Abstract
The role of commensal microbiota in enteric viral infections has been explored extensively, but the interaction between human gut microbiota (HGM) and human norovirus (HuNoV) is poorly understood. In this study, we established an HGM-Transplanted gnotobiotic (Gn) pig model of HuNoV infection and disease, using an infant stool as HGM transplant and a HuNoV GII.4/2006b strain for virus inoculation. Compared to germ-free Gn pigs, HuNoV inoculation in HGMT Gn pigs resulted in increased HuNoV shedding, characterized by significantly higher shedding titres on post inoculation day (PID) 3, 4, 6, 8 and 9, and significantly longer mean duration of virus shedding. In addition, virus titres were significantly higher in duodenum and distal ileum of HGMT Gn pigs on PID10, while comparable and transient HuNoV viremia was detected in both groups. 16S rRNA gene sequencing demonstrated that HuNoV infection dramatically altered intestinal microbiota in HGMT Gn pigs at the phylum (Proteobacteria, Firmicutes and Bacteroidetes) and genus ( Enterococcus , Bifidobacterium , Clostridium , Ruminococcus , Anaerococcus , Bacteroides and Lactobacillus ) levels. In summary, enhanced GII.4 HuNoV infection was observed in the presence of HGM, and host microbiota was susceptible to disruption upon HuNoV infection.
Keywords: Human norovirus, diarrhea, human gut microbiota, gnotobiotic pig, faecal microbiota transplantation
Introduction
Human noroviruses (HuNoVs), non-enveloped RNA viruses with a positive-sense single-stranded genome in the Caliciviridae family, are the leading cause of epidemic acute gastroenteritis around the world [1]. Annually, HuNoV infections cause 685 million illnesses and over 212 000 deaths worldwide, in which 30 % of illnesses and 25 % of deaths are in children under 5 years old [2]. HuNoV gastroenteritis has an economic cost of ~$4 billion in direct healthcare costs and ~$60 billion in loss of productivity globally [3]. Despite the tremendous burden of disease and financial cost, no vaccines or antivirals are currently available to prevent or control HuNoV infections, primarily resulting from the long absence of a readily reproducible cultivation system and a suitable small animal model [4].
The ability of commensal microbiota to enhance enteric viral infections was first demonstrated by two landmark studies using poliovirus, reovirus and mouse mammary tumour virus [5, 6]. The microbiota-driven enhancement of murine rotavirus infection was evidenced by the reduced rotavirus infectivity and diarrhea in antibiotic-treated suckling mice [7]. Similarly, antibiotic treatment reduced the acute murine norovirus (MNV) infection and prevented the persistent MNV infection in mice [8, 9], and the persistent infection could be restored by microbial colonization [9]. However, it is also known that gut microbiota can serve as a shield against pathogenic micro-organisms due to their colonization resistance and immunomodulatory functions [10]. The existence of contradictory reports suggests that the microbiota’s role in viral infections varies in regard to the individual virus and host. For example, after depletion of the gut microbiota with antibiotics, mice were more susceptible and vulnerable to multiple flaviviruses such as West Nile, Dengue and Zika virus [11].
While commensal bacteria has been found to promote MNV infections in mice [8, 9, 12], the effects of human gut microbiota (HGM) on HuNoV infectivity remain elusive. Disruption of HGM due to HuNoV infection was observed in human patients. Seven out of thirty-eight HuNoV-infected patients showed significantly decreased abundance of Bacteroidetes and increased abundance of Proteobacteria [13]. In a study analysing saliva and stool samples obtained from healthy human volunteers, lower salivary anti-HuNoV IgA titres, which is an indicator of previous exposure to HuNoV, were correlated with higher abundance of certain bacterial groups such as Ruminococcus spp. and Faecalibacterium, demonstrating a potential link between the susceptibility to HuNoV infection and HGM composition [14]. Human B lymphocytes (BJAB cell line) supported moderate in vitro HuNoV replication using an unfiltered HuNoV-positive stool sample as inoculum, whereas the filtered inoculum failed to establish HuNoV infection [8]. This cultivation system has been replicated and applied for the evaluation of a viral polymerase inhibitor [15], suggesting a stimulatory role of commensal bacteria in HuNoV infection of target cells. In efforts to dissect such microbiota-dependent infection, synthetic histo-blood group antigen (HBGA) or HBGA-expressing bacteria such as Enterobacter cloacae was identified as the helper for HuNoV infection of B cells [8]. However, our previous study using a gnotobiotic (Gn) pig model showed that E. cloacae inhibited HuNoV infection in vivo, and viral infection of B cells was not observed with or without the presence of E. cloacae [16]. In addition, bacteria were not required for efficient viral infection of human intestinal enteroids, which have been established as a novel HuNoV cultivation system for multiple GII.3 and GII.4 strains [17, 18]. These conflicting results raise new questions about the role and importance of HGM on HuNoV infection.
Neonatal Gn pigs share high similarity of gastrointestinal physiology and immune system with infants and young children, and have been widely used for the studies of pathogenesis, host immunity, and the role of microbiome/bacteria in enteric virus infections [19]. The evaluations of vaccine candidates and therapeutic agents against enteric viruses in Gn pigs have high translational implications [20–24]. In addition, Gn pigs recapitulate the hallmark features of HuNoV biology, such as natural oral route of infection, faecal viral shedding, transient viremia, and increased and prolonged infection in immunodeficient host [19, 25]. More importantly, the germ-free environment is ideal for the reconstruction of HGM in animal models. Microbiome analysis revealed that the HGM-transplanted (HGMT) Gn pigs were colonized by microbiota similar to that of the original infant donors, indicating transplant success in previous studies [26, 27]. The HGMT Gn pig model has enabled research into the effects of enteric dysbiosis and protein malnutrition on rotavirus vaccine efficacy [28, 29].
In this study, with the aim of illuminating the complex interactions between HGM and HuNoV in vivo, we first established an HGMT Gn pig model of HuNoV infection and disease. Subsequently, HuNoV-induced disease, virus shedding in faeces, and virus distribution in tissues were evaluated and compared between HGMT Gn pigs and control groups. Finally, the composition of established HGM in Gn pigs with and without HuNoV infection was analysed, respectively.
Results
HGMT Gn pig model of HuNoV infection and disease
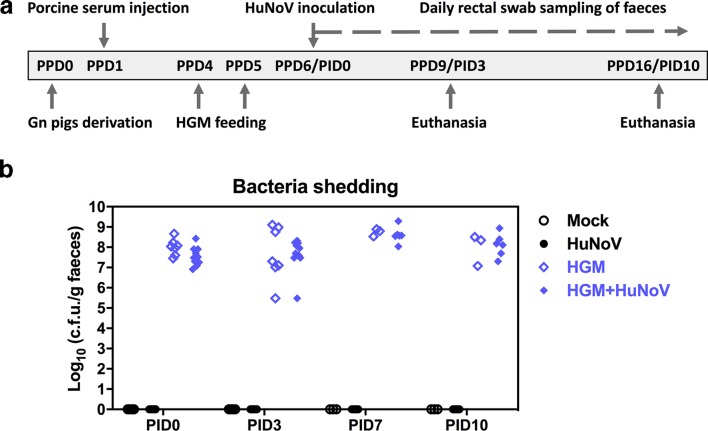
The infant stool used for transplantation in Gn pigs in this study was determined with a representative and healthy HGM. To establish and validate the HGMT Gn pig model, we tested HuNoV infection and/or HGM colonization using four treatment groups: (i) mock (n=5), naïve Gn pigs; (ii) HuNoV (n=19), Gn pigs were inoculated with HuNoV (GII.4/2006b strain); (iii) HGM (n=7), Gn pigs were colonized with HGM only; (iv) HGM+HuNoV (n=11), Gn pigs were pre-colonized with HGM prior to HuNoV inoculation (Fig. 1a). All pigs received intraperitoneal porcine serum injections on post-partum day (PPD) 1 and were euthanized on post inoculation day (PID) 3 or 10. To confirm the colonization of HGM in Gn pigs, faecal bacteria shedding was monitored after HGM feeding. Bacteria shedding was detected in all pigs in the HGM group and HGM+HuNoV group, whereas pigs in the mock group and HuNoV group remained sterile during the entire study (Fig. 1b).
Fig. 1.
Experimental design and faecal bacteria shedding. (a) Schematic representation of Gn pig study. HGM, human gut microbiota; PPD, post-partum day; PID, post inoculation day. (b) HGM colonization in Gn pigs. Concentrations of culturable aerobic bacteria were measured in serial dilution of pig faeces and enumeration of colony-forming unit (c.f.u.) grown on lysogeny broth (LB) media agar plates. Data were combined from four independent experiments and presented as individual animal data points. Sample sizes are shown in Table 1.
Increased HuNoV shedding and diarrhea in HGMT Gn pigs
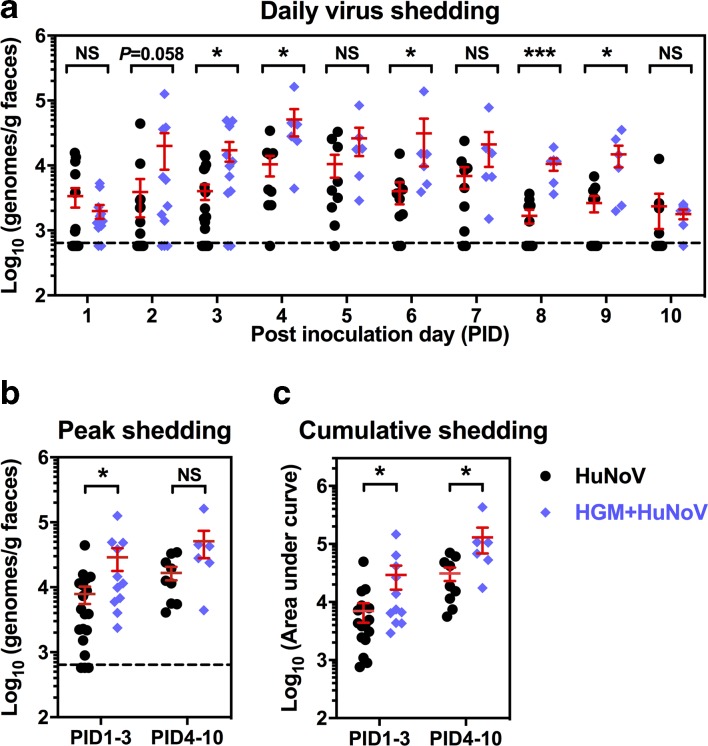
As a characteristically self-limiting enteric pathogen, HuNoV shedding in Gn pigs peaked on PID4 with or without HGM colonization (Fig. 2a). Daily faecal virus shedding increased in the HGM+HuNoV group, and statistical significance was observed on PID3, 4, 6, 8 and 9 (Fig. 2a). Compared to the HuNoV group, the peak shedding in the HGM+HuNoV group was significantly higher on PID1–3 (Fig. 2b), and the cumulative shedding in the HGM+HuNoV group was significantly higher on PID1–3 and PID4–10 (Fig. 2c). In addition, HGM+HuNoV pigs had a significantly longer mean duration of virus shedding on PID1–3 and PID4–10 (2.4 versus 1.5 days and 6.8 versus 4.9 days, respectively) (Table 1). Taken together, these data demonstrated higher HuNoV shedding in HGMT pigs, suggesting that the presence of HGM promoted HuNoV infectivity in Gn pigs.
Fig. 2.
Increased faecal HuNoV shedding in HGMT pigs. (a) Daily virus shedding was measured from PID1 to PID10 by quantitative reverse transcription (qRT) PCR to quantify HuNoV genomes in faeces. (b) Peak shedding titres during PID1 to PID3 and PID4 to PID10 in individual pigs were present. (c) Individual pigs’ cumulative shedding was shown as the area under curve based on daily virus shedding in (a). Sample sizes are indicated in Table 1. Dashed line shows the limit of detection. Data were combined from four independent experiments and presented as individual animal data points with mean±sem. Statistical significance was determined by Mann–Whitney test. NS, not significant, *P<0.05, **P<0.01.
Table 1.
Summary of clinical sign and virus shedding in Gn pigsa
|
Group |
Time |
n |
Diarrheab |
Virus shedding |
||
|---|---|---|---|---|---|---|
|
Pigs with diarrhea (%)* |
Mean duration days (semc)** |
Pigs with virus shedding (%)* |
Mean duration days (sem)** |
|||
|
Mock |
PID1-3 |
5 |
0 |
0 |
0 |
0 |
|
HuNoV |
19 |
11 (58 %)A |
0.9 (0.2)A |
16 (84 %) |
1.5 (0.2)A |
|
|
HGM |
7 |
0 |
0 |
0 |
0 |
|
|
HuNoV+HGM |
11 |
1 (9 %)B |
0.1 (0.1)B |
11 (100 %) |
2.4 (0.2)B |
|
|
Mock |
PID4-10 |
3 |
0 |
0 |
0 |
0 |
|
HuNoV |
9 |
7 (78 %) |
2.0 (0.5)A |
9 (100 %) |
4.9 (0.7)A |
|
|
HGM |
3 |
0 |
0 |
0 |
0 |
|
|
HuNoV+HGM |
6 |
6 (100 %) |
3.8 (0.7)B |
6 (100 %) |
6.8 (0.2)B |
|
a, Gn pigs were inoculated with a HuNoV GII.4 2006b variant 092895 at 6 days of age. Rectal swabs were collected daily after inoculation to determine faecal consistency scores and virus shedding.
b, Faecal consistency was scored as follows: 0, solid; 1, semisolid; 2, pasty; 3, semiliquid; and 4, liquid. Pigs with scores of or over 2 were considered with diarrhea.
c, sem, standard error of the mean.
*Fisher's exact test or **Mann–Whitney test was used for statistical analysis. Groups with significant differences (P<0.05) were indicated with letters A and B.
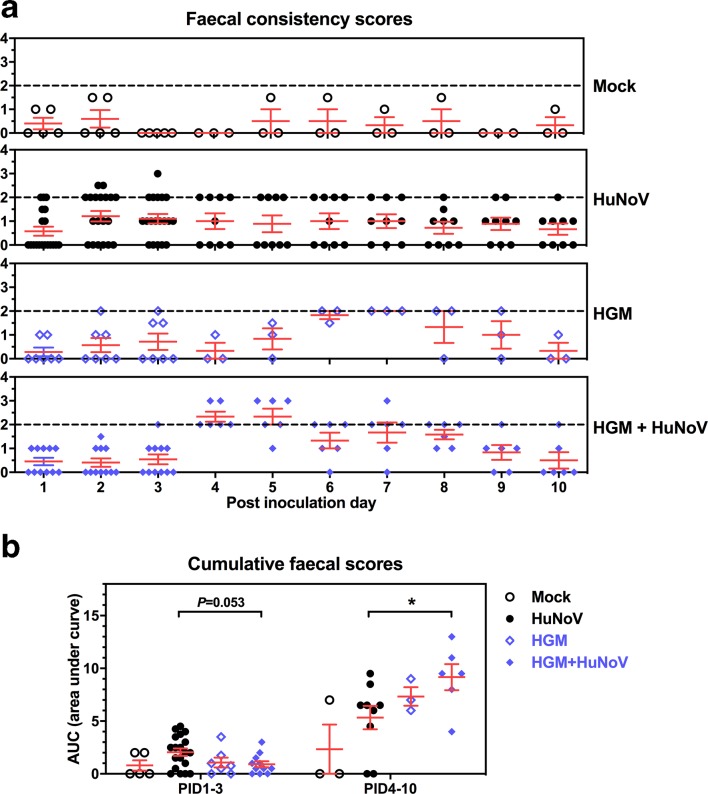
The faecal consistency was evaluated daily for all groups, the mock and HGM group had comparable scores (Fig. 3 and Table 1). Consistent with the higher virus shedding, more severe HuNoV-induced diarrhea was observed in the HGM+HuNoV group, characterized by significantly higher cumulative faecal consistency scores and mean duration of diarrhea (3.8 versus 2.0 days) on PID4–10 compared to those of the HuNoV group (Fig. 3b and Table 1). Interestingly, pigs in the HGM+HuNoV group experienced lower incidence and mean duration of diarrhea on PID1–3 (Table 1), indicating initial protection due to the pre-colonization of HGM, which might delay the occurrence of HuNoV-induced diarrhea.
Fig. 3.
Faecal consistency scores. (a) Daily faecal consistency scores after HuNoV inoculation. Faecal consistency was scored as follows: 0, solid; 1, semisolid; 2, pasty; 3, semiliquid; and 4, liquid. Dashed line shows the minimal value to be considered as diarrhea. (b) Individual pigs’ cumulative faecal scores were shown as the area under curve based on daily faecal consistency scores in (a). Data were combined from ten independent measurements and presented as individual animal data points with mean±sem. Statistical significance was determined by Mann–Whitney test, *P<0.05.
HuNoV distribution in gut tissues, blood and mononuclear cells
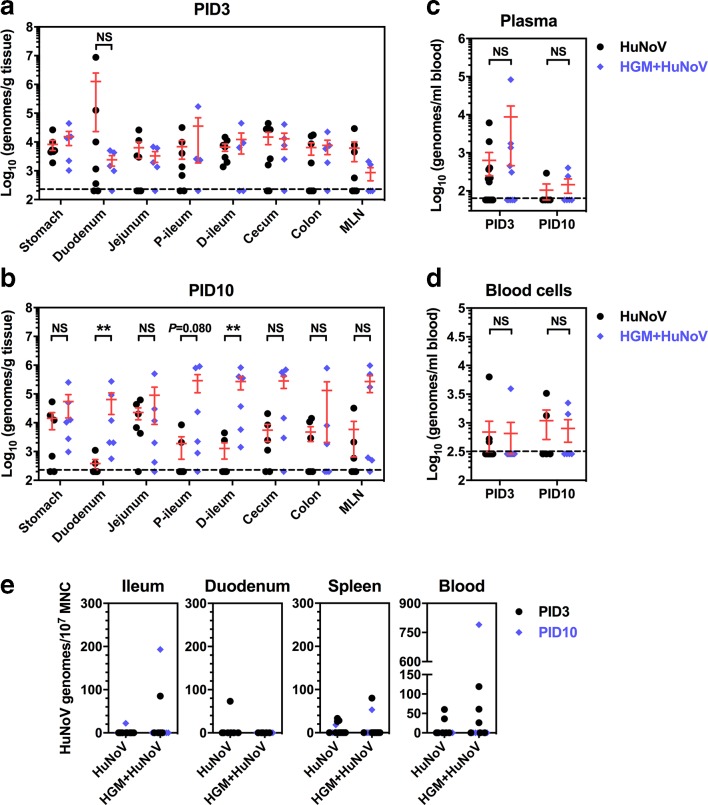
After HuNoV inoculation, pigs were euthanized at PID3 or PID10 for the collection of gut tissues and blood. Virus titres in all sections of intestine were comparable between the HuNoV group and HGM+HuNoV group on PID3 (Fig. 4a). However, virus titres were significantly higher in duodenum and distal ileum of the HGM+HuNoV group compared to the HuNoV group on PID10 (Fig. 4b). Viral genomes were detected in plasma and whole blood cells in both groups, although statistical significance was not observed (Fig. 4c and d), suggesting unaltered and transient HuNoV viremia in Gn pigs colonized with HGM. In an attempt to examine whether HuNoV could infect immune cells in the presence of HGM in Gn pigs, we performed qRT-PCR to detect viral genomes in mononuclear cells (MNC) from ileum, duodenum, spleen and blood. Although a small portion of pigs in both groups had detectable virus in MNC, the titres were generally as low as 200 genomic copies per 107 MNC (Fig. 4e).
Fig. 4.
HuNoV distribution in gut tissues, blood and MNCs. HuNoV genomes in gut tissues from pigs euthanized on PID3 (a) and PID10 (b) were quantified by qRT-PCR. P-ileum, proximal ileum; d-ileum, distal ileum. HuNoV genomes in plasma (c), whole blood cells (d), and mononuclear cells (MNCs) were quantified by qRT-PCR. (a, b) HuNoV group size: PID3 n=7, PID10 n=6. HGM+HuNoV group size: PID3 n=5, PID10 n=6. (c–e) Group sizes were shown in Table 1. Dashed line shows the limit of detection. Data were combined from four to five independent experiments and presented as individual animal data points with mean±sem. Statistical significance was determined by Mann–Whitney test. NS, not significant, *P<0.05, **P<0.01.
HuNoV infection altered intestinal microbiota in HGMT Gn pigs
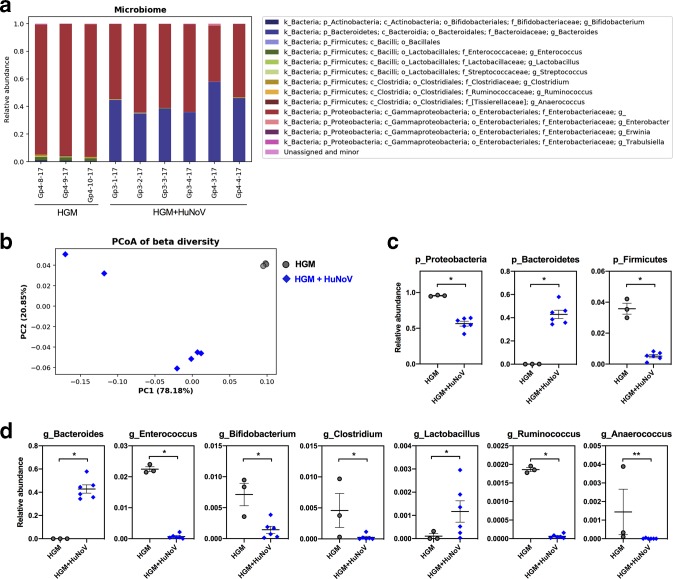
To investigate the impact of HuNoV infection on intestinal microbiota, we collected the large intestinal contents from HGMT Gn pigs euthanized on PPD16 without and with HuNoV infection (Fig. 1a), and then performed high-throughput sequencing of 16S rRNA genes. As shown by the bacterial abundance (Fig. 5a), the microbiome composition was consistent across samples in the HGM group and HGM+HuNoV group, respectively. Their beta diversity was visualized with a principal coordinate analysis (PCoA) using weighted UniFrac, which includes both sequence distance and abundance information. The results showed that HGM pig microbiota was highly similar and distinct from those of HuNoV infected pigs (Fig. 5b). Specifically, at the phylum level, Proteobacteria (95.6 % versus 56.5 %) and Firmicutes (3.6 % versus 0.5 %) significantly decreased in HuNoV-infected HGMT Gn pigs, while Bacteroidetes (0.1 % versus 42.9 %) significantly increased (Fig. 5c). At the genus level, Enterococcus , Bifidobacterium , Clostridium , Ruminococcus and Anaerococcus significantly decreased in HuNoV-infected HGMT Gn pigs, while Bacteroides and Lactobacillus significantly increased (Fig. 5d). Taken together, the variations of microbiota composition at the phylum and genus levels demonstrated that HuNoV infection dramatically altered the transplanted HGM in Gn pigs.
Fig. 5.
Microbiome composition analysis of HGMT Gn pigs. (a) Bacterial taxonomic summary showing relative abundance at the genus level. Unassigned and minor group includes genus less than 0.1 % of total community in each sample. (b) Principal coordinate analysis (PCoA) of beta diversity based on weighted UniFrac distances among HGMT Gn pigs. The first two axes that explain largest variations (PC1 and PC2) are plotted. Significantly different taxa at the phylum level (c) and at the genus level (d) between the HGM group and HGM+HuNoV group. (c, d) Data are presented as individual animal data points with mean±sem. Statistical significance was determined by Mann–Whitney test. *P<0.05, **P<0.01.
Discussion
The microbiota is indispensable for the development and maintenance of a healthy enteric immune system [10], nervous system [30] and gastrointestinal physiology [31]. The lack of maternal antibodies and gut microbiota in neonatal Gn pigs contributes to their underdeveloped mucosal immunity, predisposing these pigs to enteric pathogens [32]. However, enhanced GII.4 HuNoV infection and disease were observed in HGMT Gn pigs than that of germ-free Gn pigs in this study, indicating a favourable role of HGM in HuNoV lifecycle. Experimental HuNoV infections have not been successful in conventional pigs, presumably resulting from the well-developed mucosal immunity promoted by the naturally acquired porcine gut microbiota. Therefore, it is likely that HGM has a unique component that facilitates HuNoV infection in pigs, and such a component could be illuminated by systematic analysis, including transcriptome analysis of viral target cells and metabolome profiling of intestinal contents. Meanwhile, it is worth trying to transplant HGM in specific pathogen-free or even conventional pigs, so that HuNoV challenge study might be performed in those pig models without the need for a Gn facility.
The commensal bacterial enhancement of poliovirus infection in mice was attributed to viral binding to the bacterial outer-membrane component polysaccharides, resulting in virion thermos-stabilization and attachment to host cells [33]. HuNoV has also been shown to bind to a variety of bacteria, including a commensal bacterial species, E. cloacae [34], the representatives in HGM [35], and multiple probiotic strains [36, 37]. One underlying mechanism is the direct interaction between the viral capsid and HBGA-like carbohydrates on bacterial surface, which might also enhance HuNoV integrity when under acute heat stress [34, 38]. Both enhancement and inhibition of HuNoV P particles attachment on cells have been observed in vitro in the presence of HuNoV-binding probiotics, such as Escherichia coli Nissle 1917 and Lactobacillus casei BL23 [36]. HBGA-expressing E. cloacae has been suggested as a helper in HuNoV infection of human B cells, which is a novel HuNoV cell culture system, despite the inconsistent results in other laboratories [8, 39]. However, previous studies in Gn pigs showed that E. cloacae , Lactobacillus rhamnosus GG, and Escherichia coli Nissle 1917 exhibited inhibitory effects on HuNoV infection in vivo [16, 37], presumably resulting from bacteria-and-virus interaction and/or bacterial immunomodulatory functions. Therefore, the impact of HuNoV-binding bacteria on viral infections differs in different studies. In addition, there should be certain bacterial strains in HGM that inhibit HuNoV infection such as E. cloacae and some others enhance, resulting in the average effects of HGM, and further investigations are in great demand to differentiate the influence of different bacterial strains on HuNoV infection.
Although most enteric pathogens target intestinal epithelial cells, the presence of HuNoV antigens or virions has not been reported in clinical biopsy samples from immunocompetent humans, and the cell tropism of HuNoV has long been obscure [40–42]. Using intestinal biopsies from an immunocompromised patient cohort, HuNoV replication was observed only in enterocytes from sections of duodenum and jejunum, and the HuNoV-associated histopathological features in enterocytes were present as well [43]. Additionally, enterocytes in the stem cell-derived and nontransformed human intestinal enteroids supported the cultivation of multiple HuNoV strains [17], altogether indicating enterocytes as the primary target for HuNoV infection in vivo and in vitro. Previous studies indicated that enterocytes are the only target of HuNoV in different types of Gn pig models, including germ-free Gn pigs [44, 45], E. cloacae colonized Gn pigs [16], and RAG2/IL2RG immunodeficient Gn pigs [46]. Tuft cells have been recognized as a rare intestinal target of MNV strain CR6, and microbiota-promoted MNV-CR6 infection in mice could be partially explained by the immune-privileged tuft cells, whose proliferation could be induced by type 2 immunity [47]. Tuft cells might also be a potential HuNoV target in HGMT Gn pigs, contributing to the increased HuNoV titres in duodenum and distal ileum in the HGM+HuNoV group on PID10 (Fig. 4b), which requires further investigation.
Biased analysis showed that a minority of HuNoV-infected adults had decreased abundance of Bacteriodetes and increased abundance of Proteobacteria in their microbiota [13]. In another study analysing intestinal microbiota in children, those disruptions were not observed [48]. Under HuNoV infection in HGMT Gn pigs, significant increase of Proteobacteria and Firmicutes but decrease of Bacteroidetes were observed in the current study (Fig. 5b). It was noted that high abundance of Ruminococcus spp. was correlated with lower anti-HuNoV antibody titres and thus lower infection in humans [14]. This reverse correlation was noticed again in this study by the decreased abundance of Ruminococcus spp. after HuNoV infection. Decreased abundance of Bacteroides spp., Bifidobacterium spp. and Lactobacillus spp. were detected by RT-PCR in HuNoV-infected patients in an early study [49], but our data showed differential alterations among these bacterial families (Fig. 5c). Notably, microbiota structures differ over time in infants and young children [50], indicating that the age of HGM donor and timeline of viral infection after HGM transplantation might affect experimental outcomes.
Due to a variety of potential factors such as age, host, antibiotic usage, viral strain and initial microbial composition, it is an unsettled question whether or how HuNoV infection might affect the host microbiota and vice versa [12]. In this study, we used the well-established Gn pig system to develop an HGMT Gn pig model of HuNoV infection and disease, in which univariate analysis was performed with one infant HGM and one GII.4 strain. It is not impossible that the outcomes from the current study were HGM donor-specific and HuNoV strain-specific; future studies with multiple HGM and virus strains will shed more light on their complex interactions. In summary, the colonization of HGM in Gn pigs was associated with the enhanced GII.4 HuNoV infection, evidenced by increased virus shedding and genome titres in intestinal tissues. Significant intestinal microbiota alterations were observed under HuNoV infection in HGMT Gn pigs. To our best knowledge, this is the first in vivo evaluation on the direct effects of HGM on HuNoV infection, and our study provides a platform and guidance for future investigations of HuNoV pathogenesis, host response, antiviral and vaccine efficacies in regard to gut microbiota.
Methods
HuNoV inoculum
The HuNoV inoculum containing the GII.4/2006b variant 092895 (GenBank accession number KC990829) was prepared from a stool sample, which was obtained from a child with norovirus gastroenteritis at the Cincinnati Children’s Hospital Medical Center in 2008 [45].
HGM inoculum
The stool candidates for HGM transplantation were prepared from infant stool samples collected from León, Nicaragua, and the stool sample used in this study (ID number SV5) came from a vaginally delivered and breast-fed male infant [28, 51]. Previous analysis of SV5 showed diverse bacterial taxonomy composition and low enteropathy score [51, 52]. SV5 was confirmed negative for rotavirus, astrovirus, norovirus, sapovirus, adenovirus and Klebsiella spp. via PCR prior to oral transplantation into the Gn pigs [28]. The 5 % stool sample was washed with tenfold volume of sterile PBS to remove glycerol, centrifuged at 2000 r.p.m. for 10 min at 4 °C to pellet bacteria, and then resuspended to the original volume with sterile PBS as HGM inoculum.
Gnotobiotic pigs and treatments
Near-term Yorkshire pigs were derived via hysterectomy, maintained in Gn isolator units, and fed with sterile cow milk [32]. Neonatal Gn pigs were randomly assigned to the four treatment groups: mock (n=5), HuNoV (n=19), HGM (n=7) and HGM+HuNoV (n=11). Due to the lack of maternal antibody transfer across the porcine placenta and the deprivation of sow colostrum/milk in Gn system, Gn pigs have no maternal antibodies and thus are more susceptible to enteric pathogens than conventional pigs. In an attempt to provide immune protection (i.e. antibodies) against potentially pathogenic bacteria in the HGM transplants, all piglets received 60 ml gamma-irradiated, non-heat treated porcine serum (Rocky Mountain Biologicals) via three intraperitoneal injections at 24, 30 and 36 h post derivation. The porcine serum was screened using a luciferase immunoprecipitation system assay [53], and no antibodies against a broad range of genotype of HuNoVs, including GI.5, GI.6, GII.1, GII.2, GII.3, GII.4/MD145, GII.6, GIV.1 or GII.4/2006b were detected (data not shown). Pigs in the HGM group and HGM+HuNoV group received 450 µl HGM inoculum each day at 4 and 5 days of age. Pigs in the HuNoV group and HGM+HuNoV group were orally inoculated at 6 days of age with 2.74×104 viral RNA copies of HuNoV, the dosage was determined as 10 ID50 for neonatal pigs based on a previous study [45]. Then, 4 ml of 200 mM NaHCO3 was given 15–20 min prior to HuNoV inoculation to neutralize stomach acids. Pigs were euthanized on PID3 or PID10 for collection of blood, tissues and intestinal contents.
Assessment of faecal consistency and detection of HuNoV
Faecal consistency and virus shedding were monitored daily after HuNoV inoculation by rectal swab sampling. Faecal consistency assessment system: 0, solid; 1, semisolid; 2, pasty; 3, semiliquid; 4, liquid. HuNoV genomes in faeces, blood, mononuclear cells and tissues were detected by a one-step TaqMan qRT-PCR as described previously [46].
Microbiome analysis
Pig large intestinal contents were collected at necropsy, snap frozen and stored in liquid nitrogen. 16S rRNA amplicon sequencing was performed at the UNC Microbiome Core Facility as previously described [28]. Multiplexed paired-end fastq files were produced from the sequencing results of the Illumina MiSeq using the Illumina software BclToFastq, and then joined into a single multiplexed and single-end fastq using the tool fastq-join. Quality analysis was performed using the software FastQC. Bioinformatics analysis of bacterial 16S rRNA amplicon sequencing data was conducted using the Quantitative Insights Into Microbial Ecology (QIIME) [54]. OTU picking was performed on the quality-filtered results. Chimeric sequences were detected and removed using ChimeraSlayer. Alpha diversity and beta diversity analysis were performed on the dataset using the QIIME routines [54, 55]. Taxa of the genus level OTU with a relative abundance of 0.1 % or greater within totally community were compared between groups.
Statistics
Pigs were randomly divided into treatment groups upon derivation regardless of gender and body weight, and pigs in each group were randomly assigned for euthanasia on PID3 or PID10. For assessing faecal virus shedding and consistency scores after HuNoV infection, pigs in the PID10 subgroup contributed data to the PID3 subgroup. Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software) with different significance specified in figure legends, while only P-value<0.05 was considered as statistically significant.
Funding information
This work was supported by NIH grant R01AI089634 (to L.Y.) and Virginia-Maryland College of Veterinary Medicine (to R.A.).
Acknowledgements
We gratefully thank Xiang-Jin Meng, Xiaofeng Wang and Nanda Nanthakumar for critical discussion. We thank Kristi DeCourcy for assisting on confocal microscopy, Sherrie Clark-Deener and Kevin Pelzer for veterinary services, Karen Hall, Mariah Weiss and Jessica Park for animal care. We thank Dr Samuel Vilchez from Universidad Nacional Autonoma de Nicaragua, Leon for the collection of the HGM sample used in this study.
Author contributions
S.L., E.L.T., and L.Y. conceived the project. S.L. and E.L.T. conducted the majority of the experiments and performed data analysis. A.R., T.B., E.M. and C.M.T. conducted experiments. S.L., G.A.-A., L.Z. and M.A.A.-P. performed microbiome analysis. R.A., X.J. and S.B.-D. contributed materials. S.L. and L.Y. wrote the manuscript. All co-authors reviewed the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
The stool collection was conducted in accordance with protocols approved by the Institutional Review Boards the Cincinnati Children’s Hospital Medical Center (IRB number: 2008–1131) and the Universidad Nacional Autónoma de Nicaragua, León (IRB number: #110), informed consent from a parent or legal guardian for the study participation was obtained. All animal experiments were conducted in accordance with protocols approved by the Institutional Animal Care and Use Committee at Virginia Tech (IACUC protocols: 14–108-CVM and 17–110-CVM).
Footnotes
Abbreviations: Gn, gnotobiotic; HGM, human gut microbiota; HGMT, human gut microbiota transplanted; HuNoV, human gut microbiota transplanted; LB, lysogeny broth; MNC, mononuclear cells; MNV, murine norovirus; PID, post inoculation day; PPD, post-partum day; qRT, quantitative reverse transcription.
Reference
- 1.Atmar RL, Ramani S, Estes MK. Human noroviruses: recent advances in a 50-year history. Curr Opin Infect Dis. 2018;31:422–432. doi: 10.1097/QCO.0000000000000476. [DOI] [PubMed] [Google Scholar]
- 2.Hall AJ, Glass RI. Parashar UD: new insights into the global burden of noroviruses and opportunities for prevention. Expert Rev Vaccines. 2016:1–3. doi: 10.1080/14760584.2016.1178069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. Global economic burden of norovirus gastroenteritis. PLoS One. 2016;11:e0151219. doi: 10.1371/journal.pone.0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riddle MS, Walker RI. Status of vaccine research and development for norovirus. Vaccine. 2016;34:2895–2899. doi: 10.1016/j.vaccine.2016.03.077. [DOI] [PubMed] [Google Scholar]
- 5.Kuss SK, Best GT, Etheredge CA, Pruijssers AJ, Frierson JM, et al. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 2011;334:249–252. doi: 10.1126/science.1211057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kane M, Case LK, Kopaskie K, Kozlova A, MacDearmid C, et al. Successful transmission of a retrovirus depends on the commensal microbiota. Science. 2011;334:245–249. doi: 10.1126/science.1210718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uchiyama R, Chassaing B, Zhang B, Gewirtz AT. Antibiotic treatment suppresses rotavirus infection and enhances specific humoral immunity. J Infect Dis. 2014;210:171–182. doi: 10.1093/infdis/jiu037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones MK, Watanabe M, Zhu S, Graves CL, Keyes LR, et al. Enteric bacteria promote human and mouse norovirus infection of B cells. Science. 2014;346:755–759. doi: 10.1126/science.1257147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, et al. Commensal microbes and interferon-λ determine persistence of enteric murine norovirus infection. Science. 2015;347:266–269. doi: 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thackray LB, Handley SA, Gorman MJ, Poddar S, Bagadia P, et al. Oral antibiotic treatment of mice exacerbates the disease severity of multiple flavivirus infections. Cell Rep. 2018;22:3440–3453.:e3446. doi: 10.1016/j.celrep.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker FC, Baldridge MT. Interactions between noroviruses, the host, and the microbiota. Curr Opin Virol. 2019;37:1–9. doi: 10.1016/j.coviro.2019.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson AM, Walk ST, Taube S, Taniuchi M, Houpt ER, et al. Disruption of the human gut microbiota following norovirus infection. PLoS One. 2012;7:e48224. doi: 10.1371/journal.pone.0048224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodríguez-Díaz J, García-Mantrana I, Vila-Vicent S, Gozalbo-Rovira R, Buesa J, et al. Relevance of secretor status genotype and microbiota composition in susceptibility to rotavirus and norovirus infections in humans. Sci Rep. 2017;7:45559. doi: 10.1038/srep45559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolawole AO, Rocha-Pereira J, Elftman MD, Neyts J, Wobus CE. Inhibition of human norovirus by a viral polymerase inhibitor in the B cell culture system and in the mouse model. Antiviral Res. 2016;132:46–49. doi: 10.1016/j.antiviral.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei S, Samuel H, Twitchell E, Bui T, Ramesh A, et al. Enterobacter cloacae inhibits human norovirus infectivity in gnotobiotic pigs. Sci Rep. 2016;6:25017. doi: 10.1038/srep25017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costantini V, Morantz EK, Browne H, Ettayebi K, Zeng X-L, et al. Human norovirus replication in human intestinal enteroids as model to evaluate virus inactivation. Emerg Infect Dis. 2018;24:1453. doi: 10.3201/eid2408.180126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lei S, Twitchell E, Pathogenesis YuanL. Immunity and the role of microbiome/probiotics in enteric virus infections in humans and animal models. In: Sun J, Dudeja PK, editors. Mechanisms Underlying Host-Microbiome Interactions in Pathophysiology of Human Diseases. Boston, MA: Springer US; 2018. pp. 55–78. [Google Scholar]
- 20.Kocher J, Bui T, Giri-Rachman E, Wen K, Li G, et al. Intranasal P particle vaccine provided partial cross-variant protection against human GII.4 norovirus diarrhea in gnotobiotic pigs. J Virol. 2014;88:9728–9743. doi: 10.1128/JVI.01249-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bui T, Li G, Kim I, Wen K, Twitchell EL, et al. Effects of racecadotril on weight loss and diarrhea due to human rotavirus in neonatal gnotobiotic pigs (sus scrofa domesticus) Comp Med. 2017;67:157–164. [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, Twitchell E, Li G, Wen K, Weiss M, et al. High protective efficacy of rice bran against human rotavirus diarrhea via enhancing probiotic growth, gut barrier function, and innate immunity. Sci Rep. 2015;5:15004. doi: 10.1038/srep15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen X, Cao D, Jones RW, Hoshino Y, Yuan L. Tandem truncated rotavirus VP8* subunit protein with T cell epitope as non-replicating parenteral vaccine is highly immunogenic. Hum Vaccin Immunother. 2015;11:2483–2489. doi: 10.1080/21645515.2015.1054583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei S, Yuan L. Chapter 21 - Rice Bran Usage in Diarrhea. In: Watson RR, editor. Dietary Interventions in Gastrointestinal Diseases. Preedy VR: Academic Press; 2019. pp. 257–263. editor. [Google Scholar]
- 25.Todd K, Tripp R. Human norovirus: experimental models of infection. Viruses. 2019;11:151. doi: 10.3390/v11020151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H, Wang H, Shepherd M, Wen K, Li G, et al. Probiotics and virulent human rotavirus modulate the transplanted human gut microbiota in gnotobiotic pigs. Gut Pathog. 2014;6:39. doi: 10.1186/s13099-014-0039-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A, Vlasova AN, Deblais L, Huang HC, Wijeratne A, et al. Impact of nutrition and rotavirus infection on the infant gut microbiota in a humanized pig model. BMC Gastroenterol. 2018;18:93. doi: 10.1186/s12876-018-0810-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Twitchell EL, Tin C, Wen K, Zhang H, Becker-Dreps S, et al. Modeling human enteric dysbiosis and rotavirus immunity in gnotobiotic pigs. Gut Pathog. 2016;8:51. doi: 10.1186/s13099-016-0136-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyazaki A, Kandasamy S, Michael H, Langel SN, Paim FC, et al. Protein deficiency reduces efficacy of oral attenuated human rotavirus vaccine in a human infant fecal microbiota transplanted gnotobiotic pig model. Vaccine. 2018;36:6270–6281. doi: 10.1016/j.vaccine.2018.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Vadder F, Grasset E, Mannerås Holm L, Karsenty G, Macpherson AJ, et al. Gut microbiota regulates maturation of the adult enteric nervous system via enteric serotonin networks. Proc Natl Acad Sci USA. 2018;115:6458–6463. doi: 10.1073/pnas.1720017115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.An R, Wilms E, Masclee AAM, Smidt H, Zoetendal EG, et al. Age-Dependent changes in Gi physiology and microbiota: time to reconsider? Gut. 2018;67:2213. doi: 10.1136/gutjnl-2017-315542. [DOI] [PubMed] [Google Scholar]
- 32.Yuan L, Jobst PM, Weiss M. Gnotobiotic pigs: from establishing facility to modeling human infectious diseases. In: Schoeb TR, editor. Gnotobiotics. Eaton KA: Academic Press; 2017. pp. 349–368. editor. [Google Scholar]
- 33.Robinson CM, Jesudhasan PR, Pfeiffer JK. Bacterial lipopolysaccharide binding enhances virion stability and promotes environmental fitness of an enteric virus. Cell Host Microbe. 2014;15:36–46. doi: 10.1016/j.chom.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miura T, Sano D, Suenaga A, Yoshimura T, Fuzawa M, et al. Histo-Blood group antigen-like substances of human enteric bacteria as specific adsorbents for human noroviruses. J Virol. 2013;87:9441–9451. doi: 10.1128/JVI.01060-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almand EA, Moore MD, Outlaw J, Jaykus LA. Human norovirus binding to select bacteria representative of the human gut microbiota. PLoS One. 2017;12:e0173124. doi: 10.1371/journal.pone.0173124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubio-del-Campo A, Coll-Marqués JM, Yebra MJ, Buesa J, Pérez-Martínez G, et al. Noroviral p-particles as an in vitro model to assess the interactions of noroviruses with probiotics. PLoS One. 2014;9:e89586. doi: 10.1371/journal.pone.0089586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lei S, Ramesh A, Twitchell E, Wen K, Bui T, et al. High protective efficacy of probiotics and rice bran against human norovirus infection and diarrhea in gnotobiotic pigs. Front Microbiol. 2016;7:1699. doi: 10.3389/fmicb.2016.01699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li D, Breiman A, le Pendu J, Uyttendaele M. Binding to histo-blood group antigen-expressing bacteria protects human norovirus from acute heat stress. Front Microbiol. 2015;6:659. doi: 10.3389/fmicb.2015.00659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones MK, Grau KR, Costantini V, Kolawole AO, de Graaf M, et al. Human norovirus culture in B cells. Nat Protoc. 2015;10:1939–1947. doi: 10.1038/nprot.2015.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Agus SG, Dolin R, Wyatt RG, Tousimis AJ, Northrup RS. Acute infectious nonbacterial gastroenteritis: intestinal histopathology. histologic and enzymatic alterations during illness produced by the Norwalk agent in man. Ann Intern Med. 1973;79:18–25. doi: 10.7326/0003-4819-79-1-18. [DOI] [PubMed] [Google Scholar]
- 41.Dolin R, Levy AG, Wyatt RG, Thornhill TS, Gardner JD. Viral gastroenteritis induced by the Hawaii agent. jejunal histopathology and serologic response. Am J Med. 1975;59:761–768. doi: 10.1016/0002-9343(75)90461-1. [DOI] [PubMed] [Google Scholar]
- 42.Karst SM, Wobus CE, Goodfellow IG, Green KY. Virgin HW: Advances in norovirus biology. Cell Host Microbe. 2014;15:668–680. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karandikar UC, Crawford SE, Ajami NJ, Murakami K, Kou B, et al. Detection of human norovirus in intestinal biopsies from immunocompromised transplant patients. J Gen Virol. 2016;97:2291–2300. doi: 10.1099/jgv.0.000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheetham S, Souza M, Meulia T, Grimes S, Han MG, et al. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol. 2006;80:10372–10381. doi: 10.1128/JVI.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bui T, Kocher J, Li Y, Wen K, Li G, et al. Median infectious dose of human norovirus GII.4 in gnotobiotic pigs is decreased by simvastatin treatment and increased by age. J Gen Virol. 2013;94:2005–2016. doi: 10.1099/vir.0.054080-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei S, Ryu J, Wen K, Twitchell E, Bui T, et al. Increased and prolonged human norovirus infection in RAG2/IL2RG deficient gnotobiotic pigs with severe combined immunodeficiency. Sci Rep. 2016;6:25222. doi: 10.1038/srep25222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilen CB, Lee S, Hsieh LL, Orchard RC, Desai C, et al. Tropism for tuft cells determines immune promotion of norovirus pathogenesis. Science. 2018;360:204–208. doi: 10.1126/science.aar3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen SY, Tsai CN, Lee YS, Lin CY, Huang KY, et al. Intestinal microbiome in children with severe and complicated acute viral gastroenteritis. Sci Rep. 2017;7:46130. doi: 10.1038/srep46130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ma C, Wu X, Nawaz M, Li J, Yu P, et al. Molecular characterization of fecal microbiota in patients with viral diarrhea. Curr Microbiol. 2011;63:259–266. doi: 10.1007/s00284-011-9972-7. [DOI] [PubMed] [Google Scholar]
- 50.Stewart CJ, Ajami NJ, O'Brien JL, Hutchinson DS, Smith DP, et al. Temporal development of the gut microbiome in early childhood from the TEDDY study. Nature. 2018;562:583–588. doi: 10.1038/s41586-018-0617-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Becker-Dreps S, Vilchez S, Bucardo F, Twitchell E, Choi WS, et al. The association between fecal biomarkers of environmental enteropathy and rotavirus vaccine response in nicaraguan infants. Pediatr Infect Dis J. 2017;36:412–416. doi: 10.1097/INF.0000000000001457. [DOI] [PubMed] [Google Scholar]
- 52.Becker-Dreps S, Allali I, Monteagudo A, Vilchez S, Hudgens MG, et al. Gut microbiome composition in young nicaraguan children during diarrhea episodes and recovery. Am J Trop Med Hyg. 2015;93:1187–1193. doi: 10.4269/ajtmh.15-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tin CM, Yuan L, Dexter RJ, Parra GI, Bui T, et al. A luciferase immunoprecipitation system (lips) assay for profiling human norovirus antibodies. J Virol Methods. 2017;248:116–129. doi: 10.1016/j.jviromet.2017.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lozupone C, Hamady M, Knight R. UniFrac-an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinformatics. 2006;7:371. doi: 10.1186/1471-2105-7-371. [DOI] [PMC free article] [PubMed] [Google Scholar]