Summary
Infectious bronchitis virus (IBV) causes respiratory diseases in chickens and poses an economic threat to the poultry industry worldwide. Despite vaccine use, there have been field outbreaks of IBV in Taiwan. This study aimed to characterize the emerging IBV variants circulating in Taiwan. The analysis of the structural protein genes showed that these variants emerged through frequent recombination events among Taiwan strains, China strains, Japan strains and vaccine strains. Cross‐neutralization tests revealed that two of the variants exhibited novel serotypes. Clinicopathological assessment showed that two of the variants caused high fatality rates of 67% and 20% in one‐day‐old SPF chicks, and all the variants possessed multiorgan tropisms, including trachea, proventriculus and urogenital tissues. Furthermore, the commercial live‐attenuated Mass‐type vaccine conferred poor protection against these variants. This study identified novel genotypes, serotypes and pathotypes of emerging IBV variants circulating in Taiwan. There is an urgent need for effective countermeasures against these variant strains.
Keywords: infectious bronchitis virus, multiorgan tropism, pathotype, recombinant variants, serotype
1. INTRODUCTION
Avian infectious bronchitis is an acute, highly contagious upper respiratory tract disease of chickens. Chickens of all ages and types are susceptible to the disease, which usually causes clinical symptoms, including rales, sneezing, shakes and diarrhoea. In addition, the reproductive tract of chickens can be affected, resulting in decreased egg quality and production (Cook, Jackwood, & Jones, 2012; Jackwood et al., 2005). When young chicks are affected, damage to the reproductive tract can lead to reduced production. The virus transmits very rapidly in naive chickens and poses an economic threat to the poultry industry worldwide (Jackwood, 2012).
Infectious bronchitis virus (IBV) is a Coronaviridae family member possessing a single‐stranded positive‐sense RNA genome enclosed by an envelope. The virus is made up primarily of four structural proteins: spike glycoprotein (S), envelope protein (E), membrane glycoprotein (M) and nucleocapsid protein (N). The S protein is the most important viral protein for virus subtyping because it contains epitopes for neutralizing antibodies and thus evolves very quickly by random mutation or recombination (Lin & Chen, 2017). The protein also mediates cell attachment and virus–host membrane fusion, playing a critical role in tissue specificity. Therefore, changes in the IBV S protein can easily influence the virus phenotype (Cavanagh, 2007; Sjaak de Wit, Cook, & Heijden, 2011). In recent years, IBV variants presenting novel genotypes, serotypes or pathogenicity have been identified in China (Chen et al., 2017; Gao et al., 2016; Zhong et al., 2016; Zhou et al., 2017), Korea (Hong, Kwon, Kim, Mo, & Kim, 2012), Egypt (Zanaty et al., 2016) and Australia (Hewson et al., 2014). These variants caused different degrees of mortality in chickens in experimental inoculations.
In Taiwan, IBV was first isolated in the early 1960s; since then, the live‐attenuated Massachusetts‐type (H120) IBV vaccine has been used to prevent and control the disease (Lin, Wang, & Wang, 2005; Wang, Hsieh, & Chang, 1996). However, IBVs locally circulating in Taiwan have been found to be genetically different from all other genotypes in the world and can be divided into two groups, namely, Taiwan Group I (TW‐I) and Taiwan Group II (TW‐II) (Wang & Tsai, 1996). Because of the lack of specific vaccines against endemic strains of IBV in Taiwan, IBV infections remain a problem in the poultry industry. Since 2002, IBVs causing severe outbreaks have been isolated from the field and subsequently identified as viral variants that emerged through frequent recombination events, including strains 2992/02 (Chen, Huang, & Wang, 2009), 3374/05 and 3382/06 (Chen, Huang, & Wang, 2010), and TC3/13 and S78/14 (Tsai, Tsai, & Wang, 2016). These variants have been circulating in domestic chickens. In this study, we retrospectively characterized the genotype, serotype, pathogenicity and vaccine protection of emerging IBV variants.
2. MATERIALS AND METHODS
2.1. Virus propagation and titration
Virus propagation was performed using 10‐day‐old specific‐pathogen free (SPF) embryonated eggs (Animal Health Research Institute, Tamsui, Taiwan) via an allantoic route as previously described (Chen et al., 2009). Viral samples were inoculated in the allantoic cavity of embryos and incubated at 37°C for 72 hr. Allantoic fluid was subsequently collected and stored at −80°C until use. For virus titration, samples were diluted tenfold with sterile PBS, and each 10‐day‐old SPF egg received 0.1 ml of the diluted sample. Infection was determined by the presence of dwarfing or malformation in embryos 7 days post‐infection (dpi). Viral titres were expressed as a 50% egg infectious dose (EID50). At least five eggs were used for each dilution, and the EID50 values were calculated by the method of Reed and Muench (1938).
2.2. RNA extraction, RT‐PCR and sequencing analyses
Viral RNA was extracted from the harvested allantoic fluid by a commercial RNA extraction kit (Geneaid Biotech Ltd.) following the manufacturer's guidelines. Full S and N genes were RT‐PCR amplified and sequenced using the primers and protocols described in Lin et al. (2016). DNA sequencing was conducted by a commercial service (Tri‐I Biotech). Each nucleotide was determined from at least three identical results generated from separate PCR products. Sequences of the reference IBV strains were GenBank with the accession numbers listed in Table S1. Sequence data were compiled using the Lasergene (DNASTAR), and sequence alignments were conducted with the Clustal W method available in BioEdit software. Phylogenetic trees were constructed with the neighbour‐joining method using MEGA software (Tamura, Dudley, Nei, & Kumar, 2007), and the bootstrap values were determined from 1,000 replicates of the original data. Phylogenetic trees of the S1, S2 and N genes of IBVs were constructed, where the classification of S1 gene is according to Valastro et al. (2016). The recombinant analysis was performed using the Recombination Detection Program 4 (RDP4) software (Martin, Murrell, Golden, Khoosal, & Muhire, 2015).
2.3. Anti‐serum production and cross‐neutralization test
Anti‐sera against IBV strains were prepared in SPF chickens. Groups of three 3‐week‐old chickens were intranasally inoculated with virus at a titre of 105 EID50 and further boosted after 2 and 4 weeks with the same strain at a titre of 106 EID50 intravenously. Blood was obtained by cardiac puncture two weeks after the last inoculation. Serum was heat‐inactivated and stored at −20°C. The anti‐serum to Mass‐type H120 was acquired from Charles River Laboratories (North Franklin, CT). A cross‐neutralization test was conducted as previously published (Wang & Huang, 2000). Fourfold diluted sera were mixed with the same volume of 100 EID50 virus at room temperature for one hour. The mixtures were inoculated into 10‐day‐old SPF eggs, and the eggs were observed for survival on a daily basis. Seven days after inoculation, the eggs were opened and examined for typical lesions caused by IBV infection (dwarfing or malformation). The neutralizing titre of each serum against the homologous or heterologous virus was determined by the last serum dilution that protected 50% of the embryo. In addition, r‐values were calculated by the method described by Archetti and Horfall (1950). The antigenic (serotype) difference between two given strains was denoted as follows: r: 70%–100%, same serotype; r: 33%–70%, different subtype (minor); r: 11%–32%, different subtype (major); and r: 0%–10%, different serotype.
2.4. Experimental infections of viral variants in chickens
One‐day‐old chicks were inoculated intranasally with 106 EID50 virus; for each strain, chicks were observed daily for clinical signs and survival for 21 days (n = 5 or 6 per group). The clinical scores of IBV were interpreted according to the methods described by Avellaneda, Villegas, Jackwood, and King (1994). The clinical signs were evaluated as follows: 0 = no clinical signs; 1 = lacrimation, slight shaking, watery faeces or tracheal rales; 2 = lacrimation, presence of nasal exudate, depression, watery faeces, apparent sneezing or cough; 3 = same as 2 but stronger with severe watery faeces; and 4 = death. Mean scores of each group during the 21‐day observation period were calculated. For the tissue tropism and pathological evaluations, following infection, chicks were killed at 4, 7, 11 and 14 dpi (n = 3 or 4 each time point); the blood was collected for ELISA and tissues, including trachea, proventriculus, kidney and oviduct were collected for viral detection. Half of the harvested tissues were homogenized in tryptose phosphate broth and clarified by centrifugation. Viral detection by N gene‐based RT‐PCR was performed as described above. Another half of the tissues were further processed for immunohistochemical staining.
2.5. Immunohistochemical staining
Tissues stored in formalin were trimmed, embedded in paraffin, and cut into sections. Sections were first processed to remove the paraffin by xylene and rehydrate by ethanol. Citric buffer (10 mM, pH 6.0) was used to retrieve the viral antigens at 95°C followed by treatment with 3% hydrogen peroxide. After blocking with 1% bovine serum albumin solution, slides with sections were incubated with anti‐S1 monoclonal antibody (mAb) 2296‐B1 (prepared from S1 recombinant protein‐immunized mice) as the primary antibody (1:500) at 37°C for 40 min and an anti‐mouse IgG HRP conjugate (Jackson ImmunoResearch) as the secondary antibody. The antigens were visualized by applying a substrate of peroxidase (DAB). Finally, the slides were counterstained with haematoxylin and fixed by mounting buffer.
2.6. ELISA for detection of IBV antibodies in serum
Indirect ELISA against IBV was performed. Briefly, the IBV antigen was prepared as previously described (Chen, Wang, & Cheng, 2011). The serum antibody response was evaluated. The virus‐specific antibody response induced by the IBV strains was evaluated as previously described (Lin et al., 2016).
2.7. Commercial vaccine protection studies
To assess the protection conferred by a commercially available vaccine, groups of 10 one‐day‐old chicks were intranasally inoculated with 104 EID50 live‐attenuated H120 vaccine (Merial), the most common vaccine used in Taiwan. After 14 days, chickens were challenged with the above‐studied IBV variant strains or the Mass‐type strain as a control. Another mock group of chickens received PBS inoculation. Chickens were euthanized at 7‐ and 21‐days post‐challenge (dpc), and their pathological manifestations were evaluated. Viral shedding was also examined by collecting throat and cloacal swabs from each chicken at 7 and 21 dpc. Lesions in the trachea were evaluated as follows: 0 = no lesion; 1 = slight increase of mucin; 2 = large increase of mucin; and 3 = large increase of mucin and mucosal congestion. Lesions in the proventriculus were evaluated as follows: 0 = no lesion; 1 = slight increase in the thickness of the mucosa; 2 = large increase in the thickness of the mucosa; and 3 = large increase in the thickness of the mucosa and mucosal congestion. Lesions in the kidney were evaluated as follows: 0 = no lesions; 1 = swelling, urate visible only under stereomicroscopy; 2 = swelling with urate; and 3 = same as 2 with a large amount of urate deposition in the kidney. The lesion scores from three organs were averaged.
2.8. Statistical analysis
Data were analysed by unpaired t tests or ANOVA followed by Dunnett's multiple comparisons using GraphPad Prism (GraphPad Software). The p values smaller than .05 were considered significant.
3. RESULTS
3.1. Identification of viral variants
As listed in Table 1, five viral recombinants were previously isolated in Taiwan during 2002–2014 (Chen et al., 2009, 2010; Huang, Lee, Cheng, & Wang, 2004; Tsai et al., 2016). Among the recombinants, IBV TC3/13 and S78/14 strains were isolated from chickens and broilers native to Taiwan in 2013 and 2014, respectively. These two strains have similar genetic sequences and are grouped as Taiwan–Japan (TW–JP) recombinants. IBV 2992/02 and its homologous strain 3374/05 were previously reported to have arisen from multiple recombination events from strains from Taiwan and China; therefore, they have been defined as Taiwan–China (TW–CN) recombinants. In contrast, IBV 3382/06 was identified from broiler chickens in a poultry slaughterhouse, and this virus displays a Taiwan‐Mass‐type (TW‐Mass) recombination. In this study, the 3′ structural protein genomes of these IBV variants along with the putative parental strain, JP/Akita/92, were fully sequenced. The sequences were submitted to GenBank, and the accession numbers are shown in Table S1.
Table 1.
Viral variants identified in Taiwan
| Strain | Location | Year of isolation | Chicken type | References |
|---|---|---|---|---|
| S78/14 | Hsinchu | 2014 | Broiler | Tsai et al., (2016) |
| TC3/13 | Pingtung | 2013 | Native Taiwan chicken | Tsai et al., (2016) |
| 3382/06 | Taoyuan | 2006 | Broiler | Chen et al., (2010) |
| 3374/05 | Changhua | 2005 | Broiler | Chen et al., (2010) |
| 2992/02 | Yilan | 2002 | Broiler | Chen et al., (2009); Huang et al., (2004) |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.2. Phylogenetic analyses and recombination event of IBV variants
The phylogenetic analyses were compared among Taiwan variants and other reference strains. The S1 comparison (Figure 1a) clearly showed that the five variants clustered in different groups. IBV TC3/13 and S78/14, isolated from different geographical areas in Taiwan, share high nucleotide homology in structural proteins genes. In addition, the S1 gene sequences of these two strains were found to be clustered in the Japan group and share high identities (>94%) with the JP/Akita/92 and JP/Toyama/2000, isolated in Japan in 1992 and 2000, respectively (Mase, Tsukamoto, Imai, & Yamaguchi, 2004). In contrast to the result obtained from the S1 gene, phylogenetic trees based on additional 3’ structural proteins (S2 and N, Figure 1b and c) showed the phylogenetic closeness of TC3/13 and S78/14 to other Taiwan strains. These results suggest that TC3/13 and S78/14 were generated from the recombination of Taiwan and Japan IBV strains. We, therefore, performed the recombination analysis. RDP4 was used to display the consecutive nucleotide identity and illustrate the crossover events among the queried strain (S78/14) and the parental strains (2575/98 and Akita/92). As shown in Figure 1a,d crossover event in the 3′ of the S1 gene (nt 1,416) was suggested by the similarity plot. There was a significant difference (p < .01) between the resultant divisions of informative sites. IBV isolate S78/14 was therefore identified as an inter‐typic recombinant among the two putative parental strains. Three other variants, 2992/02, 3374/05 and 3382/06, have previously been reported to be derived from multiple recombination events with Chinese or Mass strains, and the recombination sites were located in the S1, S2, M and 5a genes (Chen et al., 2009, 2010).
Figure 1.
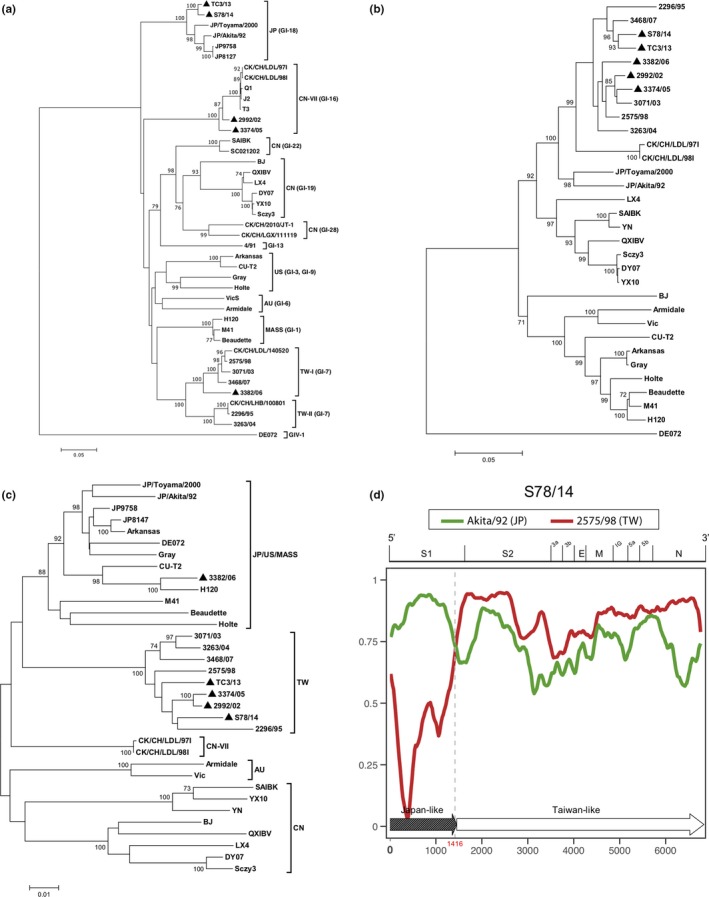
Phylogenetic analyses of Taiwan IBV isolates. Neighbour‐joining trees based on viral S1 (a), S2 (b) and N (c) gene sequences were constructed with 1,000 bootstrap replicates. Bootstrap vales higher than 70% were added to the respective nodes. The five studied Taiwan IBV variants are indicated with black triangles. AU, Australia; CN, China; TW, Taiwan; US, United States; and JP, Japan. (d) RDP4 analysis of the IBV isolate S78/14. The similarity plot displays the consecutive nucleotide identity (%) from the spike glycoprotein 1 (S1) gene to the nucleocapsid (N) gene among the queried strain (S78/14) and putative parental strains (2575/98 and Akita/92). The genomic scale is shown at the top of the plot. The recombination break point is indicated in red. The 3′ structural protein genome of IBV S78/14 was schematically assembled using Japan‐like and Taiwan‐like sequence regions. IG, inter‐genic region; E, envelope protein gene; M, membrane protein gene [Colour figure can be viewed at http://wileyonlinelibrary.com]
3.3. Novel serotypes possessed by recombinant IBV variants
To examine the antigenicity of the three viral variants, anti‐serum was cross neutralized using two well‐characterized local strains, 2575/98 (TW‐I) and 2296/95 (TW‐II), along with a vaccine strain, H120. The neutralization capacity of each anti‐serum was evaluated based on the viral infectivity in embryonated chicken eggs inoculated with a virus‐serum mixture, and the r‐value was calculated to assess the differences in the antigenicity between viruses. The r‐value of homologous virus‐serum was set as 100. The results shown in Table 2 and Table S2 demonstrated that among three viral variants, 3382/05 possess a r‐value of 68.5 with 2575/98, suggesting minor difference in the serotype shared by 3382/05 and 2575/98. Two other IBV variants, S78/14 and 2992/02, barely conferred protection against any other heterologous viruses, with r‐values no greater than 5.0. These results support the hypothesis that the IBV variants identified in Taiwan present novel serotypes that are distinct from the current two major circulating serotypes, TW‐I and TW‐II, and the Mass vaccine type.
Table 2.
Cross‐neutralization test with TW‐I, TW‐II, Mass‐type strains and three IBV recombinant variants
| IBV strain | 2575/98 (TW‐I) | 2296/95 (TW‐II) | H120 (Mass) | S78/14 (TW–JP) | 2992/02 (TW–CN) | 3382/06 (TW‐Mass) |
|---|---|---|---|---|---|---|
| 2575/98 (TW‐I) | 100 | |||||
| 2296/95 (TW‐II) | 12.0 | 100 | ||||
| H120 (Mass) | 2.6 | 12.8 | 100 | |||
| S78/14 (TW–JP) | 0.9 | 1.0 | 1.5 | 100 | ||
| 2992/02 (TW–CN) | 1.2 | 4.3 | 5.0 | 1.0 | 100 | |
| 3382/06 (TW‐Mass) | 68.5 | 2.6 | 2.1 | 0.6 | 2.0 | 100 |
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.4. Clinical and pathological assessment of chickens
To evaluate the pathogenicity of the three viral variants in chickens, SPF chickens were infected with the three viruses, and their clinical signs were recorded daily (Figure 2a and Figure 2b). Among all tested viruses, S78/14 induced chick death beginning at 7 dpi and caused the highest mortality rate of 67% at 21 dpi. IBV 2992/02 and 3382/06 caused fewer mortalities, with rates of 20% and 0%, respectively. Clinical signs such as tracheal rales and lacrimation occurred since the day following infection in all study groups of chicks except for the 3382/06 group, which showed only mild symptoms beginning at 3 dpi. More apparent respiratory signs, including sneezing and coughing, began at 5–7 dpi. Severe clinical signs, such as depression, were only present before animal death. As representative pictures shown in Figure 3a, all tracheal tissues sampled from infected chicks euthanized at 14 dpi or upon death showed substantial increases in mucin or mucosal congestion. Lesions in the proventriculus (haemorrhage and thickened mucosa) were observed in chicks infected with S78/14 and 2992/02, but not in chicks infected with 3382/06. In addition, lesions in the kidney were correlated with the outcome of death; in the S78/14 and 2992/02 groups, 4/6 and 2/5 chicks had visible urate deposits and swelling in the kidneys, which suggests that the predominant cause of lethality for strains S78/14 and 2992/02 was nephropathogenicity. Immunohistochemical staining with IBV‐specific mAb detected viruses in different tissues, including the trachea, lung, proventriculus and kidney (Figure 3b).
Figure 2.
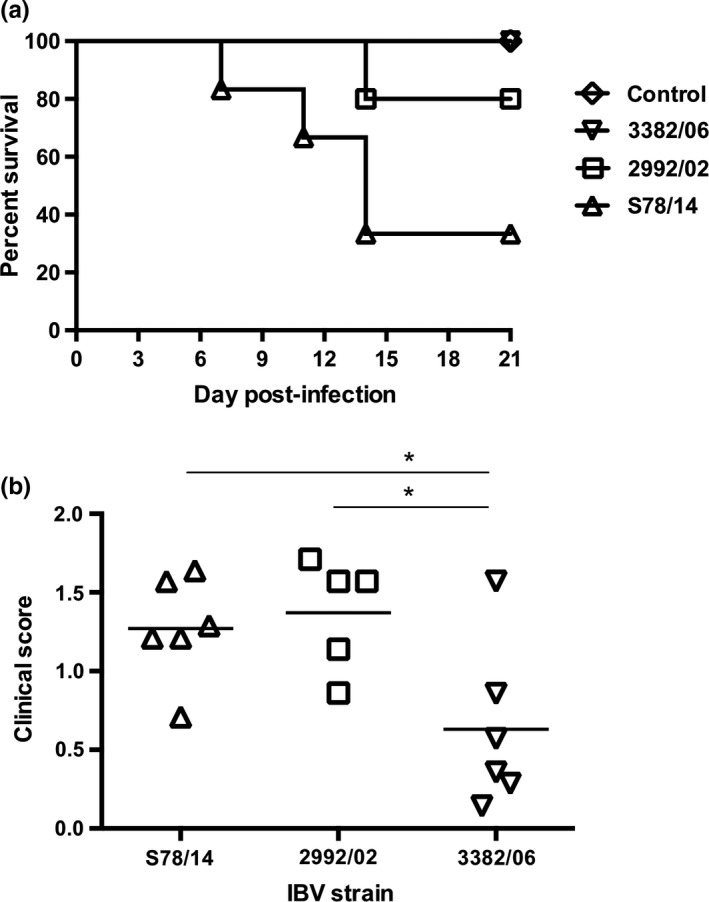
Pathogenicity of three Taiwan IBV variants in chickens. (a) One‐day‐old chicks were intranasally inoculated with 106 EID50 viruses, and their survival rates were recorded daily. (b) Following infection, the daily clinical scores were recorded. The clinical signs were evaluated as follows: 0 = no clinical signs; 1 = lacrimation, slight shaking, watery faeces or tracheal rales; 2 = lacrimation, presence of nasal exudate, depression, watery faeces, apparent sneezing or cough; 3 = same as 2 but stronger with severe watery faeces; and 4 = death. Mean scores of each group during the 21‐day observation period are indicated (n = 5 or 6 chickens per group). *p < .05
Figure 3.
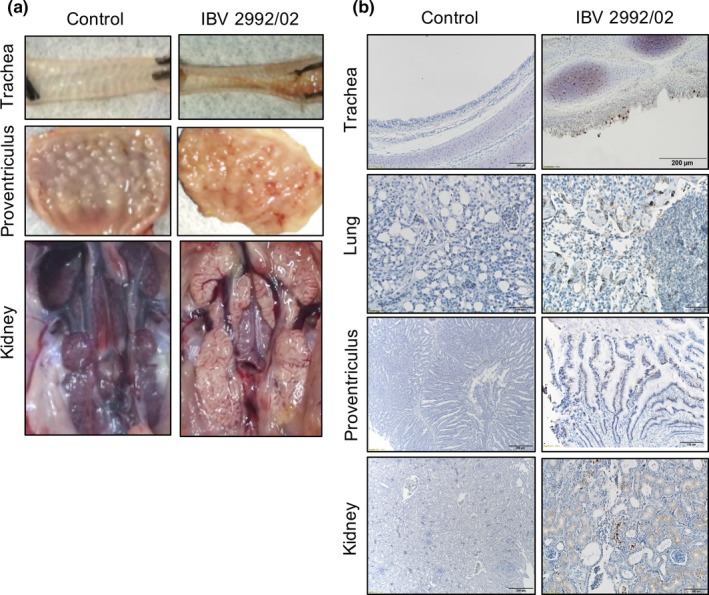
Pathological examination and immunohistochemical staining from IBV variant‐infected chickens. One‐day‐old chicks were intranasally inoculated with 106 EID50 of IBV 2992/02. Tissues were collected from IBV‐ or mock‐infected chicks at 7 dpi. (a) Gross lesions from experimentally infected chicks showed congestion and haemorrhage in trachea and proventriculus, and urate deposit in kidney. (b) Immunohistochemical staining detected virus antigens (brown colour signals) in trachea, lung, proventriculus and kidney by IBV spike protein‐specific monoclonal antibody [Colour figure can be viewed at http://wileyonlinelibrary.com]
Serum samples were also collected at different time points post‐infection; antibody responses against each strain were measured by commercial kits (Figure 4). The results showed that antibody titres against IBV of 2992/02‐ and 3382/06‐infected chicks increased significantly from 11 to 14 dpi, whereas the titres of the S78/14‐infected chicks increased moderately. Notably, the antibody titres of chicks infected with S78/14 at 11 dpi were higher than those of chicks infected with the other two selected viral strains.
Figure 4.
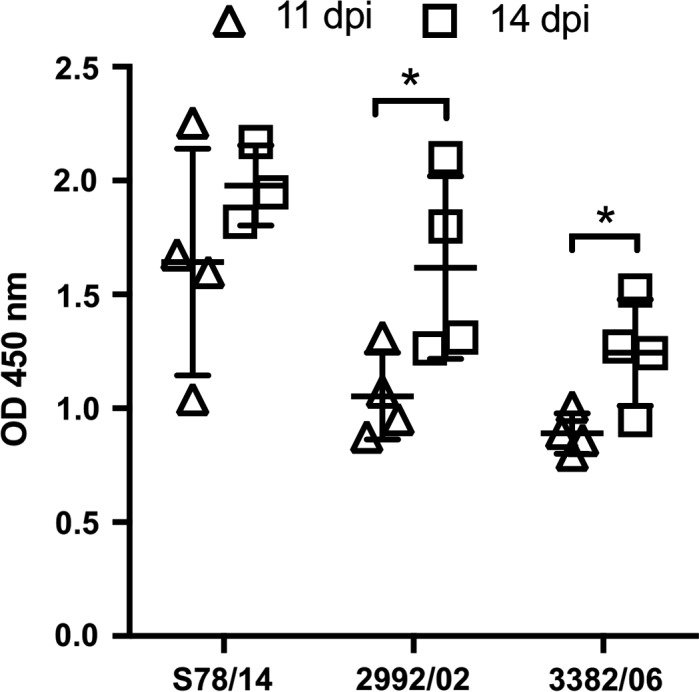
Serum antibodies of IBV variants‐infected chickens. Antibody responses post‐inoculation with different IBV strains were evaluated by ELISA. Chicken serum was collected at 11 and 14 dpi, respectively. Bars represent mean ± standard deviation (n = 3 or 4 chickens per group). *p < .05
3.5. Viral RNA distribution in infected chickens
To further study the tissue tropism of these strains, tissues from the trachea, proventriculus, kidney and oviduct were collected at 4, 7, 11 and 14 dpi, and viral RNA was examined by RT‐PCR (Table 3). Tissues from the trachea were positive in all three groups of viruses, indicating the respiratory tract tropism of the three viruses, and the viruses were readily transmitted by the respiratory route. Increased positive rates were also found in the proventriculus and kidney in the S78/14 and 2992/02 groups, whereas lower positive rates were found in the 3382/06 groups for these two organs; the rate for tissues from the kidney was especially low at 25% (4/16). Oviduct samples of the tested strains, except for 3382/06, were positive for viral RNA.
Table 3.
IBV detection by RT‐PCR in chicken organs at 4, 7, 11 and 14 days post‐infection
| Strain | Trachea (dpi) | Proventriculus (dpi) | Kidney (dpi) | Oviducta (dpi) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 7 | 11 | 14 | 4 | 7 | 11 | 14 | 4 | 7 | 11 | 14 | 4 | 7 | 11 | 14 | |
| S78/14 | 4/4 | 4/4 | 4/4 | 3/3 | 3/4 | 4/4 | 4/4 | 0/3 | 0/4 | 3/4 | 3/4 | 3/3 | 1/2 | 1/2 | n/ab | 2/2 |
| 2992/02 | 4/4 | 4/4 | 3/4 | 1/3 | 3/4 | 4/4 | 4/4 | 2/3 | 1/4 | 4/4 | 3/4 | 3/3 | 0/2 | 0/1 | 3/4 | n/a |
| 3382/06 | 4/4 | 4/4 | 4/4 | 4/4 | 3/4 | 4/4 | 2/4 | 0/4 | 0/4 | 1/4 | 1/4 | 2/4 | n/a | 0/1 | 0/1 | 0/2 |
Abbreviation: dpi, day post‐infection.
Only female chickens were examined.
n/a: female chickens not available.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
3.6. Vaccination against novel IBV isolates using the Mass vaccine
Based on the results above, the three novel IBV strains exhibited substantial antigenic distances to the Mass vaccine strain, and two of the strains exhibited virulence to chickens. Therefore, we inoculated chicks with live H120 vaccine prior to the challenge with different strains of virus to assess the protective effect of the vaccine against each studied strain. From the throat and cloacal swabs, viruses in all three groups were detected, but the positive rates declined in a temporal manner (Table 4). Among the three groups of IBV variants‐infected chickens, chickens inoculated with S78/14 had the highest levels of virus detected from two swabbed sites across two time points, with positive rates of 78% and 89% at 7 dpc in throat swabs and cloacal swabs, respectively. For the 2992/02 group, which began with slightly lower positive rates at 7 dpc in the cloacal swabs, a similar shedding pattern was observed compared to the S78/14 group. At 21 dpc, 40% and 60% of viral shedding from the throat swabs and cloacal swabs, respectively, were still observed from these two variants‐challenged groups. On the contrary, vaccination conferred better protection to 3382/06‐infected chickens, with 50 and 20% of infected animals presenting positive viral detection at 7 dpc in the throat and cloacal swabs, respectively, and viruses were undetectable at 21 dpc. The group of chickens received homologous viral challenge or mock challenge demonstrated a low or zero detection rate for virus shedding. In addition to viral shedding, the challenged chickens were killed at 7 and 21 dpc to examine the gross lesions made by viruses. As shown in Figure 5, the results from the post‐mortem necropsy demonstrated that S78/14 viruses caused more severe damage to vaccinated chickens (the highest lesion scores among five challenge groups) than both 2992/02 and 3382/06 strains caused. The 2992/02 and 3382/06 groups showed apparent lesions compared to the Mass‐type‐ and mock‐challenged group. Taken together, our data point out that the current vaccine plan using the Mass strain as an immunogen is insufficiently capable of stopping viral transmission and reducing viral pathological effects.
Table 4.
Rate of IBV detection by RT‐PCR in throat and cloacal swabs of Mass‐type immunized chickens after IBV variant challenge
| Challenge strain | Throat swab % IBV positive | Cloacal swab % IBV positive | ||
|---|---|---|---|---|
| 7 dpc | 21 dpc | 7 dpc | 21 dpc | |
| S78/14 | 78 (7/9) | 40 (2/5) | 89 (8/9) | 60 (3/5) |
| 2992/02 | 78 (7/9) | 40 (2/5) | 56 (5/9) | 60 (3/5) |
| 3382/06 | 50 (5/10) | 20 (1/5) | 0 (0/10) | 0 (0/5) |
| Mass | 20 (2/10) | 0 (0/5) | 0 (0/10) | 0 (0/5) |
| Mock | 0 (0/10) | 0 (0/5) | 0 (0/10) | 0 (0/5) |
Abbreviation: dpc, day post‐challenge.
This article is being made freely available through PubMed Central as part of the COVID-19 public health emergency response. It can be used for unrestricted research re-use and analysis in any form or by any means with acknowledgement of the original source, for the duration of the public health emergency.
Figure 5.
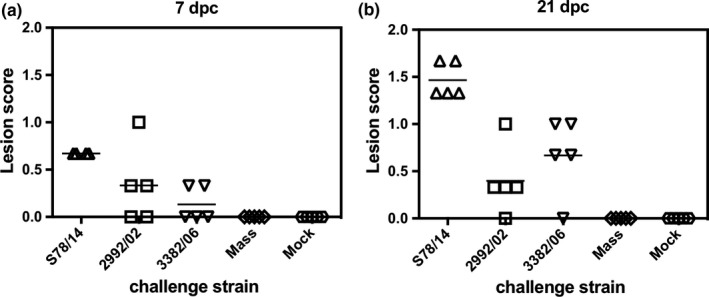
Vaccination against novel IBV isolates using the Mass vaccine. One‐day‐old chicks were intranasally inoculated with H120 live‐attenuated vaccine (104 EID50), and after 14 days, chicks were infected with S78/14, 2992/02 or 3382/06 viruses intranasally. At 7‐ and 21‐days post‐challenge (dpc), animals were killed to examine their gross lesions made by viruses. Lesions in the trachea were evaluated as follows: 0 = no lesion; 1 = slight increase of mucin; 2 = large increase of mucin; and 3 = large increase of mucin and mucosal congestion. Lesions in the proventriculus were evaluated as follows: 0 = no lesion; 1 = slight increase in the thickness of the mucosa; 2 = large increase the thickness of the mucosa; and 3 = large increase in the thickness of the mucosa and mucosal congestion. Lesions in the kidney were evaluated as follows: 0 = no lesions; 1 = swelling, urate visible only under stereomicroscopy; 2 = swelling with urate; and 3 = same as 2 with a large amount of urate deposition in the kidney. The lesion scores from trachea, proventriculus and kidney were averaged
4. DISCUSSION
This study comprehensively characterized three strains of recombinant IBVs isolated in Taiwan from 2002 to 2014. Two of the strains, S78/14 and 2992/02, exhibited serotypes distinct from classical Taiwan IBV lineages. S78/14 and 2992/02 strains were lethal to one‐day‐old chicks, with mortality rates of 67% and 20%, respectively. These two strains also caused more severe clinical signs than 3382/06, which did not kill chicks. Furthermore, an extensive nephropathic effect was observed in S78/14‐ and 2992/02‐infected chicks, while viral RNA was detected systemically in infected hosts. Finally, the results from the challenge experiments showed that the current vaccine based on the Mass strain could not stop viral transmission from host to host, and lesions made by viruses were not prevented in vaccinated chickens inoculated with the studied viruses.
Recombination in S78/14 and TC3/13 strains was first recognized through nucleotide identity searching, wherein the S1 gene sequences were found to be similar with the IBVs clustered in the JP‐I group (Mase et al., 2004), but the S2 and N gene sequences were found to be highly homologous with local viruses isolated in Taiwan. Using the RDP4 analysis, S78/14 was confirmed to emerge through recombination events between Taiwan and Japan strains; however, in the one‐day‐old chicken experimental infection, S78/14 exhibited increased lethality (67%) as compared to the 33.3% lethality caused by the TW‐I strain 2575/98 (Lin et al., 2016). Furthermore, a previous study has described the abnormal egg production of chickens caused by the infection of the Japan IBV strains JP8127 (Shieh et al., 2004), which is closely related to S78/14 in term of the S1 gene, supporting our finding on the oviduct tropism of S78/14. However, the number of examined female chickens in this study was relatively limited, and the experimental period was not long enough to observe pathological lesions. Further investigation using a defined group of female chickens is warranted to confirm this observation.
During recent years, an increasing number of TW‐like strains have been isolated from other geographical regions in Asia (Hong, Kwon, Choi, & Kim, 2017; Lin & Chen, 2017; Ma et al., 2012; Xu et al., 2016), and some of these emerging variants displayed severe necropsy lesions, including respiratory tissues, kidney, heart and bursa of Fabricius (Xu et al., 2018). Interestingly, similar age (approximately 30 days) of affected broilers was described from these reported infections and in our study. Given that most of the broiler chickens are maintained at a high density in Taiwan or other countries where commercial broiler operations are practiced, viral variants seem more likely to emerge from IBVs that co‐circulate in the farm during the breeding period. Therefore, in addition to the regular immunization, improved management and biosecurity practices may help reduce the viral emergence and control the disease outbreaks.
In contrast to other variants, the 3382/06, a recombinant IBV whose S1 protein belongs to the TW‐I, did not show lethality and oviduct tropism. The mild clinical symptoms and low virulence in chickens caused by the 3382/06 can be speculated that the recombination with the H120 vaccine strain attenuated the pathogenicity. However, to what extent the swapped genes influenced the viral virulence remains unknown. Similarly, although we have examined the structural protein genes (S to N), the link of sequence variations in other non‐structural protein genes to serotype changes observed from IBV S78/14 and 2992/02 in this study is not clear. More studies based on the reverse genetic techniques are needed to elucidate the mechanisms contributing to the altered antigenicity and virulence of a variant carrying chimeric IBV genome arrangement.
The Mass lineage virus is the current industry standard vaccine strain for IBV control. Our results from the protection experiments pointed out that chickens immunized with H120 virus failed to eliminate viral shedding after infection with the three recombinant strains. Furthermore, these viruses produced appreciable pathogenic effects in the immunized chickens, indicating that H120 is not competent as the sole immunogen to defeat current circulating recombinant IBVs. New vaccines conferring desirable cross‐protection are warranted.
In conclusions, this study reported recombinant IBVs with novel serotypes, multiorgan tropism and lethality (Table S3). These strains caused disease outbreaks in the field, and the use of commercial Mass‐type vaccine is not effective in preventing these infections. Our study indicates that more intense surveillance efforts and a review of the current IBV vaccine strategy are required.
ETHICAL APPROVAL
Experiments involving chickens were conducted at National Taiwan University under an approved Institutional Animal Care and Use Committee (IACUC) protocol (no. NTU‐103‐EL‐3). All animal experiments were carried out in accordance with the approved guidelines.
CONFLICT OF INTEREST
Authors declare that none of conflict interests exist.
AUTHOR CONTRIBUTIONS
HWC conceived and designed the experiments; YTL TCC SYL and HWC performed the experiments; YTL TCC SYL and HWC analysed the data; MM SM and TH contributed essential materials; YTL and HWC wrote the paper. All authors read the final manuscript and approved it for submission.
Supporting information
ACKNOWLEDGEMENTS
The authors thank Dr. Ching‐Ho Wang for providing the viruses. This work was supported by the Ministry of Science and Technology (103‐2321‐B‐002‐066, 104‐2321‐B‐002‐023, 105‐2321‐B‐002‐007, 106‐2311‐B‐002‐030‐MY3) and National Taiwan University.
Li Y‐T, Chen T‐C, Lin S‐Y, et al. Emerging lethal infectious bronchitis coronavirus variants with multiorgan tropism. Transbound Emerg Dis. 2020;67:884–893. 10.1111/tbed.13412
REFERENCES
- Archetti, I. , & Horsfall, F. L. Jr (1950). Persistent antigenic variation of influenza A viruses after incomplete neutralization in ovo with heterologous immune serum. Journal of Experimental Medicine, 92, 441–462. 10.1084/jem.92.5.441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avellaneda, G. E. , Villegas, P. , Jackwood, M. W. , & King, D. J. (1994). In vivo evaluation of the pathogenicity of field isolates of infectious bronchitis virus. Avian Diseases, 38, 589–597. 10.2307/1592083 [DOI] [PubMed] [Google Scholar]
- Cavanagh, D. (2007). Coronavirus avian infectious bronchitis virus. Veterinary Research, 38, 281–297. 10.1051/vetres:2006055 [DOI] [PubMed] [Google Scholar]
- Chen, H. W. , Huang, Y. P. , & Wang, C. H. (2009). Identification of Taiwan and China‐like recombinant avian infectious bronchitis viruses in Taiwan. Virus Research, 140, 121–129. 10.1016/j.virusres.2008.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. W. , Huang, Y. P. , & Wang, C. H. (2010). Identification of intertypic recombinant infectious bronchitis viruses from slaughtered chickens. Poultry Science, 89, 439–446. 10.3382/ps.2009-00322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H. W. , Wang, C. H. , & Cheng, I. C. (2011). A type‐specific blocking ELISA for the detection of infectious bronchitis virus antibody. Journal of Virological Methods, 173, 7–12. 10.1016/j.jviromet.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , Jiang, L. , Zhao, W. , Liu, L. , Zhao, Y. , Shao, Y. , … Liu, S. (2017). Identification and molecular characterization of a novel serotype infectious bronchitis virus (GI‐28) in China. Veterinary Microbiology, 198, 108–115. 10.1016/j.vetmic.2016.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, J. K. , Jackwood, M. , & Jones, R. C. (2012). The long view: 40 years of infectious bronchitis research. Avian Pathology, 41, 239–250. 10.1080/03079457.2012.680432 [DOI] [PubMed] [Google Scholar]
- Gao, M. , Wang, Q. , Zhao, W. , Chen, Y. , Zhang, T. , Han, Z. , … Liu, S. (2016). Serotype, antigenicity, and pathogenicity of a naturally recombinant TW I genotype infectious bronchitis coronavirus in China. Veterinary Microbiology, 191, 1–8. 10.1016/j.vetmic.2016.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewson, K. A. , Noormohammadi, A. H. , Devlin, J. M. , Browning, G. F. , Schultz, B. K. , & Ignjatovic, J. (2014). Evaluation of a novel strain of infectious bronchitis virus emerged as a result of spike gene recombination between two highly diverged parent strains. Avian Pathology, 43, 249–257. 10.1080/03079457.2014.914624 [DOI] [PubMed] [Google Scholar]
- Hong, S. M. , Kwon, H. J. , Choi, K. S. , & Kim, J. H. (2017). Comparative genomics of QX‐like infectious bronchitis viruses in Korea. Archives of Virology, 162, 1237–1250. 10.1007/s00705-016-3208-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, S. M. , Kwon, H. J. , Kim, I. H. , Mo, M. L. , & Kim, J. H. (2012). Comparative genomics of Korean infectious bronchitis viruses (IBVs) and an animal model to evaluate pathogenicity of IBVs to the reproductive organs. Viruses, 4, 2670–2683. 10.3390/v4112670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, Y. P. , Lee, H. C. , Cheng, M. C. , & Wang, C. H. (2004). S1 and N gene analysis of avian infectious bronchitis viruses in Taiwan. Avian Diseases, 48, 581–589. 10.1637/7186-033004R [DOI] [PubMed] [Google Scholar]
- Jackwood, M. W. (2012). Review of infectious bronchitis virus around the world. Avian Diseases, 56, 634–641. 10.1637/10227-043012-Review.1 [DOI] [PubMed] [Google Scholar]
- Jackwood, M. W. , Hilt, D. A. , Lee, C. W. , Kwon, H. M. , Callison, S. A. , Moore, K. M. , … Thayer, S. (2005). Data from 11 years of molecular typing infectious bronchitis virus field isolates. Avian Diseases, 49, 614–618. 10.1637/7389-052905R.1 [DOI] [PubMed] [Google Scholar]
- Lin, K. Y. , Wang, H. C. , & Wang, C. H. (2005). Protective effect of vaccination in chicks with local infectious bronchitis viruses against field virus challenge. Journal of Microbiology, Immunology and Infection, 38, 25–30. [PubMed] [Google Scholar]
- Lin, S. Y. , & Chen, H. W. (2017). Infectious bronchitis virus variants: Molecular analysis and pathogenicity investigation. International Journal of Molecular Sciences, 18, 2030 10.3390/ijms18102030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, S. Y. , Li, Y. T. , Chen, Y. T. , Chen, T. C. , Hu, C. J. , & Chen, H. W. (2016). Identification of an infectious bronchitis coronavirus strain exhibiting a classical genotype but altered antigenicity, pathogenicity, and innate immunity profile. Scientific Reports, 6, 37725 10.1038/srep37725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, H. , Shao, Y. , Sun, C. , Han, Z. , Liu, X. , Guo, H. , … Liu, S. (2012). Genetic diversity of avian infectious bronchitis coronavirus in recent years in China. Avian Diseases, 56, 15–28. 10.1637/9804-052011-Reg.1 [DOI] [PubMed] [Google Scholar]
- Martin, D. P. , Murrell, B. , Golden, M. , Khoosal, A. , & Muhire, B. (2015). RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution, 1, vev003 10.1093/ve/vev003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mase, M. , Tsukamoto, K. , Imai, K. , & Yamaguchi, S. (2004). Phylogenetic analysis of avian infectious bronchitis virus strains isolated in Japan. Archives of Virology, 149, 2069–2078. 10.1007/s00705-004-0369-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed, L. J. , & Muench, H. (1938). A simple method of estimating fifty per cent endpoints. The American Journal of Hygiene, 27, 493–497. [Google Scholar]
- Shieh, H. K. , Shien, J. H. , Chou, H. Y. , Shimizu, Y. , Chen, J. N. , & Chang, P. C. (2004). Complete nucleotide sequences of S1 and N genes of infectious bronchitis virus isolated in Japan and Taiwan. Journal of Veterinary Medical Science, 66, 555–558. 10.1292/jvms.66.555 [DOI] [PubMed] [Google Scholar]
- Sjaak de Wit, J. J. , Cook, J. K. , & van der Heijden, H. M. (2011). Infectious bronchitis virus variants: A review of the history, current situation and control measures. Avian Pathology, 40, 223–235. 10.1080/03079457.2011.566260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , Dudley, J. , Nei, M. , & Kumar, S. (2007). MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution, 24, 1596–1599. 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Tsai, C. T. , Tsai, H. F. , & Wang, C. H. (2016). Detection of infectious bronchitis virus strains similar to Japan in Taiwan. Journal of Veterinary Medical Science, 78, 867–871. 10.1292/jvms.15-0609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastro, V. , Holmes, E. C. , Britton, P. , Fusaro, A. , Jackwood, M. W. , Cattoli, G. , & Monne, I. (2016). S1 gene‐based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification. Infection, Genetics and Evolution, 39, 349–364. 10.1016/j.meegid.2016.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. H. , Hsieh, M. C. , & Chang, P. C. (1996). Isolation, pathogenicity, and H120 protection efficacy of infectious bronchitis viruses isolated in Taiwan. Avian Diseases, 40, 620–625. 10.2307/1592273 [DOI] [PubMed] [Google Scholar]
- Wang, C. H. , & Huang, Y. C. (2000). Relationship between serotypes and genotypes based on the hypervariable region of the S1 gene of infectious bronchitis virus. Archives of Virology, 145, 291–300. 10.1007/s007050050024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. H. , & Tsai, C. T. (1996). Genetic grouping for the isolates of avian infectious bronchitis virus in Taiwan. Archives of Virology, 141, 1677–1688. 10.1007/BF01718291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, G. , Cheng, J. L. , Ma, S. H. , Jia, W. F. , Yan, S. H. , & Zhang, G. Z. (2018). Pathogenicity differences between a newly emerged TW‐like strain and a prevalent QX‐like strain of infectious bronchitis virus. Veterinary Microbiology, 227, 20–28. 10.1016/j.vetmic.2018.10.019 [DOI] [PubMed] [Google Scholar]
- Xu, G. , Liu, X. Y. , Zhao, Y. , Chen, Y. , Zhao, J. , & Zhang, G. Z. (2016). Characterization and analysis of an infectious bronchitis virus strain isolated from southern China in 2013. Virology Journal, 13, 40 10.1186/s12985-016-0497-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanaty, A. , Naguib, M. M. , El‐Husseiny, M. H. , Mady, W. , Hagag, N. , & Arafa, A. S. (2016). The sequence of the full spike S1 glycoprotein of infectious bronchitis virus circulating in Egypt reveals evidence of intra‐genotypic recombination. Archives of Virology, 161, 3583–3587. 10.1007/s00705-016-3042-1 [DOI] [PubMed] [Google Scholar]
- Zhong, Q. , Hu, Y. X. , Jin, J. H. , Zhao, Y. , Zhao, J. , & Zhang, G. Z. (2016). Pathogenicity of virulent infectious bronchitis virus isolate YN on hen ovary and oviduct. Veterinary Microbiology, 193, 100–105. 10.1016/j.vetmic.2016.08.017 [DOI] [PubMed] [Google Scholar]
- Zhou, H. , Zhang, M. , Tian, X. , Shao, H. , Qian, K. , Ye, J. , & Qin, A. (2017). Identification of a novel recombinant virulent avian infectious bronchitis virus. Veterinary Microbiology, 199, 120–127. 10.1016/j.vetmic.2016.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


