Abstract
The stalling of ribosomes during protein synthesis results in the production of truncated polypeptides that can have deleterious effects on cells, and therefore must be eliminated. In eukaryotes, this function is carried out by a dedicated surveillance mechanism known as ribosome-associated protein quality control (RQC). The E3 ubiquitin ligase Ltn1 (Listerin in mammals) plays a key part in RQC by targeting the aberrant nascent polypeptides for proteasomal degradation. Consistent with having an important protein quality control function, mutations in Listerin cause neurodegeneration in mice. Ltn1/Listerin is part of the multi-subunit RQC complex, and recent findings have revealed that the Rqc2 subunit of this complex catalyses the formation of C-terminal alanine and threonine tails (CAT tails), which are extensions of nascent chains known to either facilitate substrate ubiquitylation and targeting for degradation or induce protein aggregation. RQC, originally described for quality control on ribosomes translating cytosolic proteins, is now known to also have a role on the surfaces of the endoplasmic reticulum and mitochondria. This Review describes our current knowledge on RQC mechanisms, highlighting key features of Ltn1/Listerin action that provide a paradigm for understanding how E3 ligases operate in protein quality control in general, and discusses how defects in this pathway may compromise cellular function and lead to disease.
Introduction
Aberrant proteins are continuously produced as a consequence of, for example, gene mutations, errors during gene expression, chemical damage or the absence of an interacting partner. As such proteins can fold improperly, lose normal function, and/or acquire new anomalous functions (such as forming toxic aggregates) cells have a variety of protein quality control mechanisms that detect their production and prevent their accumulation—in general, relying on molecular chaperones to promote correct protein folding and/or prevent protein aggregation, and on the ubiquitin-proteasome and autophagy systems for protein degradation1,2. Consistent with having a crucial role in ensuring the integrity of the proteome and cellular fitness, defects protein quality control are associated with various diseases and are a hallmark of neurodegeneration3.
Protein surveillance, which must occur with great sensitivity and precision to detect as many forms of protein aberration as possible, is carried out by a variety of specialized machineries. Studies over the past years have increased our understanding of one such surveillance process: ribosome-associated protein quality control (RQC). The RQC pathway is initiated as a result of ribosome stalling on mRNA (Box 1; Fig. 1) and targets newly-synthesized proteins for proteasomal degradation while they are still associated with the ribosomal 60S subunit. Degrading proteins at their birthplace may seem counterintuitive, as nascent chains only have the opportunity to achieve their final fold, and sometimes to engage in stabilizing interactions with other cellular components, after translation is terminated and the complete polypeptides are released from ribosomes. However, stalled ribosomes are perceived by cells of all organisms as a dangerous source of defective nascent polypeptide chains4,5. Thus, a commitment to proteolysis is made already at the ribosomal level, minimizing the potential to release toxic protein species (Fig. 1). Furthermore, this mechanism of targeting protein quality control substrates expands the code that cells utilize to recognize aberrant proteins, as it senses the state of translation rather than the folding state of the nascent chain.
Box 1: The translation cycle and causes for ribosome stalling.
Following initiation, protein synthesis proceeds through a phase of elongation before the process is terminated and the small (40S) and large (60S) ribosomal subunits are recycled for engaging in other rounds of translation (see the figure, part a). During elongation, aminoacyl-tRNAs (aa-tRNAs) are loaded onto the ribosomal A site by the GTPase eukaryotic elongation factor 1A (eEF1A). The nascent chain linked to the P-site tRNA is transferred to the aminoacyl-tRNA on the A site. With this reaction, the ribosomal subunits become rotated relative to each other, and the nascent chain-accepting tRNA adopts a ‘hybrid’ (P/A) state. The GTPase eEF2 binds to such rotated ribosomes and drives the subunits back to the classical conformation, resulting in the nascent chain-linked tRNA adopting a P-site state, while emptying the A site for the next aminoacyl-tRNA to be recruited. When translating ribosomes encounter a stop codon on the A site, the GTPase eukaryotic peptide chain release factor 3 (eRF3) loads the translation termination factor eRF1 instead of tRNA. eRF1 catalyses hydrolysis of the peptidyl-tRNA linkage to release the nascent chain and recruits the ATPase Rli1 (ABCE1 in mammals) to split the ribosomal subunits. Translation termination and ribosome recycling are coupled in eukaryotes.
During the elongation phase of translation, there can be different causes for ribosomal stalling (see the figure, part b): decoding of suboptimal codons, fluctuations in tRNA levels or tRNA modifications, chemical damage of mRNA or ribosomes, strong mRNA secondary structure, certain nascent chain sequences, immature 60S subunits that have escaped from the nucleus, or mRNAs lacking stop codons (nonstop mRNA), which can be generated by endonuclease cleavage or premature polyadenylation4,5,96. Premature cleavage and polyadenylation of mRNA within coding sequences is a naturally occurring error during gene expression, and is believed to be a prevalent source of nonstop mRNA. Unless a stop codon is generated at the junction between the truncated open reading frame and the poly(A) tail, ribosomes continue translating through the poly(A) tail, producing a truncated polypeptide that is extended with the poly(A)-encoded polyLys sequence (part b, right panel). Translation into poly(A) sequences is at present the only known mechanism to cause efficient stalling in reporter gene assays with mammalian cells97,98. Programmed ribosomal pausing can also occur in response to different signals and can have regulatory functions99. It remains unknown how cells distinguish such ribosomal pauses from more permanent and detrimental stalling that requires rescue4.
Box 1.
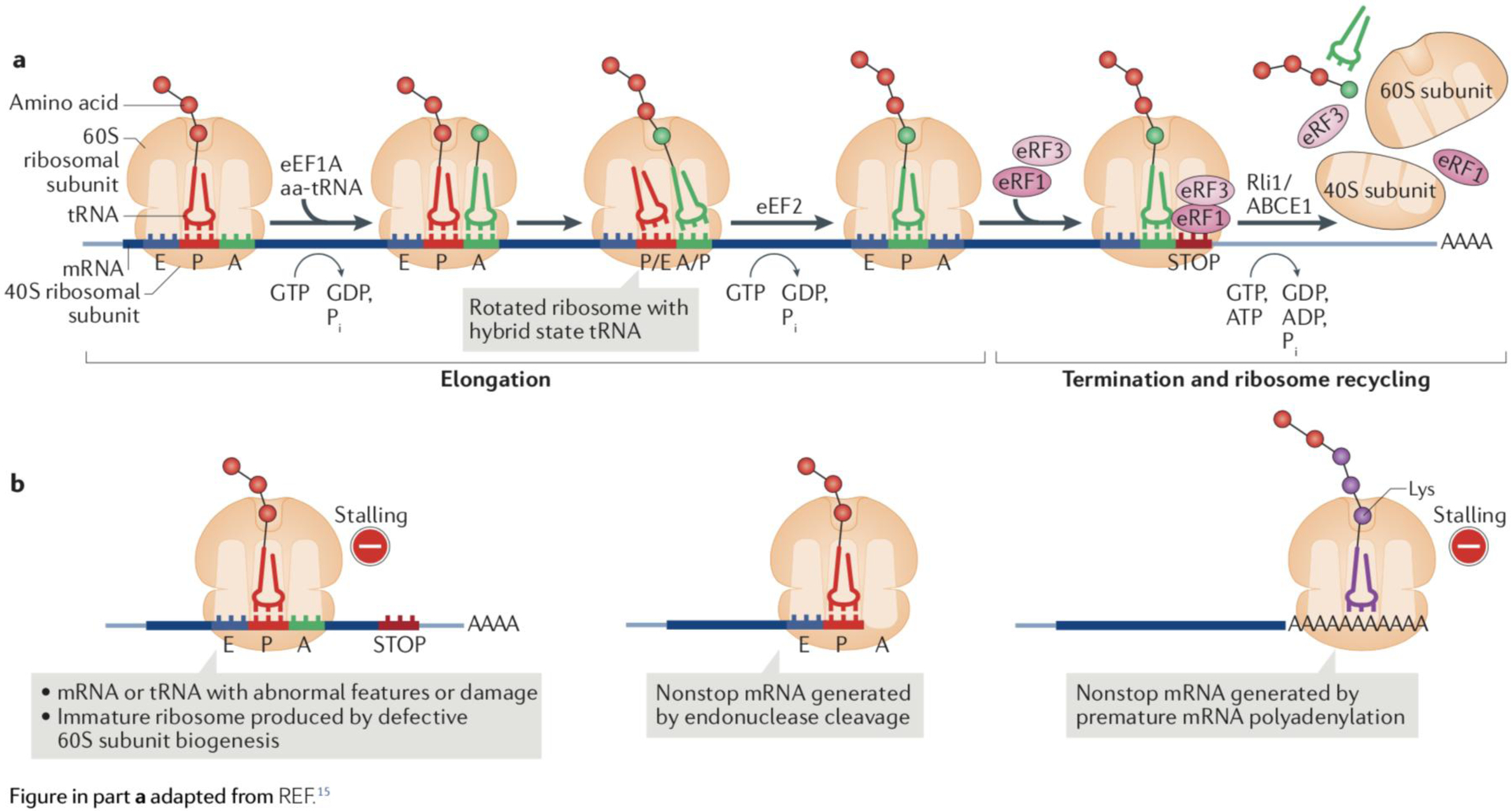
Fig. 1: The eukaryotic ribosome-associated quality control pathway.
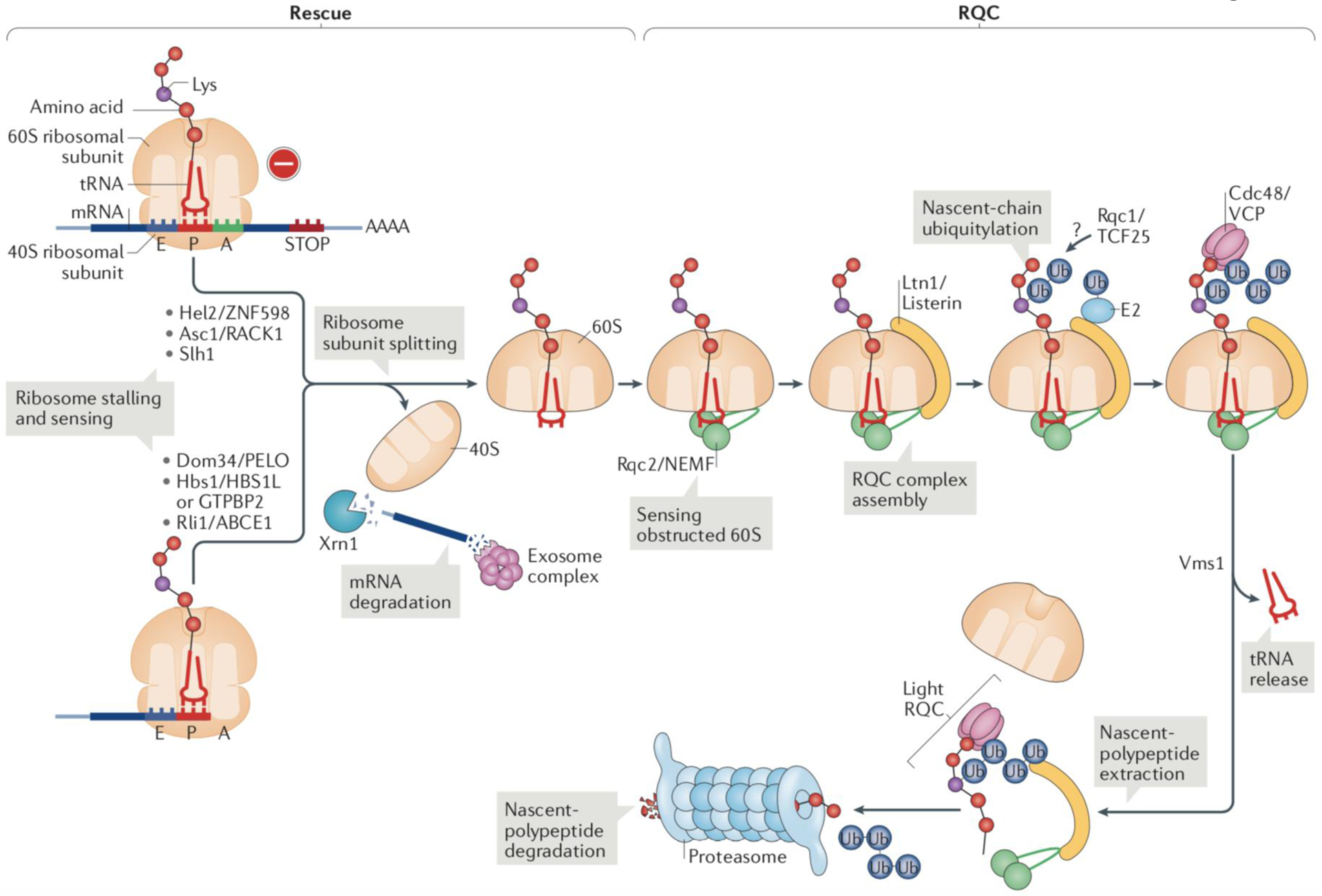
The ribosomal rescue and Ribosome-Associated Quality Control (RQC) pathways for protein surveillance consist of two sequential steps that each sense a unique defect. The first step (rescue) senses stalled ribosomes and mediates 80S subunit splitting; the second step (RQC) detects 60S subunits obstructed with peptidyl-tRNA—products of the rescue step—and promotes the resolution of this aberrant structure, to release free, translation-competent 60S ribosomal subunits and mediate proteolysis of the nascent chain.
(Left) Ribosomal rescue. Ribosomes that are stalled at the mRNA 3’ end are sensed by Hbs1 in yeast (or HBS1L and GTPBP2 in mammals) and Dom34 (pelota (PELO) in mammals). The mechanisms for the recognition of ribosomes stalled on internal mRNA sequences are poorly understood and at least in some cases involve the E3 ubiquitin-protein ligase Hel2/ZNF598. Whether Dom34/PELO also acts downstream of Hel2/ZNF598 is not known, but these two factors can target the same mRNA molecule, as ribosome stalling elicits endonuclease cleavage of the transcript4. Dom34/PELO recruits Rli1 (ATP-binding cassette protein ABCE1 in mammals), which separates the small (40S) and large (60S) ribosomal subunits (Box 1). The released truncated mRNA is degraded by the 5’−3’ exoribonuclease Xrn1 and the exosome complex, which prevents the mRNA from being translated again, as it may be defective. However, unlike eRF1, Dom34 lacks peptidyl-tRNA hydrolase activity, the peptidyl-tRNA remains bound to the 60S subunit. Although the peptidyl-tRNA−60S complex can potentially reassociate with free 40S subunits, the reassociation can be reversed by Dom34, Hbs1 and Rli113,14,115.
(Right) RQC. Resolution of the peptidyl-tRNA−60S complex is initiated by binding of the Rqc2 (NEMF in mammals) subunit of the RQC complex, which recruits and stabilizes the binding of the E3 ligase Ltn1/Listerin. At this stage, Rqc2 may synthesize CAT tails to help expose Lys residues that are buried in the ribosomal exit tunnel, to be ubiquitylated by Ltn1/Listerin (see below). The ribosome binding mode of the Rqc1 subunit (TCF25 in mammals) of the RQC complex and its exact function remain unknown. The ubiquitin chain that is polymerised by Ltn1 on the nascent polypeptides signals recruitment of the AAA ATPase Cdc48 and its cofactors. Cdc48 extracts nascent polypeptides from the 60S ribosomal subunit after they have been released from the conjugated tRNA by Vms1 (ANKZF1 in mammals). Released polypeptides can then be degraded by the proteasome. The extracted nascent polypeptides may remain associated with RQC subunits in the form of a ‘light RQC’ complex before proteasomal degradation.
The central player in RQC is the RING domain [G] E3 ubiquitin-protein ligase Ltn14,6,7, which polyubiquitylates the polypeptides obstructing the 60S ribosomal subunit to promote their degradation. Lnt1 is the yeast homologue of mammalian Listerin, and consistent with its important function in protein quality control, mutation of Listerin causes neurodegeneration in mice8. Ltn1 works closely with its cofactor Rqc2 (NEMF in mammals) as part of the multi-subunit RQC complex9,10.
This Review describes the function and mechanisms of RQC. It also highlights how this mechanistic understanding can inform on more general principles underlying E3 ligase-mediated protein surveillance; this is important, as knowledge on how E3 ligases work in protein surveillance is scarce, with the exception of endoplasmic reticulum-associated degradation (ERAD) [G]. Moreover, the Review discusses the molecular and cellular consequences of RQC dysfunction and work implicating RQC in protecting the integrity of organelles such as the endoplasmic reticulum (ER) and mitochondria. Finally, it comments on how our understanding of RQC provides a framework for hypothesis-driven research on the mechanisms underlying neurodegeneration.
Overview of the RQC pathway
Unlike co-translational ubiquitylation on elongation-competent ribosomes11,12, nascent chain proteolytic tagging in RQC takes place after stalled ribosomes have been sensed and split into the 40S and 60S subunits (the small and large ribosomal subunits, respectively)13,14 (Fig. 1, Box 1, Box 2). This so-called rescue of stalled ribosomes upstream of RQC has been recently reviewed4,15 and is not the focus of this article.
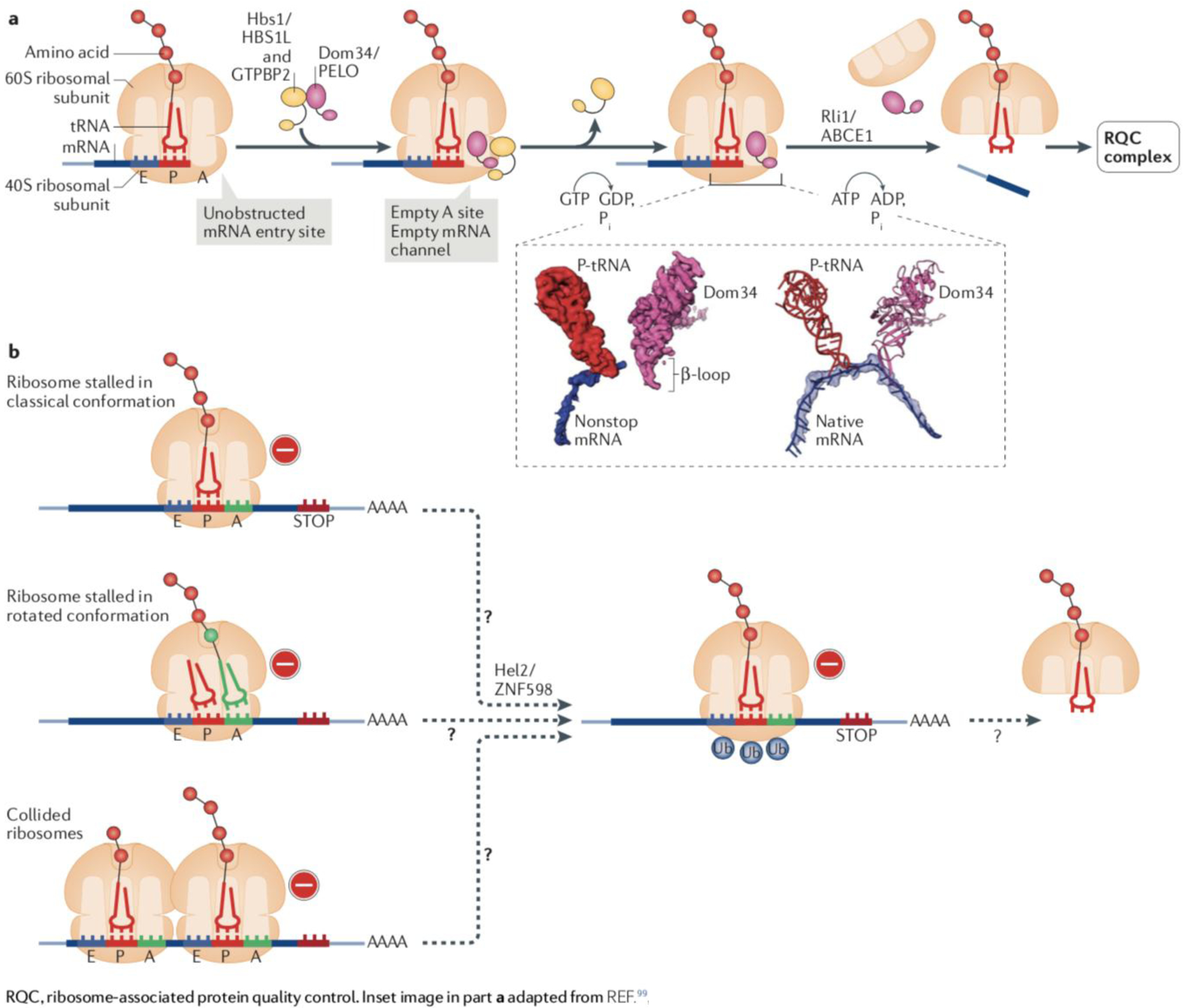
Box 2: Ribosomal stalling and rescue generate substrates for RQC.
In response to ribosomal stalling in eukaryotes, dedicated factors act to rescue and recycle the 40S and 60S ribosomal subunits. Ribosomes stalled at the 3’ end of truncated mRNAs are rescued by the concerted action of Dom34, Hbs1 and Rli1 in yeast, the mammalian orthologs of which are pelota (PELO), HBS1L and GTPBP2, and ABCE1, respectively) (see the figure, part a). The GTPase Hbs1 is a paralog of the canonical termination factor, eRF3, and Dom34 is a paralog of eRF1 (see Box 1).
The Dom34/PELO-mediated rescue reaction has been characterized at multiple levels and a recent cryo-EM structure of Dom34 bound to a ribosome stalled at the mRNA 3’ end has provided new insight into how the sensing of stalled ribosomes occurs100. The picture on the left portion of the inset (see the figure, part a) shows the segmented electron density maps for P-site tRNA (red), nonstop mRNA in (blue) and Dom34 on the ribosomal A site (pink) (adapted with permission from REF.100; ribosome not shown). Two independent events appear to detect an unobstructed mRNA entry channel in the ribosome as indicative of a ribosome being stalled at an mRNA 3’ end: the Hbs1 N-terminal domain binds to the ribosome in the vicinity of entry to the mRNA channel, whereas Dom34 binds to an empty ribosomal A site and inserts a β loop into the channel100. The picture on the right portion of the inset (see the figure, part b) shows the superposition of atomic models for P-tRNA and Dom34 with mRNA from human polysomes (native mRNA). It illustrates how the Dom34 β loop would clash with the mRNA if the latter were present.
Once Dom34/PELO is bound, Hbs1/HBS1L dissociates and, like eRF1, Dom34/PELO recruits the ATPase Rli1/ABCE1 to induce the splitting of the two ribosomal subunits. However, Dom34/PELO lacks peptidyl-tRNA hydrolase activity, leading to the release of a 60S subunit that remains obstructed with a nascent chain-tRNA conjugate.
In contrast to ribosome stalling at the mRNA 3’ end, how ribosomes that stall on internal mRNA sequences are sensed is not understood (see the figure, part b). The Zaher model proposes that the clashing of an elongating ribosome with a stalled one (collided ribosomes) produces a unique structural identifier of stalling80. An alternative model hypothesizes that stalled ribosomes may be recognized in a rotated conformation with tRNA in a hybrid state (see Box 1)101. At least in some cases, sensing of ribosomes stalled on internal mRNA sequences may be mediated by the E3 ubiquitin-protein ligase Hel2 (ZNF598 in mammals)97,101–104 (for reviews, see4,5,105). Hel2/ZNF598 catalyzes the site-specific mono- or oligo-ubiquitylation of 40S proteins in stalled ribosomes. This ubiquitin signal is not proteolytic, but rather is thought to induce the recruitment of factors required for 80S subunit splitting.
[Figure in part a adapted from ref. 15]
Box 2.
With the rescue reaction, released 40S ribosomal subunits can be recycled and the mRNA can be degraded by the 5’−3’ exoribonuclease Xrn1 and the exosome complex, preventing new rounds of translation of a transcript that is possibly aberrant, thus helping keep the levels of the encoded proteins low16,17 (Fig. 1)(see, e.g.,6,18). Another product of the rescue reaction is a 60S subunit that remains attached to an obstructing nascent chain-tRNA conjugate13,14,19 (Box 2). This aberrant 60S structure must therefore undergo further processing, and it is RQC that resolves the obstruction, while simultaneously marking the nascent chain for destruction6,9,10,20–22 (Fig. 1).
Obstructed 60S ribosomal subunits are sensed by the RQC complex subunit Rqc2 (also known as Tae2; NEMF in mammals) by binding to the 60S subunit simultaneously with the tRNA moiety of the peptidyl-tRNA that is exposed on the 60S20–22. Rqc2 then recruits Ltn1 and stabilizes the binding of Ltn1 to the 60S10,22.
Ltn1 catalyses the polymerization of poly-ubiquitin chains on the nascent polypeptide6. This ubiquitin signal recruits Cdc48 (VCP or p97 in mammals), which is an ATPase of the AAA family, and its cofactors9,10. Cdc48 recruitment also requires the Rqc1 subunit of the RQC complex, the exact function of which remains to be elucidated. Cdc48 extracts and delivers the nascent chains to the proteasome for degradation23 (Fig. 1).
Extraction by Cdc48 requires that the 60S-trapped nascent chain be released from the tRNA to which it is covalently linked (Fig. 1). The Vms1 protein (ANKZF1 in mammals) has recently been identified as a crucial factor performing this function in RQC24,25. Of note, Vms1 is a paralog of the eukaryotic translation termination factor eRF1, a peptidyl-tRNA hydrolase that releases nascent chains in canonical translation termination (Fig. 1).
Cells have built mechanisms that take over RQC when the canonical pathway does not function properly21,26–30. Notably, Rqc2 has another role in supporting Ltn1, when the nascent polypeptide sequence exposed outside the ribosome exit tunnel has no Lys residues accessible to ubiquitin modification. In this case, Rqc2 recruits charged tRNAs to extend the C-terminus of the nascent chain with Ala and Thr residues, which pushes the polypeptide out of the ribosomal exit tunnel, until a Lys residue that would otherwise be unavailable to ubiquitylation becomes exposed21,26. Moreover, in case ubiquitylation fails altogether, the C-terminal Ala and Thr tails (known as ‘CAT tails’) synthesized by Rqc2 may play a fail-safe role by mediating amyloid-like aggregation of the nascent chains28–30 (see below).
Substrate recognition and ubiquitylation
mRNAs lacking stop codons (nonstop mRNAs) encode proteins extended with polyLys, due to translation of the poly(A) tail (Box 1b). Such aberrant proteins are unstable and degraded by the proteasome31,32. At the time this phenomenon was initially described, the E3 ligase involved and the mechanism of substrate selectivity remained elusive, and contemporary paradigms suggested that the aberrant nascent chains might be released and then targeted by a cytosolic E3 ligase. However, those aberrant proteins were found to be marked for degradation by Ltn1 while still associated with the ribosome6. Ltn1 was found to be predominantly associated with the 60S ribosomal subunit and in the absence of Ltn1, nascent chains produced by ribosome stalling accumulated partially in ribosome-associated form6. These observations defined a novel pathway of protein surveillance taking place on ribosomes—now known as RQC—and have since been extended by structural biology, biochemical and molecular genetic studies.
Ltn1/Listerin is largely specialized in RQC.
E3 ligases operate in either cellular regulation or protein quality control, a functional distinction that can have implications at the mechanistic level, although some E3 ligases can perform both functions33–38(Box 3). A major difference between these two classes is that at least some regulatory E3 ligases have evolved to optimally modify a limited number of substrates, whereas quality control E3 ligases must be able to handle a wide range of substrates that can be highly heterogeneous with regard to sequence, structure and type of aberration, thus requiring mechanisms that enable broad substrate recognition and modification.
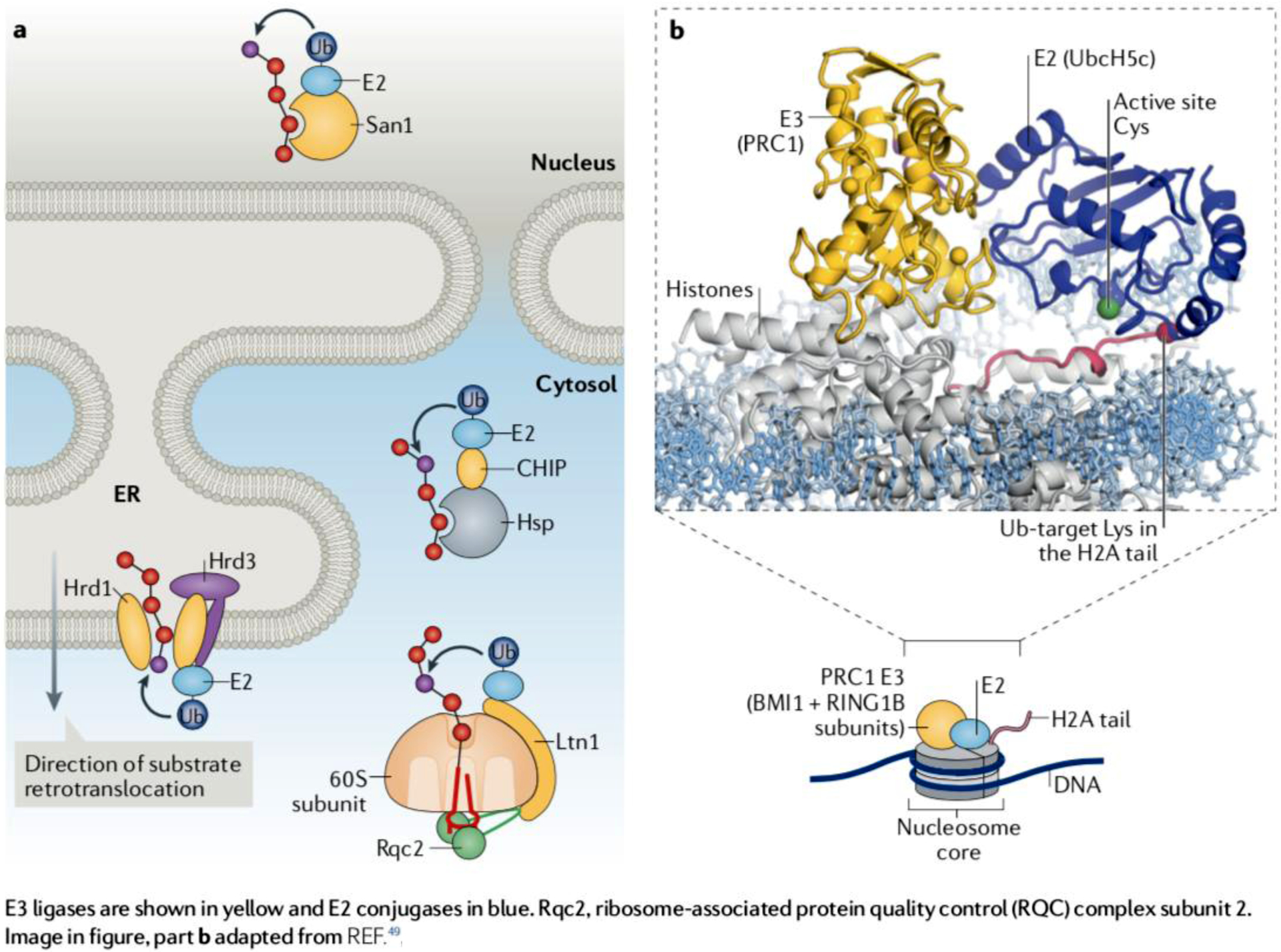
Box 3: Regulatory and protein quality control functions of E3 ligases.
E3 ligases are the components of ubiquitylation pathways that confer specificity in substrate targeting. Accordingly, the approximately 80 E3 ligases predicted to be encoded by the yeast genome and the over 650 E3 ligases encoded by the human genome are associated with a great variety of domains or motifs that bind specific substrates51,106.
In addition to substrate-binding domains, E3 ligases have a catalytic domain. 95% or so of human E3 ligases, like Ltn1/Listerin, depend on a RING domain for catalysis51 (the remaining E3 ligases have a HECT domain or operate using a HECT/RING domain hybrid mechanism55,107). The RING domain serves as binding site for E2 conjugating enzymes, whose active site Cys residue carries ubiquitin via a thioester bond. RING domains can also make direct contact with the E2-bound ubiquitin. Through these interactions, RING domain E3 ligases mediate the transfer of ubiquitin from the E2 to an acceptor residue of a target substrate, typically a Lys51,108.
E3 ligases can operate in either cellular regulation or quality control. Some have both functions, such as Huwe1, which promotes degradation of unassembled ribosomal proteins33,34 and functions in proliferation, apoptosis, DNA repair and stress responses by targeting specific substrates109 and Ubr1, which acts in cytosolic protein quality control35–38 and in the N-end rule pathway [G]110. Regulatory E3 ligases are thought to target a limited set of substrates, some of which are structurally related. c-Cbl is an example of an E3 ligase with regulatory function—it promotes the degradation of activated receptor and non-receptor tyrosine kinases thereby terminating signaling111. In contrast to regulatory E3 ligases, protein quality control E3 ligases must be able to target a great variety of unrelated substrates. For this purpose, the latter often utilize binding adapters, as illustrated in part a.
Another example of regulatory E3 ligase is the polycomb complex protein PRC1 (a heterodimer of RING1B and BMI1) which functions in epigenetic control. PRC1 binds to a nucleosome core particle and ubiquitylates the C-terminus of histone H2A. PRC1 functions with the E2 enzyme UbcH5c/UBE2D3. A structural study has provided remarkable evidence that the PRC1 RING domain acts by positioning the UbcH5c active site Cys85 right next to its specific target, Lys119 in the C-terminal tail of histone H2A49 (part b). Since regulatory E3 ligases like PRC1 act on a more limited range of substrates, they can more readily orient the reacting E2~ubiquitin and substrate acceptor residue to promote ubiquitin transfer; the main text discusses mechanisms that may allow the protein quality control E3 ligase Ltn1 to ubiquitylate diverse 60S subunit-bound substrates whose Lys residues are often improperly oriented with regard to the Ltn1-E2~ubiquitin complex.
E3 ligases are shown in yellow and E2 conjugases in blue.
Box 3.
Protein quality control E3 ligases must solve a difficult problem—the need for broad action is counteracted by the need for high sensitivity and selectivity, as their substrates can be present in low abundance (at least under normal growth conditions) relative to an excess of normal proteins. Perhaps as a result of this pressure, quality control E3 ligases have evolved some degree of specialization, which often relies on subcellular compartmentalization6,39–42. This is, e.g., the case of Ltn1, whose predominant association with the 60S subunit is linked to a specialized role in protein quality control at the ribosome6.
Ltn1/Listerin binds to the 60S subunit to recognize diverse substrates.
Mechanisms that generate aberrant proteins often lead to the exposure of hydrophobic patches that would normally be hidden inside the protein core and/or a higher-order protein complex. Sensing exposed hydrophobicity underlies the canonical mechanism of molecular chaperones, and E3 ligases can work with chaperones to target misfolded proteins. For example, the E3 ligase CHIP uses Hsp70 and Hsp90 as adaptors for substrate binding43–45 (Box 3). Alternatively, misfolded proteins can be recognized directly by E3 ligases; for example, the nuclear protein quality control E3 ligase San1 binds to substrates through its unstructured domains46 (Box 3). Ltn1/Listerin illustrates a third mechanism by which diverse aberrant proteins can be recognized by an E3 ligase, as revealed by the cryo-EM structures of RQC complexes (Fig. 2 and Box 3)—as predicted from molecular genetics and biochemistry data, Ltn1 binds to the ribosomal 60S subunit and uses it as an adaptor to target the variety of nascent chains that can be produced by translational stalling20. These observations were confirmed by reports of RQC complex structures solved at higher resolution and expanded towards the mammalian complex21,22.
Fig. 2: The function of Ltn1/Listerin in protein surveillance.
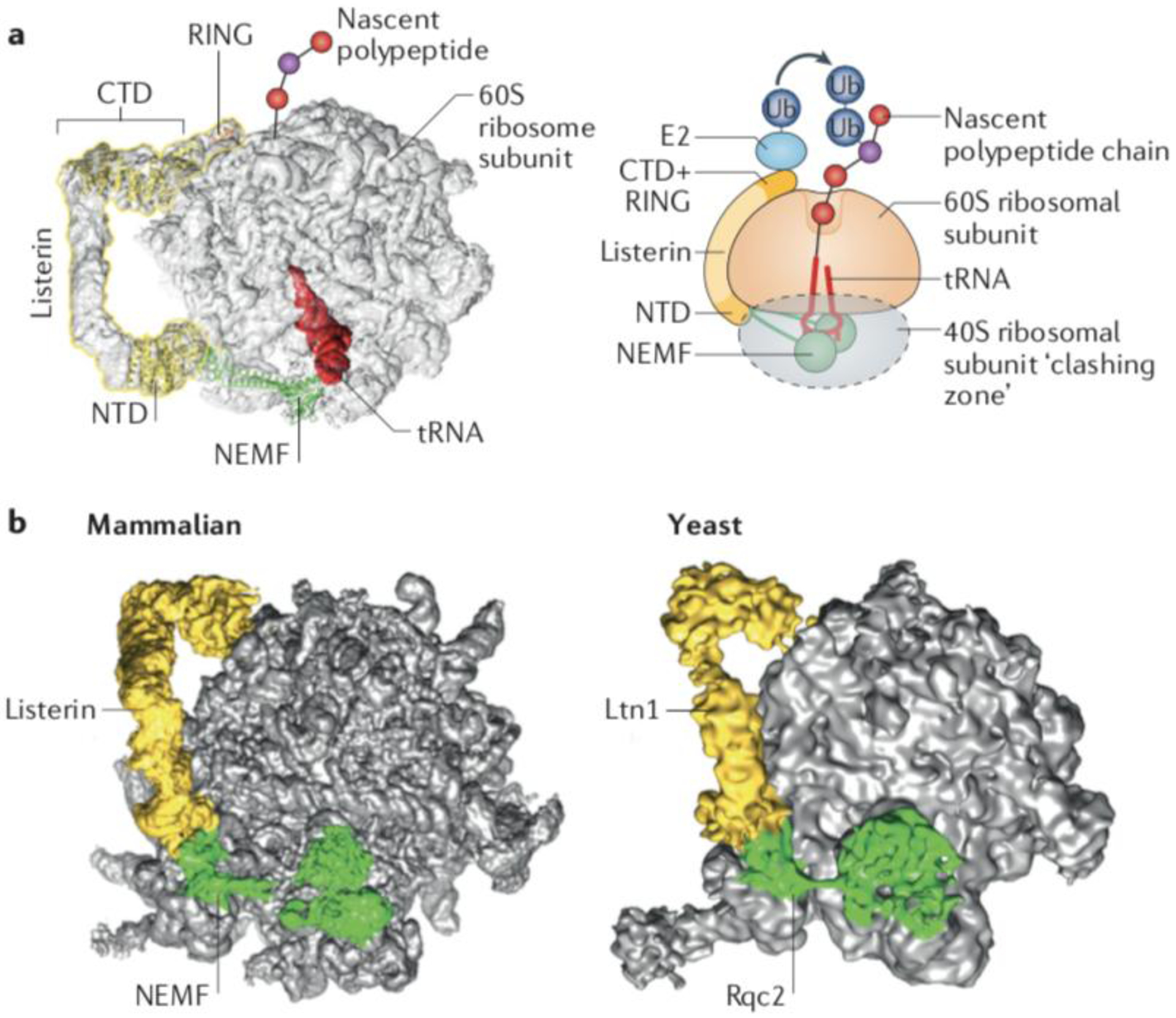
(a) Structural model of the ribosomal 60S subunit obstructed with peptidyl-tRNA and bound to the ribosomal quality control (RQC) complex subunits Listerin and NEMF. Superposition of cryo-EM volume of the RQC (EMDB 2832), molecular models of NEMF and the Listerin C-terminal Domain (CTD)22 (PDB 3J92), and the crystal structure of the Ltn1 N-terminal Domain (NTD)48 (PDB 5FG1). The cartoon on the right is a representation of the 60S subunit bond to the peptidyl-tRNA, Listerin/Ltn1 and NEMF/Rqc2. The conserved N-terminal and C-terminal domains of Ltn1, separated by a variable middle linker containing HEAT/ARM repeats are indicated. The C-terminal Ltn1 RING domain recruits E2~Ub to ubiquitylate nascent chains. The dotted grey oval indicates the space that would be occupied by the 40S subunit if present, to indicate steric clashes that would occur with Rqc2/NEMF and the Ltn1/Listerin NTD. (b) Comparison of the RQC structures from yeast (EMDB 2797)20 and mammals (EMDB 2832)22.
(Figures courtesy of H. Paternoga)
Ltn1 negatively selects against binding to normally translating ribosomes.
[Ltn1/Listerin homologs are large proteins (~150–190 kDa) conserved from yeast to humans. Sequence conservation is highest in the N-terminal domain (NTD) and C-terminal domain (CTD)6,47, which are connected via a middle linker region that has variable sequence but folds as a series of HEAT/ARM-type helical repeats in Ltn1/Listerin orthologs47 (Fig. 2). Cryo-EM and crystal structures revealed that the NTD and CTD bind directly to the 60S (Fig. 2). A subset of conserved residues in the Ltn1 NTD are part of an exposed patch48 that binds to the 60S subunit near the 40S-binding surface, whereas the CTD binds near the opening of the nascent polypeptide exit tunnel20–22. The NTD and CTD both contribute to 60S binding affinity, as suggested by the observation that mutations in either domain are detrimental to the Ltn1−60S interaction22,48 and to the function of Ltn1 in vivo48. The large size of Ltn1/Listerin and the presence of a linker region are consistent with Ltn1/Listerin functioning in the RQC complex as a monomer that has to simultaneously bind to two distal sites on the 60S subunit47 (Fig. 2).
The architecture of the Ltn1–60S complex provides the basis for a key mechanism imparting fidelity to protein surveillance. Several lines of evidence indicate that initial binding to 60S ribosomal subunits and substrate ubiquitylation by Ltn1 occur after the sensing of 80S ribosome stalling by rescue factors, and 40S and 60S ribosomal subunit separation13,14,19. The structural data are in agreement with this model as they show that Ltn1 and the 40S cannot simultaneously bind to the 60S subunit as the NTD would face extensive steric clashes with the 40S subunit if the latter were present20 (Fig. 2a). Moreover, analyses of free Ltn120,47 argue against the possibility of Ltn1 binding to 80S ribosomes even if Ltn1 was anchored to the 60S subunit via the CTD alone.
Thus, Ltn1/Listerin can select and broadly target diverse nascent chains obstructing the 60S subunit while sparing normal nascent chains in 80S translating ribosomes from becoming erroneously ubiquitylated20.
Mechanisms that enable modification of a wide variety of substrates
Solving of the structures of two regulatory RING E3 ligases (PRC1 and APC) bound to their cognate E2 ubiquitin-conjugating enzymes and substrates has shed light on the substrate modification reaction49,50. As expected of bi-substrate enzymes51, these E3 ligases have evolved to juxtapose the two reacting parts, the E2 active site Cys and the acceptor Lys in the substrate, with remarkable precision to facilitate ubiquitin transfer (Box 3). These observations raise the important question of how other E3 ligases, especially those implicated in protein surveillance, are able to modify a great diversity of substrates that are structurally unrelated and have distinct amino acid sequence and composition, and therefore have Lys residues potentially accessible for modification in different locations. The RQC system provides an example of strategies that enable tackling substrate diversity.
The CTD of Ltn1/Listerin, which includes the RING domain, binds to the 60S near the opening of the nascent chain exit tunnel. Thus, it is clear that Ltn1 positions E2~ubiquitin in the general proximity of its substrate nascent polypeptides20,22 (Fig. 2). However, further action needs to be taken to position the E2 active site Cys in close proximity to an acceptor residue in any of the enormous diversity of substrates that can become jammed on ribosomes. Potential mechanisms enabling such action are discussed here.
Although protein ubiquitylation most often occurs on Lys residues, it can also occur on other residues. The N-terminal methionine (Met1) can be modified by certain E3 ligases52,53 but it is unlikely that Ltn1 would adopt Met1 ubiquitylation as a general mechanism for substrate targeting, as the spatial location of the N-termini of polypeptides trapped in the 60S is diverse and sometimes inappropriately oriented relative to the RING domain. Moreover, in the case of 60S subunits that are blocked on organellar translocons (see below), the nascent chain N-terminus is entirely inaccessible. Alternatively, Ltn1/Listerin could modify different amino acid residues. Ubiquitylation by certain E3 ligases can occur on Cys, Ser, and Thr residues53–56, and E3 ligases that function in ERAD are thought to employ such relaxation strategy57–59. However, mutation of all Lys residues in RQC substrates markedly decreased their ubiquitylation and degradation, suggesting that Ltn1 preferentially modifies Lys26,28.
Structural flexibility can potentially facilitate access to substrate modification sites present in variable locations relative to E3 ligases. For example, members of the multi-subunit cullin-dependent E3 ligase [G] family use a variety of adaptors to target highly diverse substrates60. The RING domain is connected to the cullin subunit via a mobile linker that is unleashed through a regulated mechanism61. Furthermore, the adapter subunits can also be flexibly arranged within the architecture of the E3 ligase complex62. It is reasonable to hypothesize that structural flexibility is also important for E3 ligases involved in protein surveillance, and Ltn1 seems to be highly flexible. Ltn1 flexibility was suggested by the observation of >20 distinct conformations under negative staining and cryo-EM47 and by the poor resolution of Ltn1 in structures of the RQC complex (compared to that achieved for the 60S ribosomal subunit in the same complex)20–22. However, a requirement for flexibility in substrate modification by Ltn1 has not been demonstrated experimentally thus far.
Ltn1 has been proposed to preferentially modify Lys residues of model reporter substrates within a surprisingly short stretch of amino acids exposed immediately outside the exit tunnel26. Although it is unclear whether the same is true for endogeneous substrates, this is expected to be the case for nascent polypeptides traversing organellar translocons, as discussed below. Recent studies have uncovered a mechanism, mediated by Rqc2, that is thought to address this problem: the synthesis of CAT tails pushes out Lys residues that would otherwise be buried in the exit tunnel in a presumably stepwise manner, until one such residue is rendered accessible to ubiquitin transfer26 (discussed below). It is tempting to speculate that other E3 ligases, particularly those that function in protein quality control and target substrates in an extended conformation (such as proteins during retrotranslocation from an organelle), may likewise scan the polypeptide sequence linearly—and not only three-dimensionally—for a modification site.
Sensing of obstructed 60S subunits
The Ltn1/Listerin cofactor Rqc2/NEMF plays a crucial and more complex part in RQC than initially anticipated.
The cryo-EM structure of the endogenous yeast RQC complex indicated that Rqc2 directly senses obstructed 60S subunits20. Rqc2 recognizes aberrant proteins in a non-canonical manner as it does not bind them directly. Rqc2 binds to ribosomal proteins and rRNA on the intersubunit surface of the 60S, while simultaneously binding to the tRNA moiety of the peptidyl-tRNA residing on the 60S subunit P site. The structure thus uncovered an elegant way by which cells can detect abnormalities in macromolecules—a binary binding code that detects the juxtaposition of components (in this case a tRNA and a free 60S subunit) that should not be present together20. Higher resolution structures of yeast and mammalian RQC have confirmed and provided further detail to these observations21,22 (Fig. 2).
Rqc2, like Ltn1, binds at the inter-subunit surface of the 60S in a mutually exclusive manner with the 40S subunit (Fig. 2a, as discussed above). Consequently, Rqc2 cannot bind to a translating 80S ribosome, which confers selective degradation of substrates associated with the 60S subunit. Moreover, Rqc2 prevents dissociated 40S subunits from re-associating with obstructed 60S subunits that are generated from rescue of stalled ribosomes9,10,20–22 (Fig. 1). That is because Rqc2 is more abundant than free 40S subunits and can thus better compete for binding to the 60S subunit20,63; this is important as 40S reassociation with obstructed 60S subunits would prevent the binding of Ltn1, which is much less abundant.
Rqc2 also binds directly to Ltn120–22 (Fig. 2). The interaction is mediated by the Ltn1 NTD and the Rqc2 middle (M) domains but the exact interaction surfaces differ between yeast and mammals48. Consistent with the structural data, Rqc2 stabilized Ltn1 binding to the 60S in yeast10 and biochemical reconstitution of mammalian RQC assembly supported the notion that NEMF facilitates Listerin association with the complex22,27.
The relative abundance of RQC complex components, the ability of Rqc2/NEMF to sense obstructed 60S subunits, and its function of stabilizing and/or facilitating Ltn1/Listerin−60S interaction suggest that Rqc2/NEMF is the first RQC complex subunit to bind to the 60S. However, in the absence of Rqc2, Ltn1 is able to bind to 60S subunits and promote substrate degradation, albeit less efficiently9,10,20–22,28–30,64.
CAT tail formation and functions
The C terminus of tRNA-conjugated nascent polypeptides obstructing 60S subunits can be modified (extended) with Ala and Thr residues in a process that is independent of the mRNA and the 40S subunit, known as CATylation21. CAT tails facilitate the Ltn1/Listerin-dependent degradation of aberrant proteins but can also induce protein aggregation and stress responses (Fig. 3).
Fig. 3: CAT tail synthesis and functions.
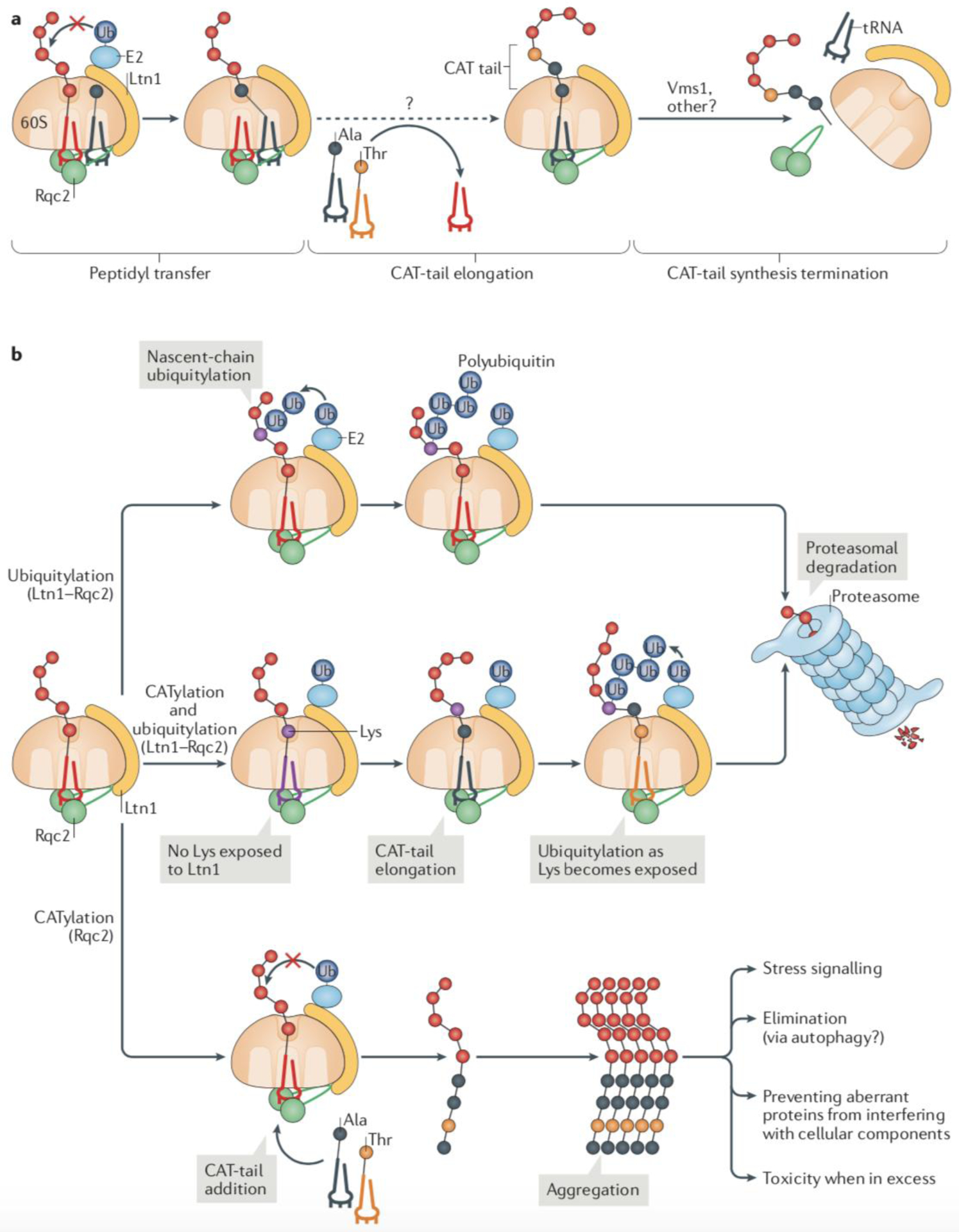
(a) The ribosome-associated protein quality control (RQC) complex subunit Rqc2 recognizes ribosomal 60S subunits that are obstructed by a peptidyl-tRNA by simultaneously binding to components of the 60S subunit and the tRNA. When ubiquitylation of the nascent polypeptide by Ltn1 fails, Rqc2 can extend the trapped nascent polypeptide with a C-terminal tail composed of Ala and Thr residues (CAT tail). The reaction is mediated by the direct recruitment of charged tRNA-Ala and tRNA-Thr by Rqc2 and occurs without an mRNA template. The Vms1 protein can terminate CAT tail synthesis by releasing the P-site tRNA and presumably promoting Rqc2 dissociation from the complex. (b) Alternative fates of RQC substrates. The canonical RQC pathway of Ltn1-mediated ubiquitylation of nascent polypeptides is kinetically preferred, provided that Lys ubiquitylation sites on nascent chains are readily accessible (top). In this pathway, Rqc2 functions in recruiting and stabilizing Ltn1 in the complex. An alternative pathway takes place when ubiquitylation is compromised (middle). In this pathway, Rqc2 catalyses the elongation of a CAT tail, which can result in the exposure of Lys residues that would otherwise be hidden in the ribosomal exit tunnel. The increased accessibility to Lys residues enables ubiquitylation by Ltn1. However, when ubiquitylation fails altogether (bottom), CAT tail-modified nascent chains form aggregates, which can have different fates and effects in cellular function, including stress signalling.
Rqc2 modifies 60S-associated nascent chains with CAT tails.
CATylation is mediated by Rqc221 (Fig. 3). Because the CATylation reaction occurs without an mRNA template, the Ala- and Thr-containing tails that it produces are of varying length and heterogeneous in sequence and in length21,28–30. CAT tail formation has been observed in response to different stalling signals, such as polyLys synthesis, mRNA truncation, or Arg CGN codons28 (see, e.g., Box 1b). It remains to be determined whether nascent polyLys peptides encoded by prematurely polyadenylated mRNA slow down the rates of CAT tail elongation, as they are thought to engage in strong electrostatic interactions with the ribosomal exit tunnel65.
Mechanisms underlying CATylation have only begun to be elucidated (Fig. 3a). While binding to the nascent chain-linked tRNA that is in the P site of the 60S subunit, Rqc2 simultaneously recruits a second tRNA molecule (tRNA-Ala or tRNA-Thr) to the A site and positions the two tRNAs close to each other to facilitate peptidyl transfer21. Accordingly, Rqc2 mutations that interfere with A-site tRNA binding, but do not affect Rqc2 binding to the 60S or Ltn1, led to a selective defect in CATylation21,28–30.
CAT tail elongation seems to be independent of the canonical eukaryotic translation elongation factors eEF1A and eEF2, as a non-hydrolyzable GTP analog did not block CATylation in an in vitro reconstitution of the reaction27. Whether any cofactors are required for elongation and whether repeated cycles of Rqc2 association and dissociation underlie chain elongation remains unknown.
Recent findings have shed some light on the termination of CAT tail synthesis24,25. These studies suggest that the Vms1 protein releases nascent chains from their linked tRNA, thereby preventing further tail elongation and presumably decreasing the binding affinity of Rqc2 towards the RQC complex. This model is consistent with the report that Vms1 antagonizes the formation of CAT tail-dependent protein aggregates (see below), and stimulates Rqc2 dissociation from the complex66.
Consequences of nascent chain CATylation.
Different functions for CAT tails have been proposed (Fig. 3)
Nascent chain aggregation.
CATylated polypeptides form stable protein aggregates and cellular inclusions that share properties with amyloids and yeast prions28–30 (Fig. 3b). Reporter constructs encoding a GFP fusion with C-terminal Ala-Thr repeats led to GFP aggregation independent of ribosome stalling, suggesting that CAT tails play a direct and sufficient role in the process28. Notably, engineered tails containing both Ala and Thr were found to have greater propensity for aggregation than tails made of only Ala or Thr28, raising the question of why the amyloid-forming Ala and Thr combination has not been selected against. One possibility is that aggregation may be an intended fail-safe function of CAT tails. Although protein aggregation is usually associated with cellular toxicity, one can speculate on the potential benefits of diverting aberrant nascent chains that fail to be ubiquitylated by Ltn1 towards aggregation. The sequestration of aberrant proteins in confined compartments, such as the aggresome [G], would reduce the likelihood of disrupting cellular functions67,68. Furthermore, the aggregation of aberrant proteins could provide alternative pathways for elimination, via proteasomal degradation, loss owing to asymmetric distribution during cell division, or autophagy (see, e.g.,30,69,70). Lastly, aggregates may elicit adaptive stress signalling, which is indeed known to take place in response to Ltn1 defects9.
Activation of stress signalling.
A universal response to perturbations in protein homeostasis is the activation of stress signaling pathways, such as that orchestrated by heat shock factor 1 (HSF1)71. In yeast, Hsf1 is normally kept inactive by interaction with chaperones. Under conditions of proteotoxic stress, chaperones are diverted to handling misfolded proteins, releasing Hsf1 and allowing this factor to activate the expression of stress genes, in particular genes encoding chaperones themselves71. Chaperone induction prevents further aggregation of misfolded proteins72,73. Notably, Hsf1 is activated in response to defective Ltn1 function and RQC being compromised9. Furthermore, under these conditions Hsf1 activation is dependent on Rqc29, suggesting that it is mediated by the formation of CAT tail aggregates (Fig. 3b), which sequester chaperones28–30.
Nascent chain proteolysis.
CAT tails can promote proteolysis by facilitating Ltn1-mediated ubiquitylation26 (Fig. 3b). Such a role is consistent with a partitioning model of ubiquitylation versus CATylation in RQC (Fig. 3b). CATylation of stalling reporters is more conspicuous in the absence of either Ltn1 or Rqc121,28–30 or in wild type cells expressing a reporter polypeptide lacking Lys residues28. A possible explanation for these observations is that nascent chain ubiquitylation and extraction is kinetically preferred over CATylation, but if acceptor Lys residues to be modified by Ltn1 cannot be readily targeted, Rqc2 is allowed time to add Ala and Thr residues to the C-terminal end of the nascent polypeptide. Given that the ribosomal tunnel covers ~35 amino acids of a nascent polypeptide74 and that CAT tails formed in the absence of Ltn1 can have as many as ~60 residues added (judged on the size of the smears formed in SDS-PAGE), repeated rounds of Ala and Thr addition would eventually expose any Lys residues buried in the tunnel to the outside (Fig. 3b). This strategy would certainly be effective for ribosomes stalled in the poly(A) tails of prematurely polyadenylated mRNA4,5 (Box 1b), as poly(A) encodes polyLys. Furthermore, computational analyses predict that CATylation would increase the fraction of RQC substrates that can be targeted by Ltn1 if ribosome stalling occurred with equal probability at each codon on the yeast transcriptome26.
The role of Rqc2-mediated CAT tail synthesis in exposing Lys residues for ubiquitylation may be especially relevant when ribosomes stall during co-translational protein import into organelles. In this case, the nascent polypeptide chain that that exits the ribosomal tunnel becomes quickly inaccessible to Ltn1 because it enters the translocon [G] on the organelle membrane and is then transferred to the organelle interior (where there is no Ltn1 or other components of the ubiquitin system) (Fig. 4). Therefore, the window of opportunity for Ltn1-dependent ubiquitylation is probably smaller in comparison to RQC on 60S subunits that are free in the cytosol.
Figure 4: RQC on the endoplasmic reticulum and mitochondrial membranes.
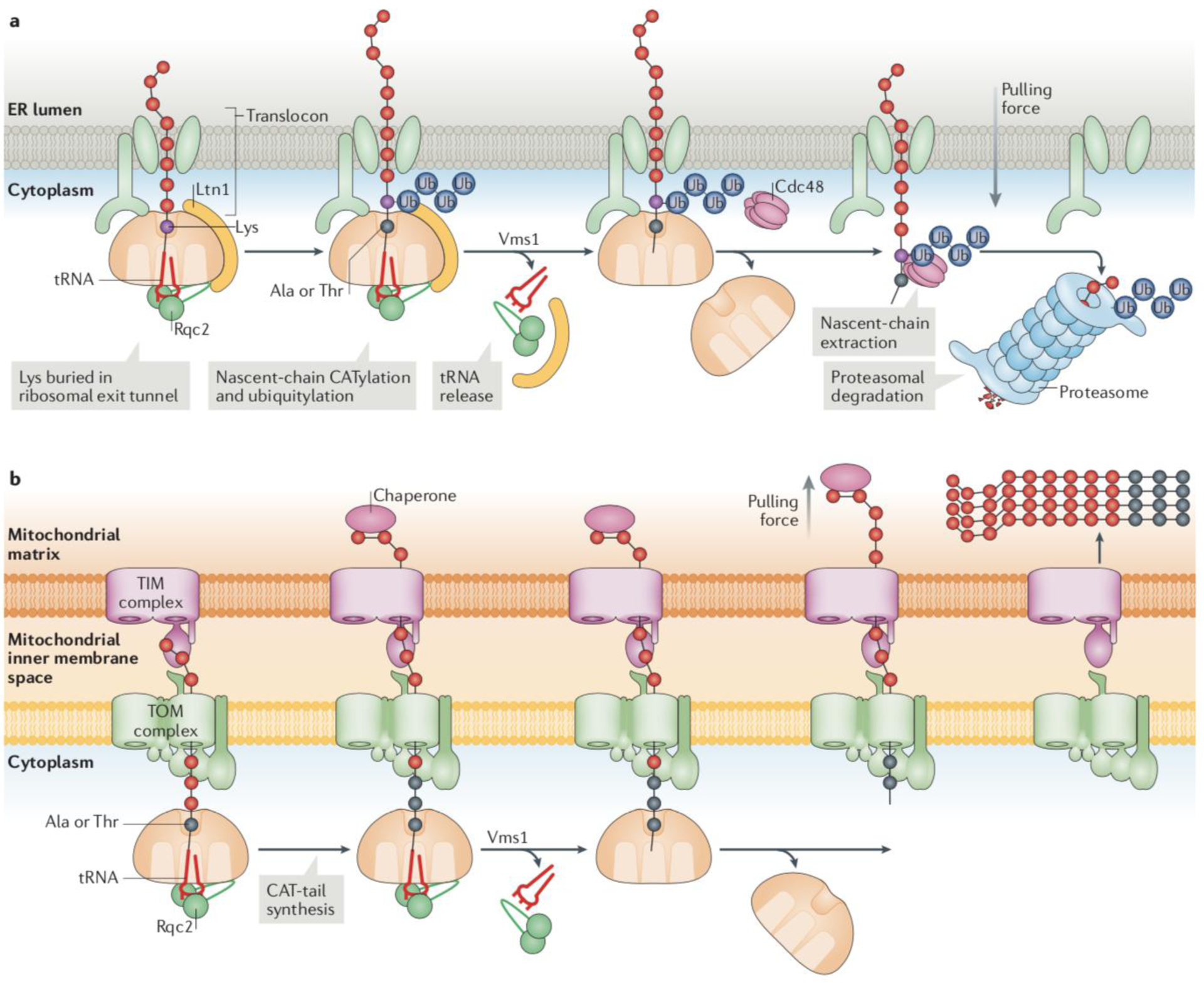
Ribosomes can stall while translating proteins destined to different subcellular compartments, such as the cytosol, mitochondria and the endoplasmic reticulum (ER). (a) ER-RQC: hypothetical model for RQC at ER-associated ribosomes. On nascent chains obstructing the 60S ribosomal subunit on organellar surfaces, Ltn1 only has limited access to Lys residues, within a short segment of cytosolic-exposed polypeptide sequence immediately outside the exit tunnel. The nascent chain segments buried in the ribosomal exit tunnel or the translocon, or inside the organelle, are not accessible to ubiquitylation. However, Lys residues hidden in the ribosomal exit tunnel can be exposed to Ltn1 through Rqc2-mediated nascent chain CATylation. Cdc48 is recruited to extract the ubiquitylated nascent chain and deliver it to the proteasome for degradation in the cytosol, after Vms1 catalyses nascent chain release from the tRNA. (b) Mito-RQC: Example of Ltn1 dysfunction on the mitochondrial surface. In the absence of nascent chain ubiquitylation, nascent chains are CATylated by Rqc2 and released into the mitochondrial matrix by Vms1. The directional pulling force by a mitochondrial chaperone (in pink) is indicated. In the matrix, moderate levels of CAT tail-dependent aggregates can be handled by the protein homeostasis machinery. However, if both Ltn1- and Vms1-mediated processes fail, CATylation proceeds for an extended period, resulting in the accumulation of larger aggregates which mitochondria may have more difficulty in eliminating, and which can interfere with the function of the organelle66.
In conclusion, the synthesis of CAT tails during RQC uncovers a fail-safe strategy that may both facilitate E3 ligase function during protein surveillance and promote alternative mechanisms that handle and dispose of aberrant proteins when the E3 ligase function is compromised (Fig. 3b). The CAT tail studies also highlight that protein quality control E3 ligases have evolved alternative solutions to modify heterogeneous substrates— whereas some E3 ligases seem to have adopted a relaxed amino acid specificity approach, Ltn1 appears to utilize a cofactor (Rqc2) to scan a polypeptide sequence one-dimentionally and unidirectionally for the presence of Lys residues.
The termination phase of RQC
The ubiquitin chains that are linked to nascent polypeptides obstructing ribosomal 60S subunits function as a signal for protein extraction from the 60S subunit both for 60S recycling for nascent polypeptide proteasomal degradation.
Vms1 releases nascent chains from linked tRNA.
The bulky tRNA that is linked to nascent chains on the opposite end of the ribosomal exit tunnel relative to the ubiquitin signal poses an impediment to nascent chain extraction. Recent studies have identified Vms1/ANKZF1 as a specific factor that functions in the release of the nascent chain from tRNA24,25 (Figs. 1 and 3a).
Vms1 interacts with the 60S subunit and RQC complex components10,24,25,66. Although Vms1 had been previously characterized as a Cdc48 cofactor, the role of this association in RQC is unclear and the Vms1 motif that binds to Cdc48 has been reported as not essential for its function in RQC in one study66.
Vms1 is conserved in eukaryotes (like other RQC components) and was found to be a paralog of eRF1 and Dom3424. Conservation includes a ribonuclease H [G] fold and a glutamine residue (Gln295) at a position comparable to that of the eRF1 catalytic Gln necessary for peptidyl-tRNA hydrolysis24,25. As predicted by these observations, both Vms1 and ANKZF1 were able to promote release of nascent chains obstructing 60S subunits in vitro, in a Gln295-dependent manner24,25. Furthermore, loss of Vms1 function or mutation of Gln295 led to the accumulation of ubiquitylated peptidyl-tRNA associated with the 60S subunit in vivo, and mutated Vms1 could not rescue growth defects caused by ribosome stalling or Ltn1 dysfunction24. Despite these relationships between eRF1 and Vms1, more recent work has suggested that, rather than functioning as a bona fide peptidyl-tRNA hydrolase, ANKZF1 acts as an endonuclease and releases the nascent chain still linked to a few nucleotides from the tRNA 3’ end75. Further studies will be required to clarify the precise mechanism of Vms1/ANKZF1 in RQC.
Thus, in contrast with canonical translation termination, where eRF1 mediates ribosome subunit splitting and nascent chain release, in RQC these functions have been uncoupled such that Dom34 promotes the splitting but nascent chains remain anchored to the 60S subunit via the tRNA until Vms1 releases them. It remains to be established whether and how the activities of Ltn1 and Vms1 are coordinated to prevent premature release of non-ubiquitylated nascent chains--while in vitro studies indicate that peptidyl-tRNA susceptibility to human ANKZF1 can be stimulated by NEMF, Listerin and ubiquitylation75, results of yeast molecular genetic analyses suggest that Ltn1 binding or nascent chain ubiquitylation are not essential for Vms1 to release nascent chains24,25,66.
It is also reasonable to speculate that the release of nascent chain from tRNA plays a part in the disassembly of the RQC complex as, although the complex is stable in vitro (C.A.P.J. unpublished observations), it must be rapidly disassembled in vivo given that there are only very few Ltn1 molecules in a cell6. By releasing the tRNA from its conjugated polypeptide, Vms1 might induce the dissociation of Rqc2 and the entire complex from the 60S subunit (see also76).
Cdc48/p97 extracts and delivers nascent chains to the proteasome.
Cdc48 (p97 or VCP in mammals) is a AAA ATPase that functions as a segregase in various ubiquitin-dependent pathways77. Cdc48 also delivers polypeptides extracted from complexes to the proteasome for degradation, which may be important to prevent their aggregation or unwanted interactions with other cellular components.
Cdc48 and its cofactors, Ufd1 and Npl4 (UFD1 and NPLOC4 in mammals), are recruited to RQC complexes in a manner that is dependent on Rqc1 and on Ltn1/Listerin-dependent ubiquitylation9,10,23(Fig. 1). Loss of Cdc48 function resulted in increased amount of RQC substrates remaining trapped in 60S subunits10,23,78. Thus, Cdc48 contributes to unobstructing 60S subunits for recycling and facilitates proteasomal degradation of the nascent chain.
Nascent chains can remain associated with RQC complex subunits following extraction by Cdc4876. These complexes, which have been called ‘light RQC’, have been proposed to function in preventing aggregation and/or facilitating proteasomal delivery of the aberrant nascent chains (Fig. 1)76.
The function of Rqc1 in the RQC complex remains poorly understood.
The Rqc1/TCF25 subunit is conserved among eukaryotes, suggesting it has an important function. Deletion of the RQC1 gene in yeast leads to accumulation of reporter substrates that become CATylated, indicating that loss of Rqc1 causes a defect in some step before or during nascent chain extraction from the 60S28–30. RQC1 deletion did not affect either the binding of Ltn1 to the RQC complex or the extent of ubiquitylation of RQC substrates. It was thus suggested that Rqc1 is required, together with the ubiquitin chains placed by Ltn1, for the recruitment of Cdc48 to the complex9,10,76 (Fig. 1). Recent biochemical analyses provide a potential mechanistic basis for this observation: ubiquitin chains linked via Lys48 of ubiquitin—which are selectively bound by Cdc48-Ufd1-Npl4 (e.g.,79)—were preferentially formed over other types of chain in presence of TCF2575. However, whether this is also true in vivo remains to be shown.
In contrast, another study found that overall nascent chain ubiquitylation levels were greatly reduced when the yeast RQC system was reconstituted in vitro using extracts of Rqc1-deficient yeast, and that this defect could be reversed by addition of recombinant Rqc1 to the extracts27. Based on these observations it was concluded that Rqc1 is required for Ltn1-mediated ubiquitylation, although alternative explanations remain possible. Thus, the role of Rqc1 in the RQC complex is controversial, and structural analyses have so far failed to provide insights on its function or mechanism of action20,21.
RQC protects organellar function
Maintaining the structure and function of organelles is key to cellular homeostasis and requires control of the quality of proteins that transit through or reside in those subcellular compartments. An important quality control step occurs during cotranslational protein import in to the organelles, via the RQC pathway.
Studies of ribosomal stalling and Ltn1-mediated protein ubiquitylation and degradation have largely used reporters encoding cytosolic proteins; however, many proteins are targeted to organelles cotranslationally and ribosome stalling can occur during this process (Fig. 4a). For example, ribosomes could stall on the ER surface while translating aberrant (e.g., prematurely polyadenylated) mRNA encoding proteins in the secretory pathway. On the surface of mitochondria, ribosome stalling could occur as a result of mRNA or ribosomal damage caused by reactive oxygen species produced by mitochondria80 (note that mitochondrial ribosomes that become stalled during translation within the organelle are handled by a distinct mechanism81). Aberrant proteins that are produced as a result of ribosome stalling during cotranslational import into organelles must be eliminated. Indeed, nonstop reporters targeted to the ER or mitochondria are unstable, and, in the absence of the Dom34 or Hbs1 rescue factors, they block the translocons (Sec61 in the ER and translocase of the outer membrane (TOM) complexes in mitochondria)82. Importantly, this blockade results in general impairment of protein import and growth defects82. Thus, ribosomal rescue protects the structure and function of organelles, which depend largely on import of nuclear-encoded proteins synthesized on their cytosolic side.
Like ribosomal rescue, the RQC pathway has been shown to function both on the ER83–85 (Fig 4b) and on mitochondria66 (Fig 4c). In the current model, ribosome stalling during cotranslational import prevents further translocation of the nascent chain. Ribosomal rescue then releases the mRNA and 40S subunits, allowing the RQC complex to assemble. Finally, Ltn1-mediated ubiquitylation promotes reverse translocation—probably via Cdc48-dependent nascent chain extraction—so that the nascent chain can be degraded in the cytosol by the proteasome.
ER-RQC.
Biochemical studies indicate that active mammalian RQC complexes can be found associated with the ER membrane, and the recruitment of RQC components to those membranes is stimulated by translational stalling85. In both yeast and mammalian cells, model substrates with diverse topologies, encoding soluble or transmembrane ER proteins, were successfully targeted for degradation by Ltn1 in a translational stalling-dependent manner83–85. Interestingly, ERAD E3 ligases apparently cannot substitute for Ltn1 function to induce the degradation of ER-stalled RQC substrates84, which illustrates the specificity of protein quality control pathways.
Structural modeling suggests that there is a gap between the 60S subunit and the translocon that is sufficiently large to allow access of the Ltn1 C-terminus to its canonical binding site on the 60S subunit, thus enabling nascent chain ubiquitylation85. Of note, the short length of nascent polypeptide sequence exposed through this gap limits the availability of Lys residues for ubiquitylation26. Although the involvement of Rqc2/NEMF and CAT tails in ER-RQC has yet to be examined, this observation provides a plausible rationale underlying a role for CATylation in exposing buried Lys residues to Ltn1 (Fig 4b).
Mito-RQC.
A link between RQC and mitochondria has been recently established. Notably, RQC was found to be required for yeast growth under conditions that promote oxidative phosphorylation metabolism66. As for RQC involving cytosolic proteins, nonstop proteins targeted to the mitochondria were degraded in a Ltn1-dependent manner, and in the absence of Ltn1, reporter polypeptides were modified with CAT tails in an Rqc2-dependent manner66. Vms1 was also implicated in the metabolism of mitochondrial nonstop proteins66. Vms1 may help unclog the 60S and the TOM complex through its ability to cleave tRNA off the 60S-trapped nascent chain24,25 (Fig. 4c).
Cellular consequences of defects in RQC
Even in the absence of any exogenously applied stress, the yeast RQC factors are found bound to the 60S and active, suggesting that the RQC pathway is constitutively active and receiving a continuous flux of substrates6,9,10,78. This is consistent with known causes of ribosome stalling upstream of RQC, such as constitutive errors in gene expression leading to premature mRNA polyadenylation. However, Ltn1 is not essential for yeast cell growth in the absence of stress. It is possible that other fail-safe mechanisms—both at the level of RQC and other machineries—can handle substrates in the absence of Ltn1 function on non-stressed conditions. Ltn1 becomes essential for growth when the amounts of substrate to be degraded increases6, or when other factors that restrain the accumulation of aberrant mRNAs or proteins become limiting10. Different mechanisms that may contribute to the growth defects observed under these conditions are discussed below.
Ribosome sequestration.
Failure to resolve obstructed 60S ribosomal subunits, leading to their sequestration in an inactive form, can potentially decrease cellular translational efficiency. This is better exemplified by studies of ribosome stalling in bacteria—in many situations, the critical function of the tmRNA/ssrA system [G] appears to be ribosomal rescue rather than proteolytic targeting86.
Accumulation of aberrant proteins and protein aggregates.
Polypeptides produced by ribosome stalling are often misfolded and can be extended with polyLys if encoded by a prematurely polyadenylated mRNA. Misfolded proteins in general, and polylysine extensions in specific, have the potential to interact with different factors or cellular components and thus interfere with cellular functions (e.g.,87). Moreover, misfolded proteins can exhibit toxic gain-of-function through formation of different types of aggregates (e.g.,1,2).
As discussed above, polypeptides produced by ribosome stalling can also be modified with CAT tails, and CAT tail-mediated aggregation may serve as a protective mechanism when those polypeptides fail to be marked with ubiquitin for degradation. However, result show that if produced in excess, CAT tail aggregates can be toxic, possibly owing to sequestration of components of the chaperone system. This has been observed, for example, following inactivation of Ltn1 alone or both Ltn1 and Vms1 by gene deletion28–30,66,88,89. Under these conditions, the growth defect could be suppressed by deletion of the RQC2 gene28–30,66.
Notably, in cells deficient for both Ltn1 and Vms1, overexpression of a nonstop reporter for mitochondrial RQC (mito-RQC; Fig. 4c) caused a more severe growth phenotype in comparison to a nonstop cytosolic reporter66; furthermore, Rqc2-dependent endogenous aggregates isolated in the absence of reporter constructs were mostly composed of mitochondrial proteins. Aggregates were also found to sequester components of the mitochondrial protein homeostasis machinery, which correlated with the collapse of mitochondrial activities66. These results suggest that mitochondrial dysfunction due to CAT tail-mediated aggregation may be a major cause of yeast growth defects in response to loss of RQC activity and raise the question why mitochondria seem to be especially sensitive to loss of Ltn1 function. One possibility is that mitochondria are not able to clear CAT tail-dependent aggregates as efficiently as the cytosol, although earlier studies with polyGln-mediated aggregation suggested the contrary90.
Organellar protein import defects.
In comparison to stalling of ribosomes producing cytosolic proteins, stalling of ribosomes on the surfaces of ER and mitochondria (Fig. 4) has at least one additional detrimental consequence. 60S subunits obstructed with nascent chain-tRNA can block the organellar import channels (translocons)66,83 and thus have the potential to impair general protein import to organelles and cause organellar dysfunction. The extent to which the toxicity resulting from ribosome stalling in organellar surfaces83,84 is due to the blockade of translocons, the aggregation of proteins within organelles, or both, requires further investigation.
Listerin mutation causes neurodegeneration
Defects in protein surveillance leading to imbalance in protein homeostasis and to the accumulation of toxic proteins are associated with various types of cellular and tissue dysfunction in metazoans, collectively referred to as proteinopathies (e.g.,91). In particular, defects in protein quality control are a hallmark of neurodegeneration, and studies of Listerin support this notion.
Whereas complete loss of Listerin function impairs mouse embryonic development, a hypomorphic mutation [G] in the Listerin non-conserved middle region (listerENU mice) allows embryonic development to proceed but later gives rise to a neurodegenerative phenotype8. Notably, a mutation in mouse GTPBP2 (a paralog of Hbs1), which functions in ribosomal rescue upstream of RQC, also gives rise to neurodegeneration92.
Simple model systems such as the yeast RQC have greatly contributed to our understanding of the molecular mechanisms of RQC and the cellular consequences of RQC dysfunction, providing insights in the possible causes underlying neurodegeneration in mammals. For example, results from those studies raise the possibility that the accumulation and/or aggregation of aberrant RQC substrates, ribosomal subunit sequestration and defective organellar protein import could all contribute to the pathophysiology of neurodegeneration. In support of this hypothesis, dysfunction of ER and mitochondria—much like defects in protein homeostasis—are known hallmarks of neurodegeneration93,94. Moreover, mutations directly affecting the function of the mitochondrial import machinery have been linked to progressive neurodegenerative syndromes95. Therefore, studying organellar dysfunction due to defective protein import, accumulation of protein aggregates, or both, will be of particular interest in the context of listerENU mice.
Conclusions and future perspectives
Our understanding of the mechanisms and functions of the RQC pathway has substantially increased in the last 8 years, since the initial report on Ltn1/Listerin function. Together, the studies highlight the complexity of RQC, and how knowledge on Ltn1/Listerin can provide new insights on general principles of E3 ligase-mediated protein surveillance (Table 1). Yet, much needs to be learned about fundamental as well as disease-related aspects of RQC. Box 4 discusses mechanistic and functional or physiological aspects of the RQC requiring further investigation.
Table 1:
Key features of RQC-mediated protein quality control
| General element of protein quality control | Corresponding characteristic for Ltn1/Listerin and RQC |
|---|---|
| Source of substrate | Ribosomal stalling; can occur without exogenous stress |
| E3 ligase localization | Mostly 60S-associated in steady state (yeast); free in the cytoplasm but 60S-bound with increased ribosome stalling (mammalian cells) |
| Nature of protein aberration | Any nascent chain trapped on the 60S subunit should in principle be a substrate of RQC, regardless of being aberrant. However, those proteins are generally expected to be misfolded due to truncation or extension. They can also be functionally defective without exposing hydrophobicity (e.g., having dominant-negative activity due to truncation) |
| Mechanism for broad substrate recognition | Ltn1 uses the 60S subunit as adapter to target 60S-trapped nascent chains |
| Mechanism for substrate specificity | Ltn1 is unable to bind to translating 80S ribosomes due to the presence of the 40S subunit |
| Ubiquitylation | Ltn1 targets Lys residues; it sometimes requires CAT tail synthesis by the Rqc2 cofactor, which enables scanning the polypeptide linearly for a modification site |
| Fate of ubiquitylated proteins | Proteasomal degradation |
| Fail-safe mechanism | CAT tail-dependent nascent chain aggregation (?); other? |
| Stress signaling | CAT tail-dependent Hsf1 activation |
| Loss of function phenotypes | Sensitivity to increased substrate production (yeast), defective growth when the need for oxidative phosphorylation is increased (yeast), embryonic lethality (Listerin knockout or RING domain deletion in mice), neurodegeneration (Listerin hypomorphic mutation in mice) |
| Other key features | Aberrant proteins are ubiquitylated shortly after synthesis and before being released to other subcellular compartments. The pathway is coupled to mRNA quality control; experimentally, it is often necessary to simultaneously inactivate both pathways to observe accumulation of aberrant protein substrates. |
Box 4: Open questions on mechanisms and functions of RQC.
An important subject of current investigation is how cells sense ribosomes stalled internally on an mRNA, and how this reaction generates RQC substrates. Understanding the former will require indentifying the factors directly involved (likely Hel2/ZNF598), and solving their structure bound to stalled ribosomes.
In the canonical RQC pathway, the sequence of events leading to Cdc48-mediated extraction remains to be understood. That includes elucidating the mechanism of action of Rqc1 and how Vms1 function is coordinated with Ltn1-mediated ubiquitylation.
Much has yet to be learned about CAT tails, including: their function; the mechanism of synthesis; why CAT tails are composed of Ala and Thr. Of note, reagents to study CAT tails in a physiological context are unavailable. Furthermore, although Rqc2 residues implicated in A-site tRNA recruitment and CAT tail synthesis are highly conserved, including in mammalian NEMF, CAT tail formation (or the formation of related tails with different composition) has so far only been described in yeast.
RQC in organellar surfaces is generally assumed to function with a large degree of similarity to RQC of cytosolic proteins, but whether subcellular compartment-specific RQC factors or mechanisms exist remains to be explored.
Research on the RQC has been mainly focused on yeast and in vitro translation systems. It is now appropriate to expand functional and mechanistic analyses towards more complex systems, such as metazoan model organisms and mammalian cells; indeed, although the RQC complex is highly conserved with regard to both components and structure, there are already indications that the yeast and mammalian complexes can have different properties. For example, the RQC complex is more difficult to isolate from mammalian cells, with its subunits apparently existing mostly in free form in equilibrium (64,112 and C.A.P.J. unpublished results);
Next to nothing is known about other direct, non-RQC functions of RQC complex components, so this is also a relevant area for future studies. An example is Ltn1’s proposed function in ribophagy113, a process that selectively degrades ribosomal 60S subunits. Under normal growth conditions, ribophagy is negatively regulated by Ltn1, via ubiquitylation of the 60S ribosomal subunit protein, Rpl25. Nitrogen starvation leads to decreased Ltn1 expression, which allows removal of the ubiquitin inhibitory signal in Rpl25 Lys74/75 by the Ubp3/Bre5 deubiquitylase, and ribophagy to be triggered. The extent to which Ltn1 function in ribophagy [G] relates to its role in RQC, and whether other RQC components are involved, remains unknown. It is also unclear how the few Ltn1 molecules/cell support ubiquitylation of the much larger Rpl25 pool;
Little is known about how RQC components are regulated. Ltn1 levels decrease drastically and rapidly upon nutrient starvation, and this occurs at least in part via auto-ubiquitylation113. Rqc1 appears to be modified by ubiquitylation as well114.
Finally, mechanistic insights derived from fundamental RQC studies need to be tested in the context of mammalian physiology and disease. For example, it will be important to determine whether mutations in RQC complex subunits other than Listerin can also result in neurodegeneration in mice, and whether the pathway is directly or indirectly implicated in human disease. These and other more advanced analyses on RQC could also benefit from the identification of endogenous substrates of the pathway, which could serve as biomarkers for studies with animal models or human disease samples.
Acknowledgements
The author thanks members of the Joazeiro laboratory for critical comments on the manuscript. I am also grateful to Tarek Hilal, Christian Spahn, Song Tan and members of the Joazeiro laboratory for preparing figures. Work in the Joazeiro laboratory is supported by a grant from the Deutsche Forschungsgemeinschaft (SFB1036) and by R01 Grants NS075719 and NS102414 from the National Institute of Neurological Disorders and Stroke (NINDS) of the NIH.
GLOSSARY
- RING domain
globular domain that characterizes the vast majority of E3 ligases and functions by recruiting E2 conjugases and sometimes additionally binding the E2-conjugated ubiquitin moiety to prime it for transfer.
- Endoplasmic reticulum-associated degradation (ERAD)
process in which aberrant proteins in the lumen or membrane of the ER are ubiquitylated by ER membrane-resident E3 ligases and retrotranslocated for degradation in the cytosol.
- Cullin-dependent E3 ligase
E3 ligase complex consisting of a RING domain subunit (Rbx1/2), a cullin subunit, and adapter proteins that link the RING-cullin subunit core to substrates.
- Aggresome
cellular inclusion containing misfolded proteins and formed in a regulated manner, typically increased under stress.
- Translocon
complex on organellar surfaces that mediates import of proteins made in the cytosol (with this term being most commonly utilized for the ER translocon).
- Ribonuclease H
family of endonuclease enzymes that cleave RNA phosphodiester bonds in an RNA-DNA duplex in a sequence-unspecific manner.
- tmRNA/ssrA
hybrid transfer-messenger RNA (tmRNA) molecule, a central component of the bacterial pathway of ribosomal rescue and protein quality control elicited in response to translational stalling.
- Hypomorphic mutation
recessive mutation that causes partial loss of gene function due to reduced activity or expression.
- N-end rule pathway
ubiquitylation pathway that targets proteins for degradation as a function of their N-terminal residue.
- Ribophagy
selective autophagy of 60S ribosomal subunits in response to nutrient starvation and requiring the Ubp3 deubiquitylating enzyme in yeast.
References
- 1.Pilla E, Schneider K & Bertolotti A Coping with Protein Quality Control Failure. Annu Rev Cell Dev Biol 33, 439–465 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Klaips CL, Jayaraj GG & Hartl FU Pathways of cellular proteostasis in aging and disease. J. Cell Biol 217, 51–63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartl FU Protein Misfolding Diseases. Annu. Rev. Biochem 86, 21–26 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Joazeiro CAP Ribosomal Stalling During Translation: Providing Substrates for Ribosome-Associated Protein Quality Control. Annu. Rev. Cell Dev. Biol 33, 343–368 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Buskirk AR & Green R Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc Lond B Biol Sci 372, 20160183(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bengtson MH & Joazeiro CA Role of a ribosome-associated E3 ubiquitin ligase in protein quality control. Nature 467, 470–473 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]; This seminal work identifies Ltn1 as an E3 ligase that marks nascent chains produced by ribosome stalling for degradation while still ribosome-associated, defining a previously unknown specialized pathway of protein quality control--RQC.
- 7.Brandman O & Hegde RS Ribosome-associated protein quality control. Nat Struct Mol Biol 23, 7–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu J et al. A mouse forward genetics screen identifies LISTERIN as an E3 ubiquitin ligase involved in neurodegeneration. Proc. Nat. Acad. Sci. USA 106, 2097–2103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study established a first link between Listerin/RQC and neurodegeneration.
- 9.Brandman O et al. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell 151, 1042–1054 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]; Studies in references 9 and 10 report the results of a powerful combination of genetic and proteomic analyses, uncovering that Ltn1 functions together with cofactors in an “RQC complex”.
- 10.Defenouillere Q et al. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proc. Nat. Acad. Sci. USA 110, 5046–5051 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Canadeo LA & Huibregtse JM Ubiquitination of newly synthesized proteins at the ribosome. Biochimie 114, 127–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duttler S, Pechmann S & Frydman J Principles of Cotranslational Ubiquitination and Quality Control at the Ribosome. Mol. Cell 50, 379–393 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shoemaker CJ, Eyler DE & Green R Dom34:Hbs1 promotes subunit dissociation and peptidyl-tRNA drop-off to initiate no-go decay. Science 330, 369–372 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pisareva VP, Skabkin MA, Hellen CU, Pestova TV & Pisarev AV Dissociation by Pelota, Hbs1 and ABCE1 of mammalian vacant 80S ribosomes and stalled elongation complexes. EMBO J 30, 1804–1817 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuller AP & Green R Roadblocks and resolutions in eukaryotic translation. Nat. Rev. Mol. Cell Biol 19, 526–541 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker CJ & Green R Translation drives mRNA quality control. Nat Struct Mol Biol 19, 594–601 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graille M & Seraphin B Surveillance pathways rescuing eukaryotic ribosomes lost in translation. Nat. Rev. Mol. Cell Biol 13, 727–735 (2012). [DOI] [PubMed] [Google Scholar]
- 18.Arribere JA & Fire AZ Nonsense mRNA suppression via nonstop decay. Elife 7, e33292(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao S, von der Malsburg K & Hegde RS Listerin-dependent nascent protein ubiquitination relies on ribosome subunit dissociation. Mol Cell 50, 637–648 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]; Further developing studies in references 13 and 14, this work reports biochemical evidence that the dissociation of stalled ribosomal subunits by rescue factors is required upstream of Listerin function.
- 20.Lyumkis D et al. Structural basis for translational surveillance by the large ribosomal subunit-associated protein quality control complex. Proc. Nat. Acad. Sci. USA 111, 15981–15986 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; Study reporting the first cryo-EM structure of the RQC complex, identifying Rqc2 as the subunit that senses obstructed 60S subunits and setting forth novel principles of substrate recognition and selectivity in protein quality control.
- 21.Shen PS et al. Protein synthesis. Rqc2p and 60S ribosomal subunits mediate mRNA-independent elongation of nascent chains. Science 347, 75–78 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work reports the surprising discovery of CAT tails and offers an underlying mechanism, mediated by Ala- and Thr-tRNA recruitment by Rqc2.
- 22.Shao S, Brown A, Santhanam B & Hegde RS Structure and assembly pathway of the ribosome quality control complex. Mol Cell 57, 433–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work describes the high-resolution cryo-EM structure of the mammalian RQC complex, and an elegant biochemical reconstitution of the complex assembly.
- 23.Verma R, Oania RS, Kolawa NJ & Deshaies RJ Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. Elife 2, e00308(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verma R et al. Vms1 and ANKZF1 peptidyl-tRNA hydrolases release nascent chains from stalled ribosomes. Nature 557, 446–451 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; Studies in references 24 and 25 report that Vms1 acts in releasing the nascent chain from tRNA in an obstructed 60S/peptidyl-tRNA complex.
- 25.Zurita Rendon O et al. Vms1p is a release factor for the ribosome-associated quality control complex. Nat Commun 9, 2197(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostova KK et al. CAT-tailing as a fail-safe mechanism for efficient degradation of stalled nascent polypeptides. Science 357, 414–417 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; References 26 and 28–30 report work elucidating consequences of the modification of nascent chains with CAT tails.
- 27.Osuna BA, Howard CJ, Kc S, Frost A & Weinberg DE In vitro analysis of RQC activities provides insights into the mechanism and function of CAT tailing. Elife 6, e27949(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yonashiro R et al. The Rqc2/Tae2 subunit of the Ribosome-Associated Quality Control (RQC) complex marks ribosome-stalled nascent polypeptide chains for aggregation. Elife 5, e11794(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choe YJ et al. Failure of RQC machinery causes protein aggregation and proteotoxic stress. Nature 531, 191–195 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Defenouillere Q et al. Rqc1 and Ltn1 Prevent C-terminal Alanine-Threonine Tail (CAT-tail)-induced Protein Aggregation by Efficient Recruitment of Cdc48 on Stalled 60S Subunits. J Biol Chem 291, 12245–12253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito-Harashima S, Kuroha K, Tatematsu T & Inada T Translation of the poly(A) tail plays crucial roles in nonstop mRNA surveillance via translation repression and protein destabilization by proteasome in yeast. Genes Dev. 21, 519–524 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson MA, Meaux S & van Hoof A A genomic screen in yeast reveals novel aspects of nonstop mRNA metabolism. Genetics 177, 773–784 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sung MK et al. A conserved quality-control pathway that mediates degradation of unassembled ribosomal proteins. Elife 5, e19105(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Anderson DE & Ye Y The HECT domain ubiquitin ligase HUWE1 targets unassembled soluble proteins for degradation. Cell Discov 2, 16040(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khosrow-Khavar F et al. The yeast ubr1 ubiquitin ligase participates in a prominent pathway that targets cytosolic thermosensitive mutants for degradation. G3 (Bethesda, Md.) 2, 619–628 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nillegoda NB et al. Ubr1 and ubr2 function in a quality control pathway for degradation of unfolded cytosolic proteins. Mol. Biol. Cell 21, 2102–2116 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heck JW, Cheung SK & Hampton RY Cytoplasmic protein quality control degradation mediated by parallel actions of the E3 ubiquitin ligases Ubr1 and San1. Proc Natl Acad Sci U S A 107, 1106–1111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eisele F & Wolf DH Degradation of misfolded protein in the cytoplasm is mediated by the ubiquitin ligase Ubr1. FEBS Lett 582, 4143–4146 (2008). [DOI] [PubMed] [Google Scholar]
- 39.Wu X & Rapoport TA Mechanistic insights into ER-associated protein degradation. Curr Opin Cell Biol 53, 22–28 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jones RD & Gardner RG Protein quality control in the nucleus. Curr Opin Cell Biol 40, 81–89 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khmelinskii A et al. Protein quality control at the inner nuclear membrane. Nature 516, 410–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Foresti O, Rodriguez-Vaello V, Funaya C & Carvalho P Quality control of inner nuclear membrane proteins by the Asi complex. Science 346, 751–755 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Taipale M et al. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 158, 434–448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murata S, Chiba T & Tanaka K CHIP: a quality-control E3 ligase collaborating with molecular chaperones. The Intl. J. Biochem. Cell Biol 35, 572–578 (2003). [DOI] [PubMed] [Google Scholar]
- 45.Kevei E, Pokrzywa W & Hoppe T Repair or destruction-an intimate liaison between ubiquitin ligases and molecular chaperones in proteostasis. FEBS Lett 591, 2616–2635 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenbaum JC et al. Disorder targets misorder in nuclear quality control degradation: a disordered ubiquitin ligase directly recognizes its misfolded substrates. Mol Cell 41, 93–106 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lyumkis D et al. Single-particle EM reveals extensive conformational variability of the Ltn1 E3 ligase. Proc. Natl. Acad. Sci. USA 110, 1702–1707 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doamekpor SK et al. Structure and function of the yeast listerin (Ltn1) conserved N-terminal domain in binding to stalled 60S ribosomal subunits. Proc Natl Acad Sci U S A 113, E4151–4160 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McGinty RK, Henrici RC & Tan S Crystal structure of the PRC1 ubiquitylation module bound to the nucleosome. Nature 514, 591–596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown NG et al. RING E3 mechanism for ubiquitin ligation to a disordered substrate visualized for human anaphase-promoting complex. Proc Natl Acad Sci U S A 112, 5272–5279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deshaies RJ & Joazeiro CA RING domain E3 ubiquitin ligases. Annu. Rev. Biochem 78, 399–434 (2009). [DOI] [PubMed] [Google Scholar]
- 52.Breitschopf K, Bengal E, Ziv T, Admon A & Ciechanover A A novel site for ubiquitination: the N-terminal residue, and not internal lysines of MyoD, is essential for conjugation and degradation of the protein. EMBO J 17, 5964–5973 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McDowell GS & Philpott A Non-canonical ubiquitylation: mechanisms and consequences. The Intl. J. Biochem. Cell Biol 45, 1833–1842 (2013). [DOI] [PubMed] [Google Scholar]
- 54.Kravtsova-Ivantsiv Y & Ciechanover A Non-canonical ubiquitin-based signals for proteasomal degradation. J. Cell Sci 125, 539–548 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Pao KC et al. Activity-based E3 ligase profiling uncovers an E3 ligase with esterification activity. Nature 556, 381–385 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Weber A et al. Sequential Poly-ubiquitylation by Specialized Conjugating Enzymes Expands the Versatility of a Quality Control Ubiquitin Ligase. Mol Cell 63, 827–839 (2016). [DOI] [PubMed] [Google Scholar]
- 57.Boban M, Ljungdahl PO & Foisner R Atypical ubiquitylation in yeast targets lysine-less Asi2 for proteasomal degradation. J Biol Chem 290, 2489–2495 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ishikura S, Weissman AM & Bonifacino JS Serine residues in the cytosolic tail of the T-cell antigen receptor alpha-chain mediate ubiquitination and endoplasmic reticulum-associated degradation of the unassembled protein. J Biol Chem 285, 23916–23924 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimizu Y, Okuda-Shimizu Y & Hendershot LM Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol Cell 40, 917–926 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skaar JR, Pagan JK & Pagano M Mechanisms and function of substrate recruitment by F-box proteins. Nat. Rev. Mol. Cell Biol 14, 369–381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Duda DM et al. Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation. Cell 134, 995–1006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fischer ES et al. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell 147, 1024–1039 (2011). [DOI] [PubMed] [Google Scholar]
- 63.Ghaemmaghami S et al. Global analysis of protein expression in yeast. Nature 425, 737–741 (2003). [DOI] [PubMed] [Google Scholar]
- 64.Shao S & Hegde RS Reconstitution of a minimal ribosome-associated ubiquitination pathway with purified factors. Mol Cell 55, 880–890 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lu J & Deutsch C Electrostatics in the ribosomal tunnel modulate chain elongation rates. J. Mol. Biol 384, 73–86 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Izawa T, Park SH, Zhao L, Hartl FU & Neupert W Cytosolic Protein Vms1 Links Ribosome Quality Control to Mitochondrial and Cellular Homeostasis. Cell 171, 890–903 e818 (2017). [DOI] [PubMed] [Google Scholar]; This is an elegant analysis first reporting consequences of RQC dysfunction regarding cytosolic ribosomes stalled while translating mitochondrial proteins (“mito-RQC”).
- 67.Sontag EM, Samant RS & Frydman J Mechanisms and Functions of Spatial Protein Quality Control. Annu. Rev. Biochem 86, 97–122 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Miller SB, Mogk A & Bukau B Spatially organized aggregation of misfolded proteins as cellular stress defense strategy. J. Mol. Biol 427, 1564–1574 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Dikic I & Elazar Z Mechanism and medical implications of mammalian autophagy. Nat. Rev. Mol. Cell Biol 19, 349–364 (2018). [DOI] [PubMed] [Google Scholar]
- 70.Spokoini R et al. Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep. 2, 738–747 (2012). [DOI] [PubMed] [Google Scholar]
- 71.Gomez-Pastor R, Burchfiel ET & Thiele DJ Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol 19, 4–19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anckar J & Sistonen L Regulation of HSF1 function in the heat stress response: implications in aging and disease. Annu. Rev. Biochem 80, 1089–1115 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Gomez-Pastor R, Burchfiel ET & Thiele DJ Regulation of heat shock transcription factors and their roles in physiology and disease. Nat. Rev. Mol. Cell Biol 19, 4–19 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nissen P, Hansen J, Ban N, Moore PB & Steitz TA The structural basis of ribosome activity in peptide bond synthesis. Science 289, 920–930 (2000). [DOI] [PubMed] [Google Scholar]
- 75.Kuroha K, Zinoviev A, Hellen CUT & Pestova TV Release of Ubiquitinated and Non-ubiquitinated Nascent Chains from Stalled Mammalian Ribosomal Complexes by ANKZF1 and Ptrh1. Mol Cell 72, 286–302 e288 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Defenouillere Q, Namane A, Mouaikel J, Jacquier A & Fromont-Racine M The ribosome-bound quality control complex remains associated to aberrant peptides during their proteasomal targeting and interacts with Tom1 to limit protein aggregation. Mol. Biol. Cell 28, 1165–1176 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Meyer H, Bug M & Bremer S Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14, 117–123 (2012). [DOI] [PubMed] [Google Scholar]
- 78.Shcherbik N, Chernova TA, Chernoff YO & Pestov DG Distinct types of translation termination generate substrates for ribosome-associated quality control. Nu. Ac. Res 44, 6840–6852 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsuchiya H et al. In Vivo Ubiquitin Linkage-type Analysis Reveals that the Cdc48-Rad23/Dsk2 Axis Contributes to K48-Linked Chain Specificity of the Proteasome. Mol Cell 66, 488–502 (2017). [DOI] [PubMed] [Google Scholar]
- 80.Simms CL & Zaher HS Ribosome Collision Is Critical for Quality Control during No-Go Decay. Mol. Cell 68, 361–373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wesolowska MT, Richter-Dennerlein R, Lightowlers RN & Chrzanowska-Lightowlers ZM Overcoming stalled translation in human mitochondria. Front Microbiol 5, 374(2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Izawa T et al. Roles of dom34:hbs1 in nonstop protein clearance from translocators for normal organelle protein influx. Cell Rep. 2, 447–453 (2012). [DOI] [PubMed] [Google Scholar]
- 83.Arakawa S et al. Quality control of nonstop membrane proteins at the ER membrane and in the cytosol. Scientific reports 6, 30795(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Crowder JJ et al. Rkr1/Ltn1 Ubiquitin Ligase-mediated Degradation of Translationally Stalled Endoplasmic Reticulum Proteins. J Biol Chem 290, 18454–18466 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.von der Malsburg K, Shao S & Hegde RS The ribosome quality control pathway can access nascent polypeptides stalled at the Sec61 translocon. Mol. Biol. Cell 26, 2168–2180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Moore SD & Sauer RT The tmRNA system for translational surveillance and ribosome rescue. Annu. Rev. Biochem 76, 101–124 (2007). [DOI] [PubMed] [Google Scholar]
- 87.Lang WH, Calloni G & Vabulas RM Polylysine is a Proteostasis Network-Engaging Structural Determinant. J Proteome Res 17, 1967–1977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Saarikangas J & Barral Y Protein aggregation as a mechanism of adaptive cellular responses. Curr. Gen 62, 711–724 (2016). [DOI] [PubMed] [Google Scholar]
- 89.Wang Y, Meriin AB, Costello CE & Sherman MY Characterization of proteins associated with polyglutamine aggregates: a novel approach towards isolation of aggregates from protein conformation disorders. Prion 1, 128–135 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rousseau E et al. Targeting expression of expanded polyglutamine proteins to the endoplasmic reticulum or mitochondria prevents their aggregation. Proc Natl Acad Sci U S A 101, 9648–9653 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schneider K & Bertolotti A Surviving protein quality control catastrophes--from cells to organisms. J. Cell Sci 128, 3861–3869 (2015). [DOI] [PubMed] [Google Scholar]
- 92.Ishimura R et al. RNA function. Ribosome stalling induced by mutation of a CNS-specific tRNA causes neurodegeneration. Science 345, 455–459 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Taylor JP, Brown RH Jr. & Cleveland DW Decoding ALS: from genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dawson TM & Dawson VL Molecular pathways of neurodegeneration in Parkinson’s disease. Science 302, 819–822 (2003). [DOI] [PubMed] [Google Scholar]
- 95.Bauer MF & Neupert W Import of proteins into mitochondria: a novel pathomechanism for progressive neurodegeneration. J Inherit Metab Dis 24, 166–180 (2001). [DOI] [PubMed] [Google Scholar]
- 96.Sarkar A et al. Preribosomes escaping from the nucleus are caught during translation by cytoplasmic quality control. Nat Struct Mol Biol 24, 1107–1115 (2017). [DOI] [PubMed] [Google Scholar]
- 97.Juszkiewicz S & Hegde RS Initiation of Quality Control during Poly(A) Translation Requires Site-Specific Ribosome Ubiquitination. Mol Cell 65, 743–750 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arthur L et al. Translational control by lysine-encoding A-rich sequences. Science advances 1, e1500154(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ito K & Chiba S Arrest peptides: cis-acting modulators of translation. Annu. Rev. Biochem 82, 171–202 (2013). [DOI] [PubMed] [Google Scholar]
- 100.Hilal T et al. Structural insights into ribosomal rescue by Dom34 and Hbs1 at near-atomic resolution. Nat Commun 7, 13521(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Matsuo Y et al. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat Commun 8, 159(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sundaramoorthy E et al. ZNF598 and RACK1 Regulate Mammalian Ribosome-Associated Quality Control Function by Mediating Regulatory 40S Ribosomal Ubiquitylation. Mol Cell 65, 751–760 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sitron CS, Park JH & Brandman O Asc1, Hel2, and Slh1 couple translation arrest to nascent chain degradation. RNA (New York, N.Y.) 23, 798–810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garzia A et al. The E3 ubiquitin ligase and RNA-binding protein ZNF598 orchestrates ribosome quality control of premature polyadenylated mRNAs. Nat Commun 8, 16056(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Simms CL, Thomas EN & Zaher HS Ribosome-based quality control of mRNA and nascent peptides. RNA 8, e1366(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li W et al. Genome-wide and functional annotation of human E3 ubiquitin ligases identifies MULAN, a mitochondrial E3 that regulates the organelle’s dynamics and signaling. PloS one 3, e1487(2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wenzel DM, Lissounov A, Brzovic PS & Klevit RE UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474, 105–108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buetow L & Huang DT Structural insights into the catalysis and regulation of E3 ubiquitin ligases. Nat. Rev. Mol. Cell Biol 17, 626–642 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kao SH, Wu HT & Wu KJ Ubiquitination by HUWE1 in tumorigenesis and beyond. J Biomed Sci 25, 67(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Varshavsky A The Ubiquitin System, Autophagy, and Regulated Protein Degradation. Annu. Rev. Biochem 86, 123–128 (2017). [DOI] [PubMed] [Google Scholar]
- 111.Joazeiro CA et al. The tyrosine kinase negative regulator c-Cbl as a RING-type, E2-dependent ubiquitin-protein ligase. Science 286, 309–312 (1999). [DOI] [PubMed] [Google Scholar]
- 112.Zuzow N et al. Mapping the mammalian ribosome quality control complex interactome using proximity labeling approaches. Mol. Biol. Cell 29, 1258–1269 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ossareh-Nazari B et al. Ubiquitylation by the Ltn1 E3 ligase protects 60S ribosomes from starvation-induced selective autophagy. J. Cell Biol 204, 909–917 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Defenouillere Q & Fromont-Racine M The ribosome-bound quality control complex: from aberrant peptide clearance to proteostasis maintenance. Curr. Gen 63, 997–1005 (2017). [DOI] [PubMed] [Google Scholar]
- 115.Shoemaker CJ & Green R Kinetic analysis reveals the ordered coupling of translation termination and ribosome recycling in yeast. Proc Natl Acad Sci U S A 108, E1392–1398 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


