ABSTRACT
The autophagy receptor for selective reticulophagy, RETREG1/FAM134B is essential for ER maintenance, and its dysfunction is associated with neuronal disorders, vascular dementia, or viral infections. The protein consists of the reticulon-homology domain (RHD) that is flanked at the N- and C-termini by an intrinsically disordered protein region (IDPR), where the C terminal IDPR carries the indispensable LC3-interacting region (LIR) motif for the interaction with LC3. The RHD of RETREG1 is presumed to play a role in membrane remodeling, but the absence of a known 3D structure of this domain so far prevented researchers from gaining mechanistic insights into how the RETREG1 RHD curves membranes, and thereby facilities reticulophagy. The recent study by Bhaskara et al., which is described in this editor’s corner article, used molecular dynamics (MD) simulations to create a structural model of the RETREG1 RHD. MD simulations along with in vitro liposome remodeling experiments reveal how the RHD domain acts on the ER membrane and, in concert with the C terminal IDPR, executes the function of RETREG1 in selective reticulophagy.
Abbreviations: ER, endoplasmic reticulum; IDPR, intrinsically disordered protein region; LIR, LC3-interacting region; MD, molecular dynamics; RHD, reticulon-homology domain; TM, transmembrane
KEYWORDS: Endoplasmic reticulum, lysosome, protein intrinsic disorder, receptor, selective autophagy, stress
Selective autophagy is an indispensable pathway for the maintenance of organelle homeostasis in cells. When a major site for protein production, the endoplasmic reticulum (ER), undergoes stress conditions, cells respond by induction of selective reticulophagy. In this process, the autophagy machinery is selectively recruited to ER-specific locations where the ER is fragmented and engulfed by the growing phagophore that ultimately closes, and becomes an autophagosome. After fusion of the autophagosome with the lysosome, the ER fragments are degraded. Successful completion of this process requires two important events, fragmentation of the ER and linking the ER fragments to the autophagy machinery. Both of these events are mediated by selective reticulophagy receptors, one of them being RETREG1.
The reticulophagy receptor RETREG1 is assumed to function primarily in ER homeostasis. RETREG1 contains a reticulon-homology domain that anchors the receptor in the ER membrane. The RHD domain is flanked on its N- and C-termini by an intrinsically disordered protein region of ~ 80 and ~ 240 amino acids, respectively. Whereas the function of the N terminal IDPR has not been assigned yet, the role of the C terminal IDPR is ascribed to linking RETREG1 to LC3. In particular, the very C terminus of this IDPR carries the LIR motif (455FELL) that binds to PE-conjugated LC3 on the phagophore membrane [1]. Thus, the C terminal IDPR of RETREG1 fulfills the function of connecting the ER, which is destined for degradation, to the autophagy machinery. The other function, fragmentation of the ER, is assumed to be carried out by the RETREG1 RHD via induction of high-membrane curvature that leads to vesiculation of the ER and subsequent engulfing by the phagophore. Due to the absence of information about the RHD structure, the molecular mechanism of membrane remodeling during RETREG1-mediated reticulophagy was so far elusive. The study by Bhaskara et al. [2], described here, gives us a new insight into this molecular mechanism.
These researchers used all-atom molecular dynamics simulations to create an initial model of each structural element that comprises the RETREG1 RHD. The structural models of these elements were then assembled into an overall structural model of this domain. The authors showed that four transmembrane (TM) helices of RHD fold into two sets of helical hairpins. Each hairpin has a polar lumenal loop of 3–4 residues. One loop links TM1 with TM2, and the other loop connects TM3 with TM4. Another helical feature of the RHD in RETREG1 are two cytoplasmic amphipathic helices (AHL and AHC) that have hydrophobic faces embedded in the lipid bilayer. These helices are derived from a disordered conformation, and thus are folded upon interaction with membrane. The first amphipathic helix (AHL) originates from the linker sequence that connects the TM1-TM2 hairpin with the TM3-TM4 hairpin, whereas the second helix (AHC) arises from the disordered C-terminal segment of the RHD. All-atom MD simulations revealed local perturbations of the lipid bilayer by the simulated molecule of the RETREG1 RHD. This is consistent with in silico curvature assays that simulate the behavior of flat bicelles after addition of the RETREG1 RHD. Bicelles undergo complete vesiculation in the presence of the RETREG1 RHD, suggesting the ability of the RHD to induce membrane curvature. Determination of the rates of vesicle formation for each structural element of the RHD (TM1-TM2, TM3-TM4, AHL, and AHC) shows that AHL and AHC are responsible for curvature induction, whereas the transmembrane hairpins are inefficient in causing bicelle vesiculation. The results obtained for AHL and AHC are consistent with the membrane-remodeling capability of lipid-inducible amphipathic helices in other proteins, for example BAR proteins [3], ATG14 [4], ATG3 [5], or Atg2 [6].
To determine if the RETREG1 RHD senses membrane curvature and what type of local curvature it prefers, Bhaskara et al. used simulations that tracked the RHD along the buckled membrane with a sinusoidal shape and local mean curvatures H(x,y) ranging from −0.05 to 0.05 nm−1. The RHD was found to prefer regions of high local curvature, specifically H(x,y) of 0.026 nm−1.
The best insight into the molecular mechanism of RETREG1 function was obtained by long simulations of the intact RHD in flat bilayers. These simulations show the overall topology of the RHD in the membrane and reveal how RETREG1 achieves membrane remodeling. The two transmembrane hairpins are embedded in the membrane in a slightly tilted position that facilitates interaction of their hydrophilic lumenal loops. On the cytosolic side, the TM hairpins are farther apart, separated by AHL. With AHC inserted in the membrane next to TM3 and TM4, the resulting structure creates a wedge shape with the disordered N and C termini protruding into the cytoplasm. This asymmetrical shape of the RETREG1 RHD strongly curves the bilayer (Figure 1A). Positive membrane curvature induced by the RHD is further enhanced by clustering of the RETREG1 RHDs. Simulations on large flat bilayers as well as on tubules show that the RHD molecules dynamically cluster, and this clustering amplifies membrane deformation. A closer visualization of a cluster containing three RHD molecules in the strongly curved bilayers reveals that this cluster forms a unique inverted-pyramid-like shape, where the lumenal loops of the six TM hairpins interact and form a narrow tip, while the six AHs are assembled into a base of the inverted pyramid on the cytosolic face of the membrane. Thus, the inverted-pyramid-like shape can be interpreted as an amplification of the wedge shape, which is adopted by each RETREG1 RHD molecule. This explains why clustering of the multiple RETREG1 RHDs enhances positive curvature that is induced, and sensed, by a single RETREG1 RHD.
Figure 1.
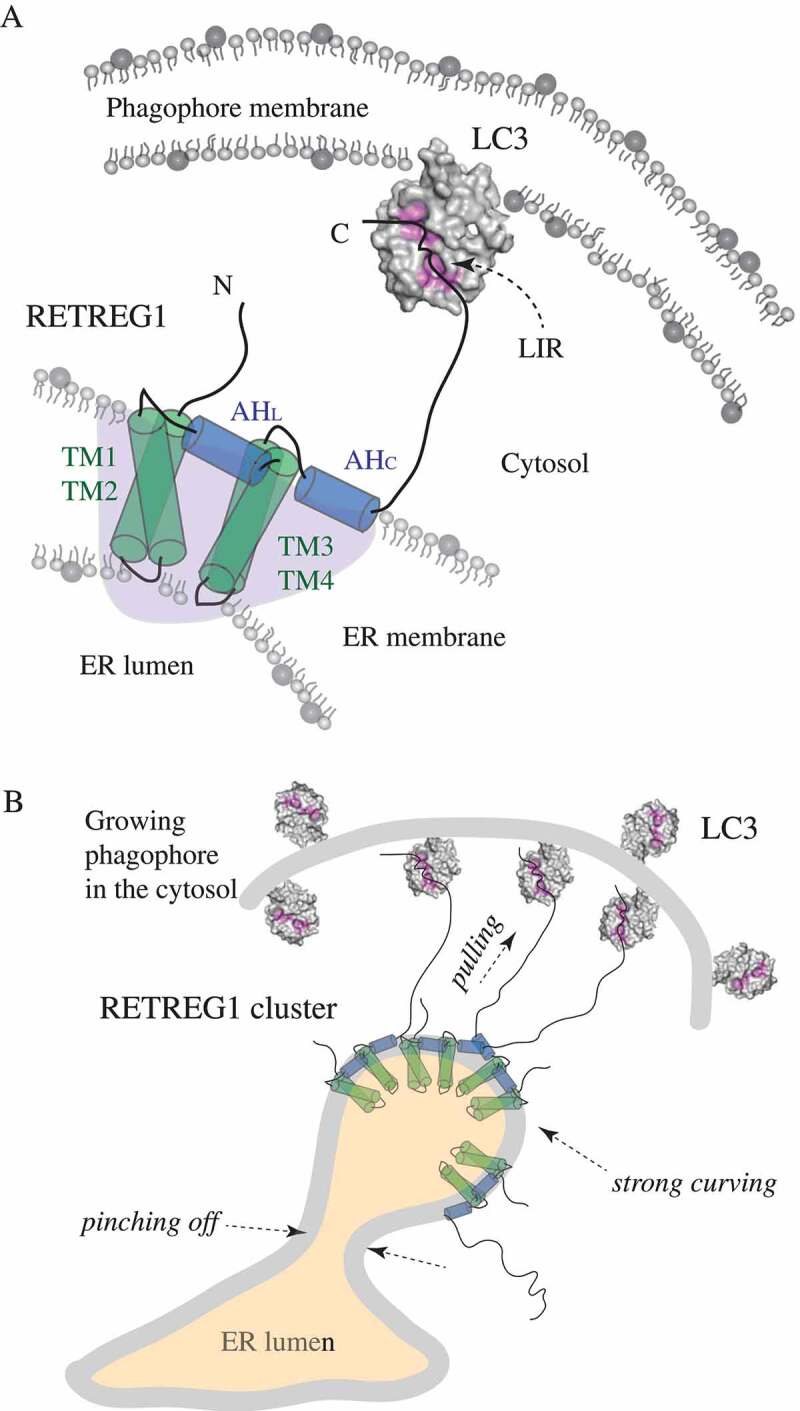
Schematic representation of the molecular mechanism of RETREG1 function. (A) A detailed zoom-in on a single molecule of the RETREG1 receptor embedded in the ER membrane via its RHD. The functional topology of the RETREG1 RHD, as revealed by MD simulations (redrawn here schematically based on ref [2].), is formed by several structural elements including the two transmembrane hairpins (TM1-TM2 and TM3-TM4) and two amphipathic helices (AHL and AHC). Together these elements adopt a wedge shape (light purple shading) of a single RHD molecule. The disordered N and C termini protrude into the cytosol. The C terminus of RETREG1 carries the LIR motif (FELL) that binds to the hydrophobic pockets (highlighted in pink) of LC3 (PDB ID: 4ZDV) that is conjugated to PE on the phagophore membrane. (B) The molecular mechanism of RETREG1 function in selective reticulophagy as proposed by Bhaskara et al. The wedge-shaped RHDs of RETREG1 cluster together and form an inverted-pyramid-like shape, which strongly curves the ER membrane. The C terminal LIR motif of each RETREG1 molecule binds to LC3 on the concave side of the phagophore. Pulling forces by the multiple long disordered C termini of RETREG1 molecules along with strong curving forces by the clustered RHDs facilitate ER vesicle budding and subsequent pinching off. This leads to fragmentation of the ER, which is necessary for engulfing by the phagophore.
To strengthen the results of MD simulations and to assess experimentally direct membrane binding and remodeling capabilities of the RHD structural elements, the authors produced the recombinant wild-type RETREG1 RHD along with several deletion constructs. Using liposome flotation assays, they showed that the TM hairpins are the most important elements for liposome binding, and at least one TM hairpin is required for stable anchoring of the RETREG1 RHD into lipid membranes. The ability of the individual RHD elements to induce curvature was probed using negative-stain transmission electron microscopy of remodeled proteoliposomes. In agreement with the results of the in silico curvature assays, the negative-stain transmission electron microscopy images show that AHL and AHC are critical for remodeling of liposomes, whereas the TM1-TM2 hairpin is not required. Interestingly, the TM3-TM4 hairpin plays a substantial role in liposome remodeling.
To assess the RETREG1 RHD structural elements in selective reticulophagy, wild-type RETREG1 and deletion mutants were evaluated for ER fragmentation in U2OS cells 24 h after transfection. ER fragmentation was monitored by immunofluorescence of HA-tagged RETREG1. As in the case of liposome binding, removal of the TM hairpins renders the reticulophagy receptor completely unable to fragment the ER, as does removal of the LIR motif. This shows that, next to the LIR motif, the intact RHD is required for ER fragmentation. This finding also explains why disease- or virus-associated dysfunction of the RETREG1 receptor abolishes RETREG1-mediated selective reticulophagy. Explicitly, sensory neuropathy is a disease caused by genetic truncation of RETREG1 at Q145 [7], and a Zika virus protease cleaves RETREG1 at R142 in order to subvert selective reticulophagy [8]. Both of these shortenings remove the TM3-TM4 hairpin along with AHL, AHC, and the LIR-containing C terminus from the intact RETREG1. Thus, the study by Bhaskara et al. provides structural insight into RETREG1 dysfunction by proposing a model for the molecular mechanism of RETREG1-mediated reticulophagy. In this model (Figure 1B), the RETREG1 RHD senses and induces regions with positive curvature in the ER membrane via its wedge shape adopted by the TM hairpins and AHs. This mediates clustering of the several RETREG1 RHDs that form together the inverted-pyramid-like structure, which further amplifies membrane deformation, and produces locally highly curved ER membranes. Along with pulling forces created by the long disordered RETREG1 C terminus bound via the LIR motif to LC3 on the phagophore, the strong membrane deformation by the RETREG1 RHD leads to vesicles budding and pinching off from the ER membrane. Proximity of the phagophore close to where the ER vesicles pinch off enables efficient engulfing of the ER fragments and subsequent degradation in autolysosomes.
Elucidation of the molecular mechanism of membrane deformation by RETREG1 was possible primarily by building a structural model of the membrane-embedded RHD using molecular and MD simulations. For proteins that need interactions with membranes for folding into their complete functional structure, visualization of their 3D architecture merely based on the amino acid sequence, that is, irrespective of inputs from lipid membranes, is much less meaningful; this approach does not yield a functional protein topology and comprehension of mutual protein-membrane effects. For example, the homology model of the RETREG1 RHD created by Phyre2, the protein fold recognition server [9], merely based on its amino acid sequence provides overall information about the existence of four transmembrane helices and some additional helical structures (Figure 2), but the functional topology of these helical elements and their behavior in the lipid bilayer remain elusive (compare Figure 1A and 2). Furthermore, most of the Phyre2 model represents a low-confidence prediction. For this reason, MD simulations are an innovative approach that can become one of the essential tools in the field of structural autophagy, which has the uneasy task of solving functional structures of many membrane-binding autophagy proteins highly enriched in disordered regions.
Figure 2.
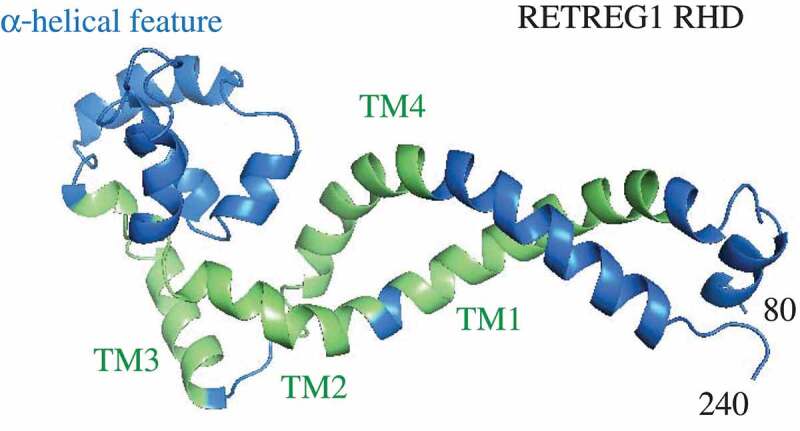
The homology model of the RETREG1 RHD (amino acid residues 80–240) predicted by Phyre2 based on the amino acid sequence obtained in the UniProt database. The α-helical feature was modeled by Phyre2 based on the Nogo-66 NMR structure (PDB ID: 2KO2). The rest of the model, including the TM segments, is a low confidence prediction. Modeling to the best of the ability of the fold recognition server cannot reveal the functional topology of RETREG1 without a direct input from lipid membranes.
Funding Statement
This work was supported by the National Institute of General Medical Sciences (GM131919), and the Protein Folding Disease FastForward Initiative, University of Michigan.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Khaminets A, Heinrich T, Mari M, et al. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015. June 18;522(7556):354–358. PubMed PMID: 26040720; English. [DOI] [PubMed] [Google Scholar]
- [2].Bhaskara RM, Grumati P, Garcia-Pardo J, et al. Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat Commun. 2019. May 30;10(1):2370. PubMed PMID: 31147549; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Bhatia VK, Madsen KL, Bolinger PY, et al. Amphipathic motifs in BAR domains are essential for membrane curvature sensing. Embo J. 2009. November 4;28(21):3303–3314. PubMed PMID: 19816406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fan W, Nassiri A, Zhong Q.. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L). Proc Natl Acad Sci U S A. 2011. May 10;108(19):7769–7774. PubMed PMID: 21518905; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Nath S, Dancourt J, Shteyn V, et al. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol. 2014. May;16(5):415–424. PubMed PMID: 24747438; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kotani T, Kirisako H, Koizumi M, et al. The Atg2-Atg18 complex tethers pre-autophagosomal membranes to the endoplasmic reticulum for autophagosome formation. Proc Natl Acad Sci U S A. 2018. October 9;115(41):10363–10368. PubMed PMID: 30254161; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kurth I, Pamminger T, Hennings JC, et al. Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet. 2009. November;41(11):1179–1181. PubMed PMID: 19838196; English. [DOI] [PubMed] [Google Scholar]
- [8].Lennemann NJ, Coyne CB.. Dengue and Zika viruses subvert reticulophagy by NS2B3-mediated cleavage of FAM134B. Autophagy. 2017. February;13(2):322–332. PubMed PMID: 28102736; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kelley LA, Mezulis S, Yates CM, et al. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015. June;10(6):845–858. PubMed PMID: 25950237; English. [DOI] [PMC free article] [PubMed] [Google Scholar]


