Abstract
Long non-coding RNAs (lncRNAs) are recognized as critical regulators of self-renewal in human cancer stem-like cells (CSCs), which are a subpopulation of cancer cells primarily responsible for the malignant features of cancer. However, most CSC-related lncRNAs remain unidentified. The results of the present study suggested that growth-arrest-specific transcript 5 (GAS5), a tumor suppressor, exhibited increased expression and was associated with malignant features in human colorectal cancer cell HCT116-derived CSCs. Phenotypic analysis indicated that GAS5 knockdown by specific siRNA significantly decreased CSC self-renewal capacity, proliferation and migration. Moreover, GAS5 knockdown sensitized CSCs to the chemotherapeutic agents 5-fluorouracil and doxorubicin by inducing apoptosis detected by Annexin V-FITC/PI double staining. Inhibition of Nodal growth differentiation factor (NODAL) signaling, which has been reported to be protected by GAS5, presented similar chemosensitivity effects to the GAS5 knockdown results. The present study also assessed the effects of GAS5 overexpression on HCT116 cells, and revealed that overexpression of GAS5 sensitized HCT116 cells to chemotherapeutic agents, which is the opposite of the effect observed in CSCs derived from HCT116 cells. Therefore, it was hypothesized that GAS5 may function as a critical factor for maintaining stemness and that it may exert protective effects on CSCs in a NODAL-dependent manner. Collectively, the results of the present study indicate that GAS5 may be a promising therapeutic target for overcoming malignant features and chemoresistance in colorectal cancer cells.
Keywords: long non-coding RNA, growth-arrest-specific transcript 5, cancer stem-like cells, chemoresistance, Nodal growth differentiation factor
Introduction
Colorectal cancer is the 3rd most common and fatal type of cancer worldwide (1); ~1.4 million cases are diagnosed annually, and mortality occurs in half these cases each year, globally (2). Chemotherapeutic treatment is considered the primary strategy for colorectal cancer; however, chemoresistance frequently develops in response to first-line chemotherapeutic agents, including 5-fluorouracil (5-FU) and doxorubicin (DOX), which is a major barrier for achieving effective therapy (3). Despite numerous studies examining the potential mechanism underlying the induction of chemoresistance, including investigations into activated NF-κB (4), induction of the transcription factor activator protein 1 (5) and activation of multiple drug resistance protein 1 signaling (6), the mechanisms remain largely unknown.
In different cancer types, including colon cancer, a subpopulation of cancer stem-like cells (CSCs) has been identified (7,8). This subpopulation is characterized by cells presenting with stem-like properties, including self-renewal and differentiation, resulting in the production of tumor cells that have a long-term ability for tumor renewal (9,10). It has also been revealed that CSCs may contribute to malignant features, such as tumorigenesis, progression, maintenance and recurrence in several types of cancer (11–13). Seymour et al (14) reported that in glioblastoma, the existence of glioma stem-like cells led to recurrence and metastasis via upregulation of the pluripotency gene, Sox2. Moreover, CSCs have been successfully enriched from several cancer cell lines using novel medium (15,16) and being identified to increase resistance of cells to several anticancer drugs (17). In pancreatic cancer-derived CSCs, it has been revealed that enriched cells presented with increased chemoresistance via regulation of epithelial-mesenchymal transition (EMT) and increased surveillance (18).
Growth-arrest-specific transcript 5 (GAS5), which was originally isolated from mouse NIH3T3 cells and is found on chromosome 1q25 (19), is one of the most common long non-coding RNAs (lncRNAs). A previous study demonstrated that GAS5 may act as a key regulator of tumor proliferation, migration and EMT in cancer cells (20). Furthermore, it has been reported that in cervical cancer, GAS5 may act as a tumor suppressor by sponging microRNA (miR)-21, which induced regulation of cisplatin resistance (21). In pancreatic cancer, GAS5 reversed EMT and induced gemcitabine sensitivity by targeting the miR-221/suppressor of cytokine signaling 3 pathway (22). It has also been reported that in clear cell renal cell carcinoma, GAS5 functioned as a competitive endogenous RNA to regulate solute carrier family 39 member 1 (SLC39A1, also termed hZIP1) expression via sponging miR-223, which resulted in chemosensitivity to cisplatin (23). Furthermore, lncRNAs are reported to be involved in the regulation of physiological processes in CSCs, including liver (24,25) and pancreatic CSCs (24,25). However, the potential role of GAS5 in CSCs derived from colorectal cancer is not fully understood.
In the present study, CSCs were successfully enriched from HCT116 cells, and their potential to maintain self-renewal capacity and to regulate the malignant features of CSCs (including proliferation, tumor formation, migration and chemoresistance) were identified. It was revealed that GAS5 exerted its protective roles via nodal growth differentiation factor (NODAL) signaling, resulting in the maintenance of CSC stemness and induction of chemoresistance. Moreover, it was demonstrated that GAS5 exerted opposing effects on CSCs derived from HCT116 cells compared with parental HCT116 cells. Collectively, the present study identified a novel role of GAS5 in regulating physiological processes in CSCs, which suggests that GAS5 may be a potential therapeutic target for colorectal cancer.
Materials and methods
Cell culture and enrichment of CSCs from HCT116 cells
Human colorectal cancer HCT116 cells were purchased from the American Type Culture Collection and stored in liquid nitrogen. Cells were cultured in DMEM (Thermo Fisher Scientific, Inc.), supplemented with 10% heat-inactivated FBS (Gibco; Thermo Fisher Scientific, Inc.) in a 5% CO2 incubator at 37°C. The medium was refreshed every 2 days, and when the confluence reached 80–90%, cells were harvested with 0.25% trypsin (Gibco; Thermo Fisher Scientific, Inc.) and passaged.
To enrich CSCs, HCT116 cells were maintained in DMEM/Ham Nutrient Mixture F-12 (1:1) supplemented with epidermal growth factor (EGF; 20 ng/ml), human fibroblast growth factor basic (hFGFb; 10 ng/ml) and 2% B27. All reagents were bought from Thermo Fisher Scientific, Inc. Every 3 days the medium was half replaced.
Serial replating assay
To detect self-renewal capacity, cells were replated at a clonal density of 1,000 cells/well in 6-well plate and cultured in DMEM/Ham Nutrient Mixture F-12 (1:1) supplemented with 2% B27, 10 ng/ml EGF and 20 ng/ml hFGFb. Every 2 days, the medium was half replaced, and after 14 days, spheres >40 µm in diameter were counted under a X71 (U-RFL-T) fluorescence microscope (Olympus Corporation; magnification, ×40). This process was repeated three times.
Western blotting
Cells were suspended in lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 0.02% NaN3, 100 µg/ml PMSF, 1 µg/ml aprotinin, 1 µg/ml pepstatin A and 1% Triton X-100. A SoniConvert® homogenizer (DocSense Biotech) was used to lyse the cells. After centrifugation at 12,000 × g for 10 min at 4°C, 20 µg total protein quantitatively measured using the bicinchoninic acid kit (Sigma-Aldrich; Merck KGaA) was resolved using SDS-PAGE on 10% gels, and was then transferred to a nitrocellulose membrane. After transferring, the membrane was blocked with 5% BSA (Beyotime Institute of Biotechnology) in PBS for 30 min at room temperature. The primary antibodies used were as follows: Rabbit monoclonal anti-Oct4 antibody (1:2,000; cat. no. ab181557), rabbit monoclonal anti-Sox2 antibody (1:2,000; cat. no. ab93689) and rabbit monoclonal anti-β-actin antibody (1:5,000; cat. no. ab179467). In addition, goat anti-rabbit IgG H&L antibody (horseradish peroxidase labeled; 1:10,000; cat. no. ab7090) was used as the secondary antibody. All antibodies were purchased from Abcam and incubated with the membrane at room temperature for 1 h, followed by three washes with PBS supplemented with 0.1% Tween-20. Blot bands were semi-quantified via densitometry with ImageJ software (version 1.52r; National Institutes of Health). β-actin was used as an internal reference. The signal was detected with ECL plus western blotting detection reagents (Pierce; Thermo Fisher Scientific, Inc.) and imaged using X-ray film.
Reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using TRIzol® (Thermo Fisher Scientific, Inc.) according to the manufacturer's instructions. Subsequently, 1 µg total RNA underwent RT using a RT kit (Thermo Fisher Scientific, Inc.) at 42°C for 1 h. After RT, cDNA was used as a template for qPCR by using SYBR™ Green PCR master mix (Thermo Fisher Scientific, Inc.), which was conducted under the following conditions: 95°C for 5 min; 35 cycles of 95°C for 10 sec; and a final extension at 60°C for 1 min. An ABI 7500 system (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used for PCR. The primers used for qPCR were as follows: GAS5, forward 5′-CGACTCCTGTGAGGTATGGTG-3′ and reverse 5′-ATCCTTCCTTGGGGACACAAC-3′; and β-actin, forward 5′-CATGTACGTTGCTATCCAGGC-3′ and reverse 5′-CTCCTTAATGTCACGCACGAT-3′. The qPCR results were analyzed and expressed relative to the threshold cycle (Cq) values, and were then converted to fold changes; all data was analyzed using 2−ΔΔCq method. A 2.0-fold change was considered to be significant (26). In total, three repeats were conducted for each sample.
Transfection with small interfering (si)RNA or GAS5 coding sequence
siRNA was used for knocking down the expression of GAS5 in CSCs derived from HCT116 cells. In total, two 21-nucleotide siRNAs were used for silencing: siGAS5-1, 5′-GCAAGCCTAACTCAAGCCA-3′; siGAS5-2, 5′-GGACCAGCTTAATGGTTCT-3′. si-negative control (siNC): CCTGAGACCAAGCCATAAC was employed as a NC. Briefly, siRNA (50 nmol) was chemically synthesized by Guangzhou RiboBio Co., Ltd. and transfected into ~1×106 cells with Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.), according to the siRNA manufacturer's protocol (Guangzhou RiboBio Co., Ltd.). After 48 h, cells were used for subsequent experimentation.
The coding sequence of GAS5 was amplified from CSCs cDNA using the followed primers: Forward, 5′-ATAGGGCTAGCTTTCGAGGTAGGAGTCGACT-3′ and reverse, 5′-ATAGGCGGCCGCGGATTGCAAAAATTTATTAA-3′. The PCR product was amplified using PlatinumTM Green Hot Start PCR Master mix (2X; Thermo Fisher Scientific, Inc.) under the following conditions: 95°C for 5 min; 35 cycles of 95°C for 10 sec, extension at 60°C for 3 min. The PCR product and pEGFP-N3 plasmid (Beijing Solarbio Science & Technology Co., Ltd.) were digested using NheI and NotI restriction enzymes (Takara Bio, Inc.). Subsequently, the digested PCR product was inserted into the pEGFP-N3 plasmid using the Quick Ligation kit (cat. no. M2200S; New England Biolabs, Inc.) and successful insertion was confirmed by sequencing which was provided by Sangon Biotech Co., Ltd. In total, 1.6 µg plasmid was transfected into ~1×106 cells with Lipofectamine® 2000, according to the manufacturer's protocol. Also, an empty vector was transfected into cells, and this was considered as the vector group. After 48 h, cells were used for further analysis.
Cell Counting Kit-8 (CCK-8) assay for cell viability
Cells (2×105/well) were seeded into 96-well plates. 0.1, 1, 3, 5 and 10 µg/ml of 5-FU, or 0.1, 0.3, 0.5, 0.7 and 1.0 µg/ml DOX was added into medium for 24-h culturing. Next, 10 µl CCK-8 solution (Beyotime Institute of Biotechnology) was added to the cell culture for a 2 h co-incubation at 37°C. Absorbance was measured at 450 nm and cell viability was expressed as optical density.
Cells (2×105/well) were seeded into 96-well plates. After 1, 2, 3 or 4-day culturing with or without SB431542, 10 µl CCK-8 solution (Beyotime Institute of Biotechnology) was added to the cell culture for a 2 h co-incubation at 37°C. The mock group was treated with the same volume of DMSO. Absorbance was measured at 450 nm and cell viability was expressed as optical density. Each assay was repeated three times.
Cell cycle analysis
The percentage of cells in each cell cycle phase was analyzed by propidium iodide (PI) staining and flow cytometry. Cells were washed three times with ice-cold PBS and a final concentration of 75% ethanol was added for fixation at 4°C for 4 h. Subsequently, 100 µg/ml Rnase (Beyotime Institute of Biotechnology) was added to each sample and incubated at 37°C for 30 min, and cells were stained with 50 µg/ml PI for 10 min at room temperature and analyzed on a FACS LSRII flow cytometer (BD Biosciences). Then data were analyzed using FlowJo software (FlowJo LLC, version 9.7.4)
Tumor formation in soft agar
Cells (2×103) were suspended in serum-free DMEM/F-12 medium and dissolved in 0.3% low-melting agarose (Sigma-Aldrich; Merck KGaA) supplemented with 20 ng/ml EGF, 10 ng/ml hFGFb, 2% B27 and 1% antibiotic-antimycotic mixture. Cells were allowed to grow for 14 days and colonies with >50 cells were counted under a X71 (U-RFL-T) fluorescence microscope (Olympus Corporation; magnification, ×40).
Transwell assay
To analyze migratory ability, 2×103 cells were plated in the upper chambers of semi-permeable Transwell inserts filled with 200 µl of SFM and 600 µl DMEM/Ham Nutrient Mixture F-12 (1:1), supplemented with 20 ng/ml EGF, 10 ng/ml hFGFb, 2% B27 and 1% antibiotic-antimycotic mixture was added to the lower chambers. After incubation at 37°C for 24 h, cells were fixed using 4% paraformaldehyde at room temperature for 10 min, and stained with 0.5% crystal violet at room temperature for 10 min. After 2–3 washes with ice-cold PBS, images were captured under a X71 (U-RFL-T) fluorescence microscope (Olympus Corporation; magnification, ×40).
Annexin V-FITC/PI double staining
Annexin V-FITC/PI double staining was performed with the Annexin V-FITC/PI apoptosis detection kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's instructions. Cells (1×106) were suspended from 6-well plate and washed with ice-cold PBS. Then cells were co-incubated with 5 µg/ml 5-FU, 0.7 µg/ml DOX or 10 µM of SB431542 for 24 h. For staining, cells were resuspended in binding buffer containing 5 µl Annexin V-FITC for 10 min in the dark at room temperature, and then 5 µl PI was added for 5 min staining at room temperature. Then, 400 µl binding buffer was added to each sample and flow cytometric analysis was performed on a FACS LSRII flow cytometer (BD Biosciences). Data were analyzed using FlowJo software (version 10.6.1; FlowJo LLC).
Statistical analysis
Data are presented as the mean ± SD, and all experiments were repeated three times independently. One-way ANOVA followed by Bonferroni's multiple comparison tests was used for comparisons among experimental groups, and was conducted using GraphPad Prism 5 software (GraphPad Software, Inc.). P<0.05 was considered to indicate a statistically significant difference.
Results
Identifying and enriching CSCs from HCT116 cells and detection of GAS5
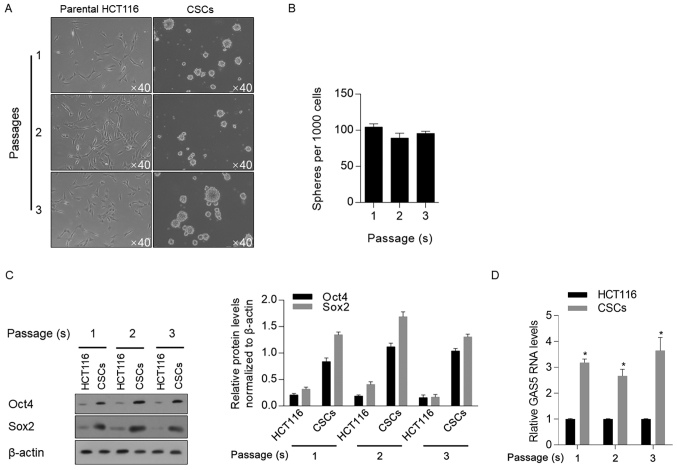
For CSC enrichment, parental HCT116 cells were cultured in serum-free medium supplemented with EGF, hFGFb and B27, as described previously (27). Cells were passaged every 10 days, and it was observed that spheres formed in every passage (Fig. 1A). When counting spheres >40 µm in diameter, no significant decrease in sphere formation ability was observed at the different passages; these data indicate the existence of a self-renewal capacity (Fig. 1B), which is a characteristic of CSCs (28).
Figure 1.
Enrichment and characterization of CSCs from HCT116 cells. (A) Sphere formation in serum-free medium was assessed and cells were passaged three times. Magnification, ×40. (B) Serial replating assay was performed and spheres >40 µm in diameter were counted. (C) Western blotting was performed to detect stem cell factors, including Oct4 and Sox2 in different passage of CSCs and parental HCT116 cells. (D) Reverse transcription-quantitative PCR was performed to measure GAS5 mRNA expression. *P<0.05 vs. HCT116 group. CSCs, cancer stem-like cells; GAS5, growth-arrest-specific transcript 5.
To further characterize the stemness of enriched cells, quantitative analyses were performed on Oct4 and Sox2, two stem cell factors that are important for the self-renewal properties of CSCs (29). An increase in the protein expression levels of Oct4 and Sox2 was identified in three passages of enriched cells compared with parental HCT116 cells (Fig. 1C). Subsequently, all three passages of CSCs were used to assess GAS5 mRNA expression; it was revealed that GAS5 was significantly upregulated in CSCs compared with parental HCT116 cells, regardless of the passage number (Fig. 1D).
GAS5 is essential for maintaining stemness in CSCs derived from HCT116 cells
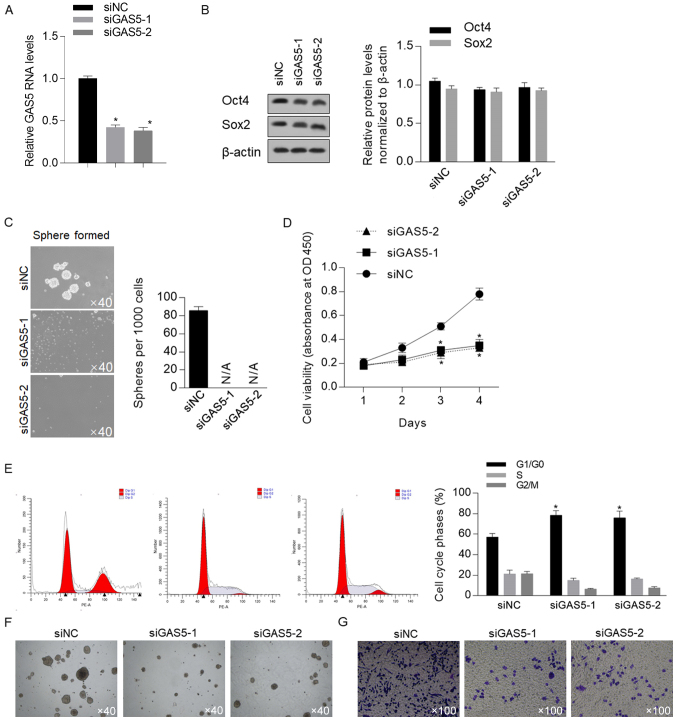
Xu et al (30) reported that in human embryonic stem cells, the presence of GAS5 was critical for controlling self-renewal capacity; therefore, the present study assessed whether GAS5 was involved in regulating stemness in CSCs derived from HCT116 cells. The efficiency of GAS5 knockdown using GAS5-targeting siRNA at different sites (siGAS5-1 and siGAS5-2; Fig. 2A) was assessed by transfection at the early passage. Western blotting demonstrated that knockdown of GAS5 did not affect Oct4 and Sox2 protein expression levels, which indicated that GAS5 may not affect the stemness of CSCs (Fig. 2B). However, the serial replating assay suggested that knockdown of GAS5 abolished sphere formation ability (Fig. 2C). Therefore, these two controversial results indicate that GAS5 may affect self-renewal capacity by regulating other physiological processes, such as proliferation, but not stem cell factors. Thus, cell viability was detected from day 1–4 after GASG5 knockdown; it was identified that proliferative capacity was significantly decreased by GAS5 knockdown (Fig. 2D). The cell phase distribution had a consistent tendency, which demonstrated that knockdown of GAS5 significantly increased the proportion of cells in G1/G0 phase and decrease the proportion of cells phase in S and G2/M, thus indicating that the cell cycle was arrested at this point. Collectively, these results suggest that knockdown of GAS5 primarily decreases cellular proliferation, which as a result inhibits sphere formation in CSCs.
Figure 2.
GAS5 inhibits cell proliferation and sphere formation in CSCs derived from HCT116 cells. (A) After introduction of siRNA to target GAS5, GAS5 mRNA expression was measured by reverse transcription-quantitative PCR. (B) Western blotting was performed to measure Oct4 and Sox2 protein expression levels after GAS5 knockdown. (C) Serial replating assay was performed after GAS5 knockdown. Magnification, ×40. (D) Cell viability was measured from day 1–4 after GAS5 knockdown. (E) Flow cytometry was performed after propidium iodide staining to determine the proportion of cells in cell cycle phases after GAS5 knockdown. After GAS5 knockdown, (F) tumor formation in soft agar and (G) Transwell assays were performed. *P<0.05 vs. siNC group. CSCs, cancer stem-like cells; GAS5, growth-arrest-specific transcript 5; NC, negative control; OD, optical density; siRNA, small interfering RNA.
The effects of GAS5 on other malignant features were also detected, including tumor formation in soft agar and cell migration. It was revealed that knockdown of GAS5 inhibited these two malignant features (Fig. 2F and G), which suggested its important role in regulating physiological processes in CSCs.
GAS5 may desensitize CSCs to chemotherapeutic agents
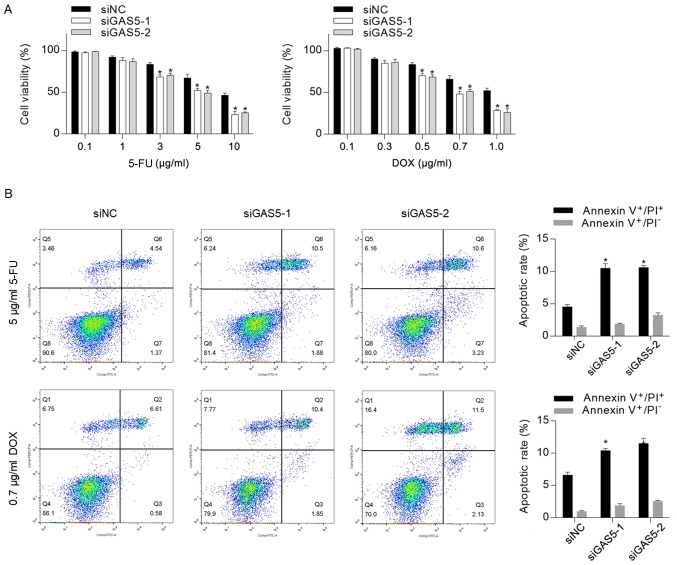
Previous studies have revealed that GAS5, which acts as a tumor suppressor, increased chemosensitivity in several types of cancer, including cervical cancer (21), ovarian cancer (31), pancreatic cancer (22) and clear cell renal cell carcinoma (23). To assess whether knockdown of GAS5 exerted a regulatory role in CSC chemosensitivity, the cytotoxicity of 5-FU and DOX to CSCs was measured, and it was identified that knockdown of GAS5 significantly sensitized CSCs to 5-FU and DOX (Fig. 3A). Moreover, 5 µg/ml 5-FU or 0.7 µg/ml DOX were supplemented into medium for 24-h treatment followed by apoptosis analysis. In line with the cytotoxicity results, it was revealed that GAS5 knockdown significantly increased chemotherapeutic treatment-induced apoptosis (Fig. 3B).
Figure 3.
Knockdown of GAS5 sensitizes CSCs to chemotherapeutic agents. (A) Cell Counting Kit-8 assay was performed to detect cytotoxicity of 5-FU and DOX to CSCs. (B) Annexin V-FITC/PI double staining was performed and analyzed by performing flow cytometry to measure apoptotic death rate. *P<0.05 vs. siNC group. 5-FU, 5-fluorouracil; CSCs, cancer stem-like cells; DOX, doxorubicin; GAS5, growth-arrest-specific transcript 5; NC, negative control; PI, propidium iodide; siRNA, small interfering RNA.
GAS5 exerts regulatory roles via regulating NODAL
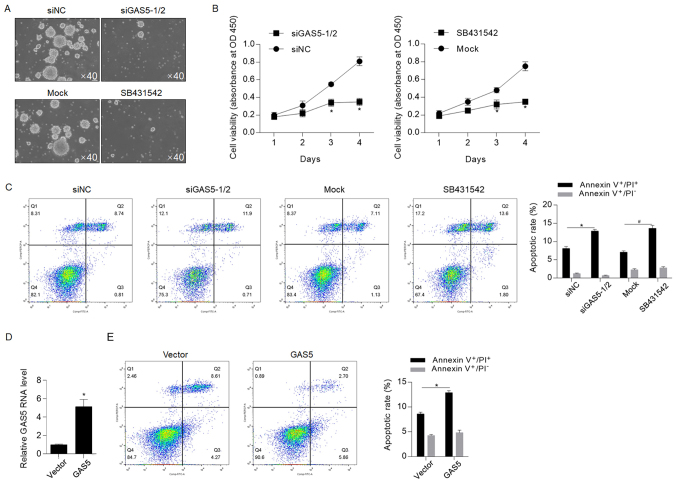
NODAL signaling has been reported to be regulated by GAS5, helping to maintain stemness (30). Therefore, the present study aimed to identify whether NODAL was involved in GAS5-mediated regulation of CSCs. The serial replating assay demonstrated that GAS5 knockdown and pretreatment with SB431542 significantly decreased self-renewal capacity in CSCs, thus indicating that both of these are important for maintaining stemness (Fig. 4A). The cell viability assay also suggested that GAS5 knockdown and inhibition of NODAL signaling via SB431542 decreased cell proliferation (Fig. 4B). To further assess whether NODAL signaling contributed to GAS5 knockdown-induced chemosensitivity, the apoptotic rate after 5-FU treatment was measured, and it was revealed that inhibition of NODAL signaling sensitized CSCs to chemotherapeutic treatment (Fig. 4C). However, this finding was inconsistent with those from previous studies, which reported the anti-tumor effects of GAS5 (21–23,31). To assess whether GAS5 exerts different effects on HCT116 cells, GAS5 was efficiently overexpressed in HCT116 cells (Fig. 4D) and apoptotic cell death was detected (Fig. 4E). Flow cytometry results indicated that, in HCT116 cells, overexpression of GAS5 sensitized cells to chemotherapy, thus suggesting that GAS5 potentially exerts opposite effects on chemoresistance in HCT116 cells and CSCs derived from HCT116.
Figure 4.
GAS5 knockdown may sensitize CSCs to chemotherapy depending on NODAL signaling. (A) After GAS5 knockdown or inhibition of NODAL, sphere formation was assessed. Magnification, ×40. Mock cells were treated with the same volume of DMSO. (B) Cell Counting Kit-8 assay was performed to detect the effects of GAS5 knockdown or inhibition of NODAL on viability. (C) Annexin V-FITC/PI double staining was performed to detect apoptotic cell death. *P<0.05 vs. siNC group. #P<0.05. vs. Mock group. (D) Transfection efficiency of GAS5 overexpression vector was detected by reverse transcription-quantitative PCR. (E) Apoptotic cell death was measured by performing Annexin V-FITC/PI double staining. *P<0.05 vs. Empty vector group. CSCs, cancer stem-like cells; GAS5, growth-arrest-specific transcript 5; NC, negative control; OD, optical density; PI, propidium iodide; siRNA, small interfering RNA.
Discussion
The present study detected the expression of GAS5 in CSCs derived from a human colorectal cancer cell line, and investigated its roles in regulating malignant features and maintaining the stemness of CSCs. While GAS5 has been reported to act as a tumor suppressor in several types of cancer (19–21), the present results suggested that GAS5 may be essential for maintaining stemness, and that it exerted critical roles in promoting the malignant features of CSCs, including proliferation, tumor formation, migration and chemoresistance.
By culturing HCT116 cells in serum-free medium supplemented with EGF, hFGFb and B27, CSCs presented a stem cell morphological phenotype and exhibited high expression levels of stem cell factors, including Oct4 and Sox2; the cells were enriched and identified by assessing their self-renewal capacity. Moreover, future studies aim to isolate CSCs from colorectal tumor tissue and investigate whether the potential role of GAS5 found in the HCT116 cell line also exists in primary cells. In the present study, compared with parental HCT116 cells, the expression of GAS5 was significantly higher in CSCs in different passages, thus indicating its potential role in maintaining stemness. Considering the relatively high expression of GAS5, its overexpression may have an effect on the malignant features and stemness of CSCs. In the present study, GAS5 expression was knocked down in CSCs, in order to investigate its potential effects. While GAS5 knockdown inhibited sphere formation of CSCs, the stem cell factors Oct4 and Sox2 were not significantly affected, which suggested that GAS5 knockdown may affect sphere formation by modifying pluripotency. Cellular proliferation and cell cycle distribution were subsequently assessed, and it was revealed that GAS5 knockdown inhibited cell proliferation via cell cycle arrest at the G1/G0 phase. In addition, the inhibitory effects of GAS5 knockdown on other malignant features were also detected, including tumor formation and migration. GAS5 has been widely reported to act as a tumor suppressor in several types of cancer cells (19–21), which is the opposite of the effects of siGAS5 on the malignant features of CSCs observed in the present study. Although there is no direct evidence of the mechanisms underlying how GAS5 modifies pluripotency, it is speculated that modification of pluripotency is the primary mechanism by which GAS5 exerts its regulatory roles in CSCs.
GAS5 has also been studied for its beneficial role in increasing chemosensitivity in several cancer cell types (21–23). In the present study, 5-FU and DOX were used to detect the effects of GAS5 knockdown on chemoresistance in CSCs; it was revealed that GAS5 knockdown increased the chemosensitivity and chemotherapeutic agent-induced apoptosis of CSCs. NODAL signaling has an important role in regulating the stem-like properties of CSCs (32,33), and a previous study revealed a critical role for NODAL signaling in the induction of chemoresistance in CSCs (33). Therefore, the present study examined whether NODAL signaling was involved in the GAS5-mediated regulation of CSCs. It was demonstrated that GAS5 knockdown and NODAL inhibition with SB431542 pretreatment decreased the self-renewal capacity and chemoresistance of cells, thus indicating that GAS5 may exert tumor-promoting effects in a NODAL-dependent manner. However, whilst the present study did not identify the exact regulatory mechanism between GAS5 and NODAL signaling, the results indicated that it may be beneficial to investigate the regulatory mechanism between GAS5 and NODAL signaling in other cancer cells. Moreover, a limitation of the present study was that the effects of GAS5 on chemosensitivity were not assessed in vivo, thus it is important to examine whether GAS5 is relevant to the regulation of malignant features in vivo.
In conclusion, the present study identified GAS5 as a factor that may promote colorectal CSC activity in cells derived from HCT116 cells. The present study investigated the effects of GAS5 on CSCs vs. parental HCT116 cells, and demonstrated that GAS5 exerted opposing effects on malignant features in CSCs compared with parental cells. Furthermore, the involvement of NODAL signaling was indicated to be essential for GAS5-mediated modification in CSCs, which is a potential key reason for the opposing effects of GAS5 in CSCs and parental HCT116 cells. However, further investigation into the interaction of GAS5 with NODAL signaling is required to understand the exact roles of GAS5 in regulating malignant features in colorectal CSCs.
Acknowledgements
The authors would like to thank Dr Huimin Shi (Sichuan University) for language editing.
Funding
The present work was supported by Scholar supporting program, Chongqing Scientific project (grant no. S20180827).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XZ and DX designed and performed all experiments, data collection and analysis. DX supervised the experiments, and was involved in writing and revision of the manuscript. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Marmol I, Sanchez-de-Diego C, Pradilla DA, Cerrada E, Rodriguez YM. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci. 2017;18:E197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 3.Papanastasopoulos P, Stebbing J. Molecular basis of 5-fluorouracil-related toxicity: Lessons from clinical practice. Anticancer Res. 2014;34:1531–1535. [PubMed] [Google Scholar]
- 4.Bharti AC, Aggarwal BB. Nuclear factor-kappa B and cancer: Its role in prevention and therapy. Biochem Pharmacol. 2002;64:883–888. doi: 10.1016/S0006-2952(02)01154-1. [DOI] [PubMed] [Google Scholar]
- 5.Huang J, Chen Y, Li J, Zhang K, Chen J, Chen D, Feng B, Song H, Feng J, Wang R, Chen L. Notch-1 confers chemoresistance in lung adenocarcinoma to Taxanes through AP-1/microRNA-451 mediated regulation of MDR-1. Mol Ther Nucleic Acids. 2016;5:e375. doi: 10.1038/mtna.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie Y, Zhong DW. AEG-1 is associated with hypoxia-induced hepatocellular carcinoma chemoresistance via regulating PI3K/AKT/HIF-1alpha/MDR-1 pathway. EXCLI J. 2016;15:745–757. doi: 10.17179/excli2016-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 8.Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, Hoey T, Gurney A, Huang EH, Simeone DM, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci USA. 2007;104:10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magee JA, Piskounova E, Morrison SJ. Cancer stem cells: Impact, heterogeneity, and uncertainty. Cancer Cell. 2012;21:283–296. doi: 10.1016/j.ccr.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clarke MF, Dick JE, Dirks PB, Eaves CJ, Jamieson CH, Jones DL, Visvader J, Weissman IL, Wahl GM. Cancer stem cells-perspectives on current status and future directions: AACR Workshop on cancer stem cells. Cancer Res. 2006;66:9339–9344. doi: 10.1158/0008-5472.CAN-06-3126. [DOI] [PubMed] [Google Scholar]
- 11.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 12.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 13.Vander Griend DJ, Karthaus WL, Dalrymple S, Meeker A, DeMarzo AM, Isaacs JT. The role of CD133 in normal human prostate stem cells and malignant cancer-initiating cells. Cancer Res. 2008;68:9703–9711. doi: 10.1158/0008-5472.CAN-08-3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seymour T, Nowak A, Kakulas F. Targeting aggressive cancer stem cells in glioblastoma. Front Oncol. 2015;5:159. doi: 10.3389/fonc.2015.00159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watanabe Y, Yoshimura K, Yoshikawa K, Tsunedomi R, Shindo Y, Matsukuma S, Maeda N, Kanekiyo S, Suzuki N, Kuramasu A, et al. A stem cell medium containing neural stimulating factor induces a pancreatic cancer stem-like cell-enriched population. Int J Oncol. 2014;45:1857–1866. doi: 10.3892/ijo.2014.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsukuma S, Yoshimura K, Ueno T, Oga A, Inoue M, Watanabe Y, Kuramasu A, Fuse M, Tsunedomi R, Nagaoka S, et al. Calreticulin is highly expressed in pancreatic cancer stem-like cells. Cancer Sci. 2016;107:1599–1609. doi: 10.1111/cas.13061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hashimoto N, Tsunedomi R, Yoshimura K, Watanabe Y, Hazama S, Oka M. Cancer stem-like sphere cells induced from de-differentiated hepatocellular carcinoma-derived cell lines possess the resistance to anti-cancer drugs. BMC Cancer. 2014;14:722. doi: 10.1186/1471-2407-14-722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang F, Ma L, Zhang Z, Liu X, Gao H, Zhuang Y, Yang P, Kornmann M, Tian X, Yang Y. Hedgehog signaling regulates epithelial-mesenchymal transition in pancreatic cancer stem-like cells. J Cancer. 2016;7:408–417. doi: 10.7150/jca.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nakamura Y, Takahashi N, Kakegawa E, Yoshida K, Ito Y, Kayano H, Niitsu N, Jinnai I, Bessho M. The GAS5 (growth arrest-specific transcript 5) gene fuses to BCL6 as a result of t(1;3)(q25;q27) in a patient with B-cell lymphoma. Cancer Genet Cytogenet. 2008;182:144–149. doi: 10.1016/j.cancergencyto.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 20.Ye K, Wang S, Zhang H, Han H, Ma B, Nan W. Long noncoding RNA GAS5 suppresses cell growth and epithelial-mesenchymal transition in osteosarcoma by regulating the miR-221/ARHI pathway. J Cell Biochem. 2017;118:4772–4781. doi: 10.1002/jcb.26145. [DOI] [PubMed] [Google Scholar]
- 21.Wen Q, Liu Y, Lyu H, Xu X, Wu Q, Liu N, Yin Q, Li J, Sheng X. Long Noncoding RNA GAS5, which acts as a tumor suppressor via microRNA 21, regulates cisplatin resistance expression in cervical cancer. Int J Gynecol Cancer. 2017;27:1096–1108. doi: 10.1097/IGC.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu B, Wu S, Ma J, Yan S, Xiao Z, Wan L, Zhang F, Shang M, Mao A. lncRNA GAS5 reverses EMT and tumor stem cell-mediated gemcitabine resistance and metastasis by targeting miR-221/SOCS3 in pancreatic cancer. Mol Ther Nucleic Acids. 2018;13:472–482. doi: 10.1016/j.omtn.2018.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dong X, Kong C, Liu X, Bi J, Li Z, Li Z, Zhu Y, Zhang Z. GAS5 functions as a ceRNA to regulate hZIP1 expression by sponging miR-223 in clear cell renal cell carcinoma. Am J Cancer Res. 2018;8:1414–1426. [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J, Fu Y, Wu J, Li J, Huang G, Qin L. The diverse mechanisms of miRNAs and lncRNAs in the maintenance of liver cancer stem cells. Biomed Res Int. 2018;2018:8686027. doi: 10.1155/2018/8686027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L, Dong P, Wang W, Huang M, Tian B. Gemcitabine treatment causes resistance and malignancy of pancreatic cancer stem-like cells via induction of lncRNA HOTAIR. Exp Ther Med. 2017;14:4773–4780. doi: 10.3892/etm.2017.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Noh KH, Kim BW, Song KH, Cho H, Lee YH, Kim JH, Chung JY, Kim JH, Hewitt SM, Seong SY, et al. Nanog signaling in cancer promotes stem-like phenotype and immune evasion. J Clin Invest. 2012;122:4077–4093. doi: 10.1172/JCI64057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gkountela S, Aceto N. Stem-like features of cancer cells on their way to metastasis. Biol Direct. 2016;11:33. doi: 10.1186/s13062-016-0135-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Singh S, Trevino J, Bora-Singhal N, Coppola D, Haura E, Altiok S, Chellappan SP. EGFR/Src/Akt signaling modulates Sox2 expression and self-renewal of stem-like side-population cells in non-small cell lung cancer. Mol Cancer. 2012;11:73. doi: 10.1186/1476-4598-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu C, Zhang Y, Wang Q, Xu Z, Jiang J, Gao Y, Gao M, Kang J, Wu M, Xiong J, et al. Long non-coding RNA GAS5 controls human embryonic stem cell self-renewal by maintaining NODAL signalling. Nat Commun. 2016;7:13287. doi: 10.1038/ncomms13287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long X, Song K, Hu H, Tian Q, Wang W, Dong Q, Yin X, Di W. Long non-coding RNA GAS5 inhibits DDP-resistance and tumor progression of epithelial ovarian cancer via GAS5-E2F4-PARP1-MAPK axis. J Exp Clin Cancer Res. 2019;38:345. doi: 10.1186/s13046-019-1329-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu LL, Chen XH, Zhang G, Liu ZC, Wu N, Wang H, Qi YF, Wang HS, Cai SH, Du J. CCL21 facilitates chemoresistance and cancer stem cell-like Properties of colorectal cancer cells through AKT/GSK-3β/Snail signals. Oxid Med Cell Longev. 2016;2016:5874127. doi: 10.1155/2016/5874127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cioffi M, Trabulo SM, Sanchez-Ripoll Y, Miranda-Lorenzo I, Lonardo E, Dorado J, Reis Vieira C, Ramirez JC, Hidalgo M, Aicher A, et al. The miR-17-92 cluster counteracts quiescence and chemoresistance in a distinct subpopulation of pancreatic cancer stem cells. Gut. 2015;64:1936–1948. doi: 10.1136/gutjnl-2014-308470. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.