Abstract
At present, the treatment of hyperuricemia is designed primarily to decrease the production of uric acid using xanthine oxidase inhibitors; however, the therapeutic effect is not satisfactory. Therefore, the key to the successful treatment of hyperuricemia is to increase the excretion of uric acid. The aim of present study was to construct uricase-expressing genetically engineered bacteria and analyze the effects of these engineered bacteria on the lowering of uric acid levels in a rat model of hyperuricemia. The uricase expression vector was constructed by gene recombination technology and transfected into Escherichia coli. The expression and activity of uricase were analyzed by SDS-PAGE analysis and Bradford assay. The water consumption, food intake, body weight, eosinophil count and intestinal histology, in addition to the levels of serum uric acid (SUA) and allantoin in the feces of the rats, were assessed. The intestinal contents of the rats were analyzed by 16S rDNA sequencing technology. The results demonstrated that uricase-expressing genetically engineered bacteria secreted active uricase. All rats exhibited a natural growth trend during the entire experiment, and the SUA of hyperuricemic rats treated with uricase-expressing engineered bacteria was significantly decreased. In conclusion, these results indicate that uricase secreted by recombinant uricase-expressing genetically engineered bacteria served an important role in decreasing SUA levels in a rat model of hyperuricemia.
Keywords: gene recombination technology, uricase-expressing genetically engineered bacteria, serum uric acid, hyperuricemia, enteric microorganism
Introduction
Uric acid is the product of purine metabolism in animals (1). Some animals decompose uric acid into allantoin by uricase and expel it from body. However, due to the lack of the uricase gene, the product of purine metabolism is excreted by humans in form of uric acid (2,3). Therefore, serum uric acid (SUA) is maintained at a high level in humans. When normal human purine metabolism is disrupted, the production and excretion of uric acid in the body are dysregulated, leading to hyperuricemia (4). The clinical diagnostic criteria for hyperuricemia are SUA >420 μmol/l in males and SUA >360 μmol/l in females (5). Recent studies have shown that following changes to the structure of the human diet, the incidence of hyperuricemia in the world is increasing (6–8), and the age of onset of hyperuricemia is decreasing (9,10).
It has been demonstrated that hyperuricemia is not only a risk factor for gout and chronic kidney disease, but also a risk factor for hypertension, hyperlipidemia, diabetes and cardiovascular disease, which are conditions that seriously affects human health (11–17). Therefore, xanthine oxidase inhibitors are commonly used in clinical practice to inhibit the synthesis of uric acid (18,19), but the effect is not as good as expected and has serious side effects (20–23). It was observed from previous studies that approximately two-thirds of uric acid in the human body is excreted by the kidneys, and one-third is excreted by intestinal microorganisms following degradation (24–28). Due to the limited water-solubility of uric acid, increasing the level of uric acid excretion in the kidneys will increase the burden on the organ and lead to renal diseases (29,30). Furthermore, recombinant uricase has been used to convert uric acid into allantoin in the human body in order to decrease the levels of uric acid, but its clinical application is limited due to its side effects, including immunogenicity of heterologous proteins (31,32). Therefore, increasing the excretion of uric acid through the intestine has become an alternative treatment strategy. In the present study, genetically engineered bacteria containing a recombinant uricase gene that could be expressed extracellularly without induction were constructed. The uricase secreted by the engineered bacteria in the intestinal tissues of a hyperuricemia rat model served a role in decreasing the levels of uric acid. The present study aimed to identify new methods for the treatment of hyperuricemia.
Materials and methods
Ethics statement
In the present study, all the experimental protocols involving animals approved by the Ethics Committee of Chongqing Medical University.
Vectors, bacterial strains and growth conditions
The plasmid containing the gene of staphylococcal protein A (spa) and uricase (UOX), pET-28a-sUOX (accession no. D32043.1) (33–36), was preserved by the Department of Biochemistry and Molecular Biology, Chongqing Medical University, and the plasmid was purchased from Beijing Beina Chuanglian Biotechnology Research Institute. The pMG36e vector was purchased from the Suzhou Bei Na Chuanglian Biological Corporation. The pUC57 vector containing the S-layer protein signal peptide gene (spo) (accession no. Z14250.1) of Lactobacillus brevis was purchased from the Chengdu Qingke Zixi Biological Company. In this study, Escherichia coli DH5α and BL-21 cells were used for cloning and expression, respectively, which were grown in LB medium (Beijing Dingguo Changsheng Biotechnology Co., Ltd.) broth containing 200 μg/ml erythromycin incubated at 37°C with 200 r/min rotation. Restriction enzymes XbaI, HindIII, SalI and T4 ligase and polymerase chain reaction (PCR) kit (catalog no. R060A) were purchased from Takara Biotechnology Co., Ltd.
Construction of recombinant plasmids pMG36e-sUOX and pMG36e-spo-uox
UOX and sUOX gene primers were designed by Primer 5.0 software (Premier Software, Inc.) using the UOX and sUOX sequences in the pET-28a-sUOX vector as templates and the corresponding restriction sites and protection base sequences were added to the primers. The primers were synthesized by Shanghai Shenggong Biological Engineering Co., Ltd. The sequences of primer are listed in Table I. The vector pet-28a-sUOX was used as a template, and corresponding target genes were amplified by PCR using primers OUX-forward (f), UOX-reverse (r), sUOX-f, sUOX-r and high-fidelity rTaq DNA polymerase (Takara Biotechnology Co., Ltd.). The PCR reaction conditions were as follows: Pre-denaturation at 94°C for 10 min, followed by 32 cycles of 94°C for 1 min, 60°C for 30 sec and 72°C for 2 min, and final extension for 10 min at 72°C. PCR products, and the pUC57 (containing the spo signal peptide) and pMG36e vectors were double digested with the corresponding restriction enzymes: The sUOX fragment and pMG36e vector were digested with restriction endonucleases XbaI and HindIII, respectively. Both ends of the UOX fragment were digested with SalI and HindIII. The spo fragment was digested with restriction endonucleases XbaI and SalI, and then ligated by T4 ligase. Next, the recombinant plasmids were introduced into E. coli DH5α and BL-21 cells. Then, these bacteria were spread onto LB agar plates containing erythromycin. Following successful culture, single colonies were selected and grown in LB liquid medium containing erythromycin.
Table I.
Target gene primer sequences.
| Primer | Primer sequence (5′-3′) | Restriction sites |
|---|---|---|
| UOX-F | GGTCGACATGTCAACAACGCTCTCATCCACC | SalI |
| UOX-R | GCGAAGCTTTTAGTGGTGGTGGTGGTGGTGCAACTTG | HindIII |
| sUOX-F | CGTCTAGAATGACTTTACAAATACATACAGGGGGTATTAATTTGAAAAAG | XbaI |
| sUOX-R | ATAAAGCTTTTAGTGGTGGTGGTGGTGGTG | HindIII |
The underlined letters indicate restriction enzyme cutting sites. UOX, uricase; sUOX, Staphylococcal Protein A + uricase; F, forward; R, reverse.
Secretion and expression of uricase
Positive clones were selected and grown in LB medium containing 200 μg/ml erythromycin, and incubated at 37°C with 200 r/min for 12 h. The bacterial solution was used to inoculate the LB medium at a ratio of 1:20 at 37°C for 200 r/min. Following agitation for 3, 6, 9 and 12 h, the supernatant of the bacterial solution was collected after centrifugation (4,000 × g, 15 min) at 4°C and concentrated by polyethylene glycol. The protein concentration was detected using the Bradford protein detection kit (Beyotime Institute of Biotechnology; cat. no. P0006). The steps were as follows: 10 μl protein standard was diluted by 0.9% NaCl to a concentration of 0.5 mg/ml. Subsequently, 0, 1, 2, 4, 8, 12, 16 or 20 μl of the protein standard was added to the standard wells of a 96-well plate, and then the standard dilution was added to make up to 20 μl. The sample (10 μl) was added to the sample wells of the 96-well plate and then made up to 20 μl with standard dilution. Then, 200 μl coomassie blue g-250 staining solution was added to each well, placed at room temperature for 5 min, and the absorbance was detected at 595 nm. Finally, the protein concentration in the samples was calculated according to the standard curve. The optimal protein expression time was determined by 10% SDS-PAGE analysis with 40 μg protein loaded per lane. Coomassie G-250 dye solution purchased from Beyotime Institute of Biotechnology was used for visualization at room temperature for 1 h.
The activity of uricase was determined, and the unit of activity of the enzyme was defined as: The amount of enzyme required to catalyze the decomposition of 1 μmol of uric acid per min at 40°C and pH 8.5. The protein content was determined using a Bradford assay.
Application of engineered bacteria in a rat model Establishment of rat model of hyperuricemia and administration of engineered bacteria
A total of 50 male SPF-grade Sprague-Dawley (SD) rats were purchased from the Animal Experimental Center of Chongqing Medical University. The rats were maintained at 24±2°C, 50–70% humidity and a 12 h light: Dark cycle (8:00 a.m.–8:00 p.m.). Rats were divided into the following 5 groups by random: Recombinant uricase-expressing genetically engineered E. coli bacteria group (E36eu); empty vector-expressing engineered E. coli bacteria group (E36e); E. coli group (E); carboxymethylcellulose suspension group (NS); and blank control (Con) group. The Con group was fed with water, the rats in the other groups were fed with 5% fructose and injected intraperitoneally with oteracil potassium (100 mg/kg/day) twice daily for 8 weeks to maintain a high uric acid state, as described previously (37–43). During the entire experiment, all rats were fed standard rat chow and were given ad libitum access to water.
The suspension of engineered bacteria (bacterial solution concentration of 0.02 g/ml) was made from carboxymethylcellulose solution. An total of 5 ml/kg/day was administered via intragastric gavage at 2:00 p.m. every day to the E36eu group (carboxymethylcellulose suspension administered with recombinant uricase E. coli genetically engineered bacteria), E36e group (carboxymethylcellulose suspension administered with empty vector E. coli engineered bacteria), E group (carboxymethylcellulose suspension administered with E. coli) and NS group (carboxymethylcellulose suspension).
Collection and processing of samples
During the experiment, the water consumption, food intake and body weight of rats were measured daily. After fasting rats for 12 h, at 8:00 a.m. every Sunday, 0.5–1.0 ml of blood was collected by tail cutting, and serum was collected by centrifugation (1,000 × g, 10 min) at 4°C.
i) Measurement of SUA levels. SUA levels were determined by enzyme colorimetry. The uric acid detection kit was purchased from Nanjing Jiancheng Technology Co., Ltd (cat. no. C012). The principle of measuring uric acid is that, in the protein-free filtrate, uric acid reduces phosphotungstic acid to generate tungsten blue, allantoin and carbon dioxide in the alkaline state. The shade of blue coloring is directly proportional to the concentration of uric acid. The kit was used as follows: A total of 0.2 ml serum, uric acid standard application solution and distilled water were added to the test tube, standard tube and blank tube respectively. Then, 2 ml tungstate protein precipitator was added to the 3 tubes. The fully mixed solutions were allowed to stand for 10 min, and then centrifuged at 1,000 × g for 5 min at 4°C. Following removal of 1.6 ml supernatant from each tube, 0.5 ml sodium tungstate reagent and phosphotungstic acid reagent were added to each tube, and the solutions were mixed well. After 10 min, the absorbance value of each tube was measured at a wavelength of 690 nm with 1 cm light diameter. A blank tube was used to calibrate the machine. Finally, the values of SUA were calculated according to the formula: SUA=OD of the test tube/(OD of the standard tube × content of the standard substance). At the end of the experiment, blood samples from the rats were used to coat slides. At room temperature, 0.5 ml Wright’s stain was added to the slides for 1 min, and then 1 ml buffer was added to mix with the Wright’s stain for 5 min. Finally, the dyeing solution was washed away by running water. Subsequently, the cells on each slide were observed at magnification, ×40, and images were captured at a magnification, ×400 from three different field of view, and eosinophil counts were performed.
ii) Histology. To obtain tissue samples for further analysis, 4% paraformaldehyde was injected into the heart of anesthetized rats (IP, 40 mg/kg sodium pentobarbital). Following perfusion, small intestine tissues were extracted, fixed in 4% paraformaldehyde at 4°C for 24 h, and sectioned to 5 μm. At room temperature, paraffin sections were dewaxed with pure xylene (twice for 5 min each), and then washed with distilled water following rehydration using high concentration to low concentration alcohol (anhydrous ethanol, twice for 5 min each; 95% alcohol, 80% alcohol, 70% alcohol, each time for 5 min). The slides were then stained using hematoxylin for 5 min at room temperature. Color separation was then conducted using acidic and ammonia solutions (1% hydrochloric acid alcohol solution, 15 sec; 0.6% ammonia water, 15 sec). The slides were rinsed with running water and placed in distilled water for 15 min. The samples were then stained with eosin for 2 min at room temperature, followed by dehydration with alcohol (95% alcohol, twice for 5 min each; anhydrous ethanol, twice for 5 min each) and then treatment with pure xylene (twice for 5 min each). They were then covered and observed using an optical microscope (magnification, ×400) and images were captured.
iii) High-throughput sequencing. The contents of the small intestine tissues of rats were stored at −80°C, and then analyzed by Chengdu Lilai Biotechnology Co., Ltd. The methods for high-throughput sequencing were as follows: The total microbiome DNA was extracted using the E.Z.N.A.® Stool DNA kit (cat. no. D4015; Omega Bio-Tek, Inc.), and target fragment libraries were constructed. The corresponding primers were designed according to the conserved region in the microbial ribosomal RNA sequence, and the sequencing universal adapters and sample specific Barcode sequences were added to the common primers, so as to amplify the variable region gene (V3+V4) or specific gene fragments of rDNA by PCR. The PCR amplification products were then detected by 2% agarose gel electrophoresis and the target fragments were recovered using the AMPure XT beads recovery kit (cat. no. A63881; Beckman Coulter, Inc.). A Qubit fluorometer was used to quantify the purified PCR products, using a cut-off value of >2 nM. Following gradient dilution of the qualified sequencing libraries, they were mixed according to the amount required for sequencing, and then modified into a single chain by NaOH for machine sequencing. The MiSeq sequencer was used for 2 × 300 bp double-end sequencing. Finally, bioinformatics analysis was performed. Bioinformatics analysis included Operational Taxonomic Units (OTU) cluster analysis, Alpha diversity analysis, Bate analysis and species analysis. The sequence for OTU cluster analysis was performed using USEARCH software (https://www.drive5.com/usearch/; version 10.0) based on 97% sequence similarity. USEARCH software was also used for Alpha diversity analysis and Beta analysis. Alpha diversity analysis includes Chao1, Observed_species, Goods_coverage, Shannon and Simpson index. And Beta diversity analysis and species analysis were performed based on the results obtained by OTU cluster analysis. Beta diversity analysis was started by calculating the distance matrix between environmental samples, which included the distance between any two samples. The sample distance matrix was clustered using the Unweighted Pair Group Method with Arithmetic Mean method (44). The generated cluster tree data file was imported into the cluster tree viewing software Figtree (version v1.4.4; http://tree.bio.ed.ac.uk/software/figtree/). Principal coordinates analysis and clustering analysis were used to decompose the community data structure naturally, and to observe the differences between samples by ordinating the samples. Then, species analysis was performed. RDP (http://rdp.cme.msu.edu/index.jsp) and NT-16S (http://ftp.ncbi.nlm.nih.gov/blast/db) databases were used for species classification and subsequent analysis. According to the OTU annotation results and the OTU abundance table of each sample, the species abundance tables of phylum and genera level were obtained, and the species composition and difference analysis of different groups were performed for different levels of species abundance table. In the analysis, Blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for classification comparison.
iv) Allantoin ELISA analysis. Fecal excrement from the rat large intestine tissues was stored in liquid nitrogen for determination of allantoin levels. A rat allantoin ELISA kit was purchased from Jiangsu Mei Biao Biological Technology Co., Ltd. (cat. no. MB-7159A). The stop solution changed the reaction color from blue to yellow, and the color intensity was measured at 450 nm using a spectrophotometer. In order to measure the concentration of allantoin in the sample, the allantoin ELISA kit used included specific calibration standards. The calibration standards were assayed concomitantly with the samples, which produced a standard curve of optical density (OD) vs. allantoin concentration. The concentration of allantoin in the samples was then determined by comparing the OD of the samples with the standard curve.
Statistical analysis
Measurement data were expressed as mean ± standard deviation. The Student’s t-test was used for comparison between the two groups (Tables II, III, IV, V and VI). One-way analysis of variance was used to compare multiple groups (Table VII). If the variances were equal, Tukey’s post hoc test was used. If the variances were not equal, Tamhane T2 test was used. GraphPad Prism (GraphPad Software, Inc.) was used to analyze the data. P<0.05 was considered to indicate a statistically significant difference.
Table II.
Changes in water consumption in rats ml/(kg/day).
| Group | Time interval, weeks | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| E36eu | 132.24±2.24D | 136.0±2.07a | 151.44±3.00a,B,c | 144.29±7.28a | 152.56±9.45 | 154.40±3.50A,d | 143.05±2.39a,c,D | 148.23±1.62A,d |
| E36e | 142.21±2.06c | 140.03±2.05A,c | 141.92±9.02a | 160.22±11.66A | 155.75±2.59a | 161.93±1.37A,B | 153.60±2.59a,c | 156.25±1.93 |
| E | 136.48±2.24 | 145.04±2.64A | 144.48±1.01a | 142.66±2.02a,b | 154.28±6.23 | 157.70±1.56A | 147.87±1.57a | 142.00±2.96 |
| NS | 136.10±1.98 | 136.70±1.94a | 141.06±2.41a | 135.90±2.04a | 153.56±0.54a | 150.17±2.16a | 157.43±7.79A | 147.86±1.72 |
| Con | 119.08±1.11 | 110.99±3.27 | 113.81±4.14 | 115.96±3.10 | 118.33±6.17 | 118.46±1.69 | 117.70±5.85 | 121.81±3.22 |
P<0.05 and
P<0.01 vs. Con group.
P<0.05 and
P<0.01 vs. NS group.
P<0.05 vs. E group.
P<0.05 and
P<0.01 vs. E36e group.
E36eu, recombinant uricase-expressing genetically engineered E. coli bacteria group; E36e, empty vector-expressing engineered E. coli bacteria group; E, E. coli group; NS, carboxymethylcellulose suspension group; Con, blank control.
Table III.
Changes of food intake in rats (g/kg/day).
| Group | Time, weeks | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| E36eu | 73.91±2.43 | 73.26±2.46c | 73.04±1.78d | 71.88±1.10c | 66.46±2.85a | 68.12±1.60 | 66.03±2.13 | 63.26±3.11 |
| E36e | 72.33±1.89 | 67.85±2.11 | 70.14±1.32 | 66.36±2.67 | 73.63±2.79 | 65.29±1.86 | 64.86±1.53 | 62.87±1.59 |
| E | 74.53±1.28 | 70.06±3.48 | 68.58±2.63 | 63.66±2.25a,b | 66.67±2.29a | 65.85±4.51 | 62.38±2.01 | 61.61±2.44 |
| NS | 70.81±2.87 | 72.43±3.00 | 70.14±1.72 | 72.35±1.12a | 64.96±3.13 | 67.47±2.45 | 64.16±3.56 | 62.13±2.59 |
| Con | 74.14±2.76 | 68.91±5.54 | 66.80±1.49 | 66.24±2.14 | 73.69±3.10 | 67.77±3.10 | 62.47±1.39 | 60.37±3.41 |
P<0.05 vs. Con group;
P<0.05 vs. NS group;
P<0.05 vs. E group;
P<0.05 vs. E36e group.
E36eu, recombinant uricase-expressing genetically engineered E. coli bacteria group; E36e, empty vector-expressing engineered E. coli bacteria group; E, E. coli group; NS, carboxymethylcellulose suspension group; Con, blank control.
Table IV.
Changes of body weight in rats (g).
| Group | Time, weeks | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| E36eu | 183.30±2.70 | 212.72±4.51 | 233.80±15.45 | 236.67±4.99 | 278.50±2.36 | 301.33±4.03 | 311.71±4.03B,d | 351.25±4.50D |
| E36e | 184.56±10.88 | 218.69±3.07 | 244.67±8.26 | 265.66±13.10 | 282.16±3.40a | 310.57±7.59 | 326.68±2.6B | 333.76±4.49b |
| E | 182.90±13.16 | 207.37±19.26 | 241.77±18.28 | 258.03±8.60 | 272.24±4.03a | 294.28±3.86 | 317.29±7.85b | 326.49±5.56a,b |
| NS | 175.17±14.48 | 208.67±4.11a | 233.23±11.15 | 254.43±9.46 | 278.67±7.36 | 312.87±20.17 | 334.51±3.40 | 351.19±4.19a |
| Con | 188.20±8.74 | 224.68±8.60 | 249.00±12.33 | 278.35±15.17 | 297.09±5.89 | 305.69±8.81 | 347.37±15.52 | 341.21±2.05 |
P<0.05 vs. Con group.
P<0.05 and
P<0.01 vs. NS group.
P<0.05 and
P<0.01 vs. E36e group.
E36eu, recombinant uricase-expressing genetically engineered E. coli bacteria group; E36e, empty vector-expressing engineered E. coli bacteria group; E, E. coli group; NS, carboxymethylcellulose suspension group; Con, blank control.
Table V.
Changes in serum uric acid levels in rats (μmol/l).
| Group | Time, weeks | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| E36eu | 384.56±14.51A | 427.61±9.42A | 428.51±22.01A | 424.90±27.69A | 405.84±32.04B,C,D | 375.98±26.08A,B,C,D | 361.49±16.71A,B,c,D | 345.45±4.27A,B,C,D |
| E36e | 380.90±15.72A | 421.91±5.10A | 414.13±13.92A | 431.92±34.73A | 448.02±22.61b | 463.79±16.23A | 451.48±33.47A,b | 450.23±3.97A,B |
| E | 386.52±17.82a | 418.41±7.01A | 428.06±20.52A | 433.95±10.89A | 446.31±6.57A,b | 443.84±35.61 | 449.12±26.87A,b | 455.95±45.75A,B |
| NS | 379.59±15.88a | 415.22±33.87A | 424.72±26.88A | 435.62±10.95A | 432.40±13.50A | 436.81±25.07A | 434.19±23.83A | 436.08±13.39A |
| Con | 203.23±10.06 | 192.70±24.04 | 189.13±4.89 | 198.34±37.87 | 195.32±13.70 | 191.35±14.59 | 198.07±46.63 | 188.83±44.66 |
P<0.05 and
P<0.01 vs. Con group.
P<0.05 and
P<0.01 vs. NS group.
P<0.05 and
P<0.01 vs. E group.
P<0.01 vs. E36e group.
E36eu, recombinant uricase-expressing genetically engineered E. coli bacteria group; E36e, empty vector-expressing engineered E. coli bacteria group; E, E. coli group; NS, carboxymethylcellulose suspension group; Con, blank control.
Table VI.
Fecal allantoin levels.
| Group | E36eu | E36e | E | NS | Con |
|---|---|---|---|---|---|
| Allantoin, μg/ml | 393.94±5.16A–D | 259.92±14.58a | 271.71±9.34 | 250.49±11.09 | 241.27±8.87 |
P<0.05 and
P<0.01 vs. Con group.
P<0.01 vs. NS group.
P<0.01 vs. E group.
P<0.01 vs. E36e group.
E36eu, recombinant uricase-expressing genetically engineered E. coli bacteria group; E36e, empty vector-expressing engineered E. coli bacteria group; E, E. coli group; NS, carboxymethylcellulose suspension group; Con, blank control.
Table VII.
Eosinophil count.
| Group | E36eu | E36e | E | NS | Con |
|---|---|---|---|---|---|
| Number | 3.80±2.42 | 4.01±0.18 | 3.63±0.12 | 2.30±0.15 | 2.08±0.17 |
E36eu, recombinant uricase-expressing genetically engineered E. coli bacteria group; E36e, empty vector-expressing engineered E. coli bacteria group; E, E. coli group; NS, carboxymethylcellulose suspension group; Con, blank control.
Results
Identification of target genes and recombinant expression vectors pMG36e-sUOX and pMG36e-spo-uox
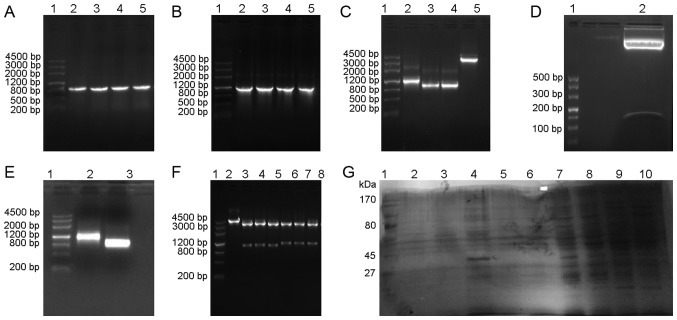
The target genes UOX, sUOX and spo exhibited target bands of 948 bp [Fig. 1A (lanes 2–5) and C (lanes 3–4)], 1,093 bp [Fig. 1B (lanes 2–5) and C (lane 2)] and 105 bp [Fig. 1D (lane 2)] by 1.0% agarose gel electrophoresis, respectively. The pMG36e plasmid exhibited a band at 3,600 bp [Fig. 1C (lane 5)] by 1.0% agarose gel electrophoresis. Plasmids of positive clones were extracted for PCR identification and restriction enzyme digestion, and were confirmed by 1.0% agarose gel electrophoresis. As demonstrated in Fig. 1, vector pMG36e was 3.6 kb [Fig. 1F (lane 2)], UOX gene was 1 kb [Fig. 1E (lane 3)], and sUOX gene was 1.1 kb [Fig. 1E (lane 2)], which coincided with the size of the target gene. Sequencing results indicated that the base sequence was complete and consistent with GenBank base sequence (45).
Figure 1.
Results of target gene amplification, identification of recombinant plasmid and expression of uricase. (A) Molecular weight marker DL4500 (lane 1) and PCR results for the UOX gene (lanes 2–5). (B) DL4500 (lane 1) and PCR results for sUOX gene analysis (lanes 2–5). (C) DL4500 (lane 1), double digestion of sUOX gene (lane 2), double digestion of UOX gene (lanes 3–4) and double digestion of pMG36e (lane 5). (D) DL500 (lane 1) and double digestion of spo gene (lane 2). (E) DL4500 (lane 1), PCR results of sUOX gene analysis in recombinant plasmid (lane 2); PCR results of UOX gene analysis in recombinant plasmid (lane 3). (F) DL4500 (lane 1), double digestion of pMG36e in recombinant plasmid (lane 2), double digestion of recombinant plasmid pMG36e-spo-uox (lanes 3–5) and double digestion of recombinant plasmid pMG36e-sUOX (lanes 6–8). (G) Protein marker 170 kDa (lane 1), protein expression of pmg36e-spo-uox in the periplasmic space, protein inclusion bodies and supernatant following 9 h culture (lanes 2–4), protein expression of pmg36e-sUOX in the periplasmic space, protein inclusion bodies and supernatant following 9 h culture (lanes 5–7) and protein expression of pmg36e-spo-uox in supernatants after 3, 6, and 12 h culture (lanes 8–10). PCR, polymerase chain reaction; UOX, uricase, sUOX, staphylococcal protein A + uricase.
Secretion expression of uricase
Following transformation of pMG36e-sUOX and pMG36e-spo-uox into E. coli BL21 cells, the expression levels of proteins were analyzed by SDS-PAGE at different culture times in the supernatant, periplasmic space and inclusion bodies. As indicated in Fig. 1G, the supernatants of the recombinant plasmids pMG36e-sUOX and pMG36e-spo-uox exhibited a band at 34 kDa [Fig. 1G (lane 4)], which was consistent with the expected molecular weight of the target protein. In addition, the expression level of pMG36e-spo-uox was increased compared with that of pMG36e-sUOX, and maximum expression was obtained at 9 h. The enzyme activity was identified to be 2.97 U/mg. Therefore, the recombinant plasmid pMG36e-spo-uox was selected for subsequent analysis in vivo.
Application of engineered bacteria in rat model Changes in water consumption, food intake, body weight and SUA of rats, and fecal allantoin
Compared with the Con group, the water consumption of the E36eu group was increased at weeks 6 and 8 (P<0.05; Table II) and increased significantly at weeks 2, 3, 4, and 7 (P<0.01; Table II), and food intake was decreased at week 5 (P<0.05; Table III). Compared with the NS group, water consumption in the E36eu group was significantly increased at week 3 (P<0.01; Table II) and body weight was significantly decreased at week 7 (P<0.01; Table IV). The changes of SUA in rats are presented in Table V. During the whole experiment, SUA in the Con group was significantly decreased compared with that in the other groups (P<0.01). Between week 5 to week 8, the levels of SUA in the E36eu group were decreased, and were significantly decreased compared with those in the NS, E and E36e groups (P<0.01). Compared with the NS group, the SUA levels in the E36e group at week 5 and the E group at weeks 5 and 7 were higher than that in the NS group (P<0.05). The SUA levels in the E36e group at weeks 7 and 8 and the E group at week 8 were significantly higher than that in the NS group (P<0.01). As observed in Table VI, the allantoin levels in the feces of the E36eu group were significantly increased compared with those of the other groups (P<0.01). These data indicate that there was no decrease in the SUA levels in the rats who were administered the empty vector E. coli engineered bacteria and E. coli. The SUA levels in the rats who were administered uricase genetically engineered bacteria was significantly decreased.
Analysis of eosinophil count and histological observation of small intestine in rats
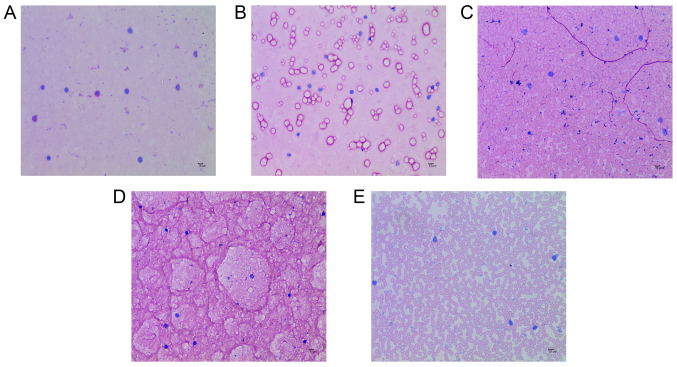
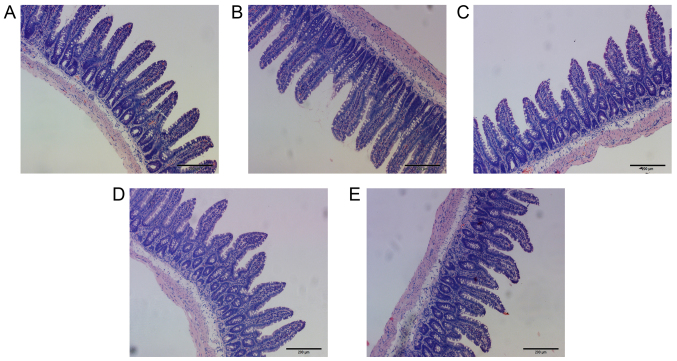
Compared with the Con group, the number of eosinophils in the E36eu, E36e, E and NS groups increased slightly, but the difference was not significant (P>0.05; Table VII and Fig. 2A–E). The structure of the intestinal mucosa was intact, epithelial cells were closely arranged and goblet cells were scattered between absorptive cells. There were no pathological changes in observed in the cells (Fig. 3A–E).
Figure 2.
Eosinophil count in each treatment group. (A) E36eu group. (B) E36e group. (C) E group. (D) NS group. (E) Con group. All images at magnification, x400. Scale bar=200 μm.
Figure 3.
Histological analysis of intestinal tissue in rats. (A) E36eu group. (B) E36e group. (C) E group. (D) NS group. (E) Con group. The tissue structure of the small intestine in each group of rats was intact. There were no pathological changes observed in the cells. All images at magnification, ×100.
Analysis of intestinal contents in rats
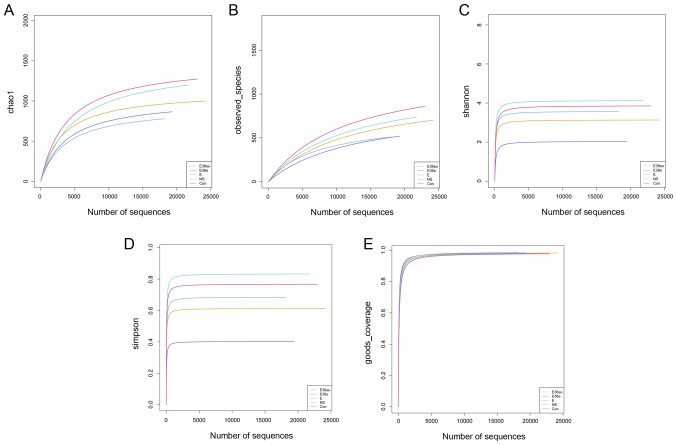
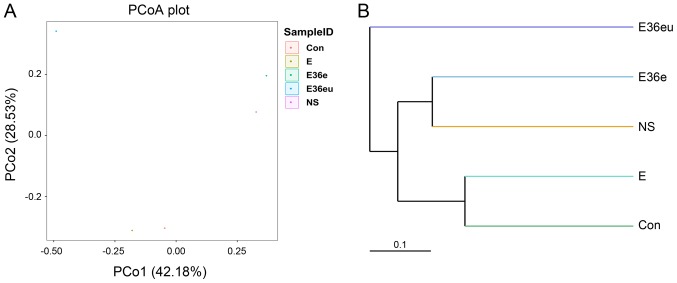
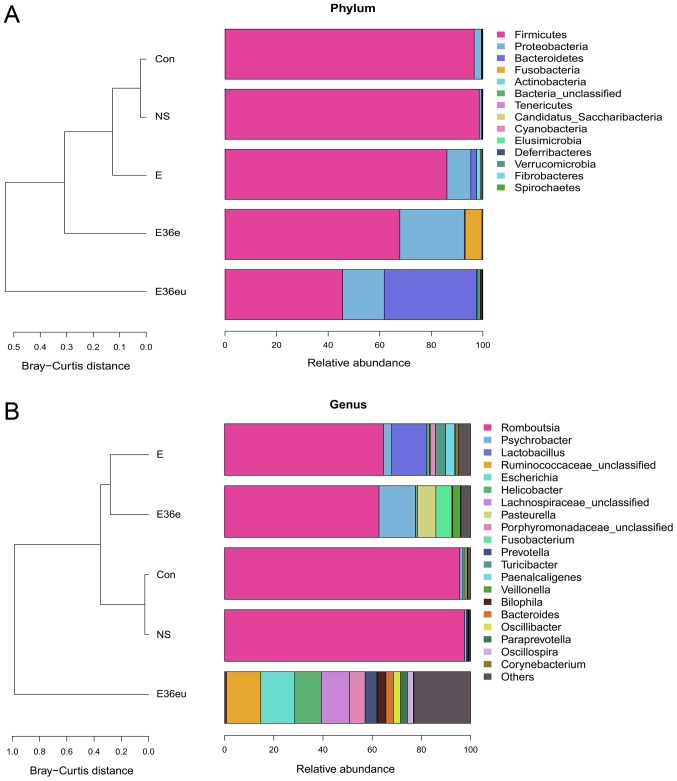
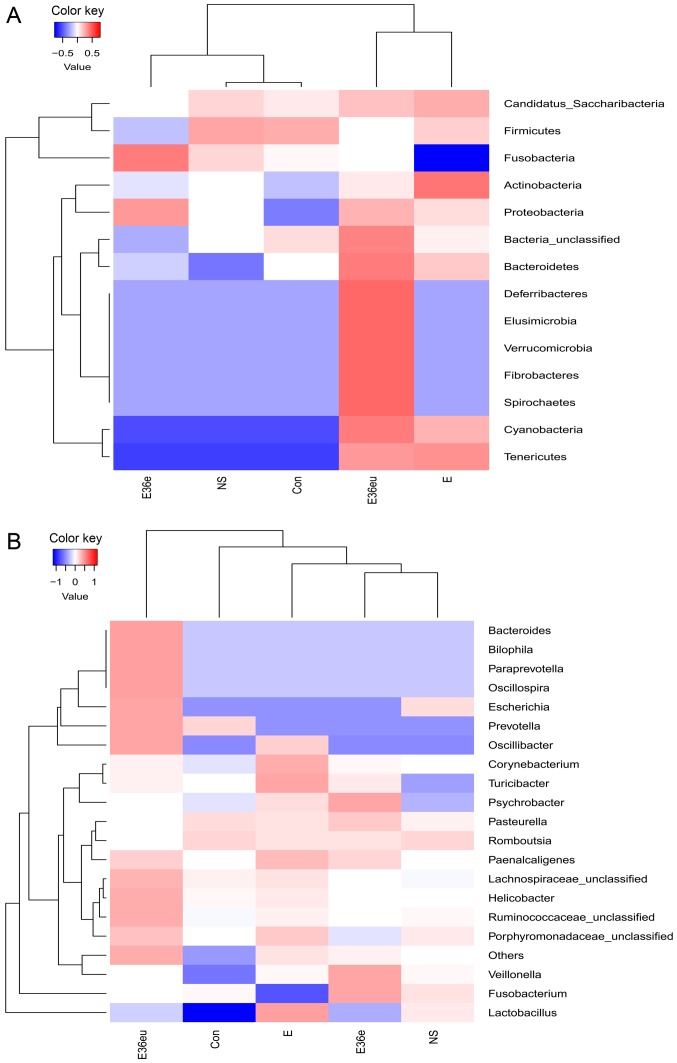
Following high-throughput sequencing, the intestinal contents of the rats were analyzed by bioinformatics, including Operational Taxonomic Units (OTU) cluster analysis, Alpha diversity analysis, Beta diversity analysis and species analysis. The results are presented in Figs. 4–7. The results of the OTU cluster analysis and Alpha diversity analysis demonstrated that the dilution curves of each group were generally smooth, indicating that the data volume of sequencing was reasonable and could include the majority of microorganisms in the samples (Fig. 4). According to the Goods_coverage index, the results of sequencing can represent all samples (Fig. 4E). According to the Observed_species, Chao1, Shannon and Simpson indexes, the species richness and evenness of the Con and NS groups were relatively high (Fig. 4A–D). The results of the Bate analysis and species analysis indicated that there were differences in species diversity between the E36eu and Con groups (Fig. 5A and B). According to the phylum level cluster analysis, intestinal flora of each group was primarily composed of Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Actinobacteria and Teneriquets (Figs. 6A and 7A). The compositions of the colonies in the Con and NS groups were similar, and the compositions of the colonies in the Con and E36eu groups were different. According to the genus level cluster analysis, composition and abundance of colonies in each group were different (Figs. 6B and 7B).
Figure 4.
OTU cluster analysis and Alpha diversity analysis of the intestinal contents in the rat model. (A) Chao1 index. (B) Observed_species index. (C) Shannon Index. (D) Simpson index. (E) Goods_coverage index.
Figure 5.
Beta analysis of the intestinal contents in the rats model. (A) PCoA analysis chart. (B) Unweighted Pair Group Method with Arithmetic Mean hierarchical cluster tree diagram. PCoA, principle coordinates analysis.
Figure 6.
Species analysis of the intestinal contents in the rat model. (A) Histogram of sample abundance by phylum. (B) Histogram of sample abundance by genus.
Figure 7.
Species analysis of the intestinal contents in the rat model. (A) Taxa heatmap of species by phylum. (B) Taxa heatmap of species by genus.
Discussion
Apart from the kidney, the intestine is the most important organ for excreting uric acid (27,46). The uric acid in the intestine primarily originates from both endogenous (SUA) and exogenous (food) sources. Endogenous uric acid is not only comprised of SUA (46,47), but also a small amount of uric acid secreted from liver (48). The penetration of SUA into the intestine occurs through passive diffusion (49), and may be associated with the organic ion transporter 10 (OAT10) in the intestinal mucosa (50). Therefore, hyperuricemia significantly increases the level of uric acid entering the intestine in this way (46). Furthermore, uric acid in the intestine may also be absorbed into the blood, which is associated with the secretory and absorptive functions of the intestinal mucosa. Under normal circumstances, microorganisms in the intestine convert some uric acid into allantoin, which is more easily soluble in water, and excreted in feces (51). The conversion of uric acid into allantoin not only decreases the level of uric acid in the intestine, but also facilitates the excretion of SUA into the intestine, which is conducive to the overall decrease of uric acid levels in the body.
Therefore, the present study successfully constructed uricase-expressing genetically engineered bacteria by gene recombination technology, and the efficacy of the engineered bacteria was verified in a hyperuricemia rat model using oteracil potassium, which is a commonly used drug in animal models that inhibits uricase activity. When administered in certain amounts, it may partially inhibit the activity of uricase due to the competitive inhibition mechanism of the drug, which results in an rapid increase in the levels of SUA in rats. However, oteracil potassium is metabolized quickly in the body; therefore, the time of injection of oteracil potassium in the animal model of the present study was different from the time of administration of the genetically engineered bacteria, so that the oteracil potassium would not affect the activity of uricase secreted by the engineered bacteria. The results indicated that the SUA levels in rats who received uricase-expressing genetically engineered bacteria decreased significantly and that the rats grew normally. In the upper part of the intestine, the number of bacteria in the lumen is relatively low, and it is generally accepted that bacterial communities increase in relative diversity and complexity in the proximal-distal direction, from duodenum through to the jejunum and ileum. Based on early molecular studies, the genus Streptococcus appears to be the dominant genus in the duodenum and jejunum (52,53). There are more gram-negative bacteria compared with gram-positive bacteria in the terminal ileum. Located next to the ileocecal valve, the environment of the colon is conducive to the growth of obligate anaerobic bacteria, and the number of bacteria increases rapidly. Normally, the flora of the small intestine is primarily distributed in the ileum. The duodenum and jejunum are relatively sterile, and the primary bacterial species are gram-positive aerobic bacteria, including Streptococcus, Staphylococcus and Lactobacillus (54). In the distal ileum, the gram-negative bacteria begin to outnumber the gram-positive bacteria. Therefore, there are few bacteria in the upper part of the small intestine, and it is difficult to collect samples. As such, the contents of the whole intestinal segment of the small intestine were collected in the present study for intestinal contents analysis. It has been demonstrated that SUA can be excreted into the intestinal lumen throughout the small intestine to the large intestine (49,50). The mucosal area of the small intestine is large, and this organ serves an important role in lowering SUA levels. In the present study, the analysis of the whole contents of the small intestine, SUA values and allantoin values in rats suggested that uricase-expressing genetically engineered bacteria colonized the intestine and served a role in lowering uric acid levels. In addition, one study has used montmorillonite to adsorb intestinal uric acid in order to decrease SUA levels (55), which is consistent with the results of the present study. However, the aforementioned study only analyzed the effect of adsorption of intestinal uric acid by montmorillonite on SUA in a short period of time, but the effect of long-term use of montmorillonite on SUA was not analyzed. Therefore, it has not been demonstrated that this method can be used for long-term treatment of hyperuricemia. Recombinant uricase has also been used to decompose uric acid in vivo, but its clinical application has been limited by side effects such as immunogenicity of heterologous proteins (31,32). Using a different method, the present study used gene recombination technology to introduce the s-layer protein signal peptide gene from Lactobacillus brevis at the N-terminal end of the UOX gene, which is the signal peptide gene that directs extracellular secretion of uricase. Therefore, uricase-expressing engineered bacteria colonized in the intestine may secrete endogenous uricase. Additionally, E. coli is one of the normal species of intestinal flora. Uricase, which is expressed through normal intestinal flora, cannot enter the blood, and even if exposed for prolonged periods, the body will not produce antibodies. The present study also contains certain limitations; although E. coli is a normal intestinal flora but a conditional pathogen, excessive use may cause some side effects, including diarrhea and inflammation.
An increasing number of studies investigating probiotics have been performed in recent years. Lactobacillus is one of the probiotics present in the human intestine, which can improve intestinal microenvironment, regulate immunity and promote nutrient absorption (56–59). Recent studies have demonstrated that probiotics also have a role in regulating abnormal brain activity (60,61). Therefore, this study group plans to transfect a recombinant UOX gene into Lactobacillus, to further analyze the practical application value and significance of engineered bacteria.
In summary, the present study described a novel approach to treat hyperuricemia through degrading uric acid by uricase secreted by intestinal microorganisms carrying a recombinant uricase gene. The uricase secreted by uricase-expressing engineered bacteria, constructed through gene recombination technology, promoted the conversion of intestinal uric acid into allantoin, which not only prevented the absorption of intestinal uric acid into the bloodstream but also accelerated the diffusion of SUA into the intestine, thereby decreasing SUA levels.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the Chongqing Science and Technology Foundation in China (grant no. cstc2016jcyjA0220) awarded to XJ.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Authors’ contributions
XL was responsible for the conceptualization of the study. LC was involved in data curation. QL performed the formal analysis. XJ was involved in funding acquisition, project administration and supervision of the project. WD was involved in reviewing the literature. YD was responsible for developing the methodology. LC, QL and WD were involved in performing experiments. XJ was involved in processing and editing the figures. LC was responsible for writing the original draft. XJ was involved in revision of the manuscript and final approval of the version to be released. All authors read and approved the final manuscript.
Ethics approval and consent to participate
In the present study, all the experimental protocols involving animals approved by the Ethics Committee of Chongqing Medical University (Chonqing, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Rock KL, Kataoka H, Lai JJ. Uric acid as a danger signal in gout and its comorbidities. Nat Rev Rheumatol. 2013;9:13–23. doi: 10.1038/nrrheum.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mallat SG, Al Kattar S, Tanios BY, Jurjus A. Hyperuricemia, hypertension, and chronic kidney disease: An emerging association. Curr Hypertens Rep. 2016;18:74. doi: 10.1007/s11906-016-0684-z. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Lario B, Macarron-Vicente J. Uric acid and evolution. Rheumatology (Oxford) 2010;49:2010–2015. doi: 10.1093/rheumatology/keq204. [DOI] [PubMed] [Google Scholar]
- 4.Mandal AK, Mount DB. The molecular physiology of uric acid homeostasis. Annu Rev Physiol. 2015;77:323–345. doi: 10.1146/annurev-physiol-021113-170343. [DOI] [PubMed] [Google Scholar]
- 5.Li Q, Li X, Wang J, Liu H, Kwong JS, Chen H, Li L, Chung SC, Shah A, Chen Y, et al. Diagnosis and treatment for hyperuricemia and gout: A systematic review of clinical practice guidelines and consensus statements. BMJ Open. 2019;9:e26677. doi: 10.1136/bmjopen-2018-026677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuo CF, Grainge MJ, Mallen C, Zhang W, Doherty M. Rising burden of gout in the UK but continuing suboptimal management: A nationwide population study. Ann Rheum Dis. 2015;74:661–667. doi: 10.1136/annrheumdis-2013-204463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smith E, Hoy D, Cross M, Merriman TR, Vos T, Buchbinder R, Woolf A, March L. The global burden of gout: Estimates from the global burden of disease 2010 study. Ann Rheum Dis. 2014;73:1470–1476. doi: 10.1136/annrheumdis-2013-204647. [DOI] [PubMed] [Google Scholar]
- 8.Elfishawi MM, Zleik N, Kvrgic Z, Michet CJ, Jr, Crowson CS, Matteson EL, Bongartz T. The rising incidence of gout and the increasing burden of comorbidities: A population-based study over 20 years. J Rheumatol. 2018;45:574–579. doi: 10.3899/jrheum.170806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiang KM, Tsay YC, Vincent NT, Yang HC, Huang YT, Chen CH, Pan WH. Is hyperuricemia, an early-onset metabolic disorder, causally associated with cardiovascular disease events in han Chinese? J Clin Med. 2019;8:E1202. doi: 10.3390/jcm8081202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seoane-Mato D, Sanchez-Piedra C, Silva-Fernandez L, Sivera F, Blanco FJ, Pérez Ruiz F, Juan-Mas A, Pego-Reigosa JM, Narváez J, Quilis Martí N, et al. Prevalence of rheumatic diseases in adult population in Spain (EPISER 2016 study): Aims and methodology. Reumatol Clin. 2019;15:90–96. doi: 10.1016/j.reuma.2017.06.009. (In English, Spanish) [DOI] [PubMed] [Google Scholar]
- 11.Liu CW, Liao PC, Chen KC, Chiu YW, Liu YH, Ke SR, Wu YW. Relationship of serum uric acid and Killip class on mortality after acute ST-segment elevation myocardial infarction and primary percutaneous coronary intervention. Int J Cardiol. 2017;226:26–33. doi: 10.1016/j.ijcard.2016.10.025. [DOI] [PubMed] [Google Scholar]
- 12.Neogi T, Jansen TL, Dalbeth N, Fransen J, Schumacher HR, Berendsen D, Brown M, Choi H, Edwards NL, Janssens HJ, et al. 2015 gout classification criteria: An American college of rheumatology/ European league against rheumatism collaborative initiative. Arthritis Rheumatol. 2015;67:2557–2568. doi: 10.1002/art.39254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roumeliotis S, Roumeliotis A, Dounousi E, Eleftheriadis T, Liakopoulos V. Dietary antioxidant supplements and uric acid in chronic kidney disease: A review. Nutrients. 2019;11 doi: 10.3390/nu11081911. pii: E1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryu S, Chang Y, Zhang Y, Kim SG, Cho J, Son HJ, Shin H, Guallar E. A cohort study of hyperuricemia in middle-aged South Korean men. Am J Epidemiol. 2012;175:133–143. doi: 10.1093/aje/kwr291. [DOI] [PubMed] [Google Scholar]
- 15.Johnson RJ, Bakris GL, Borghi C, Chonchol MB, Feldman D, Lanaspa MA, Merriman TR, Moe OW, Mount DB, Sanchez Lozada LG, et al. Hyperuricemia, acute and chronic kidney disease, hypertension, and cardiovascular disease: Report of a scientific workshop organized by the national kidney foundation. Am J Kidney Dis. 2018;71:851–865. doi: 10.1053/j.ajkd.2017.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu CW, Chen KH, Tseng CK, Chang WC, Wu YW, Hwang JJ. The dose-response effects of uric acid on the prevalence of metabolic syndrome and electrocardiographic left ventricular hypertrophy in healthy individuals. Nutr Metab Cardiovasc Dis. 2019;29:30–38. doi: 10.1016/j.numecd.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Williams B, Mancia G, Spiering W, Agabiti Rosei E, Azizi M, Burnier M, Clement DL, Coca A, de Simone G, Dominiczak A, et al. 2018 ESC/ESH Guidelines for the management of arterial hypertension. Eur Heart J. 2018;39:3021–3104. doi: 10.1093/eurheartj/ehy339. [DOI] [PubMed] [Google Scholar]
- 18.Presa M, Pérez-Ruiz F, Oyagüez I. Second-line treatment with lesinurad and allopurinol versus febuxostat for management of hyperuricemia: A cost-effectiveness analysis for Spanish patients. Clin Rheumatol. 2019;38:3521–3528. doi: 10.1007/s10067-019-04739-3. [DOI] [PubMed] [Google Scholar]
- 19.Becker MA, Schumacher HJ, Jr, Wortmann RL, MacDonald PA, Eustace D, Palo WA, Streit J, Joseph-Ridge N. Febuxostat compared with allopurinol in patients with hyperuricemia and gout. N Engl J Med. 2005;353:2450–2461. doi: 10.1056/NEJMoa050373. [DOI] [PubMed] [Google Scholar]
- 20.Liu CW, Chang WC, Lee CC, Shau WY, Hsu FS, Wang ML, Chen TC, Lo C, Hwang JJ. The net clinical benefits of febuxostat versus allopurinol in patients with gout or asymptomatic hyperuricemia-A systematic review and meta-analysis. Nutr Metab Cardiovasc Dis. 2019;29:1011–1022. doi: 10.1016/j.numecd.2019.06.016. [DOI] [PubMed] [Google Scholar]
- 21.Mu Z, Wang W, Wang J, Lv W, Chen Y, Wang F, Yu X, Wang Y, Cheng B, Wang Z. Predictors of poor response to urate-lowering therapy in patients with gout and hyperuricemia: A post-hoc analysis of a multicenter randomized trial. Clin Rheumatol. 2019;38:3511–3519. doi: 10.1007/s10067-019-04737-5. [DOI] [PubMed] [Google Scholar]
- 22.White WB, Saag KG, Becker MA, Borer JS, Gorelick PB, Whelton A, Hunt B, Castillo M, Gunawardhana L CARES Investigators. Cardiovascular safety of febuxostat or allopurinol in patients with gout. N Engl J Med. 2018;378:1200–1210. doi: 10.1056/NEJMoa1710895. [DOI] [PubMed] [Google Scholar]
- 23.Richette P, Doherty M, Pascual E, Barskova V, Becce F, Castañeda-Sanabria J, Coyfish M, Guillo S, Jansen TL, Janssens H, et al. 2016 updated EULAR evidence-based recommendations for the management of gout. Ann Rheum Dis. 2017;76:29–42. doi: 10.1136/annrheumdis-2016-209707. [DOI] [PubMed] [Google Scholar]
- 24.Xu X, Li C, Zhou P, Jiang T. Uric acid transporters hiding in the intestine. Pharm Biol. 2016;54:3151–3155. doi: 10.1080/13880209.2016.1195847. [DOI] [PubMed] [Google Scholar]
- 25.Jalal DI, Chonchol M, Chen W, Targher G. Uric acid as a target of therapy in CKD. Am J Kidney Dis. 2013;61:134–146. doi: 10.1053/j.ajkd.2012.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maiuolo J, Oppedisano F, Gratteri S, Muscoli C, Mollace V. Regulation of uric acid metabolism and excretion. Int J Cardiol. 2016;213:8–14. doi: 10.1016/j.ijcard.2015.08.109. [DOI] [PubMed] [Google Scholar]
- 27.Yun Y, Yin H, Gao Z, Li Y, Gao T, Duan J, Yang R, Dong X, Zhang L, Duan W. Intestinal tract is an important organ for lowering serum uric acid in rats. PLoS One. 2017;12:e190194. doi: 10.1371/journal.pone.0190194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matsuo H, Tsunoda T, Ooyama K, Sakiyama M, Sogo T, Takada T, Nakashima A, Nakayama A, Kawaguchi M, Higashino T, et al. Hyperuricemia in acute gastroenteritis is caused by decreased urate excretion via ABCG2. Sci Rep. 2016;6:31003. doi: 10.1038/srep31003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adejumo OA, Okaka EI, Madumezia G, Okwuonu CG, Ojogwu LI. Assessment of some cardiovascular risk factors in predialysis chronic kidney disease patients in Southern Nigeria. Niger Med J. 2015;56:394–399. doi: 10.4103/0300-1652.171616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakhaee K. Epidemiology and clinical pathophysiology of uric acid kidney stones. J Nephrol. 2014;27:241–245. doi: 10.1007/s40620-013-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamp LK, O’Donnell JL, Chapman PT. Emerging therapies in the long-term management of hyperuricaemia and gout. Intern Med J. 2007;37:258–266. doi: 10.1111/j.1445-5994.2007.01315.x. [DOI] [PubMed] [Google Scholar]
- 32.Terkeltaub R. Gout. Novel therapies for treatment of gout and hyperuricemia. Arthritis Res Ther. 2009;11:236. doi: 10.1186/ar2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cao C, Zhang H, Yang Z, Wu B. Peptide recognition, signaling and modulation of class B G protein-coupled receptors. Curr Opin Struct Biol. 2018;51:53–60. doi: 10.1016/j.sbi.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Tristan A, Benito Y, Montserret R, Boisset S, Dusserre E, Penin F, Ruggiero F, Etienne J, Lortat-Jacob H, Lina G, et al. The signal peptide of Staphylococcus aureus panton valentine leukocidin LukS component mediates increased adhesion to heparan sulfates. PLoS One. 2009;4:e5042. doi: 10.1371/journal.pone.0005042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Negi S, Pandey S, Srinivasan SM, Mohammed A, Guda C. LocSigDB: A database of protein localization signals. Database (Oxford) 20152015 doi: 10.1093/database/bav003. pii: bav003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lv XG, Shi YF, Xu X, Wei YL, Wang HM, Zhang XB, Wu JL. Oryza sativa chloroplast signal recognition particle 43 (OscpSRP43) is required for chloroplast development and photosynthesis. PLoS One. 2015;10:e143249. doi: 10.1371/journal.pone.0143249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen G, Jia P. Allopurinol decreases serum uric acid level and intestinal glucose transporter-5 expression in rats with fructose-induced hyperuricemia. Pharmacol Rep. 2016;68:782–786. doi: 10.1016/j.pharep.2016.04.014. [DOI] [PubMed] [Google Scholar]
- 38.Zhu C, Tai LL, Wan XC, Li DX, Zhao YQ, Xu Y. Comparative effects of green and black tea extracts on lowering serum uric acid in hyperuricemic mice. Pharm Biol. 2017;55:2123–2128. doi: 10.1080/13880209.2017.1377736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dalbeth N, Phipps-Green A, House ME, Gamble GD, Horne A, Stamp LK, Merriman TR. Body mass index modulates the relationship of sugar-sweetened beverage intake with serum urate concentrations and gout. Arthritis Res Ther. 2015;17:263. doi: 10.1186/s13075-015-0781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caliceti C, Calabria D, Roda A, Cicero AFG. Fructose intake, serum uric acid, and cardiometabolic disorders: A critical review. Nutrients. 2017;9 doi: 10.3390/nu9040395. pii: E395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abdulla MH, Sattar MA, Abdullah NA, Johns EJ. The effect of high-fructose intake on the vasopressor response to angiotensin II and adrenergic agonists in sprague-dawley rats. Pak J Pharm Sci. 2013;26:727–732. [PubMed] [Google Scholar]
- 42.Lecoultre V, Egli L, Theytaz F, Despland C, Schneiter P, Tappy L. Fructose-induced hyperuricemia is associated with a decreased renal uric acid excretion in humans. Diabetes Care. 2013;36:e149–e150. doi: 10.2337/dc13-0866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xilifu D, Abudula A, Rehemu N, Zhao L, Zhou X, Zhang X. Effect of rosuvastatin on hyperuricemic rats and the protective effect on endothelial dysfunction. Exp Ther Med. 2014;8:1683–1688. doi: 10.3892/etm.2014.2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.LI Y, Xu L. Improvement for unweighted pair group method with arithmetic mean and its application. J Beijing Univ Technol. 2007;33 [Google Scholar]
- 45.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2013;41(Database Issue):D36–D42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hosomi A, Nakanishi T, Fujita T, Tamai I. Extra-renal elimination of uric acid via intestinal efflux transporter BCRP/ABCG2. PLoS One. 2012;7:e30456. doi: 10.1371/journal.pone.0030456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Szczurek P, Mosiichuk N, Woliński J, Yatsenko T, Grujic D, Lozinska L, Pieszka M, Święch E, Pierzynowski SG, Goncharova K. Oral uricase eliminates blood uric acid in the hyperuricemic pig model. PLoS One. 2017;12:e179195. doi: 10.1371/journal.pone.0179195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Takada T, Ichida K, Matsuo H, Nakayama A, Murakami K, Yamanashi Y, Kasuga H, Shinomiya N, Suzuki H. ABCG2 dysfunction increases serum uric acid by decreased intestinal urate excretion. Nucleosides Nucleotides Nucleic Acids. 2014;33:275–281. doi: 10.1080/15257770.2013.854902. [DOI] [PubMed] [Google Scholar]
- 49.Bronk JR, Shaw MI. The transport of uric acid across mouse small intestine in vitro. J Physiol. 1986;378:229–239. doi: 10.1113/jphysiol.1986.sp016216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bahn A, Hagos Y, Reuter S, Balen D, Brzica H, Krick W, Burckhardt BC, Sabolic I, Burckhardt G. Identification of a new urate and high affinity nicotinate transporter, hOAT10 (SLC22A13) J Biol Chem. 2008;283:16332–16341. doi: 10.1074/jbc.M800737200. [DOI] [PubMed] [Google Scholar]
- 51.Góth L. Rasburicase therapy may cause hydrogen peroxide shock. Orv Hetil. 2008;149:1587–1590. doi: 10.1556/OH.2008.28422. (In Hungarian) [DOI] [PubMed] [Google Scholar]
- 52.Skar V, Skar AG, Bratlie J, Osnes M. Beta-glucuronidase activity in the bile of gallstone patients both with and without duodenal diverticula. Scand J Gastroenterol. 1989;24:205–212. doi: 10.3109/00365528909093038. [DOI] [PubMed] [Google Scholar]
- 53.Justesen T, Nielsen OH, Jacobsen IE, Lave J, Rasmussen SN. The normal cultivable microflora in upper jejunal fluid in healthy adults. Scand J Gastroenterol. 1984;19:279–282. doi: 10.1080/00365521.1984.12005721. [DOI] [PubMed] [Google Scholar]
- 54.Hollister EB, Gao C, Versalovic J. Compositional and functional features of the gastrointestinal microbiome and their effects on human health. Gastroenterology. 2014;146:1449–1458. doi: 10.1053/j.gastro.2014.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Y, Lin Z, Zhang B, Nie A, Bian M. Cichorium intybus L. Promotes intestinal uric acid excretion by modulating ABCG2 in experimental hyperuricemia. Nutr Metab (Lond) 2017;14:38. doi: 10.1186/s12986-017-0190-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yadav H, Lee JH, Lloyd J, Walter P, Rane SG. Beneficial metabolic effects of a probiotic via butyrate-induced GLP-1 hormone secretion. J Biol Chem. 2013;288:25088–25097. doi: 10.1074/jbc.M113.452516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Balakrishnan B, Taneja V. Microbial modulation of the gut microbiome for treating autoimmune diseases. Expert Rev Gastroenterol Hepatol. 2018;12:985–996. doi: 10.1080/17474124.2018.1517044. [DOI] [PubMed] [Google Scholar]
- 58.Rinaldi E, Consonni A, Guidesi E, Elli M, Mantegazza R, Baggi F. Gut microbiota and probiotics: Novel immune system modulators in myasthenia gravis? Ann NY Acad Sci. 2018;1413:49–58. doi: 10.1111/nyas.13567. [DOI] [PubMed] [Google Scholar]
- 59.Chang CJ, Lin TL, Tsai YL, Wu TR, Lai WF, Lu CC, Lai HC. Next generation probiotics in disease amelioration. J Food Drug Anal. 2019;27:615–622. doi: 10.1016/j.jfda.2018.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bagheri S, Heydari A, Alinaghipour A, Salami M. Effect of probiotic supplementation on seizure activity and cognitive performance in PTZ-induced chemical kindling. Epilepsy Behav. 2019;95:43–50. doi: 10.1016/j.yebeh.2019.03.038. [DOI] [PubMed] [Google Scholar]
- 61.Akkasheh G, Kashani-Poor Z, Tajabadi-Ebrahimi M, Jafari P, Akbari H, Taghizadeh M, Memarzadeh MR, Asemi Z, Esmaillzadeh A. Clinical and metabolic response to probiotic administration in patients with major depressive disorder: A randomized, double-blind, placebo-controlled trial. Nutrition. 2016;32:315–320. doi: 10.1016/j.nut.2015.09.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.