Abstract
Group A Streptococcus is a pathogen of global importance, but despite the ubiquity of group A Streptococcus infections, the relationship between infection, colonization, and immunity is still not completely understood. The M protein, encoded by the emm gene, is a major virulence factor and vaccine candidate and forms the basis of a number of classification systems. Longitudinal patterns of emm types collected from 457 Fijian schoolchildren over a 10-month period were analyzed. No evidence of tissue tropism was observed, and there was no apparent selective pressure or constraint of emm types. Patterns of emm type acquisition suggest limited, if any, modification of future infection based on infection history. Where impetigo is the dominant mode of transmission, circulating emm types either may not be constrained by ecological niches or population immunity to the M protein, or they may require several infections over a longer period of time to induce such immunity.
Keywords: Streptococcus pyogenes, emm cluster, immunity, skin infection
We examined longitudinal patterns of emm types in group A Streptococcus samples collected from Fijian schoolchildren. In a setting where impetigo is the dominant mode of transmission, we found no evidence that infection history modifies future infection.
(See the Editorial Commentary by Beall and Van Beneden, on pages 1394–7.)
Group A Streptococcus (GAS) is a pathogen of global importance, responsible for >700 million superficial infections and ≥500 000 deaths per year [1, 2]. Almost all diseases caused by GAS are most common in developing regions, from superficial conditions, such as pyoderma (including impetigo) and pharyngitis, to severe sequelae, including invasive disease, rheumatic heart disease, and poststreptococcal glomerulonephritis [1]. Despite the ubiquity of GAS, our understanding of its immunobiology and the relationship between infection, colonization, and immunity remain incomplete, hindering efforts at sustainable control, including the development of safe and effective vaccines.
The GAS M protein is a major virulence factor that elicits antibody production and enables the bacteria to inhibit phagocytosis in the absence of antibodies, making it a prime vaccine candidate [3]. This protein forms the basis of a number of classification systems for GAS: emm typing, emm patterns, and emm clusters, where emm refers to the emm gene encoding this surface M protein [4]. More than 240 emm types have been identified based on the variable N-terminus part of the protein, contributing to the complexity of epidemiologic studies [5]. Based on the structure of emm and emm-like genes in the GAS genome, emm types may be further grouped into emm patterns, referred to as A–C, D, and E [6].
Recently, emm types have been grouped into 48 emm clusters based on closely related sequences, shared structural characteristics, and similar binding capacities [7, 8]. It has been hypothesized that cross-protective immunity may occur between emm types that exist within the same emm cluster. Preliminary and limited laboratory studies have shown that in vitro cross-protection does occur within certain emm clusters in Fijian children [8]. The existence of cross-protective immunity within emm clusters could substantially aid vaccine development against this multistrain pathogen. However, it remains to be seen whether these findings translate to population-level protection.
The relationship between emm types and disease burden is important when selecting priority strains for prevention. It has long been believed that different GAS strains preferentially cause either impetigo or pharyngitis [9]. Based on a number of population-based surveys, GAS strains with the emm pattern A–C display a tropism for the throat, whereas D have a tropism for the skin and E are found in both tissue sites [6, 10]. However, the mechanism responsible for different disease manifestations remains to be identified [11]. The distributions of emm types and clinical manifestations differ between settings, with fewer emm types circulating in low-prevalence settings (typically dominated by pharyngitis) than in high-prevalence settings (typically dominated by impetigo), and with some emm types common in developed countries rarely found in developing countries [12–14].
In the current study, we investigated longitudinal data on emm types, patterns, and clusters at individual, school, and regional levels to evaluate emm immunobiology within a streptococcal disease endemic setting. We examined emm types isolated from children over time for evidence of immune protection at either the emm type or emm cluster level. First, we hypothesized that if immunity to prevalent emm types did develop, we would observe the subsequent disappearance of these emm types at individual and population levels as opportunities for further transmission diminished, followed by the appearance of new emm types to which the population had no prior immunity. Second, we also hypothesized that if immunity to emm types did develop, we would observe different emm types circulating in children infected at a single time point than in children infected at multiple time points, because immunity would limit the acquisition of emm types that could cause a subsequent infection. Third, we framed similar hypotheses in terms of emm clusters.
METHODS
Ethical Approval
This study was approved by the Fiji National Research Ethics Review Committee, the Fiji National Health Research Committee, and the University of Melbourne Human Research Ethics Committee. Written informed consent was required from participants or from a parent or guardian before collection of information.
Setting and Participants
This was a prospective longitudinal cohort study conducted in 3 schools in the Central Division of Fiji from February 2006 to November 2006. Two of the schools were rural, with all children of iTaukei (Indigenous Fijian) ethnicity, and the third school was located in Suva, with most children of Indo-Fijian ethnicity. Enrollment rates for the 3 schools were 96.4% (rural school 1), 80.6% (rural school 2), and 53.2% (urban school) [15].
For skin screening, each school was visited every 2 months over a 10-month period, with a total of 6 visits per school. Children aged 5–15 years were screened for skin sores, and swab samples were obtained from crusted or purulent sores [15]. At the first skin screening visit, children without sore throat symptoms had their throats swabbed for evidence of asymptomatic colonization [16]. Over the 10-month period, each school was visited twice per week, and throat swab samples were collected from children reporting sore throat symptoms within the preceding 7 days [16].
Laboratory Methods
Skin and throat swab samples were collected, transported, and tested for the presence of β-hemolytic colonies using standard methods [15, 16]; emm typing of the dominant β-hemolytic colony was undertaken according to US Centers for Disease Control and Prevention standard methods [15–17].
Data Preparation and Analysis
Characterization and Analysis of emm Types
Data were restricted to Lancefield group A isolates, and emm types were assigned to emm clusters per Sanderson-Smith et al [7] and analyzed by time period, school, disease type, and participant. Each skin screening visit took place during a period of approximately 2 weeks, followed by a window of about 6 weeks before the next round of skin screening visits began. The study period was therefore divided into 6 screening time periods, including 5 of approximately 2 months’ duration, each running from the date of the first skin screening associated with that visit to the day before the first skin screening associated with the next visit, and a sixth visit of approximately 2 weeks’ duration, covering the final skin screening visit. Throat isolates were assigned to the screening period within which they were collected.
Diversity and Prevalence
At the child level, the number of distinct emm types and emm clusters was tabulated against the number of positive swab samples. The Simpson index of diversity, the probability that 2 randomly selected emm types are different, was calculated for each screening period by school [18].
The total number of isolates of each emm cluster and each emm type were calculated. The prevalence of each emm type per 1000 children was calculated at each time point by dividing the number of isolates by the number of children participating in that visit, stratified to the same level. The emm clusters were ordered by the highest number of samples overall, and within each cluster, emm types were similarly ordered.
Individual-Level Exposure Responses
Children with >1 isolate of the same emm type during the study were identified and their screening, pharyngitis and emm type history reported, noting that the absence of an isolate for a screening visit may indicate that no sores were present, sores were present but no swab sample was obtained (because sores were not crusted or purulent, as specified in the study protocol), or a swab sample was obtained but GAS did not grow. For these children, we also report the emm clusters corresponding to their emm types, in the Supplementary Material.
The ordering of emm types and emm clusters from highest to lowest number of isolates was compared between 2 groups of children: those with a single positive isolate and those with multiple positive isolates. Data were prepared using Stata (release 14; StataCorp) and MATLAB 2017b (The Mathworks) software, and analyses were conducted using R software (version 3.4.4).
RESULTS
Demographics
A total of 457 children were enrolled in the study, with a minimum of 400 seen at any of the 6 skin screening visits and 80% of children seen at all of them. The number of children screened per visit was 73–80 and 160–175 for rural schools 1 and 2, and 161–202 for the urban school. All 255 children enrolled from rural schools 1 and 2 identified as iTaukei (Indigenous Fijian); the population at the urban school included of 99 Indo-Fijian children (49%), 67 (33%) iTaukei, and 36 (18%) of other ethnicities. The sexes were evenly distributed, with 229 of the 457 children female. The children’s median age was 9.9 years (interquartile range, 7.9–12.0 years).
Clinical Data
There were 451 GAS-positive isolates collected from 245 children during the study. Of these, 379 (84%) were from impetigo samples, 45 (10%) from pharyngitis samples, and 27 (6%) from asymptomatic throat colonization samples, which were collected only at the first visit. The median prevalence of GAS infection per 1000 children per screening period (excluding colonization) was highest in rural school 2 (227.5; interquartile range, 199.2–247.4), followed by rural school 1 (167.6, 135.1–237.5) and the urban school, with the lowest observed prevalence (97.0, 78.4–135.5).
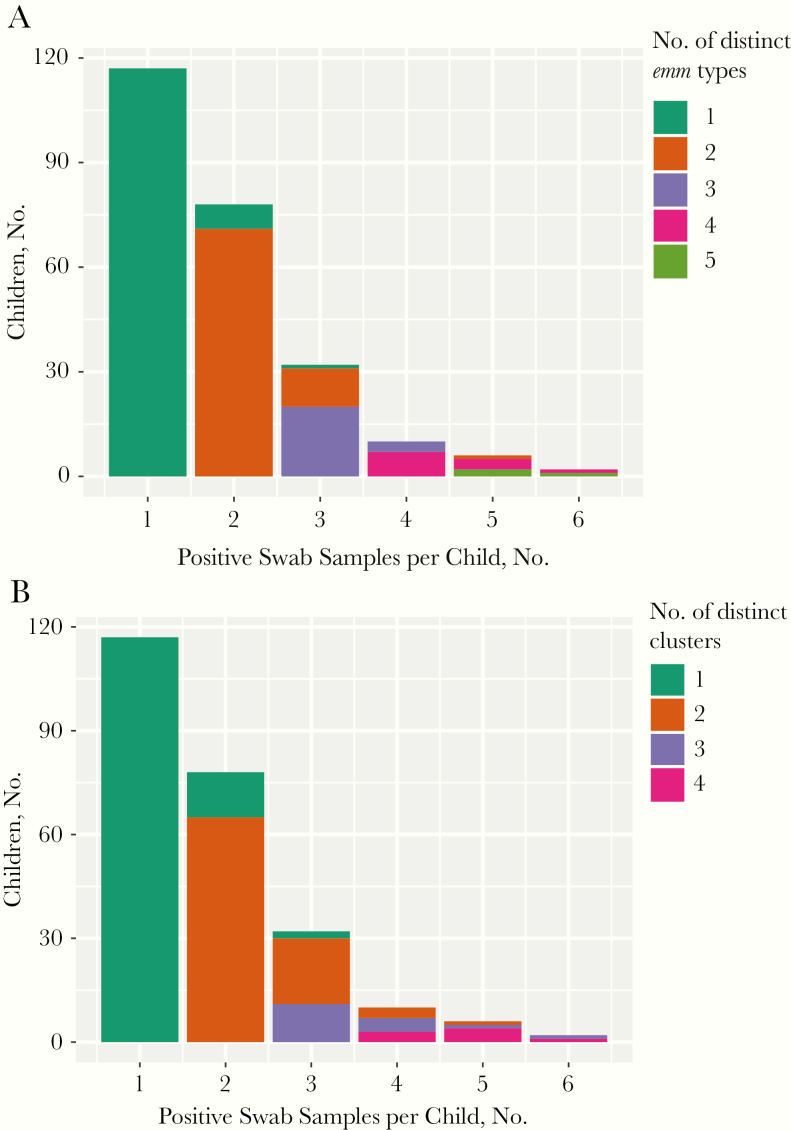
Of the 245 children with GAS-positive isolates, most (195 [80%]) had either 1 or 2 GAS-positive swab samples (Figure 1A). The highest number of GAS-positive swab samples (impetigo, pharyngitis, and asymptomatic throat colonization combined) for any child was 6, in 2 children (<1%).
Figure 1.
Numbers of group A Streptococcus–positive swab samples per child, colored according to the number of distinct emm types (A) or distinct emm clusters (B) present in these samples.
Diversity and Prevalence
The 5 most frequently observed emm types were emm70, emm33, emm25, emm93.3, and emm11, accounting for about 30% of positive isolates. This ranking held both for children infected only once and for children infected more than once. The 5 most frequently observed clusters were D4, E3, E6, E4, and E2, for children infected only once and for children infected more than once, accounting for 77% of positive isolates in both groups.
Of the 128 children with >1 GAS-positive swab sample, 100 had a different emm type and 83 had a different emm cluster isolated from each of their swab samples. Most children with 2 positive swab samples had 2 different emm clusters (65 of 78 children [83%]) rather than a single cluster. Most children with ≥3 positive swab samples had ≥1 repeated emm cluster (36 of 50 children [72%]) (Figure 1B).
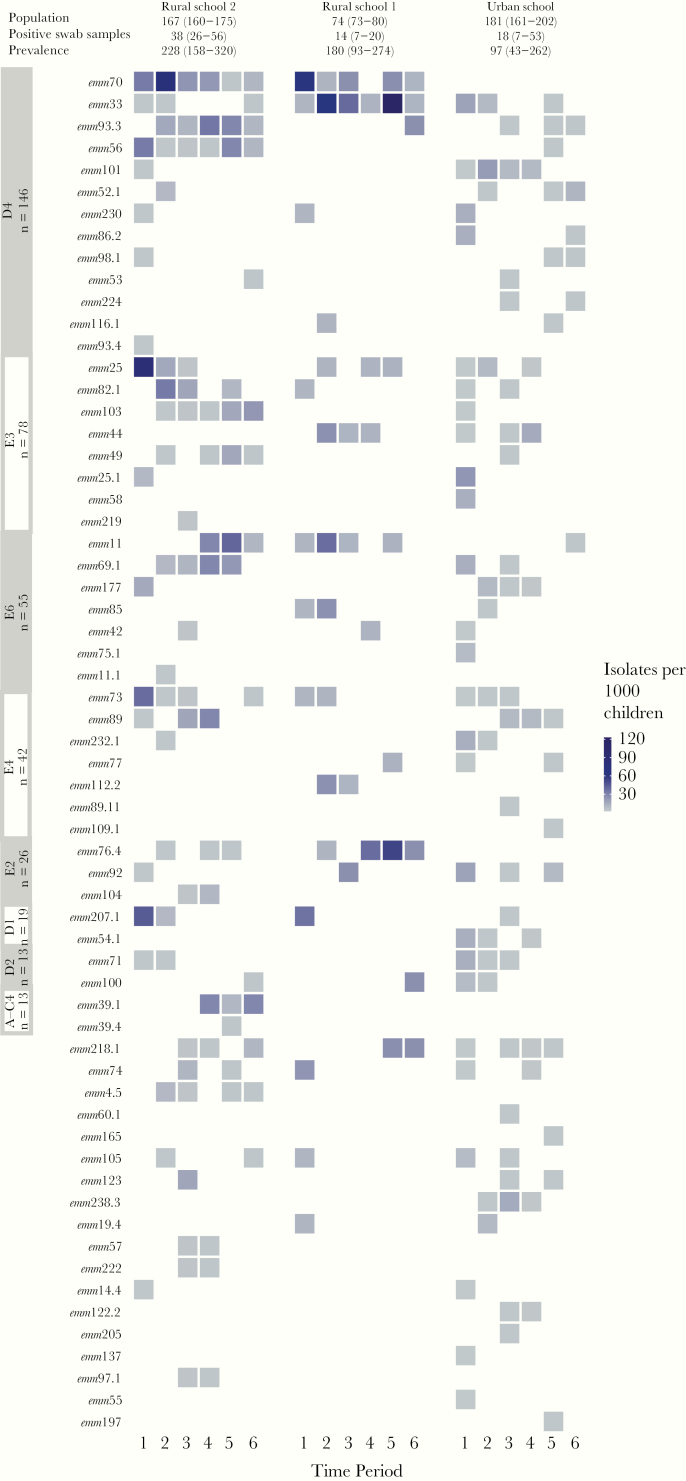
A wide variety of emm types circulated during all 6 screening periods (range, 19–37 emm types per screening period). There was no evidence of prevalent emm types disappearing, followed by the appearance of new dominant emm types (Figure 2). Rather, the most prevalent emm types were consistently present in a given setting (eg, emm70 and emm33), and less prevalent emm types were detected during only 1 or 2 screening periods. There was no evidence of competitive exclusion of emm types at the population level, with each emm cluster having several isolates circulating concurrently in the same setting. Urban and rural schools seemed to have different patterns of circulation, with emm70 highly prevalent in both rural schools throughout the entire study but completely absent from the urban school. In contrast to the rural schools, there were no dominant emm types at the urban school, and no emm types consistently isolated across all screening periods.
Figure 2.
Prevalence of emm types per 1000 children for each screening time period, grouped by cluster and stratified by school. The emm clusters (labeled on y-axis) are ordered by the overall number of isolates, and within each emm cluster, emm types are also ordered by overall number of isolates. Data for each school include the median population, number of positive swab samples, and overall prevalence, together with their ranges.
Despite differences in the overall prevalence of GAS-positive swab samples for each screening period, high diversity was observed across both rural and urban settings (Figure 2), with the Simpson index of diversity ranging from 0.77 to 0.99 (Supplementary Figure 1). No clear association was observed between prevalence and diversity, and we did not perform a formal test of association, given the small number of data points and observed differences per setting. The prevalence of infection by cluster related directly to the number of emm types categorized within each cluster. For example, the most prevalent cluster in our study was D4, which is the largest cluster with 32 emm types. The next most prevalent cluster was E3, which is the second largest cluster with 19 emm types (Supplementary Figure 2).
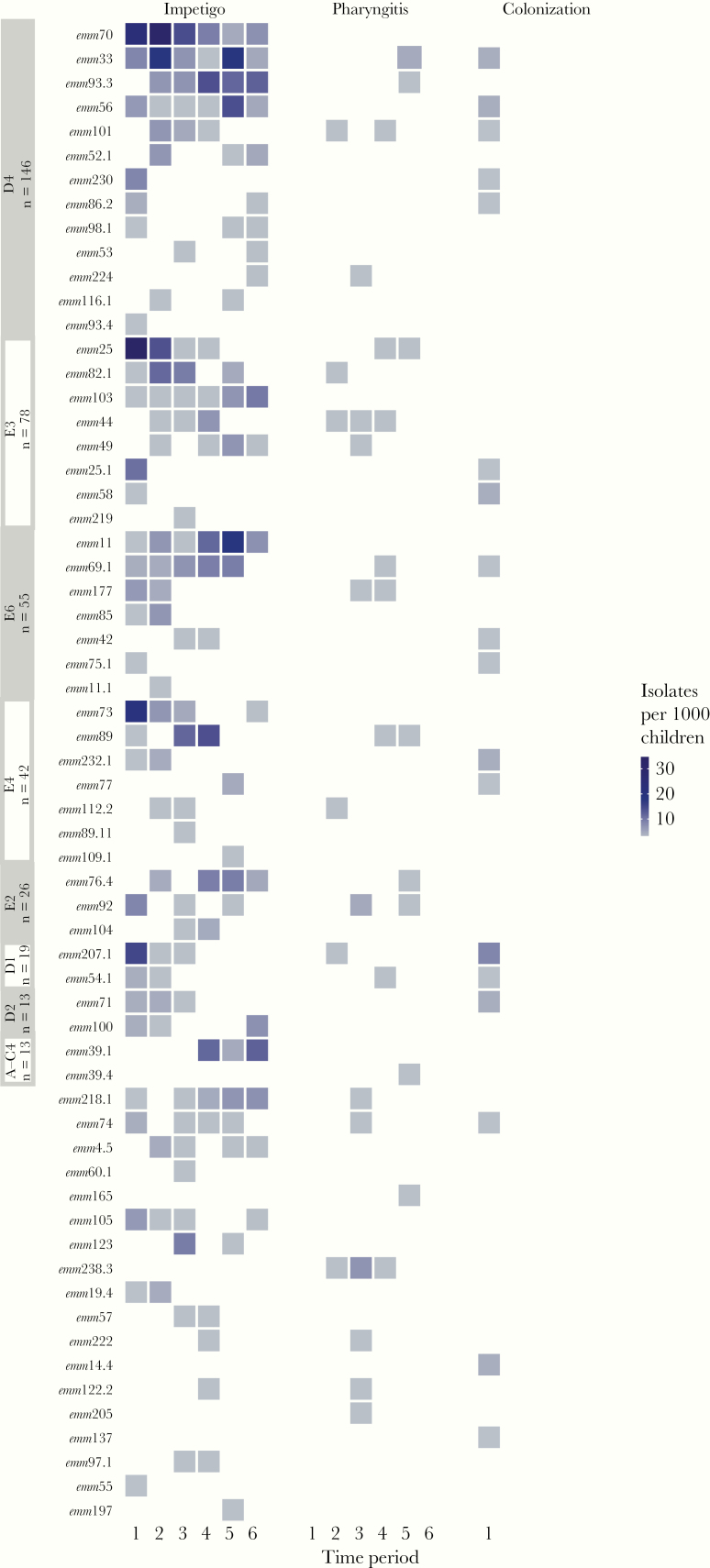
The vast majority of emm types (56 of 62 detected) were recovered from impetigo samples (Figure 3). Even for those emm types consistently present throughout the study period, isolation from pharyngitis was rare and generally not repeated over >2 consecutive screening periods (with the exceptions of emm44, emm92 emm101, and emm238.3). Throat colonization isolates collected at the first skin screening visit did not greatly increase the emm type diversity, adding only 2 emm types (emm137 and emm14.4). Whereas cluster D4, typically associated with skin infection, dominated the impetigo isolates, it was also present in pharyngitis isolates during each screening period, along with cluster E3 (Supplementary Figure 3). Throat colonization isolates collected at the first skin screening visit were mostly from the clusters with the highest prevalence of skin isolates at that time, with emm pattern D comprising 63% of the colonization isolates (Supplementary Table 1). Conversely, emm pattern A–C, typically associated with throat infection, was isolated from about 16% of pharyngitis isolates.
Figure 3.
Prevalence of emm types per 1000 children for each screening time period, grouped by cluster and stratified by specimen type (impetigo, pharyngitis, or colonization). The emm clusters (labeled on y-axis) are ordered by the overall number of isolates, and within each emm cluster, emm types are also ordered by the overall number of isolates.
Individual-Level Exposure Responses
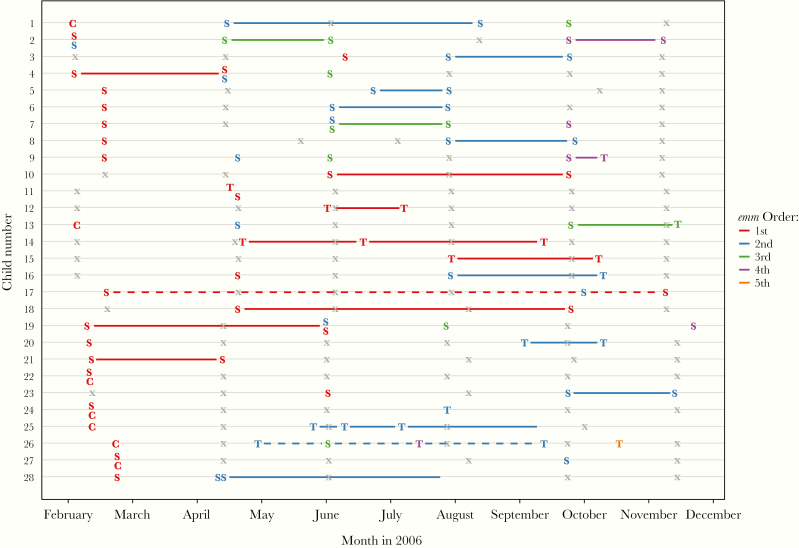
For the 28 children with repeated isolation of the same emm type, a variety of longitudinal patterns was observed (Figure 4). Four children (children 9, 11, 13, and 16) had the same emm type isolated from pharyngitis and impetigo samples at different times, and 3 (children 22, 24, and 27) had the same emm type isolated from throat colonization and impetigo samples on the same day. For skin infections, reacquisition of the same emm type after a documented skin screening without GAS infection was observed in 6 children (children 1, 10, 17, 18, 19, and 28). Two different emm types were isolated from impetigo samples obtained on the same day in 4 children (children 2, 4, 7, and 19). For these same 28 children (Supplementary Figures 4 and 5), we observed different emm types from the same cluster isolated at the same time (child 4), and at different times (children 1, 2, 4, 7, 17, and 26). In addition, 3 of the 28 children had emm types from different clusters isolated concurrently (children 2, 7, and 19).
Figure 4.
Longitudinal information for the subset of 28 children with repeated isolation of the same emm type. Solid lines connect the same emm type isolated more than once; dotted lines, different emm types isolated in the 2 samples. Colors represent unique emm types, with symbols representing the type of sample (S, skin; T, throat [with symptoms]; and C, throat [without symptoms]). Each x represents a skin screening visit attended by the child during which no group A Streptococcus (GAS) was isolated, either because no sores were present, sores were present but no swab sample was obtained, or a sample was obtained but GAS did not grow.
Discussion
Our analysis of longitudinal GAS emm types, patterns, and clusters in Fijian schoolchildren provides insights into the links between prevalence, diversity, and tissue tropism. We found no evidence of tissue tropism in this setting, with the emm types isolated from pharyngitis and throat colonization samples reflecting those isolated from impetigo samples, the dominant mode of infection in this cohort. There was no evidence of displacement of one emm type by another over time, with common emm types generally present throughout the study. There was no apparent selective pressure or constraint of emm types within clusters, with multiple emm types from each cluster circulating concurrently. Despite differences in prevalence by site and by time, high levels of diversity were observed in both urban and rural settings.
The patterns of acquisition in the small number of children with the same emm type isolated more than once suggest limited, if any, modification of future infection based on infection history in these children. It may be that the small number of children experiencing repeated infection with the same emm type indicates that most children developed immunity to the emm type that caused their infection. We cannot be certain whether the limited number of repeated infections we observed is due to immune protection or a lack of reexposure to the same emm type. Our observation that the same emm types and clusters circulated in children infected at single and multiple time points raises questions about the immunity induced by the M protein in this tropical setting and its capacity to limit the collection of emm types or clusters that could cause subsequent infections. It is important to note that our conclusions regarding individual and population immunity to GAS are based on infection patterns rather than measurement of M type–specific or other antibodies to GAS. However, measurement and understanding of the GAS immune response is not straightforward, with inconsistent development of antibodies after acquisition and a lack of evidence for a protective effect [19–21].
A particular strength of our study is the novel longitudinal analysis of emm types and clusters, with limited loss to follow-up. More than 80% of children in our cohort were seen at all 6 skin screening visits, with swab samples obtained from all crusted and purulent skin sores, providing rich data on the bacterial population. As with all studies, there are limitations. With a likely resolution time of <1 month (Adrian Marcato, The Peter Doherty Institute for Infection and Immunity, personal communication), we may have missed detecting some impetigo infections that arose and resolved within our 2-month screening period. Additional infections could add to the level of observed strain diversity, making our estimates a lower bound of diversity.
Furthermore, although children may have had as many as 2 swab samples collected from different impetigo lesions, only the dominant colony from each sample was emm typed. We may have missed detecting multiple emm types within a single lesion, again meaning that our results may underestimate overall diversity. The sporadic detection of many emm types in our age-limited population sample suggests that the relevant mixing pool is wider than schools, and infection may be readily acquired elsewhere in the community, including households. However, because only symptomatic infections were swabbed after the first skin screening visit, we were unable to investigate any relationship between throat colonization and skin infection, and these emm types may have been present asymptomatically in our cohort throughout the study. It is possible that GAS recovered from throat swab samples during pharyngitis episodes was also asymptomatic colonization. For the single time point with throat colonization data, all but 2 of the emm types found in asymptomatic children were also present in impetigo and/or pharyngitis isolates. However, without longitudinal data on asymptomatic colonization in this cohort, we cannot estimate the overall contribution of colonization to strain diversity.
The large number of concurrently circulating emm types (between 19 and 37 types per screening period) is consistent with observations from other settings with a high prevalence of impetigo [13, 22–24]. Studies in a mouse model of skin infection suggest that reinfection with the same emm type within a short period is required to stimulate type-specific immunity after skin infection [25]. If this requirement holds true for human infections, this phenomenon would be anticipated to positively select for diversity in high-prevalence settings, because a large number of circulating emm types makes reexposure to the same emm type less likely. The observation that very few children (28 of 457 [6.1%]) had repeated isolation of the same emm type within the study period suggests that there would be limited opportunity for the development of type-specific immunity, should reexposure be necessary. In an earlier study in a low-prevalence population, where colonization was the dominant source of GAS isolates and pharyngitis the dominant disease manifestation, children were equally likely to acquire the same emm type as to acquire a different emm type in a single year [26]. This observation further supports the notion that low-prevalence/low-diversity settings afford greater opportunity for the development of natural immunity, given the greater likelihood of reexposure to the same strain.
We did not observe the classic epidemiologic picture of A–C pattern strains dominating pharyngitis isolates and pattern D representing a very small percentage of pharyngitis isolates [6]. Rather, we observed patterns D and E dominant for both impetigo and pharyngitis (and pattern D dominant for colonization), consistent with studies in other high-prevalence settings [12–14, 27, 28]. Causal mechanisms for this tropism remain uncertain [6, 29], but they seem to be overwhelmed by transmission pressure in high-prevalence settings.
An in vitro study of a subset of the children in our analysis found that skin infection may elicit a functional immune response in some children with M type–specific and cross-reactive immune responses after skin infection [8]. However, results varied depending on the emm cluster analyzed, and the evidence was not strong for the D4 cluster, the most prevalent in our population [8]. Our observations of reacquisition of the same emm type or cluster during the period of our study suggest that the duration of protective immunity after infection may be short or even absent in some children. One could hypothesize that other, non–M protein, GAS antigens may be needed to protect against GAS infection owing to D4 cluster in particular, and owing to skin-associated emm types in a tropical setting in general. A GAS vaccine would definitively need to provide a substantially longer duration of protection than that we observed for some children to have an impact on prevalence.
This longitudinal study suggests that in settings where impetigo is the dominant mode of transmission, circulating emm types either may not be constrained by ecological niches or population immunity to the M protein or may require several infections over a longer period of time to induce such immunity. With limited evidence of M immunity to GAS apparent in our data, further work is needed to understand how settings have transitioned from high to low prevalence (as has happened in many developed countries), because this may provide clues for future control.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Financial support. This work was supported by the National Institute of Allergy and Infectious Diseases (grant U01AI60579) and the Australian National Health and Medical Research Council (Career Development Fellowship GNT1145033 to S. Y. C. T., Principal Research Fellowship GNT1117140 to J. M., and project grants GNT1098319 and GNT1130455).
Potential conflicts of interest. All authors: No reported conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Carapetis JR, Steer AC, Mulholland EK, Weber M. The global burden of group A streptococcal diseases. Lancet Infect Dis 2005; 5:685–94. [DOI] [PubMed] [Google Scholar]
- 2. Sanyahumbi AS, Colquhoun S, Wyber R, Carapetis JR. Global disease burden of group A Streptococcus. In: Ferretti JJ, Stevens DL, Fischetti VA, eds. Streptococcus pyogenes: basic biology to clinical manifestations. Oklahoma City, OK: The University of Oklahoma Health Sciences Center, 2017:661–704. [Google Scholar]
- 3. Fischetti VA. M protein and other surface proteins on streptococci. In: Ferretti JJ, Stevens DL, Fischetti VA, eds. Streptococcus pyogenes: basic biology to clinical manifestations. Oklahoma City, OK: The University of Oklahoma Health Sciences Center, 2017:27–54. [Google Scholar]
- 4. Bessen DE. Molecular basis of serotyping and the underlying genetic organization of Streptococcus pyogenes. In: Ferretti JJ, Stevens DL, Fischetti VA, eds. Streptococcus pyogenes: basic biology to clinical manifestations. Oklahoma City, OK: The University of Oklahoma Health Sciences Center, 2017:97–108. [Google Scholar]
- 5. Bessen DE, Smeesters PR, Beall BW. Molecular epidemiology, ecology, and evolution of group A streptococci. Microbiol Spectr 2018; 6:CPP3-0009-2018. [DOI] [PubMed] [Google Scholar]
- 6. Bessen DE. Tissue tropisms in group A Streptococcus: what virulence factors distinguish pharyngitis from impetigo strains? Curr Opin Infect Dis 2016; 29:295–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sanderson-Smith M, De Oliveira DM, Guglielmini J, et al. ; M Protein Study Group A systematic and functional classification of Streptococcus pyogenes that serves as a new tool for molecular typing and vaccine development. J Infect Dis 2014; 210:1325–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frost HR, Laho D, Sanderson-Smith ML, et al. . Immune cross-opsonization within emm clusters following group A Streptococcus skin infection: broadening the scope of type-specific immunity. Clin Infect Dis 2017; 65:1523–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wannamaker LW. Differences between streptococcal infections of the throat and of the skin (second of two parts). N Engl J Med 1970; 282:78–85. [DOI] [PubMed] [Google Scholar]
- 10. Bessen DE, Lizano S. Tissue tropisms in group A streptococcal infections. Future Microbiol 2010; 5:623–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Smeesters PR, McMillan DJ, Sriprakash KS, Georgousakis MM. Differences among group A Streptococcus epidemiological landscapes: consequences for M protein-based vaccines? Expert Rev Vaccines 2009; 8:1705–20. [DOI] [PubMed] [Google Scholar]
- 12. Tewodros W, Kronvall G. M protein gene (emm type) analysis of group A beta-hemolytic streptococci from Ethiopia reveals unique patterns. J Clin Microbiol 2005; 43:4369–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smeesters PR, Vergison A, Campos D, de Aguiar E, Miendje Deyi VY, Van Melderen L. Differences between Belgian and Brazilian group A Streptococcus epidemiologic landscape. PLoS One 2006; 1:e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sakota V, Fry AM, Lietman TM, Facklam RR, Li Z, Beall B. Genetically diverse group A streptococci from children in far-western Nepal share high genetic relatedness with isolates from other countries. J Clin Microbiol 2006; 44:2160–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Steer AC, Jenney AW, Kado J, et al. . High burden of impetigo and scabies in a tropical country. PLoS Negl Trop Dis 2009; 3:e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Steer AC, Jenney AW, Kado J, et al. . Prospective surveillance of streptococcal sore throat in a tropical country. Pediatr Infect Dis J 2009; 28:477–82. [DOI] [PubMed] [Google Scholar]
- 17. Centers for Disease Control and Prevention. M protein gene (emm) typing https://www.cdc.gov/streplab/groupa-strep/emm-background.html. Accessed 23 September 2019.
- 18. Magurran AE. Measuring biological diversity. Oxford, United Kingdom: Blackwell Science, 2004. [Google Scholar]
- 19. Lancefield RC. Persistence of type-specific antibodies in man following infection with group A streptococci. J Exp Med 1959; 110:271–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quinn RW, Vander Zwaag R, Lowry PN. Acquisition of group A streptococcal M protein antibodies. Pediatr Infect Dis 1985; 4:374–8. [DOI] [PubMed] [Google Scholar]
- 21. Hysmith ND, Kaplan EL, Cleary PP, Johnson DR, Penfound TA, Dale JB. Prospective longitudinal analysis of immune responses in pediatric subjects after pharyngeal acquisition of group A streptococci. J Pediatric Infect Dis Soc 2017; 6:187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Richardson LJ, Towers RJ, Cheng AC, et al. . Diversity of emm sequence types in group A beta-haemolytic streptococci in two remote Northern Territory Indigenous communities: implications for vaccine development. Vaccine 2010; 28:5301–5. [DOI] [PubMed] [Google Scholar]
- 23. Speers DJ, Levy A, Gichamo A, Eastwood A, Leung MJ. M protein gene (emm type) analysis of group A Streptococcus isolates recovered during an acute glomerulonephritis outbreak in northern Western Australia. Pathology 2017; 49:765–9. [DOI] [PubMed] [Google Scholar]
- 24. Steer AC, Law I, Matatolu L, Beall BW, Carapetis JR. Global emm type distribution of group A streptococci: systematic review and implications for vaccine development. Lancet Infect Dis 2009; 9:611–6. [DOI] [PubMed] [Google Scholar]
- 25. Pandey M, Ozberk V, Calcutt A, et al. . Streptococcal immunity is constrained by lack of immunological memory following a single episode of pyoderma. PLoS Pathog 2016; 12:e1006122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin JM, Green M, Barbadora KA, Wald ER. Group A streptococci among school-aged children: clinical characteristics and the carrier state. Pediatrics 2004; 114:1212–9. [DOI] [PubMed] [Google Scholar]
- 27. McDonald MI, Towers RJ, Fagan P, Carapetis JR, Currie BJ. Molecular typing of Streptococcus pyogenes from remote Aboriginal communities where rheumatic fever is common and pyoderma is the predominant streptococcal infection. Epidemiol Infect 2007; 135:1398–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Williamson DA, Smeesters PR, Steer AC, et al. . Comparative M-protein analysis of Streptococcus pyogenes from pharyngitis and skin infections in New Zealand: implications for vaccine development. BMC Infect Dis 2016; 16:561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Loh JMS, Tsai JYC, Proft T. The ability of group A Streptococcus to adhere to immortalized human skin versus throat cell lines does not reflect their predicted tissue tropism. Clin Microbiol Infect 2017; 23:677.e1–. e3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.