Abstract
Rax is one of the key transcription factors crucial for vertebrate eye development. In this study, we conducted comprehensive evolutionary analysis of Rax. We found that Bilateria and Cnidaria possess Rax, but Placozoa, Porifera, and Ctenophora do not, implying that the origin of the Rax gene dates back to the common ancestor of Cnidaria and Bilateria. The results of molecular phylogenetic and synteny analyses on Rax loci between jawed and jawless vertebrates indicate that segmental duplication of the Rax locus occurred in an early common ancestor of jawed vertebrates, resulting in two Rax paralogs in jawed vertebrates, Rax and Rax2. By analyzing 86 mammalian genomes from all four major groups of mammals, we found that at least five independent Rax2 gene loss events occurred in mammals. This study may provide novel insights into the evolution of the eye.
Keywords: eye, homeobox gene, molecular evolution, Pax6, Rax, retina
Evolution of RAX transcription factor, crucial for eye development, was comprehensively analyzed. Our analyses suggest that Rax appeared in the common ancestor of Cnidaria and Bilateria. Jawed vertebrate Rax and Rax2 might originate from segmental duplication of the ancient Rax locus. At least five independent Rax2 gene loss events occurred in mammals. This study provides novel insights into eye evolution.
Abbreviations
- JTT
Jones–Taylor–Thornton model
- Ka/Ks
nonsynonymous‐to‐synonymous substitution ratio
- LG
Le–Gascuel model
- WAG
Whelan and Goldman model
- WGD
whole‐genome duplication
Acquiring visual information from the external environment is critical for animal survival. Over the course of evolution, animals have developed different kinds of eyes, from eyespots to complex refractive and compound eyes, which allow them to respond to light stimulus [1]. Notably, vertebrates have developed camera eyes, in which the retina receives the visual input [1].
The homeobox gene superfamily encodes transcription factors with diverse functional roles [2]. For DNA recognition, these transcription factors share a 60‐amino‐acid homeodomain, which comprises a helix‐turn‐helix structure, similar to the one found in prokaryotic gene regulatory proteins [3]. Since animals, plants, and fungi possess homeobox genes, the origin of such genes preceded the divergence of these kingdoms [4]. Among these kingdoms, animal homeobox genes are the most diverse due to extensive gene duplication in the early eumetazoan lineage [5].
We previously identified the retina and anterior neural fold homeobox (Rax, also known as Rx) gene, which plays critical roles in the eye and forebrain development of vertebrate species [6, 7, 8]. Vertebrate Rax is composed of an N‐terminal octapeptide, a paired‐type homeobox, and a C‐terminal OAR motif [6]. In the early mouse embryo, Rax is expressed in the anterior neural fold [6]. Subsequently, its expression is limited to the embryonic diencephalon region, which develops into the retina and pineal gland [6]. Rax‐null mouse embryos do not form optic vesicles and exhibit the reduction of brain structures [8]. Likewise, mutations of the RAX gene were reported in human microphthalmia patients [9, 10]. In the retina, Rax plays an essential role in cell fate determination and maturation of photoreceptor cells [11, 12]. It has been shown that Pax6, another homeobox gene, plays an essential role in eye development [13]. It should be noted that, in Pax6‐null mouse embryos, optic vesicles are formed and the Rax expression is unaffected, but cell proliferation in optic vesicles is severely impaired, resulting in a defective eye structure at later developmental stages. On the other hand, the cornea and lens are not formed in Pax6‐null mouse embryos [14]. Thus, Rax is one of the critical transcription factors functioning at the initial stage of eye development, but acts independently of Pax6 [15, 16].
As a result of recent advances in DNA sequencing and computation, whole‐genome sequencing has become widely available [17]. To date, 2618 animal genomes including those of 1171 invertebrates have been sequenced, according to the NCBI Assembly database [18]. Since invertebrates diverged from vertebrates more than 500 million years ago [19], the genome sequences of invertebrates provide us with an unique opportunity to study the origin and evolution of genes [20]. Previous studies on the molecular evolution of Rax focused primarily on vertebrates [21] or on Bilateria, Cnidaria, and Placozoa [22, 23]. This may be due to the fact that the number of genomic sequences available when these studies were conducted was very limited compared to those currently available. Therefore, to investigate the origin and molecular evolution of Rax and to gain insights into the evolution of the eye, we conducted a comprehensive evolutionary analysis of Rax. To do so, we analyzed the abundant number of currently available genome sequences, including Ctenophora and Porifera in addition to Bilateria, Cnidaria, and Placozoa.
Materials and methods
Collection of Rax orthologs
To identify Rax orthologs, we downloaded NCBI Gnomon gene models of various species (https://www.ncbi.nlm.nih.gov/genome/annotation_euk/gnomon/). Gnomon gene models were constructed using comprehensive gene predictions with a combination of homology searching and ab initio modeling. For every species analyzed, we obtained protein‐coding sequences from genome sequences based on the corresponding NCBI Gnomon gene models. We obtained complete protein sets by translating these protein‐coding sequences. To identify putative Rax ortholog sequences, we performed blastp against every complete set of protein sequences using the Rax protein sequences for human (NP_038463.2), octopus (XP_014777656.1), and Pocillopora damicornis (XP_027036745.1) as query sequences with E‐values < 1e−10. In cases where Rax was not identified in the complete set of protein sequences in a species, we performed tblastn against its genome sequence using the above three query sequences with E‐values < 1e−10. All protein sequences for putative Rax orthologs were subjected to a blastp search against all human protein sequences to confirm their orthologous relationships with human RAX. To identify protein‐coding sequences of Rax from transcriptomic data, we downloaded publicly available RNA‐seq raw reads from the NCBI SRA database [24] and performed de novo transcriptome assembly using Trinity under default settings [25]. We searched for Rax putative protein‐coding sequences in the Trinity contigs using tblastn as described above. The protein sequence of Meara stichopi Rax (AVK72338.1) [26] was obtained from the NCBI nucleotide database. In every Rax ortholog, regions of the octapeptide, homeodomain, and OAR motif were defined with reference to human RAX. The complete list of accession numbers for the taxon names, genome assemblies, and Rax orthologs is provided in Tables 1 and 2. Domain organizations of genes or proteins were illustrated by Illustrator for Biological Sequences [27].
Table 1.
Rax and Rax2 orthologs in various animal species.
| Group | Taxon name | Genome or transcriptome | Reference ID | Genome assembly level | Rax | Rax2 |
|---|---|---|---|---|---|---|
| Porifera | Amphimedon queenslandica | Genome | GCA_000090795.1 | Scaffold | – | – |
| Porifera | Aplysina aerophoba | Genome | GCA_900275595.1 | Contig | – | – |
| Porifera | Sycon ciliatum | Transcriptome | ERR466755 | n.a. | – | – |
| Ctenophora | Pleurobrachia bachei | Genome | GCA_000695325.1 | Scaffold | – | – |
| Ctenophora | Mnemiopsis leidyi |
Genome Transcriptome |
GCA_000226015.1 SRR1971277 |
Scaffold | – | – |
| Ctenophora | Beroe ovata | Genome | GCA_900239995.1 | Contig | – | – |
| Placozoa | Trichoplax adhaerens | Genome | GCA_000150275.1 | Scaffold | – | – |
| Placozoa | Trichoplax | Genome | GCA_003344405.1 | Scaffold | – | – |
| Cnidaria | Tripedalia cystophora | Transcriptome | SRR8101523 | n.a. | a | – |
| Cnidaria | Nematostella vectensis | Genome | GCA_000209225.1 | Scaffold | XM_001634160.1 | – |
| Cnidaria | Stylophora pistillata | Genome | GCA_002571385.1 | Scaffold | XM_022924671.1 | – |
| Cnidaria | Pocillopora damicornis | Genome | GCA_003704095.1 | Scaffold | XM_027180944.1 | – |
| Cnidaria | Orbicella faveolata | Genome | GCA_002042975.1 | Scaffold | XM_020776224.1 | – |
| Bilateria | Drosophila melanogaster | Genome | GCA_000001215.4 | Chromosome | NM_166413.3 | – |
| Bilateria | Apis mellifera | Genome | GCA_003254395.2 | Chromosome | XM_001119966.5 | – |
| Bilateria | Caenorhabditis elegans | Genome | GCA_000002985.3 | Complete Genome | NM_059845.2 | – |
| Bilateria | Octopus bimaculoides | Genome | GCA_001194135.1 | Scaffold | XM_014922170.1 | – |
| Bilateria | Mizuhopecten yessoensis | Genome | GCA_002113885.2 | Scaffold | XM_021516578.1 | – |
| Bilateria | Meara stichopi | Transcriptome | n.a. | n.a. | KY709787.1 | – |
| Bilateria | Acanthaster planci | Genome | GCA_001949145.1 | Scaffold | XM_022243979.1 | – |
| Bilateria | Saccoglossus kowalevskii | Genome | GCA_000003605.1 | Scaffold | NM_001164903.1 | – |
| Bilateria | Branchiostoma belcheri | Genome | GCA_001625305.1 | Scaffold | XM_019761392.1 | – |
| Bilateria | Ciona intestinalis | Genome | GCA_000224145.2 | Chromosome | NM_001032511.1 | – |
| Bilateria | Eptatretus burgeri | Genome | GCA_900186335.2 | Scaffold | ENSEBUT00000011203.1 | – |
| Bilateria | Petromyzon marinus | Genome | GCA_002833325.1 | Scaffold | b | – |
| Bilateria | Callorhinchus milii | Genome | GCA_000165045.2 | Scaffold | XM_007903126.1 | XM_007908006.1 |
| Bilateria | Erpetoichthys calabaricus | Genome | GCA_900747795.2 | Chromosome | XM_028803026.1 | XM_028814691.1 |
| Bilateria | Acipenser ruthenus | Genome | GCA_004119895.1 | Scaffold | RXM93534.1 | RXM94969.1 |
| Bilateria | Lepisosteus oculatus | Genome | GCA_000242695.1 | Chromosome | XM_006627139.2 | XM_015365287.1 |
| Bilateria | Danio rerio | Genome | GCA_000002035.4 | Chromosome | NM_131227.1 | |
| Bilateria | Takifugu rubripes | Genome | GCA_901000725.2 | Chromosome | XM_029837663.1 | |
| Bilateria | Latimeria chalumnae | Genome | GCA_000225785.1 | Scaffold | XM_006005788.1 | XM_005999053.1 |
| Bilateria | Xenopus tropicalis | Genome | GCA_000004195.3 | Chromosome | XM_002936669.4 | XM_002941390.4 |
| Bilateria | Taeniopygia guttata | Genome | GCA_003957565.2 | Chromosome | NM_001243734.1 | XM_030256513.1 |
| Bilateria | Monodelphis_domestica | Genome | GCA_000002295.2 | Chromosome | XM_007487510.2 | XM_001373844.3 |
| Bilateria | Homo sapiens | Genome | GCA_000001405.27 | Chromosome | NM_013435.3 | NM_032753.3 |
| Bilateria | Mus musculus | Genome | GCA_000001635.8 | Chromosome | NM_013833.2 | – |
The putative sequence of Rax ortholog was obtained from transcriptome
The putative sequence of lamprey Rax was directly retrieved from the genomic sequence using UCSC Genome Browser.
Table 2.
Mammalian Rax and Rax2 orthologs.
| Group | Taxon name | Genome ID | Genome assembly level | Rax | Rax2 |
|---|---|---|---|---|---|
| Euarchontoglires | Homo sapiens | GCA_000001405.27 | Chromosome | NM_013435.3 | NM_032753.3 |
| Euarchontoglires | Pan troglodytes | GCA_002880755.3 | Chromosome | XM_001142510.3 | NM_001081487.1 |
| Euarchontoglires | Pan paniscus | GCA_000258655.2 | Chromosome | XM_008952703.1 | XM_008972700.2 |
| Euarchontoglires | Pongo abelii | GCA_002880775.3 | Chromosome | XM_024236038.1 | XM_009252600.2 |
| Euarchontoglires | Nomascus leucogenys | GCA_000146795.3 | Chromosome | XM_030810092.1 | XM_004091072.2 |
| Euarchontoglires | Macaca mulatta | GCA_000772875.3 | Chromosome | XM_015122061.2 | XM_002801027.3 |
| Euarchontoglires | Macaca fascicularis | GCA_000364345.1 | Chromosome | XM_005586557.2 | XM_015440231.1 |
| Euarchontoglires | Macaca nemestrina | GCA_000956065.1 | Scaffold | XM_011733358.1 | XM_011747810.2 |
| Euarchontoglires | Chlorocebus sabaeus | GCA_000409795.2 | Chromosome | XM_008013918.1 | XM_007994799.1 |
| Euarchontoglires | Papio anubis | GCA_000264685.2 | Chromosome | XM_021930179.1 | XM_003914667.1 |
| Euarchontoglires | Cercocebus atys | GCA_000955945.1 | Scaffold | XM_012069057.1 | XM_012073070.1 |
| Euarchontoglires | Theropithecus gelada | GCA_003255815.1 | Chromosome | XM_025365606.1 | XM_025368381.1 |
| Euarchontoglires | Mandrillus leucophaeus | GCA_000951045.1 | Scaffold | XM_011971742.1 | XM_011967344.1 |
| Euarchontoglires | Piliocolobus tephrosceles | GCA_002776525.2 | Scaffold | XM_023218311.1 | XM_023200859.2 |
| Euarchontoglires | Rhinopithecus bieti | GCA_001698545.1 | Scaffold | XM_017890457.1 | XM_017847249.1 |
| Euarchontoglires | Rhinopithecus roxellana | GCA_000769185.1 | Scaffold | XM_030926113.1 | XM_010367273.2 |
| Euarchontoglires | Colobus angolensis | GCA_000951035.1 | Scaffold | XM_011949591.1 | XM_011943448.1 |
| Euarchontoglires | Callithrix jacchus | GCA_000004665.1 | Chromosome | XM_002757287.2 | XM_002761582.3 |
| Euarchontoglires | Cebus capucinus | GCA_001604975.1 | Scaffold | XM_017528838.1 | XM_017507399.1 |
| Euarchontoglires | Saimiri boliviensis boliviensis | GCA_000235385.1 | Scaffold | XM_010336873.1 | XM_010349647.1 |
| Euarchontoglires | Aotus nancymaae | GCA_000952055.2 | Scaffold | XM_012446398.1 | XM_012436148.2 |
| Euarchontoglires | Otolemur garnettii | GCA_000181295.3 | Scaffold | XM_003788425.1 | XM_003788870.2 |
| Euarchontoglires | Propithecus coquereli | GCA_000956105.1 | Scaffold | XM_012651751.1 | XM_012646140.1 |
| Euarchontoglires | Microcebus murinus | GCA_000165445.3 | Chromosome | XM_012745461.1 | XM_012769405.1 |
| Euarchontoglires | Galeopterus variegatus | GCA_000696425.1 | Scaffold | XM_008580185.1 | XM_008582889.1 |
| Euarchontoglires | Tupaia chinensis | GCA_000334495.1 | Scaffold | XM_006139947.2 | XM_006171790.2 |
| Euarchontoglires | Ochotona princeps | GCA_000292845.1 | Scaffold | XM_004579462.1 | – |
| Euarchontoglires | Ictidomys tridecemlineatus | GCA_000236235.1 | Scaffold | XM_013357567.2 | XM_021730889.1 |
| Euarchontoglires | Urocitellus parryii | GCA_003426925.1 | Scaffold | XM_026393165.1 | XM_026404447.1 |
| Euarchontoglires | Marmota flaviventris | GCA_003676075.1 | Scaffold | XM_027948970.1 | XM_027952705.1 |
| Euarchontoglires | Mus musculus | GCA_000001635.8 | Chromosome | NM_013833.2 | – |
| Euarchontoglires | Rattus norvegicus | GCA_000001895.4 | Chromosome | NM_053678.1 | – |
| Euarchontoglires | Nannospalax galili | GCA_000622305.1 | Scaffold | XM_008840033.1 | – |
| Euarchontoglires | Cavia porcellus | GCA_000151735.1 | Scaffold | XM_013157082.1 | – |
| Euarchontoglires | Octodon degus | GCA_000260255.1 | Scaffold | XM_004647907.1 | – |
| Euarchontoglires | Dipodomys ordii | GCA_000151885.2 | Scaffold | XM_013018940.1 | – |
| Laurasiatheria | Panthera pardus | GCA_001857705.1 | Scaffold | XM_019463885.1 | XM_019430847.1 |
| Laurasiatheria | Felis catus | GCA_000181335.4 | Chromosome | XM_023242049.1 | XM_023243834.1 |
| Laurasiatheria | Canis lupus familiaris | GCA_000002285.2 | Chromosome | XM_022423505.1 | XM_849723.4 |
| Laurasiatheria | Vulpes vulpes | GCA_003160815.1 | Scaffold | XM_025997549.1 | XM_026019439.1 |
| Laurasiatheria | Ailuropoda melanoleuca | GCA_000004335.1 | Scaffold | XM_011231633.1 | XM_002923561.3 |
| Laurasiatheria | Ursus arctos horribilis | GCA_003584765.1 | Scaffold | XM_026495947.1 | XM_026481074.1 |
| Laurasiatheria | Ursus maritimus | GCA_000687225.1 | Scaffold | XM_008705444.1 | XM_008711252.1 |
| Laurasiatheria | Leptonychotes weddellii | GCA_000349705.1 | Scaffold | XM_006734844.1 | XM_006750976.2 |
| Laurasiatheria | Neomonachus schauinslandi | GCA_002201575.1 | Scaffold | XM_021686390.1 | XM_021705428.1 |
| Laurasiatheria | Eumetopias jubatus | GCA_004028035.1 | Scaffold | XM_028102872.1 | XM_028126063.1 |
| Laurasiatheria | Zalophus californianus | GCA_900631625.1 | Scaffold | XM_027576397.1 | XM_027587801.1 |
| Laurasiatheria | Mustela putorius furo | GCA_000215625.1 | Scaffold | XM_004745687.2 | – |
| Laurasiatheria | Manis javanica | GCA_001685135.1 | Scaffold | XM_017662163.1 | XM_017641956.1 |
| Laurasiatheria | Equus caballus | GCA_002863925.1 | Scaffold | XM_023647900.1 | XM_023644413.1 |
| Laurasiatheria | Equus asinus | GCA_001305755.1 | Scaffold | XM_014859029.1 | XM_014843046.1 |
| Laurasiatheria | Ceratotherium simum simum | GCA_000283155.1 | Scaffold | XM_004422590.2 | XM_004441310.2 |
| Laurasiatheria | Lagenorhynchus obliquidens | GCA_003676395.1 | Scaffold | XM_027118662.1 | XM_027086050.1 |
| Laurasiatheria | Orcinus orca | GCA_000331955.2 | Scaffold | XM_004268055.1 | XM_004277200.1 |
| Laurasiatheria | Lipotes vexillifer | GCA_000442215.1 | Scaffold | XM_007450117.1 | XM_007460506.1 |
| Laurasiatheria | Neophocaena asiaeorientalis asiaeorientalis | GCA_003031525.1 | Scaffold | XM_024754865.1 | XM_024745933.1 |
| Laurasiatheria | Delphinapterus leucas | GCA_002288925.2 | Scaffold | XM_022584361.2 | XM_022557349.2 |
| Laurasiatheria | Physeter catodon | GCA_002837175.1 | Scaffold | XM_024133906.1 | XM_007109282.2 |
| Laurasiatheria | Balaenoptera acutorostrata | GCA_000493695.1 | Scaffold | XM_007192060.1 | XM_007169190.1 |
| Laurasiatheria | Ovis aries | GCA_002742125.1 | Chromosome | XM_027960933.1 | XM_027969870.1 |
| Laurasiatheria | Capra hircus | GCA_001704415.1 | Chromosome | XM_018039348.1 | XM_005682656.3 |
| Laurasiatheria | Bubalus bubalis | GCA_003121395.1 | Chromosome | XM_025273363.1 | XM_006047896.2 |
| Laurasiatheria | Bison bison bison | GCA_000754665.1 | Scaffold | XM_010854825.1 | XM_010828446.1 |
| Laurasiatheria | Bos taurus | GCA_002263795.2 | Chromosome | XM_024984497.1 | NM_182653.1 |
| Laurasiatheria | Bos mutus | GCA_000298355.1 | Scaffold | XM_005911824.1 | XM_005895951.1 |
| Laurasiatheria | Bos indicus | GCA_000247795.2 | Chromosome | XM_019986442.1 | XM_019963601.1 |
| Laurasiatheria | Sus scrofa | GCA_000003025.6 | Chromosome | XM_003121712.3 | XM_005661348.3 |
| Laurasiatheria | Camelus dromedarius | GCA_000767585.1 | Scaffold | XM_031441660.1 | XM_010996050.2 |
| Laurasiatheria | Camelus bactrianus | GCA_000767855.1 | Scaffold | XM_010961429.1 | XM_010966706.1 |
| Laurasiatheria | Vicugna pacos | GCA_000164845.3 | Scaffold | XM_006205802.1 | XM_006206395.1 |
| Laurasiatheria | Miniopterus natalensis | GCA_001595765.1 | Scaffold | XM_016223487.1 | XM_016197969.1 |
| Laurasiatheria | Hipposideros armiger | GCA_001890085.1 | Scaffold | XM_019640612.1 | XM_019653323.1 |
| Laurasiatheria | Desmodus rotundus | GCA_002940915.2 | Scaffold | XM_024580521.1 | XM_024569247.1 |
| Laurasiatheria | Rhinolophus sinicus | GCA_001888835.1 | Scaffold | XM_019729367.1 | XM_019744054.1 |
| Laurasiatheria | Pteropus alecto | GCA_000325575.1 | Scaffold | XM_025048123.1 | XM_006904102.2 |
| Laurasiatheria | Pteropus vampyrus | GCA_000151845.2 | Scaffold | XM_011361137.2 | XM_011372678.2 |
| Laurasiatheria | Erinaceus europaeus | GCA_000296755.1 | Scaffold | XM_007529018.1 | – |
| Laurasiatheria | Sorex araneus | GCA_000181275.2 | Scaffold | XM_004601992.1 | – |
| Laurasiatheria | Condylura cristata | GCA_000260355.1 | Scaffold | XM_004684020.1 | XM_004688892.1 |
| Afrotheria | Loxodonta africana | GCA_000001905.1 | Scaffold | XM_003406278.1 | – |
| Afrotheria | Trichechus manatus latirostris | GCA_000243295.1 | Scaffold | XM_023733937.1 | XM_004378466.1 |
| Afrotheria | Chrysochloris asiatica | GCA_000296735.1 | Scaffold | XM_006837574.1 | XM_006869042.1 |
| Afrotheria | Echinops telfairi | GCA_000313985.1 | Scaffold | XM_004703116.1 | XM_004714381.1 |
| Afrotheria | Elephantulus edwardii | GCA_000299155.1 | Scaffold | XM_006892750.1 | XM_006897892.1 |
| Afrotheria | Orycteropus afer | GCA_000298275.1 | Scaffold | XM_007935546.1 | XM_007951029.1 |
| Xenarthra | Dasypus novemcinctus | GCA_000208655.2 | Scaffold | XM_004447204.1 | XM_004447500.3 |
Multiple sequence alignment of the octapeptide, homeodomain, or OAR motif in Rax orthologs
Amino acid sequences of the octapeptide, homeodomain, or OAR motif in Rax orthologs were aligned by clustal omega with the default parameters [28]. Resulting multiple sequence alignments were verified and visualized by jalview [29].
Molecular phylogenetic analysis of Rax and Rax2 of jawed vertebrates and Rax of jawless vertebrates
Amino acid sequences of Rax and Rax2 of jawed vertebrates and Rax of jawless vertebrates were aligned using clustal omega [28] and muscle [30] under default parameters. The multiple sequence alignment results were verified and visualized using jalview [29]. Maximum‐likelihood trees were constructed using the Poisson, Whelan and Goldman (WAG), Le–Gascuel (LG), or Jones–Taylor–Thornton (JTT) models using mega7 [31]. Neighbor‐joining trees were constructed with the Poisson, Dayhoff, or JTT models using mega7 [31]. All positions with < 90% site coverage were excluded from analyses. In other words, fewer than 10% alignment gaps or missing data were allowed at any position. The bootstrap values were estimated from 500 replicates in all analyses.
Synteny analysis of mammalian Rax and Rax2
Genome sequences and annotations were obtained from the NCBI Assembly database. In this database, the genome assembly quality is classified into four categories, in order of highest to lowest quality: complete genome, chromosome, scaffold, and contig (https://www.ncbi.nlm.nih.gov/assembly/help/). We used genome assemblies of scaffold to complete genome quality in this analysis. The arrangements of genes around Rax or Rax2 were compared between species. Even if a relatively high‐quality genome assembly is used for analysis, low‐quality regions are often included to some extent due to low sequencing read coverage or the presence of repetitive sequences [32]. Therefore, regions with fewer than three genes surrounding Rax or Rax2 were excluded. We reported the genome analysis results where Rax and Rax2 loci were identified.
Maximum‐likelihood tree construction based on mammalian Rax and Rax2 alignment
The amino acid sequences of 86 placental mammalian Rax, 75 placental mammalian Rax2, opossum Rax, and opossum Rax2 protein sequences were aligned using clustal omega under default parameters [28]. Based on this alignment, a maximum‐likelihood tree was constructed with the JTT model using MEGA7 [31]. All positions with < 90% site coverage were excluded from analyses. Bootstrap values were estimated from 500 replicates. The complete list of accession numbers for the analyzed protein sequences is provided in Table 2.
Calculation of nonsynonymous‐to‐synonymous substitution ratio
The amino acid sequences of 86 placental mammalian Rax, 75 placental mammalian Rax2, opossum Rax, and opossum Rax2 protein sequences were aligned using clustal omega under default parameters as described above [28]. Based on this alignment and the set of protein‐coding sequences, we generated codon alignment using tranalign [33]. By comparing human RAX and RAX2 with respective mammalian Rax and Rax2 orthologs, we calculated nonsynonymous‐to‐synonymous rate ratios (Ka/Ks) for each ortholog pair using the Jukes–Cantor model. All positions with < 90% site coverage in the alignment were excluded from analyses. The average Rax Ka/Ks and Rax2 Ka/Ks were compared using a Welch two‐sample t‐test. The difference was considered statistically significant if the P value was < 0.05.
Results
Identification of Rax in various animal species
To investigate the evolutionary origin of Rax, we comprehensively searched for Rax in the genomes or transcriptomes of animals that are evolutionarily distant from each other. The criteria to determine whether a Rax ortholog is present in a species are described in Materials and Methods. This analysis included genome sequences of 34 animal species: two Porifera, three Ctenophora, two Placozoa, four Cnidaria, and 23 Bilateria (Fig. 1A, Table 1). Transcriptome data from Sycon ciliatum, a Porifera organism, and Mnemiopsis leidyi, a Ctenophora organism, were also analyzed. In all Cnidaria and Bilateria analyzed, we identified Rax orthologs (Fig. 1A). In contrast, we did not identify any Rax gene in Porifera, Ctenophore, and Placozoa, which are groups phylogenetically more distant from both Bilateria and Cnidaria (Fig. 1A). Most of the Rax genes in Cnidaria and Bilateria were shown to have all of the octapeptide, homeodomain, and OAR motif (Fig. 1A). Sequence alignment analyses revealed that amino acid sequences of octapeptide, homeodomain, and OAR motif are highly conserved among these animals (Fig. 1B). These results suggest that Rax appeared in the common ancestor of Bilateria and Cnidaria, and has been highly conserved in terms of domain organization and sequence similarity over the course of evolution.
Fig. 1.
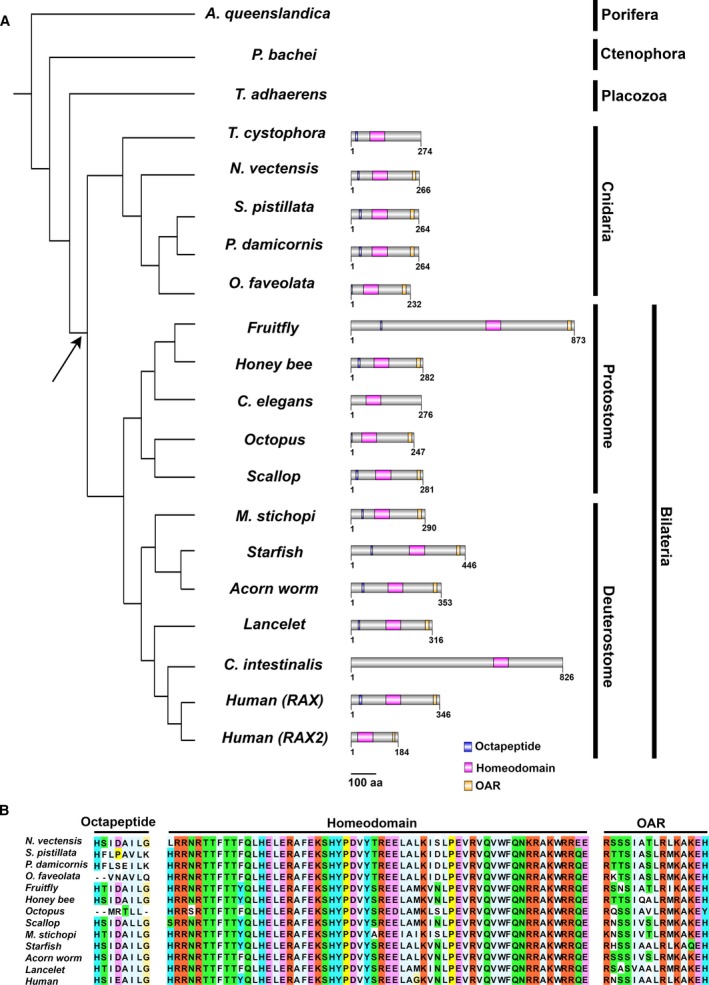
Phylogeny of animal species and their Rax genes. (A) Cladogram of representative animals and domain organizations of their Rax genes. Note the absence of Rax in Ctenophora, Porifera, and Placozoa. Blue boxes indicate octapeptides, magenta boxes indicate homeodomains, and yellow boxes indicate OAR motifs. The cladogram topology is derived from previous studies [54, 55, 56, 57]. It should be noted that the Metazoan phylogeny is controversial. The arrow indicates the presumed origin of the Rax gene. The scale bar indicates 100 amino acid residues. (B) Sequence alignments of the octapeptide, homeodomain, and OAR motif in Rax orthologs. These three domains/motifs are well conserved in Cnidaria and Bilateria. Each residue is colored according to the Clustal X residue code [58].
Phylogenetic analysis of Rax and Rax2 in jawed and jawless vertebrates
Vertebrates are divided into two major groups, jawed and jawless, depending on whether the jaw is present. Jawed vertebrates possess Rax2 in addition to Rax [12, 21]. The high sequence similarity of jawed vertebrate Rax and Rax2 suggests that they resulted from gene duplication. We analyzed the genomes of jawed and jawless vertebrate species to estimate the evolutionary timepoint when these two genes appeared. We analyzed the genomes of elephant shark, spotted gar, and coelacanth as representatives of jawed vertebrates. On the other hand, as representative of jawless vertebrates, we analyzed the genomes of lamprey and hagfish, whose genomes were recently sequenced. In jawed vertebrates, we identified both Rax and Rax2 genes (Fig. 2). The arrangement of the genes Malt1‐Rax‐Cplx4 and their paralogous counterparts was conserved in these organisms. In contrast, we identified only single Rax in both lamprey and hagfish (Fig. 2). Notably, the arrangements of genes around Rax in lamprey or hagfish were very similar, suggesting that their Rax genes are orthologous to each other (Fig. 2).
Fig. 2.
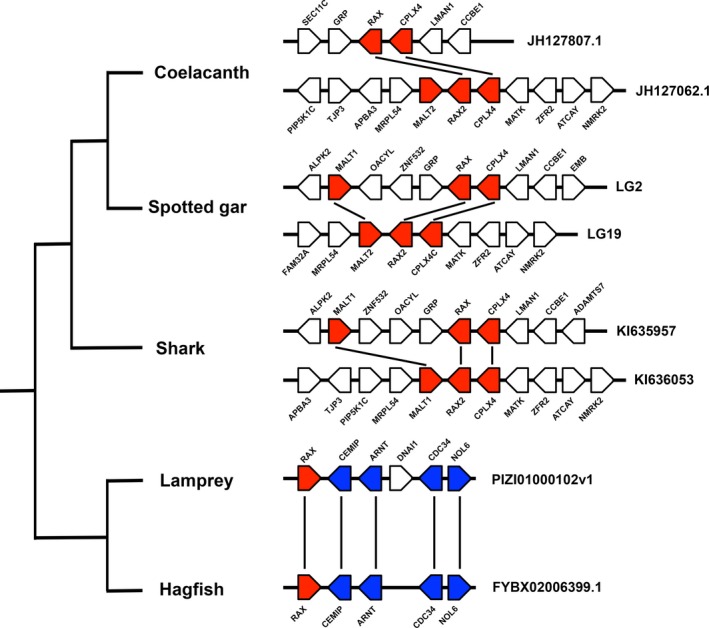
Evolution of Rax and Rax2 gene structures in vertebrates. Synteny around Rax and Rax2 genes in basal vertebrates. While jawed vertebrates possess both Rax and Rax2, jawless vertebrates possess a single Rax. Black lines indicate paralogous or orthologous relationships. Scaffold names are indicated on the right of the panel.
We next analyzed the phylogenetic relationships between jawless vertebrate Rax, jawed vertebrate Rax, and jawed vertebrate Rax2. Tetrapod Rax2 lacks the N‐terminal region, including octapeptide, and is shorter than Rax [21]. Therefore, we focused on nontetrapod vertebrate Rax2 for molecular phylogenetic analysis in order to obtain as much phylogenetic information from the sequence alignments as possible. A total of 22 protein sequences were analyzed, including seven jawed vertebrate Rax, nine jawed vertebrate Rax2, two jawless vertebrate Rax, and four invertebrate Rax. To perform robust molecular phylogenetic analysis, we used the clustal omega [28] and muscle [30] programs to generate two multiple protein sequence alignments (Fig. 3). Based on these alignments, we constructed phylogenetic trees using the maximum‐likelihood method in the character state methods and the neighbor‐joining trees in the distance matrix methods (Fig. 4, Figs S1 and S2). Maximum‐likelihood trees were constructed using the Poisson, WAG, LG, or JTT models. Neighbor‐joining trees were constructed using the Poisson, Dayhoff, or JTT models. In all cases, lamprey and hagfish Rax formed a sister group to Rax and Rax2 of jawed vertebrates (Fig. 4, Figs S1 and S2). Taken together, the current synteny analysis and molecular phylogenetic analysis suggest that Rax and Rax2 of jawed vertebrates resulted from segmental duplication of a small region containing Malt1, Rax, and Cplx4 ancestors that occurred after jawed vertebrates diverged from jawless ones.
Fig. 3.

Multiple sequence alignments of Rax orthologs. (A) Multiple sequence alignment of Rax orthologs using clustal omega. The 22 protein sequences of Rax were aligned by clustal omega [28]. (B) Multiple sequence alignment of Rax orthologs using muscle. The same set of protein sequences in (A) were aligned by muscle [30]. Each residue is colored according to the Clustal X residue code [58].
Fig. 4.
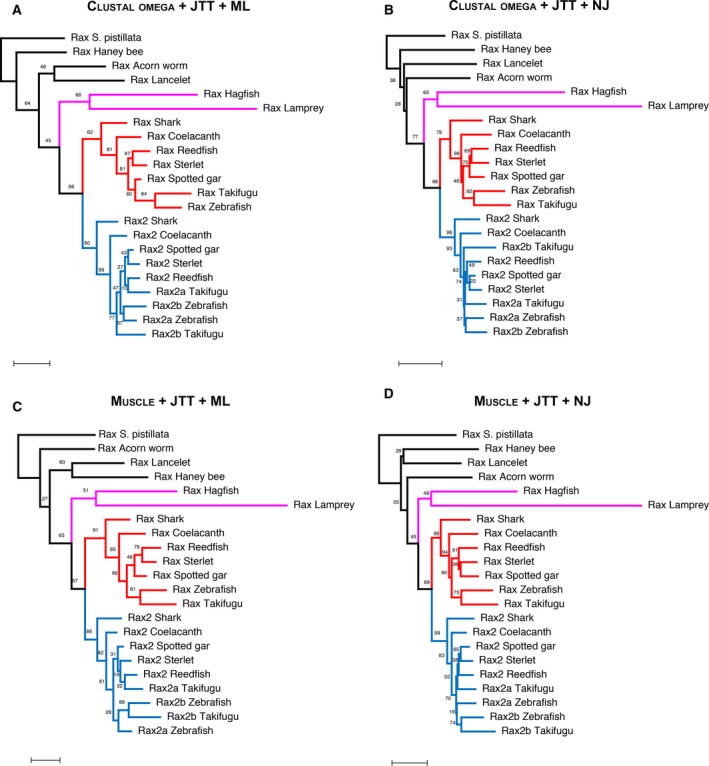
Molecular phylogenetic analysis of Rax and Rax2 in various animal species. Multiple sequence alignments were used to construct maximum‐likelihood trees from (A) clustal omega or (C) muscle and neighbor‐joining trees from (B) clustal omega or (D) muscle. In all these analyses, the JTT model was used as the amino acid substitution model. Jawed vertebrate Rax sequences are colored in red. Jawed vertebrate Rax2 sequences are colored in blue. Lamprey and hagfish Rax sequences are colored in magenta. The scale bars represent 0.2 amino acid substitutions per site. Bootstrap values are given on each node.
Comparative analysis of Rax and Rax2 gene structures in vertebrates
Tetrapod Rax2 genes were reported to lack octapeptide domains [21]. To investigate how the loss of octapeptide in tetrapods occurred, we compared the gene structures of vertebrate Rax and Rax2. We included seven vertebrates from shark to human in this analysis (Fig. 5A). In all species analyzed, Rax or Rax2 gene was composed of three exons (Fig. 5A). In both genes, octapeptides were coded in the first exon (Fig. 5B). Start codons of Rax were located on the first exons in all species (Fig. 5A). Similarly, start codons of Rax2 in shark, spotted gar, and coelacanth were located on the first exon; however, in all tetrapods analyzed, start codons of Rax2 were shifted to the second exon, resulting in the loss of octapeptides (Fig. 5A).
Fig. 5.
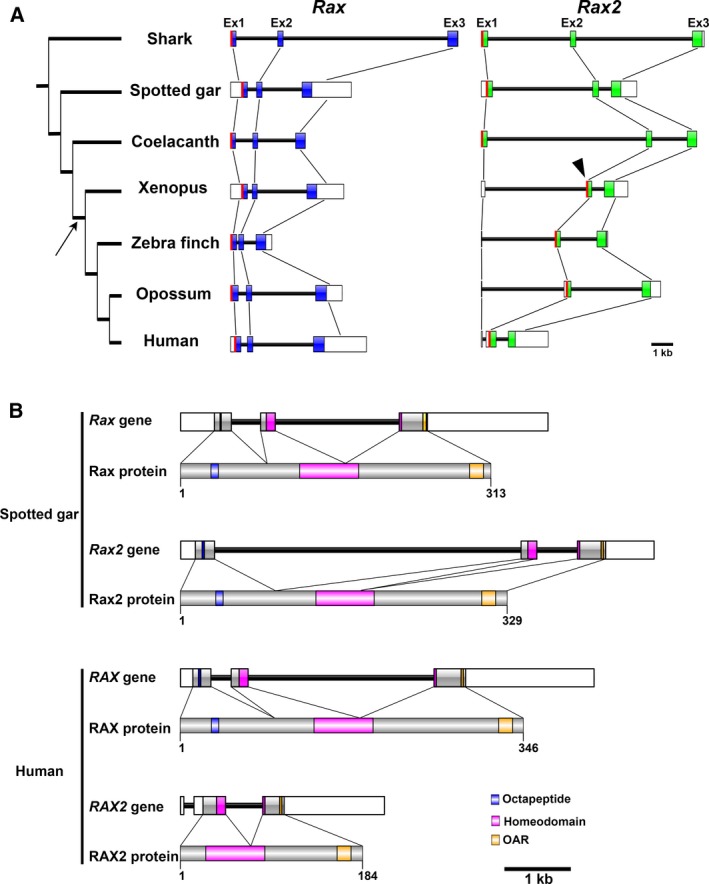
Rax and Rax2 gene structures. (A) Rax and Rax2 gene structures of seven vertebrates. All Rax and Rax2 genes are composed of three exons. Protein‐coding sequences of Rax or Rax2 are colored blue or green, respectively. In shark, spotted gar, and coelacanth, start codons of Rax2 (red line) are located on the first exon. In Xenopus, zebra finch, opossum, and human, start codons of Rax2 are shifted to the second exon (arrowhead). The arrow indicates the presumed point of Rax2 octapeptide loss. The scale bar indicates 1 kbp. (B) Rax and Rax2 gene structures of spotted gar and human. Rax and Rax2 gene structures and their respective protein domain/motif architectures are shown. Note that human RAX2 has its start codon at the second exon and lacks the octapeptide. Blue boxes indicate octapeptides, magenta boxes indicate homeodomains, and yellow boxes indicate OAR motifs. The scale bar indicates 1 kbp.
Identification of Rax2 gene loss events in mammals
Since mice are known to lack the Rax2 gene [12], we investigated whether more Rax2 gene loss events have occurred in mammals by comparative analysis of Rax and Rax2 loci in mammals. Mammals are phylogenetically divided into four major groups: Euarchontoglires, Laurasiatheria, Afrotheria, and Xenarthra (Fig. 6A). We examined both Rax and Rax2 loci in 86 mammalian genomes (Table 2). While all 86 Rax loci contained Rax genes, 11 Rax2 loci lacked the Rax2 gene (Table 2). Of the investigated Euarchontoglires, rock rabbit and six rodent species lack Rax2 (Fig. 6B, Table 2). Notably, three squirrel species possess Rax2 genes, suggesting that an ancestor of rodents and one of the rabbits independently lost Rax2 genes (Fig. 6B, Table 2). In Laurasiatheria, ferret, hedgehog, and common shrew lack Rax2 genes (Fig. 6C, Table 2). Recent studies on the Eulipotyphla phylogeny indicated that hedgehog and common shrew are the sister group to star‐nosed mole [34, 35]. Since star‐nosed mole has the Rax2 gene, a single Rax2 gene loss event appears to have occurred in the common ancestor of hedgehog and common shrew. In Afrotheria, elephant lacks the Rax2 gene (Fig. 6D, Table 2). In Xenarthra, we analyzed the armadillo genome and identified the Rax2 gene. In summary, we detected five independent gene loss events of the Rax2 gene in mammals (Fig. 6A).
Fig. 6.
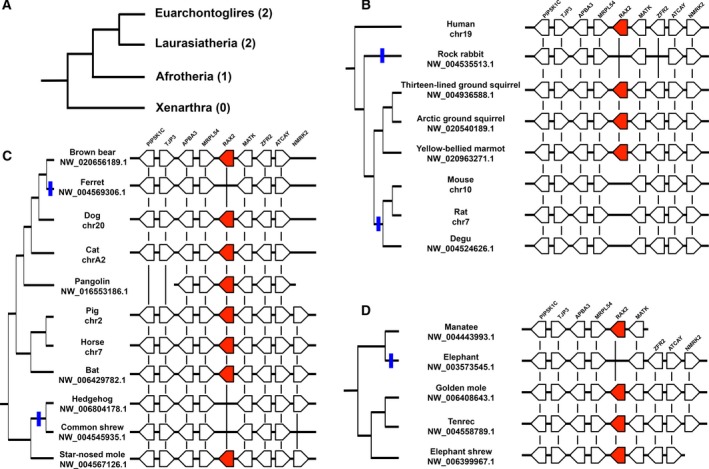
Rax2 gene losses in mammals. (A) Phylogeny of mammals and Rax2 gene loss. Mammals are divided into four major groups. Numbers of Rax2 gene loss events are given in parentheses. The topology of this cladogram is based on previous reports [59, 60, 61]. (B) Synteny of the Rax2 locus in Euarchontoglires. Rock rabbit and a subgroup of rodents, including mouse, rat, and degu, show independent Rax2 gene loss (blue lines). (C) Synteny of Rax2 locus in Laurasiatheria. There are two independent Rax2 gene loss events in Laurasiatheria (blue lines). (D) Synteny of Rax2 locus in Afrotheria. Elephant lacks the Rax2 gene (blue line). Rax2 genes are colored red. Black lines indicate orthologous relationships. Scaffold name is shown below each taxon name.
Molecular phylogenetic analysis of mammalian Rax and Rax2
In order to further analyze mammalian Rax and Rax2 evolution, we aligned all 86 Rax and 75 Rax2 protein sequences identified in the current study and constructed a maximum‐likelihood tree (Fig. 7A, Table 2). As expected, Rax and Rax2 formed a monophyletic group in the tree (Fig. 7A). The Rax2 branch lengths appeared to be longer than the Rax lengths. Therefore, to quantitatively compare the degree of amino acid substitutions, we compared the nucleotide substitutions between human RAX and RAX2 with their respective mammalian Rax and Rax2 orthologs. Then, we calculated Ka/Ks ratios for each ortholog gene pair. The Rax2 Ka/Ks values showed a broader distribution compared to Rax (Fig. 7B). The average Ka/Ks value was 0.042 for Rax and 0.070 for Rax2 (Fig. 7B), indicating that the average mammalian Rax2 Ka/Ks value was ~ 67% greater than mammalian Rax (P < 0.01, Welch two‐sample t‐test; Fig. 7B).
Fig. 7.
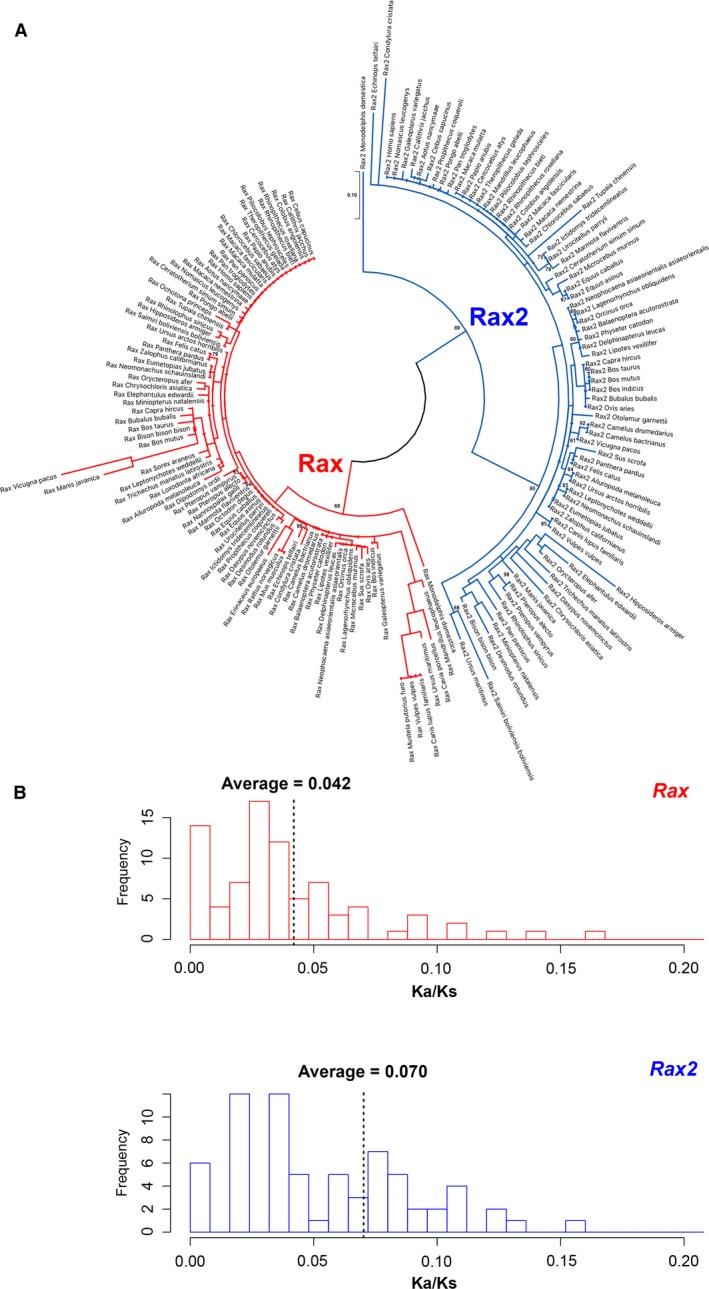
Phylogenetic analysis of mammalian Rax and Rax2. (A) A maximum‐likelihood tree of mammalian Rax and Rax2. A maximum‐likelihood tree was constructed from the amino acid sequence alignment containing Rax (red) and Rax2 (blue) from 86 placental mammals and opossum (Monodelphis domestica), a marsupial. The scale bars represent 0.1 amino acid substitutions per site. Bootstrap values > 0.6 are given on each node. (B) Ka/Ks ratio distributions of mammalian Rax (upper panel) or Rax2 (lower panel). Ka/Ks ratios comparing human RAX and RAX2 with respective mammalian Rax and Rax2 were calculated. The average Ka/Ks values are indicated by dotted lines.
Discussion
Our comprehensive analysis of Rax orthologs suggests that Rax appeared after Bilateria and Cnidaria diverged from other lineages over the course of evolution (Fig. 1A). It has remained unknown whether Rax is present in species evolutionarily distant from Bilateria and Cnidaria, probably because genome sequences of such distant species remained unavailable [23]. Fortunately, genome sequences of such distant species have recently become available, including Porifera, Ctenophora, and Placozoa, providing us with a valuable resource for comparative genomics [20]. Consistent with the previous studies, we definitively showed that Cnidaria and Bilateria possess Rax, whereas Placozoa does not (Fig. 1A) [22, 23]. Conversely, we showed that Porifera and Ctenophora may lack Rax using a comprehensive analysis of their genomes or transcriptomes (Fig. 1A, Table 1). It has been proposed that the Hox genes diversified due to rapid gene duplication before diversification of Cnidaria and Bilateria [36]. Similarly, ancestral paired‐type homeobox genes may have diverged, resulting in the appearance of the Rax gene before the diversification of Cnidaria and Bilateria. However, the divergence between Placozoa and the common ancestor of Cnidaria and Bilateria is very ancient. Therefore, our analysis cannot exclude the possibility that highly accumulated substitutions affect our Rax ortholog search results. The incompleteness of the genome assemblies should also be considered because all genome assemblies for Porifera, Ctenophora, and Placozoa are assembled at the contig or scaffold level. We also showed that the domain organization and amino acid sequences of the octapeptide, homeodomain, and OAR motif are highly conserved between cnidarian Rax and bilaterian Rax (Fig. 1B). Based on these observations, we propose that the origin of the Rax gene dates back to the common ancestor of Cnidaria and Bilateria and that Rax is highly conserved among Cnidaria and Bilateria.
From the very simple structure seen in Porifera to more complex structures, animal body plans have become more elaborate over the course of evolution [37]. Cnidaria were the first animal organisms to develop nervous systems [38]. Moreover, some Cnidaria in the medusozoan group display complex lens‐containing eyes [39, 40]. Since the current analysis suggests that Rax appeared in the common ancestor of Cnidaria and Bilateria, the evolutionary appearance of Rax might underlie the evolution of the eye in this ancestor. Functional analysis of Rax in extant Cnidaria may provide important clues to clarify the evolution of the eye.
Pax6 is a paired‐type homeobox gene [13], which is an ortholog of eyeless in flies and plays a critical role in eye formation in both flies and mammals. However, its evolutionary origin is after the divergence of Bilateria and Cnidaria [41, 42]. Although the cnidarian PaxB gene is considered to be related to Pax6 in Bilateria, the domain organization and DNA‐binding specificity of the paired domain differ between these two genes [41, 42]. In contrast, cnidarian Rax has the same domain organization as bilaterian Rax. Furthermore, amino acid sequences of the octapeptide, homeodomain, and OAR motif are highly conserved between cnidarian Rax and bilaterian Rax (Fig. 1B). Based on these observations, we propose the following scenario for the roles of Rax and Pax6 in eye evolution: Rax appeared in the common ancestor of Bilateria and Cnidaria, predating Pax6 in terms of the evolutionary origin and the involvement in eye formation; after the emergence of Pax6 in Bilateria, Rax and Pax6 began to act jointly in the eye development of Bilateria. Future evolutionary analyses of other homeobox transcription factors involved in eye formation, including Six3, Six6, and Lhx2, may deepen our understanding of the evolution of the eye [15].
The previous study examining Rax evolution analyzed earlier versions of the lamprey genome assemblies [43, 44] and identified one Rax gene [21]. They suggested that future studies may identify another Rax gene because of the incomplete nature of these genome assemblies [21]. The current study analyzed the latest assembly of the lamprey [45] and hagfish (GCA_900186335.2) genomes and identified one Rax gene in both genomes (Table 1). Moreover, our synteny analysis results suggest that the lamprey and hagfish Rax loci are orthologous (Fig. 2). Taken together, the current results further support the possibility that jawless vertebrates only possess one Rax.
Based on the results of our synteny and molecular phylogenetic analyses, we propose an alternative origin hypothesis of Rax and Rax2 in jawed vertebrates. These two genes might have resulted from segmental duplication of a small region containing Malt1, Rax, and Cplx4 ancestor genes in the common ancestor of jawed vertebrates (Fig. S3). This conclusion conflicts with the previous hypothesis that vertebrate Rax and Rax2 resulted from two rounds of whole‐genome duplication (WGD) [21]. Following two rounds of WGD that occurred at the root of vertebrates, many vertebrate genes have two to four paralogs [46]. For example, vertebrate genomes contain four Hox gene clusters or three Otx family genes: Otx2, Crx, and Otx5 [47]. Likewise, the previous study used molecular phylogenetic and synteny analyses to conclude that vertebrate Rax and Rax2 originated from two rounds of WGD that occurred in the common ancestor of vertebrates [21]. The authors conducted phylogenetic analysis including jawed vertebrate Rax, jawed vertebrate Rax2, and lamprey Rax. However, they did not include invertebrate Rax as an outgroup. Therefore, lamprey Rax can be arbitrarily assigned to Rax or Rax2. The authors assigned the lamprey Rax to Rax2 without detailing their justification [21]. Further, synteny analysis showed that the Rax and Rax2 loci were mapped to the same regions of the lancelet genome [21]. The authors used their synteny analysis results to support the conclusion that Rax and Rax2 originated from WGDs. However, segmental duplication of the ancient Rax locus could produce similar synteny analysis results (Fig. S4). Segmental duplications in vertebrate genomes are commonly observed. For example, ~ 4% of the human genome is covered by duplications, with segmental duplication accounting for up to 14% in individual chromosomes [48]. Segmental duplication is believed to occur via nonallelic homologous recombination in regions flanked by highly homologous sequences [49]. Since it is very likely that the genomes of jawed and jawless vertebrates’ common ancestor contained highly homologous duplicated sequences, such as transposable elements, segmental duplication events could occur frequently in their genome. Together, these considerations indicate that the synteny analysis results alone cannot completely exclude the possibility of segmental duplication. Moreover, we included four invertebrate Rax sequences as outgroups and used the lamprey and hagfish Rax sequences from their latest genomes [45] to perform more robust phylogenetic analyses than the previous study [21]. The current molecular phylogenetic analysis indicated that lamprey and hagfish Rax forms a sister group with Rax and Rax2 in jawed vertebrates, indicating that jawed vertebrate Rax and Rax2 originated from lineage‐specific segmental duplication events, not the WGDs (Fig. 4, Figs S1 and S2). However, since it is known that jawless vertebrates show amino acid composition biases, resolving orthology among jawless vertebrate Rax and jawed vertebrate Rax and Rax2 is challenging [50]. Therefore, it should be noted that our phylogenetic analysis results cannot exclude the possibility that Rax and Rax2 generation in jawed and jawless vertebrates is due to the two rounds of WGD as proposed in the previous study [21].
Another possible explanation for the current molecular phylogenetic analysis results regarding Rax and Rax2 of jawed vertebrates and Rax of jawless vertebrates is a delayed rediploidization after genome duplication [51]. In this model, following WGD, speciation predates rediploidization [51]. This leads to independent ohnolog divergence in sister lineages that share a common WGD event and provides ohnologs solely available for lineage‐specific adaptation [51]. It has been reported that 27.1% of ohnologs showed delayed rediploidization in salmonid fish [51]. Resolving orthology is difficult if ohnologs of interest independently diverged in sister lineages that share a common WGD. Therefore, it should be noted that the current molecular phylogenetic analysis results can also be affected by delayed rediploidization.
A comprehensive ortholog search for Rax and Rax2 in 86 mammalian genomes from all four major mammal groups found at least five independent Rax2 gene loss events (Fig. 6). These comprehensive analyses enabled us to raise an alternative explanation regarding the Rax2 loss in lagomorph and rodent (Glires). The previous study proposed that Rax2 was lost in a common ancestor of lagomorph and rodent [21]. However, they did not show the presence of Rax2 in squirrel species, which are a sister group to other rodent species [21]. In contrast, the current study demonstrated that Rax2 is present in the thirteen‐lined ground squirrel, arctic ground squirrel, and yellow‐bellied marmot (Fig. 6B). This finding suggests that Lagomorpha and Rodent independently lost Rax2.
We observed a loss of the Rax2 octapeptide in tetrapods and five independent Rax2 gene loss events in mammals (Figs 5 and 6). In contrast, no Rax gene loss events were identified in the analyzed mammalian genomes, suggesting that Rax is a highly evolutionarily conserved and functionally significant gene in mammals. Rax gene loss events may not be present in the analyzed mammalian species because it plays an essential role in central nervous system development [8]. Deletion of the Rax gene in mice, which lack the Rax2 gene, results in severe brain malformation, such as the absence of the ventral forebrain and failure of the optic vesicle to form [8]. Conversely, Rax can functionally compensate for loss of mammalian Rax2 in mice [6, 7, 11, 12]. In humans, unlike RAX2, RAX mutations are associated with symptoms affecting the whole eye. RAX mutations result in microphthalmia [9, 10], whereas RAX2 mutations are associated with cone–rod dystrophy or age‐related macular degeneration [52, 53]. Since RAX2 mutations can lead to age‐related disease, we hypothesize that the effects of RAX2 loss in some mammal lineages occur after the age of sexual maturity. Therefore, RAX2 loss has little effect on fitness. However, it also should be noted that sudden RAX2 loss cannot be compensated by RAX, as indicated by the association of RAX2 mutations with human diseases [52, 53]. Evolutionary deletion of Rax2 from mammal genomes might require gradual accumulation of amino acid substitutions. We found that the average Ka/Ks ratio of mammalian Rax2 was ~ 67% greater than that of mammalian Rax (Fig. 7B). This difference between mammalian Rax and Rax2 might partially explain why Rax2 is more defect‐prone than Rax in this animal group.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
TK and TF designed the study, performed molecular evolutionary analyses, and prepared the manuscript.
Supporting information
Fig. S1. Maximum‐likelihood trees of Rax and Rax2 in various animal species (related to Fig. 4).
Fig. S2. Neighbor‐joining trees of Rax and Rax2 in various animal species (related to Fig. 4).
Fig. S3. A hypothetical model of the origin of Rax and Rax2 in jawed vertebrates.
Fig. S4. Two possible Rax evolution scenarios.
Acknowledgements
We thank Dr. T. Chaya for helpful comments and the GIRC Computer System at Osaka University for conducting computations. This work was supported by Grant‐in‐Aid for Scientific Research (18H02593), Grant‐in‐Aid for Challenging Research (Exploratory) (T18K19427), and the Takeda Science Foundation.
References
- 1. Lamb TD, Collin SP and Pugh EN Jr (2007) Evolution of the vertebrate eye: opsins, photoreceptors, retina and eye cup. Nat Rev Neurosci 8, 960–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Burglin TR and Affolter M (2016) Homeodomain proteins: an update. Chromosoma 125, 497–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gehring WJ, Muller M, Affolter M, Percival‐Smith A, Billeter M, Qian YQ, Otting G and Wuthrich K (1990) The structure of the homeodomain and its functional implications. Trends Genet. 6, 323–329. [DOI] [PubMed] [Google Scholar]
- 4. Lappin TR, Grier DG, Thompson A and Halliday HL (2006) HOX genes: seductive science, mysterious mechanisms. Ulster Med J 75, 23–31. [PMC free article] [PubMed] [Google Scholar]
- 5. Larroux C, Fahey B, Degnan SM, Adamski M, Rokhsar DS and Degnan BM (2007) The NK homeobox gene cluster predates the origin of Hox genes. Curr Biol 17, 706–710. [DOI] [PubMed] [Google Scholar]
- 6. Furukawa T, Kozak CA and Cepko CL (1997) rax, a novel paired‐type homeobox gene, shows expression in the anterior neural fold and developing retina. Proc Natl Acad Sci USA 94, 3088–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Muranishi Y, Terada K and Furukawa T (2012) An essential role for Rax in retina and neuroendocrine system development. Dev Growth Differ 54, 341–348. [DOI] [PubMed] [Google Scholar]
- 8. Mathers PH, Grinberg A, Mahon KA and Jamrich M (1997) The Rx homeobox gene is essential for vertebrate eye development. Nature 387, 603–607. [DOI] [PubMed] [Google Scholar]
- 9. Voronina VA, Kozhemyakina EA, O'Kernick CM, Kahn ND, Wenger SL, Linberg JV, Schneider AS and Mathers PH (2004) Mutations in the human RAX homeobox gene in a patient with anophthalmia and sclerocornea. Hum Mol Genet 13, 315–322. [DOI] [PubMed] [Google Scholar]
- 10. Lequeux L, Rio M, Vigouroux A, Titeux M, Etchevers H, Malecaze F, Chassaing N and Calvas P (2008) Confirmation of RAX gene involvement in human anophthalmia. Clin Genet 74, 392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Muranishi Y, Terada K, Inoue T, Katoh K, Tsujii T, Sanuki R, Kurokawa D, Aizawa S, Tamaki Y and Furukawa T (2011) An essential role for RAX homeoprotein and NOTCH‐HES signaling in Otx2 expression in embryonic retinal photoreceptor cell fate determination. J Neurosci 31, 16792–16807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Irie S, Sanuki R, Muranishi Y, Kato K, Chaya T and Furukawa T (2015) Rax homeoprotein regulates photoreceptor cell maturation and survival in association with Crx in the postnatal mouse retina. Mol Cell Biol 35, 2583–2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Osumi N, Shinohara H, Numayama‐Tsuruta K and Maekawa M (2008) Concise review: Pax6 transcription factor contributes to both embryonic and adult neurogenesis as a multifunctional regulator. Stem Cells 26, 1663–1672. [DOI] [PubMed] [Google Scholar]
- 14. Klimova L and Kozmik Z (2014) Stage‐dependent requirement of neuroretinal Pax6 for lens and retina development. Development 141, 1292–1302. [DOI] [PubMed] [Google Scholar]
- 15. Zagozewski JL, Zhang Q, Pinto VI, Wigle JT and Eisenstat DD (2014) The role of homeobox genes in retinal development and disease. Dev Biol 393, 195–208. [DOI] [PubMed] [Google Scholar]
- 16. Zhang L, Mathers PH and Jamrich M (2000) Function of Rx, but not Pax6, is essential for the formation of retinal progenitor cells in mice. Genesis 28, 135–142. [PubMed] [Google Scholar]
- 17. Shendure J, Balasubramanian S, Church GM, Gilbert W, Rogers J, Schloss JA and Waterston RH (2017) DNA sequencing at 40: past, present and future. Nature 550, 345–353. [DOI] [PubMed] [Google Scholar]
- 18. Kitts PA, Church DM, Thibaud‐Nissen F, Choi J, Hem V, Sapojnikov V, Smith RG, Tatusova T, Xiang C, Zherikov A et al (2016) Assembly: a resource for assembled genomes at NCBI. Nucleic Acids Res 44, D73–D80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kumar S, Stecher G, Suleski M and Hedges SB (2017) TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol 34, 1812–1819. [DOI] [PubMed] [Google Scholar]
- 20. Alfoldi J and Lindblad‐Toh K (2013) Comparative genomics as a tool to understand evolution and disease. Genome Res 23, 1063–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Orquera DP and de Souza FSJ (2017) Evolution of the Rax family of developmental transcription factors in vertebrates. Mech Dev 144, 163–170. [DOI] [PubMed] [Google Scholar]
- 22. Mazza ME, Pang K, Reitzel AM, Martindale MQ and Finnerty JR (2010) A conserved cluster of three PRD‐class homeobox genes (homeobrain, rx and orthopedia) in the Cnidaria and Protostomia. Evodevo 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ryan JF, Burton PM, Mazza ME, Kwong GK, Mullikin JC and Finnerty JR (2006) The cnidarian‐bilaterian ancestor possessed at least 56 homeoboxes: evidence from the starlet sea anemone, Nematostella vectensis . Genome Biol 7, R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Leinonen R, Sugawara H, Shumway Ml; International Nucleotide Sequence Database Collaboration (2011) The sequence read archive. Nucleic Acids Res 39, D19–D21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q et al (2011) Full‐length transcriptome assembly from RNA‐Seq data without a reference genome. Nat Biotechnol 29, 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin‐Duran JM, Pang K, Borve A, Le HS, Furu A, Cannon JT, Jondelius U and Hejnol A (2018) Convergent evolution of bilaterian nerve cords. Nature 553, 45–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu W, Xie Y, Ma J, Luo X, Nie P, Zuo Z, Lahrmann U, Zhao Q, Zheng Y, Zhao Y et al (2015) IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics 31, 3359–3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sievers F and Higgins DG (2014) Clustal Omega, accurate alignment of very large numbers of sequences. Methods Mol Biol 1079, 105–116. [DOI] [PubMed] [Google Scholar]
- 29. Waterhouse AM, Procter JB, Martin DM, Clamp M and Barton GJ (2009) Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kumar S, Stecher G and Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nagarajan N and Pop M (2013) Sequence assembly demystified. Nat Rev Genet 14, 157–167. [DOI] [PubMed] [Google Scholar]
- 33. Rice P, Longden I and Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16, 276–277. [DOI] [PubMed] [Google Scholar]
- 34. Sato JJ, Ohdachi SD, Echenique‐Diaz LM, Borroto‐Paez R, Begue‐Quiala G, Delgado‐Labanino JL, Gamez‐Diez J, Alvarez‐Lemus J, Nguyen ST, Yamaguchi N et al (2016) Molecular phylogenetic analysis of nuclear genes suggests a Cenozoic over‐water dispersal origin for the Cuban solenodon. Sci Rep 6, 31173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. He K, Shinohara A, Helgen KM, Springer MS, Jiang XL and Campbell KL (2017) Talpid mole phylogeny unites shrew moles and illuminates overlooked cryptic species diversity. Mol Biol Evol 34, 78–87. [DOI] [PubMed] [Google Scholar]
- 36. Schierwater B, Kamm K, Srivastava M, Rokhsar D, Rosengarten RD and Dellaporta SL (2008) The early ANTP gene repertoire: insights from the placozoan genome. PLoS ONE 3, e2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Levin M, Anavy L, Cole AG, Winter E, Mostov N, Khair S, Senderovich N, Kovalev E, Silver DH, Feder M et al (2016) The mid‐developmental transition and the evolution of animal body plans. Nature 531, 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Holland LZ, Carvalho JE, Escriva H, Laudet V, Schubert M, Shimeld SM and Yu JK (2013) Evolution of bilaterian central nervous systems: a single origin? EvoDevo 4, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Picciani N, Kerlin JR, Sierra N, Swafford AJM, Ramirez MD, Roberts NG, Cannon JT, Daly M and Oakley TH (2018) Prolific origination of eyes in Cnidaria with co‐option of non‐visual opsins. Curr Biol 28, 2413–2419.e4. [DOI] [PubMed] [Google Scholar]
- 40. Piatigorsky J and Kozmik Z (2004) Cubozoan jellyfish: an Evo/Devo model for eyes and other sensory systems. Int J Dev Biol 48, 719–729. [DOI] [PubMed] [Google Scholar]
- 41. Kozmik Z (2008) The role of Pax genes in eye evolution. Brain Res Bull 75, 335–339. [DOI] [PubMed] [Google Scholar]
- 42. Kozmik Z (2005) Pax genes in eye development and evolution. Curr Opin Genet Dev 15, 430–438. [DOI] [PubMed] [Google Scholar]
- 43. Smith JJ, Kuraku S, Holt C, Sauka‐Spengler T, Jiang N, Campbell MS, Yandell MD, Manousaki T, Meyer A, Bloom OE et al (2013) Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat Genet 45, 415–421, 421e1‐2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mehta TK, Ravi V, Yamasaki S, Lee AP, Lian MM, Tay BH, Tohari S, Yanai S, Tay A, Brenner S et al (2013) Evidence for at least six Hox clusters in the Japanese lamprey (Lethenteron japonicum). Proc Natl Acad Sci USA 110, 16044–16049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith JJ, Timoshevskaya N, Ye C, Holt C, Keinath MC, Parker HJ, Cook ME, Hess JE, Narum SR, Lamanna F et al (2018) The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat Genet 50, 270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vandepoele K, De Vos W, Taylor JS, Meyer A and Van de Peer Y (2004) Major events in the genome evolution of vertebrates: paranome age and size differ considerably between ray‐finned fishes and land vertebrates. Proc Natl Acad Sci USA 101, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Holland PW (2013) Evolution of homeobox genes. Wiley Interdiscip Rev Dev Biol 2, 31–45. [DOI] [PubMed] [Google Scholar]
- 48. Zhang L, Lu HH, Chung WY, Yang J and Li WH (2005) Patterns of segmental duplication in the human genome. Mol Biol Evol 22, 135–141. [DOI] [PubMed] [Google Scholar]
- 49. Sharp AJ, Locke DP, McGrath SD, Cheng Z, Bailey JA, Vallente RU, Pertz LM, Clark RA, Schwartz S, Segraves R et al (2005) Segmental duplications and copy‐number variation in the human genome. Am J Hum Genet 77, 78–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qiu H, Hildebrand F, Kuraku S and Meyer A (2011) Unresolved orthology and peculiar coding sequence properties of lamprey genes: the KCNA gene family as test case. BMC Genom 12, 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Robertson FM, Gundappa MK, Grammes F, Hvidsten TR, Redmond AK, Lien S, Martin SAM, Holland PWH, Sandve SR and Macqueen DJ (2017) Lineage‐specific rediploidization is a mechanism to explain time‐lags between genome duplication and evolutionary diversification. Genome Biol 18, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wang QL, Chen S, Esumi N, Swain PK, Haines HS, Peng G, Melia BM, McIntosh I, Heckenlively JR, Jacobson SG et al (2004) QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum Mol Genet 13, 1025–1040. [DOI] [PubMed] [Google Scholar]
- 53. Yang P, Chiang PW, Weleber RG and Pennesi ME (2015) Autosomal dominant retinal dystrophy with electronegative waveform associated with a novel RAX2 mutation. JAMA Ophthalmol 133, 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pisani D, Pett W, Dohrmann M, Feuda R, Rota‐Stabelli O, Philippe H, Lartillot N and Worheide G (2015) Genomic data do not support comb jellies as the sister group to all other animals. Proc Natl Acad Sci USA 112, 15402–15407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Feuda R, Dohrmann M, Pett W, Philippe H, Rota‐Stabelli O, Lartillot N, Worheide G and Pisani D (2017) Improved modeling of compositional heterogeneity supports sponges as sister to all other animals. Curr Biol 27, 3864–3870.e4. [DOI] [PubMed] [Google Scholar]
- 56. Philippe H, Poustka AJ, Chiodin M, Hoff KJ, Dessimoz C, Tomiczek B, Schiffer PH, Muller S, Domman D, Horn M et al (2019) Mitigating anticipated effects of systematic errors supports sister‐group relationship between xenacoelomorpha and ambulacraria. Curr Biol 29, 1818–1826.e6. [DOI] [PubMed] [Google Scholar]
- 57. Moroz LL, Kocot KM, Citarella MR, Dosung S, Norekian TP, Povolotskaya IS, Grigorenko AP, Dailey C, Berezikov E, Buckley KM et al (2014) The ctenophore genome and the evolutionary origins of neural systems. Nature 510, 109–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F and Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Meredith RW, Janecka JE, Gatesy J, Ryder OA, Fisher CA, Teeling EC, Goodbla A, Eizirik E, Simao TL, Stadler T et al (2011) Impacts of the cretaceous terrestrial revolution and KPg extinction on mammal diversification. Science 334, 521–524. [DOI] [PubMed] [Google Scholar]
- 60. O'Leary MA, Bloch JI, Flynn JJ, Gaudin TJ, Giallombardo A, Giannini NP, Goldberg SL, Kraatz BP, Luo ZX, Meng J et al (2013) The placental mammal ancestor and the post‐K‐Pg radiation of placentals. Science 339, 662–667. [DOI] [PubMed] [Google Scholar]
- 61. Zhao T and Schranz ME (2019) Network‐based microsynteny analysis identifies major differences and genomic outliers in mammalian and angiosperm genomes. Proc Natl Acad Sci USA 116, 2165–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Maximum‐likelihood trees of Rax and Rax2 in various animal species (related to Fig. 4).
Fig. S2. Neighbor‐joining trees of Rax and Rax2 in various animal species (related to Fig. 4).
Fig. S3. A hypothetical model of the origin of Rax and Rax2 in jawed vertebrates.
Fig. S4. Two possible Rax evolution scenarios.


