Abstract
Microbial biofilms are ubiquitous in drinking water systems, yet our understanding of drinking water biofilms lags behind our understanding of those in other environments. Here, a six-member model bacterial community was used to identify the interactions and individual contributions of each species to community biofilm formation. These bacteria were isolated from the International Space Station potable water system and include Cupriavidus metallidurans , Chryseobacterium gleum , Ralstonia insidiosa, Ralstonia pickettii, Methylorubrum (Methylobacterium) populi and Sphingomonas paucimobilis , but all six species are common members of terrestrial potable water systems. Using reconstituted assemblages, from pairs to all 6 members, community biofilm formation was observed to be robust to the absence of any single species and only removal of the C. gleum / S. paucimobilis pair, out of all 15 possible 2-species subtractions, led to loss of community biofilm formation. In conjunction with these findings, dual-species biofilm formation assays supported the view that the contribution of C. gleum to community biofilm formation was dependent on synergistic biofilm formation with either R. insidiosa or C. metallidurans . These data support a model of multiple, partially redundant species interactions to generate robustness in biofilm formation. A bacteriophage and multiple predatory bacteria were used to test the resilience of the community to the removal of individual members in situ, but the combination of precise and substantial depletion of a single target species was not achievable. We propose that this assemblage can be used as a tractable model to understand the molecular bases of the interactions described here and to decipher other functions of drinking water biofilms.
Keywords: tap water, bacterial ecology, species interactions, robustness, resilience
Introduction
Simplified microbial communities are important tools to study interactions between species and understand the species contributing to various community processes. Such studies have been useful in understanding the contributions of individual species to community functions in a variety of environments, including the mammalian oral cavity, gut and infected lungs, plant nectaries and roots, marine environments and microbially impacted food products, and these interactions play important roles, including promoting antimicrobial tolerance [1–11]. In many of these models the types of microbial interactions are conceptually similar but differ in the specific mechanisms driving the interactions and the identities of the members that participate. We are interested in a less well-studied community: the bacteria that compose the biofilms in potable water plumbing, particularly in the terminal portions of the plumbing (faucets, showerheads, etc.). The goal in this study was to use a simplified subset of our previously described model potable water bacterial community [12] to characterize the contributions of individual species to biofilm formation.
Plumbing biofilms are a universal aspect of potable water systems [13], inescapable even after leaving Earth, as demonstrated by the drinking water biofilm issues on board all long-term manned spaceflight platforms to date [14–18]. These communities play important roles in water quality and pipe corrosion and are the reservoir of a number of opportunistic pathogens, including Legionella pneumophila , non-tuberculosis mycobacteria, Stenotrophomonas maltophilia and Pseudomonas aeruginosa , among others [19–22]. Many compositional studies have been conducted on the bacterial community in terminal plumbing fixtures and generally show a similar set of species, predominately Gram-negative and dominated by Methylorubrum/Methylobacterium, Sphingomonas and Ralstonia species (reviewed in [13]).
Here we use a six-species bacterial community composed of bacteria isolated from the potable water reclamation system of the International Space Station (ISS) as a model potable water community. The component species, Cupriavidus metallidurans , Chryseobacterium gleum , Ralstonia insidiosa, Ralstonia pickettii, Methylorubrum populi and Sphingomonas paucimobilis , are common and abundant constituents of drinking water systems worldwide and could function as a model community to study bacterial interactions in the potable water environment. In this study, we have manipulated the species compositions of the community to understand the effects of community composition on biofilm formation within this assemblage. We show that community biofilm formation is not affected by single-species removal and that only one of the possible combinations of two-species removal results in near-complete ablation of the community biofilm. These findings and follow-up experiments led us to propose a model where S. paucimobilis and C. gleum can independently contribute to community biofilm formation, with C. gleum ’s contribution dependent upon synergistic biofilm formation with either one of two other community members.
Methods
Bacterial strains and maintenance conditions
C. metallidurans 101480065–2, C. gleum 113330055–2, R. insidiosa 130770013–1, M. populi 122620021–1, S. paucimobilis 121220007–2 and R. pickettii 113330051–2 were isolated from the ISS potable water delivery system (except for C. gleum , isolated from the Russian SVO-ZV) and identified by the Microbiology Laboratory at the NASA Johnson Space Center (Houston, TX, USA) [16]. The strain numbers listed after each species are for the strain database at the Johnson Space Center, and strains may be requested directly using these designations for all species described herein. We specify our Methylorubrum ( Methylobacterium ) strain as M. populi for the closest named species based on 16S sequencing and partial genome sequencing phylogenetic assessment, but it also has close similarity to M. fujiwaensis. Future whole-genome sequencing may suggest different nomenclature for this strain. Methylorubrum nomenclature for the genus is based on a recent taxonomic reassessment [23]. All bacteria were stored in 20 % glycerol stocks at −80 °C and were recovered on R2A plates at 30 °C.
This particular community and its membership were chosen for a few reasons. First, these species are common in potable water systems (multiple metagenomic studies reviewed in [13]) and represent a subset of the major members in the ISS potable water system [16]. Second, this community simplifies our previously described potable water community [12] by removal of the species present in the lowest proportions, which will aid in future omics analyses, and inclusion of M. populi. M. populi was added to better mimic tap water communities and to expand the metabolic repertoire of the model community, particularly in low-nutrient conditions, where the single carbon compound metabolism of Methylorubrum is predicted to be important.
For all experiments below, the R2A plates were used as a source of inoculation into MRLP media (2/3 modified MOPS media [24], 1/3 R2B, 1/50th LB, with 10 mM additional sodium pyruvate; in composition, this medium comprises 26.4 mM MOPS, 2.64 mM tricine, 6.28 mM NH4Cl, 34.7 mM NaCl, 0.35 mM MgCl2, 0.19 mM K2SO4, 1.57 mM K2HPO4, 10.91 mM sodium pyruvate, 0.94 mM glucose, 32 µM MgSO4, 21.1 µM CaCl2, 6 µM FeSO4, 5.28 µM FeCl2, 0.33 g l−1 peptone, 0.27 g l−1 yeast extract, 0.20 g l−1 tryptone, 0.17 g l−1 soluble starch, 0.66X MOPS medium micronutrients) and grown overnight in 3 ml culture volume at 30 °C with orbital shaking of angled glass tubes. MRLP was generated empirically during the identification of media formulations that allowed pregrowth of Methylorubrum in primarily unaggregated form, which was therefore amenable to quantification by OD600 and dilution plating.
Community initiation and biofilm quantification
The overnight MRLP-grown cultures were used as the starting source for biofilm experiments. The optical density of each species was determined and normalized to generate an initial OD600 of 0.05 for each community member. This means that communities with fewer members have a lower total starting OD600, but we have found that proportional initiation (increasing input to normalize the total starting OD600) did not measurably alter biofilm formation. As we previously reported, the colony-forming units (c.f.u.) ml−1 values at an OD600 of 0.05 for each species with the exception of C. gleum were within 0.4 log10 units of each other, centred around 1E8 c.f.u. ml−1. This variance is close to the technical error level of our spot-based serial dilution plating (roughly 0.3 log10 in control counts). The c.f.u. ml−1 for C. gleum was 0.4 log10 units below the mean of the other species. Thus, while equal cell densities were used, this does not lead to equal cell numbers, particularly for C. gleum .
After pipetting 150 µl of the various species mixtures into flexible 96-well dishes, the dishes were placed into humidified chambers and incubated under static conditions (no shaking) at 30 °C for 24–96 h. To measure biofilm formation, the crystal violet staining protocol was used as described by O’Toole and colleagues [25]. Biofilms were quantified by dissolving the crystal violet with 10 % acetic acid and measuring absorbance at 550 nm.
Assessing community compositions
To determine static community composition, cells were gathered by resuspending the contents of each well via vigorous pipetting and scraping the well walls with a pipette tip. As measured by crystal violet staining, <1 % of the biomass remained after this treatment. We used a previously reported differential plating technique to quantify the c.f.u. for each species [12]. We rarely identified Methylorubrum colonies during plating, although we can detect DNA and RNA. Given the ability of Methylorubrum to form tenacious biofilms and self-aggregates, we are not particularly surprised by our inability to recover colonies from this organism. In the context of biofilm formation, Methylorubrum does not appear to contribute substantively to any of the phenoytypes we observe, and so, while future studies may require Methylorubrum enumeration to support conclusions, we do not think it a requirement here.
For the phage treatments, community composition was determined just by plating on R2A plates and binning colonies by colour, yellow for S. paucimobilis , orange for C. gleum , and lumping all others together simply as white colonies. We have seen no effect of proximity between colonies of the same or different species altering colony colour or morphology on the media described here.
Sphingomonas paucimobilis phage isolation, characterization, sequencing and biofilm treatment
A number of water sources were screened to identify a S. paucimobilis -specific plaque-forming phage and only one was isolated (designated ΦScott), isolated from Lake Champlain water (Burlington, VT, USA). The phage was collected from the initial plaque by elution into 100 µl of 10 mM Tris pH 7.4 and filtration through a 0.22 µm PVDF filter and amplified from this initial isolation by adding 10 µl of the original ΦScott stock to 50 ml R2B inoculated with 1 ml of S. paucimobilis overnight culture. This was incubated overnight at 30 °C. Cells and debris were collected by centrifugation and the supernatant was filtered through a 0.22 µm filter. Plaque-forming units (p.f.u.) were determined by plating serial dilutions of each phage prep onto a lawn of S. paucimobilis .
High-titre ΦScott preparation was spotted onto formvar/carbon-coated nickel 200 mesh grids made hydrophilic with peroxide treatment, and the sample was left on the grid for 1 min. Excess liquid was wicked off and the grid rinsed on five sequential drops of ultrapure water before staining with 2 % uranyl acetate for 60 s. Excess stain was wicked off and the grid air-dried in the dark. Phage were imaged by transmission electron microscopy (TEM; JEM-1400, JEOL USA, Inc., Peabody, MA, USA) at 80kV.
The ΦScott DNA was isolated by treating 5 ml of filtered supernatant (described above) with 10 µl DNase I to eliminate DNA from lysed bacteria. After DNase I treatment, the enriched phage DNA was isolated by phenol : chloroform : isoamyl alcohol fractionation and ethanol precipitation in the presence of sodium acetate. Genome sequencing was performed using the Illumina HiSeq 1500 from a Nextera library on a single-read flow cell. A total of 518 929 sequences were collected. The quality of the raw sequencing data was assessed using FastQC (v. 0.11.6). Adapters and low-quality sequences were removed using Trim Galore! (v. 0.5.0), removing Illumina adapters and clipping 6 bp from the leading and trailing ends to a minimum length of 36 bp. Post-trimming quality was again assessed via FastQC prior to mapping and assembly. Reads mapping to the host genome, S. paucimobilis , were filtered from the dataset to reduce assembly errors. Reads were mapped using Bowtie 2 (v. 2.3.4.1) [26], leaving 499 904 sequences for bacteriophage genome assembly with an average length of 145 bp and GC content of 56 %.
Reads were assembled using Unicycler (v. 0.4.6) [27], which generated a single contig that was 43 077 bp in length, with a GC content of 57 % and ~1600-fold coverage. The ΦScott genome was annotated using DNAMaster (v. 5.0.2) [28]. Putative coding sequences (CDSs) were identified by both GeneMark and Glimmer to assess coding potential, and CDSs were queried against the National Center for Biotechnology Information (NCBI) nr nucleotide database for annotation. Evidence for gene calls was assessed using additional information from PECAAN and RAST [29], leveraging synteny and similarity to known bacteriophage genomes to assess confidence in gene calls. Predicted terminators and tRNAs were annotated using ARNold [30] and tRNAscan-SE [31], respectively, with eight predicted terminator-like hairpins and no tRNAs. A total of 51 coding sequences were predicted, of which 22 could be assigned functions and 3 were conserved proteins among similar bacteriophages. The sequence is available via the NCBI (BioSample ID SAMN09703972).
To test whether phage-dependent depletion of S. paucimobilis could phenocopy complete loss of S. paucimobilis from the community, static communities were treated with ~20 000 p.f.u. (2 µl of ~10 000 p.f.u. ml−1) of ΦScott either at initiation of the community, 0 h, or 24 h post-inoculation. Biofilm quantification and species composition were determined as described above.
Predation experiments
The predatory bacteria used for this study were Micavibrio aeruginosavorus ARL-13 [32] and Bdellovibrio bacteriovorus strains HD100 (ATCC 15356) and 109J (ATCC 15143). Predatory bacteria were cultured as described previously, using Escherichia coli WM3064 as prey [33–35]. Predator stock lysates were made by co-culturing the predators with prey cells at 30 °C in HEPES buffer (25 mM HEPES supplemented with 3 mM MgCl2 and 2 mM CaCl2). Following 24 or 48 h of incubation, for B. bacteriovorus and M. aeruginosavorus, respectively, cultures were filtered to remove residual prey (harvested predators). To evaluate the susceptibility of drinking water bacterial species to predation, co-cultures were prepared by adding 0.25 ml of HEPES-washed prey cells (∼0.5−1×109 c.f.u. ml−1) to 0.25 ml of harvested predators (∼0.5−1×109 p.f.u. ml−1 for B. bacteriovorus and ∼0.5−1×108 p.f.u. ml−1 for M. aeruginosavorus) and 2.0 ml HEPES medium. Predation was measured by the change in prey population enumerated by dilution plating during a 72 h incubation period and compared to a predator-free control [34, 35].
Statistical analysis and data visualization
All data visualization and statistical analyses were conducted in GraphPad Prism using one-way or two-way analysis of variance (ANOVA) with Tukey, Sidak, Bonferroni, or Dunnett’s post-testing as described in the individual figures. Values of P below 0.05 were considered statistically significant. Data are presented, when possible, with bars representing the mean and experimental replicates are shown as individual overlying data points. Where individual data points are not shown, error bars represent the standard deviation.
Results
The contribution of individual species to community biofilm formation
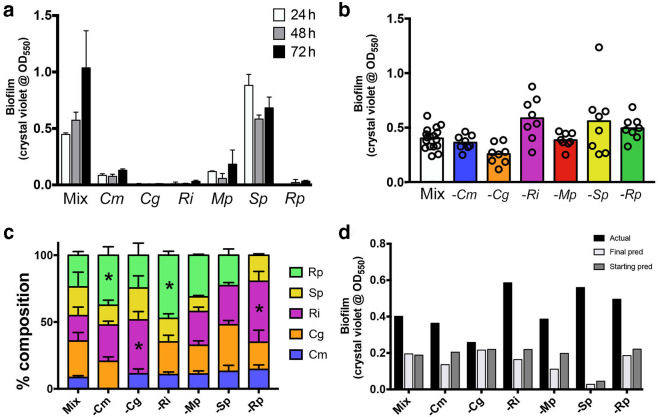
A mixed community comprising all six species readily formed a biofilm that increased over time, while only S. paucimobilis was capable of forming a consistently strong biofilm on its own (Fig. 1a). While S. paucimobilis could form a biofilm alone, removal of this species from the community did not significantly alter biofilm formation (Fig. 1b), and S. paucimobilis made up approximately 20 % of the c.f.u. in the full community (Fig. 1c). Likewise, removal of any single community member did not change biofilm formation in a statistically significant manner (Fig. 1b), although species removal did impact on the proportion of species in the community (Fig. 1c). The biofilm formation of each community was not simply a sum of the individual biofilm-forming potential of its members, whether calculated based on the starting c.f.u. or the c.f.u. at the termination of the assay (Fig. 1d, see the Methods section for specifics of the predicted biofilm calculations). These data suggested that interactions within the community could provide resistance to changes in the initial species compositions to support biofilm formation, a property termed robustness in the field of ecology.
Fig. 1.
Biofilm formation related to species composition in the model drinking water community. (a) Biofilm formation increases over time for the full community; only S. paucimobilis forms substantial biofilms alone. (b) Biofilm formation for the full community and each community lacking one member. While there are trends, none of the groups are significantly different from the full community. (c) Species composition of the five species measured by c.f.u. counts. There is a strong interaction between changes in species compositions and the different community compositions by two-way ANOVA (P<0.0001), even when controlling for the missing members. Statistically significant changes in a species’ % composition compared to the full community was assessed using Dunnett’s post-test after the two-way ANOVA, with changes significant at P<0.01 being noted with an asterisk (*). (d) Comparison of actual biofilm means [from panel (b)] to the predicted biofilm formation using the compositions at the start or end of the experiment with the biofilm formation by each species on its own [panel (a)]. All data are from at least three independent biological experiments, each with at least two technical replicates per experiment. Mix, complete community; Cm, Cupriavidus metallidurans ; Cg, Chryseobacterium gleum ; Ri, Ralstonia insidiosa ; Mp, Methylorubrum populi ; Sp, Sphingomonas paucimobilis ; Rp, Ralstonia pickettii ; pred, prediction. ‘−’ denotes that that community member has been left out.
The impact of removing all combinations of two species on community biofilm formation
The robustness of the community biofilm to loss of individual members (Fig. 1) supported the hypothesis that multiple redundant biofilm formation pathways could occur within this community. To identify these redundant pathways, the biofilm-forming capability of species assemblages that were missing all possible combinations of two species was assessed (Fig. 2). The removal of a few different pairs of species moderately affected community biofilm formation, whereas concurrent removal of C. gleum and S. paucimobilis reduced biofilm formation by ∼95 %, suggesting that these two organisms participated in (at least two) redundant biofilm formation pathways.
Fig. 2.
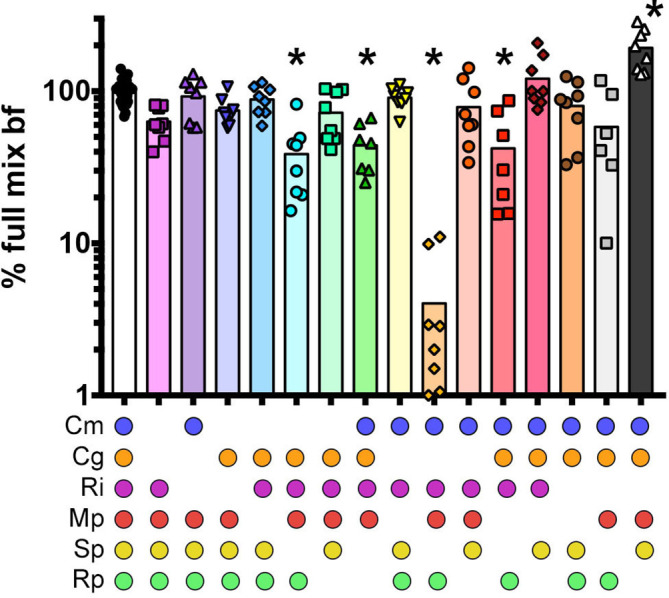
Impact of removing two species on community biofilm formation. Biofilm formation was normalized to the full community, which was set at 100 %. The data are from at least three biological experiments each with at least two technical replicates per experiment. The data were analysed by one-way ANOVA with a Dunnett’s post-test with statistical significance (P<0.01) indicated with an asterisk (*). Cm, C. metallidurans ; Cg, C. gleum ; Ri, R. insidiosa ; Mp, M. populi ; Sp, S. paucimobilis ; Rp, R. pickettii .
Characterization of dual-species biofilm formation
Loss of community biofilm formation was only noted when both S. paucimobilis and C. gleum were not present (Fig. 2), suggesting that biofilm enhancing interactions were occurring that included one or both of these species in conjunction with other species in the community. To characterize these interactions, biofilm formation for every possible two-member interaction was quantified (Fig. 3). Co-growth of S. paucimobilis with C. metallidurans , M. populi , or R. pickettii showed mild stimulation of biofilm formation above the predicted value (sum of individual biofilm formation values proportional to starting cell numbers). However, much more striking was the synergistic biofilm formation shown during co-growth of C. gleum with either R. insidiosa or C. metallidurans .
Fig. 3.
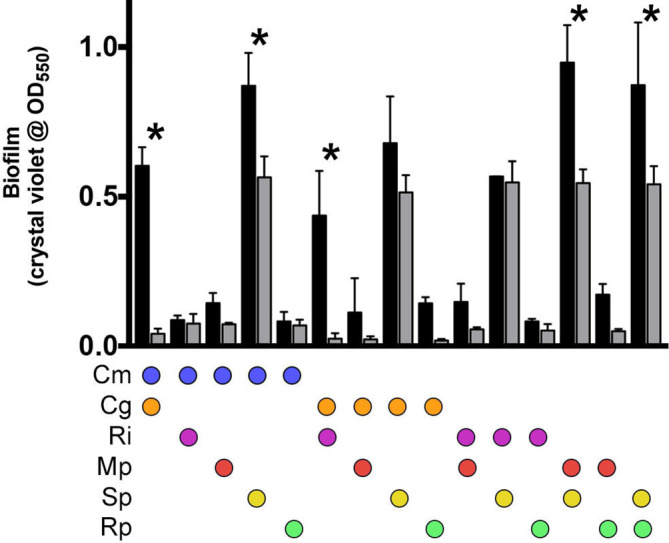
Biofilm formation between all two-species combinations from the six-member community. Black bars represent the means of the experimentally measured biofilm formation between these two members and the grey bars represent the means of the predicted biofilm formation based on individual species biofilm formation starting at equal proportions. The error bars represent the standard deviation. Analysis was performed using two-way ANOVA with Sidak’s post-test to compare actual measures with predicted measures within each two-species mix (*, P<0.01). Cm, C. metallidurans ; Cg, C. gleum ; Ri, R. insidiosa ; Mp, M. populi ; Sp, S. paucimobilis ; Rp, R. pickettii .
Testing the contributions of predicted interactions in community biofilm formation
The data above suggest a model where there are two main drivers of biofilm formation in this community: (i) an S. paucimobilis -dependent one and (ii) another that is dependent on C. gleum interaction with either R. insidiosa or C. metallidurans (Fig. 4a). This model predicts that losing R. insidiosa and C. metallidurans would be equivalent to losing C. gleum . This model was tested by measuring the biofilm formation of an assemblage lacking R. insidiosa and C. metallidurans in the presence or absence of S. paucimobilis . The absence of R. insidiosa and C. metallidurans partially phenocopied the loss of C. gleum (Fig. 4b), in that their loss only substantially reduces biofilm formation in the absence of S. paucimobilis , supporting the interaction model (Fig. 4a). This model is not fully predictive and suggests there are other unknown contributions. In particular, the grey arrow in Fig. 4a represents the observation that when C. metallidurans , R. insidiosa and S. paucimobilis are absent, there is more biofilm formation than is seen with the loss of C. gleum and S. paucimobilis (Fig. 4b). This may represent an interaction with C. gleum by one of the remaining species that is exposed upon removal of the other three bacteria.
Fig. 4.
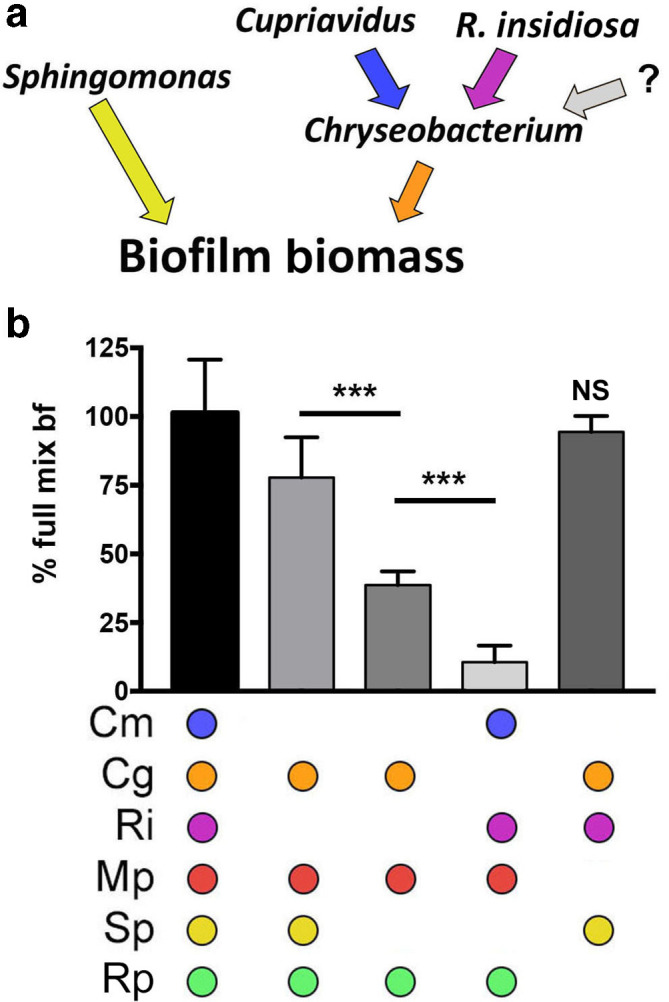
Species interaction model and explicit test for functionality within the community. (a) Model of positive species contributions to biofilm formation based on the data from Figs 1–3. (b) Test of necessity or sufficiency of community members for total biofilm formation as visualized and quantified by crystal violet. The error bars represent the standard deviation. Analysis was performed using one-way ANOVA with Tukey’s post-test comparing all pairs (***, P<0.001) with comparisons noted by bars or reported compared to the full mix. Cm, C. metallidurans ; Cg, C. gleum ; Ri, R. insidiosa ; Mp, M. populi ; Sp, S. paucimobilis ; Rp, R. pickettii ; ns, not significant. ‘?’ denotes an unknown combination of species.
Impact of S. paucimobilis -specific phage on community biofilm formation
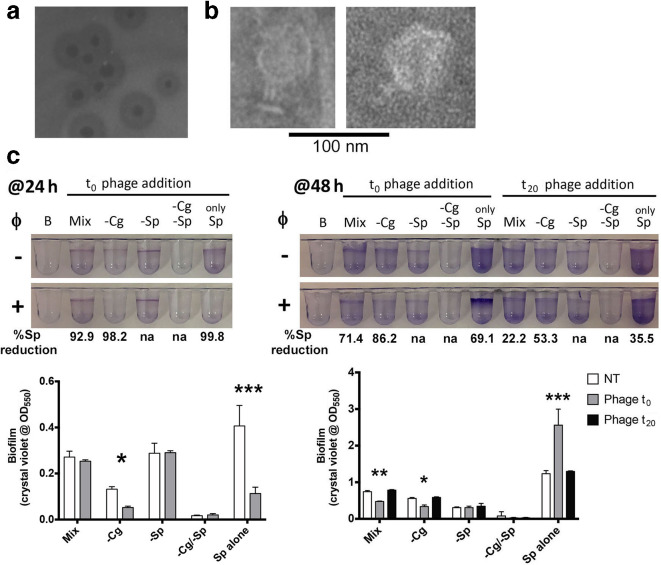
The model community described above is amenable to experiments where one or more members are omitted from the beginning, which is quite different from the depletion of a member after the biofilm has been formed. In an effort to generate tools for this model, we set out to identify reagents that would allow us to deplete a key species in situ. No tested antibiotic allowed specific removal of only one member, thus the first step was isolation of a S. paucimobilis -specific temperate phage from Lake Champlain water (Burlington, VT, USA), which was designated ΦScott.
ΦScott generated plaques with well-defined lysogenic halos (Fig. 5a) and showed a morphology characteristic of a short-tailed phage (Fig. 5b). High-coverage (~1600-fold) short-read sequencing enabled the assembly of a complete 43 kbp genome for ΦScott. The closest matches by gene sequence are in the family P odoviridae . The ΦScott genome is available through he NCBI (GenBank BioSample ID SAMN09703972).
Fig. 5.
Treatment of bacterial biofilms with the S. paucimobilis phage ΦScott. (a) Image of plaque formation on a lawn of S. paucimobilis . Note the wide halos of lysogens. (b) TEM imaging from a high-titre ΦScott preparation. (c) Biofilm formation visualized and quantified with the presence (+) and absence (−) of ΦScott treatment. Reduction in S. paucimobilis c.f.u. reported below crystal violet images normalized to non-treated values. Quantified biofilm formation data analysed using two-way ANOVA with Sidak’s multiple comparison testing for the 24 h time point (testing non-treated vs phage-treated) and with Dunnett’s post-test for the 48 h time point (testing t 0 and t 20 phage, each versus non-treated), with significance denoted by *, P<0.05; **, P<0.01; ***, P<0.001. Mix, complete community; Cg, C. gleum ; Sp, S. paucimobilis ; NT, non-treated.’−’ denotes that that community member has been left out.
A high-titre ΦScott preparation was used as a tool to deplete S. paucimobilis from the mixed-species assemblages. ΦScott readily lysed >99.99 % of S. paucimobilis in shaking planktonic culture (data not shown) and ~99.8 % of S. paucimobilis in static culture conditions (Fig. 5c), but it was less efficient in the mixed assemblage, showing a ~93 % reduction when added concurrently with bacterial inoculation (Fig. 5c). As seen for S. paucimobilis removal experiments (Figs 1 and 2), the application of ΦScott at the start of the experiment (t 0) led to reduced biofilm formation in S. paucimobilis -alone culture and phenocopied he absence of S. paucimobilis at 24 h of biofilm formation for mixed cultures. However, treatment of pre-existing biofilms resulted in no substantial biofilm removal, which correlated with poor lysis rates (20–50 % c.f.u. reduction) for S. paucimobilis in these communities (Fig. 5c). Interestingly, the S. paucimobilis lysogens that survive the initial lysis in the S. paucimobilis -alone condition eventually generate more biofilm biomass than is seen in the absence of ΦScott (Fig. 5c).
The poor lysis of S. paucimobilis and failure to impact on biofilm biomass in preformed biofilms leads to two possible interpretations: (i) the amount of killing observed in these conditions (20–50 %) is not sufficient to perturb the S. paucimobilis -dependent pathway of the community biofilm, or (ii) that amount of killing is sufficient, but S. paucimobilis is only required for the early stages of the community biofilm assembly and is dispensable at later time points.
Characterization of predatory bacteria predation on community members
Specific phage treatment did not demonstrate the capacity to allow species removal from a preformed biofilm, which led to testing the susceptibility of the community members to three different predatory bacteria. When co-cultured in suspension, there was no measurable predation on M. populi and R. insidiosa, as well as the two critical community members S. paucimobilis and C. gleum (Fig. 6). Similar results were seen when predation assays were conducted on established biofilms of S. paucimobilis , and C. gleum (data not shown). However, both Bdellovibrio strains were able to prey on C. metallidurans as well as R. pickettii . Thus, as with phage treatment, the removal of a single species from pre-formed biofilms could not be achieved with the tested predatory bacteria.
Fig. 6.
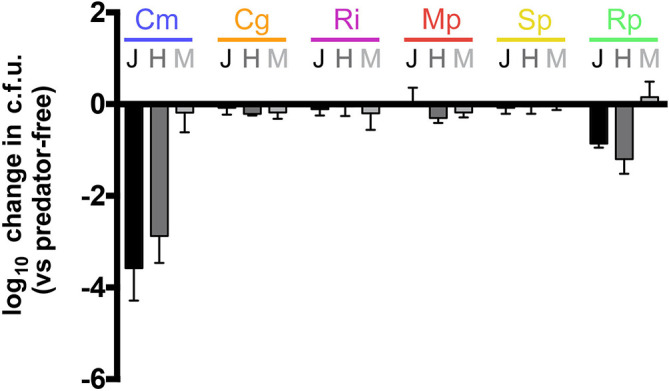
Quantification of bacterial predation in planktonic culture by predatory bacteria. Co-cultures were prepared by adding prey cells to harvested predator cells. Values represent maximal population change (log10) of each species, compared to a predator-free control, following predation. Each experiment was conducted at least three times and the error bars represent the standard deviation. Cm, C. metallidurans ; Cg, C. gleum ; Ri, R. insidiosa ; Mp, M. populi ; Sp, S. paucimobilis ; Rp, R. pickettii ; J, Bdellovibrio bacteriovorous 109J; H, Bdellovibrio bacteriovorous HD100; M, Micavibrio aeruginosavorus.
Discussion
In this study we sought to understand the species interactions that drive biofilm formation in a model drinking water community. The species contributing to biofilm formation in this six-species community were determined, demonstrating that S. paucimobilis and C. gleum were necessary (no biofilm if both were removed), but either was dispensable if one was present. The contribution of C. gleum required either R. insidiosa or C. metallidurans, but not both (Fig. 4), and dual-species interactions suggest that both R. insidiosa – C. gleum and C. metallidurans – C. gleum interactions are positively synergistic for biofilm formation (Fig. 3). Combined with species removal data, these findings suggest that C. gleum -dependent synergistic biofilm formation is a critical portion of biofilm formation and its robustness to alternative community membership, supporting an important role for positive microbial interactions in this community [36]. Importantly, at this point, spatial distribution was not assessed, although we assume that community composition would alter organization within the biofilms [37]. Additionally, the choice of coexisting species (from the same niche) likely enhanced our capacity to identify interspecies biofilm interactions [38]. The identification of interactions and the relative stability and reproducibility of the population proportions is not necessarily surprising, as many others have demonstrated in recreated natural or fully synthetic communities that a stable, or at least predictably fluctuating, community is arrived at very quickly [39, 40]. To bypass the large differences in growth rate between some members of this community and to drive the community to nutrient limitation and predicted stability rapidly, we chose to use very high inocula at equal initial optical densities. The final concentration of cells in each static system was approximately 1/3 of the maximal carrying capacity of the system.
Single-species removal in this system did not alter biofilm formation in a statistically significant way, but there was a trend to decreased biofilm with loss of C. gleum (Fig. 1b). Interestingly, the biofilm formation of the community missing C. gleum was closest to that predicted by the biofilm formation capacities of each remaining species (predicted values, Fig. 1d). One caveat to the predicted values is an inability to accurately assess c.f.u. for Methylorubrum (formerly Methylobacterium [23]) in this system due to poor correlation of plating with biomass for this species (inability to disrupt clumps). For these cases, it was assumed that 10 % of the c.f.u. in the system were Methylorubrum (0 % in the −Mp group). We are working on molecular methods to address this shortcoming, but have found incomplete lysis of biofilm-grown cells even under very harsh conditions, which makes the molecular analysis less straightforward. One final observation of the single-species removals is that R. insidiosa removal leads to R. pickettii expansion and vice versa, but while R. pickettii seems to take over the growth niche of R. insidiosa , it does not appear to participate in community biofilm formation. Specifically, R. pickettii cannot substitute for R. insidiosa in the C. gleum synergistic interaction.
When each combination of two species was left out of the starting inoculum, removal of C. gleum and S. paucimobilis was the only combination to result in nearly complete loss of biofilm formation. Moderate reduction in biofilm formation was seen for the absence of C. metallidurans / S. paucimobilis , R. pickettii / S. paucimobilis and M. populi / S. paucimobilis (all showing a reduction of between 40 and 60%). Conversely, removal of R. insidiosa / R. pickettii led to a statistically significant increase in biofilm formation, although this may be of limited biological importance, being roughly a 10 % increase. S. paucimobilis was absent in all instances where two-species removal led to reduced biofilm formation, suggesting that the strong biofilm formation capacity of S. paucimobilis (Fig. 1a) is an important contributor to the community biofilm when other perturbations are present, even though the absence of S. paucimobilis alone was not sufficient to cause reduced biofilm formation (Fig. 1b). An interesting question regarding these interactions, and the dual-species interactions described below, is how specific they are for the individual strains used in this model. For instance, we do not know if other C. gleum and S. paucimobilis strains could take the place of these ISS isolates in this model system. It would be interesting to determine the extent of strain impact, and also use their absence to identify other drinking water community members that could serve a similar function.
The literature covering dual-species interactions is extensive, demonstrating every variety of interaction, from wholly antagonistic to mutually dependent, with many shown to be synergistic [41]. One goal of this study was to understand the role of the pair-wise interactions driving biofilm formation. In addition to moderately synergistic interactions between C. metallidurans / S. paucimobilis , M. populi / S. paucimobilis and R. pickettii / S. paucimobilis , we identified two very strong synergistic interactions that were dependent on C. gleum , in which C. gleum / R. insidiosa and C. gleum / C. metallidurans mixes form much greater biofilm than predicted from either species alone. The role of dual species synergy has been noted in many other communities. From a functional viewpoint, the examples of synergistic biofilm in this model could be the result of competition-induced biofilm formation [42], niche construction by one member for the other [43] that may even include synergistic syntrophy [36], or contact-dependent coaggregation [44, 45]. Many dual-species biofilm increases are dependent on coaggregation, including bacteria from drinking water communities [46, 47]. Simoes and colleagues [46, 47] demonstrated inter-genus coaggregation interactions, particularly driven by an Acinetobacter species, which they propose plays a keystone role similar to that of Fusobacterium and Prevotella in dental biofilms. We have specifically tested coaggregation between our synergistic biofilm-forming species and detected none by eye or microscopically using the methods of Simoes (data not shown). Thus, while it is possible our synergistic species bind to each other within biofilms, they appear not to express the specific carbohydrate/lectin pairs necessary for rapid coaggregation.
One of the members in our community that can form synergistic biofilms with C. gleum is R. insidiosa . This bacterium has been implicated as a ‘bridge’ species, increasing biofilm formation and promoting pathogen integration into industrial and drinking water biofilms [48–51]. In one case, R. insidiosa promoted Listeria monocytogenes biofilm formation, even though it formed spatially segregated clusters within the dual-species biofilm [48]. In this study, R. insidiosa specifically interacts with C. gleum to promote biofilm formation, and C. metallidurans functions in a similar fashion. It remains to be seen whether C. gleum is playing a partial bridging role or perhaps C. gleum can stimulate R. insidiosa and/or C. gleum to function as bridges in the community. Biofilm formation synergy between Chryseobacterium sp. and Pseudomonas putida , isolated from agricultural soil, was previously observed [52], although the mechanism by which Chryseobacterium promotes such synergy is currently unknown.
To better understand the stability of biofilm formation to perturbations on an established community, a specific perturbation was tested to deplete a key community member in situ, starting with a S. paucimobilis phage. However, ΦScott was not effective at substantially depleting S. paucimobilis once a biofilm had formed, even when Sphingomonas was the only organism present (Fig. 5). Therefore, while the addition of ΦScott at t 0 phenocopied he removal of S. paucimobilis from the community, phage addition at 24 h did not result in either biofilm reduction in the absence of C. gleum , or in substantial depletion of S. paucimobilis c.f.u. Additionally, the S. paucimobilis that regrew after t 0 phage treatment were nearly all lysogens, while many of those isolated at 48 h after the 24 h phage treatment were not lysogens as assessed by their susceptibility to the phage during dilution (i.e. low dilution wells showed full lysis due to remaining phage under these conditions, something not seen when phage was added at t 0 and cells sampled 24 or 48 h later). We have not further characterized the lysogenic phenotypes or phage–bacteria interactions for ΦScott and S. paucimobilis . While predatory bacteria have generally been shown to be adept at predation within biofilms [53, 54], none of the predatory bacteria were able to effectively prey specifically on a biofilm-critical member of this community (Fig. 6). Thus, the biological or chemical tools necessary to specifically test resilience in this community after biofilm formation are not currently available.
In this study, biofilm formation by a six-member potable water bacterial community was examined and shown to be robust to the absence of individual members. Manipulation of community membership at inoculation was used to determine interactions between members that impact on community biofilm formation. These studies resulted in a simplified biofilm promoting network (Fig. 4) that describes the two pathways that contribute to the robustness of community biofilm formation, although when the primary drivers of biofilm are removed, there does appear to be some compensatory activity from the remaining members (Fig. 4, grey arrow). This community differs from other model drinking water assemblages and the constituent species appear to interact in a different manner. Therefore, this model is complementary to existing models, and we propose its utility for dissection of the molecular interactions within drinking water biofilm communities. It is important to highlight the limitations of this study. This study used a single, simultaneous inoculation into a static system with no nutrient replenishment. Thus, flow effects, the relative contributions of biofilm and planktonic members within the static system, and the effects of community member priority (order of addition) on the interactions were not examined. Many of these issues have been carefully studied in other model systems and this drinking water community can function as an independent model to examine these processes.
Funding information
This study was supported by NASA cooperative agreement NNX16ZHA001C to M. J. W., T32 HL076122 fellowship support of G. G. W., graduate student rotation support for A. T. and A. F. by the University of Vermont Cell, Molecular, and Biomedical Sciences (CMB) Program, and an undergraduate summer fellowship to M. B. supported by the Vermont Genetics Network P20 GM103449. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Acknowledgements
We’d like to thank C. Mark Ott, PhD from the NASA Johnson Space Center for the bacterial strains in this study. Electron microscopy was performed at the Microscopy Imaging Center at the University of Vermont and sequencing of the bacteriophage genome was conducted at the Vermont Integrative Genomic Resource at the University of Vermont.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Edited by: D. Grainger and S. P Diggle
References
- 1.Tucker CM, Fukami T. Environmental variability counteracts priority effects to facilitate species coexistence: evidence from nectar microbes. P Roy Soc B-Biol Sci. 1778;2014:281. doi: 10.1098/rspb.2013.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mark Welch JL, Hasegawa Y, McNulty NP, Gordon JI, Borisy GG. Spatial organization of a model 15-member human gut microbiota established in gnotobiotic mice. Proc Natl Acad Sci U S A. 2017;114:E9105–E9114. doi: 10.1073/pnas.1711596114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Limoli DH, Whitfield GB, Kitao T, Ivey ML, Davis MR, et al. Pseudomonas aeruginosa Alginate Overproduction Promotes Coexistence with Staphylococcus aureus in a Model of Cystic Fibrosis Respiratory Infection. MBio. 2017;8:e00186-17. doi: 10.1128/mBio.00186-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niu B, Paulson JN, Zheng X, Kolter R. Simplified and representative bacterial community of maize roots. Proc Natl Acad Sci USA. 2017;114:E2450–E2459. doi: 10.1073/pnas.1616148114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolfe BE, Button JE, Santarelli M, Dutton RJ. Cheese RIND communities provide tractable systems for in situ and in vitro studies of microbial diversity. Cell. 2014;158:422–433. doi: 10.1016/j.cell.2014.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kolenbrander PE, Palmer RJ, Rickard AH, Jakubovics NS, Chalmers NI, et al. Bacterial interactions and successions during plaque development. Periodontol 2000. 2006;42:47–79. doi: 10.1111/j.1600-0757.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 7.Kviatkovski I, Minz D. A member of the Rhodobacteraceae promotes initial biofilm formation via the secretion of extracellular factor(s) Aquat Microb Ecol. 2015;75:155–167. doi: 10.3354/ame01754. [DOI] [Google Scholar]
- 8.Vandeplassche E, Coenye T, Crabbé A. Developing selective media for quantification of multispecies biofilms following antibiotic treatment. PLoS One. 2017;12:e0187540. doi: 10.1371/journal.pone.0187540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer RJ, Shah N, Valm A, Paster B, Dewhirst F, et al. Interbacterial adhesion networks within early oral biofilms of single human hosts. Appl Environ Microbiol. 2017;83 doi: 10.1128/AEM.00407-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burmølle M, Ren D, Bjarnsholt T, Sørensen SJ. Interactions in multispecies biofilms: do they actually matter? Trends Microbiol. 2014;22:84–91. doi: 10.1016/j.tim.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 11.Crabbé A, Jensen Peter Østrup, Bjarnsholt T, Coenye T. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol. 2019 doi: 10.1016/j.tim.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 12.Willsey GG, Wargo MJ. Extracellular lipase and protease production from a model drinking water bacterial community is functionally robust to absence of individual members. PLoS One. 2015;10:e0143617. doi: 10.1371/journal.pone.0143617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berry D, Xi C, Raskin L. Microbial ecology of drinking water distribution systems. Curr Opin Biotechnol. 2006;17:297–302. doi: 10.1016/j.copbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Novikova N, De Boever P, Poddubko S, Deshevaya E, Polikarpov N, et al. Survey of environmental biocontamination on board the International space Station. Res Microbiol. 2006;157:5–12. doi: 10.1016/j.resmic.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 15.La Duc MT, Sumner R, Pierson D, Venkat P, Venkateswaran K. Evidence of pathogenic microbes in the International space Station drinking water: reason for concern? Habitation. 2004;10:39–48. doi: 10.3727/154296604774808883. [DOI] [PubMed] [Google Scholar]
- 16.Castro VA, Thrasher AN, Healy M, Ott CM, Pierson DL. Microbial characterization during the early habitation of the International space Station. Microb Ecol. 2004;47:119–126. doi: 10.1007/s00248-003-1030-y. [DOI] [PubMed] [Google Scholar]
- 17.Song B, Leff LG. Identification and characterization of bacterial isolates from the miR space Station. Microbiol Res. 2005;160:111–117. doi: 10.1016/j.micres.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 18.Roman MC, Minton-Summers S. Assessment of biofilm formation in the International space Station water recovery and management system. Life Support Biosph Sci. 1998;5:45–51. [PubMed] [Google Scholar]
- 19.Falkinham JO, Hilborn ED, Arduino MJ, Pruden A, Edwards MA. Epidemiology and ecology of opportunistic premise plumbing pathogens: Legionella pneumophila, Mycobacterium avium, and Pseudomonas aeruginosa . Environ Health Perspect. 2015;123:749–758. doi: 10.1289/ehp.1408692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haig S-J, Kotlarz N, LiPuma JJ, Raskin L. A High-Throughput Approach for Identification of Nontuberculous Mycobacteria in Drinking Water Reveals Relationship between Water Age and Mycobacterium avium . MBio. 2018;9:e02354-17. doi: 10.1128/mBio.02354-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hardalo C, Edberg SC. Pseudomonas aeruginosa: assessment of risk from drinking water. Crit Rev Microbiol. 1997;23:47–75. doi: 10.3109/10408419709115130. [DOI] [PubMed] [Google Scholar]
- 22.van der Wielen PWJJ, van der Kooij D. Nontuberculous mycobacteria, fungi, and opportunistic pathogens in unchlorinated drinking water in the Netherlands. Appl Environ Microbiol. 2013;79:825–834. doi: 10.1128/AEM.02748-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Green PN, Ardley JK. Review of the genus Methylobacterium and closely related organisms: a proposal that some Methylobacterium species be reclassified into a new genus, Methylorubrum gen. nov. Int J Syst Evol Microbiol. 2018;68:2727–2748. doi: 10.1099/ijsem.0.002856. [DOI] [PubMed] [Google Scholar]
- 24.Labauve AE, Wargo MJ. Growth and laboratory maintenance of Pseudomonas aeruginosa. Curr Protoc Microbiol. 2012;1 doi: 10.1002/9780471729259.mc06e01s25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merritt JH, Kadori DE, O'Toole GA. Growing and Analyzing Static Biofilms. Current Protocols in Microbiology . Hoboken, NJ: J. Wiley & Sons. 2005:1–17. doi: 10.1002/9780471729259.mc01b01s00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pope WH, Jacobs-Sera D. Annotation of bacteriophage genome sequences using DNA master: an overview. Methods in molecular biology (Clifton, NJ. 2018;1681:217–229. doi: 10.1007/978-1-4939-7343-9_16. [DOI] [PubMed] [Google Scholar]
- 29.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, et al. The RAST server: rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naville M, Ghuillot-Gaudeffroy A, Marchais A, Gautheret D. Arnold: a web tool for the prediction of rho-independent transcription terminators. RNA Biol. 2011;8:11–13. doi: 10.4161/rna.8.1.13346. [DOI] [PubMed] [Google Scholar]
- 31.Lowe TM, Chan PP. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:W54–W57. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Z, Kadouri DE, Wu M. Genomic insights into an obligate epibiotic bacterial predator: Micavibrio aeruginosavorus ARL-13. BMC Genomics. 2011;12:453. doi: 10.1186/1471-2164-12-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dashiff A, Junka RA, Libera M, Kadouri DE. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol. 2011;110:431–444. doi: 10.1111/j.1365-2672.2010.04900.x. [DOI] [PubMed] [Google Scholar]
- 34.Kadouri DE, To K, Shanks RMQ, Doi Y. Predatory bacteria: a potential ally against multidrug-resistant gram-negative pathogens. PLoS One. 2013;8:e63397. doi: 10.1371/journal.pone.0063397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanks RMQ, Davra VR, Romanowski EG, Brothers KM, Stella NA, et al. An eye to a kill: using predatory bacteria to control gram-negative pathogens associated with ocular infections. PLoS One. 2013;8:e66723. doi: 10.1371/journal.pone.0066723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schink B. Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek. 2002;81:257–261. doi: 10.1023/A:1020579004534. [DOI] [PubMed] [Google Scholar]
- 37.Liu W, Røder HL, Madsen JS, Bjarnsholt T, Sørensen SJ, et al. Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front Microbiol. 2016;7:1366. doi: 10.3389/fmicb.2016.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Madsen JS, Røder HL, Russel J, Sørensen H, Burmølle M, et al. Coexistence facilitates interspecific biofilm formation in complex microbial communities. Environ Microbiol. 2016;18:2565–2574. doi: 10.1111/1462-2920.13335. [DOI] [PubMed] [Google Scholar]
- 39.Vanwonterghem I, Jensen PD, Dennis PG, Hugenholtz P, Rabaey K, et al. Deterministic processes guide long-term synchronised population dynamics in replicate anaerobic digesters. Isme J. 2014;8:2015–2028. doi: 10.1038/ismej.2014.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stenuit B, Agathos SN. Deciphering microbial community robustness through synthetic ecology and molecular systems synecology. Curr Opin Biotechnol. 2015;33:305–317. doi: 10.1016/j.copbio.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Ren D, Madsen JS, Sørensen SJ, Burmølle M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. Isme J. 2015;9:81–89. doi: 10.1038/ismej.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliveira NM, Oliveria NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, et al. Biofilm formation as a response to ecological competition. PLoS Biol. 2015;13:e1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Callahan BJ, Fukami T, Fisher DS. Rapid evolution of adaptive niche construction in experimental microbial populations. Evolution. 2014;68:3307–3316. doi: 10.1111/evo.12512. [DOI] [PubMed] [Google Scholar]
- 44.Rickard AH, Gilbert P, High NJ, Kolenbrander PE, Handley PS. Bacterial coaggregation: an integral process in the development of multi-species biofilms. Trends Microbiol. 2003;11:94–100. doi: 10.1016/S0966-842X(02)00034-3. [DOI] [PubMed] [Google Scholar]
- 45.Buswell CM, Herlihy YM, Marsh PD, Keevil CW, Leach SA. Coaggregation amongst aquatic biofilm bacteria. J Appl Microbiol. 1997;83:477–484. doi: 10.1046/j.1365-2672.1997.00260.x. [DOI] [Google Scholar]
- 46.Simões LC, Simões M, Vieira MJ. Intergeneric coaggregation among drinking water bacteria: evidence of a role for Acinetobacter calcoaceticus as a bridging bacterium. Appl Environ Microbiol. 2008;74:1259–1263. doi: 10.1128/AEM.01747-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simões LC, Simões M, Vieira MJ. Biofilm interactions between distinct bacterial genera isolated from drinking water. Appl Environ Microbiol. 2007;73:6192–6200. doi: 10.1128/AEM.00837-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guo A, Xu Y, Mowery J, Nagy A, Bauchan G, et al. Ralstonia insidiosa induces cell aggregation of Listeria monocytogenes. Food Control. 2016;67:303–309. doi: 10.1016/j.foodcont.2016.03.006. [DOI] [Google Scholar]
- 49.Liu NT, Nou X, Lefcourt AM, Shelton DR, Lo YM. Dual-species biofilm formation by Escherichia coli O157:H7 and environmental bacteria isolated from fresh-cut processing facilities. Int J Food Microbiol. 2014;171:15–20. doi: 10.1016/j.ijfoodmicro.2013.11.007. [DOI] [PubMed] [Google Scholar]
- 50.Liu NT, Nou X, Bauchan GR, Murphy C, Lefcourt AM, et al. Effects of environmental parameters on the dual-species biofilms formed by Escherichia coli O157:H7 and Ralstonia insidiosa, a strong biofilm producer isolated from a fresh-cut produce processing plant. J Food Prot. 2015;78:121–127. doi: 10.4315/0362-028X.JFP-14-302. [DOI] [PubMed] [Google Scholar]
- 51.Liu NT, Bauchan GR, Francoeur CB, Shelton DR, Lo YM, et al. Ralstonia insidiosa serves as bridges in biofilm formation by foodborne pathogens Listeria monocytogenes, Salmonella enterica, and enterohemorrhagic Escherichia coli. Food Control. 2016;65:14–20. doi: 10.1016/j.foodcont.2016.01.004. [DOI] [Google Scholar]
- 52.Burmølle M, Hansen LH, Sørensen SJ. Establishment and early succession of a multispecies biofilm composed of soil bacteria. Microb Ecol. 2007;54:352–362. doi: 10.1007/s00248-007-9222-5. [DOI] [PubMed] [Google Scholar]
- 53.Kadouri D, O'Toole GA. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol. 2005;71:4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kadouri D, Venzon NC, O'Toole GA. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol. 2007;73:605–614. doi: 10.1128/AEM.01893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]