Abstract
The filamentous fungus Penicillium chrysogenum Q176 secretes the antimicrobial proteins (AMPs) PAF and PAFB, which share a compact disulfide-bond mediated, β-fold structure rendering them highly stable. These two AMPs effectively inhibit the growth of human pathogenic fungi in micromolar concentrations and exhibit antiviral potential without causing cytotoxic effects on mammalian cells in vitro and in vivo.
The antifungal mechanism of action of both AMPs is closely linked to - but not solely dependent on - the lipid composition of the fungal cell membrane and requires a strictly regulated protein uptake into the cell, indicating that PAF and PAFB are not canonical membrane active proteins. Variations in their antifungal spectrum and their killing dynamics point towards a divergent mode of action related to their physicochemical properties and surface charge distribution.
In this review, we relate characteristic features of PAF and PAFB to the current knowledge about other AMPs of different sources. In addition, we present original data that have never been published before to substantiate our assumptions and provide evidences that help to explain and understand better the mechanistic function of PAF and PAFB. Finally, we underline the promising potential of PAF and PAFB as future antifungal therapeutics.
Keywords: Antimicrobial proteins and peptides, Penicillium chrysogenum, β-Fold structure, γ-Core, Fungal membrane lipids, Endocytosis, Apoptosis
Graphical abstract
Highlights
-
•
Penicillium chrysogenum secretes the small, cysteine-rich proteins PAF and PAFB.
-
•
Both exhibit antifungal activity, but with differences in their mode of action.
-
•
Structure, membrane interaction and cellular uptake determine their function.
-
•
PAF and PAFB are well tolerated by mammalian cells.
-
•
They promise applicability in medicine, plant protection and food industry.
1. Introduction
Approximately one billion people are suffering from severe fungal diseases which cause around 1.5 million deaths per year [1]. Ninety percent of all deaths are a consequence of fungal infections caused by species of the genera: Aspergillus, Candida and Cryptococcus [2]. The treatment of these pathogens is hampered by the limited number of available antifungal drugs belonging mainly to the classes of azoles, echinocandins and polyenes [3]. The extensive use of antimycotics stimulates the development of resistance towards these agents in many fungal pathogens [4]. Also, in agriculture the control of plant diseases caused by filamentous fungi pose severe problems as species like Fusarium are responsible for enormous crop losses every year [5]. Therefore, there is a great urgency to develop alternative antifungal drugs and treatment strategies.
Filamentous fungi produce a wide spectrum of antimicrobial proteins (AMPs) that serve as defense and/or signaling molecules for their hosts [[6], [7], [8]]. Such AMPs are promising candidates for the development of new therapeutic compounds. These small (~5.6–6.6 kDa), cysteine-rich and amphipathic proteins are secreted into the culture supernatant and can be easily purified via ion-exchange chromatography due to their positive net charge. Therefore, large scale industrial production of AMPs would be easy and inexpensive [9,10].
AMPs of fungal origin are highly active against pathogenic filamentous fungal species e.g. the human pathogen Aspergillus fumigatus or the plant pathogens Botrytis spp. and Fusarium spp. [8]. Additionally, some show anti-yeast activity and inhibit the growth of pathogenic yeasts like Candida albicans at low micromolar [μM] concentrations [[11], [12], [13]].
In this review, we focus on the two AMPs derived from the well-known penicillin producing ascomycete Penicillium chrysogenum Q176: the P. chrysogenum antifungal protein (PAF) and the P. chrysogenum antifungal protein B (PAFB). These two AMPs do not only inhibit the growth of filamentous fungi and yeasts but also possess antiviral potential without exhibiting any cytotoxic or hemolytic activity on mammalian cells [11]. A detailed knowledge about the mechanistic function of PAF and PAFB is an important prerequisite for their exploitation and also related biomolecules in industrial drug design and the development of new anti-infective treatment strategies for medical and agricultural application. Here, we highlight and discuss the similarities and differences of these two P. chrysogenum AMPs regarding structure, antimicrobial spectrum and mode of action.
2. Phylogenetic aspects
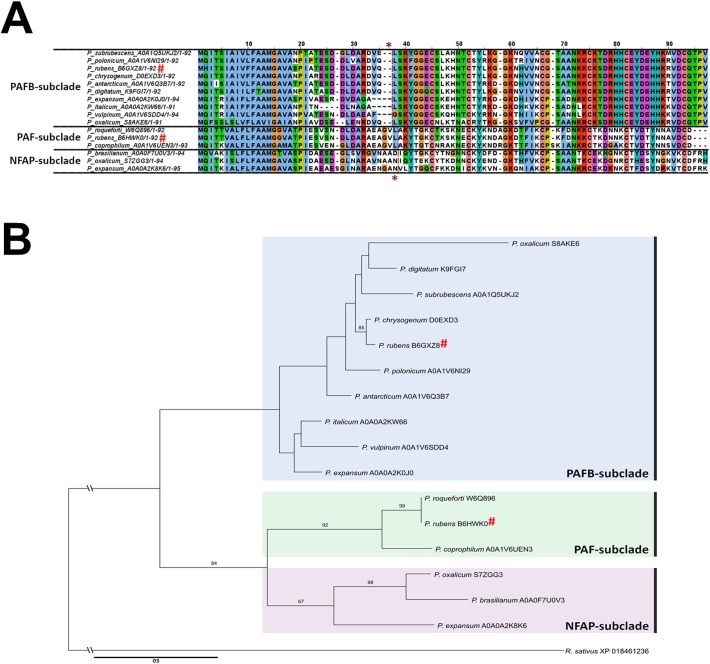
Screening of genomic databases revealed that AMPs with potential antifungal activity are widespread among filamentous ascomycetes, especially in the class Eurotiomycetes (specifically the genera Aspergillus, Monascus, Penicillium, Paecilomyces) [14] and Sordariomycetes (containing the genus Fusarium) [15]. A detailed phylogenetic analysis revealed that the so far isolated small, cysteine-rich antifungal AMPs from Eurotiomycetes and their putative and predicted homologs can be divided into four well-separated clades. These are named in this review according to their first characterized representatives: the clade of (i) Aspergillus giganteus antifungal protein (AFP) [16]; (ii) P. chrysogenum antifungal protein (PAF) [17]; (iii) Penicillium brevicompactum “bubble protein” (BP) [18] and (iv) Neosartorya (Aspergillus) fischeri antifungal protein 2 (NFAP2) [13]. Interestingly, some ascomycetous isolates are able to produce more than one AMP belonging to different clades [14]; e.g. the Penicillium rubens Wisconsin 54-1255 strain (PAF and BP-clade) and N. fischeri NRRL 181 (PAF-, BP-, and NFAP2-clade). Notably, the PAF and PAFB producing P. chrysogenum Q176 [11,17] is the original ancestor of the industrially exploited Wisconsin strains [19]. This analysis also revealed that the PAF-clade can be further divided into four subclades [14], containing (1) the Aspergillus niger antifungal protein (AnAFP) [12] and its putative homologs; (2) the PAF-like proteins; (3) the N. (A.) fischeri antifungal protein (NFAP) [20] and respective Penicillium spp. homologs; as well as (4) the Penicillium protein PAFB [11] and other closely related proteins. A recent mining of the UniProt database [21] for PAF-like Penicillium AMPs resulted in several putative representatives with 34–88% amino acid (aa) identity (Fig. 1A) and distinct physicochemical features (Table 1 ). Based on the primary structure they are clearly separated into three different subclades, representing PAF-, PAFB-, and NFAP-like proteins, but no AnAFP-similar ones. The well-supported separation of Penicillium PAFBs from PAFs in this phylogenetic tree proposes differences in protein structure and antifungal mode of action, which is further discussed below (Fig. 1B).
Fig. 1.
(A) Clustal W multiple alignment of the putative PAF-like antifungal AMPs from Penicillium spp. from the UniProt database [21]. Alignment was generated by BioEdit [22] and visualized by Jalview 2.11.0. [23]. The cleavage of the preprosequence and the first aa of the mature protein is marked by brown dotted line and asterisks. After the species name the accession number of the respective AMP (see Table 1) is indicated. The Clustal X default color scheme was applied (http://www.jalview.org/help/html/colourSchemes/clustal.html). (B) Maximum-likelihood (ML) tree of putative PAF-like AMPs from Penicillium spp. from UniProt database [21]. The alignment of the full-length proteins was generated with PRANK v. 140,110 [24] with default settings for phylogenetic analysis. ML analysis was performed with RAxML v. 8.2.10 [25] under the GAMMA distributed rate heterogeneity empirical frequencies model with DCmut substitution matrix in 1000 through bootstrap replicates. ML bootstrap values >60% are shown next to branches. After the species name the accession number of the respective AMP (see Table 1) is indicated. “#” marks PAF and PAFB from P. rubens Wisconsin 54-1255, which is the descendent strain of P. chrysogenum Q176 [19].
Table 1.
Putative PAF-like AMP homologs of Penicillium spp. from UniProt database [21], and their physicochemical properties analyzed in silico.§
| Species | UniProt ID | MW$ [Da] |
pI | Net charge (pH 7) | GRAVY⁎ | No. positively charged aa |
|---|---|---|---|---|---|---|
| PAF-subclade | ||||||
| P. roqueforti | W6Q896 | 6134.96 | 9.09 | +4.7 | −1.335 | 13 |
| P. rubens | B6HWK0 | 6250.05 | 8.93 | +4.7 | −1.375 | 13 |
| P. coprophilum | A0A1V6UEN3 | 6272.10 | 9.28 | +6.7 | −1.166 | 13 |
| PAFB-subclade | ||||||
| P. subrubescens | A0A1Q5UKJ2 | 6464.30 | 8.29 | +2.7 | −0.781 | 12 |
| P. polonicum | A0A1V6NI29 | 6591.44 | 9.22 | +6.9 | −1.153 | 17 |
| P. rubens | B6GXZ8 | 6500.32 | 8.83 | +5.2 | −1.031 | 16 |
| P. chrysogenum | D0EXD3 | 6500.32 | 8.83 | +5.2 | −1.031 | 16 |
| P. antarcticum | A0A1V6Q3B7 | 6621.33 | 8.29 | +3.2 | −1.284 | 16 |
| P. digitatum | K9FGI7 | 6575.39 | 9.06 | +5.9 | −1.000 | 15 |
| P. expansum | A0A0A2K0J0 | 6592.28 | 7.80 | +2.2 | −1.570 | 17 |
| P. italicum | A0A0A2KW66 | 6718.48 | 8.27 | +3.2 | −1.538 | 18 |
| P. vulpinum | A0A1V6SDD4 | 6685.55 | 9.14 | +7.2 | −1.467 | 18 |
| P. oxalicum | S8AKE6 | 6528.37 | 9.02 | +5.7 | −1.031 | 15 |
| NFAP-subclade | ||||||
| P. brasilianum | A0A0F7U0V3 | 6510.26 | 8.96 | +5.2 | −1.172 | 12 |
| P. oxalicum | S7ZGG3 | 6638.31 | 8.27 | +2.5 | −1.435 | 13 |
| P. expansum | A0A0A2K8K6 | 6644.70 | 9.48 | +8.7 | −1.081 | 15 |
ExPASy ProtParam tool [26].
MW: Molecular Weight.
GRAVY: Grand Average of Hydropathy.
Protein Calculator v3.4 (The Scripps Research Institute; http://protcalc.sourceforge.net/) was applied for calculations of physicochemical properties.
3. Antimicrobial and antiviral activity
PAF and PAFB exhibit growth inhibitory activity against numerous pathogenic molds, yeasts and fungal model organisms, whereas both proteins were found inactive against bacteria such as the Gram-negative Escherichia coli or the Gram-positive Bacillus subtilis in the concentration range tested (up to 32 μM) [11,27]. Pathogenic fungi that are highly susceptible towards low doses of PAF and PAFB are the human pathogens: A. fumigatus, A. niger, Trichophyton spp. and Candida spp., as well as the model fungi Neurospora crassa and Saccharomyces cerevisiae. These strains are similarly sensitive towards both proteins as their growth is inhibited in the presence of 0.25–4 μM PAF or PAFB, respectively (Table 2 ).
Table 2.
Activity of PAF and PAFB on fungi.
| Organisms | IC90 [μM]⁎ |
References | |
|---|---|---|---|
| PAF | PAFB | ||
| Filamentous fungi | |||
| Aspergillus fumigatus§ | 1 | 0.25 | [11] |
| Aspergillus niger§ | 0.25 | 0.50 | [11] |
| Aspergillus terreus§ | 32 | 1 | [11] |
| Neosartorya fischeri§ | 0.50 | 1 | This study |
| Neurospora crassa# | 0.06 | 0.12 | [11] |
| Penicillium chrysogenum§ | >32 | 0.50 | [11] |
| Trichophyton rubrum+ | 0.25 | 0.50 | [11] |
| Trichophyton mentagrophytes+ | 0.02 | 0.30 | This study |
| Yeasts | |||
| Candida albicans | 4 | 1 | [11] |
| Candida glabrata | 2.5 | 0.60 | [28] |
| Candida krusei | 5 | 0.60 | [28] |
| Candida parapsilosis | 2.5 | 0.60 | [28] |
| Saccharomyces cerevisiae | 2 | 1 | [11] |
IC90, AMP concentration inhibiting growth ≥90%. The antifungal activity was tested in a microdilution broth assay by inoculating conidia (104/mL) or yeast cells (104/mL) with increasing concentrations of PAF or PAFB.
0.1 × potato dextrose broth (PDB) medium 48 h, 25–30 °C.
0.2 × Vogel's medium 32 h at 25 °C. +0.1 × PDB medium 8 days, 37 °C; Yeasts were grown for 24–48 h at 30 °C in 0.05–0.1 × PDB.
Notably, differences exist in the spectrum of microorganisms targeted by these two related proteins. The clinically relevant and amphotericin B-resistant human pathogen Aspergillus terreus is highly susceptible towards low doses of PAFB (1 μM) whereas a high concentration of PAF (32 μM) is required for growth inhibition [11,29]. The same holds true for the producing mold P. chrysogenum itself, whose growth is inhibited at 0.5 μM PAFB. In contrast, the highest PAF concentration tested (32 μM) was not sufficient to achieve the same growth inhibition [11]. Notably, the AMP encoding genes paf and pafB are differently regulated in P. chrysogenum, paf being induced under nutrient limitation whereas pafB being expressed under excess nutrient availability [30]. The difference in the gene expression pattern and in the AMP susceptibility of the host strengthen our hypothesis that PAF and PAFB cover additional functions, possibly related to signaling in growth and development regulation in P. chrysogenum [30,31].
However, in sensitive target fungi PAF acts in a dose-dependent manner and causes a linear growth reduction with increasing protein concentrations as documented with the fungal model N. crassa (Fig. 2 ). PAFB, instead, does not significantly alter the fungal proliferation within a wide concentration range until the protein amount is reached that effectively inhibits growth by 90% (IC90) (Fig. 2). The differences in the antimicrobial spectrum as well as in the dynamics of antifungal activity suggest different modes of interaction of PAF and PAFB with the fungal target cell.
Fig. 2.
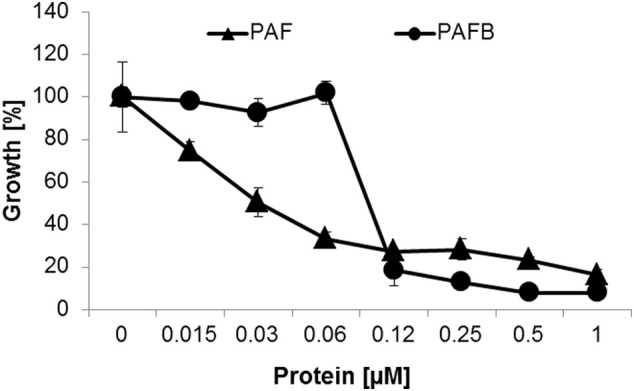
Growth inhibition of N. crassa by increasing concentrations of PAF and PAFB. The antifungal activity was tested in a microdilution broth assay by inoculating conidia (104/mL) with increasing concentrations of PAF or PAFB in undiluted Vogel's medium and incubation for 30 h at 25 °C without shaking. Growth was determined spectrophotometrically (FLUOstar Omega, BMG Labtech) measuring the optical density at λ = 620 nm. Values represent the mean ± SD (n = 3, technical triplicates) growth (%) in the presence of AMPs in comparison to the untreated control which was set to be 100%. The result of one representative experiment of two biological repeats is shown.
The risk of resistance development in fungal pathogens is greatly reduced with fungicidal agents [32]. Notably, PAF and PAFB show fungicidal potential on the wide-spread opportunistic human pathogen C. albicans killing planktonic cells and inhibiting biofilms (Fig. 3 ) [11,14].
Fig. 3.
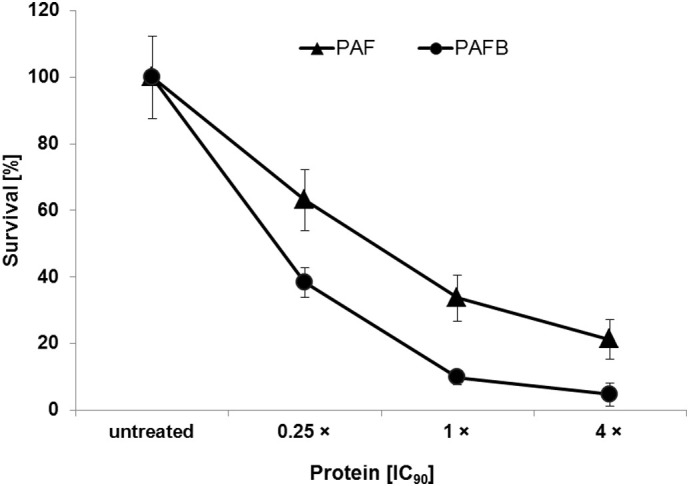
Killing of C. albicans exposed to PAF and PAFB. Planktonic cells (104/mL) were mixed with 0.25 ×, 1 × and 4 × the IC90 of PAF or PAFB in 0.1 × PDB and incubated for 6 h at 30 °C. Samples with appropriate dilutions were plated on 0.1 × PDB agar and the colony number was determined after 24 h of incubation at 30 °C. Values represent the mean ± SD (n = 3, technical triplicates) colony number (%) in the presence of AMPs in comparison to the number of colonies from the untreated control at time point 0 h, which was set to be 100%. The result of one representative experiment of two biological repeats is shown.
In addition to their antifungal activity, the P. chrysogenum AMPs also possess antiviral potential. Both proteins diminished the cytopathogenic effects of the Human Coronavirus in human cervix carcinoma cells [11]. To our best knowledge, this was the first report on the antiviral potential of small, cysteine-rich and cationic proteins from fungal origin. Investigations are in progress to study in more detail the antiviral spectrum and unravel the mechanism of action that impedes virus activity.
4. Structure-function relation of PAF and PAFB
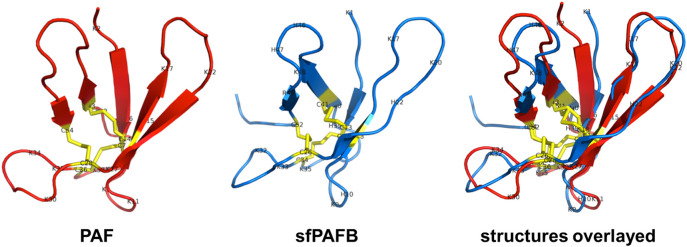
Understanding the mechanistic mode of action of AMPs requires detailed knowledge of their structural features. PAF and PAFB are produced as 92 aa long preproproteins. Both proteins contain a signal (pre)sequence that directs their secretion and a prosequence, which is suggested to prevent premature protein folding and activity [33]. A high degree of homology (56% aa identity) can be found within the preprosequence of PAF and PAFB. The pre- and prosequence are removed before or during protein secretion into the supernatant. The mature PAF and PAFB consist of 55 or 58 aa, respectively, with a high content of positively charged residues: PAF contains 13 lysines leading to a net charge of +4.7 (at pH 7), whereas PAFB contains 16 positively charged aa: eight lysines, two arginines and six histidines, which renders this protein slightly higher positively charged (+5.2 at pH 7) than PAF (Table 3 ). Importantly, N-terminal shorter variants of PAFB exist, which differ from the full-length protein by the lack of one (lysine) or two (serine/lysine) aa. The shortest N-terminal variant of PAFB (sfPAFB) served to determine its solution structure (Fig. 4 ) [11].
Table 3.
Amino acid sequences of P. chrysogenum AMPs and AMP variants and their physicochemical properties analyzed in silico.§
| Proteins and variants | MW$ [Da] |
pI | Net charge at pH 7 |
GRAVY* | No. positively charged aa | References |
|---|---|---|---|---|---|---|
| PAF wt | 6250.05 | 8.93 | +4.7 | −1.375 | 13 | [17] |
| AKYTGKCTKSKNECKYKNDAGKDTFIKCPKFDNKKCTKDNNKCTVDTYNNAVDCD | ||||||
| PAFopt | 6290.20 | 9.30 | +7.7 | −1.438 | 15 | [14] |
| PAFD19S | 6222.03 | 9.09 | +5.7 | −1.325 | 13 | [38] |
| PAFD53SD55S | 6194.02 | 9.22 | +6.7 | −1.276 | 13 | [39] |
| PAFY48Q | 6215.00 | 8.95 | +4.7 | −1.415 | 13 | [9] |
| PAFF31N | 6216.97 | 8.93 | +4.7 | −1.489 | 13 | [9] |
| PAFK9A | 6192.95 | 8.77 | +3.7 | −1.271 | 12 | [34] |
| PAFK35A | 6192.95 | 8.77 | +3.7 | −1.271 | 12 | [34] |
| PAFK38A | 6192.95 | 8.77 | +3.7 | −1.271 | 12 | [34] |
| PAFB wt | 6500.32 | 8.83 | +5.2 | −1.031 | 16 | [11] |
| LSKFGGECSLKHNTCTYLKGGKNHVVNCGSAANKKCKSDRHHCEYDEHHKRVDCQTPV | ||||||
| PAFBopt | 6546.52 | 9.51 | +9.9 | −1.236 | 19 | This study |
ExPASy ProtParam tool [26].
MW: Molecular Weight.
GRAVY: Grand Average of Hydropathy.
Protein Calculator v3.4 (The Scripps Research Institute; http://protcalc.sourceforge.net/) was applied for calculation of physicochemical properties. Red letters indicate aa exchanges.
Fig. 4.
Structure of PAF (PDB ID: 2mhv, red) and sfPAFB (PDB ID: 2nc2, blue) and structural alignment of both proteins. sfPAFB is a truncated variant of PAFB, shorter with two aa residues at the N-terminus. Antiparallel β-strands represented by arrows form two overlapping β-sheets. Consecutive strands are connected by short turns or longer loop regions. The positions of cysteines and cationic residues are indicated in one letter code. Disulfide bonds are marked in yellow. PyMol 1.4.1 (Schrödinger, Inc.) software was used for structure alignment.
For both AMPs the solution structures have been solved by nuclear magnetic resonance (NMR) spectroscopy [11,34,35], and for PAF the crystal structures complexed with sulfonato-calixarenes reflect the NMR structure (PDB ID: 2mhv) [36]. Compared to the NMR structures that are based on proton-proton distance measurements only, the X-ray structures more accurately define disulfide conformations. Indeed, the PAF X-ray structures (PDB ID: PAF-sclx4, 6 ha6; PAF-sclx6, 6hah; PAF-sclx8, 6hah) [37] exhibited lower energy disulfide conformers than the 2mhv NMR structure [35]. Similar to other members of the PAF cluster, PAF and PAFB adopt a Greek key super-secondary structure and five antiparallel β-strands, including the fifth amphipathic strand, are connected by four slightly flexible loops. The strands form two orthogonally packed β-sheets leading to a β-barrel topology, rendering the tertiary structure of PAF and PAFB very similar despite low sequence similarity (35.2%) (Fig. 4) [11]. Both proteins contain six cysteine residues, that form three intramolecular disulfide bridges with a common “abcabc” pattern [11,34]. This strongly stabilized structure with a central hydrophobic core around the disulfides results in resistance against adverse environmental conditions, e.g. proteolytic degradation, high temperature, chemical denaturants and extreme pH range [34] (PAFB; unpublished data). In PAFB, the side chains of Tyr 43 and Tyr 15 equipped with other apolar residues also contribute to the hydrophobic core. In the sfPAFB (PDB ID: 2nc2) the strands are connected by three small loops (1, 2, 4) and one large loop (3) forming the β-turns. The conformation of loop 3 shows significant differences between PAF and sfPAFB. The central part of loop 3 in sfPAFB includes three conserved lysines (Lys 36, 37, 39), and equipped with the N-terminal Lys 9, they contribute to a contiguous positively charged surface. Still, PAFB does not have a “belt” of aligned lysines as compared to PAF (Lys 6, 15, 42) and has a different electrostatic surface potential [11]. The loops exhibit a limited flexibility that might allow the interaction with so far unidentified fungal target molecules, e.g. membrane lipids and proteins. A breakthrough for the possibility of structural analysis of the P. chrysogenum AMPs was the development of a P. chrysogenum-based expression system. This tool uses the strong promoter of the paf-gene to produce cysteine-rich and cationic AMPs in a P. chrysogenum paf deletion strain (Δpaf), which provides the secretion of the recombinant proteins in high yields into the supernatant and the correct disulfide-bonding and protein folding. The use of defined minimal medium in combination with 15N and/or 13C isotopes allows uniform labelling of the proteins and easy one-step purification by ion-exchange chromatography avoiding impurities that hamper NMR-based analysis [9]. The functional importance of the loop sequences was demonstrated by creating the recombinant PAF variants PAFD19S, PAFF31N and PAFY48Q, in which distinct aa in loop 2 (position 19), loop 3 (position 31) and loop 4 (position 48) were substituted, respectively [9,38] (Table 3). All analyzed variants retained an overall 3D structure similar to PAF but lost antifungal activity (see Chapter 5). This shows that AMPs are finely tuned for their activity and selectivity, and neither the overall fold, nor the gross physical properties of a given sequence warrant biological impact.
4.1. The γ-core motif - functional or structural determinant?
PAF and PAFB contain a ten-aa long conserved consensus sequence in loop 1, the so-called gamma(γ)-core motif [11,14]. This motif is present in many disulfide-stabilized AMPs of prokaryotes and eukaryotes. When folded, it resembles the Greek letter “γ” and it is composed of the primary structure GXCX3-9C, for which a central role in AMP function has been reported [14,40]. Synthetic peptides derived from specific plant defensins that span this motif exhibit antimicrobial activity per se [41].
In agreement with these reports, a positively charged peptide (Pγ) (net charge +3.9 at pH 7) spanning the γ-core of PAF showed anti-Candida activity per se, whereby exhibiting reduced activity by 2.5-fold (IC90 10 μM) compared to PAF (IC90 4 μM) (Table 2, Table 4 ). Interestingly, the substitution of neutral aa in Pγ by positively charged and less hydrophobic ones resulted in the peptide Pγopt with eight-fold increased efficacy (IC90 1.3 μM) than Pγ and four times higher efficacy than PAF (Table 4) [14]. The improvement in antifungal activity could also be transferred to the full-length PAF when the aa in loop 1 were substituted by the same residues as those used for designing Pγopt. The resulting PAFopt had a four-fold higher anti-Candida activity than the native protein (Table 2, Table 3) [14]. Both, Pγopt and PAFopt counteracted Candida-biofilm formation more effectively than the unmodified peptide and protein [14]. These results underline the important role of the γ-core motif for the antifungal activity of PAF. They further demonstrate that the antifungal efficacy of a synthetic γ-core peptide and of the full-length AMP can be improved by rational design, increasing the net charge and the hydrophilicity in this conserved motif by specific aa exchanges.
Table 4.
In silico predicted physicochemical properties of γ-core motives derived from PAF and PAFB and their anti-Candida efficacy.
| Synthetic peptide | γ-core sequence§ | Net charge at pH 7# |
GRAVY$ | IC90 [μM]⁎ C. albicans |
References |
|---|---|---|---|---|---|
| Pγ | +1.8 | −1.56 | 10 | [14] | |
| Pγopt | +4.8 | −1.91 | 1.3 | [14] | |
| PBγ | +0.1 | −0.72 | >400 | [28] | |
| PBγopt | +4.8 | −1.91 | 1.3 | [28] |
Peptides were synthesized with –SH group of the cysteines being reduced. Red letters indicate aa exchanges.
Protein Calculator v3.4 (The Scripps Research Institute; http://protcalc.sourceforge.net/) was applied for net charge calculation.
GRAVY: Grand Average of Hydropathy.
IC90, AMP concentration inhibiting growth ≥90%.
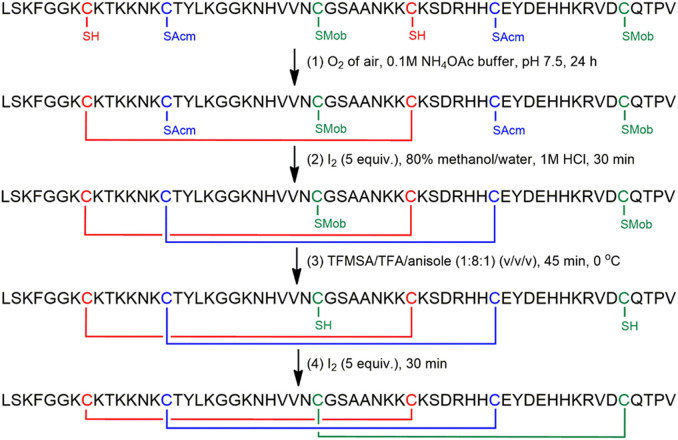
However, this observation is not necessarily true for all AMPs carrying a γ-core motif. A synthetic peptide spanning the γ-core of PAFB (PBγ) exhibited no activity against C. albicans when applied at concentrations of up to 400 μM [28]. It has to be noted here that the PBγ is nearly neutral (net charge +0.1 at pH 7) and less hydrophilic compared to Pγ. This could possibly explain its inactivity. An “optimized” version of PBγ (PBγopt) was generated by the exchange of distinct aa to positively charged and hydrophilic ones, leading to an increase in the net charge and to a reduction of the grand average of hydropathy (GRAVY). PBγopt exhibited anti-Candida activity at an IC90 of 1.3 μM (Table 4), which is similar to the IC90 of the full-length PAFB (Table 2). These results again underline that a high positive net charge and hydrophilicity result in a markedly higher antifungal efficacy. In contrast, our attempt to improve the PAFB efficacy by generating a PAFB variant with the primary structure of PBγopt in its γ-core was unsuccessful. The recombinant protein PAFBopt was readily degraded when expressed with the P. chrysogenum-based expression system [28]. It is likely that the γ-core of PAFB contributes to protein structure and stability. The exchange of certain aa in this sensitive region could have resulted in a different folding of the mutated protein PAFBopt and rendered it more susceptible for proteolysis. Therefore, PAFBopt was prepared stepwise, using microwave-assisted solid-phase peptide synthesis and fluorenylmethoxycarbonyl (Fmoc) chemistry. To ensure the naturally occurring “abcabc” pattern, disulfide bridges were formed in a regio-selective manner (Fig. 5 ) [42]. Sulfhydryl (SH) groups of the three pairs of cysteines were protected by trityl (Trt), acetamidomethyl (Acm) and methoxybenzyl (Mob) groups. Partially protected intermediates were isolated from the reaction mixtures after each step by reversed-phase high performance liquid chromatography (RP-HPLC). At the end of the synthesis, purification of PAFBopt was also performed by RP-HPLC.
Fig. 5.
Chemical synthesis of PAFBopt by regio-selective disulfide bond formation. The synthesized protein was detached from the solid support and protecting groups of the aa except Acm and Mob were removed by a trifluoroacetic acid (TFA)/water/dithiothreitol (95:5:3) (v/v/w) cleavage cocktail. The first disulfide bond was formed with O2 of air in a pH 7.5 buffer (1). Iodine treatment cleaved Acm and oxidized free thiols of the second pair of cysteines in one step (2). After cleavage of Mob by a trifluoromethanesulfonic acid (TFMSA)/TFA/anisole mixture (3), iodine was used to form the third disulfide bond (4).
Unexpectedly, the synthetic PAFBopt exhibited no improvement; rather a severe reduction in antifungal efficacy was observed when compared to that of the native PAFB. PAFBopt acted at a slightly increased IC90 (2 μM) against C. albicans when compared to PAFB and showed no activity against A. fumigatus (IC90 > 400 μM), which underlines a role in structure and stability rather than a functional role of the γ-core in PAFB. To address the hypothesis that the PAFB γ-core contributes to protein stability, we exposed PAFB and the synthetic PAFBopt to conditioned cell-free supernatants of 48 h cultures of C. albicans and A. fumigatus. The treated AMPs were then size fractionated on SDS-polyacrylamide gels to visualize any proteolytic degradation that might have occurred during fungal incubation. Indeed, PAFBopt was readily degraded when exposed to the cell-free A. fumigatus supernatant, whereas only a mild degradation - if at all - was observed with samples exposed to the C. albicans supernatant (Fig. 6 ). Obviously, A. fumigatus secretes a different set of proteases into the culture broth than C. albicans. The result explains the loss of activity of PAFBopt against A. fumigatus. PAFB, in contrast, showed no signs of proteolytic degradation when incubated with the supernatant of any of these fungal cultures, which underlines its higher stability against proteolysis (Fig. 6).
Fig. 6.

Stability testing of PAFB wt and PAFBopt in cell-free supernatants of A. fumigatus and C. albicans. Spores or yeast cells (104/mL) were grown for 48 h in 0.05 × PDB medium as stationary cultures at 37 °C and 30 °C, respectively. The hyphae and yeast cells were removed by centrifugation and 100 μL of the cell free supernatant were transferred to an Eppendorf tube together with 100 μg/mL of PAFB wt and PAFBopt. The samples were incubated for 1 h at 30 °C (C. albicans supernatant) and 37 °C (A. fumigatus supernatant). The proteins incubated in water served as controls (control). Samples were loaded in 10 μL aliquots (corresponding to 1 μg protein per sample) on a 18% (w/v) SDS polyacrylamide gel and size fractionated. Silver staining was performed to visualize the proteins.
According to other reports, the role of the γ-core motif indeed differs between distinct AMPs. For example, peptides spanning the γ-core of AMPs from Penicillium digitatum (AfpB) or N. fischeri (NFAP2) had no antifungal activity indicating that this motif is not important for the antifungal mode of action of these AMPs [43,44]. Similarly, a peptide spanning the γ-core of the plant defensin MsDef1 from Medicago sativa was inactive against Fusarium graminearum, whereas the γ-core peptide derived from the plant defensin MtDef4 from Medicago truncatula inhibited the growth of this plant pathogen at very low concentrations [41]. Notably, these two γ-core peptides significantly differ from each other with regard to primary structure and net charge, e.g. MtDef4 showing a higher positive net charge (+4.8 at pH 7) than MsDef1 (+0.9 at pH 7) [41].
Interestingly, the γ-core motifs in PAFB, AfpB and NFAP2 all have a lower positive net charge as compared to PAF. Functional mapping of AfpB [43,44] and NFAP2 [44] assigned an antifungal role to protein parts other than this specific motif. Thus, the γ-core motifs of AfpB and NFAP2 were proposed to be structurally important for folding and protein stability [43,44]. We therefore conclude that the γ-core motif fulfills different functions in the P. chrysogenum AMPs, contributing to folding and stability in PAFB and antimicrobial activity in PAF.
5. Mode of action
5.1. The medium composition determines fungal susceptibility
PAF and PAFB exhibit anti-yeast activity only in 0.1 × PDB (Table 2, Table 5 ), whereas no growth inhibitory potential was detected in other media tested, e.g. diluted yeast-extract peptone dextrose (0.1 × YPD) medium (Table 5). This could be explained with compounds of the medium that may (i) directly interact with the AMPs and reduce their activity; (ii) inhibit the interaction of the AMP with the fungal target molecule by competing for the binding site; or (iii) influence the physiology of the fungal target cell by regulating the presence/activity of interaction molecules located for example on the cell surface (cell wall, membrane). In a control experiment we tested the susceptibility of the PAF and PAFB sensitive filamentous ascomycete A. niger in 0.1 × PDB and 0.1 × YPD. Both AMPs were active in the two media, which excludes the inactivation by media compounds binding to the proteins (data not shown). Instead, the accessibility, presence and/or activity of susceptibility determinants for PAF and PAFB in fungi might be influenced by the medium composition.
Table 5.
Susceptibility of C. albicans towards PAF and PAFB in different media.
IC90, AMP concentration inhibiting growth ≥90%; Yeast cells (104/mL) were grown in the presence of increasing AMP concentrations (0–32 μM) for 40 h at 30 °C in 0.1 × PDB or 0.1 × YPD, respectively. Values represent % growth in the presence of AMPs in comparison to the untreated control which was set to be 100% growth. All values are given as mean ± SD (n = 3).
As it is true for many AMPs from different origin [38,[45], [46], [47]], the antifungal activity of PAF and PAFB is strongly influenced by the ionic strength of the medium. Among monovalent and divalent ions, Ca2+ is the most effective ion that interferes with the activity of these two AMPs. High extracellular Ca2+ reduce the antifungal activity of both AMPs in a concentration dependent manner [39,48] (PAFB, unpublished data). This raised the question whether Ca2+-binding modulates AMP activity. Recently we could show that the C-terminal residues Asp53 and Asp55 in PAF indeed specifically bind Ca2+, but this protein-ion interaction does not affect the antifungal activity [39]. The relevance of the C-terminal Ca2+-binding remains unknown. Notably, low Ca2+ concentrations in the medium are necessary for the growth inhibitory activity of PAF, because Ca2+ deprivation by the extracellular Ca2+-specific chelator BAPTA-AM counteracts PAF function [48,49].
5.2. The uptake of PAF and PAFB is a prerequisite for fungal cell death
Little is known about the role of the fungal cell wall in AMP function. The PAF/PAFB related AMP AFP from A. giganteus was shown to interact with the fungal cell wall first and then with the cell membrane [50]. This is supported by a strong in vitro binding capacity of AFP to chitin and the activation of the protein kinase C (PKC)-dependent cell wall integrity pathway as compensatory response in sensitive fungi exposed to AFP [34,51,52]. In contrast, the structural properties of PAF did not suggest any chitin binding ability [34]. Further studies revealed that PAF negatively interferes with PKC/mitogen-activated protein kinase (Mpk) signaling and does not activate the cell wall integrity pathway [53]. To substantiate these findings, in vitro activity assays with N. crassa using culture medium supplemented with β-1,3-glucan (a major cell wall polysaccharide in filamentous fungi) or glucosamine (building units of chitin) did not hamper the antifungal activity of PAF and PAFB (unpublished data). Therefore, the polysaccharide components of the cell wall do not seem to be the primary binding targets for these two P. chrysogenumderived proteins and their retention by the cell wall is unlikely.
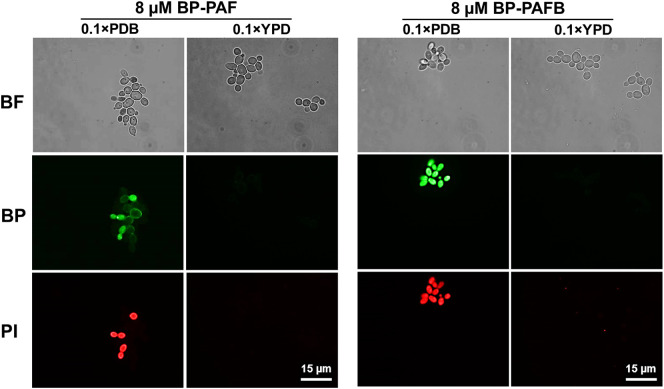
Our studies further revealed that the binding of PAF and PAFB to the fungal membrane is not sufficient to induce cell death, but that these AMPs need to be taken up into the fungal cell. This can be visualized by PAF and PAFB labelled with the fluorescent dye BODIPY (BP-PAF and BP-PAFB) [11,38]. Only those cells that have internalized the AMPs also appear positively stained with the cell death dye propidium iodide (Fig. 7 ).
Fig. 7.
Cellular localization of PAF and PAFB and cell death induction in C. albicans in different media. Yeast cells were shaken at 200 rpm for 3 h at 30 °C in 0.1 × PDB or 0.1 × YPD in the presence of 8 μM BODIPY-labelled PAF and PAFB. Co-staining with 5 μg/mL propidium iodide was performed for 10 min before imaging. Images were taken with the same exposition time (1.500 ms). BF, Brightfield; BP, BODIPY-labelled proteins; PI, Propidium iodide.
The uptake of PAF and PAFB is a regulated mechanism, which can be inhibited by reducing the metabolism or by blocking oxidative phosphorylation of the fungal cell. Both suggest an endocytic uptake in sensitive fungi [11,54]. This mechanism was investigated in more detail with BP-PAF. Conidia bind PAF in the outer layers without internalizing the protein, but as soon as they germinate, the protein is taken up and kills the growing hyphae in a concentration and time dependent manner [38]. This indicates that only actively growing fungal cells dispose of PAF susceptibility determinants for its internalization. Most probably, PAF is retained in the conidial cell wall because of its more complex and different cell wall composition compared to that of hyphal cells [55]. In contrast, the binding of PAF and PAFB to the fungal cell is impaired and no protein uptake occurs (i) in AMP resistant fungal strains [54,56]; (ii) when sensitive fungi are exposed to inactive PAF variants [38] or (iii) the medium composition affects the efficacy of the proteins as described in Chapter 5.1. (Fig. 7) [11,56]. In all these cases the cells are not killed, as evidenced by the lack of a propidium iodide positive signal.
After internalization, PAF and PAFB are compartmentalized, most probably in vacuoles, which are part of the endocytic pathway. In contrast to the A. giganteus AFP [57] or the plant defensin Psd1 from Pisum sativum [58], none of the P. chrysogenum AMPs localize in the fungal nuclei [34] (PAFB, data not shown). As long as the proteins remain in these compartments, the fungal cells show no signs of cell death. Fungal killing happens when the proteins localize in the cytoplasm, which coincides with the membrane disintegration and uptake of the otherwise membrane impermeable dye propidium iodide [56]. Finally, the damage of the fungal plasma membrane seems to be a secondary effect induced by PAF and PAFB. The mechanistic mode of action strongly resembles that described for the small, synthetic antifungal hexapeptide PAF26 [59,60] and specific plant defensins [61,62].
5.3. PAF and PAFB are not canonical membrane-active AMPs
Many antifungal proteins, such as plants defensins, were reported to interact with lipids of fungal plasma membranes [63]. Thereby, the electrostatic interaction of these cationic proteins with the anionic fungal membrane lipids is suggested. In agreement with this assumption, the substitution of lysines (Lys 9, 35, 38) by alanines reduced the antifungal activity of PAF against A. niger, most probably because the positive net charge was lowered in the protein variants PAFK9A, PAFK35A and PAFK38A (Table 3) [34].
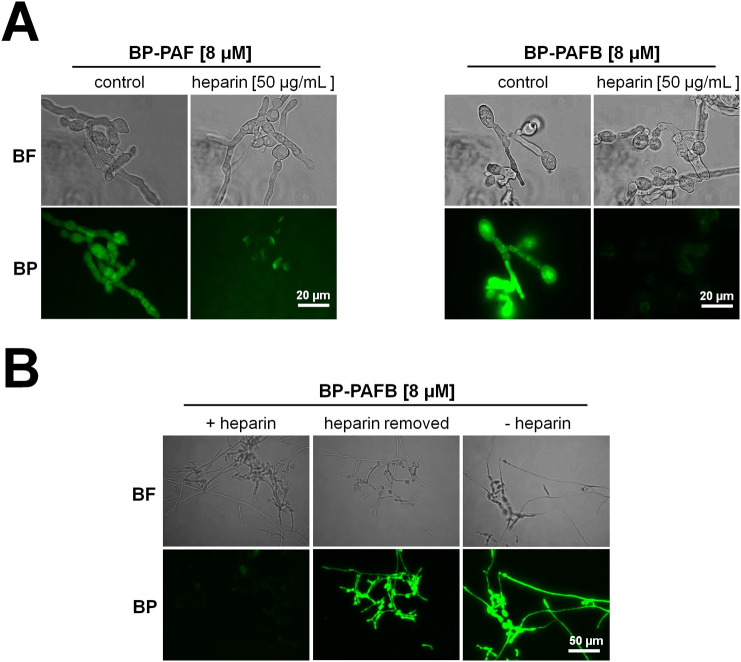
To further confirm the importance of an electrostatic interaction of PAF and PAFB with the fungal cell membrane, broth microdilution assays were performed with N. crassa exposed to PAF or PAFB in combination with the polysaccharide heparin. Heparin is a linear polysaccharide consisting of repetitive units of pyranosyluronic acid and glucosamine residues. Due to the high content of sulfo and carboxyl groups, heparin has an average of 2.7 negative charges per disaccharide. Therefore, the interaction of heparin with the cationic aa of proteins, such as lysines or arginines possibly organized in clusters on the protein surface, is assumed to be electrostatically driven [64]. To test this hypothesis, we expected heparin to bind to PAF and PAFB and mask their positive charge, resulting in the loss of function. Indeed, heparin increased the IC90 value of PAF four-fold and that of PAFB even more than 30-fold (Table 6 ). Additionally, fluorescent microscopy revealed a significantly reduced interaction (binding and uptake) of both BODIPY-labelled proteins with the fungal cell in the presence of heparin (Fig. 8A). In a control experiment, heparin was added to N. crassa germlings and then removed by washing the grown hyphae three-times with fresh medium before exposure to BP-PAFB (Fig. 8B). The microscopic evaluation revealed that PAFB retained its antifungal activity. This control experiment confirmed our assumption that heparin binds to positively charged AMPs but does not mask susceptibility determinants of N. crassa (Fig. 8B).
Table 6.
Susceptibility of N. crassa towards PAF and PAFB in the presence of heparin.
| Compound | IC90 [μM]⁎ |
|---|---|
| PAF | 0.06 |
| 50 μg/mL heparin | n.d. |
| PAF + 50 μg/mL heparin | 0.25 |
| PAFB | 0.12 |
| 50 μg/mL heparin | n.d. |
| PAFB +50 μg/mL heparin | 4 |
IC90, AMP concentration inhibiting growth ≥90%; The antifungal activity of PAF and PAFB in the presence of heparin was tested in a microdilution broth assay. N. crassa conidia (104/mL) were grown in 0.2 × Vogel's medium under increasing PAF (0–0.25 μM) or PAFB (0–4 μM) concentrations combined with heparin (50 μg/mL) for 30 h at 25 °C. n.d. not determined as no growth inhibition is observed.
Fig. 8.
Susceptibility of N. crassa towards PAF and PAFB in the presence of heparin. (A) Uptake of PAF and PAFB: N. crassa conidia (5 × 105/mL) were grown for 4 h in 0.2 × Vogel's medium (25 °C, shaking at 200 rpm). 8 μM BODIPY-labelled PAF (BP-PAF) and PAFB (BP-PAFB) were added alone or in combination with 50 μg/mL heparin to the germlings and samples were further incubated for 2.5 h at 25 °C until microscopic evaluation. (B) Control experiments: N. crassa conidia (5 × 105/mL) were grown for 4 h in 0.2 × Vogel's medium in the presence of 50 μg/mL heparin or without heparin. Then, all samples were washed three-times with fresh medium and 8 μM BP-PAFB was added to the germlings alone (heparin removed and - heparin) or in combination with 50 μg/mL heparin (+ heparin). The samples were further incubated for 2.5 h at 25 °C until microscopic evaluation. Images were taken with the same exposition time (1.500 ms). BF, Brightfield; BP, BODIPY-labelled proteins.
Zeta-potential measurements with large unilamellar vesicles (LUVs) generated from membrane lipid extracts derived from N. crassa confirmed the binding capacity of PAF and PAFB with the anionic fungal membrane lipids via electrostatic interaction, whereby the affinity of PAFB for LUVs was higher than that of PAF, most probably because of its overall higher positive net charge and/or the presence of a positively charged surface area (see Chapter 4) [56]. In contrast, none of the two AMPs were able to neutralize the negative Zeta-potential of LUVs derived from a N. crassa mutant defective in glycosylation of ceramides (Δgcs), proposing sugar moieties to be important for high affinity binding [56]. This parallels with in silico molecular dynamics simulation data of the A. giganteus AFP that propose the formation of stable salt bridges and hydrogen bonds between the negatively charged glycosylinositol phosphorylceramides in the fungal membrane with the positively charged protein [65].
In addition to the electrostatic attraction of PAF and PAFB by LUVs generated from fungal membranes, the interaction of these two AMPs seems to necessitate additional binding partners (e.g. receptors in the membrane and/or intracellular molecules). This is corroborated by the fact that PAF, but not PAFB, acts in a glucosylceramide dependent manner as demonstrated by the susceptibility testing of N. crassa mutants defective in distinct enzymatic steps of this synthesis pathway [56]. Furthermore, PAF variants carrying aa substitutions that neither change the net charge nor influence the overall solution structure, e.g. PAFF31N and PAFY48Q, lost their antifungal activity [9]. This was also found to be the case for the mutant PAFD19S with similar PAF structure, but increased net charge (+5.7 at pH 7) [38]. We therefore favor the assumption that the glucosylceramide depletion affects the correct sorting, distribution or activity of lipids and/or membrane proteins that could be putative interaction molecules of PAF. Furthermore, ceramides function as second messengers in fungal signal transduction pathways responsive to environmental stimuli. Therefore, we cannot exclude that the observed phenotypes resulted from deregulated signaling in the N. crassa mutants in response to PAF [66].
The killing mechanism of many AMPs that target the microbial plasma membrane starts with the interaction of the AMPs with the lipid matrix followed by the subsequent permeabilization or disintegration of the membrane [67]. This has been related to numerous AMPs, most of them with antibacterial function [68].
So far, all our studies indicated that fungal killing by PAF and PAFB does not occur through membrane disruption in the first place. Leakage experiments using LUVs composed of the major phospholipid constituents of eukaryotic membranes, palmitoyl-oleoyl-phosphatidylcholine (POPC) and mixtures of POPC/phosphatidylinositol, POPC/ergosterol, POPC/ceramide and POPC/sphingomyelin, were performed according to [69] to investigate the membrane disruption potential of PAF and PAFB.
These assays, however, revealed that PAFB does not cause membrane leakage, independent from the lipid composition of the LUVs. PAF induced only 10–20% leakage at a lipid to peptide ratio of 4:1 and 2:1 in POPC vesicles and in all other tested lipid combinations, respectively (data not shown). However, these protein concentrations were dramatically higher than the concentration range normally applied with classical membrane-disruptive AMPs (molar ratios lipid:peptide = 400:1 to 25:1; [67]), though the degree of disruption strongly depends on the mechanistic mode of action. As in all tested lipid vesicles, POPC was present, suggesting that PAF interacted with PC. In order to reappraise the observed leakage assay data, differential scanning calorimetry (DSC) was carried out using dipalmitoyl phosphatidylcholine (DPPC) as described before [67]. This method enables to observe phase transition (Tm) behavior of phospholipid membranes, which represents a very sensitive method for the detection of any interaction with AMPs [70]. Upon incubation of DPPC with PAFB at a lipid to peptide ratio of 25:1, a slight shift of Tm from 41.2 °C to 41.4 °C accompanied by a 20–30% increase in enthalpy was observed (Fig. 9 ). According to DSC data analysis described and studied by Lohner 2016 [70], an increased phase transition temperature and enthalpy points towards stabilization of the gel phase due to higher lipid packing and ordering. Amongst others, this can be a consequence of shielding the surface charge or dehydration of the phospholipid head groups without damaging the membrane. In the case of PAF, the opposite behavior was observed: a Tm decrease from 41.2 °C to 40.8 °C without changes in enthalpy (Fig. 9). The decrease in Tm refers to destabilization of the membranes and this may explain the low leakage induced by PAF. Furthermore, the pre-phase transition temperature of DPPC at 35 °C was shifted to 33 °C in the presence of PAF. The pre-phase transition temperature is particularly sensitive to interaction of peptides with the lipid matrix [70].
Fig. 9.
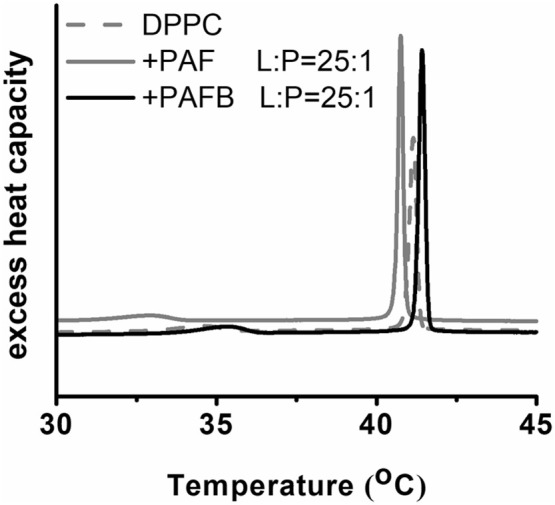
Impact of PAF and PAFB on the thermotrophic behavior. DSC scan of DPPC in absence and presence of PAF and PAFB as observed by heating scans. The concentration of PAF and PAFB is indicated in relation to lipid to peptide (L:P) molar ratio of 25:1.
In summary, the mild effects on membrane stabilization or destabilization may explain no or negligible leakage or peptide-induced temporal reorganization of the membrane, which is in contrast to the damage induced by canonical membrane-active peptides. For comparison, the antimicrobial peptide OP-145 induced 20–30% leakage of POPC vesicles at a lipid to peptide ratio of 25:1–12.5:1 [67]. In this concentration range, DSC recorded DPPC phase transition was almost lost and DPPC membranes were completely destroyed in a detergent-like manner as confirmed via X-ray techniques by the presence of small disk like particles [67]. Thus, the results obtained with PAF/PAFB and model membranes do not support any strong membrane disruptive activities.
The fungal membrane lipid composition can have a dramatic effect on the susceptibility of fungi to PAF and PAFB. N. crassa mutants defective in the synthesis of the sphingolipid glucosylceramide are resistant to PAF, whereas the susceptibility to PAFB is not affected by the respective gene deletions [56]. However, it remains largely elusive to which extent the treatment with these two compounds impacts the membrane lipid composition itself. Application of PAF and PAFB in doses corresponding to 4 × IC90 did not cause significant alteration in class-wise normalized lipid profiles. Instead, a significantly elevated ceramide content could be observed in PAFB treated cells, while there was a consistent trend towards a reduction of total cardiolipin, phosphatidylcholine and phosphatidylethanolamine content in N. crassa (Fig. 10 ). While the latter effect could potentially be explained by the inhibition of growth caused by PAF and PAFB activity, the specific increase on ceramide levels showed that the interaction between antifungal proteins and this lipid class can be a key to understand the exact molecular mechanism of action responsible for their potency, which should be addressed in detail in future studies.
Fig. 10.
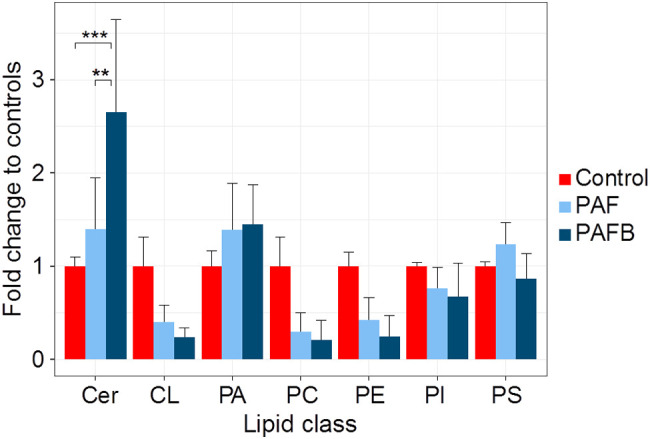
Impact of PAF and PAFB treatment on the phospholipid composition of N. crassa. Conidia (107) were inoculated in 20 mL Vogel's medium for 18 h (25 °C, shaking at 200 rpm). Hyphae were then treated with 4 × IC90 of PAF (6 μM) and PAFB (3 μM) for additional 3 h until harvesting for total lipid extraction and phospholipidomics analysis. Sample preparation and LC-MS/MS measurement were performed as described in [56] in negative ESI mode. Different lipid features were quantified, normalized to total protein content and summed per class (Cer: ceramide; CL: cardiolipin; PA: phosphatidic acid; PC: phosphatidylcholine; PE: phosphatidylethanolamine; PI: phosphatidylinositol; PS: phosphatidylserine). The obtained values were normalized to the respective mean control values. CLs, PCs and PEs show a trend to be reduced by PAF and PAFB treatment, while the total ceramide content was significantly increased when treated with PAFB compared to PAF and the controls. Values are shown as mean ± SD (n = 3). Normality of fold changes was tested with Shapiro-Wilk test class wise (p > 0.05, Holm multiple testing adjustment). Homogeneity of variances of the changes by class and treatment was assessed by Levene's test (p = 0.638). Fold changes were found significantly associated with treatment and lipid class, F(12, 42) = 5.517, p < 0.001. Post-hoc Tukey HSD analysis was applied, and p-values were adjusted for multiple testing (**: p = 0.01; ***: p = 0.001).
Thus, the results confirm our observations that the interaction of PAF and PAFB with the fungal cell membrane does not result in killing of the sensitive organism by membrane disruption in the first place.
5.4. The fungal killing mechanism is complex and regulated
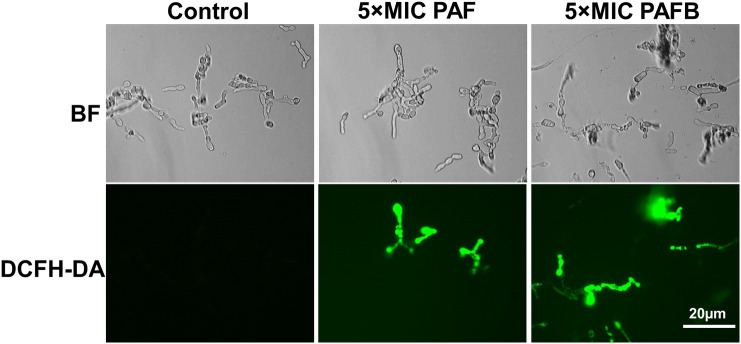
Instead, we propose a more complex and regulated mechanism of action based on our detailed investigations with PAF. An immediate response upon exposure of A. nidulans to this AMP includes the hyperpolarization of the plasma membrane at the hyphal tips and the efflux of potassium ions into the supernatant [71]. In N. crassa and A. niger a fast influx of extracellular Ca2+ was detected resulting in a sustained elevation of the cytoplasmic Ca2+ concentration [38,48,49]. The induction of intracellular reactive oxygen species (ROS) was detected in PAF- and PAFB-treated fungi by applying an indicator for oxidative radicals (2′,7′-dichlorofluorescein diacetate) (Fig. 11 ) [8,45]. The permeabilization of the fungal membrane for ions, the deregulation of the intracellular Ca2+ homeostasis and the elevation of the ROS burden in the fungal cell are recognized triggers for apoptotic cell death. Indeed, apoptotic markers, like phosphatidylserine exposure on the cell surface, DNA strand breaks, mitochondrial disintegration and membrane damage, were detected in A. nidulans exposed to PAF [71].
Fig. 11.
ROS induction by PAF and PAFB in N. crassa. Conidia (5 × 105/mL) were grown for 4.5 h in 0.2 × Vogel's medium at 25 °C as stationary cultures. Germlings were treated with 5 × IC90 PAF or 5 × IC90 PAFB for 1.5 h, respectively. ROS production was detected by incubating the samples with 10 μM of the fluorescent dye 2′,7′-dichlorofluorescin diacetate for 30 min. Images were taken with the same exposition time (1.500 ms). BF, Brightfield; DCFH-DA, 2′,7′-dichlorofluorescin diacetate.
In conclusion, the growth inhibitory and fungicidal effect of PAF and PAFB is a complex process that elicits different timely regulated responses in the fungal target cell. The identification of the exact mechanism of interaction with the fungal plasma membrane and the specific intracellular interaction molecule(s) of both P. chrysogenum AMPs still requires our utmost attention.
6. Impact on mammalian hosts
To guarantee a safe applicability of AMPs to treat fungal infections in humans and plants, any detrimental and cytotoxic effects have to be excluded. For PAF and PAFB neither cytotoxic effects on human epithelial cells L132 nor hemolytic activity on sheep erythrocytes were observed [11,14]. The in vitro cytotoxicity of PAF has been more extensively studied in the past than that of PAFB. PAF caused no detectable cytotoxic effects on endothelial cells from the umbilical vein and only had minor pro-inflammatory activity as assessed by the quantification of interleukin (IL)-6, IL-8, and TNF-α production. Furthermore, no modification of voltage-gated K+ channels of neurons, skeletal muscle fibers and astrocytes were detected [72]. Similar results were obtained for the A. giganteus AFP [73].
Experiments were carried out to investigate the harmlessness of PAF in an in vivo murine model and to give insight in its safe applicability. These studies revealed no evident change in the histology of mouse tissues and skin after PAF administration [74]. Furthermore, Palicz and co-workers (2016) showed in a follow-up study that PAF was similarly effective as amphotericin B and mildly prolonged the survival of a mouse model suffering from invasive pulmonary aspergillosis when administered intraperitoneally. Notably, the efficacy could be significantly increased when PAF was applied in combination with amphotericin B [75].
Another example further underlines the most promising applicability of AMPs from filamentous fungi. The N. fischeri NFAP2 inhibits the growth of Candida spp. without harming primary human skin cells. It effectively reduced the fungal burden of a fluconazole-resistant C. albicans strain in a murine vulvovaginitis infection model when applied alone. Similar to the findings with PAF, the efficacy could also be further improved in this model by combining NFAP2 with fluconazole [76]. Other examples for the combinatorial efficacy of AMPs with licensed drugs, e.g. caspofungin, against C. albicans biofilms are the plant defensin HsAFP1 from Heuchera sanguinea and the radish defensins RsAFP1 and RsAFP2 [77,78].
These results promise low - if at all - side effects of AMPs from filamentous fungi and a safe applicability as mono- or polytherapeutic agents with acceptable compatibility for the host.
7. Future perspectives
Up to date none of the small, cationic, antifungal proteins from ascomycetes have been applied biotechnologically or therapeutically in large scale. However, their growth inhibitory and fungicidal activity against human and plant pathogenic fungi as well as their efficacy against Candida biofilm formation renders them most promising biomolecules for the development of new antifungal treatment strategies. By the use of our recently developed P. chrysogenum-based expression system AMPs can be secreted in high abundance into the supernatant of easily fermentable molds [9,[79], [80], [81], [82]]. The secretion and accumulation of the proteins in the culture broth of fungi grown in mineral minimal medium enables fast and low-cost protein purification. This system also allows the production of recombinant AMP variants with improved efficacy that are created by rational design [14]. In this respect PAF and PAFB represent valuable model biomolecules that could inspire the research of AMPs from other filamentous ascomycetes that still await their identification.
The high stability of PAF and PAFB at high temperatures, against protease degradation and within a wide pH range renders them suitable therapeutics for use in clinical treatments, plant and food protection [34,83] (PAFB, unpublished data).
Especially a topical application to treat superficial fungal infections caused by Candida spp. or dermatophytes could be promising, whereby the combination with licensed drugs such as fluconazole proved to be highly effective in inhibiting the growth of Microsporum spp. and Trichophyton spp. [84]. Another possible application could be the constitutive expression of AMPs in transgenic plants, which would contribute to an improved plant “immune system” and defense towards plant pathogenic fungi. Adding preservatives to food is a common way to prevent food spoilage and poisoning of food products by fungi. As naturally secreted compounds, AMPs such as PAF or PAFB could be advantageous options as new food preservatives.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors want to thank Karl Lohner for the helpful discussion and Sándor Kocsubé for advices in the phylogenetic analysis. Research has been funded to F.M. by the Austrian Science Fund (FWF P25894-B20, I1644-B20, I3132-B21), and to L.G. by the Postdoctoral Excellence Program (PD 131340), the bilateral Austrian-Hungarian Joint Research Project (ANN 131341) of the Hungarian National Research, Development and Innovation (NKFI) Office. Present work of L.G. is supported by ÚNKP-19-4 New National Excellence Program of the Ministry for Innovation and Technology. L.G. has been supported by the János Bolyai Research Scholarship of the Hungarian Academy of Sciences. Structural research was supported by the EU and co-financed by the European Regional Development Fund under the projects GINOP-2.3.2-15-2016-00008 to G.B. and GINOP-2.3.3-15-2016-00004 (access to 700 MHz NMR facilities). Synthetic work was supported by GINOP-2.3.2-15-2016-00014 and 20391-3/2018/FEKUSTRAT of the Hungarian National Research, Development and Innovation (NKFI) Office, and by TUDFO/47138-1/2019-ITM FIKP of the Ministry for Innovation and Technology (ITM).
References
- 1.Tudela J.L.R., Denning D.W. Recovery from serious fungal infections should be realisable for everyone. Lancet Infect. Dis. 2017;17:1111–1113. doi: 10.1016/S1473-3099(17)30319-5. [DOI] [PubMed] [Google Scholar]
- 2.Brown G.D., Denning D.W., Gow N.A., Levitz S.M., Netea M.G., White T.C. Hidden killers: human fungal infections. Sci. Transl. Med. 2012;4 doi: 10.1126/scitranslmed.3004404. [DOI] [PubMed] [Google Scholar]
- 3.Gupte M., Kulkarni P., Ganguli B.N. Antifungal antibiotics. Appl. Microbiol. Biotechnol. 2002;58:46–57. doi: 10.1007/s002530100822. [DOI] [PubMed] [Google Scholar]
- 4.Barker K.S., Rogers P.D. Recent insights into the mechanisms of antifungal resistance. Curr. Infect. Dis. Rep. 2006;8:449–456. doi: 10.1007/s11908-006-0019-3. [DOI] [PubMed] [Google Scholar]
- 5.Edwards S.G. Influence of agricultural practices on fusarium infection of cereals and subsequent contamination of grain by trichothecene mycotoxins. Toxicol. Lett. 2004;153:29–35. doi: 10.1016/j.toxlet.2004.04.022. [DOI] [PubMed] [Google Scholar]
- 6.Hegedűs N., Marx F. Antifungal proteins: more than antimicrobials? Fungal Biol. Rev. 2013;26:132–145. doi: 10.1016/j.fbr.2012.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer V., Jung S. Antifungal peptides of the AFP family revisited: are these cannibal toxins? Microorganisms. 2018;6:50. doi: 10.3390/microorganisms6020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marx F. Small, basic antifungal proteins secreted from filamentous ascomycetes: a comparative study regarding expression, structure, function and potential application. Appl. Microbiol. Biotechnol. 2004;65:133–142. doi: 10.1007/s00253-004-1600-z. [DOI] [PubMed] [Google Scholar]
- 9.Sonderegger C., Galgóczy L., Garrigues S., Fizil Á., Borics A., Manzanares P., Hegedűs N., Huber A., Marcos J.F., Batta G., Marx F. A Penicillium chrysogenum-based expression system for the production of small, cysteine-rich antifungal proteins for structural and functional analyses. Microb. Cell Factories. 2016;15:192. doi: 10.1186/s12934-016-0586-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marx F., Binder U., Leiter E., Pocsi I. The Penicillium chrysogenum antifungal protein PAF, a promising tool for the development of new antifungal therapies and fungal cell biology studies. Cell. Mol. Life Sci. 2008;65:445–454. doi: 10.1007/s00018-007-7364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber A., Hajdu D., Bratschun-Khan D., Gáspári Z., Varbanov M., Philippot S., Fizil Á., Czajlik A., Kele Z., Sonderegger C., Galgóczy L., Bodor A., Marx F., Batta G. New antimicrobial potential and structural properties of PAFB: a cationic, cysteine-rich protein from Penicillium chrysogenum Q176. Sci. Rep. 2018;8:1751. doi: 10.1038/s41598-018-20002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee D.G., Shin S.Y., Maeng C.Y., Jin Z.Z., Kim K.L., Hahm K.S. Isolation and characterization of a novel antifungal peptide from Aspergillus niger. Biochem. Bioph. Res. Commun. 1999;263:646–651. doi: 10.1006/bbrc.1999.1428. [DOI] [PubMed] [Google Scholar]
- 13.Tóth L., Kele Z., Borics A., Nagy L.G., Váradi G., Virágh M., Takó M., Vágvölgyi C., Galgóczy L. NFAP2, a novel cysteine-rich anti-yeast protein from Neosartorya fischeri NRRL 181: isolation and characterization. AMB Express. 2016;6:75. doi: 10.1186/s13568-016-0250-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonderegger C., Váradi G., Galgóczy L., Kocsubé S., Posch W., Borics A., Dubrac S., Tóth G.K., Wilflingseder D., Marx F. The evolutionary conserved γ-core motif influences the anti-Candida activity of the Penicillium chrysogenum antifungal protein PAF. Front. Microbiol. 2018;9:1655. doi: 10.3389/fmicb.2018.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galgóczy L., Virágh M., Kovács L., Tóth B., Papp T., Vágvölgyi C. Antifungal peptides homologous to the Penicillium chrysogenum antifungal protein (PAF) are widespread among Fusaria. Peptides. 2013;39:131–137. doi: 10.1016/j.peptides.2012.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Wnendt S., Ulbrich N., Stahl U. Molecular cloning, sequence analysis and expression of the gene encoding an antifungal-protein from Aspergillus giganteus. Curr. Genet. 1994;25:519–523. doi: 10.1007/bf00351672. [DOI] [PubMed] [Google Scholar]
- 17.Marx F., Haas H., Reindl M., Stöffler G., Lottspeich F., Redl B. Cloning, structural organization and regulation of expression of the Penicillium chrysogenum paf gene encoding an abundantly secreted protein with antifungal activity. Gene. 1995;167:167–171. doi: 10.1016/0378-1119(95)00701-6. [DOI] [PubMed] [Google Scholar]
- 18.Olsen J.G., Flensburg C., Olsen O., Bricogne G., Henriksen A. Solving the structure of the bubble protein using the anomalous sulfur signal from single-crystal in-house Cu Kalpha diffraction data only. Acta Crystallogr. D. Biol. Crystallogp. 2004;60:250–255. doi: 10.1107/S0907444903025927. [DOI] [PubMed] [Google Scholar]
- 19.Jami M.S., Barreiro C., García-Estrada C., Martín J.F. Proteome analysis of the penicillin producer Penicillium chrysogenum: characterization of protein changes during the industrial strain improvement. Mol. Cell. Proteomics. 2010;9:1182–1198. doi: 10.1074/mcp.M900327-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovács L., Virágh M., Takó M., Papp T., Vágvöelgyi C., Galgóczy L. Isolation and characterization of Neosartorya fischeri antifungal protein (NFAP) Peptides. 2011;32:1724–1731. doi: 10.1016/j.peptides.2011.06.022. [DOI] [PubMed] [Google Scholar]
- 21.UniProt Consortium UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall T.A. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999;41:95–98. doi: 10.1021/bk-1999-0734.ch008. [DOI] [Google Scholar]
- 23.Waterhouse A.M., Procter J.B., Martin D.M., Clamp M., Barton G.J. Jalview version 2-a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Löytynoja A., Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 2010;11:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasteiger E. Protein identification and analysis tools on the ExPASy server. In: Walker J.M., editor. The Proteomics Protocols Handbook. Humana Press; 2005. pp. 571–607. [DOI] [Google Scholar]
- 27.Galgóczy L., Yap A., Marx F. Cysteine-rich antifungal proteins from filamentous fungi are promising bioactive natural compounds in anti-Candida therapy. Isr. J. Chem. 2019;59:360–370. doi: 10.1002/ijch.201800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yap A. 2019. The Role of the γ-Core Motif in the Structure and Finction of the Antifungal Proteins PAF and PAFB of Penicillium chrysogenum. Master thesis. [Google Scholar]
- 29.Lass-Flörl C. Treatment of infections due to Aspergillus terreus species complex. J. Fungi (Basel) 2018;4 doi: 10.3390/jof4030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huber A., Lerchster H., Marx F. Nutrient excess triggers the expression of the Penicillium chrysogenum antifungal protein PAFB. Microorganisms. 2019;7 doi: 10.3390/microorganisms7120654. pii:E654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hegedűs N., Sigl C., Zadra I., Pócsi I., Marx F. The paf gene product modulates asexual development in Penicillium chrysogenum. J. Basic Microbiol. 2011;51:253–262. doi: 10.1002/jobm.201000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kumar A., Zarychanski R., Pisipati A., Kumar A., Kethireddy S., Bow E.J. Fungicidal versus fungistatic therapy of invasive Candida infection in non-neutropenic adults: a meta-analysis. Mycology. 2018;9:116–128. doi: 10.1080/21501203.2017.1421592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marx F., Salvenmoser W., Kaiserer L., Graessle S., Weiler-Görz R., Zadra I., Oberparleiter C. Proper folding of the antifungal protein PAF is required for optimal activity. Res. Microbiol. 2005;156:35–46. doi: 10.1016/j.resmic.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Batta G., Barna T., Gáspári Z., Sándor S., Kövér K.E., Binder U., Sarg B., Kaiserer L., Chhillar A.K., Eigentler A., Leiter E., Hegedűs N., Pócsi I., Lindner H., Marx F. Functional aspects of the solution structure and dynamics of PAF-a highly-stable antifungal protein from Penicillium chrysogenum. FEBS J. 2009;276:2875–2890. doi: 10.1111/j.1742-4658.2009.07011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fizil Á., Gáspári Z., Barna T., Marx F., Batta G. “Invisible” conformers of an antifungal disulfide protein revealed by constrained cold and heat unfolding, CEST-NMR experiments, and molecular dynamics calculations. Chemistry. 2015;21:5136–5144. doi: 10.1002/chem.201404879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alex J.M., Rennie M.L., Engilberge S., Lehoczki G., Dorottya H., Fizil Á., Batta G., Crowley P.B. Calixarene-mediated assembly of a small antifungal protein. IUCrJ. 2019;6:238–247. doi: 10.1107/S2052252519000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Váradi G., Tóth G.K., Batta G. Structure and synthesis of antifungal disulfide β-strand proteins from filamentous fungi. Microorganisms. 2018;7:5. doi: 10.3390/microorganisms7010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sonderegger C., Fizil Á., Burtscher L., Hajdu D., Muñoz A., Gáspári Z., Read N.D., Batta G., Marx F. D19S mutation of the cationic, cysteine-rich protein PAF: novel insights into its structural dynamics, thermal unfolding and antifungal function. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fizil Á., Sonderegger C., Czajlik A., Fekete A., Komáromi I., Hajdu D., Marx F., Batta G. Calcium binding of the antifungal protein PAF: structure, dynamics and function aspects by NMR and MD simulations. PLoS One. 2018;13 doi: 10.1371/journal.pone.0204825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yount N.Y., Yeaman M.R. Multidimensional signatures in antimicrobial peptides. Proc. Natl. Acad. Sci. U. S. A. 2004;101:7363–7368. doi: 10.1073/pnas.0401567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sagaram U.S., Pandurangi R., Kaur J., Smith T.J., Shah D.M. Structure-activity determinants in antifungal plant defensins MsDef1 and MtDef4 with different modes of action against Fusarium graminearum. PLoS One. 2011;6 doi: 10.1371/journal.pone.0018550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kellenberger C., Hietter H., Luu B. Regioselective formation of the three disulfide bonds of a 35-residue insect peptide. Pept. Res. 1995;8:321–327. [PubMed] [Google Scholar]
- 43.Garrigues S., Gandía M., Borics A., Marx F., Manzanares P., Marcos J.F. Mapping and identification of antifungal peptides in the putative antifungal protein AfpB from the filamentous fungus Penicillium digitatum. Front. Microbiol. 2017;8:592. doi: 10.3389/fmicb.2017.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tóth L., Váradi G., Borics A., Batta G., Kele Z., Vendrinszky Á., Tóth R., Ficze H., Tóth G.K., Vágvölgyi C., Marx F., Galgóczy L. Anti-Candidal activity and functional mapping of recombinant and synthetic Neosartorya fischeri antifungal protein 2 (NFAP2) Front. Microbiol. 2018;9:393. doi: 10.3389/fmicb.2018.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaiserer L., Oberparleiter C., Weiler-Görz R., Burgstaller W., Leiter E., Marx F. Characterization of the Penicillium chrysogenum antifungal protein PAF. Arch. Microbiol. 2003;180:204–210. doi: 10.1007/s00203-003-0578-8. [DOI] [PubMed] [Google Scholar]
- 46.Virágh M., Vörös D., Kele Z., Kovács L., Fizil Á., Lakatos G., Maróti G., Batta G., Vágvölgyi C., Galgóczy L. Production of a defensin-like antifungal protein NFAP from Neosartorya fischeri in Pichia pastoris and its antifungal activity against filamentous fungal isolates from human infections. Protein Expr. Purif. 2014;94:79–84. doi: 10.1016/j.pep.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 47.Vriens K., Cammue B.P., Thevissen K. Antifungal plant defensins: mechanisms of action and production. Molecules. 2014;19:12280–12303. doi: 10.3390/molecules190812280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Binder U., Chu M., Read N.D., Marx F. The antifungal activity of the Penicillium chrysogenum protein PAF disrupts calcium homeostasis in Neurospora crassa. Eukaryot. Cell. 2010;9:1374–1382. doi: 10.1128/EC.00050-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Binder U., Bencina M., Fizil Á., Batta G., Chhillar A.K., Marx F. Protein kinase A signaling and calcium ions are major players in PAF mediated toxicity against Aspergillus niger. FEBS Lett. 2015;589:1266–1271. doi: 10.1016/j.febslet.2015.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Theis T., Stahl U. Antifungal proteins: targets, mechanisms and prospective applications. Cell. Mol. Life Sci. 2004;61:437–455. doi: 10.1007/s00018-003-3231-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meyer V. A small protein that fights fungi: AFP as a new promising antifungal agent of biotechnological value. Appl. Microbiol. Biotechnol. 2008;78:17–28. doi: 10.1007/s00253-007-1291-3. [DOI] [PubMed] [Google Scholar]
- 52.Hagen S., Marx F., Ram A.F., Meyer V. The antifungal protein AFP from Aspergillus giganteus inhibits chitin synthesis in sensitive fungi. Appl. Environ. Microbiol. 2007;73:2128–2134. doi: 10.1128/AEM.02497-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Binder U., Oberparleiter C., Meyer V., Marx F. The antifungal protein PAF interferes with PKC/MPK and cAMP/PKA signalling of Aspergillus nidulans. Mol. Microbiol. 2010;75:294–307. doi: 10.1111/j.1365-2958.2009.06936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oberparleiter C., Kaiserer L., Haas H., Ladurner P., Andratsch M., Marx F. Active internalization of the Penicillium chrysogenum antifungal protein PAF in sensitive aspergilli. Antimicrob. Agents Chemother. 2003;47:3598–3601. doi: 10.1128/aac.47.11.3598-3601.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Latgé J.P. 30 years of battling the cell wall. Med. Mycol. 2017;55:4–9. doi: 10.1093/mmy/myw076. [DOI] [PubMed] [Google Scholar]
- 56.Huber A., Oemer G., Malanovic N., Lohner K., Kovács L., Salvenmoser W., Zschocke J., Keller M.A., Marx F. Membrane sphingolipids regulate the fitness and antifungal protein susceptibility of Neurospora crassa. Front. Microbiol. 2019;10:605. doi: 10.3389/fmicb.2019.00605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Del Pozo A.M., Lacadena V., Mancheno J.M., Olmo N., Onaderra M., Gavilanes J.G. The antifungal protein AFP of Aspergillus giganteus is an oligonucleotide/oligosaccharide binding (OB) fold-containing protein that produces condensation of DNA. J. Biol. Chem. 2002;277:46179–46183. doi: 10.1074/jbc.M207472200. [DOI] [PubMed] [Google Scholar]
- 58.Lobo D.S., Pereira I.B., Fragel-Madeira L., Medeiros L.N., Cabral L.M., Faria J., Bellio M., Campos R.C., Linden R., Kurtenbach E. Antifungal Pisum sativum defensin 1 interacts with Neurospora crassa cyclin F related to the cell cycle. Biochemistry. 2007;46:987–996. doi: 10.1021/bi061441j. [DOI] [PubMed] [Google Scholar]
- 59.Muñoz A., Marcos J.F., Read N.D. Concentration-dependent mechanisms of cell penetration and killing by the de novo designed antifungal hexapeptide PAF26. Mol. Microbiol. 2012;85:89–106. doi: 10.1111/j.1365-2958.2012.08091.x. [DOI] [PubMed] [Google Scholar]
- 60.Muñoz A., Harries E., Contreras-Valenzuela A., Carmona L., Read N.D., Marcos J.F. Two functional motifs define the interaction, internalization and toxicity of the cell-penetrating antifungal peptide PAF26 on fungal cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cools T.L., Struyfs C., Cammue B.P., Thevissen K. Antifungal plant defensins: increased insight in their mode of action as a basis for their use to combat fungal infections. Future Microbiol. 2017;12:441–454. doi: 10.2217/fmb-2016-0181. [DOI] [PubMed] [Google Scholar]
- 62.El-Mounadi K., Islam K.T., Hernández-Ortiz P., Read N.D., Shah D.M. Antifungal mechanisms of a plant defensin MtDef4 are not conserved between ascomycete fungi Neurospora crassa and Fusarium graminearum. Mol. Microbial. 2016;100:542–559. doi: 10.1111/mmi.13333. [DOI] [PubMed] [Google Scholar]
- 63.Thevissen K., Ferket K.K., Francois I.E., Cammue B.P. Interactions of antifungal plant defensins with fungal membrane components. Peptides. 2003;24:1705–1712. doi: 10.1016/j.peptides.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 64.Muñoz E.M., Linhardt R.J. Heparin-binding domains in vascular biology. Arterioscler. Thromb. Vasc. Biol. 2004;24:1549–1557. doi: 10.1161/01.ATV.0000137189.22999.3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Utesch T., de Miguel Catalina A., Schattenberg C., Paege N., Schmieder P., Krause E., Miao Y., McCammon J.A., Meyer V., Jung S., Mroginski M.A. A computational modeling approach predicts interaction of the antifungal protein AFP from Aspergillus giganteus with fungal membranes via its β-core motif. mSphere. 2018;3 doi: 10.1128/mSphere.00377-18. e0037718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Santos F.C., Lobo G.M., Fernandes A.S., Videira A., de Almeida R.F.M. Changes in the biophysical properties of the cell membrane are involved in the response of Neurospora crassa to staurosporine. Front. Physiol. 2018;9:1375. doi: 10.3389/fphys.2018.01375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malanovic N., Leber R., Schmuck M., Kriechbaum M., Cordfunke R.A., Drijfhout J.W., de Breij A., Nibbering P.H., Kolb D., Lohner K. Phospholipid-driven differences determine the action of the synthetic antimicrobial peptide OP-145 on gram-positive bacterial and mammalian membrane model systems. Biochim. Biophys. Acta. 2015;1848:2437–2447. doi: 10.1016/j.bbamem.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 68.Malanovic N., Lohner K. Antimicrobial peptides targeting gram-positive bacteria. Pharmaceuticals (Basel) 2016;9:59. doi: 10.3390/ph9030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zweytick D., Deutsch G., Andrä J., Blondelle S.E., Vollmer E., Jerala R., Lohner K. Studies on lactoferricin-derived Escherichia coli membrane-active peptides reveal differences in the mechanism of N-acylated versus nonacylated peptides. J. Biol. Chem. 2011;286:21266–21276. doi: 10.1074/jbc.M110.195412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lohner K. DSC studies on the modulation of membrane lipid polymorphism and domain organization by antimicrobial peptides. In: Bastos M., editor. Biocalorimetry. Taylor & Francis Group; 2016. pp. 169–190. [DOI] [Google Scholar]
- 71.Leiter E., Szappanos H., Oberparleiter C., Kaiserer L., Csernoch L., Pusztahelyi T., Emri T., Pócsi I., Salvenmoser W., Marx F. Antifungal protein PAF severely affects the integrity of the plasma membrane of Aspergillus nidulans and induces an apoptosis-like phenotype. Antimicrob. Agents Chemother. 2005;49:2445–2453. doi: 10.1128/AAC.49.6.2445-2453.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szappanos H., Szigeti G.P., Pál B., Rusznák Z., Szucs G., Rajnavölgyi E., Balla J., Balla G., Nagy E., Leiter E., Pócsi I., Marx F., Csernoch L. The Penicillium chrysogenum-derived antifungal peptide shows no toxic effects on mammalian cells in the intended therapeutic concentration. Naunyn Schmiedeberg’s Arch. Pharmacol. 2005;371:122–132. doi: 10.1007/s00210-004-1013-7. [DOI] [PubMed] [Google Scholar]
- 73.Szappanos H., Szigeti G.P., Pál B., Rusznák Z., Szucs G., Rajnavölgyi E., Balla J., Balla G., Nagy E., Leiter E., Pócsi I., Hagen S., Meyer V., Csernoch L. The antifungal protein AFP secreted by Aspergillus giganteus does not cause detrimental effects on certain mammalian cells. Peptides. 2006;27:1717–1725. doi: 10.1016/j.peptides.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 74.Palicz Z., Jenes A., Gáll T., Miszti-Blasius K., Kollár S., Kovács I., Emri M., Márián T., Leiter E., Pócsi I., Csösz E., Kalló G., Hegedűs C., Virág L., Csernoch L., Szentesi P. In vivo application of a small molecular weight antifungal protein of Penicillium chrysogenum (PAF) Toxicol. Appl. Pharmacol. 2013;269:8–16. doi: 10.1016/j.taap.2013.02.014. [DOI] [PubMed] [Google Scholar]
- 75.Palicz Z., Gáll T., Leiter E., Kollár S., Kovács I., Miszti-Blasius K., Pócsi I., Csernoch L., Szentesi P. Application of a low molecular weight antifungal protein from Penicillium chrysogenum (PAF) to treat pulmonary aspergillosis in mice. Emerg. Microbes Infec. 2016;5 doi: 10.1038/emi.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kovács R., Holzknecht J., Hargitai Z., Papp C., Farkas A., Borics A., Tóth L., Váradi G., Tóth G.K., Kovács I., Dubrac S., Majoros L., Marx F., Galgóczy L. In vivo applicability of Neosartorya fischeri antifungal protein 2 (NFAP2) in treatment of vulvovaginal candidiasis. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.01777-18. e 01777-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vriens K., Cools T.L., Harvey P.J., Craik D.J., Braem A., Vleugels J., De Coninck B., Cammue B.P., Thevissen K. The radish defensins RsAFP1 and RsAFP2 act synergistically with caspofungin against Candida albicans biofilms. Peptides. 2016;75:71–79. doi: 10.1016/j.peptides.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 78.Vriens K., Cools T.L., Harvey P.J., Craik D.J., Spincemaille P., Cassiman D., Braem A., Vleugels J., Nibbering P.H., Drijfhout J.W., De Coninck B., Cammue B.P., Thevissen K. Synergistic activity of the plant defensin HsAFP1 and caspofungin against Candida albicans biofilms and planktonic cultures. PLoS One. 2015;10 doi: 10.1371/journal.pone.0132701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Garrigues S., Gandía M., Popa C., Borics A., Marx F., Coca M., Marcos J.F., Manzanares P. Efficient production and characterization of the novel and highly active antifungal protein AfpB from Penicillium digitatum. Sci. Rep. 2017;7:14663. doi: 10.1038/s41598-017-15277-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Heredero M., Garrigues S., Gandía M., Marcos J.F., Manzanares P. Rational design and biotechnological production of novel AfpB-PAF26 chimeric antifungal proteins. Microorganisms. 2018;6:106. doi: 10.3390/microorganisms6040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garrigues S., Gandía M., Castillo L., Coca M., Marx F., Marcos J.F., Manzanares P. Three antifungal proteins from Penicillium expansum: different patterns of production and antifungal activity. Front. Microbiol. 2018;9:2370. doi: 10.3389/fmicb.2018.02370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hajdu D., Huber A., Czajlik A., Tóth L., Kele Z., Kocsubé S., Fizil Á., Marx F., Galgóczy L., Batta G. Solution structure and novel insights into phylogeny and mode of action of the Neosartorya (Aspergillus) fischeri antifungal protein (NFAP) Int. J. Biol. Macromol. 2019;129:511–522. doi: 10.1016/j.ijbiomac.2019.02.016. [DOI] [PubMed] [Google Scholar]
- 83.Delgado J., Acosta R., Rodríguez-Martín A., Bermúdez E., Núnez F., Asensio M.A. Growth inhibition and stability of PgAFP from Penicillium chrysogenum against fungi common on dry-ripened meat products. Int. J. Food Microbiol. 2015;205:23–29. doi: 10.1016/j.ijfoodmicro.2015.03.029. [DOI] [PubMed] [Google Scholar]
- 84.Galgóczy L., Papp T., Pócsi I., Hegedűs N., Vágvölgyi C. In vitro activity of Penicillium chrysogenum antifungal protein (PAF) and its combination with fluconazole against different dermatophytes. Antonie Van Leeuwenhoek. 2008;94:463–470. doi: 10.1007/s10482-008-9263-x. [DOI] [PubMed] [Google Scholar]