Abstract
Key to onset and progression of periodontitis is a complex relationship between oral bacteria and the host. The organisms most associated with severe periodontitis are the periodontal pathogens of the red complex: Tannerella forsythia , Treponema denticola and Porphyromonas gingivalis . These organisms express sialidases, which cleave sialic acid from host glycoproteins, and contribute to disease through various mechanisms. Here, we expressed and purified recombinant P. gingivalis sialidase SiaPG (PG_0352) and characterized its activity on a number of substrates, including host sialoglycoproteins and highlighting the inability to cleave diacetylated sialic acids – a phenomenon overcome by the NanS sialate-esterase from T. forsythia . Indeed SiaPG required NanS to maximize sialic acid harvesting from heavily O-acetylated substrates such as bovine salivary mucin, hinting at the possibility of interspecies cooperation in sialic acid release from host sources by these members of the oral microbiota. Activity of SiaPG and P. gingivalis was inhibited using the commercially available chemotherapeutic zanamivir, indicating its potential as a virulence inhibitor, which also inhibited sialic acid release from mucin, and was capable of inhibiting biofilm formation of P. gingivalis on oral glycoprotein sources. Zanamivir also inhibited attachment and invasion of oral epithelial cells by P. gingivalis and other periodontal pathogens, both in monospecies but also in multispecies infection experiments, indicating potential to suppress host–pathogen interactions of a mixed microbial community. This study broadens our understanding of the multifarious roles of bacterial sialidases in virulence, and indicates that their inhibition with chemotherapeutics could be a promising strategy for periodontitis therapy.
Keywords: Sialidase, host-pathogen interactions, biofilm, glycoprotein, periodontitis, gingivitis
Introduction
The black-pigmented, Gram-negative, asaccharolytic anaerobe Porphyromonas gingivalis has been the focus of much research due to its role in periodontitis where it is often referred to as a ‘keystone pathogen’ [1]. P. gingivalis exists as a small part of the microbiome, which sees a population shift to include a greater proportion of certain bacterial species, a state now referred to as dysbiosis [2]. In periodontal disease this population shift was observed in the 1990s, with certain organisms considered particularly important to periodontal disease. Tannerella forsythia , P. gingivalis and Treponema denticola form the so-called red complex, which is strongly associated with disease [3], and with the advent of next-generation sequencing their presence as a signature of periodontal disease has been well established, along with other organisms, which seem to play a causative role in periodontitis [2, 4]. During treatment of periodontitis, systemic antibiotics such as azithromycin can be administered as an adjunct to mechanical removal of the subgingival plaque [5]. Despite this a proportion of patients poorly respond to periodontal therapy, and in these patients (post-treatment) the levels of periodontal pathogens are higher than in patients who respond to treatment, or in healthy controls [6]. This difficulty in treatment has led to increased interest in development of an ‘anti-virulence approach’ to target the virulence factors of specific pathogens or groups of pathogens, including those responsible for periodontitis. This has been seen in the case of the gingipains of P. gingivalis , with several plant extracts [7, 8], and short peptides [9] shown to reduce gingipain activity, with in vitro studies showing reductions in P. gingivalis biofilm formation [8], and disruption of bacterial nutrient acquisition leading to decreased P. gingivalis growth [9]. Plant extracts that inhibit gingipains have also shown disruption of host– P. gingivalis interactions; attachment and invasion of host cells, and cytokine production [7]. Moreover, a recent study showed that treatment with a potent small molecule gingipain inhibitor in mice reduced P. gingivalis infection in the brain and further blocked Aβ1-42 production and neurodegenerative Alzheimer disease pathology [10].
Another virulence factor of periodontal pathogens including P. gingivalis are sialidases (also called neuraminidases), enzymes frequently expressed by host-dwelling organisms, which cleave terminal sialic acid – a family of 9-carbon sugars – from host sialoglycans. In humans, the most common sialic acid is N-acetylneuraminic acid (Neu5Ac), and is usually linked by its second carbon to the third or sixth carbon of the underlying sugar (α2–3- or α2–6-linked sialic acid). Sialidases are well-established virulence factors in the gut and nasopharyngeal pathogens Vibrio cholerae and Streptococcus pneumoniae , where they contribute to colonization, persistence and ultimately disease [11, 12]. Interestingly, all three periodontal red-complex pathogens express a sialidase; PG_0352 (termed SiaPG here) in P. gingivalis , NanH in T. forsythia and TDE0471 in T. denticola . Other periodontal pathogens also possess mechanisms to process sialic acid in the absence of sialidase expression, such as some Fusobacterium nucleatum spp., which possess neuraminate lyase for catabolism of sialic acid [13]. It is thought that acquisition of sialic acid by organisms that do not possess sialidases is due to the overall action of the microbial community of which they are part of. Additionally, periodontal organisms tentatively associated with health such as Veillonella spp. [14] and the recently characterized Tannerella HOT (oral taxon BU063) [15] do not possess sialidases. The apparent importance of sialidases for periodontal pathogens has been investigated in studies of sialidase knockout. T. forsythia nanH sialidase mutants displayed decreased attachment to oral epithelial cells compared to their parent strain [16]. Different strains of P. gingivalis have also been made deficient in SiaPG [17, 18], although the exact implications for virulence are less clear because the sialidase mutants display defects in capsule formation. Mutant strains do show decreased association with host cells, while the virulence of ΔsiaPG mutants in a mouse abscess model is also decreased relative to the parent strain, where subcutaneous injections of wild-type P. gingivalis (strain W83) resulted in the formation of abscesses and ultimately lethal infection that were not seen in sialidase-deficient P. gingivalis [17]. Sialidase-deficient mutants of T. denticola have also been studied, and these display reduced complement evasion and decreased virulence in a mouse abscess model [19].
Given the importance of sialidases for periodontal pathogen–host interactions and subsequent virulence, abrogation of sialidase activity using sialidase inhibitors might exert anti-virulence effects or be detrimental to the lifecycle of these organisms. This is particularly pertinent given recent evidence that levels of sialidase transcript (T. forsythia nanH gene) are elevated in periodontal disease [20]. Similarly, sialidase enzyme activity is raised in the gingival crevicular fluid of patients with periodontal disease and this high sialidase activity is an indicator of poor responsiveness to standard treatment [21].
Sialidase inhibitors are undergoing extensive development and some are available as a treatment for influenza, with the sialidase inhibitors oseltamivir and zanamivir licensed for use globally. Sialidase inhibition has been investigated in T. forsythia , where the inhibitor oseltamivir was shown to disrupt biofilm formation on host sialoglycan substrates [16]. Considering the above, we set out to further characterize the sialidase of P. gingivalis , performing a biochemical characterization and estabilishing an activity profile while also examining the potential of sialidase inhibitors to modulate pathogenesis of P. gingivalis alone and in combination with a range of other periodontal pathogens with the commercially available sialidase inhibitor zanamivir.
Methods
Bacterial strains, human cell lines and culture conditions
T. forsythia strains used here were the WT type strain ATCC 43037, supplied by William Wade, Queen Mary University of London, UK, and the sialidase-deficient nanH mutant (TFΔnanH) [22]. P. gingivalis strains used here were the WT strains ATCC 33277, 381 (Pg381), and a sialidase-deficient mutant of Pg381 (Pg381ΔsiaPG). F. nucleatum strain NCTC 25586 (subspecies nucleatum) was also used in this study, supplied by William Wade. All strains were cultured at 37 °C under anaerobic conditions (10 % CO2, 10 % H2, 80 % N2) in a Don–Whitley mini-macs anaerobic cabinet. P. gingivalis and F. nucleatum strains were cultured on Fastidious Anaerobe Agar (Lab M, Lancashire, UK) supplemented with 5 % (v/v) oxalated horse blood (Thermo Fisher Scientific, UK), for 2–3 days before harvesting for use in experiments. T. forsythia strains were cultured on Fastidious Anaerobe Agar (Lab M, UK) supplemented with 5 % (v/v) oxalated Horse Blood (Thermo Fisher Scientific), 10 µg ml−1 N-acetylneuraminic acid (NAM, Sigma Aldrich, UK), and 25 µg ml−1 Gentamicin (Sigma-Aldrich) for 3–7 days before harvesting for use in experiments. In addition, TF∆nanH and Pg381∆SiaPG were intermittently subcultured in the presence of 10 µg ml−1 erythromycin to ensure maintenance of the antibiotic resistance cassette marker responsible for sialidase inactivation. Immortalized human oral keratinocytes (OKF6/Tert2) Dickson et al. [23] were kindly provided by Dr J. Rheinwald (Harvard Medical School, Cambridge, MA) and were grown in keratinocyte-serum-free media supplemented with defined growth supplements (DKSFM, Fisher Scientific, Loughborough, UK). The Oral Squamous Cell Carcinoma (OSCC) cell line H357 [24, 25 ] – a generous gift from Professor S. Prime, University of Bristol, UK – was grown in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10 % (v/v) FBS, 2 mM l-glutamine and 100 units ml−1 Penicillin-Streptomycin (Sigma Aldrich). Cells were incubated at 37 ˚C, 5 % CO2 when not undergoing media changes or passaging. Cells were grown to 70–90 % confluence and media was changed every 2–4 days. OBA-9 cells were maintained in keratinocyte basal medium KGM-2 supplemented with epidermal growth factor (EGF), bovine pituitary extract, epinephrine, transferrin, hydrocortisone and insulin, as per the manufacturer’s recommendations (Lonza). Cells were used after reaching near confluence.
Cloning, expression and purification of recombinant proteins
NanH and NanS were cloned, expressed and purified as previously described [26, 27]. A nucleotide sequence encoding SiaPG (PG_0352, based on the genome of strain ATCC 33277) without the secretion signal sequence (supplemental text), was codon optimized for expression in Escherichia coli , and synthesized commercially (ThermoFisher). The sequence also contained restriction sites for NdeI and XhoI at its 5′ and 3′ ends, enabling it to be ligated into pET vectors. Restriction digest and ligation was performed using NEB buffers, restriction enzymes and T4 ligase, according to the manufacturer’s protocols. SiaPG was ligated into pET21a, and ultimately transformed into chemically competent E. coli BL21 (origami B). Expression and purification of recombinant SiaPG was performed as previously described for NanH [27].
Construction of a P. gingivalis siaPG deletion mutant and genetic complementation
For generating a sialidase deficient mutant, the siaPG gene (PG0352) in P. gingivalis 381 strain was inactivated by an allelic replacement strategy. The primers used in the construction and complementation strategy are listed in Table 1. Briefly, a DNA fragment containing PG0352 coding region and flanking sequences was PCR amplified with primers 1 and 2 using the P. gingivalis 381 gDNA. This product was then used as a template to amplify the upstream and downstream regions of PG0352 with primer sets 1 and 3 (for 5′ end) and 2 and 5 (for 3′ end), respectively. In parallel, the ermF gene (797 bp) was amplified from the plasmid pVA2198 with primers 4 and 6. Primers 3, 4, 5 and 6 contain overlapping sequences for ermF and PG0352 to allow generation of a fusion fragment via overlap PCR. Finally, PCR products of PG0352 with flanking regions and ermF gene were combined in an overlap PCR reaction using primers 1 and 2. The overlap PCR product was sequenced to confirm the correct fusion of fragments and then transformed into P. gingivalis 381 by electroporation as previously described [28]. Transformants were plated on TSB-blood agar plates containing 10 µg ml−1 of erythromycin and incubated at 37 °C anaerobically for 10 days. Following incubation, erythromycin-resistant colonies were screened by PCR and Southern blotting. One representative deletion mutant as confirmed by PCR and Southern blotting (data not shown) and showing no sialidase activity was selected and designated as Pg381ΔsiaPG. For complementation of Pg381ΔsiaPG, in trans complementation with the siaPG gene on the self-replicating shuttle plasmid pT-COW was employed as we have performed previously [28]. Briefly, the siaPG gene fragment amplified by PCR with the primers FWD BamHI and REV SalI (Table 1) was digested with BamHI and SalI and ligated into BamHI and SalI digestd pT-COW plasmid. The resulting plasmid, pT-PG352C was transformed into Pg381ΔsiaPG by tri-parental conjugation involving with Pg381ΔsiaPG, E. coli carrying shuttle vector pT-PG352C and E. coli carrying helper plasmid pRK231 4,5. The transformants were selected on TSB-blood agar plate containing 100 µg ml−1 of gentamicin (Invitrogen), 10 µg ml−1 of erythromycin (Sigma) and 1 µg ml−1 of tetracycline (Sigma). The completed clones with sialidase expression were saved and one random clone designated ΔsiaPG + served as a representative.
Table 1.
List of primers used in this study
|
Primers for siaPG deletion mutant | |
|---|---|
|
Primer |
Sequence (5′>3′) |
|
1 |
TACGGCATCGCGGTTTTGA |
|
2 |
TCGCCATAGAATACAGGATAAGC |
|
3 |
gggcaatttcttttttgtcatTGAAAACTATTTTATACCATTTTGGGA |
|
4 |
TCCCAAAATGGTATAAAATAGTTTTCAatgacaaaaaagaaattgccc |
|
5 |
aaaaatttcatccttcgtagGAATAGTGCTTTTTTTATCGAGTTTTTC |
|
6 |
GAAAAACTCGATAAAAAAAGCACTATTCctacgaaggatgaaattttt |
|
Sequences in upper case correspond to siaPG and lower case to ermF. | |
|
Primers for siaPG complementation | |
|
FWD BamHI |
CGCGGATCCG taatacgactcactatagg |
|
REV SalI |
TTGACGTCGACGCGTctagttattgctcagcggtgg |
|
Endonuclease sites are underlined. | |
Inhibition of whole periodontal pathogens and purified sialidases by zanamivir
Zanamivir was provided by GlaxoSmithKline (GSK, Weybridge, UK). Whole T. forsythia or P. gingivalis , or purified sialidases 2.5 nM NanH or 5 nM rSiaPG were incubated in the presence of 0.1 mM MU-NANA, in 50 mM sodium phosphate 200 mM NaCl, pH 7.4 (to mimic host physiological conditions), in the presence of different concentrations of zanamivir (0–10 mM). Sialidase inhibition was expressed as the percentage change in fluorescence at a given concentration of inhibitor, relative to fluorescence in the absence of inhibitor. Sialidase inhibition (%) was plotted against log[inhibitor], and the variable slope model applied to obtain the IC50 of zanamivir for SiaPG and NanH in Graphpad Prism 7, using the equation: Y=100/[1+10 (LogIC50-X)×HillSlope]. Y=sialidase activity relative to no inhibitor condition (%), X=log[inhibitor] and Hillslope=steepness of the curve.
Biochemistry of SiaPG: Determination of optimum pH, reaction kinetics with MUNANA and sialyllactose, and sialic acid release from mucin in concert with NanS from T. forsythia .
Whole P. gingivalis and purified SiaPG were tested for pH optima in a similar manner to T. forsythia and its sialidase [27]. For whole cell assays, bacteria were resuspended from agar plates, washed 3× in PBS, pH 7.4 (PBS, Sigma Aldrich) and centrifugation at 10 000 g for 2 min, and resuspended to an OD600 of 0.05 in appropriate reaction buffers [20 mM sodium citrate–citric acid (pH 3.0–6.4), sodium phosphate mono-basic/dibasic (pH 6.8–8.8) or sodium carbonate–sodium bicarbonate (pH 9.2–10.5)]. For pH optimum derivation for purified SiaPG, enzyme was used at 2.5 nM. In both cases, cells or enzyme were incubated with 100 µM MU-NANA and quenched with addition of 100 mM sodium carbonate buffer, pH 10.5, at a volume ratio of 1 : 1.5 (reaction: sodium carbonate), and sialidase activity quantified by measuring 4-MU fluorescence (λex=350 nm; λem=450 nm). Whole P. gingivalis assays were performed over 1–3 h at 37 °C, incubation with SiaPG was performed over 1–3 min at room temperature (~20 °C).
For derivation of Michaelis–Menten kinetics for SiaPG, 2.5 nM enzyme was incubated with varying concentrations of MU-NANA and again quenched. A standard curve of the fluorescence signal of 4-MU at defined concentrations was also obtained, enabling sialidase activity to be expressed as 4-MU released µmol min−1 mg−1 SiaPG. Similar conditions were used for derivation of kinetic parameters with 3/6-siallyl-lactose, except no quenching was required and the levels of released sialic acid using a modified Thiobarbituric acid (TBA) assay. In this case reactions were stopped at time points over 1–5 min by commencement of the TBA assay: in total, 50 µl of these reactions was added to 25 µl of 25 mM sodium periodate (Sigma–Aldrich) in 60 mM H2SO4 (Thermo Fisher) and incubated for 30 min at 37 °C. This oxidation step was stopped by the addition of 20 µl of 2 % (w/v) sodium meta-arsenite (Sigma–Aldrich) in 500 mM HCl. Then, 47 µl of this reaction was added to 100 µl of 100 mM TBA, pH 9.0, and incubated at 95 °C for 7.5 min, resulting in the thiol-labelling of free sialic acid. Upon centrifugation at 1500 g for 5 min, the pink chromophore in the clarified supernatant was spectrophotometrically quantified at A549. A standard curve of known sialic acid (Neu5Ac, Carbosynth) concentrations was used to calculate sialic acid release in µmol min−1 mg−1 SiaPG. Finally, release of Neu5Ac from mucin was assessed using BSM and the TBA assay. PBS+6 µM BSM was incubated with combinations of 100 nM rSiaPG, 0.5 mM zanamivir, and/or the sialate-O-acetylesterase NanS from T. forsythia (100 nM), and incubated for 30 min at 37 °C, before the reaction was stopped and the TBA assay commenced as described above.
Analysis of procainamide-labelled glycans
SLeX and FA2G2S2 (A2F) glycans (Ludger) underwent procainamide labelling (see below), as glycans were released from recombinant human EPO as previously described [26]. Briefly, human EPO was expressed in Chinese hamster ovary (CHO) cells (a gift from Antonio Vallin, Center for Molecular Immunology, La Habana, Cuba) and released using peptide N-glycosidase F (PNGase F, E-PNG01; Ludger). EPO (in 17.5 µl) was denatured at 100 °C for 5 mins with the addition of 6.25 µl 2 % (w/v) SDS, and 1 M 2-mercaptoethanol, then incubated at 37 °C for 16 h with 1 µl PNGase F and 1.25 µl 15 % (w/v) Triton X-100. Released N-glycans were fluorescently labelled with 2-aminobenzamide (2-AB) as described previously [29] using a LT-KPROC-96 kit (Ludger). The released glycans were incubated with labelling reagents for 3 h at 65 °C. The 2-AB-labelled glycans were cleaned up using LudgerClean S Cartridges (Ludger), then incubated with 1 µl of 1 mg ml−1 SiaPG in a final volume of 10 µl (50 mM sodium acetate buffer, pH 5.5) for 16 h at 37 °C. Glycans were cleaned-up and SiaPG removed using a LC-PROC-96 kit (Ludger) procainamide-labelled glycans (Ludger) were analysed by UHPLC-MS/MS. Here, 25 µl of each sample was injected into a Waters ACQUITY UPLC Glycan BEH Amide column (2.1×150 mm, 1.7 µm particle size, 130 Å pore size) at 40 °C on a Dionex Ultimate 3000 UHPLC instrument with a fluorescence detector (λex=310 nm, λem=370 nm) attached to a Bruker Amazon Speed ETD. Mobile phase A was a 50 mM ammonium formate solution (pH 4.4) and mobile phase B was neat acetonitrile. Analyte separation was accomplished by a gradient running from 76–51 % mobile phase B over 70 min at a flow rate of 0.4 ml min−1. The Amazon Speed was operated in the positive sensitivity mode using the following settings: source temperature, 180 C; gas flow, 4 L min−1; capillary voltage, 4500 V; ICC target, 200 000; maximum accumulation time, 50.00 ms; rolling average, 2; number of precursor ions selected, 3; scan mode, enhanced resolution; mass range scanned, 400 to 1700. Data was analysed using Bruker Compass Data Analysis software v4.1 and glycan diagrams made using GlycoWorkbench v2.0. Glycan compositions were elucidated based on MS2 fragmentation.
Lectin staining for cell surface sialic acid-immunofluorescence microscopy
H357 cells were seeded at a density of 1.5×105 cells ml−1 into the wells of a 24-well tissue culture plate, which contained sterile glass coverslips (BDH). These were incubated at 37 °C, 5 % CO2 for 18 h. Treatment with SiaPG and NanH was performed by washing 2× with PBS, then incubated with 200 nM of each sialidase for 30 min at 37 °C. Cells then underwent sialic acid staining with lectins from Sambucus nigra (FITC conjugated SNA, Vector Labs, specific for α2–6 Neu5Ac linkages), Maackia amuriensis (Biotinylated MAA, Vector Labs, specific for α2–3 Neu5Ac linkages). After treatment, cells were washed twice with 500 µl PBS followed by application of lectins: 4 µg ml−1 SNA or 8 µg ml−1 MAA, for 30 min at 37 °C, 5 % CO2. Cells were washed twice with 500 µl PBS, and conditions containing MAA underwent a second incubation with 2 µg ml−1 Texas Red-Streptavidin, for 30 min at 37 °C. Stained cells were washed three times with 500 µl PBS and fixed with 500 µl 2 % (w/v) paraformaldehyde for 15 min at 37 °C. Coverslips with fixed cells were removed from wells and mounted onto glass slides with ProLong Gold Antifade mount (containing DAPI, ThermoFisher Scientific). Mounted cells were incubated for at least 18 h and visualized within 1 week. Imaging was performed with an Axiovert 200M fluorescence microscope (Zeiss) and associated Axiovert software (Zeiss). Images were processed using Fiji-imageJ, Software [30]. Fluorescence background subtraction was performed for all images using the same parameters for each fluorescence colour channel in a given experiment.
Biofilm assays
P. gingivalis was cultured in the wells of a 96-well tissue culture plate (poly-lysine coated, Greiner): bacteria from an agar plate were resuspended to an OD600 of 0.05 in Tryptic Soy Broth (TSB; Sigma-Aldrich) supplemented with 2 % (w/v) yeast extract, 1 µg ml−1 vitamin K, 5 µg ml−1 hemin, 50 µg ml−1 gentamicin, and either 6 mM Neu5Ac or no additional supplements. In conditions testing host glycoproteins, 100 µl of 6 µM BSM, 100 % (v/v) pooled human saliva or FBS was added to plate wells and left to coat the wells by incubation at 4 °C overnight, removed and wells washed with PBS once. In conditions testing the effect of sialidase inhibition, 10 mM zanamivir was included. All media was equilibrated overnight in culture conditions, namely, 37 ˚C under anaerobic atmosphere (10 % CO2, 10 % H2, 80 % N2). Biofilms were cultured for 5 days. To quantify planktonic or total growth, all conditions underwent OD600 measurement using a Tecan Infinite M200 plate reader. Quantification was carried out by either crystal violet staining or manual counting of organisms. Crystal violet solution (0.1 % w/v) was added to each well and incubated at room temperature for 30 min. After incubation, crystal violet was removed and wells were gently washed three–four times with PBS before visualization using light microscopy or extraction with 80 : 20 ethanol: acetone. For manual counting, biofilms were gently washed twice using PBS to remove planktonic cells, and vigorously resuspended in PBS and serial diluted where appropriate. Bacteria were enumerated by counting using a Helber chamber (Hawksley) under phase contrast microscopy, 400× magnification.
Antibiotic protection assays
H357 or OKF6 cells were seeded into the wells of a 24-well tissue culture plate at a density of 1.2×105–2×105 cells and incubated for 24–48 h. Media were removed, the cells in one well were detached by trypsinization and counted by haemocytometry, to determine the number of cells per well. Remaining cells were incubated for 1 h in media supplemented with 2 % (w/v) bovine serum albumin (BSA), then washed twice with PBS. Bacteria were quantified using a Helber chamber (Hawksley) under phase contrast microscopy, 400× magnification. Bacterial suspensions were diluted in culture media (with no FBS or antibiotics) to a ratio of 1 : 100 host cells: bacteria, and for inhibitor testing conditions 10 mM zanamivir was included. These suspensions were incubated with the host cells at 37 °C, 5 % CO2 for 1.5 h, or in empty wells containing no host cells – constituting a bacterial ‘viability’ control. After incubation, the wells were washed twice with PBS, and either harvested (see below) – forming the ‘total association’ condition, or media with 200 µg ml−1 metronidazole was added, then incubated at 37 °C, 5 % CO2 for 1 h, forming the ‘invasion’ conditions. To harvest bacteria, wells were washed three times with PBS, lysed with deionized water and scraped with a pipette tip for 1 min. Sufficient lysis was checked by microscopy (this method reliably resulted in >99 % lysis). Harvested bacterial suspensions underwent serial dilutions and were plated onto agar by Miles–Misra methodology. After incubation and colony counting, the number of bacteria associated with host cells was determined and expressed as a percentage of viable bacteria that were associated with and had invaded the OKF6 cells. By subtracting invaded organisms from associated, the number of ‘attached’ bacteria was obtained.
Assessment of zanamivir cytotoxicity
Quantifying metabolism as an indicator of cell viability was performed using 3-(4,5-dimethylthiazol-2-yl)−2,5-diphenyltetrazolium Bromide (MTT). Overall, 25 000 OKF6 cells in DKSFM were seeded into the wells of a 96-well tissue culture plate (Greiner), and incubated for 24 h at 37 °C, 5 % CO2. Cells were then washed twice with PBS, and media supplemented with 1 mg ml−1 MTT, and 0, 2.5, 5, 7.5 or 10 mM zanamivir was added. Cells were incubated for 2.5 h at 37 °C, 5 % CO2. MTT-supplemented media was removed, and cells were washed twice using PBS. Formazan crystals were solubilized using isopropanol+0.125 % (v/v) HCl, and quantified by measuring absorbance at 540 nm, reference at 630 nm using a Tecan Infinite M200 plate reader. A commercially available lactate dehydrogenase (LDH) assay, Cytotox 96 (Promega) was also used to assess cell membrane permeability as a measure of viability, according to manufacturers’ instructions, with cells seeded as described for MTT assays.
Results
SiaPG is a highly active, broad specificity sialidase, and desialylates host glycans
The coding sequence of SiaPG without secretion signal sequence was synthesized as a codon-optimized fragment and cloned into a pET21a plasmid. Expression was performed in E. coli BL21 (origami B), and purified via nickel affinity chromatography to high homogeneity (Fig. S1, available with the online version of this article). The commonly used fluorogenic 4-methylumbeliferyl-N-acetyl neuraminic acid (MUNANA) substrate was used to assess sialidase activity with liberated 4-methylumbelliferone (4-MU) measured with fluorescence change per minute used to quantify sialidase activity (Fig. 1a). We observed that SiaPG has optimal activity under acidic conditions, with highest activity observed at a pH of 5.2–5.6. However, it retained 40–50 % activity between pH 4.4 and 7.2, and ~20–30 % retained even at pH 4 and 8. Whole bacterial sialidase activity of P. gingivalis was also assessed, finding that ≥~70 % maximal activity was retained in the pH range 4.8–8.0, with the highest activity observed at pH 6.8, (Fig. 1b). The stronger activity of whole P. gingivalis sialidase in a wider pH range compared to the sialidase protein may be due to a buffering or stabilizing effect of cell membranes or other components and is relevant given that subgingival GCF pH is in the neutral range.
Fig. 1.
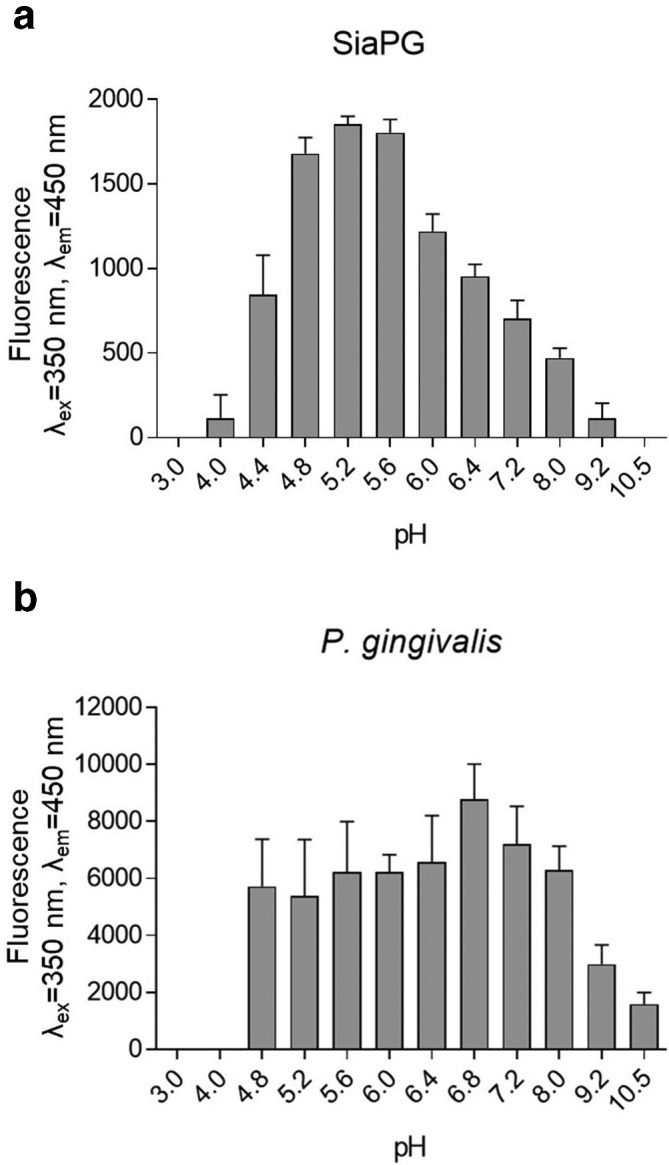
pH optima of P. gingivalis whole cell sialidase activity and purified recombinant SiaPG. MUNANA was incubated with SiaPG for 1 min or P. gingivalis (ATCC 33277) for 1 h, in a variety of buffers with variable pH. Reactions were halted and the pH equalised by addition of an excess of sodium carbonate-bicarbonate buffer, pH 10.5. Sialidase activity catalysed the production of 4-MU from MU-NANA, which was quantified by measuring fluorescence of the reactions at excitation and emission wavelengths of 350 and 450 nm. (a) SiaPG (b) P. gingivalis . Data shown represent the mean of one experiment where each condition was repeated three times. Error bars, sem.
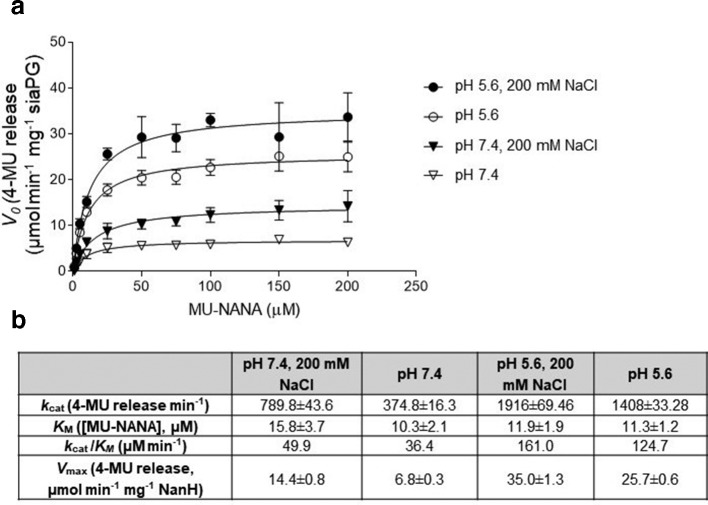
We also performed kinetics experiments to enable comparison of SiaPG to other sialidases. Given the pH variations in the pathogens’ ecological niche, i.e. the conditions the sialidases would encounter in the host, we evaluated sialidase activity at SiaPG-optimum (pH 5.6) as well as physiological pH (7.4) that would be present in the mouth, and in the presence and absence of additional salt (Fig. 2). SiaPG had the greatest catalytic efficiency (k cat/KM) at pH 5.6 in the presence of salt (161.0 µM min−1 with 200 mM NaCl, 124.7 µM min−1 without NaCl). At pH 7.4, salt had a similar impact on catalytic efficiency of SiaPG (49.9 µM min−1 with NaCl, 36.4 µM min−1 without NaCl). These values are broadly in line with those previously reported for the NanH sialidase of T. forsythia [27], though salt had the opposite effect on NanH catalytic efficiency.
Fig. 2.
Reaction kinetics of MUNANA and SiaPG under different conditions. Variable concentrations of MUNANA were exposed to SiaPG, under different pH and salinity conditions. Reactions were quenched by addition of pH 10.5 buffer at 1, 2, and 3 min, and the rate of 4-MU release determined by application of a 4-MU standard curve. (a) Michaelis–Menten plot, rate of 4-MU release (V0, µmol MU released min−1 mg−1 SiaPG), plotted against [MUNANA] (µM) using Prism 7 (GraphPad). Error bars, sd (b) Table summarizing Michaelis–Menten reaction kinetics and catalytic efficiency of SiaPG and MU-NANA, the table includes the k cat (4-MU release min−1), and k cat/KM (µM min−1). Data shown represent the mean (±sd) of one experiment, where each condition was repeated three times per experiment.
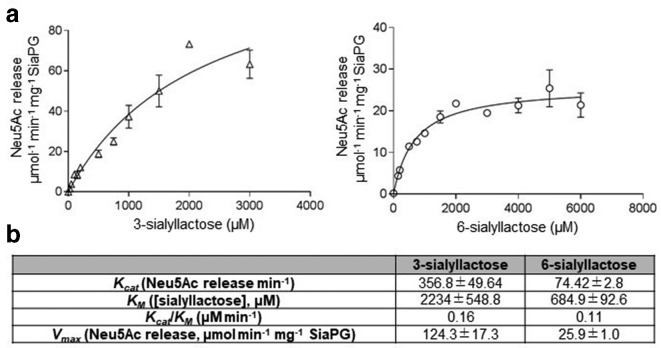
In addition to MUNANA, we assessed the ability of SiaPG to cleave 2–3′- and 2–6′-linked sialic acid, initially with the trisaccharide substrate sialyllactose (3- and 6-sialyllactose), using a TBA assay as previously described for NanH. SiaPG was capable of cleaving both linkage types (Fig. 3), and although it had a much higher affinity (lower KM) for the 2,6 linkage than 2,3, its reaction velocity with the 2,3 version means that it has a~50 % higher catalytic efficiencies (Kcat/KM) for the 3-sialyllactose-linked sugars.
Fig. 3.
Reaction kinetics of 3- or 6-sialyllactose and SiaPG under different conditions. Variable concentrations of sialyllactose were exposed to SiaPG, under conditions mimicking physiological (pH 7.4 200 mM NaCl). Reactions were quenched by commencing the thiobarbituric acid assay at 1, 2 and 3 min, and the rate of Neu5Ac release determined by application of a Neu5Ac standard curve. (a) Michaelis–Menten plot, rate of 4-MU release (V0, µmol MU released min−1 mg−1 SiaPG), plotted against [3- or 6-sialyllactose] (µM) using Prism 7 (GraphPad). Error bars, sem. (b) Table summarizing Michaelis–Menten reaction kinetics and catalytic efficiency of SiaPG and MU-NANA, the table includes the k cat (4-MU release min−1), and k cat/KM (µM min−1). Data shown represent the mean (±sd) of one experiment, where each condition was repeated three times per experiment.
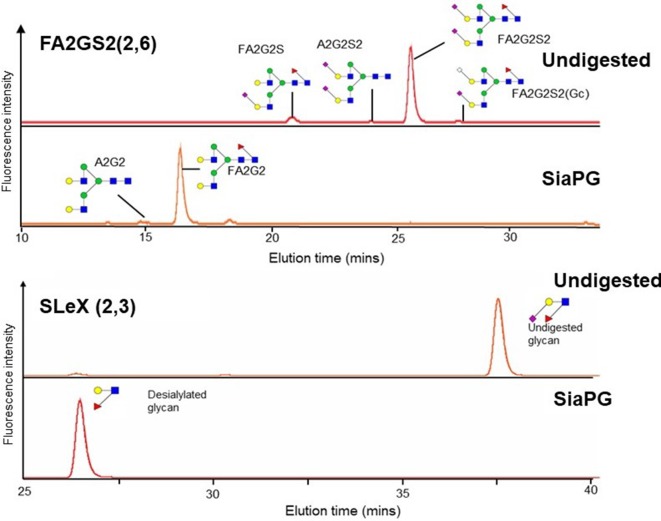
Desialylation of more complex and physiologically relevant glycans by SiaPG was also assessed using HPLC and mass spectrometry. SLeX is an oligosaccharide containing α2–3-linked sialic acid, and is present in numerous host niches – on the plasma membrane of various cell types and at the termini of secreted glycans. SLeX may also play a role in P. gingivalis pathogenesis [31]. We exposed soluble SLeX to SiaPG, subjected the reaction to HPLC, and found that SLeX was efficiently desialylated by SiaPG (Fig. 4a). Cleavage of α2–6-linked sialic acid by SiaPG was tested using the host-relevant FA2G2S2 (also known as A2F) glycan, a two-branched polysaccharide chain where both branches terminate with a sialic acid, which is present on a number of secreted glycoproteins including IgG. SiaPG was again capable of desialylating FA2G2S2 (Fig. 4b). Notably, other peaks were observed in the chromatogram for FA2G2S2, which are likely to correspond to differently sialylated versions of the FA2G2S2 glycan, and these also appeared to be desialylated – notably the mammalian NeuGc versions. It would be interesting to test the activity of SiaPG against such NeuGc-linked glycoconjugates, which the human host does not synthesize but obtains from dietary sources.
Fig. 4.
Desialylation of host-relevant glycans by SiaPG. UHPLC chromatogram showing elution of FA2G2S2 and SLeX with and without digestion by SiaPG. FA2G2S2 contains α2–6-linked Neu5Ac. Some contaminating glycans can be observed in the undigested sample, but these also appear to be desialylated. Gc=N-glycolylneuraminic acid. SLeX contains α2–3-linked Neu5Ac.
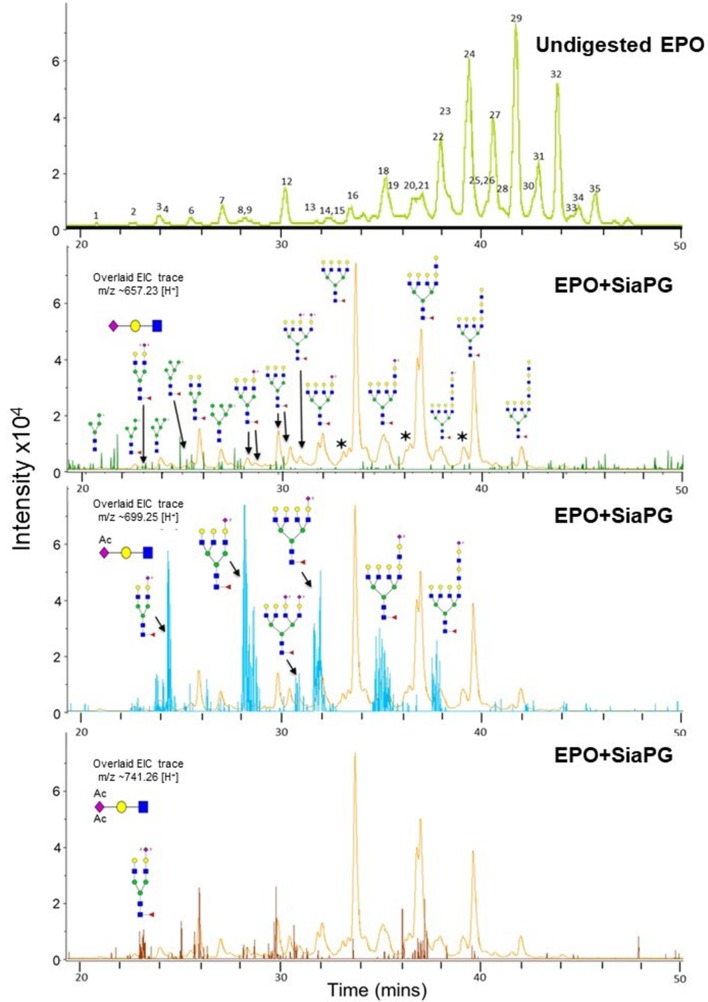
Finally, we wanted to test desialylation of glycans derived from a complex sialoglycoprotein by SiaPG. To this end, we chose the host glycoprotein erythropoietin (EPO) due to its possession of multiple bi-, tri- and tetra-branched glycans, the presence of α2–3 and α2–6 sialic acid linkages, and because some of its NeuAc is mono or di-O-acetylated (Neu5,9Ac, and perhaps tri-acetylated Neu5,8,9Ac). EPO was digested with SiaPG, N-glycans were procainamide labelled, and these underwent UHPLC-MS/MS (Fig. 5) . As expected, the UHPLC chromatogram of undigested EPO showed the presence of multiple peaks corresponding to various EPO glycans, further details and structures of these are shown in Table S1. Following digestion with SiaPG, we observed a marked change in the chromatogram. The remaining peaks were investigated by MS/MS, providing evidence that SiaPG was able to cleave the sialic acid from a wide-range of glycan structures. However, this analysis also revealed the inability of SiaPG to cleave diacetylated (or triacetylated) sialic acids (Fig. 5, lower two panels). These analyses showed that extracted ion chromatogram (EIC) corresponding to the MS2 ion for Neu5Ac trisaccharide (GlcNAc-Gal-Neu5Ac, m/z=~657.25) was not detected above background levels, while EIC corresponding to di- and tri-acetylated sialic acid trisaccharides (GlcNAc-Gal-Neu5,9Ac and GlcNAc-Gal-Neu5,8,9Ac, m/z=~699.25 and ~741.26) were detected in high abundance (high peak intensity) – i.e. had been left uncleaved by SiaPG; a result expected given our previous studies on NanH [26].
Fig. 5.
SiaPG cleaves α2-3 and α2-6 linked Neu5Ac from erythropoietin (EPO), though diacetylated sialic acid is retained. EPO was digested by SiaPG for 18 hours at 37 °C, and subjected to UHPLC-MS/MS. A deviation of MS2 ion m/z of±0.2 was permitted during spectral assignment. Sialylated structures may possess different isomers to those depicted (i.e. it is uncertain which terminal galactose residue(s) in the branched chains are sialylated). The top panel shows the UHPLC chromatogram for undigested EPO, the numbers indicate different EPO glycans, structures and further details of which are shown in Table S1. Lower panels show the UHPLC chromatogram for SiaPG digested EPO (orange), with three overlays showing MS1 ions which when fragmented produced MS2 ions with m/z corresponding to the precursor ion for Neu5Ac trisaccharide (GlcNAc-Gal-Neu5Ac, m/z=~657.25, green overlay),diactylated sialic acid trisaccharide (GlcNAc-Gal-Neu5,9Ac, m/z=~699.25, blue overlay, middle), and triacetylated sialic acid trisaccharide (GlcNAc-Gal-Neu5,8,9Ac, m/z=~741.25,brown overlay, lower).
SiaPG releases Neu5Ac from mucins, an activity enhanced by the sialate-O-acetylesterase NanS from T. forsythia
To explore activity of SiaPG further we next tested its activity with a glycoprotein substrate relevant in the oral cavity – salivary mucin. Notably the commonly used bovine submaxillary mucin (BSM) used in these experiments contains many terminal sialic acids that have the second O-acetyl group, i.e. contain Neu5,9Ac [32]. Our previous data on NanH [26] and SiaPG (this study) indicate terminal Neu5,9Ac is resistant to cleavage by periodontal sialidases. To counter this, some bacteria express sialate-O-acetylesterases, such as NanS from T. forsythia , which remove the second acetylation (forming Neu5Ac), and rendering the sialic acid susceptible to sialidase cleavage. Indeed, we previously demonstrated that T. forsythia NanS enhances NanH sialidase activity on Neu5,9Ac terminal sugars and may act to aid sialidases from other species in harversting sialic acid [26]. To test the hypothesis that T. forsythia may act to enhance SiaPG activity we performed sialic acid release assays on BSM. The data showed that NanS enhanced sialic acid release by SiaPG from BSM by ~2.5-fold (from 302 to 746 pmol min−1) (Fig. 6).
Fig. 6.
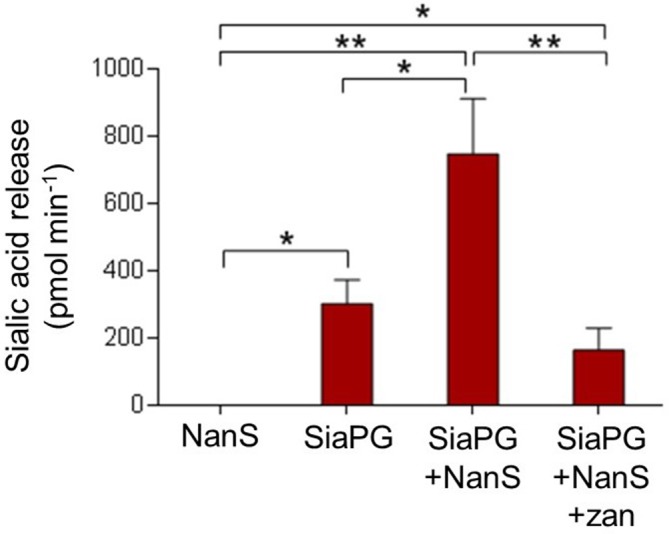
Enzyme synergy in sialic acid release from BSM, and inhibition by zanamivir. BSM was incubated with either NanS, SiaPG, or NanS+SiaPG, in thepresence or absence of 0.5 mM zanamivir, before undergoing the TBA assay to assess sialic acid release. Data based on the mean of three experiments, each condition was tested in triplicate during each experiment. Error Bars, sem. Significance determined by one-way ANOVA with repeated measures, with Bonferroni correction for multiple comparisons (*P=<0.05, **P=<0.01).
Zanamivir more efficiently inhibits sialidase activity of SiaPG and whole P. gingivalis than sialidase activity of T. forsythia NanH and whole T. forsythia
As outlined, one aim of this study was to assess the ability of a safe and FDA-approved drug in its ability to potentially reduce virulence of the keystone periodontal pathogen, P. gingivalis . Therefore we assessed the impact of zanamivir on the activity of whole P. gingivalis and another periodontal pathogen, T. forsythia , as well as their purified sialidases using the MUNANA assay as described above. Both pathogens and purified sialidases displayed decreased sialidase activity as zanamivir concentration was increased (Fig. 7). The decrease in activity was far more drastic for P. gingivalis , with a decrease in activity of ~70 % in the presence of 10 mM zanamivir, compared to T. forsythia, which only showed a decrease of ~25 % (Fig. 7a). This apparent difference in efficacy for the two sialidases was confirmed by establishing the IC50 of zanamivir for purified SiaPG and NanH (Fig. 7b), where zanamivir displayed a greater inhibitory effect on SiaPG (369 µM) than NanH (6130 µM). These data were further enhanced when we observed that zanamivir also inhibited sialidase activity in the context of removal of sialic acid from mucin (Fig. 6). There was no effect on NanS sialate-O-acetylesterase activity alone (data not shown).
Fig. 7.
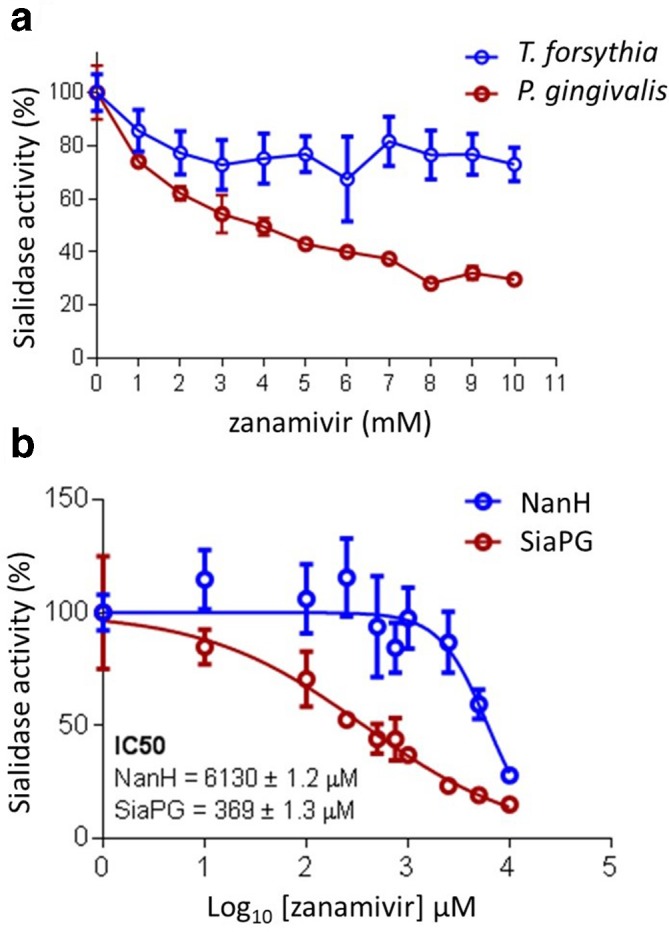
Inhibition of periodontal pathogen sialidase activity by zanamivir. MUNANA was exposed to periodontal pathogens or purified sialidases. Sialidase activitywas expressed as the difference in 4-MU fluorescence relative to conditionswith no inhibitor. (a) MUNANA was exposed to T. forsythia or P. gingivalis in the presence of zanamivir for 1 and 4 hrs, respectively. Data represent themean of two experimental repeats, where each condition was performed three times per experiment. Error bars, sd. (b) MUNANA was exposed to SiaPG and NanHin the presence or absence of zanamivir (1-10 mM, 1 mM increments), and the IC50 of zanamivir for both enzymes obtained using the variable slope model in Graphpad Prism 7. Data represent the mean of three experimental repeats, where each condition was repeated three times. Error bars, sem.
P. gingivalis biofilms on host sialoglycans are sialidase-dependent and disrupted by zanamivir
P. gingivalis was cultured in microtitre plates, where well surfaces were coated with mucin (BSM), saliva (pooled human saliva) or serum (FCS) in the presence or absence of 10 mM zanamivir. Total growth (Fig. 8a) of P. gingivalis in the mucin-coated condition was significantly reduced in the presence of zanamivir, from OD600 0.21 to 0.11 (P=<0.001), i.e. zanamivir reduced total growth of P. gingivalis in mucin-coated conditions by approximately half. In the presence of the other glycoprotein-coated conditions (saliva and serum) there was a trend towards small reductions in total growth in the presence of zanamivir but these did not reach statistical significance. Biofilm formation (normalized to total growth-optical density) of P. gingivalis (Fig. 8b) was also reduced when grown on mucin-coated surfaces (30–40 % reduction, P=<0.01), indicating that biofilm formation on these surfaces was inhibited by zanamivir. Notably we also saw a consistent small reduction on serum, but total growth (OD600) standardized to biofilm formation (bacteria ml−1) of cultures grown on human saliva-coated surfaces was too variable to make conclusions.
Fig. 8.
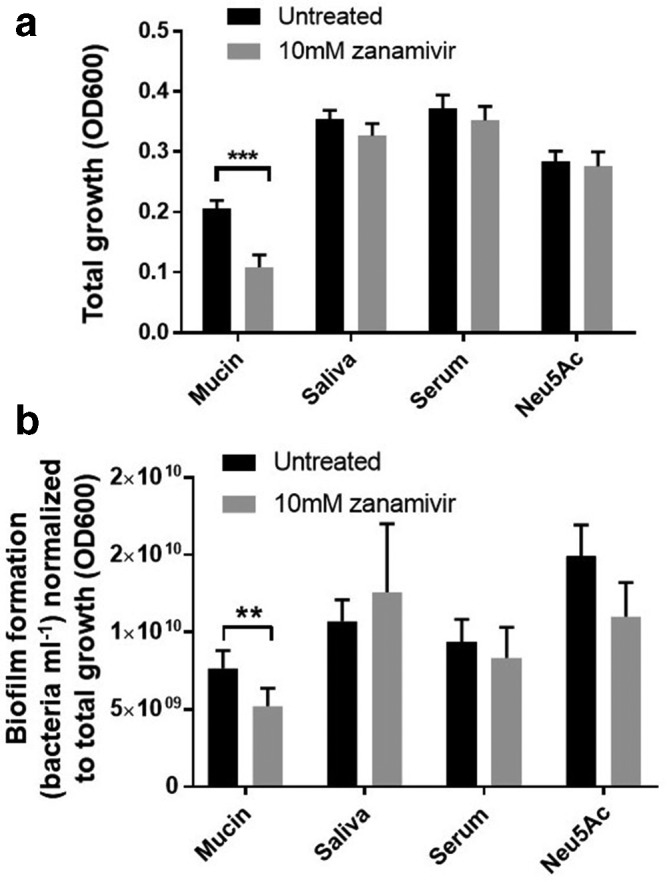
Zanamivir inhibits P. gingivalis growth and biofilm formation on sialoglycoproteins. Bacteria were cultured for 5 days at 37 °C anaerobically, in the presence of Neu5Ac, or on surfaces coated with the glycoproteins mucin, saliva or serum. All conditions were performed in the presence or absence of zanamivir. (a) Total growth; OD600 of the culture was used to quantify bacteria in both biofilm and planktonic states. (b) Biofilm formation, quantified by resuspension of biofilms and counting the number of bacteria under microscopy. All data shown represent the mean of three biological repeats, where conditions were tested three times per experiment. Error bars, sem. Significance determined by t-test, **P=>0.01, ***P=>0.005.
In support of our evidence that sialidase is important when P. gingivalis grows on mucin-coated surfaces (as would often be the case in vivo), we also tested the effect of sialidase activity in this regard in a related P. gingivalis strain – 381 (Pg381). For this purpose, a siaPG deletion mutant of P. gingivalis 381 and its complemented strain were constructed (Pg381ΔsiaPG and ΔsiaPG+). Pg381ΔsiaPG showed no sialidase activity as judged from lack of fluorescence on incubation with MUNANA, while the silaidase activity was restored in the complemented strain ΔsiaPG+ (Fig. S2). In biofilm assays, we observed a twofold reduction in biofilm growth in Pg381ΔsiaPG (relative to the WT) on mucin-coated surfaces, with this phenotype complemented by provision of the SiaPG gene in the mutant (ΔsiaPG+, Fig. 9). These data expand our view of the role of sialidase to a range of P. gingivalis strains and suggest it may be true across this species.
Fig. 9.
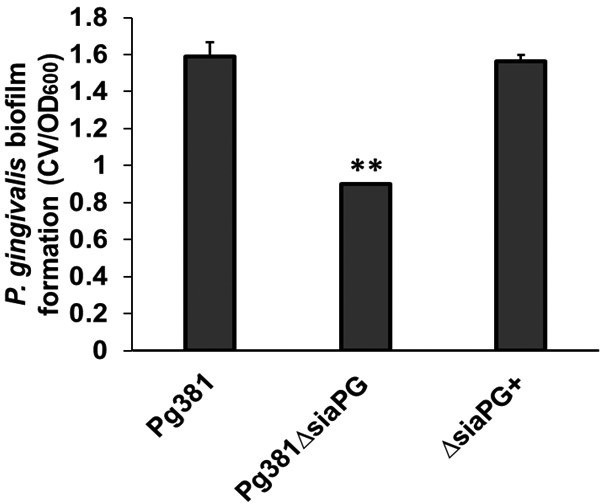
Biofilm formation by P. gingivalis 381, sialidase-deficient Pg381ΔsiaPG, and SiaPG complemented strain ΔsiaPG+. P. gingivalis strains were resuspended to an OD600 0.5, and seeded into microtitre plates precoated with mucin (BSM), and biofilms were cultured for 24 h at 37 °C anaerobically. Data represent the mean of four replicates per condition. Error bars, sd, statistical significance determined by unpaired t-test compared to the WT strain, **P=<0.01.
SiaPG releases 3- and 6-linked sialic acid from oral epithelial cells and is inhibited by zanamivir
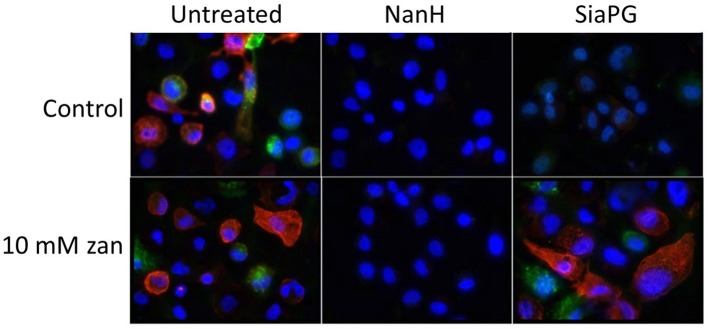
Studies of mutant strains and purified enzymes have shown that the well-studied T. forsythia NanH can desialate oral epithelial cells [22, 27], while SiaPG mutant strains have been used to highlight P. gingivalis sialidase-mediated release of sialic acid from erythrocytes [18]. In order to test the effect of SiaPG on physiologically relevant oral epithelial cells, we stained the OSCC line H357 with the lectins SNA and MAA enabling fluorescent imaging of cell surface α2–3- and α2–6-linked sialic acid. Exposure of epithelial cells to SiaPG (and T. forsythia NanH) was successful in desialylating cell surfaces (this had been shown previously for NanH [22]), while 10 mM zanamivir inhibited desialylation in the case of SiaPG, but not NanH (Figs 10, and S3).
Fig. 10.
Purified sialidases desialylate oral epithelial cell surfaces, and in the case of SiaPG this is inhibited by zanamivir. Cells were stained with lectins for α2–3- and α2–6-linked sialic acid in red and green, respectively. Prior to staining, cells were treated with NanH and SiaPG in PBS, in the presence or absence of 10 mM zanamivir (zan), as indicated. All images were visualized using the same microscopy and image-processing parameters (fluorescence intensity, exposure time and background subtraction). Images were captured in three fields of view, and this was repeated in three separate experiments. Images shown are representative of each condition. Separate colour channels are displayed in the Supplementary Material (Fig. S3).
Zanamivir inhibits periodontal pathogen–host-cell association during monospecies infection
One established aspect of P. gingivalis virulence is its ability to invade and reside within oral epithelial cells. To this end, we performed antibiotic protection assays to establish whether zanamivir had any potential effect on host-cell interaction and invasion. However, the effects of zanamivir on bacterial and human cell viability had to be established. To this end, oral epithelial cells were subjected to 10 mM zanamivir under antibiotic protection assay conditions, illustrating no effect of zanamivir on either membrane integrity (assayed by LDH) or cellular activity/viability (via MTT assay) (Fig. S4). Similarly, we incubated P. gingivalis , T. forsythia and F. nucleatum with zanamivir under antibiotic protection assay conditions (2.5 h in tissue culture medium), before obtaining c.f.u. counts, and revealed no detrimental effect of zanamivir on viability of any of these species (Fig. S5).
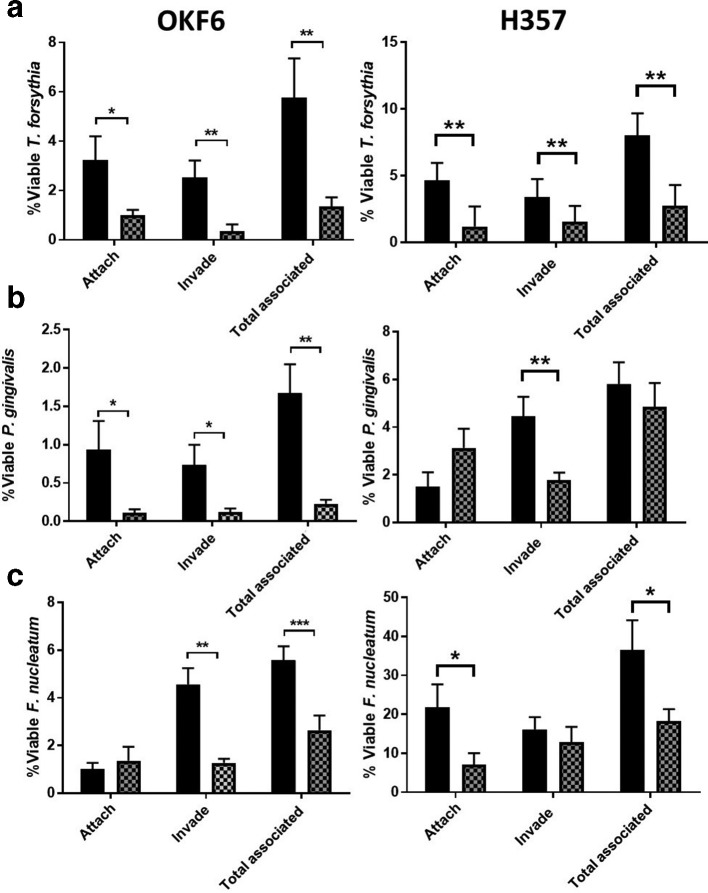
Next we performed antibiotic protection assays on two oral epithelial cell lines, namely the OSCC cell line H357 – a frequently used model for oral epithelium–bacteria interactions in our laboratory [33, 34] and normal immortalized oral epithelial cell line OKF6 [23, 35]. In addition to P. gingivalis we also tested the effect of zanamivir on two other key periodontal pathogens – namely T. forsythia and F. nucleatum subsp. nucleatum . For all three pathogens, in both cell lines, zanamivir significantly reduced one or more aspects of host–bacteria association in both cell lines (Fig. 11). For T. forsythia , attachment, invasion and total association (the sum of attached and invaded bacteria) were all significantly reduced in both cell lines; from 3.2 to 1.0 %, 2.5 to 0.3 %, and 5.8 to 1.4 % in OKF6 cells, and 4.6 to 1.2 %, 3.4 to 1.6 % and 8.0 to 2.8 % in H357 cells (Fig. 11a). For P. gingivalis , attachment, invasion and total association were significantly reduced in OKF6 cells; from 0.9 to 0.1 %, 0.7 to 0.1 %, and 1.7 to 0.2 %, however in the H357 cell line, only invasion was significantly reduced, from 4.5 to 1.8 % (Fig. 11b). Finally, for F. nucleatum , invasion and total association were significantly reduced in OKF6 cells; from 4.6 to 1.3 %, and 5.6 to 2.6 %. For H357 cells attachment and total association were significantly reduced 21.8 to 7.2 %, and 36.6 to 18.2 % (Fig. 11c).
Fig. 11.
The effect of zanamivir on attachment and invasion of epithelial cells during monospecies infection. Antibiotic protection assays on OKF6 and H357 cell lines were performed in the presence or absence of 10 mM zanamivir (thatched and black bars, respectively) with either (a) T. forsythia , (b) P. gingivalis or (c) F. nucleatum . Bacterial attachment, invasion and total association with host cells was normalized to the number of bacteria that were used to infect each condition that had survived the duration of the assay (the percentage of viable bacteria). Data represent the mean from three independent experimental repeats, and each condition was repeated in triplicate during each experiment. Error bars, sem, Significance determined by paired t-test, *P=<0.05, **
To support the notion that sialidase activity is important for bacteria–host-cell association, rather than disruption of other processes by the presence of zanamivir, the human gingival cell line OBA-9 was exposed to P. gingivalis 381 (Pg381) and a siaPG-deficient mutant (Pg381ΔsiaPG). Exposure to the latter strain was performed without or with exogenous SiaPG . Pg381ΔsiaPG displayed significantly decreased association with host cells relative to the parent Pg381, which was restored by the presence of exogenous SiaPG in a concentration-dependent manner (Fig. S6). While 100 µg ml−1 exogenous recombinant SiaPG resulted in partial restoration of host–bacteria association of Pg381ΔsiaPG (WT; 0.50 %, Pg381ΔsiaPG ; 0.15 %, Pg381ΔsiaPG+100 µg SiaPG ; 0.26), 200 µg ml−1 SiaPG resulted in complete restoration of host–bacterial association Pg381 (0.58 %).
Zanamivir inhibits attachment and invasion of oral epithelial cells during infection with multiple periodontal pathogens
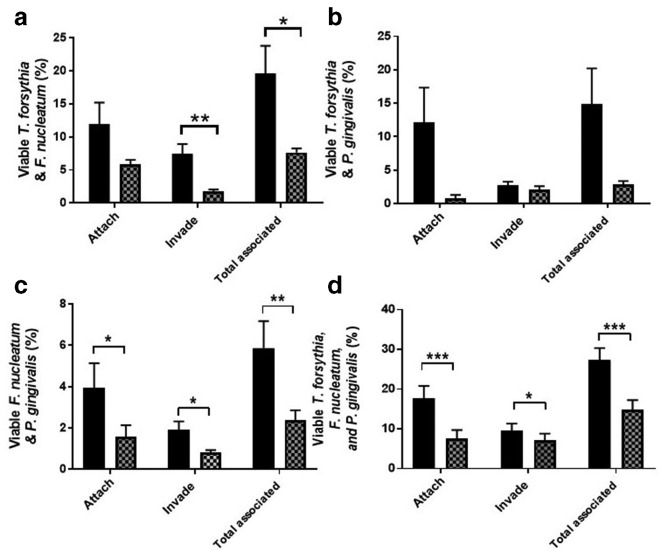
To at least partially mimic the in vivo situation, where none of these species exist in isolation, we performed antibiotic protection assays where multiple periodontal pathogens were used to infect cells. This is particularly relevant during host-cell association, since periodontal pathogens have been shown to act in synergy during attachment and invasion [36–38]. m.o.i. was maintained at 1 : 100 host cells: bacteria, so for infections using two species, the ratio was 1 : 50 : 50 (host cells: species 1: species 2), and for infections with all three species, the ratio was 1 : 33 : 33 : 33 (host cells: species 1: species 2: species 3). T. forsythia, P. gingivalis and F. nucleatum were used in a variety of combinations to infect epithelial cells. In addition to quantifying the numbers of all bacteria – not distinguishing between species during an assay – the levels of each individual species in a given assay could be enumerated by colony counting on agar plates, since the colony morphologies of T. forsythia , F. nucleatum and P. gingivalis are distinct from each other (Fig. S7). All possible combinations were tested in this way. For ease of discussion, only the total number of bacteria in each combination are described here, with complete results for each individual species provided in Figs S8–S11. For T. forsythia and F. nucleatum co-infection, zanamivir significantly reduced both invasion and total association of bacteria with host cells, from 7.4 to 1.8 %, and 19.6 to 7.6 % (P<0.01 and P<0.05), respectively (Fig. 12a). For T. forsythia and P. gingivalis co-infection, the variation between experiments meant that none of the reductions were statistically significant (where P<0.05, Fig. 12b), i.e. attachment was reduced from 12.1 to 0.8 % (P=0.076), invasion from 2.8 to 1.4 % (P=0.25), and total association from 14.4 to 2.5 % (P=0.062). For F. nucleatum and P. gingivalis , zanamivir significantly reduced attachment, invasion and total association of bacteria with host cells, from 1.9 to 0.8 % (P=<0.05), 4.0 to 1.56 % (P=<0.05) and 5.9 to 2.4 % (P=<0.01), respectively (Fig. 12c). Finally, all three species (i.e. T. forsythia, F. nucleatum and P. gingivalis ) were infected into the cells as a three-species community. Here, strikingly, zanamivir significantly reduced attachment, invasion and total association of bacteria with host cells, from 17.7 to 7.7 % (P=<0.001), 9.6 to 7.15 %, (P=<0.05) and 26.14 to 14.6 % (P=<0.001), respectively (Fig. 12d); indicating this approach might have an impact on cellular interaction in vivo.
Fig. 12.
Effect of zanamivir on attachment and invasion of epithelial cells during multispecies infection. Antibiotic protection assays were performed in the presence or absence of 10 mM zanamivir (thatched and black bars, respectively) with different combinations of T. forsythia, F. nucleatum and P. gingivalis to establish host-cell association in each instance. (a) T. forsythia+F. nucleatum. (b) T. forsythia +P. gingivalis. (c) F. nucleatum+P. gingivalis. (d) T. forsythia, F. nucleatum and P. gingivalis . Bacterial attachment, invasion and total association with host cells was normalized to the number of bacteria that were used to infect each condition that had survived the duration of the assay (the percentage of viable bacteria). Zanamivir=10 mM zanamivir present during host-cell exposure to bacteria. Data represent the mean from three experimental repeats, and each condition was repeated in triplicate during each experiment. Error bars, sem, Significance determined by paired t-test, *P=<0.05, **P=<0.01. ***P=<0.001.
Discussion
In this study, we characterized the activity of the sialidase (SiaPG) from P. gingivalis on a number of glycan and proteinaceous substrates, including relevant host sialoglycans SleX, FA2G2S2 and EPO, and established activity across a range of pH values. During these studies, it became apparent that SiaPG could cleave both α2–3- and α2–6-linked Neu5Ac, but not diacetylated Neu5,9Ac. These data also agreed with low sialic acid release from the heavily diacetylated BSM protein, with this activity being enhanced by coincubation with an enzyme capable of removing O-acetyl groups on sialic acid residues – namely the NanS sialate-O-acetylesterase from the fellow oral bacterium T. forsythia [26]. We hypothesize that in vivo NanS enhances the ability of P. gingivalis to cleave sialic acid from glycoproteins – although given the fact that P. gingivalis does not produce its own esterase or utilize sialic acid for nutrition we can only postulate that somehow removal of sialic acid opens up sites for adhesion to oral surfaces, or might modulate immune responses mediated by Siglecs (sialic acid-binding immunoglobulin-like lectins) or allow P. gingivalis to access underlying sugars or the protein component of the glycoproteins themselves, and may explain synergies between these organisms observed by others [39–41].
We tested the ability of zanamivir to inhibit SiaPG and the previously characterized NanH sialidase from another red-complex pathogen, T. forsythia . While the inhibition (IC50) of SiaPG activity on MUNANA was determined to be within the micromolar range (~350 µM), this was not the case for T. forsythia , which required >6 mM zanamivir to reduce its activity by half. A similar trend was also seen for whole bacteria. The relatively low efficacy of zanamivir for some bacterial sialidases is observed in other studies where sialidases from the pathogens S. pneumoniae and Gardnerella vaginalis are apparently only inhibited to limited extents [42, 43]. Nevertheless, here we showed that P. gingivalis sialidase was effectively inhibited by zanamivir. At present we have no mechanistic explanation for the variation between the two sialidases, but postulate it may result from structural differences, despite both possessing typical catalytic GH33 domains with predicted six-blade propeller structures the sialidases only share 23 % amino acid sequence identity.
Given this, and the ability of SiaPG to cleave sialic acid from complex glycans (FA2G2S2, EPO), we assessed the ability of SiaPG to remove sialoglycans from oral epithelial cells, illustrating removal in both cases and highlighting the broad glycan specificity of this enzyme and the ability of zanamivir to affect this process. As shown previously, NanH was also capable of desialylating oral epithelial cells [22].
Considering the ability of zanamivir to inhibit P. gingivalis sialidase activity, in vitro virulence models were carried out. It has been shown previously that NanH is key during biofilm formation in T. forsythia , and that sialidase inhibitors are also detrimental to T. forsythia biofilms [44]. We observed that sialidase inhibitors decreased P. gingivalis biofilm formation on host glycoprotein sources, significantly on saliva and mucin. In addition, biofilm formation of sialidase-deficient P. gingivalis on mucin was reduced compared to its parent, and restored upon complementation. Biofilms of sialidase-deficient P. gingivalis are also reduced on plastic surfaces [17]. There are various mechanisms by which sialidase activity may contribute to P. gingivalis biofilm formation. Possibilities include decreased proteolysis and subsequent nutrient deficiency or an inability to attach to cleaved proteins, since sialylation may protect against proteolytic activity [45]. Interference with the P. gingivalis capsule is also possible, sialidase expression appears to be important for P. gingivalis capsular synthesis, mutants deficient in SiaPG show reduced capsule formation [17, 18], and yet loss of the capsule has been shown to enhance biofilm formation [46], thus, we might expect zanamivir (or sialidase knockout) to enhance biofilm formation, but here we observed the opposite, possibly because non-capsulated (fimbriated) strains were tested. In any case, we cannot rule out the possibility that sialidase inhibition may interfere with capsular structure or interactions in the case of P. gingivalis strains, which are encapsulated. Finally, maturation and processing of gingipains – proteases central to P. gingivalis virulence and asacharolytic nutrition – is also altered in sialidase-deficient mutants [18]. Zanamivir may impact biofilm formation through any, or all, of these mechanisms. It would be particularly interesting to assess the impact of zanamivir on mixed-species biofilms, and in addition to direct impacts on sialic acid-mediated interactions and any indirect effects that may result from sialidase inhibition. For example, it has been previously shown that lysine gingipains (Kgp) of P. gingivalis mediate T. forsythia levels in mixed-species biofilms [47], and given that sialylation may influence protease access to protein backbones, inhibition of P. gingivalis sialidase may also impact the activity of Kgp, subsequently reproducing the detrimental effect of Kgp deficiency on T. forsythia abundance. Similarly, it has been noted that T. forsythia surface S-layer glycans play a potential role in localization with P. gingivalis within polymicrobial biofilms [48]. These glycans contain terminal nonulosonic analogues of sialic acid (pseudaminic and legianaminic acids) and it is possible that zanamivir may influence these interactions as a glycan mimic.
Given that periodontal pathogen sialidases have been shown to be important during host-cell association [17, 18, 22, 44] and zanamivir inhibited host-cell surface desialylation by SiaPG, but not T. forsythia NanH, it might be expected that zanamivir would decrease attachment and invasion of host cells by P. gingivalis but not T. forsythia . Surprisingly, host-cell association was greatly inhibited in the case of both organisms, and in the sialidase-negative F. nucleatum [13]. Of note here is that while our data re-emphasize the role of sialidase in the equivalent P. gingivalis capsulated W83 strain [18], indicating that this phenotype is widespread in P. gingivalis, it would be of interest to establish whether zanamavir has the same effect on capsulated strains. It is also possible that other periodontal pathogen virulence factors are inhibited by zanamivir. For example, the adhesin FadA is highly conserved among oral fusobacteria, and considered important for association with oral epithelial cells [49] and has been shown to bind cell surface E-selectin, whose native ligand is a sialoglycan (sialyl lewis A/X), so perhaps a sialic acid analogue might interfere with FadA-host-cell surface interactions.
Although the monospecies antibiotic protection assays hinted at the usefulness of zanamivir as an anti-virulence therapeutic, periodontitis is mediated by a dysbiotic polymicrobial community. Therefore, we performed mixed-species antibiotic protection assays with combinations of all three of the periodontal pathogens from the monospecies antibiotic protection assays. Importantly, zanamivir influenced invasion in the triple-species infection model with F. nucleatum , P. gingivalis and T. forsythia . Despite evidence of synergy in invasive capability reported in the literature [36, 38, 50] we were surprised zanamivir effected all species in these assays, given that F. nucleatum has no sialidase and T. forsythia sialidase is only weakly inhibited by zanamivir. One explanation might be that only a modest reduction in sialidase activity can reduce invasion for T. forsythia . However, zanamivir acts on host sialidases, in particular Neu1 and Neu3, which contribute to the immune response to bacteria via TLRs, where host sialidases are mobilized to the plasma membrane where they activate TLRs 2-, 3- and 4- [51–53], and zanamivir (and other sialidase inhibitors) has been shown to inhibit TLR activation by LPS [54]. Inhibition of host-cell sialidases by zanamivir may therefore affect the cell-surface charge and glycosylation state of receptors involved in bacterial adhesion and invasion. Indeed our data may suggest that there are host and bacterial sialidase-dependent events that not only warrant further study but make a strong case for the potential of these inhibitors to affect periodontal disease pathogenesis, since they may have dual action on host and bacterial processes.
Conclusion
In summary, we further characterized the sialidase of P. gingivalis , and provided evidence advocating for the development of sialidase inhibitors as therapeutics for periodontitis – and other diseases – where bacterial sialidases play key roles in virulence. Zanamivir was capable of inhibiting sialidase activity and virulence mechanisms of P. gingivalis , most prominently host-cell association. However, our study raises several questions and directions for further work, most notably that the host as well as bacterial sialidase may influence interactions more than currently understood. Ultimately, sialidase inhibition represents a potential novel therapy for periodontal and other diseases, and merits further investigation. Finally, zanamivir is a safe and efficacious drug for treatment of influenza, but clearly only inhibits bacterial sialidases relatively weakly (µM–mM range). However, its effectiveness in this study suggests that the development of more potent and selective inhibitors of bacterial sialidase should be a focus in upcoming years as they may hold potential as a safe and efficacious treatment in periodontal and other diseases.
Supplementary Data
Funding information
A.F. and M.S. were funded by a BBSRC iCASE studentships (BB/K501098/1, BB/M01570X/1), while C.P. was funded by a grant from the Dunhill Medical Trust to G.S. (DMT-R185/0211). We acknowledge the funding form NIH grant DE14749 to A.S. and William M. Feagans Endowed Chair Research Fund, University at Buffalo School of Dental Medicine to K.H.
Author contributions
A.F., G.S., J.P. and D.B. devised the concept of the work. A.F., M.S., P.U., D.S., K.H. and C.P. performed aspects of lab analysis. A.F., G.S. and A.S. drafted and wrote the manuscript. All authors contributed to editing of the paper.
Conflicts of interest
We declare no conflicts of interest, besides the provision of zanamivir by GlaxoSmithKline (GSK), Stevenage, UK, and A. F. was sponsored by a BBSRC iCASE studentship (BB/K501098/1) in partnership with GSK.
Footnotes
Abbreviations: 2-AB, 2-aminobenzamide; BSA, Bovine serum albumin; BSM, Bovine submaxillary mucin; CHO, Chinese hamster ovary; EGF, Epidermal Growth Factor; EIC, Extracted ion chromatogram; EPO, Erythropoietin; FCS, Fetal calf serum; LDH, Lactate dehydrogenase; MAA, Lectin from Maackia amuriensis; MTT, 3-(4,5-D dimethylthiazol-2-yl)−2,5-D diphenyltetrazolium Bromide; 4-MU, Methylumbelliferone; MU-NANA, Methylumbelliferone-N-acetylneruaminic acid; NAM, N-acetylmuramic acid; PBS, Phosphate buffer saline; PNGase, Peptide N-glycosidase; sd, Standard deviation; SDS, Sodium dodecyl sulfate; sem, Standard error of the mean; SiaPG, Sialidase from Porphyromonas gingivalis; SLeA, Sialyl Lewis A; SLeX, Sialyl Lewis X; SNA, Lectin from Sambucus nigra; TBA, Thiobarbituric acid.
Eleven supplementary figures and one supplementary table are available with the online version of this article.
Edited by: D. Grainger and M. Welch
References
- 1.Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012;10:717–725. doi: 10.1038/nrmicro2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meuric V, Le Gall-David S, Boyer E, Acuña-Amador L, Martin B, et al. Signature of microbial dysbiosis in periodontitis. Appl Environ Microbiol. 2017;83:1–13. doi: 10.1128/AEM.00462-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Socransky SS, Cugini MA, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051X.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 4.Ximénez-Fyvie La, Haffajee AD, Socransky SS. Comparison of the microbiota of supra- and subgingival plaque in health and periodontitis. J Clin Periodontol. 2000;27:648–657. doi: 10.1034/j.1600-051x.2000.027009648.x. [DOI] [PubMed] [Google Scholar]
- 5.Muniz FWMG, de Oliveira CC, de Sousa Carvalho R, Moreira MMSM, de Moraes MEA, et al. Azithromycin: a new concept in adjuvant treatment of periodontitis. Eur J Pharmacol. 2013;705:135–139. doi: 10.1016/j.ejphar.2013.02.044. [DOI] [PubMed] [Google Scholar]
- 6.Colombo APV, Boches SK, Cotton SL, Goodson JM, Kent R, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Löhr G, Beikler T, Hensel A. Inhibition of in vitro adhesion and virulence of Porphyromonas gingivalis by aqueous extract and polysaccharides from Rhododendron ferrugineum L. a new way for prophylaxis of periodontitis? Fitoterapia. 2015;107:105–113. doi: 10.1016/j.fitote.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 8.Kariu T, Nakao R, Ikeda T, Nakashima K, Potempa J, et al. Inhibition of gingipains and Porphyromonas gingivalis growth and biofilm formation by prenyl flavonoids. J Periodontal Res. 2017;52:89–96. doi: 10.1111/jre.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huq NL, Seers CA, Toh ECY, Dashper SG, Slakeski N, et al. Propeptide-mediated inhibition of cognate gingipain proteinases. PLoS One. 2013;8:e65447. doi: 10.1371/journal.pone.0065447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominy SS, Lynch C, Ermini F, Benedyk M, Marczyk A, et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 2019;5:eaau3333. doi: 10.1126/sciadv.aau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker D, Soong G, Planet P, Brower J, Ratner AJ, et al. The nana neuraminidase of Streptococcus pneumoniae is involved in biofilm formation. Infect Immun. 2009;77:3722–3730. doi: 10.1128/IAI.00228-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Galen JE, Ketley JM, Fasano A, Richardson SH, Wasserman SS, et al. Role of Vibrio cholerae neuraminidase in the function of cholera toxin. Infect Immun. 1992;60:406–415. doi: 10.1128/iai.60.2.406-415.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stafford G, Roy S, Honma K, Sharma A. Sialic acid, periodontal pathogens and Tannerella forsythia: stick around and enjoy the feast! Mol Oral Microbiol. 2012;27:11–22. doi: 10.1111/j.2041-1014.2011.00630.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palmer RJ. Composition and development of oral bacterial communities. Periodontol 2000. 2014;64:20–39. doi: 10.1111/j.1600-0757.2012.00453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey AM, Ansbro K, Kamble NS, Pham TK, Stafford GP. Characterisation and pure culture of putative health-associated oral bacterium BU063 (Tannerella sp. HOT-286) reveals presence of a potentially novel glycosylated S-layer. FEMS Microbiol Lett. 2018;365:1–8. doi: 10.1093/femsle/fny180. [DOI] [PubMed] [Google Scholar]
- 16.Roy S, Honma K, Douglas CWI, Sharma A, Stafford GP, et al. Role of sialidase in glycoprotein utilization by Tannerella forsythia . Microbiology. 2011;157:3195–3202. doi: 10.1099/mic.0.052498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li C, Kurniyati HB, Hu B, Bian J, Sun J, et al. Abrogation of neuraminidase reduces biofilm formation, capsule biosynthesis, and virulence of Porphyromonas gingivalis . Infect Immun. 2012;80:3–13. doi: 10.1128/IAI.05773-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aruni W, Vanterpool E, Osbourne D, Roy F, Muthiah A, et al. Sialidase and sialoglycoproteases can modulate virulence in Porphyromonas gingivalis. Infect Immun. 2011;79:2779–2791. doi: 10.1128/IAI.00106-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurniyati K, Zhang W, Zhang K, Li C. A surface-exposed neuraminidase affects complement resistance and virulence of the oral spirochaete Treponema denticola . Mol Microbiol. 2013;89:842–856. doi: 10.1111/mmi.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duran-Pinedo AE, Chen T, Teles R, Starr JR, Wang X, et al. Community-Wide transcriptome of the oral microbiome in subjects with and without periodontitis. Isme J. 2014;8:1659–1672. doi: 10.1038/ismej.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gul SS, Griffiths GS, Stafford GP, Al-Zubidi MI, Rawlinson A, et al. Investigation of a novel predictive biomarker profile for the outcome of periodontal treatment. J Periodontol. 2017;88:1135. doi: 10.1902/jop.2017.170187. [DOI] [PubMed] [Google Scholar]
- 22.Honma K, Mishima E, Sharma A. Role of Tannerella forsythia NanH sialidase in epithelial cell attachment. Infect Immun. 2011;79:393–401. doi: 10.1128/IAI.00629-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dickson MA, Hahn WC, Ino Y, Ronfard V, Wu JY, et al. Human keratinocytes that express hTERT and also bypass a p16(INK4a)-enforced mechanism that limits life span become immortal yet retain normal growth and differentiation characteristics. Mol Cell Biol. 2000;20:1436–1447. doi: 10.1128/MCB.20.4.1436-1447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas GJ, Lewis MP, Whawell SA, Russell A, Sheppard D, et al. Expression of the alphavbeta6 integrin promotes migration and invasion in squamous carcinoma cells. J Invest Dermatol. 2001;117:67–73. doi: 10.1046/j.0022-202x.2001.01379.x. [DOI] [PubMed] [Google Scholar]
- 25.Sugiyama M, Speight PM, Prime SS, Watt FM. Comparison of integrin expression and terminal differentiation capacity in cell lines derived from oral squamous cell carcinomas. Carcinogenesis. 1993;14:2171–2176. doi: 10.1093/carcin/14.10.2171. [DOI] [PubMed] [Google Scholar]
- 26.Phansopa C, Kozak RP, Liew LP, Frey AM, Farmilo T, et al. Characterization of a sialate-O-acetylesterase (NanS) from the oral pathogen Tannerella forsythia that enhances sialic acid release by NanH, its cognate sialidase. Biochem J. 2015;472:157–167. doi: 10.1042/BJ20150388. [DOI] [PubMed] [Google Scholar]
- 27.Frey AM, Satur MJ, Phansopa C, Parker JL, Bradshaw D, et al. Evidence for a novel carbohydrate binding module (CBM) of Tannerella forsythia NanH sialidase, key to interactions at the host-pathogen interface. Biochem J. 2018;20170592:BCJ20170592. doi: 10.1042/BCJ20170592. [DOI] [PubMed] [Google Scholar]
- 28.Honma K, Mishima E, Inagaki S, Sharma A. The oxyR homologue in Tannerella forsythia regulates expression of oxidative stress responses and biofilm formation. Microbiology. 2009;155:1912–1922. doi: 10.1099/mic.0.027920-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Royle L, Mattu TS, Hart E, Langridge JI, Merry AH, et al. An analytical and structural database provides a strategy for sequencing O-glycans from microgram quantities of glycoproteins. Anal Biochem. 2002;304:70–90. doi: 10.1006/abio.2002.5619. [DOI] [PubMed] [Google Scholar]
- 30.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komatsu T, Nagano K, Sugiura S, Hagiwara M, Tanigawa N, et al. E-Selectin mediates Porphyromonas gingivalis adherence to human endothelial cells. Infect Immun. 2012;80:2570–2576. doi: 10.1128/IAI.06098-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varki A, Diaz S. A neuraminidase from Streptococcus sanguis that can release O-acetylated sialic acids. J Biol Chem. 1983;258:12465–12471. [PubMed] [Google Scholar]
- 33.Suwannakul S, Stafford GP, Whawell SA, Douglas CWI. Identification of bistable populations of Porphyromonas gingivalis that differ in epithelial cell invasion. Microbiology. 2010;156:3052–3064. doi: 10.1099/mic.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 34.Al-Taweel FB, Douglas CWI, Whawell SA. The periodontal pathogen Porphyromonas gingivalis preferentially interacts with oral epithelial cells in S phase of the cell cycle. Infect Immun. 2016;84:1966–1974. doi: 10.1128/IAI.00111-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naylor KL, Widziolek M, Hunt S, Conolly M, Hicks M, et al. Role of OmpA2 surface regions of Porphyromonas gingivalis in host – pathogen interactions with oral epithelial cells. Microbiol Open. 2017:1–11. doi: 10.1002/mbo3.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito A, Kokubu E, Inagaki S, Imamura K, Kita D, et al. Porphyromonas gingivalis entry into gingival epithelial cells modulated by Fusobacterium nucleatum is dependent on lipid rafts. Microb Pathog. 2012;53:234–242. doi: 10.1016/j.micpath.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by "Tannerella forsythia". Infect Immun. 2006;74:5023–5028. doi: 10.1128/IAI.00062-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kirschbaum M, Schultze-Mosgau S, Pfister W, Eick S. Mixture of periodontopathogenic bacteria influences interaction with KB cells. Anaerobe. 2010;16:461–468. doi: 10.1016/j.anaerobe.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 39.Sharma A, Inagaki S, Sigurdson W, Kuramitsu HK. Synergy between Tannerella forsythia and Fusobacterium nucleatum in biofilm formation. Oral Microbiol Immunol. 2005;20:39–42. doi: 10.1111/j.1399-302X.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 40.Settem RP, El-Hassan AT, Honma K, Stafford GP, Sharma A. Fusobacterium nucleatum and Tannerella forsythia induce synergistic alveolar bone loss in a mouse periodontitis model. Infect Immun. 2012;80:2436–2443. doi: 10.1128/IAI.06276-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan KH, Seers CA, Dashper SG, Mitchell HL, Pyke JS, et al. Porphyromonas gingivalis and Treponema denticola exhibit metabolic symbioses. PLoS Pathog. 2014;10:e1003955. doi: 10.1371/journal.ppat.1003955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Govinden G, Parker JL, Naylor KL, Frey AM, Anumba DOC, et al. Inhibition of sialidase activity and cellular invasion by the bacterial vaginosis pathogen Gardnerella vaginalis . Arch Microbiol. 2018;200:1129–1133. doi: 10.1007/s00203-018-1520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walther E, Richter M, Xu Z, Kramer C, von Grafenstein S, et al. Antipneumococcal activity of neuraminidase inhibiting artocarpin. Int J Med Microbiol. 2015;305:289–297. doi: 10.1016/j.ijmm.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roy S, Honma K, Douglas CWI, Sharma A, Stafford GP. Role of sialidase in glycoprotein utilization by Tannerella forsythia. Microbiology. 2011;157:3195–3202. doi: 10.1099/mic.0.052498-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lewis WG, Robinson LS, Perry J, Bick JL, Peipert JF, et al. Hydrolysis of secreted sialoglycoprotein immunoglobulin A (IgA) in ex vivo and biochemical models of bacterial vaginosis. J Biol Chem. 2012;287:2079–2089. doi: 10.1074/jbc.M111.278135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davey ME, Duncan MJ. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis. J Bacteriol. 2006;188:5510–5523. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bao K, Belibasakis GN, Thurnheer T, Aduse-Opoku J, Curtis MA, et al. Role of Porphyromonas gingivalis gingipains in multi-species biofilm formation. BMC Microbiol. 2014;14:1–8. doi: 10.1186/s12866-014-0258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bloch S, Thurnheer T, Murakami Y, Belibasakis GN, Schäffer C, et al. Behavior of two Tannerella forsythia strains and their cell surface mutants in multispecies oral biofilms. Mol Oral Microbiol. 2017;32:404–418. doi: 10.1111/omi.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han YW, Ikegami A, Rajanna C, Kawsar HI, Zhou Y, et al. Identification and characterization of a novel adhesin unique to oral fusobacteria. J Bacteriol. 2005;187:5330–5340. doi: 10.1128/JB.187.15.5330-5340.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Saito A, Inagaki S, Ishihara K. Differential ability of periodontopathic bacteria to modulate invasion of human gingival epithelial cells by Porphyromonas gingivalis . Microb Pathog. 2009;47:329–333. doi: 10.1016/j.micpath.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 51.Stamatos NM, Carubelli I, van de Vlekkert D, Bonten EJ, Papini N, et al. Lps-Induced cytokine production in human dendritic cells is regulated by sialidase activity. J Leukoc Biol. 2010;88:1227–1239. doi: 10.1189/jlb.1209776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hata K, Koseki K, Yamaguchi K, Moriya S, Suzuki Y, et al. Limited inhibitory effects of oseltamivir and zanamivir on human sialidases. Antimicrob Agents Chemother. 2008;52:3484–3491. doi: 10.1128/AAC.00344-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Amith SR, Jayanth P, Franchuk S, Finlay T, Seyrantepe V, et al. Neu1 desialylation of sialyl α-2,3-linked β-galactosyl residues of Toll-like receptor 4 is essential for receptor activation and cellular signaling. Cell Signal. 2010;22:314–324. doi: 10.1016/j.cellsig.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 54.Amith SR, Jayanth P, Franchuk S, Siddiqui S, Seyrantepe V, et al. Dependence of pathogen molecule-induced Toll-like receptor activation and cell function on NEU1 sialidase. Glycoconj J. 2009;26:1197–1212. doi: 10.1007/s10719-009-9239-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.