogenizing 100 mg of mouse tissue or up to 5 million cells into 1 ml of DMEM, and then clarifying through centrifugation at 1200 r.p.m. in 4 °C. The supernatant from the co-culture or clarified tissue/cell lysate was used to infect the 60–80 % confluent MEF monolayers in six-well plates followed by agarose overlay, as previously described [30]. The cells were maintained in high glucose DMEM with 10 % FBS and observed by inverted-phase microscopy (5 days) for cytopathic effects characteristic of CMV infection. The plaques were visualized by crystal violet staining. Assay results were expressed as the number of p.f.u, of MCMV per milliliter of supernatant.
Flow cytometric cell staining
Primary cells from mouse BM and/or from cell culture were washed twice with PBS before further processing for staining and flow cytometry. Cell viability was assayed by staining with Fixable Viability Dye eFluor 455UV or eFluor 506 (Affymetrix eBioscience) in azide- and protein-free PBS for 30 min in the dark at room temperature (RT). It is crucial to gate and sort for viable cells. Cells were washed with 1×PBS and processed for flow-cytometry staining. For all surface antigen staining, cells were incubated in 1 μg Fc Block (BD Bioscience) for 10 min at RT, and then stained with respective panels of pre-titrated antibodies shown in Tables S1 and S2 (available in the online version of this article), in the dark for 30 min at 4 °C in staining buffer (Biolegend). Cells were then washed twice with PBS+2 % BSA wash buffer followed by flow-cytometry analysis and/or cell sorting. Antibodies used for cell-surface marker staining for immunophenotyping analysis of differentiated BMCs are listed in Table S3, those used for BMC sorting are listed in Table S4. Compensation was performed using appropriate single-colour controls prepared from AbC Total antibody Compensation Beads Kit (ThermoFisher Scientific) per the manufacturer’s instructions. Flow-cytometry cell analysis and sorting were performed on instruments located at the Robert H. Lurie Comprehensive Cancer Center-Flow Cytometry Core Facility at the Chicago campus of Northwestern University. Immunophenotyping analysis experiments were carried out on BD LSRFortessa 6 Laser 18-parameter SORP (Special Order Research Product, BD Immunocytometry Systems) analyser, equipped with 355, 405, 488, 552, 640 and 690 nm lasers. And BMC sorting was carried out using a BD FACSAria 5-laser 16-parameter SORP cell sorter, equipped with 355, 405, 488, 561 and 640 nm lasers. All data collection and sorting were performed using BD FACS Diva software (BD Biosicences), and data analysis performed using FlowJo software (Tree Star).
Nucleic acid extraction
Total BMCs isolated from mice and various cell fractions from the cell-sorting experiments were snap frozen in liquid nitrogen and then transferred to a −80 °C freezer. DNA was extracted from the total BMCs using Purgene Core Kit A (QIAGEN), and from sorted BMCs using QIAamp DNA MicroKit (QIAGEN) according to the manufacturer’s instructions. Total RNA was isolated and purified with TRIzol Plus RNA purification kit (Thermofisher Scientific). On-column DNase I digestion was performed during RNA purification. DNA and RNA were quantified by absorbance at 260 nm on a NanoDROP LITE spectrophotometer (ThermoFisher Scientific).
Reverse transcription (RT)
Oligo(dT)-primed cDNA was generated from 5 μg of total RNA by using AffinityScript Multiple Temperature cDNA Synthesis Kit (Agilent Technologies) in a 20 μl reaction. Reverse transcriptase was inactivated at 70 °С for 15 min, samples cooled to 37 °С, and treated for 30 min with 0.15 unit μl−1 RNaseH and 1 μl RNase Cocktail (ThermoFisher Scientific). Subsequently, the cDNA was purified with a QIAquick PCR Purification Kit (Qiagen, Valencia, CA, USA), and quantified using a NanoDROP LITE spectrophotometer (ThermoFisher Scientific).
Real-time PCR analysis
All of the real-time PCR reactions were performed using Taqman Gene Expression Master Mix and ABI 7500 Fast System. Standard Curve (Absolute Quantitation) Assay, and regular 7500 mode were selected. Reactions contained a 900 nM of Taqman primers, and a 250 nM of Taqman MGB probes. Each sample was analysed in triplicate. The thermal cycling conditions were 2 min at 50 °С, 10 min at 95 °С, followed by 50 cycles of 95 °С for 15 s, 60 °С for 1 min.
Quantification of MCMV DNA and RNA by real-time PCR
For absolute quantification of MCMV DNA copy number in mouse BMCs, a standard curve was generated by serial dilution of plasmid pIE111, which contains the MIEP and immediate early (IE) genes (17) in genomic DNA isolated from BMCs of uninfected mice, such that 13.5 µl of the template contained 105, 104, 103, 102 or 20 copies of pIE111 in 800 ng cellular DNA. Similarly, a standard curve for mouse eukaryotic translation elongation factor 1 alpha 1 (mEF1α) gene was generated by serial dilution of plasmid pDRIVE-mEF1α (InvivoGene), which contains the promoter region of mEF1α, such that the 13.5 µl template contained 106, 105, 104, 103 or 102 copies of pDRIVE-mEF1α in 800 ng sheared salmon sperm DNA. In total, 800 ng of sample genomic DNA for each reaction was used as a template for PCR in a 30 µl reaction. Overall, 800 ng of genomic DNA from uninfected mouse kidneys was used as a non-template control (NTC) for detection of non-specific amplification. The primers and probes used to detect MCMV MIEP and mEF1α promoter DNA were as previously described (36). The target DNA copy number in each sample was calculated based on the standard curve generated in each individual assay. Viral DNA copy number per million cells was determined by dividing the average copy number of MCMV DNA by the mEF1α copy number in the same sample and multiplying by 2×106.
Quantification of MCMV IE1, IE3, M112 and M100 mRNA was performed using the relative standard curve method as described in the Applied Biosystems support materials ‘Guide to performing relative quantitation of gene expression using real-time quantitative PCR’ (www6.appliedbiosystems.com/support/tutorials/pdf/performing_rq_gene_exp_rtpcr.pdf) and the primers and probes were as previously described [36]. Serially diluted cDNA derived from RNA isolated from kidneys at 7 days post-infection (p.i.) was used to generate the standard curve. Then, 18 ng of sample cDNA was added to a 20 µl reaction for amplification of virus cDNA and the same amount of cDNA from uninfected mouse kidneys was used as a NTC. The abundance of the target cDNA in each sample was determined according to a standard curve produced in each run. Mouse Eef2 RNA was employed as an internal control. The relative quantity of MCMV RNA in the sample was determined by dividing the average quantity of MCMV mRNA by that of Eef2, as determined from the standard curve. The sequences of primers and Taqman MGB probe for mouse Eef2 mRNA were described previously [36].
Statistical analysis
A two-tailed Student’s t-test was used to determine statistical significance. P-values≤0.05 were considered significant.
Results
Kinetics of acute MCMV infection in BM
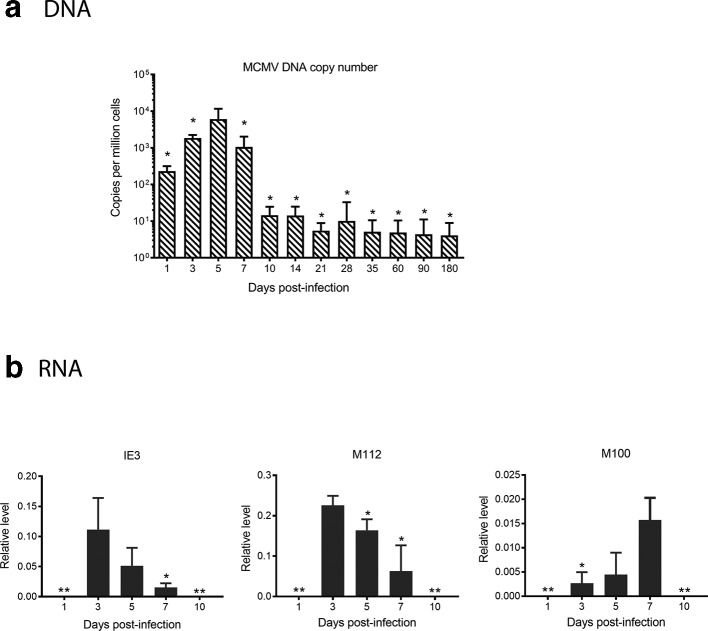
In order to characterize latency of MCMV in BM, it is essential to determine the timing of replication and gene transcription after acute infection. Using quantitative real-time PCR (qPCR), we performed absolute quantification of viral DNA in BM at different time points p.i. MCMV DNA was detectable in the BM within 1 day p.i., peaked at day 5 and then started to decline at day 7 and was stably low at day 10 p.i. (Fig. 1a).
Fig. 1.
Kinetics of viral DNA copy number and RNA expression in bone marrow of MCMV-infected mice. Three-week-old BALB/c mice were acutely infected IP (intraperitoneal injection) with Smith virus and the BM was harvested at different time points. (a) Absolute quantification of MCMV DNA copy number, determined as described in Methods. Data are expressed as the average copy number per million cells plus standard deviation after normalization against cellular gene mEF1α promoter region. N=10 for all time points; *P≤0.05 compared to the peak value (day 5). (b) Relative quantification of viral RNA was performed as described in Methods. Results are shown as the average quantity plus standard deviation after normalization against cellular gene Eef2 RNA expression. N=10 for day 3 and day 7; N=14 for day 5; **P≤0.01 compared to the peak of expression.
Similar to other herpesviruses, CMV genes are transcribed in a sequential manner during productive infection, and are classified into three categories: immediate early (IE), early (E) and late (L) genes. IE genes are the first set of genes transcribed upon infection, the products of which induce early gene expression followed by viral DNA replication and then late gene expression. Products of late genes are needed for packaging progeny virus. Based on this background information, we examined transcription of representative genes in each of these subclasses of IE (IE3), E (M112) and L (M100) by RT-qPCR. The first detectable time point for viral RNA expression was day 3 p.i. Expression of both IE3 and M112 was highest at day 3, and gradually decreased at day 5 and day 7. In contrast, expression of M100 gradually increased from day 3 through day 7. RNA expression from all three classes of viral genes was below the detection limit at day 10 although viral DNA remained detectable (Fig. 1b). While this observation does not necessarily conclude that the bone marrow is latent at this time point, it demonstrates that replication is diminished below detectability [37].
Haematopoietic cells isolated from latently infected mouse BM demonstrate evidence of reactivation in vitro after treatment with GM-CSF/IL-4 followed by LPS
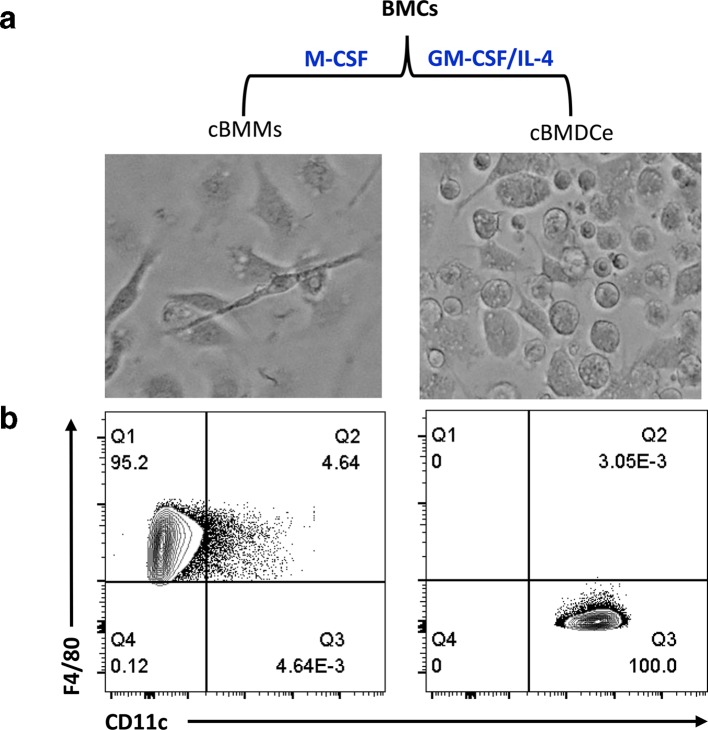
Latency is defined as a state of infection in which viral DNA is present in the cells, although lytic gene expression and production of infectious virus particles are absent. Additionally, the latent virus has the ability to be reactivated under certain conditions [38]. No infectious viral particle was detected in various organs including BM from BALB/c mice 3 months p.i., and explant of spleens from these mice resulted in production of viral progeny, which demonstrates that latency is established in these mice (Fig. S1). Thus throughout the studies described in this manuscript, we used BALB/c mice 3 months p.i. as latently infected mice. In order to prove that the MCMV DNA present in mouse BM haematopoietic cells indeed represents truly latent viral genomes, we need to also demonstrate its ability to undergo reactivation. Multiple studies have demonstrated that reactivation from latency of HCMV in haematopoietic cells is intrinsically tied to differentiation. While remaining latent in CD34+ HSPCs and monocytes, reactivation occurs when these cells further differentiate into mature macrophages or DCs [7, 10–14]. To test whether MCMV follows a similar pattern of reactivation in mouse BM haematopoietic cells, BM haematopoietic cells were induced to differentiate to cBMMs in the presence of M-CSF or cBMDCe cells in the presence of GM-CSF and IL-4. As expected, cBMMs adherent to the surface of bacterial grade petri dishes in M-CSF containing medium culture, were positive for macrophage cell-surface markers CD11b, F4/80, CD115, CD64 and MerTK, with less than 5 % of these cells expressing CD11c (left panel of Figs 2 and S2a. cBMDCe cells isolated from non-adherent or loosely adherent fractions of GM-CSF/IL-4 containing medium culture demonstrated a surface-marker profile of CD11b+CD11c+CD24+mPDCA1 F4/80- CD115-, demonstrating enrichment for DCs (right panel (Figs 2 and S3a). Furthermore, 24 h post LPS treatment, both cBMMs/LPS+ and cBMDCe/LPS+ showed upregulation of maturation markers MHCII, CD80, CD86 and CD40 (Figss S2b and S3b).
Fig. 2.
Differentiation of BMCs into cBMMs and cBMDCe cells. BMCs isolated from BALB/c mice 3 months p.i. with Smith virus were cultured in medium containing M-CSF or GM-CSF/IL-4 for 9 days followed by harvesting for imaging and immune-phenotyping. (a) Schematic representation of differentiation regimens, and morphology of cBMMs and cBMDCe cells. Pictures of the cBMMs (left) and cBMDCe (right) were taken with an EVOS Digital inverted microscope using objective 20×. (b) Phenotype of representative of M-CSF or GM-CSF/IL-4 BMC culture on day 9 by FACS. Surface markers F4/80 and CD11c were analysed on CD45+CD11b+ live cells in each culture condition.
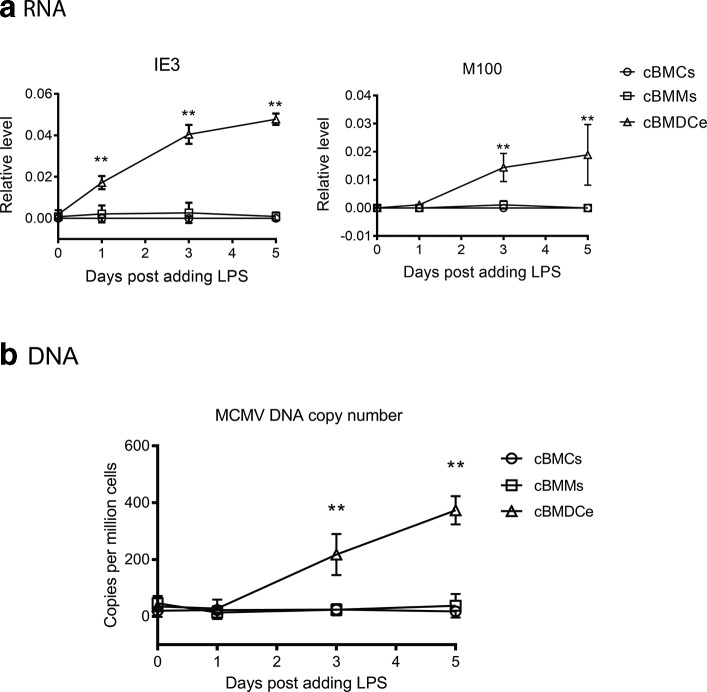
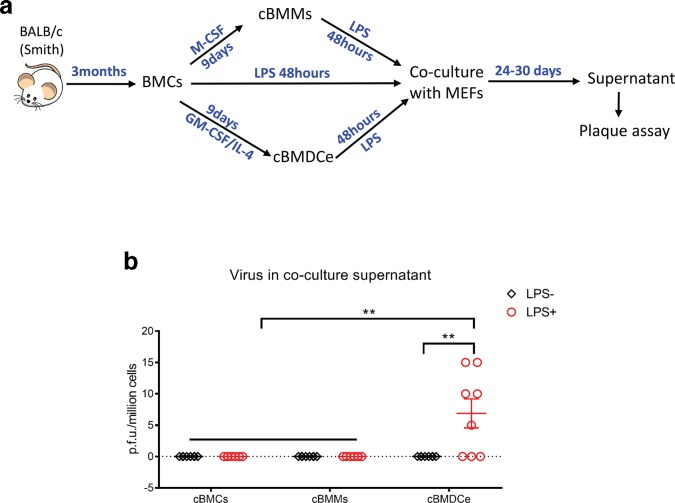
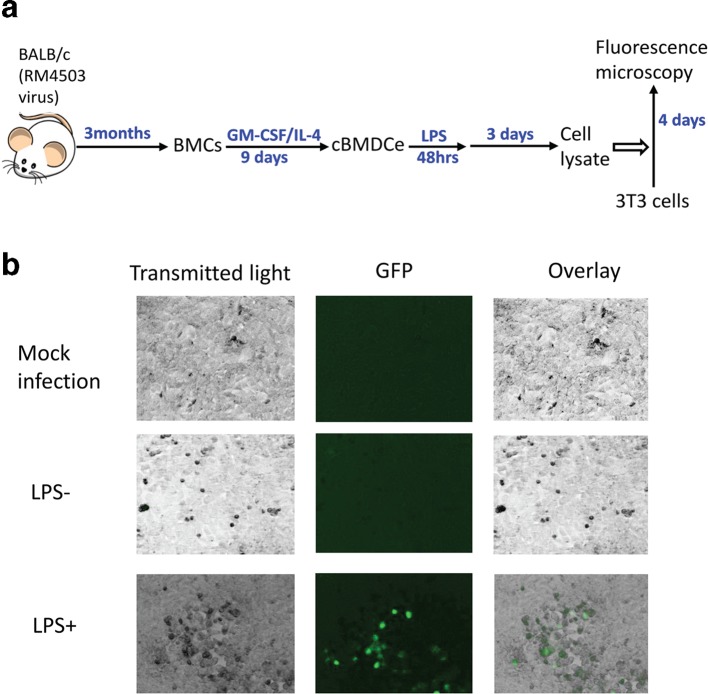
We performed a time-course study to examine the impact of LPS treatment of cBMMs and cBMDCe cells on viral gene expression and DNA replication. When BMCs that were cultured (cBMCs) in STF medium (BMC maintenance medium) were treated with LPS, we observed neither detectable expression of viral genes nor an increase in viral DNA copy number (Fig. 3). In contrast, LPS treatment led to expression of viral IE gene (IE3) and L gene (M100), and increases of viral DNA abundance in cBMDCe cells but not in cBMMs (Fig. 3). The gold standard for reactivation of MCMV is detection of infectious viral particles. After several attempts to detect MCMV particles in the supernatant of both differentiated and undifferentiated BMC culture with plaque assays failed, we reasoned that the quantity of progeny virus might be below the detection threshold of the plaque assay, and that amplification of progeny virus might be necessary for detection. Therefore, 48 h following LPS treatment, the cBMCs, cBMMs or cBMDCe cells were co-cultured with mouse embryo fibroblasts (MEFs) for up to 30 days (Fig. 4a), and an assessment for cytopathic effect (CPE) under the microscope was made every 3 days. The supernatant (conditioned medium) was collected when CPE was observed, typically between day 24 and day 30, and subjected to plaque assay. If no CPE was observed by day 30, supernatant samples were collected and subjected to plaque assay, and the co-cultures were terminated. While no p.f.u. were detected in any supernatant samples from the co-culture of cBMCs/LPS+ and cBMMs/LPS+ or with any of the three populations in the absence of LPS, p.f.u. were detected in five out of eight supernatant samples from co-culture of cBMDCe/LPS+ (Fig. 4b). In parallel with these co-cultures using the Smith strain of MCMV, we also used a recombinant virus RM4503 with an enhanced green florescence protein (EGFP) expression cassette inserted in IE2 region [31]. Five days following LPS treatment of cBMDCe from BALB/c mice 3 months p.i. with RM4503, cell lysates were placed on MCMV-permissive NIH3T3 cells. EGFP fluorescence was detected in the 3T3 cells 4 days after exposure to the cell lysate from cBMDCe/LPS+ but not cBMDCe/LPS- (Fig. 5). No fluorescence was detected from the mock-infected 3T3 cells, indicating that EGFP detection reflected infection by the RM4503 virus rather than auto-fluorescence. These results are concordant with the plaque assay results, and support the observation that differentiation of BM haematopoietic cells to cBMDCe/LPS+ leads to the detection of reactivation of latent MCMV.
Fig. 3.
LPS treatment of cBMDCe but not cBMMs leads to viral gene expression and increase in viral DNA copy number. BMCs isolated from BALB/c mice 3 months p.i. with Smith virus were cultured in BMC maintenance medium (cBMCs), M-CSF containing medium (cBMMs) or GM-CSF/IL-4 containing medium (cBMDCe) for 9 days, and subject to 100 ng ml−1 LPS treatment. (a) Viral RNA expression in different time points post LPS treatment. RT-qPCR on the RNA from cBMCs, cBMMs or cBMDCe collected at different time points upon LPS treatment. Data are presented at relative level (arbitrary number) after normalization against cellular Eef2 RNA. (b) Absolute quantification of MCMV DNA copy number. DNA was extracted from cBMCs/LPS, cBMMs/LPS or cBMDCe/LPS at different time points after adding LPS, and subject to qPCR with the Taqman primers and probe specific to the MIEP region of the viral genome. Data values are presented as copy number per million cells after normalization against cellular gene mEFα. Data values are expressed as means±sd. N=6 for all time points. Statistics were performed using multiple individual Student's t-test. *P-value≤0.05; **P-value≤0.01.
Fig. 4.
LPS treatment of cBMDCe cells but not cBMMs from latently infected mouse BM leads to production of infectious viral particles. (a) Schematic representation of experimental design. BMCs from BALB/c mice 3 months p.i. with Smith virus were cultured in M-CSF or GM-CSF/IL-4 containing medium for 9 days giving rise to cultured bone-marrow-derived macrophages (cBMMs) and cultured bone-marrow-derived dendritic cell-enriched BMCs (cBMDCe) respectively, and were subsequently pulsed with (/LPS+) or without (/LPS-) 100 ng ml−1 LPS for 48 h, and then co-cultured with MEFs for 24–30 days. Supernatant was collected from the co-culture and subject to plaque assay when a CPE was observed, which typically occurred between day 24 and day 30. If no CPE was observed by day 30, the supernatant samples were collected, and the co-cultures were terminated. (b) The titre of virus in the supernatant samples of co-culture. The results are expressed as mean±sem of p.f.u. per million cells. Multiple individual Student’s t-tests were used for statistical analysis. **P≤0.01.
Fig. 5.
Detection (GFP) of infectious virus progeny in the lysate of cBMDCe treated with LPS. (a) Schematic representation of experimental design. BMCs were isolated from BALB/c mice 3 months p.i. with RM4503 mutant MCMV, which expresses EGFP driven by HCMV MIEP, cultured for 9 days with GM-CSF/IL-4 containing medium, and pulsed with LPS for 48 h. Three days later, cell lysates made with treated with, or not treated with LPS were used to infect NIH3T3 cells. Four days p.i., the 3T3 cells were inspected under EVOS Digital Inverted Microscope for the expression of GFP. (b) Effect of LPS treatment on detection of GFP expression in LPS-treated cell lysates on NIH3T3 cells. Mock infection: 3T3 cells treated cell culture medium; LPS-: 3T3 cells treated with lysate of cBMDCe/LPS-; LPS+: 3T3 cells infected with lysate of cBMDCe/LPS+.
MCMV has broad cell tropism in the BM during acute infection, but can only establish detectable latency in a few specific myeloid cell types and HSPCs.
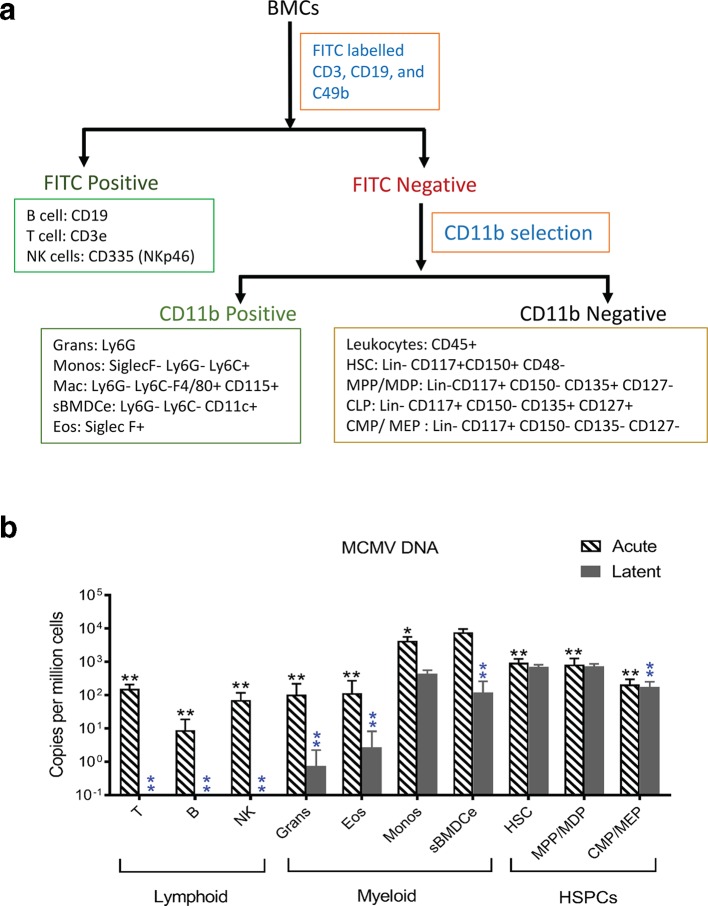
Determining a cell-specific reservoir of latent CMV is a prerequisite for understanding latency and reactivation. We therefore sought to phenotype the BM haematopoietic cells that harbour viral DNA in latently infected mouse BM. Other than red blood cells (RBCs), haematopoietic cells in BM of adult mice can be roughly categorized into three classes: HSPCs, myeloid cells and lymphoid cells. HSPCs are phenotypically characterized as Lin-CD117+, and composed of a heterogeneous cell fractions. These include haematopoietic stem cells (HSCs), multipotent progenitors (MPPs) and oligopotent progenitors (OPPs), which in turn include common myeloid progenitor cells (CMPs), common lymphoid progenitors (CLPs), megakaryocyte-erythrocyte progenitors (MEPs), granulocyte-monocyte progenitors (GMPs) and macrophage-DC progenitors (MDPs) [39]. Myeloid cells are phenotypically defined by the expression of the CD11b surface marker and include monocytes, granulocytes, eosinophils, macrophages and DCs. Lymphoid cells include B cells, T cells, and NK cells [40]. With this cellular landscape in mind, we designed a strategy for sorting BM haematopoietic cells to suit our purpose (Fig. 6a). BMCs were initially separated into three fractions using magnetic beads: FITC positive (lymphoid-enriched fraction, based on FITC conjugated antibodies specific for CD3, CD19 and CD49b), FITC negative and CD11b positive (myeloid-enriched fraction), and FITC and CD11b double negative (HSPC-enriched fraction). Each of these three cell fractions was then stained with antibody panels (Table S3), and sorted by flow cytometry. Using the gating strategy shown in Fig. S3, ten major subsets of cells were collected (Fig. S4). This sorting procedure was applied to BMCs from acutely infected (day 5 p.i.) and latently infected mice (3 months p.i.), and genomic DNA samples from these sorted BMCs (sBMCs) were used to quantify MCMV DNA via qPCR. In acutely infected BM, viral DNA was detectable in all sorted cell types, with the highest copy number in sorted (CD11c+ CD11b+ SiglecF- Ly6G- Ly6C-) DC-enriched BMCs (sBMDCe) (Fig. 6b). In contrast, in latent infection viral DNA was only detectable in fewer types of cells, including monocytes, sorted bone marrow dendritic cell-enriched (sBMDCe) cells, HSCs, MPP/MDPs and CMP/MEPs, with the highest copy numbers in HSCs and MPPs/MDPs (Fig. 6b). These results suggest that MCMV has broad tropism in the BM haematopoietic system in acute infection, but can only establish detectable latency in a few specific cell types including monocytes, sBMDCe and HSPCs following natural infection.
Fig. 6.
Identification of cell types that harbour viral DNA in haematopoietic BMCs in acutely and latently infected mice. Freshly isolated BMCs from acutely infected (day 5 p.i.) and latently infected mouse BM (3 months p.i.) were initially separated into three fractions using magnetic beads as described in Methods. Each of these three cell fractions was then stained with antibody panels (Table S3), and sorted by flow cytometry. Using this gating strategy (Fig. S3), ten major subsets of cells were collected. Genomic DNA samples from these sorted BMCs (sBMCs) were used to quantify MCMV DNA via qPCR. (a) Sorting strategy of haematopoietic BMCs. BMCs were firstly separated into FITC positive (lymphoid cells) and FITC negative (non-lymphoid cells) populations with mouse FITC positive selection magnetic beads, then the FITC negative cells were separated with CD11b selection magnetic beads into CD11b+ (myeloid cells) and CD11b- (HSPCs) populations. These three populations of cells were stained with antibodies listed in Table S4, and were sorted based on the combination of cell-surface markers shown in Fig. S4. (b) Quantification of viral DNA in sorted BMCs. DNA was extracted from each population of cells, and subject to qPCR with Taqman primers and probe specific to the MIEP region of the MCMV genome. The quantity of the MCMV genome was normalized against cellular gene mEF1α, and then converted to copy number per million cells. Values for naïve mice were set to zero and infected mice shown relative to naïve mice. BMCs from five mice were pooled and considered a biological replicate indicated by ‘N’. For infected mice N≥3, for uninfected controls N≥2. Data are presented as mean±sd. Multiple Student's t-tests were performed. *P≤0.05 compared with the peak; **P≤0.01 compared with the peak. Black and blue asterisks represent the P-values for acute and late infection, respectively. T: T cells; B: B cells; NK: natural killer cells; Grans: Ganulocytes; Eos: Eosinophils; Monos: monocytes; sBMDCe: sorted dendritic cell-enriched BMCs; Mac: Macrophages; HSCs: haematopoietic stem cells; HSPCs: haematopoietic stem cell and progenitor cells; MPP: multipotent progenitor cells; CLP: common lymphoid progenitor cells; CMP: common myeloid progenitor cells; MDP: macrophage-DC progenitors; MEP: megakaryocyte-erythrocyte progenitor;
CD11c+ cells, monocytes and HPSCs constitute the main cellular reservoirs of latent MCMV in BM; in vitro viral reactivation from CD11c+ cells is possible
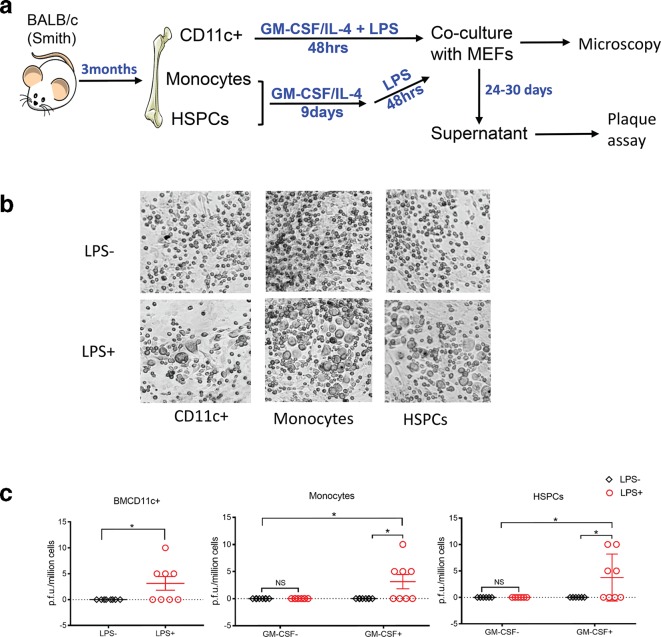
We wanted to determine whether specific types of BMCs identified by the sorting experiments above as harbouring detectable MCMV DNA in latently infected mice truly represent latent viral reservoir capable of reactivation. Therefore, informed by the data from our cultured differentiation experiments described above, we performed in vitro differentiation assays as described in the culture system above on bead-enriched cell populations. As shown in Fig. 7a, CD11c+ cells from latently infected mouse BM were cultured in GM-CSF/IL-4 containing medium for 2 days in the presence (BMcCD11c+/LPS+) or absence (BMcCD11c+/LPS-) of LPS, and then co-cultured with MEFs. CPE was observed after 24 days of co-culture (Fig. 7a and b) and p.f.u. were detected in four out of eight supernatant co-culture samples (Fig. 7c) in BMcCD11c+/LPS+, but not in (BMcCD11c+/LPS-). These results provide further evidence that treatment with LPS causes reactivation in DC-enriched CD11c +BMCs that harbour latent MCMV. Moreover, MEF co-culture of LPS-treated but not LPS-untreated cells derived from monocytes or HSPCs also resulted in CPE and p.f.u. (Fig. 7b and c), whereas direct co-culture of LPS-treated or LPS-untreated monocytes or HSPCs from latently infected mice demonstrated neither CPE nor PFUs by 30 days (Fig. 7c). These data indicate that DC-enriched CD11c+ cells, monocytes and HSPCs represent authentic cellular sites of MCMV latency and reactivation.
Fig. 7.
CD11c+ cells, monocytes and HPSCs are the main cellular reservoirs of latent MCMV in BM; exposure of CD11c+ cells to GM-CSF/IL-4 followed by treatment with LPS is associated with reactivation in vitro. (a) Experimental regimen. CD11c+ cells, monocytes and HSPCs were each independently purified from BMCs isolated from BALB/c mice 3 months p.i. with Smith virus using MACS beads. CD11c+ cells were enriched from BMCs with CD11c microbeads. CD11c+ cells were cultured in GM-CSF/IL-4 containing medium followed by treatment or no treatment with a 100 ng ml−1 LPS pulse for 48 h. These cells were co-cultured with MEFs, and inspected under microscope for CPE. The conditioned medium (supernatant) was collected for plaque assay when CPE was observed, which usually appear between 24 and 30 days. If no CPE was observed by day 30, the supernatant was also collected for plaque assay, and the co-culture was terminated. (b) CPE was observed under the microscope in the MEF co-cultures. (c) Plaque assay and viral titre in the supernatant samples of the co-cultures. The titre of the virus was expressed as p.f.u./million cells. N=8 for all of the groups. Multiple individual Student's t-tests were performed. *P≤0.05.
Discussion
Human BM is an important target for HCMV infection. Acute infection of HCMV often causes altered haematopoietic profiles, and latent infection of HSPCs can lead to graft failure, and to CMV transmission and infection in haematopoietic cell-transplant (HCT) recipients [2, 41]. MCMV has been used extensively as a model to study the pathophysiology, latency and reactivation of CMV in vivo [20, 21, 42]. MCMV infection can cause haematologic abnormalities similar to HCMV [27]. However, while MCMV DNA has been detected in BM of latently infected mice via PCR–in situ hybridization (PISH), the specific cells involved in MCMV infection and latency in the BM have not been defined [30]. Moreover, while in vitro models have demonstrated an important role for HSPCs in HCMV latency and reactivation, the role of BMCs in general, and of BM-derived HSPCs in particular remains largely unexplored in MCMV latency and reactivation.
The purpose of this study was to assess the potential for establishing an in vitro model of MCMV reactivation from latency. We sought to identify the major myeloid cell subsets based on broad immunophenotyping that might eventually lead to more minor subsets requiring more in-depth high-parameter immunophenotyping. Our focus was to design sorting strategies to identify which cell fractions were responsible for harbouring MCMV latency in the BM, by enriching for cell population known to harbour latency in HCMV. Using this approach, we have found that cell fractions enriched for CD11+ cells constitute an important site of latency and reactivation and further work will involve more in-depth identification of specific fractions.
Data presented in this study confirm that latent MCMV DNA can be detected in the BM following acute infection of mice [26, 30]. This study also establishes for the first time that reactivation of MCMV occurs following differentiation of BM haematopoietic cells to DC-enriched fractions following treatment with LPS. A previous study suggested that latently infected donors did not transmit virus to immunocompromised HCT recipients. Therefore, these authors concluded that MCMV had not established latency in BM haematopoietic cells [43]. However, while latency was established following natural infection in our study, in their study mice subjected to myelo-ablative conditioning via total body γ-irradiation (TBI) were infected with MCMV through HCT, and BMCs from these mice were then used in a second HCT [43]. It has been previously reported that apoptosis of haematopoietic cells occurs as early as 6 h post TBI [44], and that the density of BMCs decreases 67 % within day 1, and 86 % by day 3, with only partial reconstitution by 14 days [45]. Therefore, the peak of detectable MCMV in the BM following acute infection (5 days) would potentially precede engraftment of transplanted haematopoietic cells (10 days) [46], suggesting that the BM haematopoietic cells used for the second HCT in their study were not latently infected. While we were unable to find negative studies in the literature, we are aware of several studies dating back at least two decades that failed to demonstrate MCMV in latent infection. This was recently confirmed by Edward Mocarski (personal communication 7/20/2019). However, we were previously able to detect latent MCMV in BM using in-situ PCR [30], and our study now shows clear evidence that BM cells become acutely and latently infected.
HCMV latency and reactivation in haematopoietic cell lineages is well established. There is a consensus view that terminal differentiation of latently infected HSPCs and monocytes into macrophages or DCs followed by LPS-mediated maturation results in expression of lytic genes and production of progeny virus in vitro [10, 12, 13, 47, 48]. To test the possibility of a similar differentiation-driven reactivation of MCMV in HSPCs, we induced culture-driven differentiation of BMCs to cBMMs and cBMDCe cells respectively, followed by LPS treatment. While LPS treatment resulted in expression of maturation markers in both cBMMs and cBMDCe, detectable MCMV reactivation occurred in cBMDCe/LPS+, but not cBMMs/LPS+. In addition, we exposed DC-enriched freshly isolated BMCD11c+cells to a similar differentiation and maturation regimen and observed MCMV reactivation in that system as well. LPS treatment of GM-CSF-differentiated BMCs leads to rapid activation of canonical NF-kB RelA/p50 and AP-1, and production of pro-inflammatory cytokines including TNF-α, IL-6, IL-1p70 and IL-23. In contrast, M-CSF differentiation followed by LPS treatment elicits a delayed activation of NF-kB p50 and AP-1 and preferential production of IL-10 and CCL-2, suggesting that induction of M-CSF and GM-CSF differentiation result in differential cellular and molecular environments [49]. Also, human monocytes can be differentiated into M1 (pro-inflammatory) and M2 (anti-inflammatory) macrophages by GM-CSF and M-CSF respectively, and 17 % of differentially regulated genes by these CSFs are common across species. Cytokine profiles, observed following GM-CSF-induced BMDC differentiation in mice resemble M1 polarized macrophages in humans, whereas those observed following M-CSF-induced BMM differentiation in mice resemble M2 polarized macrophages in humans [50]. These previous observations provide a plausible mechanistic explanation for the differential reactivation observed in cBMDCe cells vs cBMMs following treatment with LPS.
We observed p.f.u. in only a proportion of cBMDCe/LPS+ and BMcCD11c+/LPS+MEF and co-cultures. A recurring challenge encountered in studying both HCMV and MCMV latency and reactivation revolves around the low viral genome copy number per cell, compounded by the low frequency of latently infected cells. This technical challenge is also evident in studies involving the BM haematopoietic system. There are approximately 10 copies of viral DNA per million BMCs from latently infected mice (Fig. 1a); the frequency of latently infected cells would therefore be 1/100 000 cells if each latently infected cell harbours 1 copy of latent viral genome. In the course of differentiation of BMCs into DC-enriched fractions, the viral DNA is diluted owing to the proliferation of the BMCs, which further lowers the frequency of latently infected cells. Thus, differentiation-associated reactivation of CMV is an extremely rare event, which makes detection difficult. One approach to overcome this technical hurdle is to enrich for cells known to be latently infected. To identify this population of cells, we phenotyped and sorted BMCs into ten major subpopulations and purified each of them using a fluorescence activated cell sorter (FACS) and examined them for the presence of viral DNA using qPCR. Using a strategy designed specifically for staining and sorting murine BM haematopoietic cells, we showed that MCMV demonstrates broad cellular tropism during acute infection although to varied degrees. In contrast, latent infection was limited to myeloid mononuclear cells (monocytes and DC-enriched CD11c+ cells) and HSPCs (HSCs, MPP/MDPs and CMP/MEPs).
We were surprised to find that MCMV DNA was detected in T, B and NK cells in the BM of acutely infected mice albeit at low levels (Fig. 6b). Further, though only low level IE RNA and no E or L RNA was detected in these cells (Fig. S5). These lymphoid cells were devoid of viral DNA in latently infected BM, suggesting that these cells were transiently and abortively infected or alternatively took up virus passively. It has previously been shown that HCMV DNA is present in peripheral blood lymphocytes in some of the patients without recovery of infectious viral with limited viral gene expression [2, 51]. Also, a small proportion of T, B and NK cells from human blood can be infected by HCMV in vitro, but only IE gene products are expressed [52].
Our data show that when compared to other cell types, monocytes and sBMDCe cells harboured the highest copy number of viral DNA in the BM of acutely infected mice. We also show that HSPCs from the BM of latently infected mice contain some of the highest levels of viral DNA, suggesting they serve as a major haematopoietic reservoir for latent MCMV (Fig. 6b). While it is plausible that acutely infected monocytes may develop into latently infected monocytes, it is also likely that instead, these participate in blood-borne dissemination, while acutely infected precursors give rise to latently infected monocytes and sBMDCe cells in the BM. This conceptual construct is consistent with previous observations of the role of peripheral blood monocytes and DCs in disseminating virus during the acute phase of MCMV infection [53–55], as well as with the detection of latent virus in these cell types in the peripheral blood [8, 56, 57]. In addition, latently infected cells similar to our sBMDCe cells capable of reactivation have been previously derived following acute infection of common precursor cells [58–63]. Thus, taken together, our data on the cellular profiles observed for both acute and latent MCMV infection in the murine BM haematopoietic system appear consistent with a number of observations in HCMV models [6, 7, 52, 64, 65].
This study has certain limitations mostly revolving around the low frequency and copy numbers resulting in the requirement of a large numbers of cells and in difficult detection thresholds. While our sorting and staining strategy has allowed for some degree of enrichment of latently infected BMCs, more robust approaches targeted at single populations of latently infected cells remains highly desirable. Our study may help pave the way for single-cell, multi-parameter studies in the future.
In summary, we have defined the kinetics and cellular profiles of primary MCMV infection and establishment of latency in the BM. We have also demonstrated differentiation-driven MCMV reactivation in cBMDCe cellular populations following treatment with LPS in latent mice that were infected in vivo. This in vitro model of MCMV reactivation of naturally occurring latency that has the potential to contribute significantly to our understanding of the molecular events operative in CMV reactivation. These findings complement observations from in vitro HCMV studies, and further validate the use of in vivo murine models to study CMV latency and reactivation.
Supplementary Data
Funding information
This work is supported by National Institutes of Health grants R01 AI112911-01 and P01 AI112522-01 to Michael M. Abecassis from the National Institutes of Allergy and Infectious Diseases.
Acknowledgement
Flow-cytometry analysis of cell-surface markers and the fluorescence-activated cell sorting (FACS) were performed in Robert H. Lurie Comprehensive Cancer Center-Flow Cytometry Core Facility at the Chicago campus of Northwestern University.
Author contributions
M. M. A., conceptualized this research project, and provided financial support and leadership. X. F. L., and M. M. A., conceived and designed experiments, and prepared the initial draft. X. F. L., S. S., F. A. E., S.Y., D. A. A., L. Q., Z. Z., L. A. V. and A. I., performed experiments and helped interpret data. X. F. L., T. H. S., and S. S., analysed the data. Q. C., T. H. S., and S. S. did some editing to the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
All experimental procedures with animals were done according to the standards and guidelines from the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC) and the Office of Laboratory Animal Welfare (OLAW) of National Institutes of Health (NIH), USA. Northwestern University has an Animal Welfare Assurance on file with the Office of Laboratory Animal Welfare (A3283-01). Northwestern University conducts its reviews in accordance with United States Public Health Service (USPHS) regulations and applicable federal and local laws. The composition of the IACUC meets the requirements of the USPHS policy and the Animal Welfare Act Regulations. The animal protocols (protocol numbers IS00001005 and IS00000472, respectively) used in this study were reviewed and approved by Institutional Animal Care and Use Committee (IACUC) of Northwestern University.
Footnotes
Abbreviations: Angptl3, Angiopoietin-like protein3; BM, bone marrow; BMCs, bone marrow cells; cBMCs, cultured bone marrow cells; cBMDCe, cultured bone marrow-derived dendritic cell-enriched; cBMMs, cultured bone marrow-derived macrophages; CFU, colony-forming unit; CFU-GM, CFU-granulocyte and macrophage; CFU-S, CFU-spleen; CLPs, common lymphoid progenitors; CMPs, common myeloid progenitors; CMV, cytomegalovirus; DCs, dendritic cells; DMEM, Dulbecco’s Modified Eagle’s Medium; Eos, Eosinophils; FGF1, fibroblast growth factor; GM-CSF, Granulocyte-macrophage colony stimulating factor; Grans, Granulocytes; HCMV, human cytomegalovirus; HCT, Haematopoietic cell-transplant; HSCs, haematopoietic stem cells; HSPCs, haematopoietic stem cells and progenitor cells; IE, Immediate early; IGF2, Insulin-like growth factor2; IGFBP2, Insulin-like growth factor-binding protein2; IL-4, Interleukin-4; IP, Intraperitoneal; Lin, lineage marker; LPS, Lipopolysaccharide; MCMV, murine cytomegalovirus; M-CSF, Macrophage colony-stimulating factor; MDPs, Macrophage-DC progenitors; MEFs, Mouse embryo fibroblasts; MEPs, Megakaryocyte-erythrocyte progenitors; MIEP, Major immediate enhancer and promoter region; Monos, Monocytes; MPPs, Multipopent progenitor cells; OPPs, oligopotent progenitors; p.f.u., Plaque-forming unit; p.i., Post infection; PISH, PCR-in situ hybridization; qPCR, Quantitative real-time PCR; RBCs, Red blood cells; sBMCs, Sorted bone marrow cells; sBMDCe, Sorted DC-enriched BMCs; SCF, stem-cell factor; SFEM, StemSpan serum-free expansion medium; TBI, Total body γ-irradiation; TPO, Throbopoietin.
Four supplementary tables and five supplementary figures are available with the online version of this article.
References
- 1.Ariza-Heredia EJ, Nesher L, Chemaly RF. Chemaly, Cytomegalovirus diseases after hematopoietic stem cell transplantation: a mini-review. Cancer Lett. 2014;342:1–8. doi: 10.1016/j.canlet.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Sing GK, Ruscetti FW. The role of human Cytomegalovirus in haematological diseases. Baillieres Clin Haematol. 1995;8:149–163. doi: 10.1016/S0950-3536(05)80236-7. [DOI] [PubMed] [Google Scholar]
- 3.Movassagh M, Gozlan J, Senechal B, Baillou C, Petit JC, et al. Direct infection of CD34+ progenitor cells by human cytomegalovirus: evidence for inhibition of hematopoiesis and viral replication. Blood. 1996;88:1277–1283. [PubMed] [Google Scholar]
- 4.Simmons P, Kaushansky K, Torok-Storb B. Mechanisms of cytomegalovirus-mediated myelosuppression: perturbation of stromal cell function versus direct infection of myeloid cells. Proc Natl Acad Sci USA. 1990;87:1386–1390. doi: 10.1073/pnas.87.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Laer D, Meyer-Koenig U, Serr A, Finke J, Kanz L, et al. Detection of cytomegalovirus DNA in CD34+ cells from blood and bone marrow. Blood. 1995;86:4086–4090. [PubMed] [Google Scholar]
- 6.Mendelson M, Monard S, Sissons P, Sinclair J. Detection of endogenous human cytomegalovirus in CD34+ bone marrow progenitors. J Gen Virol. 1996;77:3099–3102. doi: 10.1099/0022-1317-77-12-3099. [DOI] [PubMed] [Google Scholar]
- 7.Khaiboullina SF, Maciejewski JP, Crapnell K, Spallone PA, Dean Stock A, et al. Human cytomegalovirus persists in myeloid progenitors and is passed to the myeloid progeny in a latent form. Br J Haematol. 2004;126:410–417. doi: 10.1111/j.1365-2141.2004.05056.x. [DOI] [PubMed] [Google Scholar]
- 8.Taylor-Wiedeman J, Sissons JG, Borysiewicz LK, Sinclair JH. Monocytes are a major site of persistence of human cytomegalovirus in peripheral blood mononuclear cells. J Gen Virol. 1991;72:2059–2064. doi: 10.1099/0022-1317-72-9-2059. [DOI] [PubMed] [Google Scholar]
- 9.Taylor-Wiedeman J, Hayhurst GP, Sissons JGP, Sinclair JH. Polymorphonuclear cells are not sites of persistence of human cytomegalovirus in healthy individuals. J Gen Virol. 1993;74:265–268. doi: 10.1099/0022-1317-74-2-265. [DOI] [PubMed] [Google Scholar]
- 10.Reeves MB, Compton T. Inhibition of inflammatory interleukin-6 activity via extracellular signal-regulated kinase-mitogen-activated protein kinase signaling antagonizes human cytomegalovirus reactivation from dendritic cells. J Virol. 2011;85:12750–12758. doi: 10.1128/JVI.05878-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reeves MB, MacAry PA, Lehner PJ, Sissons JGP, Sinclair JH, et al. Latency, chromatin remodeling, and reactivation of human cytomegalovirus in the dendritic cells of healthy carriers. Proc Natl Acad Sci USA. 2005;102:4140–4145. doi: 10.1073/pnas.0408994102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Söderberg-Nauclér C, Streblow DN, Fish KN, Allan-Yorke J, Smith PP, et al. Reactivation of latent human cytomegalovirus in CD14(+) monocytes is differentiation dependent. J Virol. 2001;75:7543–7554. doi: 10.1128/JVI.75.16.7543-7554.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor-Wiedeman J, Sissons P, Sinclair J. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J Virol. 1994;68:1597–1604. doi: 10.1128/jvi.68.3.1597-1604.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang MM, Kew VG, Jestice K, Wills MR, Reeves MB, et al. Efficient human cytomegalovirus reactivation is maturation dependent in the langerhans dendritic cell lineage and can be studied using a CD14+ +experimental latency model. J Virol. 2012;86:8507–8515. doi: 10.1128/JVI.00598-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keil GM, Ebeling-Keil A, Koszinowski UH. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol. 1987;61:526–533. doi: 10.1128/jvi.61.2.526-533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keil GM, Ebeling-Keil A, Koszinowski UH. Sequence and structural organization of murine cytomegalovirus immediate-early gene 1. J Virol. 1987;61:1901–1908. doi: 10.1128/jvi.61.6.1901-1908.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messerle M, Bühler B, Keil GM, Koszinowski UH. Structural organization, expression, and functional characterization of the murine cytomegalovirus immediate-early gene 3. J Virol. 1992;66:27–36. doi: 10.1128/jvi.66.1.27-36.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mocarski E. Cytomegaloviruses and their replicatio. In: David MK, Howley PM, editors. Fields Virology. Philadelphia: Lippincott Williams & Wilkins; 2001. pp. 2629–2673. [Google Scholar]
- 19.Lawson CM, O'Donoghue HL, Reed WD. Mouse cytomegalovirus infection induces antibodies which cross-react with virus and cardiac myosin: a model for the study of molecular mimicry in the pathogenesis of viral myocarditis. Immunology. 1992;75:513–519. [PMC free article] [PubMed] [Google Scholar]
- 20.Hummel M, Abecassis MM. A model for reactivation of CMV from latency. J Clin Virol. 2002;25:123–136. doi: 10.1016/S1386-6532(02)00088-4. [DOI] [PubMed] [Google Scholar]
- 21.Melnick M, Mocarski ES, Abichaker G, Huang J, Jaskoll T, et al. Cytomegalovirus-induced embryopathology: mouse submandibular salivary gland epithelial-mesenchymal ontogeny as a model. BMC Dev Biol. 2006;6:42. doi: 10.1186/1471-213X-6-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juanjuan C, Yan F, Li C, Haizhi L, Ling W, et al. Murine model for congenital CMV infection and hearing impairment. Virol J. 2011;8:70. doi: 10.1186/1743-422X-8-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cekinovic D, Lisnic VJ, Jonjic S. Rodent models of congenital cytomegalovirus infection. Methods Mol Biol. 2014;1119:289–310. doi: 10.1007/978-1-62703-788-4_16. [DOI] [PubMed] [Google Scholar]
- 24.Oduro JD, Redeker A, Lemmermann NAW, Ebermann L, Marandu TF, et al. Murine cytomegalovirus (CMV) infection via the intranasal route offers a robust model of immunity upon mucosal CMV infection. J Gen Virol. 2016;97:185–195. doi: 10.1099/jgv.0.000339. [DOI] [PubMed] [Google Scholar]
- 25.Duppach J, Francois S, Joedicke JJ, Dittmer U, Kraft ARM, et al. Expanded regulatory T cells in chronically friend retrovirus-infected mice suppress immunity to a murine cytomegalovirus superinfection. J Virol. 2014;88:13892–13896. doi: 10.1128/JVI.01941-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bale JF, O'Neil ME, Giller R, Perlman S, Koszinowski U, et al. Murine cytomegalovirus genomic material in marrow cells: relation to altered leukocyte counts during sublethal infection of mice. J Infect Dis. 1987;155:207–212. doi: 10.1093/infdis/155.2.207. [DOI] [PubMed] [Google Scholar]
- 27.Gibbons AE, Price P, Shellam GR. Analysis of hematopoietic stem and progenitor cell populations in cytomegalovirus-infected mice. Blood. 1995;86:473–481. [PubMed] [Google Scholar]
- 28.Mori T, Ando K, Tanaka K, Ikeda Y, Koga Y, et al. Fas-mediated apoptosis of the hematopoietic progenitor cells in mice infected with murine cytomegalovirus. Blood. 1997;89:3565–3573. [PubMed] [Google Scholar]
- 29.Pollock JL, Presti RM, Paetzold S, Virgin HW. Latent murine cytomegalovirus infection in macrophages. Virology. 1997;227:168–179. doi: 10.1006/viro.1996.8303. [DOI] [PubMed] [Google Scholar]
- 30.Koffron AJ, Hummel M, Patterson BK, Yan S, Kaufman DB, et al. Cellular localization of latent murine cytomegalovirus. J Virol. 1998;72:95–103. doi: 10.1128/jvi.72.1.95-103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van Den Pol AN, Mocarski E, Saederup N, Vieira J, Meier TJ, et al. Cytomegalovirus cell tropism, replication, and gene transfer in brain. J Neurosci. 1999;19:10948–10965. doi: 10.1523/JNEUROSCI.19-24-10948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brizić I, Lisnić B, Brune W, Hengel H, Jonjić S. Cytomegalovirus Infection: Mouse Model. Curr Protoc Immunol. 2018;122:e51. doi: 10.1002/cpim.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng J, Umikawa M, Zhang S, Huynh H, Silvany R, et al. Ex vivo expanded hematopoietic stem cells overcome the MHC barrier in allogeneic transplantation. Cell Stem Cell. 2011;9:119–130. doi: 10.1016/j.stem.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Assouvie A, Daley-Bauer LP, Rousselet G. Growing murine bone marrow-derived macrophages. Methods Mol Biol. 1784;2018:29–33. doi: 10.1007/978-1-4939-7837-3_3. [DOI] [PubMed] [Google Scholar]
- 35.Lutz MB, Kukutsch N, Ogilvie AL, Rössner S, Koch F, et al. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J Immunol Methods. 1999;223:77–92. doi: 10.1016/S0022-1759(98)00204-X. [DOI] [PubMed] [Google Scholar]
- 36.Liu XF, Yan S, Abecassis M, Hummel M. Biphasic recruitment of transcriptional repressors to the murine cytomegalovirus major immediate-early promoter during the course of infection in vivo . J Virol. 2010;84:3631–3643. doi: 10.1128/JVI.02380-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mercer JA, Spector DH. Pathogenesis of acute murine cytomegalovirus infection in resistant and susceptible strains of mice. J Virol. 1986;57:497–504. doi: 10.1128/jvi.57.2.497-504.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roizman B, Sears AE. An inquiry into the mechanisms of herpes simplex virus latency. Annu Rev Microbiol. 1987;41:543–571. doi: 10.1146/annurev.mi.41.100187.002551. [DOI] [PubMed] [Google Scholar]
- 39.Wang LD, Wagers AJ. Dynamic niches in the origination and differentiation of haematopoietic stem cells. Nat Rev Mol Cell Biol. 2011;12:643–655. doi: 10.1038/nrm3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Francke A, Herold J, Weinert S, Strasser RH, Braun-Dullaeus RC, et al. Generation of mature murine monocytes from heterogeneous bone marrow and description of their properties. J Histochem Cytochem. 2011;59:813–825. doi: 10.1369/0022155411416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pergam SA, Xie H, Sandhu R, Pollack M, Smith J, et al. Efficiency and risk factors for CMV transmission in seronegative hematopoietic stem cell recipients. Biol Blood Marrow Transplant. 2012;18:1391–1400. doi: 10.1016/j.bbmt.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roback JD, Su L, Newman JL, Saakadze N, Lezhava LJ, et al. Transfusion-transmitted cytomegalovirus (CMV) infections in a murine model: characterization of CMV-infected donor mice. Transfusion. 2006;46:889–895. doi: 10.1111/j.1537-2995.2006.00820.x. [DOI] [PubMed] [Google Scholar]
- 43.Seckert CK, Renzaho A, Reddehase MJ, Grzimek NKA. Hematopoietic stem cell transplantation with latently infected donors does not transmit virus to immunocompromised recipients in the murine model of cytomegalovirus infection. Med Microbiol Immunol. 2008;197:251–259. doi: 10.1007/s00430-008-0094-1. [DOI] [PubMed] [Google Scholar]
- 44.Peng R, Wang D, Wang B, Xia G, Li Y, et al. Apoptosis of hemopoietic cells in irradiated mouse bone marrow. J Environ Pathol Toxicol Oncol. 1999;18:305–308. [PubMed] [Google Scholar]
- 45.Turner RT, Iwaniec UT, Wong CP, Lindenmaier LB, Wagner LA, et al. Acute exposure to high dose γ-radiation results in transient activation of bone lining cells. Bone. 2013;57:164–173. doi: 10.1016/j.bone.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiesmann A, Spangrude GJ. Marrow engraftment of hematopoietic stem and progenitor cells is independent of Galphai-coupled chemokine receptors. Exp Hematol. 1999;27:946–955. doi: 10.1016/S0301-472X(99)00029-6. [DOI] [PubMed] [Google Scholar]
- 47.Reeves MB, Lehner PJ, Sissons JGP, Sinclair JH. An in vitro model for the regulation of human cytomegalovirus latency and reactivation in dendritic cells by chromatin remodelling. J Gen Virol. 2005;86:2949–2954. doi: 10.1099/vir.0.81161-0. [DOI] [PubMed] [Google Scholar]
- 48.Hargett D, Shenk TE. Experimental human cytomegalovirus latency in CD14+ monocytes. Proc Natl Acad Sci USA. 2010;107:20039–20044. doi: 10.1073/pnas.1014509107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fleetwood AJ, Lawrence T, Hamilton JA, Cook AD. Granulocyte-macrophage colony-stimulating factor (CSF) and macrophage CSF-dependent macrophage phenotypes display differences in cytokine profiles and transcription factor activities: implications for CSF blockade in inflammation. J Immunol. 2007;178:5245–5252. doi: 10.4049/jimmunol.178.8.5245. [DOI] [PubMed] [Google Scholar]
- 50.Lacey DC, Achuthan A, Fleetwood AJ, Dinh H, Roiniotis J, et al. Defining GM-CSF- and Macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol. 2012;188:5752–5765. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
- 51.Schrier RD, Nelson JA, Oldstone MB. Detection of human cytomegalovirus in peripheral blood lymphocytes in a natural infection. Science. 1985;230:1048–1051. doi: 10.1126/science.2997930. [DOI] [PubMed] [Google Scholar]
- 52.Rice GP, Schrier RD, Oldstone MB. Cytomegalovirus infects human lymphocytes and monocytes: virus expression is restricted to immediate-early gene products. Proc Natl Acad Sci USA. 1984;81:6134–6138. doi: 10.1073/pnas.81.19.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoddart CA, Cardin RD, Boname JM, Manning WC, Abenes GB, et al. Peripheral blood mononuclear phagocytes mediate dissemination of murine cytomegalovirus. J Virol. 1994;68:6243–6253. doi: 10.1128/jvi.68.10.6243-6253.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Daley-Bauer LP, Roback LJ, Wynn GM, Mocarski ES. Cytomegalovirus hijacks CX3CR1(hi) patrolling monocytes as immune-privileged vehicles for dissemination in mice. Cell Host Microbe. 2014;15:351–362. doi: 10.1016/j.chom.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Farrell HE, Bruce K, Lawler C, Oliveira M, Cardin R, et al. Murine cytomegalovirus spreads by dendritic cell recirculation. mBio. 2017;8:e01264–17. doi: 10.1128/mBio.01264-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mitchell BM, Leung A, Stevens JG. Murine cytomegalovirus DNA in peripheral blood of latently infected mice is detectable only in monocytes and polymorphonuclear leukocytes. Virology. 1996;223:198–207. doi: 10.1006/viro.1996.0468. [DOI] [PubMed] [Google Scholar]
- 57.Bolovan-Fritts CA, Mocarski ES, Wiedeman JA. Peripheral blood CD14(+) cells from healthy subjects carry a circular conformation of latent cytomegalovirus genome. Blood. 1999;93:394–398. [PubMed] [Google Scholar]
- 58.Goodrum FD, Jordan CT, High K, Shenk T. Human cytomegalovirus gene expression during infection of primary hematopoietic progenitor cells: a model for latency. Proc Natl Acad Sci USA. 2002;99:16255–16260. doi: 10.1073/pnas.252630899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Connor CM, Murphy EA. A myeloid progenitor cell line capable of supporting human cytomegalovirus latency and reactivation, resulting in infectious progeny. J Virol. 2012;86:9854–9865. doi: 10.1128/JVI.01278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goodrum F, Jordan CT, Terhune SS, High K, Shenk T, et al. Differential outcomes of human cytomegalovirus infection in primitive hematopoietic cell subpopulations. Blood. 2004;104:687–695. doi: 10.1182/blood-2003-12-4344. [DOI] [PubMed] [Google Scholar]
- 61.Rauwel B, Jang SM, Cassano M, Kapopoulou A, Barde I, et al. Release of human cytomegalovirus from latency by a KAP1/TRIM28 phosphorylation switch. elife. 2015;4 doi: 10.7554/eLife.06068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu D, Pan C, Sheng J, Liang H, Bian Z, et al. Human cytomegalovirus reprogrammes haematopoietic progenitor cells into immunosuppressive monocytes to achieve latency. Nat Microbiol. 2018;3:503–513. doi: 10.1038/s41564-018-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goodrum F, Reeves M, Sinclair J, High K, Shenk T, et al. Human cytomegalovirus sequences expressed in latently infected individuals promote a latent infection in vitro. Blood. 2007;110:937–945. doi: 10.1182/blood-2007-01-070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minton EJ, Tysoe C, Sinclair JH, Sissons JG. Human cytomegalovirus infection of the monocyte/macrophage lineage in bone marrow. J Virol. 1994;68:4017–4021. doi: 10.1128/jvi.68.6.4017-4021.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hahn G, Jores R, Mocarski ES. Cytomegalovirus remains latent in a common precursor of dendritic and myeloid cells. Proc Natl Acad Sci USA. 1998;95:3937–3942. doi: 10.1073/pnas.95.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.