Summary:
We explored the potential for autophagy to regulate budding yeast meiosis. Following pre-meiotic DNA replication, we blocked autophagy by chemical inhibition of Atg1 kinase or engineered degradation of Atg14 and observed homologous chromosome segregation followed by sister chromatid separation; cells then underwent additional rounds of spindle formation and disassembly without DNA re-replication leading to aberrant chromosome segregation. Analysis of cell cycle regulators revealed that autophagy inhibition prevents meiosis II-specific expression of Clb3 and leads to the aberrant persistence of Clb1 and Cdc5, two substrates of a meiotic ubiquitin ligase activated by Ama1. Lastly, we found that during meiosis II autophagy degrades Rim4, an amyloid-like translational repressor whose timed clearance regulates protein production from its mRNA targets, which include CLB3 and AMA1. Strikingly, engineered Clb3 or Ama1 production restored meiotic termination in the absence of autophagy. Thus, autophagy destroys a master regulator of meiotic gene expression to enable irreversible meiotic exit.
Keywords: meiosis, autophagy, Atg1, Atg14, Rim4, Clb3, Ama1, APC/C, sporulation, translational repression
eTOC
Wang et al. report that autophagy promotes meiotic termination by degrading Rim4, an amyloid-like translational repressor whose timed clearance in meiosis II regulates protein production from its mRNA targets. In the absence of autophagy, cells fail to exit meiosis and undergo additional rounds of chromosome segregation after meiosis II.
Introduction
Macroautophagy (herein called autophagy) is a highly-conserved cellular degradation process that removes protein aggregates, damaged organelles, and other potentially toxic structures (Yin et al., 2016). During autophagy, cytoplasmic material becomes encapsulated into double-membrane vesicles (autophagosomes) that subsequently fuse with lysosomes, thus delivering their cargo for degradation. Autophagy was originally discovered as a starvation response that recycles macromolecules to maintain energy stores and biosynthesis of essential components. Beyond its role as a stress response, autophagy has emerged in recent years as a versatile degradation mechanism for controlling key cellular events during different stages of sexual reproduction and early stages of development (Yin et al., 2016). For example, following oocyte fertilization in mice, flies, and worms, autophagy selectively targets paternal mitochondria for destruction to enable maternal inheritance of mtDNA (Politi et al., 2014; Rojansky et al., 2016; Sato and Sato, 2011). Autophagy is also responsible for targeted elimination of P granule ribonucleoproteins during early embryonic cell divisions in worms to help restrict primordial germline specification (Zhang et al., 2009). Lastly, preimplantation development in early mouse embryos is arrested by mutations that disrupt autophagy (Tsukamoto et al., 2008).
During sexual reproduction in budding yeast, starvation generates a key signal for pre-meiotic DNA replication in diploid cells. This meiosis-initiating event is followed by two divisions in which homologous chromosomes segregate (meiosis I) and sister chromatids separate (meiosis II) to enable haploid gamete formation by an overlapping sporulation program (Neiman, 2011). Autophagy mutants lacking key autophagy-related factors (Atg) are unable to form gametes because they fail to undergo the meiotic divisions in response to starvation (Enyenihi and Saunders, 2003; Piekarska et al., 2010; Sarkar et al., 2014; Straub et al., 1997; Wen et al., 2016). We hypothesized that autophagy plays an additional, regulatory role during yeast meiotic divisions. This idea was inspired by evidence from a genome-wide analysis of translation during meiosis I and II that showed enhanced synthesis of Atg8 (Brar et al., 2012), a factor whose abundance is known to be size limiting for autophagosomes (Xie et al., 2008).
Here, we have found that inactivation of two key components of the autophagy machinery – Atg1 and Atg14 – during prophase I results in a deregulation of cell cycle events in which chromosomes continue to segregate following meiosis II and gametogenesis is aborted. Analysis of several cell cycle regulators led us to discover that with Atg1 inhibition, there was a defect in autophagic degradation of Rim4, an amyloid-like translational repressor of many mRNAs during meiosis II (Berchowitz et al., 2013). Strikingly, the effects of Atg1 inhibition on meiosis were suppressed by engineered protein expression of two Rim4 mRNA targets, the cyclin Clb3 and Ama1, a meiosis-specific activator of the APC/C ubiquitin ligase. These results provide a mechanistic explanation for how autophagy drives exit from meiosis.
Results
Autophagy inhibition results in aberrant cycles of additional spindle formation and breakdown following meiosis II
As deletion of core autophagy genes is known to preclude entry into the meiotic divisions (Enyenihi and Saunders, 2003; Piekarska et al., 2010; Sarkar et al., 2014; Straub et al., 1997; Wen et al., 2016), we used chemical genetics to inhibit the Atg1 kinase following completion of pre-meiotic DNA replication. This approach relied on a mutation in the Atg1 ATP-binding site known to enable robust inhibition of autophagy by 1-NM-PP1, a cell-permeable ATP analog (Blethrow et al., 2004; Kamada et al., 2000; Kamber et al., 2015). We introduced Atg1-as into a system for inducible expression of NDT80 (NDT80-in), a middle meiosis transcription factor required to initiate meiosis I (Benjamin et al., 2003; Carlile and Amon, 2008). NDT80-in is controlled by the GAL1 promoter and Gal4-ER transcription factor, which is inactive without β-estradiol causing prophase I arrest. Addition of β-estradiol to arrested cells activates Gal4-ER to drive NDT80 expression, leading to the onset of synchronous meiotic divisions (Figure 1A, Supplemental Figure S1A).
Figure 1: Autophagy inhibition causes additional rounds of spindle formation and breakdown after meiosis II.
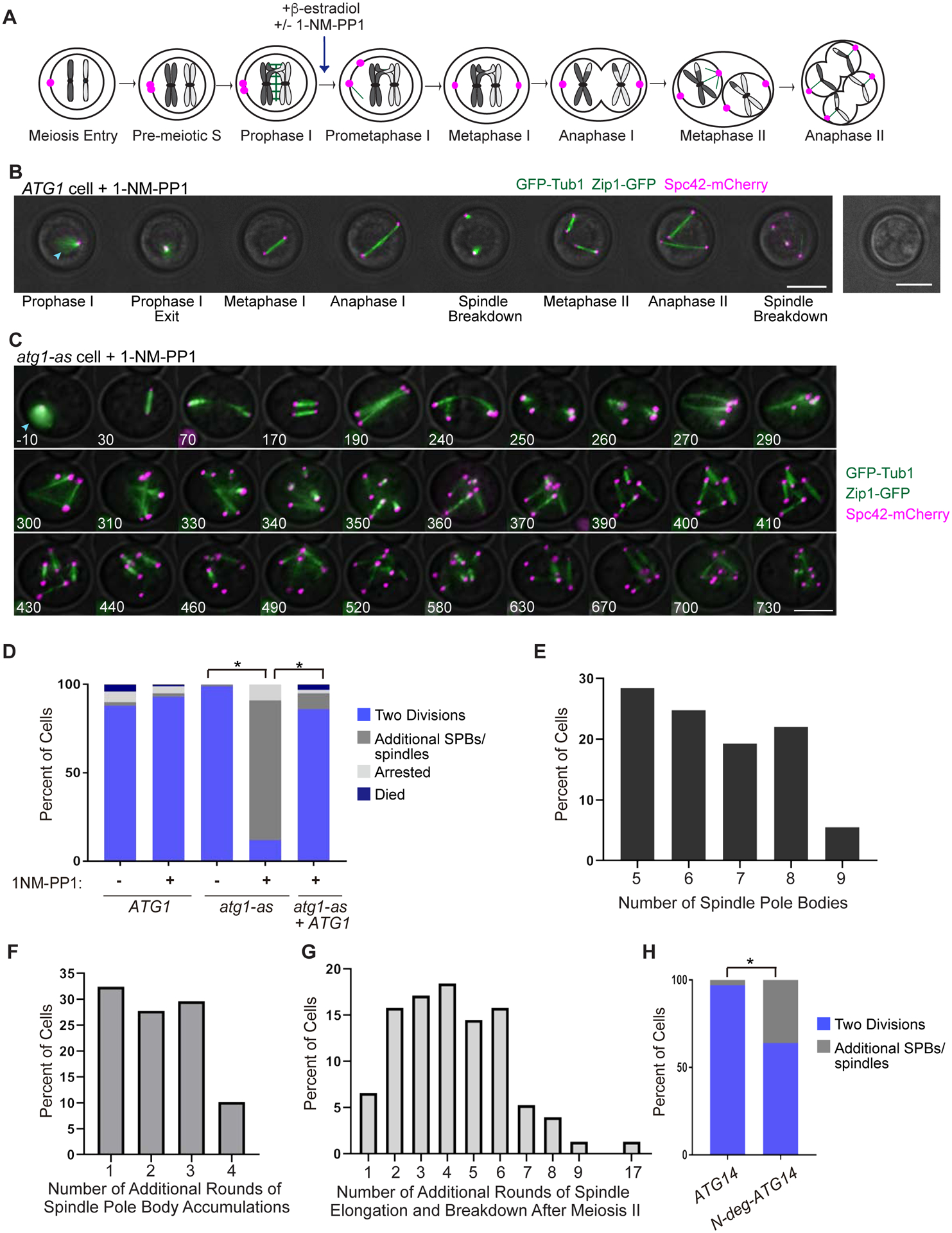
A) Cartoon showing cell cycle stages of budding yeast meiosis with synchronization by NDT80-in and release from prophase I arrest by β-estradiol addition. Unless indicated otherwise, 1-NM-PP1 was always added to meiotic cells at the time of arrest release. B) Representative time-lapse images of a wildtype (ATG1) cell undergoing synchronized meiosis in the presence of 1-NM-PP1. Also pictured, a brightfield image of spores at the end of the time-lapse. Blue arrowhead shows Zip1-GFP. Scale Bar: 5μm. C) Representative time-lapse images of an atg1-as cell undergoing synchronized meiosis in the presence of 1-NM-PP1. Blue arrowhead shows Zip1-GFP. Scale Bar: 5μm. D) Percent of wildtype, atg1-as, and atg1-as + ATG1 cells that underwent the indicated synchronized meiotic outcome in the absence or presence of 1-NM-PP1. The atg1-as + ATG1 cells have a transgenic copy of ATG1 under the control of a β-estradiol-inducible promoter (McIsaac et al., 2014). Only cells that were in prophase I (with Zip1-GFP) at the time of arrest release were analyzed. Asterisk represents statistically significant differences (n = >100 cells and 3 independent experiments for each genotype; p <.01, rx Contingency Tables). E) Graph of cells that had additional SPBs showing the percent of cells with the indicated number of additional SPBs (n=100 cells). F) Graph of cells that had additional SPBs, showing the percent of cells with the indicated number of additional rounds of SPB accumulations (n=100 cells). G) Graph of cells that had additional rounds of spindle formation, elongation, and breakdown after meiosis II, showing the percent of cells with the indicated number of additional rounds (n=100 cells). H) Percent of ATG14 NDT80-in and N-deg-ATG14 NDT80-in cells that underwent the indicated synchronized meiotic outcome. Note that both genetic backgrounds have a β-estradiol-inducible TEV protease that enables activation of the N-degron in the latter strain (Taxis and Knop, 2012). Only cells that were in prophase I (with Zip1-GFP) at the time of arrest release were analyzed. Asterisk represents statistically significant differences (n = >100 cells for each; p <.01, Fisher’s exact test).
To determine if Atg1 inhibition affects progression through the meiotic divisions, we used time-lapse microscopy to monitor the distribution of three proteins: Spc42-mCherry, a component of the spindle pole body (SPB), the yeast equivalent of the centrosome; GFP-Tub1, an alpha-tubulin that incorporates into microtubules; and Zip1-GFP, a component of the synaptonemal complex that associates with chromosomes during prophase I (Tsuchiya et al., 2011; Tsuchiya and Lacefield, 2013). Although Zip1-GFP and GFP-Tub1 are both tagged with GFP, they are easily distinguishable both morphologically and temporally, as the synaptonemal complex disassembles at the end of prophase I prior to spindle assembly. Previous work using these labeled components revealed the following series of events upon release from prophase I arrest (Tsuchiya et al., 2014). First, Zip1 disappears and the two SPBs separate from one another. Next, the spindle elongates in anaphase I to separate homologous chromosomes, and then disassembles. The SPBs then duplicate between meiosis I and meiosis II to promote assembly of two meiosis II spindles. Lastly, in anaphase II, both spindles undergo a round of elongation and disassembly to separate sister chromatids.
Our microscopy analysis of two negative controls – the untreated Atg1-as cells and 1-NM-PP1-treated wild-type ATG1 cells – revealed the expected sequence of meiotic cell cycle events culminating in the formation of four terminal haploid progeny packaged into spores (Figure 1B, Video S1, Supplemental Figure S1B). By contrast, 1-NM-PP1 treatment of Atg1-as cells led to somewhat slower progression through meiosis I and meiosis II, followed by the more striking formation of supernumerary SPBs that mediated additional rounds of spindle formation and breakdown (Figure 1C–D, Supplemental Figure S1C, Video S2). Cells accumulated between 5–9 SPBs over time and underwent between 1–17 additional aberrant cycles of spindle elongation through spindle breakdown (Figure 1E–G). The additional cycles highly varied in duration, but on average were similar to the durations of meiosis I and meiosis II (Supplemental Figure S1C, S1E). We confirmed that this phenotype was in fact the consequence of Atg1 kinase loss by achieving rescue using a transgenic wild type ATG1 (Figure 1D).
Atg1 kinase inhibition could prevent normal termination of meiosis by either inhibiting autophagy or, in principle, by a moonlighting function. We found two lines of evidence supporting the former possibility. First, we directly measured autophagy flux during meiosis using GFP-Atg8, a lipidated component of the autophagosome membrane that is processed to GFP in the vacuole (yeast lysosome) (Meiling-Wesse et al., 2002; Torggler et al., 2017). Following cell release from a Ndt80-in prophase I arrest, we detected enhanced autophagy flux at the later stages of both meiosis I and II (Supplemental Figure S2A–C). Second, we appended a conditional N-end degron to Atg14 (deg-Atg14) (Taxis et al., 2009), a core component of an autophagy-specific phosphatidylinositol 3-kinase complex that functions downstream of Atg1 (Kametaka et al., 1998; Kihara et al., 2001). Inducing deg-Atg14 degradation at the time of release from prophase I arrest resulted in 35% of cells with additional spindles and SPBs after meiosis II (Figure 1H). The relatively lower penetrance of this meiotic phenotype matches the weaker effect on GFP-Atg8 processing of inducible Atg14 degradation compared to chemical inhibition of Atg1 (Supplemental Figure S2D). In sum, our data argue that autophagic protein degradation is at the core of a novel regulatory mechanism for terminating meiosis.
Autophagy inhibition leads to a defect in pro-spore membrane formation
Meiosis II in yeast is coupled to a sporulation program that brings about the engulfment of haploid nuclei and surrounding cytoplasm by a specialized prospore membrane (Neiman, 2011). To better understand how Atg1-as inhibition results in a severe defect in spore formation (Figure 2A), we monitored recruitment of prospore nucleation factors to SPBs during meiosis II. Spo21-GFP, Mpc54-GFP, and Spo74-GFP localized normally to SPBs and continued to associate with the additional SPBs assembled following meiosis II (Figure 2B–D). Notably, while we observed some prospore membrane formation using the t-SNARE mKate-Spo2051−90 as a marker, these structures appeared grossly aberrant by comparison to uniform oval membranes that surrounded haploid nuclei in control cells (Figure 2E–F).
Figure 2: Autophagy inhibition disrupts gametogenesis.
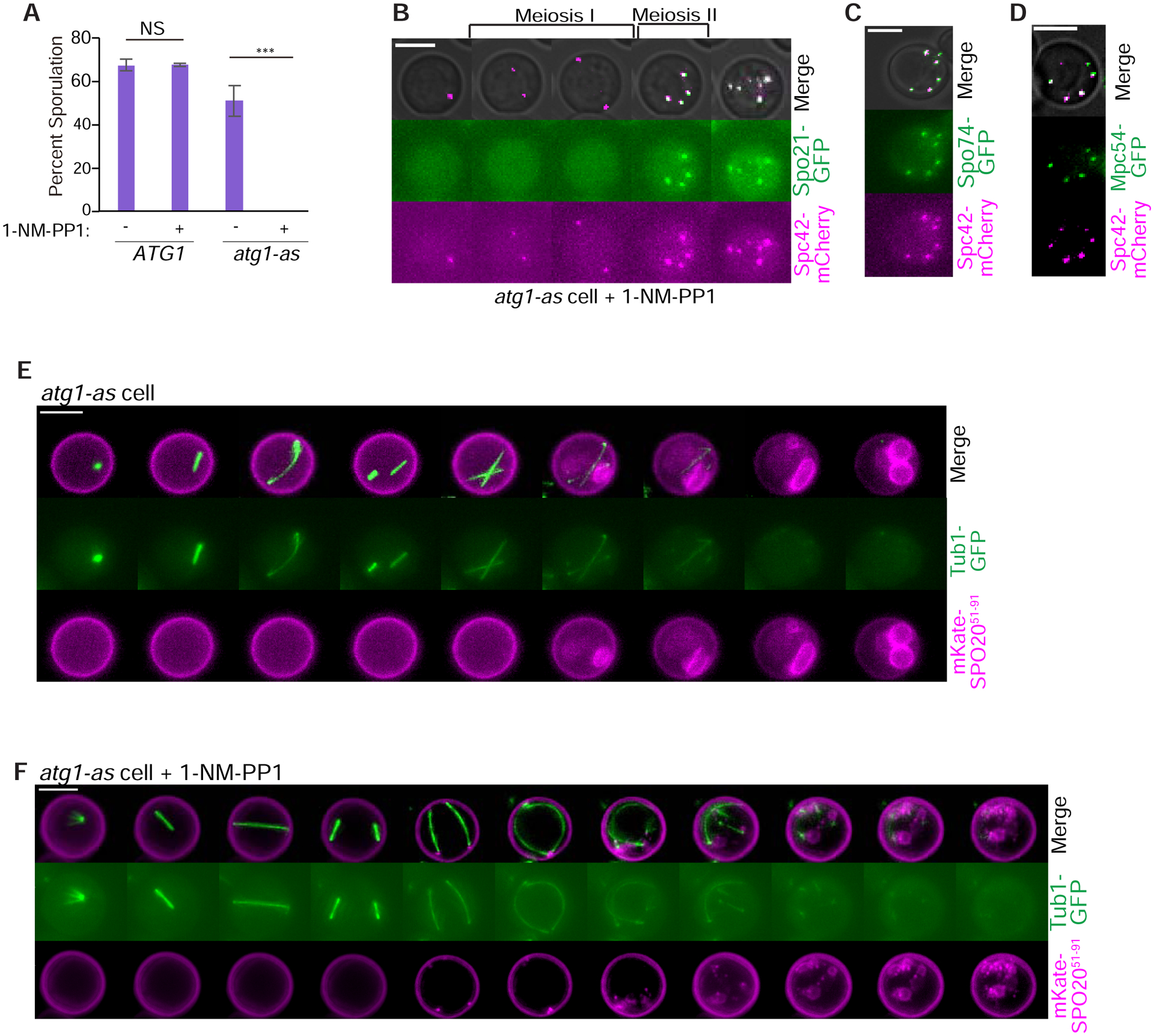
A) Graph of percentage of wildtype and atg1-as cells (without Ndt80-IN) that sporulate with and without 1-NM-PP1 inhibitor added 10 hours after introduction into sporulation medium. NS = not significant. Asterisk indicates a statistically significant difference (n > 200 cells per strain; p<0.001; t test). B) Representative time-lapse images of an atg1-as cell undergoing synchronized meiosis in the presence of 1-NM-PP1 and β-estradiol addition. Scale Bar: 5μm. C, D) Similar to post-meiosis II images in part B but with additional fluorescently-labeled SPB components, as indicated. E-F) Representative time-lapse images of atg1-as cells undergoing synchronized meiosis in the absence (E) and presence (F) of 1-NM-PP1. Prospore membranes are marked with mKate-Spo2051–90. Scale Bars: 5μm.
Next, we asked if the primary meiosis phenotype caused by autophagy inhibition could be a sequelae of a more proximal defect in prospore membrane formation. To test this idea, we imaged spo20Δ cells but did not observe any additional cycles of spindle formation and breakdown after meiosis II (Figure S3A). 7% of spo20Δ cells displayed an extra Spc42-mCherry signal after meiosis II, which might correspond to non-functional SPB fragments, as no additional spindles formed (Figure S3B). Taken together, these data argue that abortive prospore membrane biogenesis, while a product of autophagy inhibition, is unlikely to cause additional SPBs and spindles after meiosis II per se.
Autophagy inhibition results in abnormal rounds of chromosome segregation after meiosis II
To directly visualize what effect autophagy inhibition has on chromosome segregation following meiosis II, we first tracked the meiotic fate of DNA labeled with histone HTB2-mCherry. Four similar-sized DNA masses formed in all cells at the end of meiosis II in untreated Atg1-as cells (n=100 cells; Figure 3A). In 1-NM-PP1-treated cells, however, the four DNA masses that formed at the end of meiosis II became further fragmented by subsequent rounds of spindle formation, elongation, and disassembly (64% of treated cells; n = 100 cells; Figure 3B–C). To determine if DNA replication occurred in between the additional rounds of chromosome segregation, we heterozygously tagged a single chromosome III with a LacO array in a diploid parent expressing LacI-GFP. Untreated cells displayed a normal pattern of meiotic chromosome segregation (ie. one round of DNA replication, followed by two divisions) that resulted in one GFP focus in two of the four DNA masses (Figure 3D). Virtually all 1-NM-PP1 treated cells that underwent additional rounds of chromosome segregation continued to show only two GFP foci (Figure 3E; n=100 cells). These results demonstrate that the additional spindles we saw following meiosis II led to anomalous chromosome segregation in the absence of DNA re-replication.
Figure 3: Autophagy inhibition causes additional, aberrant rounds of chromosome segregation following meiosis II.
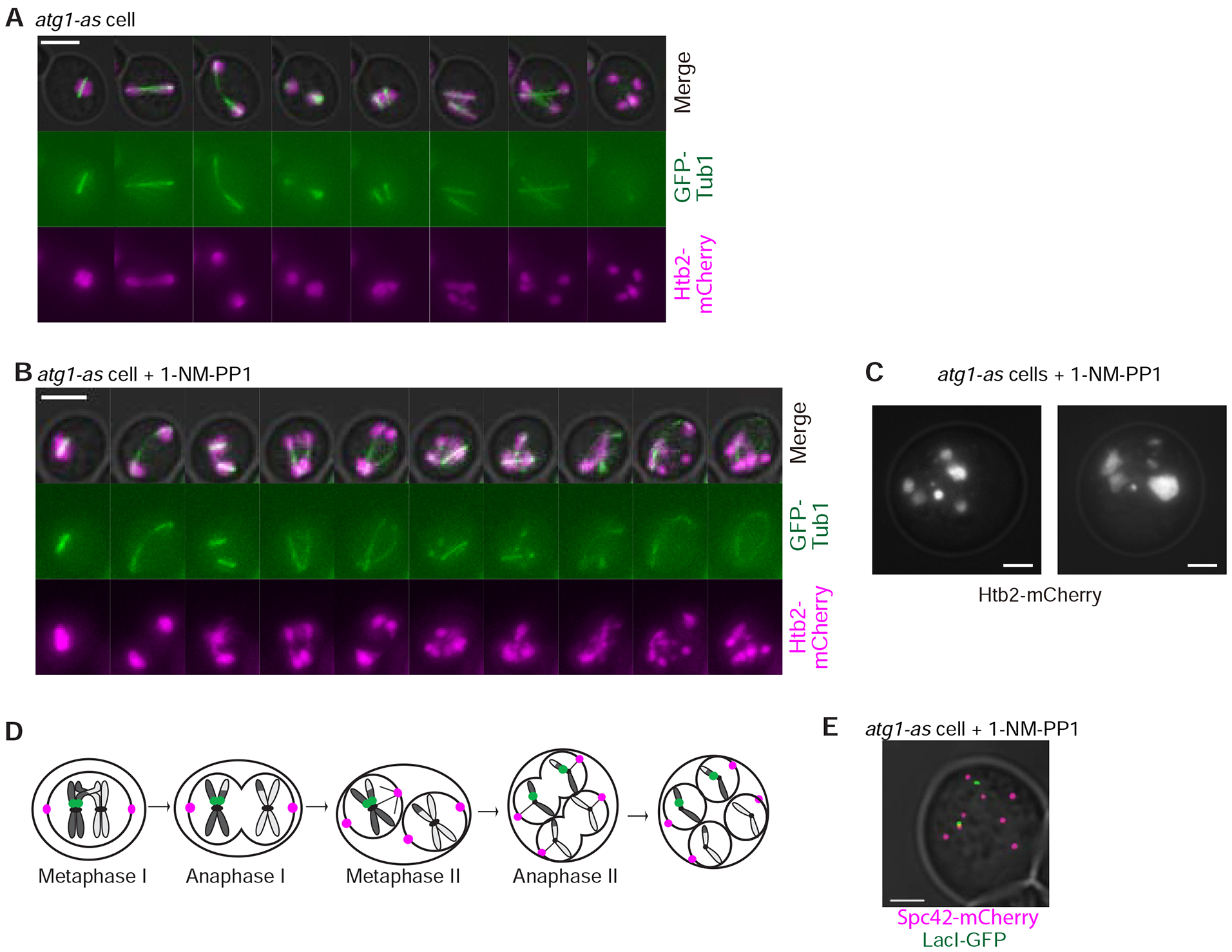
A-B) Representative time-lapse images of atg1-as cells undergoing synchronized meiosis in the absence (A) and presence of 1-NM-PP1 (B). Scale Bars: 5μm. C) Additional images of cells from part B showing different sized chromatin masses after cells underwent additional, post-meiosis II rounds of chromosome segregation. Scale bar: 2μm. D) Cartoon of a normal meiotic division with one chromosome III tagged with LacO-LacI-GFP. E) Representative image of an 1-NM-PP1-treated atg1-as cell that underwent additional, post-meiosis II rounds of chromosome segregation. LacI-GFP recognizes LacO on one chromosome III. Scale Bars: 2μm.
Evidence that autophagy inhibition indirectly disrupts diverse cell cycle regulatory steps by a direct effect on Rim4 degradation
Cycles of SPB duplication, spindle formation and disassembly during normal meiosis are driven by oscillations in meiotic CDK activity, which are terminated at the end of meiosis II (Marston and Amon, 2004). To determine if our spindle phenotype is associated with additional oscillations in CDK activity following meiosis II, we examined the effect of autophagy inhibition on Cdc14 localization. This phosphatase is normally released from the nucleolus during anaphase I to reverse CDK phosphorylations before being rapidly re-sequestered into the nucleolus in preparation for another round of release during anaphase II (Figure 4A)(Buonomo et al., 2003; Marston et al., 2003). In 64% of cells that underwent additional rounds of chromosome segregation, Atg1-as inhibition induced between one and three additional rounds of Cdc14 release from the nucleolus after meiosis II (n = 100 cells; Figure 4B–C, Supplemental Figure S3C). As the release and re-sequestration of Cdc14 could have occurred within the 10-min imaging interval, these data may underreport the phenotype.
Figure 4: Autophagy inhibition affects diverse cell cycle regulators.
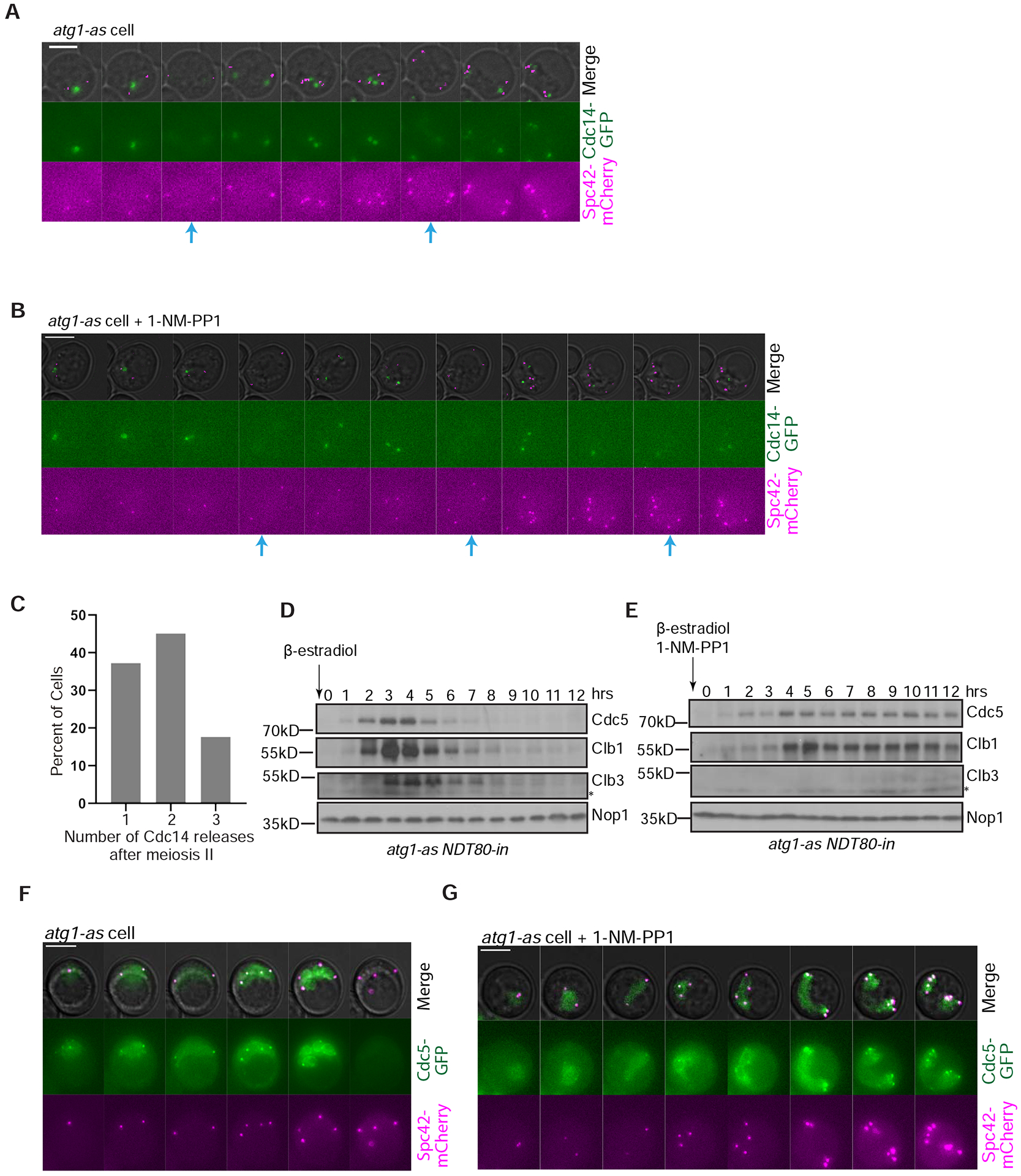
A,B) Representative time-lapse images of an atg1-as cell undergoing synchronized meiosis in the absence (A) and presence (B) of 1-NM-PP1. Blue arrows show Cdc14 release from the nucleolus. Scale Bar: 5μm. C) Graph showing the percent of cells in part B with the indicated number of Cdc14 releases after meiosis II (n=50 cells). D,E) Western blotting analysis of the indicated cell cycle regulators following release from prophase I arrest in the absence (D) and presence of 1-NM-PP1 (E). Nop1 is a nucleolar protein used as a loading control. F,G) Representative time-lapse images of atg1-as cells undergoing synchronized meiosis in the absence (A) and presence of 1-NM-PP1 (B). Scale Bars: 5μm.
Next, we analyzed several cell cycle regulators that are meiosis II-specific or involved in meiosis exit for clues to the underlying mechanism behind the phenotype we observed. Normally, expression of the B-type cyclin Clb1 drives CDK activity during meiosis I and stays present in meiosis II; but, CDK-Clb1 activity is superseded by CDK-Clb3 activity for sister chromatid separation in meiosis II, with Clb3 as the meiosis II-specific cyclin (Carlile and Amon, 2008; Miller et al., 2012). Clb3 undergoes proteasomal degradation at the end of meiosis, a process driven by the preceding activation of the APC/C ubiquitin ligase complex by Ama1 (APCAma1). Lastly, we analyzed Cdc5 because this polo kinase localizes to the nucleus and SPBs during meiosis I and II and, similar to Clb3, its clearance during meiotic exit is mediated by APCAma1 (Arguello-Miranda et al., 2017; Okaz et al., 2012). We confirmed by western blotting and microscopy that these temporal changes in cyclin and Cdc5 expression occurred as expected in untreated Atg1-as cells (Figure 4D,F). However, with Atg1-as inhibition, Clb1 rose normally during meiosis I, was present normally in meiosis II, but then persisted throughout the additional rounds of chromosome segregation; Clb3 levels failed to increase throughout the time course; and, Cdc5 levels persisted throughout the additional rounds of chromosome segregation (Figure 4E,G).
How could an autophagic degradation mechanism regulate the levels of such diverse cell cycle regulators? The production of Clb3 is directly controlled by Rim4, a translational repressor that forms amyloid-like aggregates, which sequester CLB3 mRNA and other transcripts important for meiosis II and sporulation (Berchowitz et al., 2013; Berchowitz et al., 2015). Intriguingly, AMA1 mRNA is among these latter Rim4 targets, which provides a parsimonious explanation for the observed temporal coincidence of its translational repression with that of the more established CLB3 as a Rim4 target (Berchowitz et al., 2013; Brar et al., 2012). At the onset of meiosis II, Rim4 is phosphorylated and degraded to enable translation of its target mRNAs. We hypothesized that Atg1-as inhibition interferes with normal Rim4 destruction by autophagy, thus leading to constitutive CLB3 and AMA1 mRNA translational repression, with the latter mechanism accounting for the apparent defect in APCAma1-mediated clearance of Clb1 and Cdc5. To test this idea, we looked for evidence of Rim4-GFP processing to yield a stable, cleaved GFP product in the vacuole, which is a commonly used metric for autophagic flux of GFP fusion proteins (Meiling-Wesse et al., 2002). Indeed, we observed that Rim4-GFP in untreated Atg1-as cells was completely converted to cleaved GFP following release from prophase I arrest (Figure 5A–B). Importantly, treatment of cells with 1-NM-PP1 decreased Rim4-GFP processing, thus resulting in the persistence of full-length Rim4-GFP along with a residual amount of cleaved GFP already present prior to treatment and from incomplete blockage of autophagy by 1-NM-PP1. We validated these findings by microscopy analysis, which showed that 1-NM-PP1 blocks a 40% decrease in cytosolic Rim4-GFP fluorescence seen in untreated cells as they transition from meiosis I to meiosis II (Figure 5C–E). These results suggest that loss of autophagy prevents Rim4 degradation and leads to defective translational control of several meiosis II-specific cell cycle regulators.
Figure 5: Autophagic processing of Rim4-GFP and phenotypic suppression by engineered production of Rim4 targets.
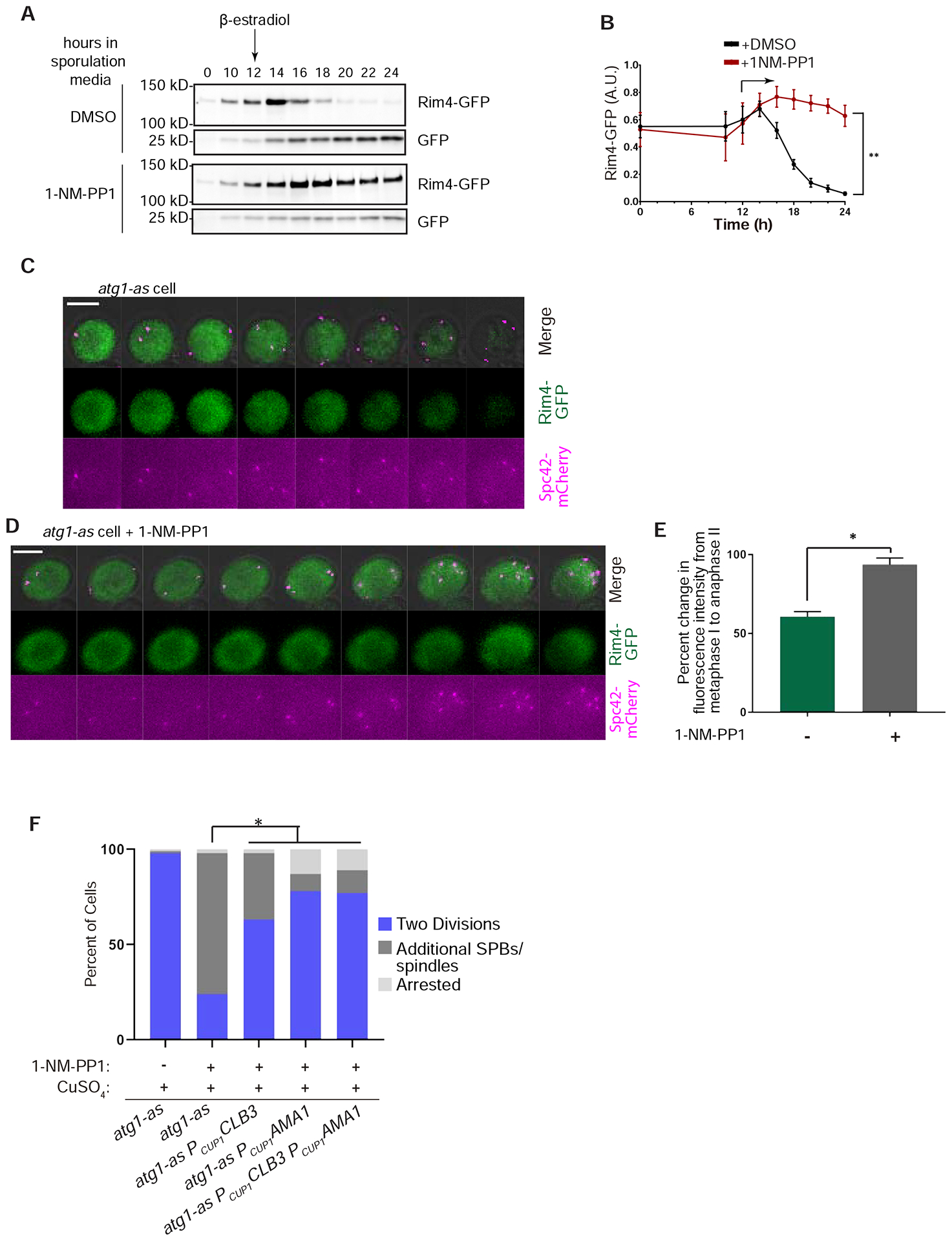
A) Representative Western blotting analysis of GFP in lysates derived from RIM4-GFP atg1-as NDT80-in cells undergoing synchronized meiosis under mock (DMSO) or 1-NM-PP1 treatment. B) Quantification of Western blotting data from three independent experiments collected as in part A. Rim4-GFP is normalized by total GFP (Rim4-GFP + GFP). Statistical significance was determined by a 2-way ANOVA. * represents statistical significance (p<0.01) C, D) Representative time-lapse images of atg1-as cells (without NDT80-in) undergoing meiosis in the absence (C) and presence of 1-NM-PP1 (D). Scale Bar: 5μm. E) GFP fluorescence intensity measurements were collected from atg1-as RIM4-GFP cells undergoing meiosis in the absence or presence of 1-NM-PP1, as indicated (n=35 cells from 3 independent experiments). Shown is the average and standard deviation of percent decrease in fluorescence intensity between metaphase I and anaphase II. * represents statistical significance (p<.001; rX contingency table). F) Percent of atg1-as, atg1-as PCUP1CLB3, atg1-as PCUP1AMA1, and atg1-as PCUP1CLB3 PCUP1AMA1 cells that undergo the indicated synchronized meiotic outcome in the presence of 1-NM-PP1. Also shown are data from control atg1-as cells in which autophagy was not inhibited. Copper sulfate was added to all cells at the time of arrest release. Only cells that were in prophase I at the time of arrest release were analyzed. Asterisk represents statistically significant differences (n = >100 cells for each genotype; p <.001, rx Contingency Tables).
Engineered expression of Rim4 mRNA targets restores normal meiotic exit in the absence of autophagy
Lastly, we asked if we could suppress the phenotypes of Atg1-as inhibition by engineering expression of Clb3 and Ama1. For technical reasons, we could not directly monitor Ama1 expression so we used the levels of APCAma1 substrates Cdc5 and Clb3 as proxies. Replacing the promoter and 5’UTR, of CLB3 and AMA1 with the corresponding regions of the CUP1 gene (PCUP1CLB3 or PCUP1AMA1) enabled us to induce Clb3 production and apparent APCAma1 activity by addition of copper sulfate (Supplemental Figure S4A–C) (Carlile and Amon, 2008). Both PCUP1CLB3 atg1-as cells and PCUP1AMA1 atg1-as cells treated with copper and 1-NM-PP1 still had a somewhat delayed meiosis I and meiosis II, similar to control atg1-as cells (Supplemental Figure S4D). Strikingly, this treatment resulted in only 34% of PCUP1CLB3 atg1-as cells and 9% of PCUP1AMA1 atg1-as cells undergoing additional rounds of SPB duplication and spindle formation (compared to 74% of control atg1-as cells) (Figure 4H). We did not observe further phenotypic suppression when we introduced PCUP1CLB3 into PCUP1AMA1 atg1-as cells. These results strongly argue that autophagy ensures the timed production of Rim4 targets during meiosis II, with Clb3 and Ama1 as two key limiting factors, to enable meiotic termination.
Discussion
Autophagy of protein aggregates, including certain amyloids, is known to be an important branch of protein quality control (Khaminets et al., 2016). Our work broadly illustrates that destruction of an amyloid by autophagy in the context of a complex gene expression program can also be a regulatory mechanism. Specifically, autophagy of the yeast amyloid-like protein Rim4 during meiosis II ensures the timely synthesis of several cell cycle regulators that are translationally repressed by Rim4. From a historical perspective, the meiotic cell cycle has been arrested at virtually every point by interfering with the function of diverse genes; our work adds to this mutant roster a unique phenotype in which cells undergo additional rounds of SPB duplication, spindle assembly, and chromosome segregation following meiosis II when autophagy is inhibited. Instead of resulting in germ cell proliferation, however, this unregulated, runaway process causes cell death due to abnormal chromosome segregation and abortive gametogenesis.
Very little is known about mechanisms that generate two and only two meiotic divisions by inducing cell cycle exit following meiosis II. Previous work has highlighted the importance of proteasomal degradation in meiotic exit. For example, in Arabidopsis thaliana, the TDM1 protein is likely an anaphase promoting complex (APC/C) component that is inactive in meiosis I but required for meiosis II, meiotic exit, and the appropriate number of meiotic divisions (Cifuentes et al., 2016; Cromer et al., 2012; Glover et al., 1998; Ross et al., 1997). A dominant mutant allele of TDM1 results in meiotic exit after meiosis I (Cifuentes et al., 2016). And, in budding yeast, the APC bound to its meiosis-specific activator Ama1 (APCAma1) has an important role in meiotic termination by targeting cell cycle regulators for degradation at the end of meiosis II (Arguello-Miranda et al., 2017). Our work argues that autophagy indirectly promotes APCAma1 activity via Rim4 translational control of AMA1 mRNA. However, cells lacking Ama1 fail to form spores but unlike autophagy mutants do not form additional SPBs and spindles after meiosis II (Cooper et al., 2000). Thus, we propose that our mutant phenotype is minimally the combined product of reduced APCAma1 activity and mis-regulation of other Rim4 targets or autophagy substrates.
Yeast meiosis takes place under starvation conditions that potently induce autophagy by activating the Atg1 kinase. At least in starved vegetative cells, preferential substrate degradation by autophagy has been shown to depend on the presence of receptor proteins that mediate physical interactions between substrates and the autophagosome membrane (Gatica et al., 2018; Johansen and Lamark, 2019; Kirkin, 2019). In addition, receptor-bound substrates are capable of directly activating Atg1 to locally induce their own autophagic engulfment (Kamber et al., 2015; Ravenhill et al., 2019; Torggler et al., 2016; Turco et al., 2019; Vargas et al., 2019). These mechanisms for substrate engagement can be controlled by kinases to provide an additional layer of regulation (Farre and Subramani, 2016). Intriguingly, phosphorylation of Rim4 by the Cdk-like kinase Ime2 has recently been shown to play a role in Rim4 clearance during meiosis (Carpenter et al., 2018). This raises the exciting possibility for a new mechanistic precedent in which a master cell cycle kinase gates substrate access to the autophagic machinery.
STAR Methods
LEAD CONTACT AND MATERIALS AVAILABILITY:
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Soni Lacefield (sonil@indiana.edu). All unique budding yeast strains and plasmids generated in this study are available upon request without restriction.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Budding Yeast Strains and Manipulations
Saccharomyces cerevisiae strains are derivatives of W303 (ade2–1 his3–11,15 leu2–3,112 trp1–1 ura3–1 can1–100). Strains used in this study can be found in Table S1. Standard PCR-based methods were used for tagging genes, deleting genes, and swapping promoters. In this method, constructs with markers/tags were amplified by PCR and then transformed into the budding yeast strain (Janke et al., 2004; Longtine et al., 1998; Sheff and Thorn, 2004). All manipulations were verified by PCR. The gatekeeper residue change (M102G) used to create an analog sensitive (as) version of Atg1 (Atg1-as) has been previously described and crossed into all of the strains used in this study (Blethrow et al., 2004; Kamber et al., 2015). The atg1-as mutation was confirmed by sequencing. The strains with GAL-NDT80 GAL4-ER were constructed by replacing the endogenous NDT80 promoter with the inducible GAL1,10 promoter using PCR off of a construct and then transformation (GAL-NDT80) in strains also expressing a Gal4-estrogen receptor fusion protein (GPD1promoted-GAL4.ER inserted at URA3 locus) (Carlile and Amon, 2008). The NDT80 transcription from the GAL1,10 promoter was induced by the addition of 1 μM ß-estradiol to the medium. Strains with a β-estradiol-inducible GFP-ATG8 transgene were made by homologous recombination of a Z4EV expression cassette with a Z4EV-driven (ZD) promoter (McIsaac et al., 2014) upstream of the GFP-ATG8 open reading frame at the LEU2 locus. Zip1 was tagged with GFP using a construct that was previously generated and described (Scherthan et al., 2007). Plasmids with GFP-TUB1, LACO, and CUP1prGFP-LACI were integrated into the genome (Straight et al., 1997). 2μ and CEN plasmids with SPO21-GFP, SPO74-GFP, and mKateSPO2051−91 were gifts from A. Neiman and were transformed into yeast (Mathieson et al., 2010; Neiman, 2011; Nickas et al., 2003).
Media
The following media were used in this work: YPD (2% peptone, 1% yeast extract, 2% glucose), YPA (2% peptone, 1% yeast extract, 2% potassium acetate), SD (0.67% yeast nitrogen base, 2% glucose, auxotrophic amino acids and vitamins), YNA (0.125% yeast extract, 2% potassium acetate), and standard sporulation medium SPM (0.6%−1% potassium acetate, pH=8.5). SD-dropout medium was made with dropout stock powder lacking histidine, tryptophan, leucine, uracil and/or methionine. Starvation experiments were performed in synthetic minimal medium lacking nitrogen, SD-N (0.17% yeast nitrogen base without amino acids, 2% glucose).
METHOD DETAILS
Sporulation, vegetative growth, and Synchronization by Ndt80 Arrest/Release
To induce meiosis for time-lapse imaging, cells were grown in YPD for 24hrs at 30°C, diluted 1/40 into YPA, grown at 30°C for 12hrs and then washed twice in water and transferred into SPM at 25°C. For biochemical experiments requiring synchronization, strains were grown on YPG (3% glycerol) plates at 30°C for 2 days. A single colony was picked, spread on YPD plates, and grown until cells formed a lawn (~24 hours). Next, cells were suspended in YPA to OD600 = 0.3 and grown overnight (16~18 hours). Cells were then pelleted, washed with water twice and resuspended in SPM to a final OD600 =3.0. To assess the percent spore formation, cells were counted after 48 hours in sporulation medium. At least 100 cells for each strain were counted under bright field OLYMPUS microscope (BX40, 40x objective).
For the vegetative growth experiment to validate the GFP-Atg8/Atg1-as system, yeast cells were cultured in SD medium to log phase (OD=0.8~1.0) and were then switched to SD-N medium for the indicated times (Feng et al., 2015; Klionsky et al., 2016).
For synchronizing NDT80-in cells, following incubation in SPM for 12 hours, strains containing GAL-NDT80 GAL4-ER were released from the prophase I arrest by addition of 1μM β-estradiol (10mM stock in ethanol, Sigma E2758–1G). To inhibit Atg1-as, 3–5μM of 1-NM-PP1, was added (10mM stock in DMSO). For induction of PCUP1AMA1 and PCUP1CLB3, 50μM copper sulfate was added one hour after beta-estradiol addition.
Inducible degradation of Atg14
We used a previously described degron sequence to tag the N-terminus of ATG14 at its endogenous locus (N-deg-ATG14) (Taxis and Knop, 2012). The construct was amplified by PCR and transformed into the yeast strains. The tag was verified by PCR. In this inducible degradation system, cleavage of the (latent) degron by the tobacco etch virus (TEV) protease exposes an active N-end rule degron that can enable rapid destruction of the tagged protein by the ubiquitin/proteasome system. TEV protease expression was controlled by the GAL1,10 promoter in the NDT80-in genetic background to initiate activation of the degradation system with 1μM β-estradiol, concomitantly with prophase I release.
Time-lapse Imaging
Time-lapse imaging was performed by loading cells onto a coverslip attached to a chamber. Cells were adhered to the coverslip with concanavalin A (Sigma, 1mg/ml in phosphate-buffered saline). Pre-conditioned SPM was added to the chamber. Cells were imaged on three microscopes. First, we used a Nikon Ti-E inverted microscope equipped with a 60X oil objective (PlanApo 1.4NA), a Lambda 10–3 optical filter changer and smartshutter (Sutter instrument), GFP and mCherry filters (Chroma Technology), and Cool-SNAPHQ2 CCD camera (Photometrics). Z-stacks of 5×1.2 μm sections were acquired in 10-minute intervals for 12–15hrs with exposure times of 60–70 ms for brightfield and 700–900 ms for GFP and mCherry. Z-stacks were combined into a single maximum intensity projection with NIS-Elements software (Nikon). Second, we used a DeltaVision Elite equipped with an Olympus 60X oil objective (PlanApo 1.4 NA), a sCMOS Edge 5.5 camera, filters for GFP/FITC and mCherry/Alexa594, and SoftWorx imaging software. For mCherry, we use 5% transmittance and 400ms exposures. For GFP, we use 2% transmittance and 200ms exposure. Z-stacks of 5×1.2 μm sections were acquired in 10-minute intervals for 12–15hrs. Third, we used a DeltaVision personal DV equipped with an Olympus 60X oil objective (PlanApo 1.4 NA), a CoolSNAPHQ2 CCD camera, and filters for GFP/FITC and mCherry/Alexa594, and SoftWorx imaging software. For mCherry, we use 5% transmittance and 400ms exposures. For GFP, we use 5% transmittance and 300ms exposure. Z-stacks of 5×1.2 μm sections were acquired in 10-minute intervals for 12–15hrs.Z-stacks of 5×1.2 μm sections were acquired in 10-minute intervals for 12–15hrs.
Cell lysate preparation and Western blotting analysis
For the autophagy assay experiments, measuring GFP-Atg8 and Rim4-GFP degradation, yeast cells were put in sporulation medium, as described above. 1 OD600 of Yeast cells were collected, resuspended and boiled at 95°C for 5 min in 100μL 2× SDS-PAGE sample Buffer (62.5 mM Tris-HCl, pH=6.8; 2% SDS; 0.05% BPB; 10% Glycerol; 5% 2-Mercaptoethanol; 1× Protease Inhibitor Cocktail (Roche, 11873580001), 10 mM PMSF), and spun down at 14,000 rpm for 5 min. Supernatant samples were resolved by SDS-PAGE (70 min at 195V) using 4–20% Criterion™ TGX Stain-Free™ gel (Bio-Rad, 5678095) and electroblotted onto nitrocellulose membranes (Bio-Rad, 1620115) using the Tran-blot SD semi-dry transfer cell (Bio-Rad, 1703940). After blocking with 5% milk, antibodies were applied for immunoblotting.
For Western blotting analysis of cyclins and Cdc5, 5mls of a meiosis culture were collected at each timepoint and spun down. 5mls of 10% trichloroacetic acids (TCA) was added on ice for 10mins. The cells were spun down, TCA removed, and 1ml of acetone was added and cells were vortexed. The cells were spun down and acetone removed. The addition on TCA and acetone was repeated twice. Tubes were left with caps open to dry in the hood for 2–3 hrs. For the protein extraction, 200μl of protein breakage buffer (60mM Tris pH7.5, 1.2mM EDTA pH8.0, 3.3mM DTT, protease inhibitor tablet) was added along with 200μl of glass beads. Cells were broken on vortex six times in 1min pulses, with 1min on ice in between. 100μl of 3×SDS buffer was added and tubes were boiled for 5mins. The supernatant was resolved on a SDS PAGE at 150 volts for 1 hr and transferred onto PVDF membrane. The membranes were blocked with 5% milk in TBST (Clb1, Cdc5, Nop1) or 2% BSA in TBST with 0.5M NaCl (Clb3).
Antibodies used in this work include: Monoclonal anti-GFP mouse IgG (Roche, 11814460001, 1:5000), monoclonal anti-mCherry Rat IgG (Thermo Fisher Scientific, M11217, 1:5000), polyclonal anti-Hexokinase 1 Rabbit IgG (United States Biological, 169073, 1:10000), polyclonal anti-Clb3 rabbit (generous gift of A. Rudner, 1:2,000), monoclonal anti-Nop1 mouse (EnCor, MCA-28F2, 1:10,000), polyclonal anti-Clb1 goat (Santa Cruz Biotechnology, sc-7647, 1:1,000), polyclonal anti-Cdc5 goat (Santa Cruz Biotechnology, sc-6733, 1:1,000). ECL-anti-rabbit HRP IgG (GE Healthcare, NA9340V, 1:5,000), ECL-antimouse IgG HRP (GE Healthcare, NA9310V, 1:5,000), anti-goat IgG HRP (R&D systems, HAF109, 1:5,000). StarBright® B700 labeled goat anti-mouse IgG secondary antibody (Bio-Rad, 12004158, 1:5000), Alexa Fluor® 488 labeled goat anti-rabbit IgG secondary antibody (Thermo Fisher Scientific, A11034, 1:5000), Cy3 labeled goat anti-rat IgG secondary antibody (Thermo Fisher Scientific, A10522, 1:5000). For detection and quantitative analysis of the autophagy assays, the western blot images were captured by ChemiDoc™ MP imaging system (Bio-Rad, 12003154), and analyzed using Image Lab™ (Ver. 6.0.1) software (Bio-Rad). For the timecourses of cyclins and Cdc5, the chemiluminescence was captured on autoradiography film (MidSci, XC59X).
Autophagy flux analysis by GFP-Atg8 processing
Autophagy was monitored with a GFP-Atg8 cleavage assay (Klionsky et al., 2016). Samples were isolated at indicated timepoints and the proteins were isolated as indicated in the section above. Cleavage by vacuolar protease mediated conversion of GFP-Atg8 to free GFP was detected by IB using GFP antibody and quantified using Image Lab™ (Ver. 6.0.1) software (Bio-Rad). To facilitate analysis of autophagy flux during the meiotic divisions, we engineered production of GFP-Atg8 from a β-estradiol-inducible transgene with addition of 1μM β-estradiol. The transgene was integrated into the genome. Flux was defined as 2(GFPn+1 − GFPn)/(GFP-Atg8n+1 + GFP-Atg8n) where n and n+1 represent signals from two sequential time points.
Determination of meiotic cell progression by Immunofluorescence
Immunofluorescence was performed to determine meiotic cell progression in Supplemental Figure S2. Specifically, after Ndt80 was induced with β-estradiol, cells were harvested every 30 minutes and fixed by pre-cold 3.7% formaldehyde. Fixed cells were washed with 0.1M KPi (4.84 g/L Potassium phosphate dibasic; 9.83 g/L Potassium phosphate monobasic; PH=6.4) buffer and 1.2M sorbitol citrate solution, and then digested by 1.4mg/ml 20T Zymolyase (amsbio, 120491) and Glusulase (PerkinElmer, NEE154001EA). Cells were spread on microscope slides and blotted with anti-tubulin Alexa Fluor® 594 antibody (Abcam, dilution 1:200) for 2 hours, treated with Prolong gold antifade reagent containing DAPI after washing, and covered with a coverslip. Spindle morphologies were then classified. Specifically, Metaphase I cells were defined as cells with a single DAPI mass spanned by a thick, short and bipolar meiotic spindle (approximately 2–3μm in length). Anaphase I cells were defined as cells with two parts of distinct (though not always separated) DAPI masses, and a single long spindle that spans both DAPI masses. Metaphase II cells were defined as cells with two separate DAPI masses with each spanned by a bipolar, thick and short meiotic spindle. Anaphase II cells were defined as cells with four distinct (though not always separated) parts of DAPI masses with two long spindles (usually crossed). At least 100 cells were counted at each time point, to determine the percentage of cells at each meiotic cell stages.
QUANTIFICATION AND STATISTICAL ANALYSIS
For the Rim4-GFP quantification of fluorescence intensity, images were first captured with time lapse microscopy on the Nikon TiE. The fluorescence intensity was measured in ImageJ (NIH). The Z-stacks were summed (sum slices) and a region of interest was chosen inside the cytosol, avoiding the vacuole (as seen in the brightfield image) when the cell was at metaphase I and anaphase II (as monitored by SPB separation). A background intensity was measured next to the cell. The background fluorescence intensity was subtracted from the GFP intensity in the cell. That number in anaphase II was subtracted from the intensity in metaphase I (as staged by spindle length).
Western blot images for the autophagy assays were captured by ChemiDoc™ MP imaging system (Bio-Rad, 12003154), and analyzed using Image Lab™ (Ver. 6.0.1) software (Bio-Rad).
Number of cells counted (n), number of repeats, p values, and the statistical tests performed to determine significance are documented in the figure legends. Statistical tests, rX Contingency Tables, Fisher’s exact test, and 2-way ANOVA were performed using the software program GraphPad Prism 7.03 (Graphpad).
DATA AND CODE AVAILABILITY
The paper does not generate any new code. The original source data for the current study have not been deposited in a public repository due to the extent and size of image files, but are available from the corresponding authors upon request.
ADDITIONAL RESOURCES
No additional resources.
Supplementary Material
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Monoclonal anti-GFP mouse IgG (1:5,000) | Roche | 11814460001 |
| Monoclonal anti-mCherry rat IgG (1:5,000) | Thermo Fisher Scientific | M11217 |
| Polyclonal anti-hexokinase 1 Rabbit IgG (1:10,000) | United States Biological | 169073 |
| Monoclonal anti-Nop1 mouse (1:10,000) | Encor | MCA-28F2 |
| Polyclonal anti-Clb3 rabbit (1:2,000) | Gift from A. Rudner | N/A |
| Polyclonal anti-Clb1 goat (1:1,000) | Santa Cruz Biotechnology | Sc-7647 |
| Polyclonal anti-Cdc5 goat (1:1,000) | Santa Cruz Biotechnology | Sc-6733 |
| ECL-anti-rabbit HRP IgG (1:5,000) | GE Healthcare | NA9340V |
| ECL-antimouse IgG HRP (1:5,000) | GE Healthcare | NA9310V |
| anti-goat IgG HRP (1:5,000) | R&D systems | HAF109 |
| StarBright® B700 labeled goat anti-mouse IgG secondary antibody (1:5000) | Bio-Rad | 2004158 |
| Alexa Fluor® 488 labeled goat anti-rabbit IgG secondary (1:5000) | Thermo Fisher Scientific | A11034 |
| Cy3 labeled goat anti-rat IgG secondary (1:5000) | Thermo Fisher Scientific | A10522 |
| Anti-tubulin Alexa Fluor 594 (1:200) | Abcam | ab195884 |
| Bacterial and Virus Strains | ||
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Bacto Peptone | BD Biosciences | Cat#211820 |
| Bacto Yeast Extract | BD Biosciences | Cat#212720 |
| Yeast Nitrogen Bases without amino acids | Difco Laboraties | Cat#291920 |
| Glucose/Dextrose | Fisher BioReagents | Cat#D16–10 |
| Potassium Acetate | Fisher BioReagents | Cat#P171–500 |
| Adenine Hemisulfate Salt | Sigma | Cat#A9126 |
| L-Tryptophan | Sigma | Cat#T0254 |
| Beta-estradiol | Sigma | Cat#E2758 |
| 1-NM-PP1 | Gift of E. O’Shea and A. Sejr Hansen | N/A |
| Glycerol | VWR Chemicals | Cat#56-81-5 |
| Copper Sulfate | Sigma | Cat#7758-99-8 |
| 4–20% Criterion™ TGX Stain-Free™ gel | Bio-Rad | Cat#5678095 |
| Tris-HCl | Sigma | Cat#76-03-9 |
| Sodium Dodecyl Sulfate | Bio-Rad | Cat#1610301 |
| BPB | Sigma | Cat#B0126 |
| 2-Mercaptoethanol | Sigma | Cat#M3148 |
| Protease Inhibitor Cocktail | Roche | Cat#11873580001 |
| PMSF | Sigma | Cat#78830 |
| Trichloroacetic acid | Sigma | Cat#76-03-9 |
| EDTA | Sigma | Cat#6381-92-6 |
| DTT | Bio-Rad | Cat#161–0611 |
| Protease inhibitor tablet | Roche | Cat#04906845001 |
| Tween | Sigma | Cat#9005-64-5 |
| Bovine Serum Albumin | Sigma | Cat#9048-46-8 |
| Sodium Chloride | EMD | Cat#7647-14-5 |
| Formaldehyde | Fisher BioReagents | Cat#BP531–500 |
| Potassium Phosphate Dibasic | Fisher BioReagents | Cat#BP363–500 |
| Potassium Phosphate Monobasic | Sigma | Cat#P5655–1 KG |
| Sorbitol Citrate | VWR Chemicals | Cat#JTV045–7 |
| Glusulase | Perkin Elmer | Cat#NEE154001EA |
| Prolong gold antifade with DAPI | Thermo Fisher Scientific | Cat#P36941 |
| Zymolyase (20T) | Amsbio | Cat#120491 |
| Critical Commercial Assays | ||
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| See Table S1 | Lacefield, Wang, and Denic Labs | N/A |
| Oligonucleotides | ||
| See Table S2 | Integrated DNA Technologies | N/A |
| Recombinant DNA | ||
| PZEVATG1:LEU2 | This paper | |
| PTUB1-GFP-TUB1:LEU2 | Gift of A. Murray | Straight et al., 1997 |
| PHIS3-GFP-TUB1:HIS3 | Gift of A. Murray | Straight et al., 1997 |
| PCUP1-GFP-LacI:HIS3 | Gift of A. Murray | Straight et al., 1997 |
| PZIP1-GFP-ZIP1:URA3 | Gift of D. Kaback | Scherthan et al., 2007 |
| PGPD1-GAL4(848).ER:URA3 | Gift of A. Amon | Carlile et al., 2008 |
| PGAL1,10-TEV:HphMX | Gift of M. Knop | Taxis and Knop, 2012 |
| pSPO21-GFP:TRP1:CEN | Gift of A. Neiman | Mathieson et al., 2010 |
| pMPC54-GFP:TRP1:2μm | Gift of A. Neiman | Mathieson et al., 2010 |
| pSPO74-GFP:TRP1:2μm | Gift of A. Neiman | Nickas et al.,, 2003 |
| pmKATE-SPO2051–91:HIS3:2um | Gift of A. Neiman | Neiman, A, 2011 |
| PZ4EV-GFP-ATG8:LEU2 | This study | N/A |
| Software and Algorithms | ||
| Image Lab™ (Version 6.0.1) | Bio-Rad | https://www.bio-rad.com/en-us/product/image-lab-software |
| Image J | National Institutes of Health (Public Domain) | https://imagej.nih.gov/ij/ |
| NIS Elements Viewer v4.20.00 (Build972) LO, 32 bit | Nikon | https://www.nikoninstruments.com/Products/Software/NIS-Elements-Advanced-Research/NIS-Elements-Viewer |
| GraphPad Prism 7.03 | GraphPad Software, Inc | https://www.graphpad.com/ |
| Other | ||
Highlights.
Autophagy is required for meiotic exit
Autophagy prevents additional rounds of chromosome segregation after meiosis II
Autophagy degrades the amyloid-like translational repressor Rim4 in meiosis II
Inhibition of autophagy leads to aberrant persistence of meiotic regulators
Acknowledgements:
We thank Jim Powers and the Indiana University Light Microscopy and Imaging Center for imaging support. We thank Aaron Neiman and Adam Rudner for reagents. We thank A. Murray, F. Solomon, B. Levine, and members of the Lacefield lab for comments on the manuscript. This work was supported by a grants from the NIH to SL (GM105755) and VD (GM127136) and by funding from Nancy Cain and Jeffrey A. Marcus Scholar in Medical Research, in honor of Dr. Bill S. Vowell, to FW. Authors declare no competing interests.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
The authors have nothing to declare.
References
- Arguello-Miranda O, Zagoriy I, Mengoli V, Rojas J, Jonak K, Oz T, Graf P, and Zachariae W (2017). Casein Kinase 1 Coordinates Cohesin Cleavage, Gametogenesis, and Exit from M Phase in Meiosis II. Developmental cell 40, 37–52. [DOI] [PubMed] [Google Scholar]
- Benjamin KR, Zhang C, Shokat KM, and Herskowitz I (2003). Control of landmark events in meiosis by the CDK Cdc28 and the meiosis-specific kinase Ime2. Genes Dev 17, 1524–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Gajadhar AS, van Werven FJ, De Rosa AA, Samoylova ML, Brar GA, Xu Y, Xiao C, Futcher B, Weissman JS, et al. (2013). A developmentally regulated translational control pathway establishes the meiotic chromosome segregation pattern. Genes Dev 27, 2147–2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Kabachinski G, Walker MR, Carlile TM, Gilbert WV, Schwartz TU, and Amon A (2015). Regulated Formation of an Amyloid-like Translational Repressor Governs Gametogenesis. Cell 163, 406–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blethrow J, Zhang C, Shokat KM, and Weiss EL (2004). Design and use of analog-sensitive protein kinases. Curr Protoc Mol Biol Chapter 18, Unit 18 11. [DOI] [PubMed] [Google Scholar]
- Brar GA, Yassour M, Friedman N, Regev A, Ingolia NT, and Weissman JS (2012). High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science 335, 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buonomo SB, Rabitsch KP, Fuchs J, Gruber S, Sullivan M, Uhlmann F, Petronczki M, Toth A, and Nasmyth K (2003). Division of the nucleolus and its release of CDC14 during anaphase of meiosis I depends on separase, SPO12, and SLK19. Developmental cell 4, 727–739. [DOI] [PubMed] [Google Scholar]
- Carlile TM, and Amon A (2008). Meiosis I is established through division-specific translational control of a cyclin. Cell 133, 280–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter K, Bell RB, Yunus J, Amon A, and Berchowitz LE (2018). Phosphorylation-Mediated Clearance of Amyloid-like Assemblies in Meiosis. Developmental cell 45, 392–405 e396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cifuentes M, Jolivet S, Cromer L, Harashima H, Bulankova P, Renne C, Crismani W, Nomura Y, Nakagami H, Sugimoto K, et al. (2016). TDM1 Regulation Determines the Number of Meiotic Divisions. PLoS Genet 12, e1005856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper KF, Mallory MJ, Egeland DB, Jarnik M, and Strich R (2000). Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc Natl Acad Sci U S A 97, 14548–14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromer L, Heyman J, Touati S, Harashima H, Araou E, Girard C, Horlow C, Wassmann K, Schnittger A, De Veylder L, et al. (2012). OSD1 promotes meiotic progression via APC/C inhibition and forms a regulatory network with TDM and CYCA1;2/TAM. PLoS Genet 8, e1002865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enyenihi AH, and Saunders WS (2003). Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics 163, 47–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farre JC, and Subramani S (2016). Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol 17, 537–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Wu T, Dan X, Chen Y, Li L, Chen S, Miao D, Deng H, Gong X, and Yu L (2015). Phosphorylation of Atg31 is required for autophagy. Protein Cell 6, 288–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatica D, Lahiri V, and Klionsky DJ (2018). Cargo recognition and degradation by selective autophagy. Nature cell biology 20, 233–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover J, Grelon M, Craig S, Chaudhury A, and Dennis E (1998). Cloning and characterization of MS5 from Arabidopsis: a gene critical in male meiosis. Plant J 15, 345–356. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, et al. (2004). A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962. [DOI] [PubMed] [Google Scholar]
- Johansen T, and Lamark T (2019). Selective Autophagy: ATG8 Family Proteins, LIR Motifs and Cargo Receptors. J Mol Biol. [DOI] [PubMed] [Google Scholar]
- Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, and Ohsumi Y (2000). Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol 150, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamber RA, Shoemaker CJ, and Denic V (2015). Receptor-Bound Targets of Selective Autophagy Use a Scaffold Protein to Activate the Atg1 Kinase. Mol Cell 59, 372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S, Okano T, Ohsumi M, and Ohsumi Y (1998). Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J Biol Chem 273, 22284–22291. [DOI] [PubMed] [Google Scholar]
- Khaminets A, Behl C, and Dikic I (2016). Ubiquitin-Dependent And Independent Signals In Selective Autophagy. Trends Cell Biol 26, 6–16. [DOI] [PubMed] [Google Scholar]
- Kihara A, Noda T, Ishihara N, and Ohsumi Y (2001). Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V (2019). History of the Selective Autophagy Research: How Did It Begin and Where Does It Stand Today? J Mol Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, et al. (2016). Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, and Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Marston AL, and Amon A (2004). Meiosis: cell-cycle controls shuffle and deal. Nat Rev Mol Cell Biol 5, 983–997. [DOI] [PubMed] [Google Scholar]
- Marston AL, Lee BH, and Amon A (2003). The Cdc14 phosphatase and the FEAR network control meiotic spindle disassembly and chromosome segregation. Developmental cell 4, 711–726. [DOI] [PubMed] [Google Scholar]
- Mathieson EM, Schwartz C, and Neiman AM (2010). Membrane assembly modulates the stability of the meiotic spindle-pole body. J Cell Sci 123, 2481–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIsaac RS, Gibney PA, Chandran SS, Benjamin KR, and Botstein D (2014). Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res 42, e48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiling-Wesse K, Barth H, and Thumm M (2002). Ccz1p/Aut11p/Cvt16p is essential for autophagy and the cvt pathway. FEBS Lett 526, 71–76. [DOI] [PubMed] [Google Scholar]
- Miller MP, Unal E, Brar GA, and Amon A (2012). Meiosis I chromosome segregation is established through regulation of microtubule-kinetochore interactions. Elife 1, e00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM (2011). Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189, 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas ME, Schwartz C, and Neiman AM (2003). Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot Cell 2, 431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaz E, Arguello-Miranda O, Bogdanova A, Vinod PK, Lipp JJ, Markova Z, Zagoriy I, Novak B, and Zachariae W (2012). Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell 151, 603–618. [DOI] [PubMed] [Google Scholar]
- Piekarska I, Kucharczyk R, Mickowska B, Rytka J, and Rempola B (2010). Mutants of the Saccharomyces cerevisiae VPS genes CCZ1 and YPT7 are blocked in different stages of sporulation. Eur J Cell Biol 89, 780–787. [DOI] [PubMed] [Google Scholar]
- Politi Y, Gal L, Kalifa Y, Ravid L, Elazar Z, and Arama E (2014). Paternal mitochondrial destruction after fertilization is mediated by a common endocytic and autophagic pathway in Drosophila. Developmental cell 29, 305–320. [DOI] [PubMed] [Google Scholar]
- Ravenhill BJ, Boyle KB, von Muhlinen N, Ellison CJ, Masson GR, Otten EG, Foeglein A, Williams R, and Randow F (2019). The Cargo Receptor NDP52 Initiates Selective Autophagy by Recruiting the ULK Complex to Cytosol-Invading Bacteria. Mol Cell 74, 320–329 e326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojansky R, Cha MY, and Chan DC (2016). Elimination of paternal mitochondria in mouse embryos occurs through autophagic degradation dependent on PARKIN and MUL1. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KJ, Fransz P, Armstrong SJ, Vizir I, Mulligan B, Franklin FC, and Jones GH (1997). Cytological characterization of four meiotic mutants of Arabidopsis isolated from T-DNA-transformed lines. Chromosome Res 5, 551–559. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Dalgaard JZ, Millar JB, and Arumugam P (2014). The Rim15-endosulfine-PP2ACdc55 signalling module regulates entry into gametogenesis and quiescence via distinct mechanisms in budding yeast. PLoS Genet 10, e1004456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, and Sato K (2011). Degradation of paternal mitochondria by fertilization-triggered autophagy in C. elegans embryos. Science 334, 1141–1144. [DOI] [PubMed] [Google Scholar]
- Scherthan H, Wang H, Adelfalk C, White EJ, Cowan C, Cande WZ, and Kaback DB (2007). Chromosome mobility during meiotic prophase in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 104, 16934–16939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheff MA, and Thorn KS (2004). Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast 21, 661–670. [DOI] [PubMed] [Google Scholar]
- Straight AF, Marshall WF, Sedat JW, and Murray AW (1997). Mitosis in living budding yeast: anaphase A but no metaphase plate. Science 277, 574–578. [DOI] [PubMed] [Google Scholar]
- Straub M, Bredschneider M, and Thumm M (1997). AUT3, a serine/threonine kinase gene, is essential for autophagocytosis in Saccharomyces cerevisiae. J Bacteriol 179, 3875–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, and Knop M (2012). TIPI: TEV protease-mediated induction of protein instability. Methods Mol Biol 832, 611–626. [DOI] [PubMed] [Google Scholar]
- Taxis C, Stier G, Spadaccini R, and Knop M (2009). Efficient protein depletion by genetically controlled deprotection of a dormant N-degron. Mol Syst Biol 5, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torggler R, Papinski D, Brach T, Bas L, Schuschnig M, Pfaffenwimmer T, Rohringer S, Matzhold T, Schweida D, Brezovich A, et al. (2016). Two Independent Pathways within Selective Autophagy Converge to Activate Atg1 Kinase at the Vacuole. Mol Cell 64, 221–235. [DOI] [PubMed] [Google Scholar]
- Torggler R, Papinski D, and Kraft C (2017). Assays to Monitor Autophagy in Saccharomyces cerevisiae. Cells 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya D, Gonzalez C, and Lacefield S (2011). The spindle checkpoint protein Mad2 regulates APC/C activity during prometaphase and metaphase of meiosis I in Saccharomyces cerevisiae. Mol Biol Cell 22, 2848–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya D, and Lacefield S (2013). Cdk1 modulation ensures the coordination of cell-cycle events during the switch from meiotic prophase to mitosis. Current biology : CB 23, 1505–1513. [DOI] [PubMed] [Google Scholar]
- Tsuchiya D, Yang Y, and Lacefield S (2014). Positive feedback of NDT80 expression ensures irreversible meiotic commitment in budding yeast. PLoS Genet 10, e1004398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto S, Kuma A, Murakami M, Kishi C, Yamamoto A, and Mizushima N (2008). Autophagy is essential for preimplantation development of mouse embryos. Science 321, 117–120. [DOI] [PubMed] [Google Scholar]
- Turco E, Fracchiolla D, and Martens S (2019). Recruitment and Activation of the ULK1/Atg1 Kinase Complex in Selective Autophagy. J Mol Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas JNS, Wang C, Bunker E, Hao L, Maric D, Schiavo G, Randow F, and Youle RJ (2019). Spatiotemporal Control of ULK1 Activation by NDP52 and TBK1 during Selective Autophagy. Mol Cell 74, 347–362 e346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen FP, Guo YS, Hu Y, Liu WX, Wang Q, Wang YT, Yu HY, Tang CM, Yang J, Zhou T, et al. (2016). Distinct temporal requirements for autophagy and the proteasome in yeast meiosis. Autophagy 12, 671–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Nair U, and Klionsky DJ (2008). Atg8 controls phagophore expansion during autophagosome formation. Mol Biol Cell 19, 3290–3298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z, Pascual C, and Klionsky DJ (2016). Autophagy: machinery and regulation. Microb Cell 3, 588–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. (2009). SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136, 308–321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The paper does not generate any new code. The original source data for the current study have not been deposited in a public repository due to the extent and size of image files, but are available from the corresponding authors upon request.


