Abstract
The contribution of N-acetylneuraminate scavenging to the nutrition of Mycoplasma alligatoris was examined. The wild-type grew substantially faster (P<0.01) than the mutant strains that were unable either to liberate (extracellular NanI− mutants) or to catabolize (NanA− mutants) N-acetylneuraminate from glycoconjugates in minimal SP-4 medium supplemented only with serum, but the growth of sialidase-negative mutants could not be restored to wild-type rate simply by adding unconjugated sialic acid to the culture medium. In 1 : 1 growth competition assays the wild-type was recovered in >99-fold excess of a sialidase-negative mutant after co-culture on pulmonary fibroblasts in serum-free RPMI 1640 medium, even with supplemental glucose. The advantage of nutrient scavenging via this mechanism in a complex glycan-rich environment may help to balance the expected selective disadvantage conferred by the pathogenic effects of mycoplasmal sialidase in an infected host.
Keywords: Mycoplasma, sialidase, nutrition, growth
N-acetylneuraminate scavenging is presumed to contribute to the nutrition of many pathogenic and commensal bacteria [1–3], but this has been directly measured in surprisingly few species besides Escherichia coli and never in one so metabolically minimalist as a mycoplasma [2–9]. In contrast to the usually subtle mycoplasmosis, Mycoplasma alligatoris causes acute lethal primary infection of susceptible hosts. A comparative genomics approach to explaining the evolutionary origin and mechanisms of that unusual virulence revealed that, amid a clade of harmless or comparatively much lower-virulence species, M. alligatoris alone encodes a full suite of ‘spreading factors’, including sialidase, galactosidase, hexosaminidase, hyaluronidase and glucuronidase, which could fuel its metabolism while simultaneously promoting fulminant necrotizing disease [10, 11]. Having first demonstrated a role for sialidase in its virulence [12, 13], through the present work we sought to assess one potential link between virulence and pathogen fitness by documenting the contribution of sialic acid scavenging to the nutrition of this species.
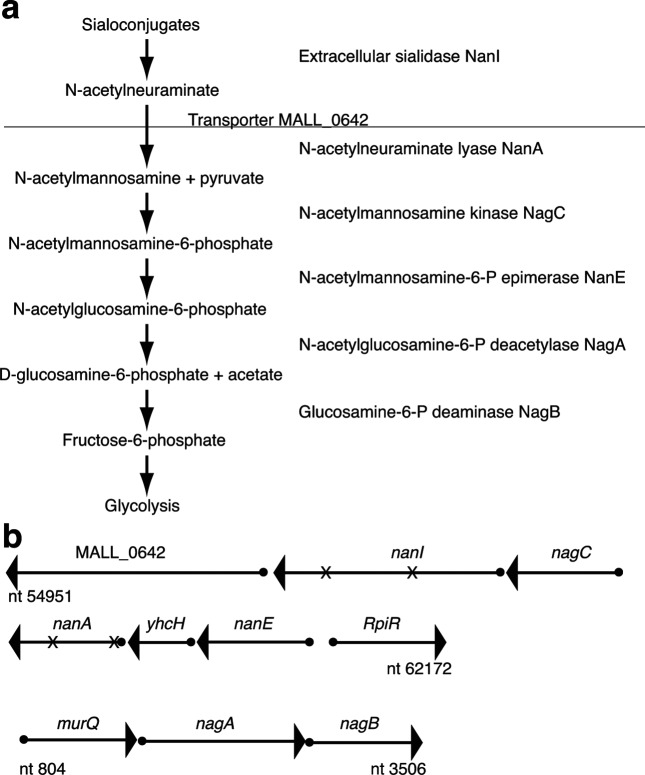
The genome of fast-growing M. alligatoris encodes two paralogues of sialidase NanI that exhibit 39 % aa identity and 55 % aa similarity to each other: MALL_0755, which is predicted to have a 34-aa secretory signal peptide, and MALL_0641, which lacks evident means of being secreted [14]. A complete pathway for unconjugated N-acetylneuraminate uptake and catabolism is also annotated. Contiguous genes encoding an SSS-family N-acetylneuraminate transporter (MALL_0642) [15, 16], N-acetylneuraminate lyase NanA (MALL_0639), N-acetylmannosamine kinase NagC (MALL_0640), N-acetylmannosamine-6-phosphate epimerase NanE (MALL_0637), the putative sugar isomerase YhcH (MALL_0638) [17] and the cytosolic allele of sialidase (MALL_0641) occur adjacent to a predicted RpiR-family transcriptional regulator of phosphosugar metabolism (MALL_0636). At a second locus, the N-acetylglucosamine-6-phosphate deacetylase nagA and glucosamine-6-phosphate deaminase nagB genes are contiguous with murQ, an N-acetylmuramic acid-6-phosphate lyase involved in amino sugar scavenging from peptidoglycan acquired from the environment [18]. The predicted extracellular sialidase gene of M. alligatoris is unlinked from other elements of the pathway. Collectively, those enzymes could liberate N-acetylneuraminate from sialoconjugates of the host or other microbes and funnel it as a nutrient to glycolysis (Fig. 1) [19, 20]. To test the prediction that extracellular sialidase contributes to the rapid growth rate of this species, we examined the growth of M. alligatoris knockout mutants that are unable either to liberate (NanI− mutants) or to catabolize (NanA− mutants) sialic acid in free saccharide-poor, complex glycan-rich environments.
Fig. 1.
Predicted capacity for N-acetylneuraminate scavenging by Mycoplasma alligatoris . (a) The horizontal line represents the interface between extracellular and intracellular processes (modified from [28]). (b) A cytosolic paralogue of sialidase NanI (MALL_0641) plus the N-acetylneuraminate transporter MALL_0642 and catabolic enzymes are encoded in two separate loci, while the gene encoding an extracellular sialidase NanI paralogue (MALL_0755) is unlinked from other elements of the pathway. Transposon insertion sites in independent mutants of the cytosolic sialidase gene nanI and the N-acetylneuraminate gene nanA are indicated by an X. The nucleotide numbering refers to GenBank assembly ASM17837v1.
Stock cultures of wild-type M. alligatoris strain A21JP2T were maintained in complete SP-4 broth medium including 0.3 mM glucose and 20 % v/v foetal bovine serum (FBS), pH 7.6–7.8. Site-specific gene knockout methods were ineffective in M. alligatoris , so a library of random knockout mutants was created by transposon insertion using the mini-Tn4001tetM plasmid pTF20 [21]. Briefly, the cell pellet from a 15 ml broth culture in the mid-log growth phase was washed 2× and resuspended in 60 µl of electroporation buffer (8 mM HEPES, 0.27 M sucrose) before being transformed with 20 µg of pTF20 DNA. After incubation on ice for 15 min, the suspension was pulsed in a pre-chilled electroporation cuvette (2.5 kV, 100 Ω, 25 µF). The suspension was then combined with 1 ml of chilled SP-4 broth and incubated at 25 °C for 10 min, and then at 30 °C for 3 h, before inoculation onto SP-4 agar including 10 µg ml−1 tetracycline. The precise point of transposon insertion was mapped for ~1600 random tetR-selected clones using direct genomic DNA sequencing via extension from primers complementary to the IS256 arms (pTF20LH_out 5′-GCT GAA ACT AAG CCC TAA AAG TAC CC-3′ and pTF20RH_out 5′-TGA GCG AGG AAG CGG AAG AG-3′) and blast comparisons of those sequences to the M. alligatoris genome. The mutant strains examined in the present study are listed in Table 1.
Table 1. Transposon insertion mapping of the strains examined in this study.
| Clone | Mutated gene | Enzyme | tetM insertion point | Phenotype |
|---|---|---|---|---|
| TNP1C23 | MALL_0755 | Extracellular sialidase | nt 1530 of 1764 | Extracellular NanI− |
| TNP1C556 | MALL_0755 | Extracellular sialidase | In dcma | Extracellular NanI− |
| TNP1C121 | MALL_0641 | Cytosolic sialidase | nt 609 of 1536 | Extracellular NanI+ |
| TNP1C420 | MALL_0641 | Cytosolic sialidase | nt 1209 of 1536 | Extracellular NanI+ |
| TP2C209 | MALL_0639 | N-acetylneuraminate lyase | nt 11 of 885 | Cytosolic NanA− |
| TP2C325 | MALL_0639 | N-acetylneuraminate lyase | nt 596 of 885 | Cytosolic NanA− |
a, isogenic tetM insertion mapped to cytosine-5-methyltransferase gene dcm (MALL_0186) while extracellular sialidase-negative phenotype was conferred by spontaneous substitution mutations in MALL_0755.
The sialidase-negative phenotype of transformed clones TNP1C23 and TNP1C556 was first recognized from their inability to hydrolyze the fluorogenic substrate 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid during the initial high-throughput phenotypic screening of intact cells [22]. It was subsequently shown that an isogenic tetM insertion occurred following nt 1530 of the 1764-nt allele of the extracellular sialidase in clone TNP1C23, whereas spontaneous substitution mutations SKDGGKTW to PKDDRKTW at aa residues 320–327 contributing to the NanI catalytic site were responsible for the phenotype of clone TNP1C556. In that clone the tetM insertion was in the DNA cytosine-5-methyltransferase gene dcm (MALL_0186), the modification partner of a predicted NgoBV type II restriction endonuclease (MALL_0185), so the insert was unlikely to have any polar effects on N-acetylneuraminate catabolism, the cell cycle, or growth in vitro [23]. The cytosolic sialidase activity of the clones TNP1C23 and TNP1C556 remained detectable when using the same fluorogenic substrate, but only after cell lysis by three cycles of rapid freeze for 30 s in a dry ice/70 % v/v ethanol bath followed by thaw for 1 min at 60 °C. Insertions in the cytosolic allele did not affect the extracellular sialidase activity of intact cells during phenotypic screening of the representative clones TNP1C121 and TNP1C420, which resolved prior ambiguity regarding gene copy number and the cellular compartmentalization of the two NanI paralogues in M. alligatoris [10].
Isogenic tetM insertions in the 885-nt N-acetylneuraminate lyase gene nanA (MALL_0639) occurred following nt 11 in clone TP2C209 and nt 596 in clone TP2C325. The lyase-negative phenotype of those mutants was confirmed using an assay for the N-acetylmannosamine liberated following the incubation of N-acetylneuraminic acid with lysed M. alligatoris cells [24]. The reaction mixture contained 16 µg of cytosolic protein and 32 mM N-acetylneuraminic acid (cat. no. 855650, Sigma-Aldrich) in 200 mM potassium phosphate buffer, pH 7.4. After 5 h of incubation at 30 °C, the wild-type and irrelevant knockout controls produced an intense chromogenic reaction at 540 nm when combined with p-dimethyl-aminobenzaldehyde, while the M. alligatoris lyase knockout mutants and control Mycoplasma crocodyli MP145T cells lacking the N-acetylneuraminate lyase gene [10] never produced any colour.
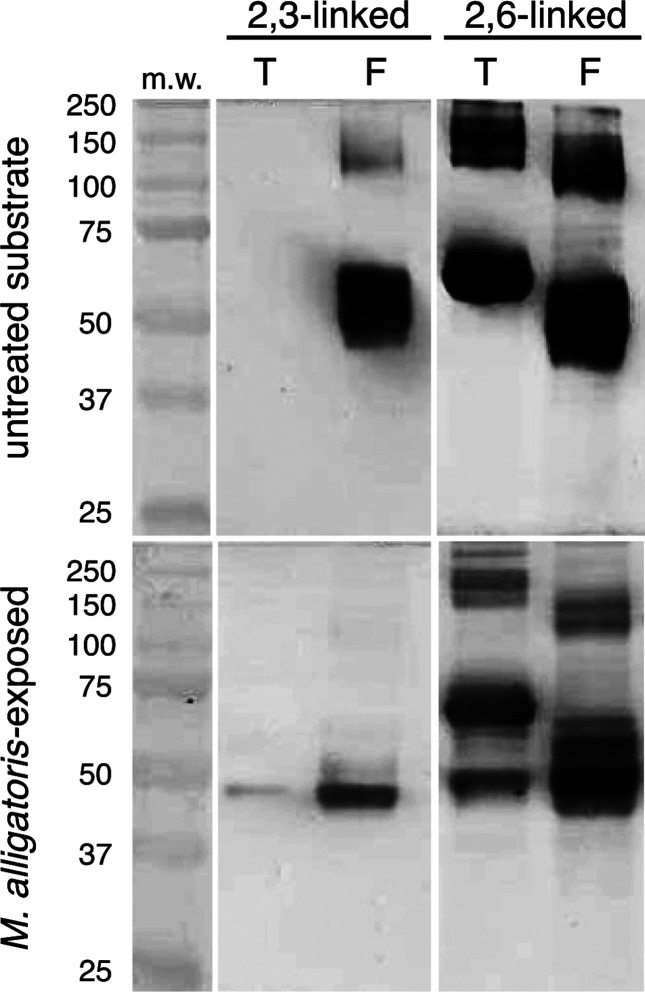
The glycosidic linkage preference of the extracellular sialidase [25] was determined using digoxigenin-labelled Maackia amurensis and Sambucus nigra lectins in a Western blot format (cat. no. 11210238001, Roche) following the incubation of fetuin and transferrin serum glycoprotein standards (5 µg per reaction) with ~2.5×108 washed wild-type M. alligatoris cells for 48 h at 30 °C in glucose-free RPMI 1640 tissue culture medium. In contrast to the sialidase allele encoded by Mycoplasma gallisepticum and Mycoplasma synoviae [26–28], the extracellular M. alligatoris sialidase depleted terminal α-2,3-linked sialic acid from subterminal galactose more efficiently than it depleted α-2,6-linked residues (Fig. 2).
Fig. 2.
Glycosidic linkage specificity of Mycoplasma alligatoris extracellular sialidase. Western blots of transferrin (T) and fetuin (F) glycoprotein substrates probed with Mackia amurensis (MAA) or Sambucus nigra (SNA) lectin, showing preferential depletion of terminal α-2,3-linked versus α-2,6-linked sialic acid by washed wild-type M. alligatoris cells. Transferrin has exclusively 2,6-linked sialogalactosyl residues, while fetuin has both 2,3- and 2,6-linked residues. MAA recognizes terminal 2,3-linked sialic acid, while SNA recognizes terminal 2,6-linked sialic acid.
The growth of the extracellular sialidase-negative and lyase-negative mutants was measured individually in SP-4 mycoplasma broth with FBS only (minimal medium), complete SP-4 broth including 0.3 mM glucose, or minimal medium plus 0.3 mM unconjugated N-acetylneuraminic acid. The serum provided ~1 mM conjugated sialic acid, >90 % in the form of N-acetylneuraminic acid (~75 % as glycoprotein and ~25 % as glycolipid) [29]. Starting with duplicate 5 ml cultures of each strain [initial density=1×106 colony-forming units (c.f.u.) ml−1), 100 µl aliquots were withdrawn over a time course from 0 to 24 h, and their serial 10-fold dilutions were inoculated onto complete SP-4 agar for colony counting after 4 days of incubation to determine changes in culture density. Separate aliquots were withdrawn at each time point to determine changes in pH. A repeated-measures ANOVA including the main effects of phenotype and timepoint, with Dunnett’s post hoc comparisons to wild-type controls when the main effects were significant (P<0.05), was performed using JMP v7.02 (SAS).
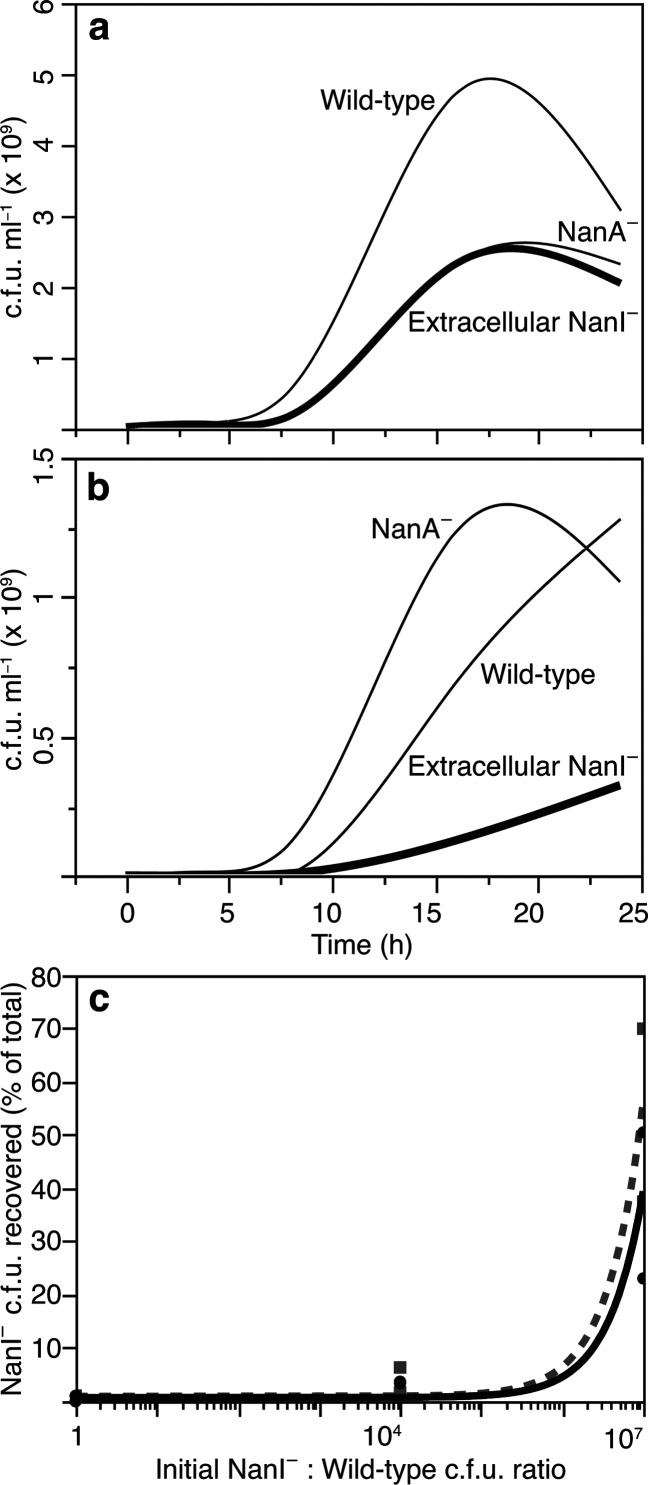
There were no statistically significant differences in the growth curves (Δc.f.u. ml−1) or acid excretion (ΔpH) among the phenotypes measured in complete SP-4 medium, confirming that all stocks were viable and that the mutant strains had no defect in growth relative to their wild-type parent strain. All grew more slowly (P<0.01) and excreted less acid (P<0.01) in unsupplemented minimal medium than in complete SP-4, but wild-type M. alligatoris grew significantly faster (P<0.01 by 8 h) in minimal medium than the strains that were unable to liberate (TNP1C23 and TNP1C556) or catabolize (TP2C209 and TP2C325) N-acetylneuraminate from serum glycans (Fig. 3a). The wild-type exponential growth rate constant (k) was 0.47 h−1, and the doubling time (g) was 1.5 h between 4 and 8 h timepoints, while the mutants grew exponentially between 8 and 16 h (mean k=0.32 h−1, g=2.2 h for extracellular sialidase-negative strains and k=0.34 h−1, g=2.0 h for lyase-negative strains). The cleavage of terminal sialic acid from antennary serum glycans likely exposed subterminal galactose and glucosamine for subsequent liberation as additional nutrients by the sequential action of M. alligatoris ’ galactosidase LacZ (MALL_0716) and hexosaminidase NagH (MALL_0841) [10, 30], a second-order effect of sialidase that is consistent with the slight differences in the k and g of the mutants. However, their similarly reduced growth rate versus the wild-type was evidence that the primary nutritional benefit was acquiring N-acetylneuraminate rather than galactose or glucosamine when glucose availability was restricted.
Fig. 3.
Contribution of N-acetylneuraminate scavenging to the growth of Mycoplasma alligatoris . The smoothed curves represent the mean number of colony-forming units (c.f.u.) of two independent clones of each mutant phenotype. (a) Knockouts that were unable to liberate (extracellular Nanl−) or catabolize (NanA−) N-acetylneuraminate grew more slowly than the wild-type in minimal SP-4 medium supplemented only with serum (Dunnett’s P<0.01 by 8 h). (b) All genotypes grew ~twofold more slowly in 0.3 mM N-acetylneuraminate-supplemented SP-4 medium with serum than in unsupplemented medium with serum (P<0.05). Presumptive toxicity was evident by 16 h in the NanA− knockouts. (c) In direct growth competition assays, the fraction of NanI− colonies recovered after co-culture on fibroblast monolayers in glucose-free medium (solid line) inoculated with mutant TNP1C23 mixed 1 : 1, 104 : 1 or 107 : 1 with the wild-type was 0.5, 4.0 and 55.6 %, respectively; inoculation in those ratios yielded only 0.5, 3.5 and 42.6 % NanI− colonies even in glucose-supplemented medium (dashed line).
All of the genotypes grew ~twofold more slowly (P<0.05) in minimal medium supplemented with unconjugated N-acetylneuraminate versus serum alone (Fig. 3b). This unexplained effect, also observed with E. coli , Streptococcus pneumoniae and Capnocytophaga canimorsus , but not with Vibrio spp. [1, 7, 24], may be related to differential kinetics of free saccharide uptake or to an intracellular overload of N-acetylneuraminate, which by 16 h became toxic to lyase-negative mutants (Fig. 3b) [4, 9, 24, 30, 31].
In direct growth competition assays, clone TNP1C23 duplicates were mixed in ratios of 1 : 1, 104 : 1, or 107 : 1 c.f.u. of wild-type M. alligatoris and co-cultured for 24 h on pulmonary fibroblast monolayers [27] as the sole source of complex nutrients in serum-free RPMI 1640 medium either with or without 11 mM glucose. Serial dilutions of culture supernatant were then inoculated onto complete SP-4 agar for mycoplasma colony counting. All colonies were individually expanded in broth, and then their sialidase phenotype was determined by fluorometry as described above [22]. Inoculation in those ratios yielded only 0.5, 4.0 and 55.6 % sialidase-negative colonies, respectively, from glucose-free medium, and 0.5, 3.5 and 42.6 % sialidase-negative colonies even in glucose-supplemented medium (Fig. 3c), which is evidence that extracellular sialidase may also play an important non-nutritional role in the natural ecology of M. alligatoris , as it does in M. synoviae [28]. From these findings we conclude that the advantageous capacity to scavenge nutrients via extracellular sialidase in a complex glycan-rich environment may help to balance the potential selective disadvantage conferred by the cytopathic effects of sialidase in an infected host [3, 11–13].
Funding information
Funded by Public Health Service grants R01GM076584 and R01GM076584-S1 from the National Institute of General Medical Sciences (D. R. B. and J. I. G.).
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: ANOVA, analysis of variance; tetM, tetracycline-resistance protein encoded by gene tetM.
Edited by: J. Stülke
References
- 1.Almagro-Moreno S, Boyd EF. Insights into the evolution of sialic acid catabolism among bacteria. BMC Evol Biol. 2009;9:118. doi: 10.1186/1471-2148-9-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Almagro-Moreno S, Boyd EF. Bacterial catabolism of nonulosonic (sialic) acid and fitness in the gut. Gut Microbes. 2010;1:45–50. doi: 10.4161/gmic.1.1.10386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vimr ER. Unified theory of bacterial sialometabolism: how and why bacteria metabolize host sialic acids. ISRN Microbiol. 2013;2013:1–26. doi: 10.1155/2013/816713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vimr ER, Troy FA. Identification of an inducible catabolic system for sialic acids (nan) in Escherichia coli . J Bacteriol. 1985;164:845–853. doi: 10.1128/jb.164.2.845-853.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Honma K, Ruscitto A, Frey AM, Stafford GP, Sharma A. Sialic acid transporter NanT participates in Tannerella forsythia biofilm formation and survival on epithelial cells. Microb Pathog. 2016;94:12–20. doi: 10.1016/j.micpath.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, McClane BA. NanI sialidase can support the growth and survival of Clostridium perfringens strain F4969 in the presence of sialyated host macromolecules (Mucin) or Caco-2 cells. Infect Immun. 2018;86:e00547-17. doi: 10.1128/IAI.00547-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mally M, Shin H, Paroz C, Landmann R, Cornelis GR. Capnocytophaga canimorsus: a human pathogen feeding at the surface of epithelial cells and phagocytes. PLoS Pathog. 2008;4:e1000164. doi: 10.1371/journal.ppat.1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roy S, Honma K, Douglas CW, Sharma A, Stafford GP. Role of sialidase in glycoprotein utilization by Tannerella forsythia . Microbiology. 2011;157:3195–3202. doi: 10.1099/mic.0.052498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown DR, Farmerie WG, May M, Benders GA, Durkin AS, et al. Genome sequences of Mycoplasma alligatoris A21JP2T and Mycoplasma crocodyli MP145T . J Bacteriol. 2011;193:2892–2893. doi: 10.1128/JB.00309-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown DR, Zacher LA, Farmerie WG. Spreading factors of Mycoplasma alligatoris, a flesh-eating mycoplasma. J Bacteriol. 2004;186:3922–3927. doi: 10.1128/JB.186.12.3922-3927.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hunt ME, Brown DR. Mycoplasma alligatoris infection promotes CD95 (FasR) expression and apoptosis of primary cardiac fibroblasts. Clin Diagn Lab Immunol. 2005;12:1370–1377. doi: 10.1128/CDLI.12.12.1370-1377.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunt ME, Brown DR. Role of sialidase in Mycoplasma alligatoris-induced pulmonary fibroblast apoptosis. Vet Microbiol. 2007;121:73–82. doi: 10.1016/j.vetmic.2006.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Severi E, Hood DW, Thomas GH. Sialic acid utilization by bacterial pathogens. Microbiology. 2007;153:2817–2822. doi: 10.1099/mic.0.2007/009480-0. [DOI] [PubMed] [Google Scholar]
- 16.Severi E, Hosie AH, Hawkhead JA, Thomas GH. Characterization of a novel sialic acid transporter of the sodium solute symporter (SSS) family and in vivo comparison with known bacterial sialic acid transporters. FEMS Microbiol Lett. 2010;304:47–54. doi: 10.1111/j.1574-6968.2009.01881.x. [DOI] [PubMed] [Google Scholar]
- 17.Teplyakov A, Obmolova G, Toedt J, Galperin MY, Gilliland GL. Crystal structure of the bacterial YhcH protein indicates a role in sialic acid catabolism. J Bacteriol. 2005;187:5520–5527. doi: 10.1128/JB.187.16.5520-5527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jaeger T, Mayer C. N-acetylmuramic acid 6-phosphate lyases (MurNAc etherases): role in cell wall metabolism, distribution, structure, and mechanism. Cell Mol Life Sci. 2008;65:928–939. doi: 10.1007/s00018-007-7399-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Corfield T. Bacterial sialidases–roles in pathogenicity and nutrition. Glycobiology. 1992;2:509–521. doi: 10.1093/glycob/2.6.509. [DOI] [PubMed] [Google Scholar]
- 20.Schauer R. Achievements and challenges of sialic acid research. Glycoconj J. 2000;17:485–499. doi: 10.1023/A:1011062223612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dybvig K, Zuhua C, Lao P, Jordan DS, French CT, et al. Genome of Mycoplasma arthritidis . Infect Immun. 2008;76:4000–4008. doi: 10.1128/IAI.00516-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.May M, Brown DR. Secreted sialidase activity of canine mycoplasmas. Vet Microbiol. 2009;137:380–383. doi: 10.1016/j.vetmic.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Løbner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Curr Opin Microbiol. 2005;8:154–160. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 24.Jeong HG, Oh MH, Kim BS, Lee MY, Han HJ, et al. The capability of catabolic utilization of N-acetylneuraminic acid, a sialic acid, is essential for Vibrio vulnificus pathogenesis. Infect Immun. 2009;77:3209–3217. doi: 10.1128/IAI.00109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Corfield AP, Higa H, Paulson JC, Schauer R. The specificity of viral and bacterial sialidases for alpha(2-3)- and alpha(2-6)-linked sialic acids in glycoproteins. Biochim Biophys Acta. 1983;744:121–126. doi: 10.1016/0167-4838(83)90080-8. [DOI] [PubMed] [Google Scholar]
- 26.Sethi KK, Müller HE. Neuraminidase activity in Mycoplasma gallisepticum . Infect Immun. 1972;5:260–262. doi: 10.1128/iai.5.2.260-262.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berčič RL, Cizelj I, Dušanić D, Narat M, Zorman-Rojs O, et al. Neuraminidase of Mycoplasma synoviae desialylates heavy chain of the chicken immunoglobulin G and glycoproteins of chicken tracheal mucus. Avian Pathol. 2011;40:299–308. doi: 10.1080/03079457.2011.565311. [DOI] [PubMed] [Google Scholar]
- 28.May M, Brown DR. Genetic variation in sialidase and linkage to N-acetylneuraminate catabolism in Mycoplasma synoviae . Microb Pathog. 2008;45:38–44. doi: 10.1016/j.micpath.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sherblom AP, Bharathan S, Hall PJ, Smagula RM, Moody CE, et al. Bovine serum sialic acid: age-related changes in type and content. Int J Biochem. 1988;20:1177–1183. doi: 10.1016/0020-711X(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 30.Marion C, Burnaugh AM, Woodiga SA, King SJ. Sialic acid transport contributes to pneumococcal colonization. Infect Immun. 2011;79:1262–1269. doi: 10.1128/IAI.00832-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim BS, Hwang J, Kim MH, Choi SH. Cooperative regulation of the Vibrio vulnificus nan gene cluster by NanR protein, cAMP receptor protein, and N-acetylmannosamine 6-phosphate. J Biol Chem. 2011;286:40889–40899. doi: 10.1074/jbc.M111.300988. [DOI] [PMC free article] [PubMed] [Google Scholar]