Abstract
Knee osteoarthritis (KOA) is a major cause of leg disability in the elderly population. Recently, the expression levels of circulating microRNA (miRNA) let-7e have been reported to be significantly reduced in KOA. The aims of the present study were to assess the feasibility of let-7e as a serum marker for detecting KOA and to explore the underlying mechanisms of its involvement. Based on previous studies and bioinformatics analysis, let-7e may regulate apoptosis and autophagy of articular chondrocytes. A total of 10 patients with KOA and 10 patients with trauma without KOA were recruited to examine the levels of let-7e in peripheral blood. Subsequently, KOA rat models were established, and the levels of let-7e in the cartilage and serum were examined, the expression of apoptotic proteins and autophagy-related proteins in the cartilage were investigated, and apoptotic and autophagic activities of primary cultured chondrocytes were also detected. In patients with KOA, let-7e levels in the peripheral serum were significantly decreased compared with the control group, and this result was confirmed in the peripheral serum and cartilage of KOA rats. In addition, the expression levels of proteins involved in the apoptotic pathway were increased in the cartilage of KOA rats, and apoptotic activity was increased. The expression of autophagy-related proteins beclin 1 and microtubule associated protein 1 light chain 3 β (LC3B) II/LC3BI in the articular cartilage of KOA rats was lower compared with the controls, and autophagy was decreased. Si-Miao-San (SMS) treatment restored the expression of let-7e and reversed the changes in apoptosis and autophagy. Therefore, the present study provided additional evidence that circulating let-7e may be a potential serum biomarker for the diagnosis and treatment of KOA. Elevated apoptosis levels and decreased autophagy levels of cartilage tissue are involved in KOA, and treatment with SMS may reverse these effects.
Keywords: let-7e, knee osteoarthritis, autophagy, apoptosis, Si-Miao-San
Introduction
With the aging of the global population, osteoarthritis (OA) is having an increasing impact on the quality of life and is becoming an increasing burden on society. At present, 50% of the elderly population (≥75 years) are diagnosed with OA (1). In the US population, the age- and body mass index (BMI)-adjusted prevalence of knee pain and symptomatic knee OA (KOA) has increased ~2-fold in women and 3-fold in men over the past 20 years (2). Therefore, the prevention and treatment of KOA is an important topic requiring urgent attention from researchers and practitioners.
Lethal-7 (let-7) gene is one of the earliest discovered microRNAs (miRNAs) (3). To date, 10 mature let-7 family members have been discovered in the human body, formed by 13 precursors, and let-7e is a member of this family (4). Altered levels of let-7e are associated with a number of diseases. As a tumor suppressor microRNA, the plasma levels of let-7e are downregulated in malignant germ cell tumors, retinoblastoma, and esophageal adenocarcinoma (5–7). Let-7e is also associated with cardiovascular diseases; let-7e expression levels were significantly upregulated in patients with hypertension (8) and ischemic stroke during the acute stage (9), and downregulated in patients with ventricular septal defect (10). In addition, let-7e is associated with the number of metabolic syndrome traits (11), and the expression levels of let-7e are significantly increased in patients with Hashimoto’s disease (12).
In 2014, a large-sample prospective study performed in Germany revealed that the levels of let-7e in circulation were significantly lower in patients with OA, independent of age, sex and BMI factors (13). miRNA let-7e has been suggested as a potential serum marker for the diagnosis of OA and predicting the risk and severity of the disease (13). However, additional studies are required to support the link between let-7e and KOA, and to uncover the underlying mechanisms between let-7e and KOA.
The pathology of KOA is complex and involves numerous factors, including genetic predisposition (14,15), altered mechanical loading (16) and inflammation (17). Although the key processes leading to OA remain unclear, the death of chondrocytes and the loss of extracellular matrix are considered to be important features of the degeneration of articular cartilage in OA (18). Articular chondrocytes synthesize and secrete extracellular matrix, and their apoptosis serves a key role in the pathogenesis of OA (19,20). Under certain circumstances, cells adapt to environmental pressures and survive through autophagy, and the apoptotic effect is attenuated (21,22). Studies have demonstrated that chondrocyte autophagy exerts a protective effect on normal chondrocytes, and increasing the levels of autophagy in chondrocytes can alleviate OA (23,24). In view of the pivotal role of apoptosis and autophagy in the pathogenesis of KOA, let-7e may be involved in KOA by dysregulating apoptosis and autophagy of chondrocytes.
Si-Miao-San (SMS) is a classic treatment prescribed for the treatment of KOA as a traditional Chinese medicine, which was initially recorded in the ‘Cheng Fang Bian Du’ written by Zhang Bingcheng >100 years ago. SMS is composed of Atractylodes, Phellodendron, Achyranthes and Coix seed; these herbs exhibit anti-inflammatory activity, thus, KOA may be treated with SMS (25). SMS is a safe and effective treatment that has been demonstrated to alleviate KOA by inhibiting cartilage matrix degradation (26,27).
The present study aimed to determine the possibility of using miRNA let-7e as a serum marker for the diagnosis of KOA, explore the underlying mechanism of let-7e function and determine whether SMS alleviated KOA through the regulation of apoptosis and autophagy.
Materials and methods
Collection of serum samples from patients
The involvement of patients in the present study was approved by The Ethics Committee of The First Affiliated Hospital of Zhejiang Chinese Medicine University (Hangzhou, China) and patients provided informed written consent prior to participation. A total of 10 patients underwent artificial knee joint replacement due to KOA at The First Affiliated Hospital of Zhejiang Chinese Medicine University between September 2014 and February 2015, and were included in this study as the KOA group. The diagnosis of KOA was based on the symptoms, physical examination and X-ray or magnetic resonance image (MRI) examinations. For the control group, 10 age-matched patients suffering from trauma without KOA during the same period were recruited. X-ray or MRI was performed to exclude the presence of KOA. The median age was 69 (64–74) years in the KOA group and 66.5 (60–77) years in the control group. Age, sex and body mass index (BMI) of the patients were comparable between the two groups (Table I). Blood samples were obtained from all patients at the time of admittance. The blood was clotted and centrifuged (4°C and 1,000 × g for 10 min), and the liquid component (serum) was transferred to a clean tube and stored at −80°C until further use.
Table I.
Summary of patient characteristics.
| Characteristic | Control (n=10) | KOA (n=10) | P-value |
|---|---|---|---|
| Age, years | 67.50±1.80 | 69.20±0.94 | 0.41 |
| Sex (male), n (%) | 5 (50%) | 3 (30%) | 0.24 |
| Height, cm | 162.20±2.57 | 160.30±2.20 | 0.58 |
| Weight, kg | 68.22±1.96 | 66.68±2.24 | 0.61 |
| BMI, kg/m2 | 25.97±0.67 | 26.02±0.91 | 0.97 |
| Tobacco consumption, n (%) | 4 (40%) | 3 (30%) | 0.32 |
Data are presented as mean ± SD or n (%). BMI, body mass index.
Establishment of an animal model of KOA
The animal protocols were reviewed and approved by The Animal Care and Use Committee of Zhejiang Chinese Medicine University. Male Sprague-Dawley rats (8 weeks old) were obtained from the Experimental Animal Center, Medical Science Academy of Zhejiang Province (Hangzhou, China) and were divided into three groups: Control, KOA and SMS (n=9 per group). Rats were anaesthetized by intraperitoneal injection of 10% chloral hydrate (400 mg/kg). For the KOA and SMS groups, the knee joint cavity of right hind limb was exposed following a medial longitudinal capsular incision. The anterior cruciate ligament was exposed and transected, and the medial meniscus was removed. The patella was relocated, the medial capsular incision was sutured, and the skin was closed (28–30).
After 6 weeks, histopathological examination was performed on one rat in each group to evaluate the KOA model. Subsequently, rats in the SMS group were intragastrically administered 1 ml SMS concentrated solution (4.3 g/kg body weight), and rats in the control and KOA groups were administered 1 ml normal saline. After daily gavage for 4 weeks, the rats were sacrificed by cervical dislocation after intraperitoneal injection of 10% chloral hydrate (400 mg/kg). Whole blood specimens were collected from the retro-orbital sinus, and serum was separated as described above and stored at −80°C. Whole knee joints were dissected into three parts; one part each was used for routine histology and cell culture, and the other part of was washed in PBS and stored at −80°C.
Histopathology of the knee joint
Dissected knee joints were fixed in 10% paraformaldehyde (pH 7.4) at room temperature for 24 h, flushed with water for 30 min and decalcified in EDTA at room temperature for 4–6 weeks. The decalcification solution was replaced weekly. Subsequently, the joints were rinsed with water for 30 min, dehydrated with different concentrations of ethanol (50–100%), transparentized with xylene, embedded in paraffin and sectioned at 5 μm. The histological sections were stained with hematoxylin for 10 min and eosin for 3–5 min at room temperature and assessed by light microscopy at ×50–400 magnification.
Reverse transcription-quantitative (RT-q) PCR
For cartilage, PureLink® miRNA Isolation kit (cat. no. K1570-01; Thermo Fisher Scientific, Inc.) was used to purify total RNA, including miRNA, from cartilage tissue. Reverse transcription was performed using SuperScript™ III Reverse Transcriptase (cat. no. 18080085; Thermo Fisher Scientific, Inc.) and hsa-let-7e Real-Time RT-PCR Detection and U6 Calibration kit (cat. no. orb220270; Biorbyt, Ltd.) in a 20 μl volume containing 100 ng miRNA, 1 μl dNTPs (10 mM), 1 μl miRNA let-7e or U6 stem-loop primer (2 μM), 1 μl 0.1 M DTT, 4 μl 5X First-Strand Buffer, 1 μl RNase Inhibitor (40 U/μl), 1 μl SuperScipt III RTase (200 U/μl) and RNase-Free Water. The reverse transcription temperature protocol was 25°C for 5 min, 50°C for 15 min and 85°C for 5 min.
For serum, a miRNeasy Serum/Plasma kit (cat. no. 217184; Qiagen, Inc.) and a miRNeasy Serum/Plasma Spike-In Control kit (cat. no. 219610; Qiagen, Inc.) were used to purify cell-free total RNA, including miRNA, from the serum. Reverse transcription was performed using SuperScript™ III Reverse Transcriptase and hsa-let-7e Real-Time RT-PCR Detection and cel-mir-39-3p Calibration kit in a 20 μl system containing 100 ng miRNA, 1 μl dNTPs (10 mM), 1 μl miRNA let-7e or cel-mir-39 stem-loop primer (2 μM), 1 μl 0.1 M DTT, 4 μl 5X First-Strand Buffer, 1 μl RNase Inhibitor (40 U/μl), 1 μl SuperScipt III RTase (200 U/μl) and RNase-Free Water. The reverse transcription temperature protocol was 25°C for 5 min, 50°C for 15 min and 85°C for 5 min.
PowerUp™ SYBR® Green Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) was used to detect the expression of miRNA let-7e. The master mix for one reaction included 8 μl sterile distilled water, 10 μl PowerUp SYBR® Green Master mix, 0.5 μl miR-let-7e (10 μM)/U6 specific primer set or miR-let-7e (10 μM)/Cel-miR-39 specific primer set (10 μM) included in the above kit; 1 μl sample cDNA was added to the respective wells. CFX384 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.) was used to run the PCR with one cycle of 95°C for 1 min and 40 cycles of 95°C for 15 sec and 60°C for 25 sec. The experiment was repeated three times. The relative mRNA expression levels were calculated using the 2−ΔΔCq method (31).
Bioinformatics analysis of miRNA-mRNA networks associated with OA
To evaluate the general expression profile of genes in OA, previously published gene expression datasets were analyzed by array and high throughput sequencing. The gene expression data of GSE114007 (32) and GSE117999 (33) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). GSE114007 compared the genome-wide molecular profile of 20 patients with OA and 18 controls based on the GPL11154 (Illumina HiSeq 2000) and GPL18573 (Illumina NextSeq 500) platforms. GSE117999 compared the transcriptomes of articular cartilage of 10 patients with OA and 10 controls based on the platform of GPL20844 (Agilent-072363 SurePrint G3 Human GE version 3 8×60K Microarray). NetworkAnalyst (http://www.networkanalyst.ca) and Cytoscape version 3.5.1 (https://cytoscape.org/download.php) were used to identify significantly differentially expressed genes (DEGs), construct miRNA-mRNA networks and visualize the results.
Unsupervised hierarchical clustering using Ward’s method was performed to cluster the genes and samples (34). The significantly DEGs were mapped to miRNA-gene interaction data obtained from TarBase (http://microrna.gr/tarbase) and miRTarBase (mirtarbase.mbc.nctu.edu.tw) (35,36). Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were identified in target genes with P≤0.05 using the ClueGo version 2.3.5 plug-in in Cytoscape. Pan-Cancer analysis of starBase (starbase.sysu.edu.cn/panCancer.php) was used to predict the co-expression between let-7e, apoptosis-related proteins and autophagy-related proteins with regression analysis.
Western blotting
Total proteins were extracted from knee tissues using T-PER Tissue Protein Extraction Reagent (cat. no. 78510; Thermo Fisher Scientific, Inc.) and Halt Protease and Phosphatase Inhibitor Cocktail (cat. no. 78440; Thermo Fisher Scientific, Inc.). The protein concentration in the supernatant was determined using a Bicinchoninic Acid Protein Assay kit (Beyotime Institute of Biotechnology). A total of 50 μg proteins were loaded per lane onto a 10% SDS-gel and resolved using SDS-PAGE. The separated samples were transferred to nitrocellulose membranes, blocked with TBS + Tween-20 (TBST) containing 5% skimmed milk powder at room temperature for 1 h and incubated with primary antibodies (target proteins and internal reference are listed in Table II) overnight at 4°C. Following washing with TBST for 10 min twice and TBS for 10 min, the membranes were incubated with the secondary antibody (Table II) for 1 h at room temperature and visualized using SuperSignal® West Dura Extended Duration Substrate (Thermo Fisher Scientific, Inc.). The experiment was repeated three times and densitometry analysis was performed with ImageJ software version 1.52 (National Institutes of Health) relative to the respective GAPDH bands in each sample.
Table II.
Antibodies used for western blotting.
| Antibody | Supplier | Cat. no. | Dilution | Molecular weight, kDa |
|---|---|---|---|---|
| LC3B | CST | 4108 | 1:1,000 | 14; 16 |
| Beclin 1 | Abcam | ab62557 | 1:1,000 | 52 |
| p-FADD | Sigma | SAB4504752 | 1:500 | 23 |
| FADD | Abcam | ab24533 | 1:500 | 28 |
| Cleaved caspase 3 | Abcam | ab2302 | 1:200 | 17 |
| Pro-caspase 3 | Abcam | ab90437 | 1:1,000 | 32 |
| GAPDH (internal reference) | Abcam | ab181602 | 1:10,000 | 36 |
| Goat anti-Mouse IgG (H+L) Secondary antibody | Thermo | 31160 | 1:5,000 | |
| Goat anti-Rabbit IgG (H+L) Secondary antibody | Thermo | 31210 | 1:5,000 |
CST, Cell Signaling Technology, Inc.; Sigma, Sigma Aldrich; Merck KGaA; Thermo, Thermo Fisher Scientific, Inc.
Primary culture of chondrocytes
Cartilage from rats was removed and rinsed twice with PBS, cut into 1 mm3 pieces, rinsed twice with PBS and digested with 0.25% trypsin solution for 30 min at 37°C with 5% CO2. The trypsin solution was aspirated, and 0.2% collagenase II solution was added and incubated at 37°C for 3 h. The suspension was collected, centrifuged and resuspended three times. Finally, DMEM (Gibco; Thermo Fisher Scientific, Inc.) containing 20% FBS (Sigma-Aldrich; Merck KGaA) and 1% penicillin and streptomycin (Beijing Solarbio Science & Technology Co., Ltd.) was added, and the cells were plated in a culture flask.
Apoptosis assay
Apoptosis analysis of chondrocytes was performed using an Annexin V-FITC/propidium iodide (PI) apoptosis kit (cat. no. AP101; Multi Sciences, Inc.), and apoptosis was detected using a BD Accuri C6 flow cytometer (BD Biosciences) with a FITC detection channel (FL1) for Annexin V-FITC and a PE detection channel (FL2) for PI. A total of 1–5×105 cells were transferred to a centrifuge tube, centrifuged at 110 × g at room temperature for 5 min and resuspended in 5-fold Binding Buffer diluted with deionized water. A total of 5 μl Annexin V-FITC and 10 μl PI were added to each centrifuge tube, gently vortexed, incubated at room temperature for 5 min in the dark and analyzed by the flow cytometer. FITC and PI staining were low (FITC-/PI-) in normal living cells and distributed in the lower left area (LL) of the flow cytometry dot plots; early apoptotic cells were strongly stained with FITC but not PI (FITC+/PI-), and were located in the lower right region of the plots (RL); in late apoptotic cells, FITC and PI staining were strong (FITC+/PI+) and were distributed in the upper right region (RU) of the plots. The sum of the percentages of early and late apoptotic cells represented the total apoptotic rate (%).
Autophagy assays
Autophagy of chondrocytes was detected using a CYTO-ID® Autophagy Detection kit 2.0 (cat. no. ENZ-51031-K200; Enzo Life Sciences). A total of 1–5×105 chondrocytes in the exponential phase were transferred to a centrifuge tube, centrifuged at room temperature at 110 × g for 5 min, washed with 1X assay buffer included in the kit and resuspended in 0.25 ml 1X assay buffer included in the kit; 0.25 ml CYTO-ID green staining solution diluted with 1X assay buffer supplemented with 5% FBS was added and incubated at room temperature in the dark for 30 min with gentle pipetting to disperse the cells and ensure equal staining. Following centrifugation and washing with the 1X assay buffer, the cells were resuspended in the 1X assay buffer and immediately analyzed using flow cytometry with a FITC detection channel (FL1). A drop of cell suspension was added to a glass microscope slide and a coverslip was placed on top. The cells on the slide were visualized using a confocal microscope at ×400 magnification. Green fluorescence represented the autophagosome and blue fluorescence represented the nucleus, which was stained with DAPI.
Statistical analysis
Statistical analysis was performed using SPSS version 19.0 (IBM Corp.). A two-tailed Student’s t-test and one-way ANOVA were used to compare the quantified data. Tukey’s post-hoc pairwise comparisons were conducted when significant terms were present. Fisher’s exact test was performed to compare the categorical data. Data are presented as the mean ± standard deviation. P<0.05 was considered to indicate a statistically significant difference.
Results
Let-7e expression is decreased in the serum of patients with KOA
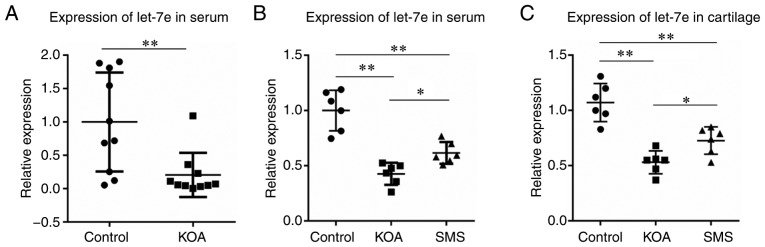
The relative expression levels of let-7e were calculated using 2−ΔΔCq method with spike-in control cel-miR-39 as the normalization control. As presented in Fig. 1A, the expression levels of let-7e in the peripheral serum of the KOA group was ~20% of that in the control group (P<0.01).
Figure 1.
mRNA expression of let-7e in patients with KOA and a rat model of KOA. (A) Expression of let-7e in the serum of patients with KOA and controls. (B) Expression of let-7e in the serum of a rat model of KOA. (C) Expression of let-7e in the cartilage of a rat model of KOA. *P<0.05, **P<0.01. SMS, Si-Miao-San; KOA, knee osteoarthritis.
Establishment of a KOA rat model
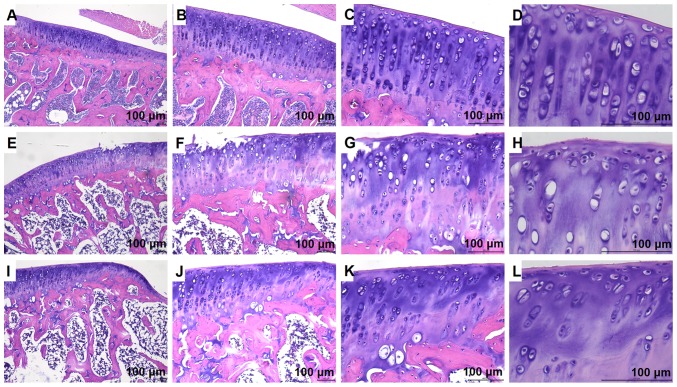
To further examine the role of let-7e in OA and the underlying mechanism, a KOA rat model was constructed by surgery. The microscopic morphological images of the cartilage tissue of the knee joints of different treatment groups were observed (Fig. 2). In the control group, the surface of the cartilage was smooth and intact with no noticeable cracks, the chondrocytes were normal in shape and arranged neatly, the cartilage lacunae were oval and regular with 2–8 cells distributed inside, and the tide line was clear and intact (Fig. 2A–D). In the KOA model group, the surface of the cartilage was less smooth, and edge defects were present in some of the visual fields, the tide line was blurred, irregular or missing, cartilage cells were arranged in a disordered manner, and cartilage lacunae were sparsely distributed (Fig. 2E–H). Compared with the KOA group, the morphology of the cartilage tissue in the SMS group was notably improved; the smoothness of the cartilage surface was increased, the tide line was clearer, and the cartilage lacunae were more regularly arranged (Fig. 2I–L).
Figure 2.
Hematoxylin and eosin staining of the knee joint cartilage tissues in the different treatment groups. (A, B, C and D) Control group. (E, F, G and H) knee osteoarthritis group. (I, J, K and L) Si-Miao-San group.
Let-7e expression is decreased in cartilage and peripheral serum in the KOA rat model
The relative expression levels of let-7e were calculated using the 2−ΔΔCq method with U6 used as the normalization control. As presented in Fig. 1B, in peripheral serum, the expression of let-7e in the KOA group was significantly decreased compared with the control group (P<0.01), and the decrease in let-7e expression was partially reversed in the animals treated with SMS (P<0.05), although it was still lower compared with the control group (P<0.01). The levels of let-7e in the cartilage tissue were examined, and similar results were observed (Fig. 1C); the expression of let-7e in the cartilage of the KOA group was significantly decreased compared with the control group (P<0.01), and the reduction was partially reversed in the SMS group (P<0.05), but was still lower compared with the control group (P<0.01).
Dysregulated mRNA and miRNA expression in OA
A total of 30 OA and 28 non-OA cases were included in the bioinformatics analysis, which identified 592 DEGs by a random effect model with a significance level of P=0.01. Unsupervised hierarchical clustering using Ward’s method was performed to cluster the genes and samples. As presented in Fig. 3, OA samples and controls were classified separately. The whole heatmap including all 592 DEGs is included in Fig. S1. The significantly DEGs were mapped to miRNA-gene interaction data obtained from the TarBase and miRTarBase. A sub-network including 425 seeds, 2,604 nodes and 9,265 edges was constructed, and let-7e was included in this network, supporting its involvement in the onset of OA.
Figure 3.
Heat map of differentially expressed genes in the cartilage of patients with OA. The hierarchical cluster was performed using NetworkAnalyst. The top two rows represent the classification of samples and datasets; red squares represent non-OA samples, blue squares represent OA samples, purple squares indicate data from GSE117999 and green squares indicate data from GSE114007. The columns represent 58 cartilage samples and the rows represent a part of the heatmap of differentially expressed mRNAs and microRNAs. Colored pixels represent changes in the expression levels: Green, downregulated; black, no change; red, upregulated. OA, osteoarthritis.
KEGG pathway analysis was performed to identify the significant pathways of the identified DEGs. Enriched KEGG pathways included ‘Small cell lung cancer’, ‘p53 signaling pathway’, ‘Apoptosis’ and ‘Amyotrophic lateral sclerosis (ALS)’. Considering that previous studies had demonstrated the role of apoptosis in OA (19,20), the focus of the present study was on apoptosis-associated pathways.
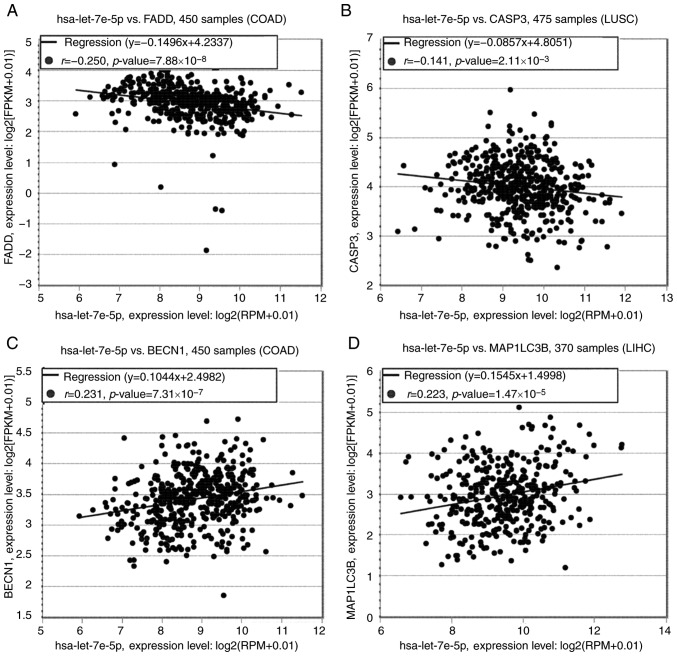
Based on previous studies (19,20,23,24) and bioinformatic analysis, let-7e may function through regulating apoptotic and autophagic activity of articular chondrocytes. Pan-Cancer analysis of starBase (starbase.sysu.edu.cn/panCancer.php) was used to predict the co-expression among let-7e, apoptosis- and autophagy-related proteins. As no data regarding chondrocytes were available, the cell lines with the highest significance in the Pan-Cancer analysis were used instead. Fig. 4 demonstrates the associations between let-7e and FAS-associated death domain (FADD), caspase-3 (CASP3), beclin 1 (BECN1) and microtubule-associated protein 1 light chain 3 β (LC3B) in different types of cancer, including colon adenocarcinoma, lung squamous cell carcinoma and liver hepatocellular carcinoma.
Figure 4.
Putative co-expression of let-7e with FADD, CASP3, Belclin-1 and LC3B. (A and B) Significant negative associations were observed between (A) let-7e and FADD in COAD and (B) let-7e and CASP3 in LUSC. (C and D) Significant positive associations were observed between (C) let-7e and Belclin-1 in COAD and (D) let-7e and LC3B in LIHC. COAD, colon adenocarcinoma; LUSC, lung squamous cell carcinoma; LIHC, liver hepatocellular carcinoma; FADD, FAS-associated death domain; CASP3, caspase-3; LC3B, microtubule-associated protein 1 light chain 3 β.
Expression of apoptotic proteins is increased, whereas expression of autophagy-related proteins is decreased in the KOA rat model
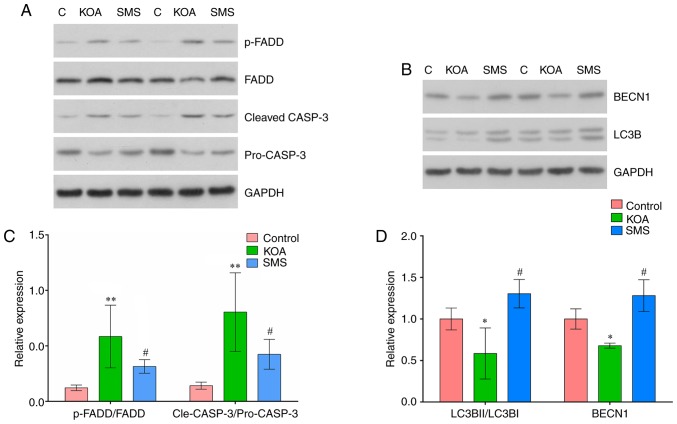
The expression of apoptotic proteins FADD and CASP3 in the cartilage were examined using western blotting. As presented in Fig. 5B, the ratio of p-FADD/FADD was significantly increased in the KOA group compared with the control group (P<0.01), and treatment with SMS significantly reduced the ratio of p-FADD/FADD compared with the KOA group (P<0.05). The ratio of cleaved-CASP3/pro-CASP3 was significantly increased in the KOA group compared with the control group (P<0.01), and treatment with SMS significantly reduced the ratio of cleaved-CASP3/pro-CASP3 compared with the KOA group (P<0.05).
Figure 5.
Expression of apoptotic and autophagy-related proteins in the cartilage of the rat KOA model. (A and C) Expression of apoptotic proteins FADD, p-FADD, pro-CASP3 and cleaved CASP3 in the cartilage. (B and D) Expression of autophagy-related proteins BECN1 and LC3B in the cartilage. *P<0.05 vs. C group; **P<0.01 vs. C group; #P<0.05 vs. KOA group. KOA, knee osteoarthritis; SMS, Si-Miao-San; FADD, FAS-associated death domain; p-, phosphorylated; CASP3, caspase-3; BECN1, beclin 1; LC3B, microtubule-associated protein 1 light chain 3 β.
The expression of autophagy-associated proteins LC3B and BECN1 are presented in Fig. 5D. The ratio of LC3BII/LC3BI was reduced in the KOA group compared with the control group (P<0.01). In rats treated with SMS, the LC3BII/LC3BI ratio was significantly increased compared with the KOA group (P<0.01). Compared with the control group, BECN1 expression was significantly reduced in the KOA group (P<0.01) and increased in rats treated with SMS (P<0.01).
Apoptotic activity is increased in the chondrocytes of KOA rats
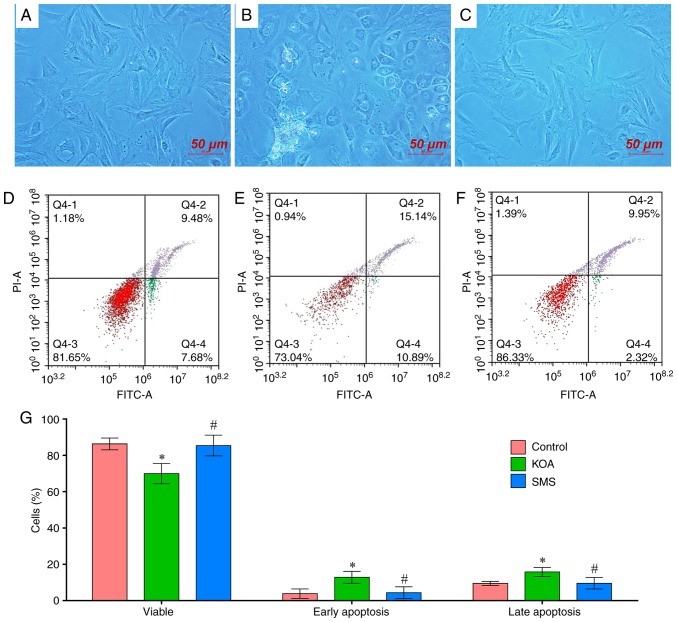
Primary culture of chondrocytes was performed using chondrocytes isolated from the rat knee cartilage, as presented in Fig. 6. In the control group, the chondrocytes were fusiform or spindle-shaped, with a small proportion exhibiting an oval-shaped morphology (Fig. 6A). In the KOA group, the chondrocytes were typically round or elliptical, with a small proportion of fusiform or spindle-shaped cells. The nuclei were large, a number of cells possessed vacuoles, and some cells died during cell culture. (Fig. 6B). Compared with the KOA group, the overall morphology was significantly improved in the chondrocytes obtained from SMS-treated rats, where the majority of the cells were spindle-shaped, although the chondrocytes of the SMS group still exhibited reduced activity and a lower number of cells compared with the control group (Fig. 6C).
Figure 6.
Primary culture and apoptotic analysis of chondrocytes from rat knee cartilage. (A and D) Control group. (B and E) KOA group. (C and F) SMS group. (G) Quantitative analysis of the apoptotic rates. *P<0.05 vs. Control; #P<0.05 vs. KOA. KOA, knee osteoarthritis; SMS, Si-Miao-San.
The chondrocytes from the control, KOA and SMS groups were stained with Annexin-V-FITC/PI and apoptosis was assessed using flow cytometry. Compared with the control group, the proportion of viable cells in the KOA group was significantly lower, and the proportion of early and late apoptotic cells was increased (P<0.01; Fig. 6D, E and G). In the chondrocytes obtained from rats treated with SMS, the proportion of live cells was significantly increased compared with the KOA group, and the proportion of early and late apoptotic cells was reduced (P<0.01; Fig. 6F and G).
Autophagic activity is decreased in the chondrocytes from the KOA rat model
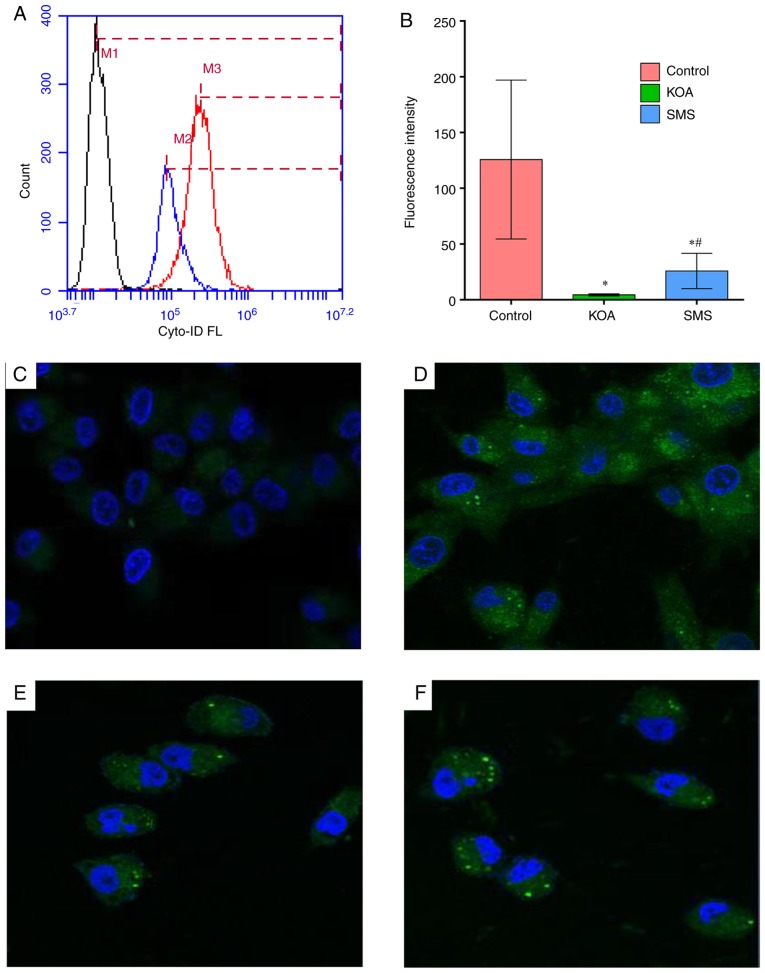
The chondrocytes from the three groups of rats were stained with an ENZO kit and analyzed using flow cytometry and confocal microscopy. The autophagosomes exhibited green fluorescence; the chondrocytes in the control group exhibited medium levels of fluorescence intensity (Fig. 7A, B and D). In the KOA group, fluorescence intensity was significantly decreased compared with the control group (P<0.01), suggesting that autophagic activity was decreased (Fig. 7E). Compared with the KOA group, the fluorescence intensity of the SMS group significantly increased, suggesting that autophagic activity was increased, although it was still significantly lower compared with the control group (Fig. 7F). No green fluorescence was observed in the blank control group (Fig. 7C).
Figure 7.
Autophagy analysis of chondrocytes from rat knee cartilage. (A and B) Autophagy analysis based on flow cytometry. M1, KOA group; M2, SMS group; M3, control group. (C–F) Autophagy analysis using confocal microscopy. Confocal microscopy of (C) blank control, (D) control, (E) KOA and (F) SMS group. *P<0.05 vs. control; #P<0.05 vs. KOA. KOA, knee osteoarthritis; SMS, Si-Miao-San.
Discussion
At present, the diagnosis of KOA is based on clinical and imaging findings. The identification of effective serum markers for diagnosis may potentially improve the rapidity and accuracy of diagnoses. Therefore, studies in the past decade have focused on the levels of circulating miRNAs. In 2014, circulating let-7e was reported to be decreased in patients with OA, and lower let-7e levels were associated with an increased risk of arthroplasty due to severe OA (13). Subsequently, a series of miRNAs have been demonstrated to be abnormally expressed in OA, such as miR-140-30, mir-33b-3p, miR-671-3p, let-7e, miR-9, miR-27, miR-34a, miR-140, miR-146a, miR-558, and miR-602 (37–39). However, the results of these studies vary widely. In the present study, in patients with KOA, let-7e levels in the peripheral blood were downregulated. In addition, the expression levels of let-7e in the serum and the cartilage of the knee in the KOA rat model were significantly decreased compared with the control group.
Beyer et al (13) demonstrated that let-7e levels were a negative predictor for total joint arthroplasty. The expression of let-7e in severe osteoarthritis cases was 0.66 of that in the control group; unadjusted Cox regression analysis revealed that let-7e was inversely associated with severe knee and hip osteoarthritis (13). After adjustment for sex, age and BMI, the results remained consistent with an adjusted HR of 0.75 (95% CI, 0.58–0.95; P=0.019) (13). In the present study, serum let-7e level in the KOA group decreased to ~20% of that in control group. Generally, the two studies achieved consistent results, although in the study by Beyer et al (13), patients with severe OA of the knee and the hip joints were included, whereas in present study only included patients with KOA. Based on the previous and the present study, miRNA let-7e may serve as a promising candidate for the diagnosis of KOA. However, as the sample size of the present study was small, future studies are required with larger sample sizes from multiple centers to explore the clinical value of this indicator.
The role of let-7e in the occurrence of KOA remains unclear. Let-7e regulates key molecules involved in apoptotic signaling pathways in neurons (40,41), breast cells (42) and hemocytes (43). In a neuron-like cell line PC12, transfection of let-7e decreased Casp3 expression and apoptosis, whereas co-transfection of anti-let-7e significantly alleviated the effects of let-7e, indicating that let-7e may protect neurons against apoptosis by negatively regulating the expression of CASP3 (40). However, the effect of let-7e on chondrocyte apoptosis is unclear. In the present study, the expression levels of let-7e were decreased in the cartilage and peripheral blood of patients with KOA, which was accompanied by an increase in apoptotic activity and decreased autophagic activity, suggesting that let-7e may be involved in the pathology of KOA through the regulation of apoptosis and autophagy.
In the present study, expression levels of various members of apoptotic signaling pathways were increased in the cartilage of KOA rats, as was the rate of apoptosis. Treatment with SMS reduced apoptotic activity and alleviated KOA. It has been reported that the incidence of apoptosis in chondrocytes in patients with OA is significantly higher compared with normal articular chondrocytes (44), and the degree of apoptosis is associated with the degree of articular cartilage destruction (45). Pro-inflammatory cytokines tumor necrosis factor-α (TNF-α) and interleukin 1β (IL-1β) reduce the anabolic metabolism of chondrocytes, inducing apoptosis and increasing the occurrence of OA (46). A large sample microarray study performed in the Netherlands revealed that the expression of molecules involved in a series of apoptotic signaling pathways in hemocytes of patients with OA were increased, including FADD and CASP3 (47). A study based on a rabbit knee arthritis model demonstrated that SMS inhibited the levels of IL-1β and TNF-α and effectively inhibited the inflammatory response (48). Modified SMS reduced the expression of IL-6 and increased the expression of basic fibroblast growth factor, thus reducing the degradation of cartilage matrix (49). In our previous study, SMS treatment significantly reduced mRNA expression levels of FADD and CASP3 in the articular cartilage of KOA rats (50). The present study confirmed the importance of chondrocyte apoptosis in the pathogenesis of OA, and SMS inhibited chondrocyte apoptosis.
The results of the present study demonstrated that the expression of autophagy-related proteins in the articular cartilage of KOA rats was lower compared with that in the controls, and autophagic activity was decreased; SMS treatment reversed the alterations in autophagic activity. At present, it is hypothesized that cell autophagy is different from apoptosis and necrosis of cells and inhibits the apoptosis of damaged chondrocytes, which may be an important mechanism for chondrocyte self-protection (51). A previous study has reported that the expression of Beclin1 and LC3 in OA cartilage and chondrocytes is reduced, and the expression levels of the associated apoptotic signaling factors are increased (23). Rapamycin enhances the autophagic activity of articular chondrocytes, thus preventing the death of chondrocytes in a mouse model of KOA and reducing the severity of OA (24). Studies using rat models of OA have demonstrated that the activity of autophagy pathways is inhibited in the cartilage from the OA group compared with a non-OA control and may be involved in the pathogenesis of OA cartilage (50,52,53). These studies suggested that the activation of chondrocyte autophagy may inhibit apoptosis, which may be a novel mechanism for delaying and controlling the progression of KOA. SMS may exert its therapeutic role through this mechanism.
The present study has several limitations. Firstly, as the sample size of this study was small, the significant reduction of let-7e in the KOA group was not robust enough; additional studies with larger sample sizes are required to validate this conclusion. Secondly, to ascertain whether let-7e is involved in the pathogenesis of KOA, further rescue experiments are required to discern whether let-7e induces the pathogenesis or its levels are increased following the onset of KOA. Thirdly, the present study was designed to explore the mechanisms underlying the involvement of let-7e; further experiments are required to confirm the association between let-7e and elevated apoptotic and decreased autophagic activity of the cartilage tissue. In addition, the present study did not provide direct evidence that SMS reversed KOA by regulating let-7e and apoptosis-related proteins. Further experiments are required to confirm this cause-effect relationship.
In conclusion, in the peripheral blood of patients with KOA, the expression of let-7e was significantly decreased, as well as in peripheral serum and cartilage of the knee in a rat KOA model. The expression of apoptotic pathway proteins and apoptotic activity was increased in the cartilage of KOA rats, whereas the expression of autophagy-related proteins and autophagic activity was decreased in the articular cartilage of KOA rats compared with the control group. SMS treatment reversed the changes in the apoptotic and autophagic activity. Therefore, the present study supported the hypothesis that circulating let-7e may serve as a potential serum biomarker for the diagnosis and treatment of KOA. Elevated apoptotic activity and decreased autophagic activity of cartilage tissues may be involved in KOA, and treatment with SMS may reverse these effects.
Supplementary Information
Acknowledgements
Not applicable.
Abbreviations
- KOA
knee osteoarthritis
- SMS
Si-Miao-San
- OA
osteoarthritis
- let-7
lethal-7
- BMI
body mass index
- DEG
differentially expressed gene
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- FADD
FAS-associated death domain
- CASP3
caspase-3
- LC3B
microtubule-associated protein 1 light chain 3 β
Funding
The present study was supported by grants from the Zhejiang Provincial Science and Technology Plan of Traditional Chinese Medicine (grant no. 2017ZQ013), the Medical and Health Science and Technology Plan of Zhejiang Province (grant no. 2017KY078), the Zhejiang Provincial Natural Science Foundation (grant no. LY17H040005) and the National Natural Science Foundation of China (grant nos. 81871176 and 81701461).
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors’ contributions
JMS conceived and designed the study. LF, CF, CXW, DYX, JJC, JFH and PLT performed the experiments. LF and CF analyzed the data. JMS and CF organized and wrote the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
All human studies were approved by the Ethics Committee of The First Affiliated Hospital of Zhejiang Chinese Medicine University, and written informed consent was obtained from each participant. All animal studies were approved by the Animal Care and Use Committee of Zhejiang Chinese Medicine University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Jordan JM, Helmick CG, Renner JB, Luta G, Dragomir AD, Woodard J, Fang F, Schwartz TA, Abbate LM, Callahan LF, et al. Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African Americans and Caucasians: The Johnston County Osteoarthritis Project. J Rheumatol. 2007;34:172–180. [PubMed] [Google Scholar]
- 2.Nguyen US, Zhang Y, Zhu Y, Niu J, Zhang B, Felson DT. Increasing prevalence of knee pain and symptomatic knee osteoarthritis: Survey and cohort data. Ann Intern Med. 2011;155:725–732. doi: 10.7326/0003-4819-155-11-201112060-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao BW, Zhou LF, Liu YL, Wan SM, Gao ZX. Evolution of fish Let-7 MicroRNAs and their expression correlated to growth development in blunt snout bream. Int J Mol Sci. 2017;18 doi: 10.3390/ijms18030646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roush S, Slack FJ. The let-7 family of microRNAs. Trends Cell Biol. 2008;18:505–516. doi: 10.1016/j.tcb.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 5.Liu SS, Wang YS, Sun YF, Miao LX, Wang J, Li YS, Liu HY, Liu QL. Plasma microRNA-320, microRNA-let-7e and microRNA-21 as novel potential biomarkers for the detection of retinoblastoma. Biomed Rep. 2014;2:424–428. doi: 10.3892/br.2014.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murray MJ, Saini HK, Siegler CA, Hanning JE, Barker EM, van Dongen S, Ward DM, Raby KL, Groves IJ, Scarpini CG, et al. LIN28 expression in malignant germ cell tumors downregulates let-7 and increases oncogene levels. Cancer Res. 2013;73:4872–4884. doi: 10.1158/0008-5472.CAN-12-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chiam K, Wang T, Watson DI, Mayne GC, Irvine TS, Bright T, Smith L, White IA, Bowen JM, Keefe D, et al. Circulating serum exosomal miRNAs as potential biomarkers for esophageal adenocarcinoma. J Gastrointest Surg. 2015;19:1208–1215. doi: 10.1007/s11605-015-2829-9. [DOI] [PubMed] [Google Scholar]
- 8.Cengiz M, Karatas OF, Koparir E, Yavuzer S, Ali C, Yavuzer H, Kirat E, Karter Y, Ozen M. Differential expression of hypertension-associated microRNAs in the plasma of patients with white coat hypertension. Medicine (Baltimore) 2015;94:e693. doi: 10.1097/MD.0000000000000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peng G, Yuan Y, Wu S, He F, Hu Y, Luo B. MicroRNA let-7e is a potential circulating biomarker of acute stage ischemic stroke. Transl Stroke Res. 2015;6:437–445. doi: 10.1007/s12975-015-0422-x. [DOI] [PubMed] [Google Scholar]
- 10.Li D, Ji L, Liu L, Liu Y, Hou H, Yu K, Sun Q, Zhao Z. Characterization of circulating microRNA expression in patients with a ventricular septal defect. PLoS One. 2014;9:e106318. doi: 10.1371/journal.pone.0106318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krause BJ, Carrasco-Wong I, Dominguez A, Arnaiz P, Farías M, Barja S, Mardones F, Casanello P. Micro-RNAs Let7e and 126 in plasma as markers of metabolic dysfunction in 10 to 12 years old children. PLoS One. 2015;10:e0128140. doi: 10.1371/journal.pone.0128140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kagawa T, Watanabe M, Inoue N, Otsu H, Saeki M, Katsumata Y, Takuse Y, Iwatani Y. Increases of microRNA let-7e in peripheral blood mononuclear cells in Hashimoto’s disease. Endocr J. 2016;63:375–380. doi: 10.1507/endocrj.EJ15-0577. [DOI] [PubMed] [Google Scholar]
- 13.Beyer C, Zampetaki A, Lin NY, Kleyer A, Perricone C, Iagnocco A, Distler A, Langley SR, Gelse K, Sesselmann S, et al. Signature of circulating microRNAs in osteoarthritis. Ann Rheum Dis. 2015;74:e85. doi: 10.1136/annrheumdis-2013-204698. [DOI] [PubMed] [Google Scholar]
- 14.Weidauer L, Beare T, Binkley T, Minett M, Specker B. Longitudinal growth and pQCT measures in hutterite children and grandchildren are associated with prevalence of hip or knee replacement resulting from osteoarthritis in parents and grandparents. Clin Orthop Relat Res. 2018;476:1093–1103. doi: 10.1007/s11999.0000000000000197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.den Hollander W, Boer CG, Hart DJ, Yau MS, Ramos YFM, Metrustry S, Broer L, Deelen J, Cupples LA, Rivadeneira F, et al. Genome-wide association and functional studies identify a role for matrix Gla protein in osteoarthritis of the hand. Ann Rheum Dis. 2017;76:2046–2053. doi: 10.1136/annrheumdis-2017-211214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wellsandt E, Khandha A, Manal K, Axe MJ, Buchanan TS, Snyder-Mackler L. Predictors of knee joint loading after anterior cruciate ligament reconstruction. J Orthop Res. 2017;35:651–656. doi: 10.1002/jor.23408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JR, Yoo JJ, Kim HA. Therapeutics in osteoarthritis based on an understanding of its molecular pathogenesis. Int J Mol Sci. 2018;19 doi: 10.3390/ijms19030674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Funato S, Yasuhara R, Yoshimura K, Miyamoto Y, Kaneko K, Suzawa T, Chikazu D, Mishima K, Baba K, Kamijo R. Extracellular matrix loss in chondrocytes after exposure to interleukin-1β in NADPH oxidase-dependent manner. Cell Tissue Res. 2017;368:135–144. doi: 10.1007/s00441-016-2551-2. [DOI] [PubMed] [Google Scholar]
- 19.Rose J, Soder S, Skhirtladze C, Schmitz N, Gebhard PM, Sesselmann S, Aigner T. DNA damage, discoordinated gene expression and cellular senescence in osteoarthritic chondrocytes. Osteoarthritis Cartilage. 2012;20:1020–1028. doi: 10.1016/j.joca.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Hosseinzadeh A, Kamrava SK, Joghataei MT, Darabi R, Shakeri-Zadeh A, Shahriari M, Reiter RJ, Ghaznavi H, Mehrzadi S. Apoptosis signaling pathways in osteoarthritis and possible protective role of melatonin. J Pineal Res. 2016;61:411–425. doi: 10.1111/jpi.12362. [DOI] [PubMed] [Google Scholar]
- 21.Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: Crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- 22.Kroemer G, Mariño G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caramés B, Taniguchi N, Otsuki S, Blanco FJ, Lotz M. Autophagy is a protective mechanism in normal cartilage, and its aging-related loss is linked with cell death and osteoarthritis. Arthritis Rheum. 2010;62:791–801. doi: 10.1002/art.27305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caramés B, Hasegawa A, Taniguchi N, Miyaki S, Blanco FJ, Lotz M. Autophagy activation by rapamycin reduces severity of experimental osteoarthritis. Ann Rheum Dis. 2012;71:575–581. doi: 10.1136/annrheumdis-2011-200557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fougere B. Si-Miao-San for chronic inflammatory conditions. Proceedings of the 40th Annual IVAS and 15th Annual ItVAS International Congress on Veterinary Acupuncture; Florence, Italy. 2014. p. 257. [Google Scholar]
- 26.Xu Y, Liu Q, Liu ZL, Lim L, Chen WH, Lin N. Treatment with SiMiaoFang, an anti-arthritis chinese herbal formula, inhibits cartilage matrix degradation in osteoarthritis rat model. Rejuvenation Res. 2013;16:364–376. doi: 10.1089/rej.2013.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen B, Zhan H, Marszalek J, Chung M, Lin X, Zhang M, Pang J, Wang C. Traditional Chinese medications for knee osteoarthritis pain: A meta-analysis of randomized controlled trials. Am J Chin Med. 2016;44:677–703. doi: 10.1142/S0192415X16500373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayami T, Pickarski M, Zhuo Y, Wesolowski GA, Rodan GA, Duong LT. Characterization of articular cartilage and subchondral bone changes in the rat anterior cruciate ligament transection and meniscectomized models of osteoarthritis. Bone. 2006;38:234–243. doi: 10.1016/j.bone.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 29.Rogart JN, Barrach HJ, Chichester CO. Articular collagen degradation in the Hulth-Telhag model of osteoarthritis. Osteoarthritis Cartilage. 1999;7:539–547. doi: 10.1053/joca.1999.0258. [DOI] [PubMed] [Google Scholar]
- 30.Hulth A, Lindberg L, Telhag H. Experimental osteoarthritis in rabbits. Preliminary report. Acta Orthop Scand. 1970;41:522–530. doi: 10.3109/17453677008991540. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Fisch KM, Gamini R, Alvarez-Garcia O, Akagi R, Saito M, Muramatsu Y, Sasho T, Koziol JA, Su AI, Lotz MK. Identification of transcription factors responsible for dysregulated networks in human osteoarthritis cartilage by global gene expression analysis. Osteoarthritis Cartilage. 2018;26:1531–1538. doi: 10.1016/j.joca.2018.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brophy RH, Zhang B, Cai L, Wright RW, Sandell LJ, Rai MF. Transcriptome comparison of meniscus from patients with and without osteoarthritis. Osteoarthritis Cartilage. 2018;26:422–432. doi: 10.1016/j.joca.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scian MJ, Maluf DG, David KG, Archer KJ, Suh JL, Wolen AR, Mba MU, Massey HD, King AL, Gehr T, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant. 2011;11:2110–2122. doi: 10.1111/j.1600-6143.2011.03666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: A functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou CH, Shrestha S, Yang CD, Chang NW, Lin YL, Liao KW, Huang WC, Sun TH, Tu SJ, Lee WH, et al. miRTarBase update 2018: A resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nugent M. MicroRNAs: Exploring new horizons in osteoarthritis. Osteoarthritis Cartilage. 2016;24:573–580. doi: 10.1016/j.joca.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 38.Ntoumou E, Tzetis M, Braoudaki M, Lambrou G, Poulou M, Malizos K, Stefanou N, Anastasopoulou L, Tsezou A. Serum microRNA array analysis identifies miR-140-3p, miR-33b-3p and miR-671-3p as potential osteoarthritis biomarkers involved in metabolic processes. Clin Epigenetics. 2017;9:127. doi: 10.1186/s13148-017-0428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seeliger C, Balmayor ER, van Griensven M. miRNAs related to skeletal diseases. Stem Cells Dev. 2016;25:1261–1281. doi: 10.1089/scd.2016.0133. [DOI] [PubMed] [Google Scholar]
- 40.Peng G, Yuan Y, He Q, Wu W, Luo BY. MicroRNA let-7e regulates the expression of caspase-3 during apoptosis of PC12 cells following anoxia/reoxygenation injury. Brain Res Bull. 2011;86:272–276. doi: 10.1016/j.brainresbull.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 41.Gong W, Zheng J, Liu X, Ma J, Liu Y, Xue Y. Knockdown of NEAT1 restrained the malignant progression of glioma stem cells by activating microRNA let-7e. Oncotarget. 2016;7:62208–62223. doi: 10.18632/oncotarget.11403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aure MR, Leivonen SK, Fleischer T, Zhu Q, Overgaard J, Alsner J, Tramm T, Louhimo R, Alnæs GI, Perälä M, et al. Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors. Genome Biol. 2013;14:R126. doi: 10.1186/gb-2013-14-11-r126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferreira AF, Moura LG, Tojal I, Ambrósio L, Pinto-Simões B, Hamerschlak N, Calin GA, Ivan C, Covas DT, Kashima S, Castro FA. ApoptomiRs expression modulated by BCR-ABL is linked to CML progression and imatinib resistance. Blood Cells Mol Dis. 2014;53:47–55. doi: 10.1016/j.bcmd.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 44.Héraud F, Héraud A, Harmand MF. Apoptosis in normal and osteoarthritic human articular cartilage. Ann Rheum Dis. 2000;59:959–965. doi: 10.1136/ard.59.12.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thomas CM, Fuller CJ, Whittles CE, Sharif M. Chondrocyte death by apoptosis is associated with the initiation and severity of articular cartilage degradation. Int J Rheum Dis. 2011;14:191–198. doi: 10.1111/j.1756-185X.2010.01578.x. [DOI] [PubMed] [Google Scholar]
- 46.Rahmati M, Mobasheri A, Mozafari M. Inflammatory mediators in osteoarthritis: A critical review of the state-of-the-art, current prospects, and future challenges. Bone. 2016;85:81–90. doi: 10.1016/j.bone.2016.01.019. [DOI] [PubMed] [Google Scholar]
- 47.Ramos YF, Bos SD, Lakenberg N, Böhringer S, den Hollander WJ, Kloppenburg M, Slagboom PE, Meulenbelt I. Genes expressed in blood link osteoarthritis with apoptotic pathways. Ann Rheum Dis. 2014;73:1844–1853. doi: 10.1136/annrheumdis-2013-203405. [DOI] [PubMed] [Google Scholar]
- 48.Li HY, Meng DF, Su JG. Si Miao San intervention rabbit knee joint inflammation traumatic synovial fluid IL-1β and TNF-α level of experiments. Chin J Tradit Chin Med. 2013;28:3043–3045. (In Chinese) [Google Scholar]
- 49.Zhao P, Wang L, Li H, Yu J. Effects on IL-6 and BFGF of the rabbits’ knee arthritises by modified Simiao power. China J Chin Med. 2015;30:1166–1169. (In Chinese) [Google Scholar]
- 50.Shen JM, Feng L, Chen J, Wu Y, Yu J. Effect of Simiaosan on apoptosis and autophagy of chondrocyte in treating knee osteoarthritis in rats. New Chin Med. 2017;49:12–15. (In Chinese) [Google Scholar]
- 51.Zhang Q, Lai S, Hou X, Cao W, Zhang Y, Zhang Z. Protective effects of PI3K/Akt signal pathway induced cell autophagy in rat knee joint cartilage injury. Am J Transl Res. 2018;10:762–770. [PMC free article] [PubMed] [Google Scholar]
- 52.Wu Z, Luan Z, Zhang X, Zou K, Ma S, Yang Z, Feng W, He M, Jiang L, Li J, Yao J. Chondro-protective effects of polydatin in osteoarthritis through its effect on restoring dysregulated autophagy via modulating MAPK, and PI3K/Akt signaling pathways. Sci Rep. 2019;9:13906. doi: 10.1038/s41598-019-50471-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53.Wang F, Liu J, Chen X, Zheng X, Qu N, Zhang B, Xia C. IL-1β receptor antagonist (IL-1Ra) combined with autophagy inducer (TAT-Beclin1) is an effective alternative for attenuating extracellular matrix degradation in rat and human osteoarthritis chondrocytes. Arthritis Res Ther. 2019;21:171. doi: 10.1186/s13075-019-1952-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.