Abstract
Patients with metastatic gastric cancer (GC) have a poor prognosis; however, the molecular mechanism of GC metastasis remains unclear. Here, we employed bioinformatics to systematically screen the metastasis‐associated genes and found that the levels of basal cell adhesion molecule (BCAM) were significantly increased in GC tissues from patients with metastasis, as compared to those without metastasis. The upregulation of BCAM was also significantly associated with a shorter survival time. Depletion of BCAM inhibited GC cell migration and invasion. Knockout (KO) of BCAM by the CRISPR/Cas9 system reduced the invasion and metastasis of GC cells. To explore the mechanism of BCAM upregulation, we identified a previously uncharacterized BCAM sense lncRNA that spanned from exon 6 to intron 6 of BCAM, and named it as BCAM‐associated long noncoding RNA (BAN). Knockdown of BAN inhibited BCAM expression at both mRNA and protein levels. Knockdown of BAN suppressed GC cell migration and invasion, which was effectively rescued by ectopic expression of BCAM. Further clinical data showed that BAN upregulation was associated with GC metastasis and poor prognosis. Importantly, BAN expression was also significantly associated with that of BCAM in GC tissues. Taken together, these results indicate that increased expression of BCAM and its sense lncRNA BAN promote GC cell invasion and metastasis, and are associated with poor prognosis of GC patients.
Keywords: BCAM, gastric cancer, lncRNA, metastasis, prognosis
We found that the levels of BCAM and its sense lncRNA BAN were increased in gastric cancer (GC) tissues with metastasis. Knockdown of BAN inhibited BCAM expression at both mRNA and protein levels. Moreover, increased expression of BCAM and its sense lncRNA BAN promote GC cell invasion and metastasis, and are associated with poor prognosis of GC patients.
Abbreviations
- ACRG
Asian cancer research group
- AJCC
American Joint Committee on Cancer
- BAN
BCAM‐associated lncRNA
- BCAM
basal cell adhesion molecule
- FACS
fluorescence‐activated cell sorting
- GAPDH
glyceraldehyde‐3‐phosphate dehydrogenase
- GC
gastric cancer
- lncRNA
long noncoding RNA
- MTT
3‐(4,5‐Dimethylthiazol‐2‐yl)‐2,5‐diphenyltetrazolium bromide
- qRT‐PCR
quantitative reverse transcription PCR
- ROC
receiver operator characteristic
- TCGA
the cancer genome atlas
1. Introduction
Gastric cancer (GC) is the fifth most frequently diagnosed cancer and is the third leading cause of cancer‐related deaths worldwide (Bray et al., 2018). Due to the lack of obvious symptoms and biomarkers for early‐stage GC, ~ 40% of GC patients present with metastasis at the time of diagnosis (Bernards et al., 2013). Moreover, the overall survival of GC patients with metastasis is poor, with an about 5% of the 5‐year survival rate (Bernards et al., 2013). Thus, it is in an emergency to clarify the molecular mechanisms of GC metastasis.
Basal cell adhesion molecule (BCAM), also known as Lutheran, is widely expressed in various tissues and is involved in many biological processes, such as cell adhesion, migration, and invasion (Bartolini et al., 2016; Campbell et al., 1994; De Grandis et al., 2013; El Nemer et al., 1998; Gauthier et al., 2005; Hines et al., 2003; Kikkawa and Miner, 2005; Kikkawa et al., 2013; Parsons et al., 1995). Emerging studies have shown that BCAM plays an important role in tumor progression, including skin tumors, hepatocellular carcinoma, colorectal cancer, and bladder cancer (Bartolini et al., 2016; Campbell et al., 1994; Chang et al., 2017; Drewniok et al., 2004; Kikkawa et al., 2013). However, the role of BCAM in GC progression is still unclear.
Long noncoding RNA, more than 200 nt in length, are transcripts without protein‐coding capacity (Derrien et al., 2012; Djebali et al., 2012; Ma et al., 2013). Increasing data demonstrate that lncRNA regulate gene expression through diverse mechanisms, including gene activation and suppression, chromatin modification and remodeling, splicing and translation modulation, acting as miRNA sponges, and small RNA precursors (Ponting et al., 2009; Spitale et al., 2011; Wang and Chang, 2011; Wilusz et al., 2009). Accumulating evidence has shown that lncRNA play key roles in the formation and progression of many cancers, including GC (Gutschner and Diederichs, 2012; Spizzo et al., 2012; Xie et al., 2016). lncRNA are important regulators of cell proliferation, apoptosis, migration, and differentiation, and dysregulated lncRNA result in tumor growth, invasion, and metastasis (Gupta et al., 2010). Emerging data demonstrate that lncRNA, including ANRIL, FENDRR, GAS5, GHET1, GMAN, MALAT1, and PVT1, are involved in GC progression (Kong et al., 2015; Sun et al., 2014; Tripathi et al., 2010; Xie et al., 2016; Xu et al., 2014; Yang et al., 2014; Zhang et al., 2014; Zhuo et al., 2019). Our recent study shows that GMAN, upregulated in GC tissues, is associated with metastasis and promotes the expression of ephrin A1 (Zhuo et al., 2019).
In this study, we found that BCAM expression was significantly correlated with GC metastasis and poor prognosis. KO of BCAM suppressed GC cell invasion and metastasis. Furthermore, we identified a previously undescribed gene BCAM‐associated lncRNA (BAN) as a sense lncRNA of BCAM, which was also associated with GC metastasis and poor prognosis. Knockdown of BAN not only inhibited BCAM expression, but also suppressed GC cell invasion, which was successfully rescued by ectopic expression of BCAM. Thus, our data suggest that BCAM and its sense lncRNA BAN play a crucial role in GC metastasis.
2. Materials and methods
2.1. Bioinformatics analysis
RNA‐seq data of The Cancer Genome Atlas (TCGA) cohort were downloaded from the Genomic Data Commons data portal (url) (Cancer Genome Atlas Research, 2014). R DESeq2 package was used to find genes with differential expression level between GC tissues with distant metastasis and those without metastasis, and the genes with FDR under 0.05 and expression fold change over 1.8 were considered as significantly upregulated genes (Love et al., 2014). Microarrays of Asian Cancer Research Group (ACRG) cohort were obtained from Gene Expression Omnibus database (url) (Cristescu et al., 2015). The raw CEL files were normalized with the RMA algorithm using Custom chip Definition Files mapping to official Gene Symbol (Brainarray v.22 http://brainarray.mbni.med.umich.edu/) (Manhong et al., 2005). Notably, the averaged expression was calculated for genes with multiple targeted probes. R limma package was used to find the differentially expressed genes between GC tissues with distant metastasis and those without metastasis, and the genes with FDR under 0.05 and expression fold change over 1.8 were considered as significantly upregulated genes (Ritchie et al., 2015). Univariate Cox regression analysis was applied to identify the survival‐related genes in TCGA and ACRG cohorts, and genes with P value under 0.05 and Z‐score over 0 were considered as adversely prognostic.
2.2. Human tissue samples
All GC tissue samples were obtained from GC patients undergoing gastrectomy with informed consent. Zhejiang cohort (n = 64) samples were collected from Sir Run Run Shaw Hospital, Zhejiang University School of Medicine (Hangzhou, China) and Zhejiang Cancer Hospital (Hangzhou, China). Among them, 11 pairs of GC tissues were from patients with distant metastasis and age‐ and sex‐matched patients without metastasis. Ethical consent was granted from the Ethical Committee Review Board of Zhejiang University School of Medicine. The study methodologies conformed to the standards set by the Declaration of Helsinki.
2.3. Cell culture
Human GC cell line BGC‐823 was obtained from the Chinese Academy of Sciences (Jiao et al., 2014). Human GC cell line SGC‐7901 was obtained from Beijing Cancer Hospital (Xing et al., 2012). BGC‐823 and SGC‐7901 cells were maintained in RPMI‐1640 medium supplemented with 10% FBS (Gibco BRL, Grand Island, NY, USA) with 5% CO2. All cell lines were routinely tested negative for mycoplasma.
2.4. RNA extraction and quantitative RT‐PCR
Total RNA was extracted from human tissue samples and cultured cells, respectively, using the TRIzol™ Reagent (Invitrogen, Carlsbad, CA, USA) following the manufacture’s protocol. The concentration and quality of RNA were determined with a NanoDrop spectrophotometer (NanoDrop Technologies, Thermo Fisher Scientific, Waltham, MA, USA) and gel analysis. Reverse transcription reactions were carried out using High‐Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA, USA). LightCycler® 480 Probes Master (Roche, Basel, Switzerland) was used to evaluate the expression of BCAM and BAN in human tissue samples and cultured cells. The relative expression of BCAM and BAN was calculated using glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) as the endogenous control to normalize the data. The sequences of the primers used are as follows: BCAM, sense 5′‐GTGCTTTCCTTACCTCTAA‐3′ antisense 5′‐GTAGGTGCCATTGGAATC‐3′ and probe 5′‐AGTCGTGAACTGCTCCGTGC‐3′; GAPDH, sense 5′‐GGACCTGACCTGCCGTCTAG‐3′, antisense 5′‐TAGCCCAGGATGCCCTTAG‐3′, and probe 5′‐CCTCCGACGCCTGCTTCACC‐ACCT‐3′; BAN, sense 5′‐GACTCTTGACCTATACTCTTAG‐3′, antisense 5′‐TACGGGTCATAGGTTTCA‐3′, and probe 5′‐CAACCTCTGAACTCTGGCACTC‐3′.
2.5. Vector construction, siRNA, and transfection
Full‐length human BCAM and BAN were amplified from the cDNA of BGC‐823 cells and were cloned into pCS2 (+) and pcDNA3.1 vectors, respectively. Both plasmids were confirmed by DNA sequencing. SGC‐7901 cells were then transfected with an empty vector or the BCAM‐expressing plasmid using Lipofectamine™ 2000 (Invitrogen). BGC‐823 cells were transfected with siRNA for BCAM or BAN using lipofectamine™ RNAiMAX (Invitrogen). siRNA corresponding to the following sequences for BCAM or BAN silencing were synthesized by GenePharma: 5′‐CAACGUGUUUGCAAAGCCATT‐3′ for siBCAM‐1, 5′‐CUGUCGCUCAGUUCUAUCATT‐3′ for siBCAM‐2, 5′‐CUCUGGCACUCAGAAUAAUTT‐3′ for siBAN‐1, and 5′‐GUUUAUGACUAAAUGGUGCTT‐3′ for siBAN‐2.
2.6. SDS/PAGE and immunoblot
Cells were lysed using RIPA protein extraction reagent (Beyotime, Shanghai, China) supplemented with a protease inhibitor cocktail (Roche). The cell lysates were separated by SDS/PAGE and were transferred onto a poly(vinylidene difluoride) membrane. The membranes were incubated with 5% BSA. The proteins were detected using an anti‐BCAM monoclonal antibody (1 : 1000; Abcam, Cambridge, MA, USA), anti‐GAPDH (1 : 1000; Sigma‐Aldrich, San Francisco, CA, USA), and anti‐ACTIN (1 : 1000; Sigma‐Aldrich).
2.7. MTT assay
The 3‐(4, 5‐dimethylthiazol‐2‐yl)‐2, 5‐diphenyl‐tetrazoliumbromide (MTT) assay was performed at 0, 24, 48, and 72 h post‐transfection. The cells in the 96‐well culture were incubated with MTT (5 mg·mL−1, 20 μL) for 4 h. After that, 150 μL of DMSO was added and resuspended until the cysts were completely dissolved. The absorbances of samples were measured with a spectrophotometer at 490 nm. Each assay was performed in triplicate and was independently repeated three times.
2.8. Colony formation assay
For the colony formation assay, transfected cells (n = 500) were placed in 6‐well plates. After 2 weeks, the cells were fixed with 4% paraformaldehyde and were stained with 0.5% crystal violet in 20% EtOH for 15 min. Visible colonies were photographed and counted by imagej software (NIH, Bethesda, MD, USA). Each assay was performed in triplicate and was independently repeated three times.
2.9. FACS assay
A fluorescence‐activated cell sorting (FACS) assay was performed to analyze the cell cycle distribution. The cells were collected and fixed with 70% EtOH overnight. Then, the fixed cells were treated with propidium iodide and subjected to cell cycle distribution analysis using a flow cytometer (Beckman, Brea, CA, USA).
2.10. Transwell assay
For the transwell assay, 5 × 104 cells were placed in the top chamber in medium with 1% FBS, and the medium supplemented with 20% FBS was filled in the lower chamber and served as a chemoattractant. After incubation, the cells on the lower surface of the membrane were stained with crystal violet and were counted by imagej software. Each assay was performed in triplicate and was independently repeated three times.
2.11. Matrigel invasion assay
For the invasion assay, 5 × 104 cells were placed in the top chamber with a Matrigel‐coated membrane (24‐well insert; pore size, 8 mm; BD Biosciences, New York, NY, USA) in medium with 1% FBS, and medium supplemented with 20% FBS was filled in the lower chamber and used as a chemoattractant. After incubation, the cells on the lower surface of the membrane were stained with crystal violet and were counted by imagej software. Each assay was performed in triplicate and was independently repeated three times.
2.12. Knockout of BCAM
The pX330 vector (Addgene plasmid 42230) was a gift from F. Zhang. Plasmids expressing hCas9 and sgRNA for BCAM were prepared by ligating oligos into the BbsI site of pX330. The sequences used for sgRNA are as follows: sense: 5′‐ ACCGCATGGAGCCCCCGGACGCAC‐3′, and antisense: 5′‐ AACGTGCGTCCGGGGGCTCCATGC‐3′. This plasmid was designated pX330‐BCAM. Then, the plasmid was introduced into BGC‐823 cells and treated with puromycin at 48 h after transfection. After 48 h, the cells were placed into 96‐well plates at the concentration of 1 cell/well. Single colonies were picked and validated by genotyping and immunoblot analysis.
2.13. Tumor metastasis model
Nude mice (6–8 weeks old) were maintained under SPF conditions with individually ventilated cages in the Animal Facility of Zhejiang University. The spleens of the mice were inoculated with 106 BGC‐823 cells. Three weeks later, the livers were harvested, and external areas of metastatic masses were quantified. Animal experiments were approved by the Institutional Animal Care and Use Committee of Zhejiang University.
2.14. Statistical analysis
The significance of the differences between groups was estimated by the Student’s t‐test or χ2 test as appropriate. P < 0.05 was considered to be statistically significant. Kaplan–Meier method with the log‐rank test was adopted to evaluate the overall survival of different groups. Univariate and multivariate Cox proportional hazards models were performed using R survival package. Receiver operating characteristic and prediction error (PE) curves were produced using the survcomp and PEC package, respectively. All the above analysis was conducted using r software (version 3.4.4, http://cran.r-project.org).
3. Results
3.1. BCAM upregulation is associated with GC metastasis and poor prognosis
To systematically screen GC metastasis‐associated genes, we first performed differential expression analysis between GC tissues with distant metastasis and those without metastasis in two cohorts from TCGA and ACRG datasets. Seventeen genes were found to be upregulated in GC tissues with metastasis (P < 0.05). These genes were further filtered by analysis of association with the shorter survival times of GC patients (P < 0.05). A Venn diagram revealed that 12 overlapping genes were upregulated in GC tissues with metastasis and associated with poor prognosis (Fig. 1A). BCAM was among the genes with lowest P value (Fig. 1B). Further paired statistical analysis confirmed that BCAM was significantly upregulated in GC tissues with metastasis compared to those without metastasis (Fig. 1C,D). BCAM upregulation was associated with poor prognosis of GC patients in both TCGA and ACRG cohorts (Fig. 1E,F). Multivariate Cox analysis showed that high expression of BCAM was independently associated with reduced overall survival time from TCGA cohort, but not ACRG cohort (Fig. 1G,H).
Figure 1.
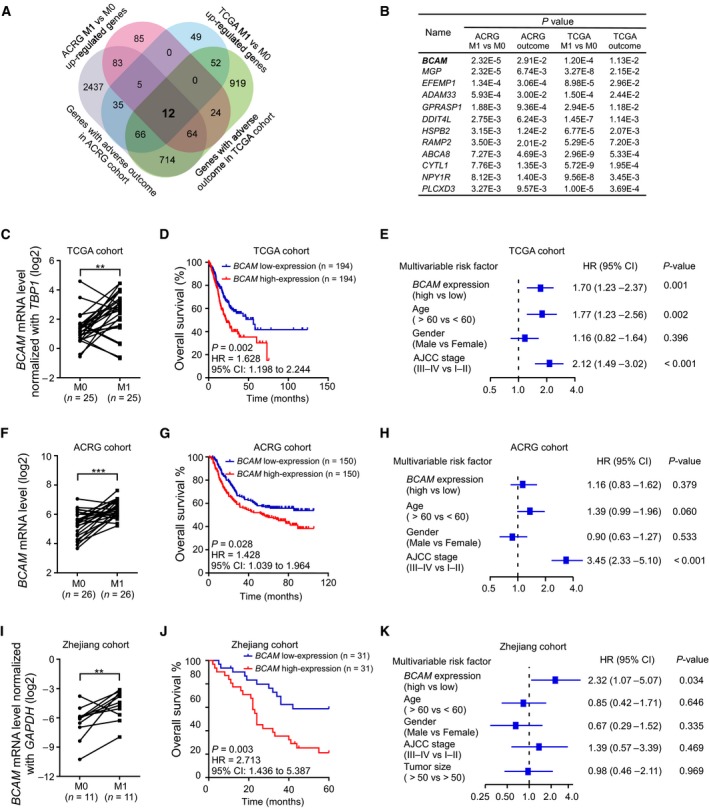
BCAM upregulation is associated with GC metastasis and poor prognosis. (A) The Venn diagram illustrating the number of upregulated genes in GC tissues with distant metastasis that had a reverse outcome in both TCGA and ACRG cohorts. (B) Twelve genes that were upregulated in metastatic GC tissues and were positively associated with a poor prognosis for GC patients were listed. (C) The relative expression of BCAM in 25 GC tissues with distant metastasis (M1) compared to that in age‐ and sex‐matched GC tissues without metastasis (M0) from the TCGA dataset. The results are presented as log2 FPKM values normalized to that of TBP1. **P < 0.01. (D) The relative expression of BCAM in 26 GC tissues with distant metastasis (M1) compared to that in age‐ and sex‐matched GC tissues without metastasis (M0) from the ACRG cohort. ***P < 0.001. (E) Kaplan–Meier survival curve analysis between patients with BCAM‐high expression group (n = 194) and BCAM‐low expression group (n = 194) in TCGA cohort. The GC patients were classified into BCAM‐high or BCAM‐low expression groups according to the median value. P = 0.002, HR = 1.628, 95% CI: 1.198–2.244. (F) Kaplan–Meier survival curve analysis between patients with BCAM‐high expression group (n = 150) and BCAM‐low expression group (n = 150) in the ACRG cohort. The GC patients were classified into BCAM‐high or BCAM‐low expression groups according to the median value. P = 0.028, HR = 1.428, 95% CI: 1.039–1.964. (G) The forest plot depicted the multivariable Cox analysis results of BCAM in the TCGA cohort. All the bars correspond to 95% confidence intervals. (H) The forest plot depicted the multivariable Cox analysis results of BCAM in the ACRG cohort. All the bars correspond to 95% confidence intervals. (I) Quantitative RT‐PCR was performed to analyze the relative expression of BCAM expression in 11 GC tissues with distant metastasis (M1) compared to that in age‐ and sex‐matched GC tissues without metastasis (M0). The results are presented as fold changes based on log2 values normalized to GAPDH. **P < 0.01. (J) Kaplan–Meier survival curve analysis between patients with BCAM‐high expression group (n = 31) and BCAM‐low expression group (n = 31) in the Zhejiang cohort. The GC patients were classified into BCAM‐high or BCAM‐low expression groups according to the median value. P = 0.003, HR = 2.713, 95% CI: 1.436–5.387. All the bars correspond to 95% confidence intervals. (K) The forest plot depicted the multivariable Cox analysis results of BCAM in the Zhejiang cohort. All the bars correspond to 95% confidence intervals.
To verify these bioinformatics results, we collected 62 GC tissues from Sir Run Run Shaw Hospital, Zhejiang University School of Medicine and Zhejiang Cancer Hospital with informed consent (Zhejiang cohort). Quantitative real‐time RT‐PCR (qRT‐PCR) analysis revealed that the levels of BCAM were significantly increased in GC tissues with metastasis compared to those in tissues without metastasis (Fig. 1I). Importantly, Kaplan–Meier curve analysis of this cohort (n = 62) showed that the upregulation of BCAM was significantly associated with the poor overall survival of GC patients (Fig. 1J). Further multivariate Cox analysis confirmed that BCAM expression was an independent predictor for predicting clinical outcome of GC patients (Fig. 1K).
3.2. Knockdown of BCAM suppresses GC cell migration and invasion
To investigate the potential role of BCAM in GC cells, we first performed qRT‐PCR analysis and found that BCAM was upregulated at high levels in AGS, MKN‐45, and BGC‐823 cells, and low levels in SGC‐7901, MKN‐74, MGC80‐3, and HGC‐27 cells (Fig. S1A). Then, we depleted BCAM expression by introducing specific siRNA into BGC‐823 cells. The knockdown efficiency was confirmed by a western blot analysis (Fig. 2A). Neither the MTT assay nor the clone formation assay showed that knockdown of BCAM had a significant effect on GC cell proliferation (Fig. 2B,C). FACS analysis showed that silencing BCAM had no significant effect on GC cell cycle progression (Fig. 2D). However, the transwell migration assay and Matrigel invasion assay revealed that knockdown of BCAM dramatically suppressed GC cell migration and invasion (Fig. 2E,F). Additionally, the migration assay and invasion assay using AGS GC cell line showed the same results (Fig. 2G,H).
Figure 2.
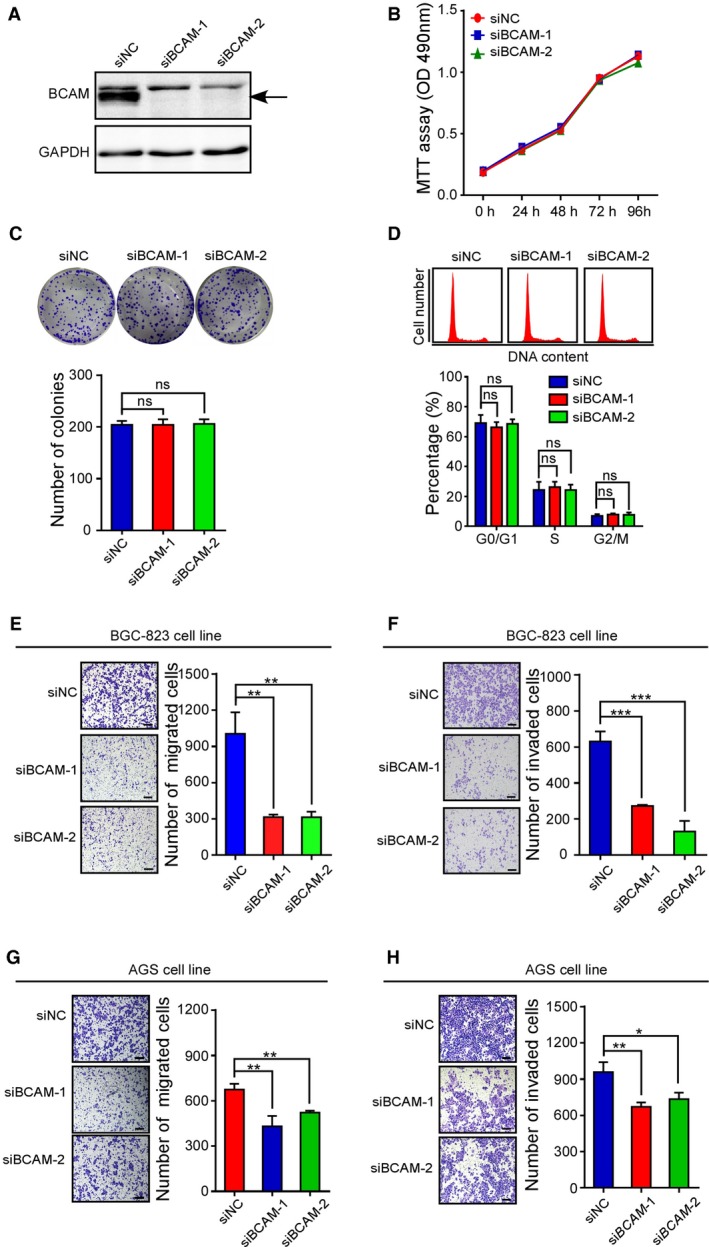
Knockdown of BCAM expression suppresses GC cell migration and invasion. (A) Immunoblot analysis of the BCAM expression levels following the treatment of BGC‐823 cells with scrambled siRNA and siBCAM. (B–D) MTT assay (B), colony‐forming growth assay (C), and cell cycle (D) analysis were performed using BGC‐823 cells with the indicated treatment. Colonies were captured and counted. The bar chart represents the percentage of cells in G0/G1, S, or G2/M phase, as indicated. (E, F) Transwell migration (E) and Matrigel invasion assays (F) were carried out using GC cells with the indicated treatments. (G, H) Transwell migration (G) and Matrigel invasion assays (H) were carried out using AGS cells with the indicated treatments. Migrated and invaded cells were counted. Scar bar, 100 μm. Experiments were performed in triplicate. Data are presented as the means ± SDs; ns, no significance; *P < 0.05, **P < 0.01, ***P < 0.001.
3.3. Ectopic expression of BCAM promotes GC cell migration and invasion
Next, we overexpressed BCAM expression by introducing pCS2‐BCAM into SGC‐7901 cells. The overexpression efficiency was confirmed by a western blot analysis (Fig. 3A). Consistent with the knockdown of BCAM, the results showed that ectopic expression of BCAM had no significant effect on SGC‐7901 cell proliferation or cell cycle progression (Fig. 3B–D). However, the migration assay and invasion assay showed that ectopic expression of BCAM significantly enhanced GC cell migration and invasion (Fig. 3E,F). Taken together, these data indicate that BCAM is involved in promoting GC cell migration and invasion.
Figure 3.
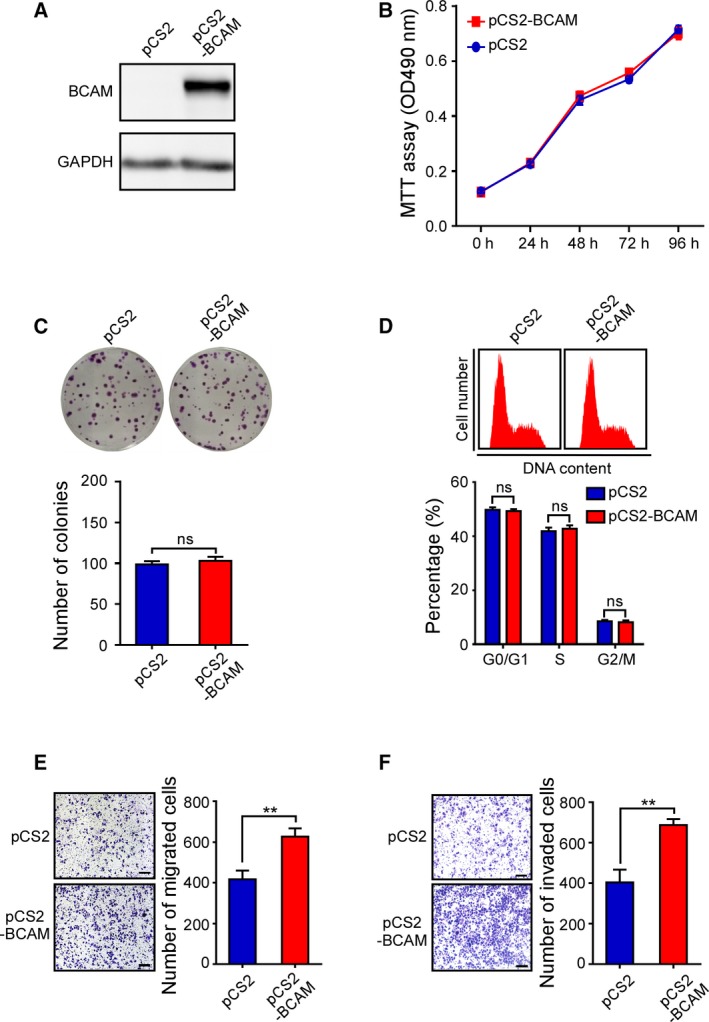
Ectopic expression of BCAM promotes GC cell migration and invasion. (A) Immunoblot analysis of BCAM expression levels following the treatment of SGC‐7901 cells with an empty vector and pCS2‐BCAM. (B–D) SGC‐7901 cells transfected with the indicated plasmids were processed for MTT assay (B), colony‐forming assay (C), and cell cycle analysis (D). The bar chart for cell cycle represents the percentage of cells in the G0/G1, S, or G2/M phase, as indicated. (E, F) Transwell migration (E) and Matrigel invasion assays (F) were carried out using GC cells with the indicated treatments. Migrated and invaded cells were counted. Scar bar, 100 μm. Experiments were performed in triplicate. Data are presented as the means ± SDs; ns, no significance; **P < 0.01.
3.4. Knockout of BCAM by the CRISPR/Cas9 system reduces GC cell metastasis in a mouse model
To further explore the role of BCAM in GC, we used the CRISPR/Cas9 system to knock out the BCAM gene in BGC‐823 cells and generated two BCAM KO subclones (Fig. 4A). The KO efficiency was confirmed by western blotting (Fig. 4B). Similar to the knockdown of BCAM, BCAM KO had no significant effect on cell proliferation, colony formation, or cell cycle distribution (Fig. 4C–E). However, the transwell migration assay and Matrigel invasion assay showed that KO of BCAM significantly inhibited GC cell migration and invasion (Fig. 4F,G).
Figure 4.

KO of BCAM by CRISPR/Cas9 system reduces GC cell metastasis in a mouse model. (A) The BCAM KO BGC‐823 cells were produced by the CRISPR/Cas9 system. The schematic diagram of the mutation in BCAM locus by CRISPR/Cas9 technique was shown. (B) Immunoblot analysis of the BCAM levels in wild‐type or BCAM KO BGC‐823 cells. (C‐G) MTT assay (C), colony‐forming growth assay (D), cell cycle analysis (E), transwell migration (F), and Matrigel invasion analysis (G) of wild‐type or BCAM KO BGC‐823 cells. The bar chart for cell cycle represents the percentage of cells in G0/G1, S, or G2/M phase. (H–J) Mice were intrasplenically injected with wild‐type or BCAM KO BGC‐823 cells and were subjected to liver metastasis analysis. Representative gross liver (H) and H&E‐stained liver sections (I) from mice were shown. Scar bars, 5 mm (H); Scar bars, 100 μm (I). The liver metastatic nodules were counted (J). Data are presented as the means ± SDs; ns, no significance. *P < 0.05. **P < 0.01. ***P < 0.001.
To investigate the potential role of BCAM in GC metastasis in a mouse model, BCAM KO cells or wild‐type cells were intrasplenically injected into nude mice, and liver metastases were measured after 6 weeks. The mice inoculated with wild‐type cells developed severe liver metastases, while the injection of BCAM KO cells robustly reduced the number of liver metastatic nodules (Fig. 4H–J). Taken together, these results suggest that BCAM plays a critical role in GC metastasis.
3.5. BCAM expression is modulated by its associated lncRNA BAN
The important role of BCAM in GC prompted us to explore the upstream mechanisms in modulating BCAM expression. We analyzed the BCAM gene locus and found a previously uncharacterized BCAM sense lncRNA, which we called BCAM‐associated long noncoding RNA (BAN) (GenBank access ID: AY927517). BAN, which spanned from exon 6 and intron 6 of the BCAM gene, is a transcript with a length of 486 nt (Fig. 5A). The coding potential calculator and coding potential assessment tool algorithms predicted that BAN was a noncoding RNA (Fig. 5B). BAN was upregulated at high levels in AGS, MKN‐45, BGC‐823, MKN‐74, and MGC80‐3 cells, and low levels in SGC‐7901 and HGC‐27 cells (Fig. S1B). Consistent with RNA‐FISH assay, nuclear and cytosolic fraction analysis showed that BAN was localized both in cytoplasm and nucleus (Fig. S2A,B). lncRNA have been documented to play versatile roles in modulating gene regulation (Sun et al., 2017; Ulitsky and Bartel, 2013). Indeed, knockdown of BAN evidently suppressed BCAM expression at both the mRNA and protein levels (Fig. 5C–E), while the depletion of BCAM had no significant effect on BAN expression (Fig. 5F,G). Moreover, we performed mRNA stability assay and protein degradation assay to investigate how BAN could promote BCAM expression. The results showed that knockdown of BAN significantly decreased the half‐life of BCAM mRNA transcripts (Fig. 5H) and BCAM protein (Fig. S3).
Figure 5.
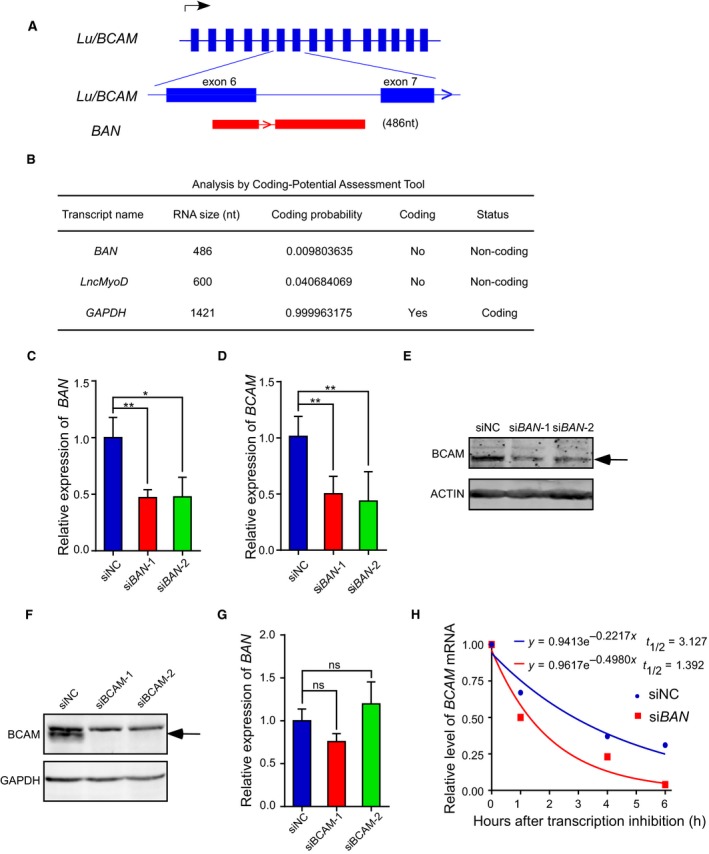
BCAM expression is modulated by its associated sense lncRNA BAN. (A) A schematic diagram of the BCAM gene locus showing the position of the lncRNA BAN within the sixth exon and sixth intron of BCAM. Arrows indicate the direction of transcription. (B) The coding potential assessment tool predicted the coding potential of BAN. The lncRNA lncMyoD and the protein‐coding gene GAPDH were also shown. (C) Quantitative RT‐PCR analysis of BAN expression levels in BGC‐823 cells transfected with scrambled siRNA and siBAN. (D, E) BGC‐823 cells transfected with the indicated siRNA were subjected to qRT‐PCR analysis (D) or immunoblot analysis (E) of BCAM expression. (F) Immunoblot analysis of BCAM expression levels following the treatment of BGC‐823 cells with scrambled siRNA and siBCAM. (G) Quantitative RT‐PCR analysis of BAN expression levels in BGC‐823 cells transfected with the indicated siRNA. (H) The effects of BAN knockdown on the half‐life (t 1/2) of BCAM mRNA. Data are presented as the means ± SDs; ns, no significance. *P < 0.05. **P < 0.01.
3.6. BCAM is involved in BAN‐mediated invasive activity of GC cells
To explore the potential role of BAN in GC cell migration and invasion in vitro, we carried out loss‐ and gain‐of‐function experiments, respectively, by introducing either siRNA specific for BAN or pcDNA3.1‐BAN into GC cells. The transwell migration assay and Matrigel invasion assay showed that knockdown of BAN significantly inhibited GC cell migration and invasion (Fig. 6A–C). Ectopic expression of BAN in SGC‐7901 cells dramatically enhanced cell migration and invasion (Fig. 6D–F). It is reasonable to propose that BCAM may be involved in BAN‐mediated invasive activity of GC cells after knockdown of BAN, BGC‐823 cells were transfected with pCS2‐BCAM. Ectopic expression of BCAM rescued the decreased cell invasion ability caused by knockdown of BAN (Fig. 6G,H). The invasion assay suggests that the cotransfection could partially rescue BAN RNAi‐decreased GC cell invasion in BGC‐823 cells. These data indicate that BAN regulated GC cell migration and invasion by modulating BCAM expression.
Figure 6.
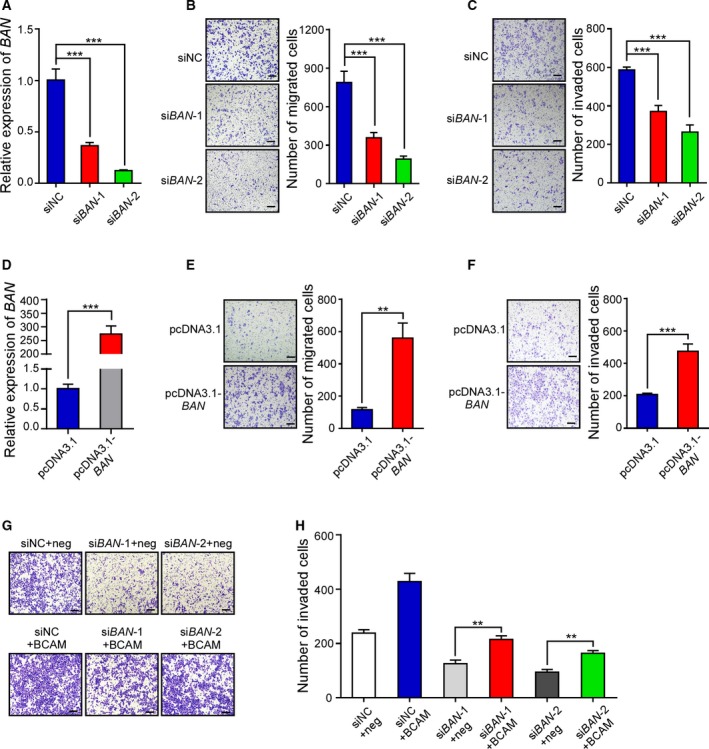
BCAM is involved in BAN‐mediated invasive activity of GC cells. (A–C) BGC‐823 cells transfected with the indicated siRNA were subjected to qRT‐PCR analysis (A), transwell migration (B), and Matrigel invasion assays (C). (D–F) SGC‐7901 cells transfected with the indicated plasmids were subjected to qRT‐PCR analysis (D), transwell migration (E), and Matrigel invasion assay (F). (G, H) After knockdown of BAN, BGC‐823 cells were transfected with pCS2‐BCAM. Ectopic expression of BCAM rescued the decrease in GC cell invasion induced by the knockdown of BAN. Scar bar, 100 μm. Experiments were performed in triplicate. Data are presented as the means ± SDs. **P < 0.01. ***P < 0.001.
3.7. BAN upregulation is associated with GC metastasis and poor prognosis
To explore the association between BAN and GC metastasis, we examined BAN expression in GC tissues with metastasis compared to those in tissues without metastasis. Our qRT‐PCR analysis revealed that increased expression of BAN was significantly associated with GC metastasis and poor prognosis in the Zhejiang cohort (Fig. 7A,B). Multivariate Cox analysis further revealed that BAN expression was an independent predictor for assessing the prognosis of GC patients (Fig. 7C). Importantly, we also found that BAN expression levels were positively correlated with that of BCAM in GC tissues from the Zhejiang cohort (Fig. 7D,E). Kaplan–Meier curve analysis showed that high expression of both BCAM and BAN was correlated with the worse prognosis of GC patients, and GC patients with low expression levels of both BCAM and BAN had relatively longer survival time (Fig. 7F). To evaluate the potential clinical value of BCAM and BAN for prognosis, we computed their accuracy by PE curves compared with the American Joint Committee on Cancer (AJCC) staging system. Our data displayed that the prediction of GC prognosis using the combination of BAN, BCAM, and AJCC stage had the lowest predicting error in the Zhejiang cohort (Fig. 7G). Furthermore, the time‐dependent receiver operator characteristic (ROC) curve at 5 years showed that the area under the curve of BAN and BCAM combining with AJCC stage was higher than that of any other factors (Fig. 7H). Taken together, these data indicate that the combination of BAN, BCAM, and AJCC stage is more precise in predicting clinical outcome of GC patients.
Figure 7.
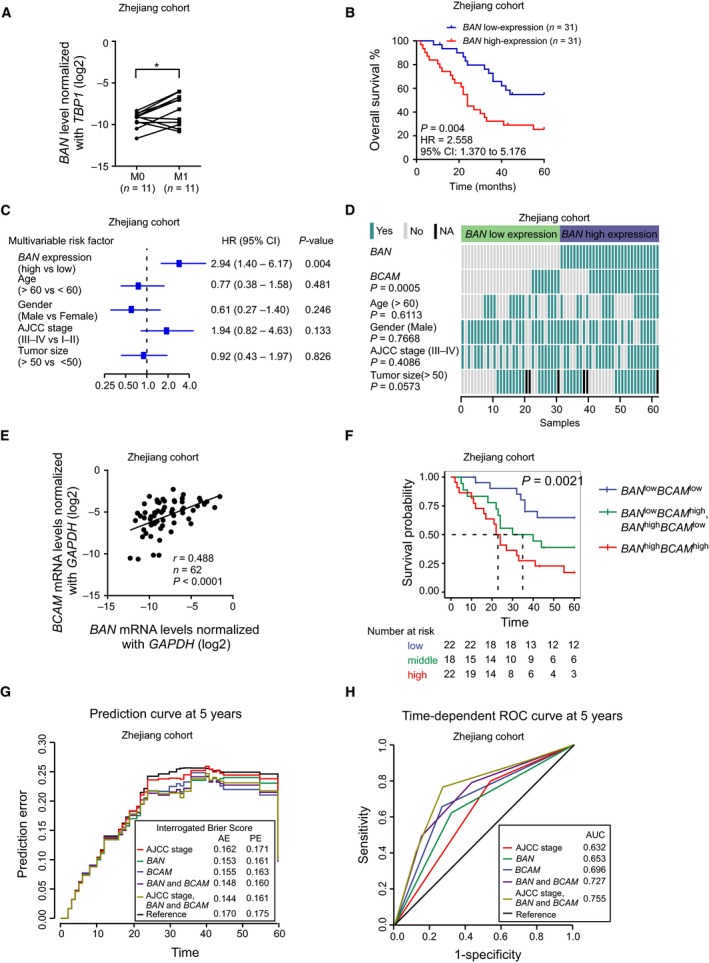
BAN upregulation is associated with GC metastasis and poor prognosis. (A) The relative expression of BAN expression in 11 GC tissues with distant metastasis (M1) compared to age‐ and sex‐matched GC tissues without metastasis (M0) in the Zhejiang cohort. (B) Kaplan–Meier survival curve analysis between patients with BAN‐high expression group and BAN‐low expression in the Zhejiang cohort. The GC patients were classified into BAN‐high or BAN‐low expression groups according to the median value. P = 0.004, HR = 2.558, 95% CI: 1.370–5.176. (C) The forest plot depicted the multivariable Cox analysis results of BAN in the Zhejiang cohort. All the bars correspond to 95% confidence intervals. (D) The heatmap illustrated the association of BAN expression, BCAM expression, and different clinical characteristics in the Zhejiang cohort. The GC patients were classified into BAN‐high and BAN‐low expression groups according to the median value. Statistical significance was performed by the χ2 test. (E) The correlation of BCAM mRNA and BAN expression levels in GC tissues was analyzed by qRT‐PCR. r = 0.488, n = 62, P < 0.0001. (F) Kaplan–Meier survival curve analysis between low (BCAM‐low and BAN‐low expression group, n = 22), middle (BCAM‐high and BAN‐low expression, or BCAM‐low and BAN‐high expression group, n = 18) and high (BCAM‐high and BAN‐high expression group, n = 22) groups in the Zhejiang cohort. The GC patients were classified into low, middle, and high groups according to the median value of BCAM and BAN. Number at risk table for BCAM and BAN expression can also be seen below the plot; P value was calculated using the log‐rank test. (G) PE curves of the different predictors in the Zhejiang cohort. Apparent error and 10‐fold cross‐validated cumulative PE at 5 years were computed using Kaplan–Meier estimation as reference. (H) Time‐dependent ROC curve analysis at 5 years of different predictors in the Zhejiang cohort.
4. Discussion
Since the mechanism of GC metastasis is still not fully understood, there is always a lack of effective GC metastasis treatment strategies and prognostic markers (Song et al., 2017). Here, we systematically screened the key genes involved in GC metastasis and the prognosis of GC patients. We found that BCAM and its sense lncRNA BAN were significantly increased in GC tissues with metastasis and correlated with the reduced survival time of GC patients. BCAM promoted GC metastasis and was regulated by BAN. Moreover, our ROC analysis showed that both BAN and BCAM integrating with the AJCC staging might be a better prognostic predictor for GC patients than that of the only AJCC staging.
Recent studies revealed that miR‐199a‐5p and the 14‐3‐3beta‐FBI1/Akirin2 complex were involved in the regulation of BCAM expression. In a previous report, miR‐199a‐5p was found to repress BCAM expression by directly targeting its 3′UTR in human keratinocytes (Kim et al., 2015). BCAM was also suggested as a target gene of the oncogenic 14‐3‐3beta‐FBI1/Akirin2 complex (Akiyama et al., 2013). The 14‐3‐3beta‐FBI1/Akirin2 complex bound to the BCAM promoter and repressed BCAM transcription. In our study, we found that an uncharacterized lncRNA BAN, which is located in the genomic locus of BCAM, modulated BCAM expression. The knockdown of BAN suppressed BCAM expression at both the mRNA and protein levels. Our previous study reported that ephrin A1 expression was modulated by its sense lncRNA GMAN, which promoted the translational expression of ephrin A1 by competitively binding its antisense RNA GMAN‐AS (Zhuo et al., 2019). It is unknown whether there is also an antisense RNA or miRNA related to BAN and BCAM. Moreover, knockdown of BAN reduced the stability of BCAM mRNA transcripts and BCAM protein. Considering the localization of BAN both in cytoplasm and nucleus, it is possible that there may exist different mechanisms for BAN to modulate BCAM expression. However, the detail molecular mechanisms were needed to be further investigated.
The previous study has shown that BCAM plays a functional role in the metastatic spreading of KRAS‐mutant colorectal cancer. Inhibition of BCAM impaired adhesion of KRAS‐mutant colorectal cancer cells specifically to endothelial cells (Bartolini et al., 2016). The exact mechanism of BCAM regulating GC invasion and metastasis should be further explored. However, the KO of BCAM significantly reduced GC cell metastasis in a mouse model. These data suggest that BCAM may act as a promising target for GC metastasis treatment.
In our study, BCAM and BAN were significantly upregulated in GC tissues with metastasis and associated with poor prognosis of GC patients. BCAM and BAN may be independent prognostic factors for GC patients according to multivariate Cox analysis. Furthermore, integrating the expression of BCAM and BAN with AJCC staging showed more sensitivity and specificity in predicting GC prognosis based on both PC and ROC analysis. Taken together, our data indicate that BCAM and BAN might be used as prognostic biomarkers for GC patients in clinical practice. Undoubtedly, the possibility of BCAM and BAN for the prognostic prediction and treatment of GC needs to be further explored.
5. Conclusions
This study reveals that the upregulation of BCAM and its sense RNA BAN are significantly associated with GC metastasis and a shorter survival time of GC patients. KO of BCAM reduced GC cell invasion and metastasis. Knockdown of BAN not only inhibits BCAM expression, but also inhibits the migration and invasion of GC cells, which is effectively rescued by the ectopic expression of BCAM. Our data indicate that BCAM and BAN might be prognostic biomarkers for GC patients.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
JJ, WZ, and TZ designed the experiments and interpreted the results, and performed research, and wrote the paper. JJ, ZH, KC, DG, XR, QS, YD, YL, SX, JD SL, and JW performed experiments. JJ, SX, ZH, DG, XR, YL WC, and WZ contributed valuable discussions on the project and participated in the manuscript preparation. TZ and WZ supervised the entire project. All authors approved the final version of the manuscript.
Supporting information
Fig. S1. The expression of BCAM and BAN in gastric cancer cell lines.
Fig. S2. The localization of BAN in gastric cancer cells.
Fig. S3. The effects of BAN knockdown on the half‐life (t1/2) of BCAM protein.
Data S1 . Supplementary Methods.
Acknowledgements
We thank Zhaocai Zhou for providing the BGC‐803 cells and Youyong Lu for providing the SGC‐7901 GC cell lines. This work was supported by the National Natural Science Foundation of China (91740205, 31771540, 31620103911, 81972276, 31571446, and 31301149), Natural Scientific Foundation of Zhejiang Province, China (LYY19H310011, LY18H160019, LQ13H160013), and Fundamental Research Funds for the Central Universities (2017QNA7005).
Contributor Information
Wei Zhuo, Email: 0012049@zju.edu.cn.
Tianhua Zhou, Email: tzhou@zju.edu.cn.
References
- Akiyama H, Iwahana Y, Suda M, Yoshimura A, Kogai H, Nagashima A, Ohtsuka H, Komiya Y and Tashiro F (2013) The FBI1/Akirin2 target gene, BCAM, acts as a suppressive oncogene. PLoS ONE 8, e78716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolini A, Cardaci S, Lamba S, Oddo D, Marchio C, Cassoni P, Amoreo CA, Corti G, Testori A, Bussolino F et al (2016) BCAM and LAMA5 mediate the recognition between tumor cells and the endothelium in the metastatic spreading of KRAS‐mutant colorectal cancer. Clin Cancer Res 22, 4923–4933. [DOI] [PubMed] [Google Scholar]
- Bernards N, Creemers GJ, Nieuwenhuijzen GA, Bosscha K, Pruijt JF and Lemmens VE (2013) No improvement in median survival for patients with metastatic gastric cancer despite increased use of chemotherapy. Ann Oncol 24, 3056–3060. [DOI] [PubMed] [Google Scholar]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA and Jemal A (2018) Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 68, 394–424. [DOI] [PubMed] [Google Scholar]
- Campbell IG, Foulkes WD, Senger G, Trowsdale J, Garin‐Chesa P and Rettig WJ (1994) Molecular cloning of the B‐CAM cell surface glycoprotein of epithelial cancers: a novel member of the immunoglobulin superfamily. Can Res 54, 5761–5765. [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014) Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang HY, Chang HM, Wu TJ, Chaing CY, Tzai TS, Cheng HL, Raghavaraju G, Chow NH and Liu HS (2017) The role of Lutheran/basal cell adhesion molecule in human bladder carcinogenesis. J Biomed Sci 24, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS, Liu J, Yue YG, Wang J, Yu K et al (2015) Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 21, 449–456. [DOI] [PubMed] [Google Scholar]
- De Grandis M, Cambot M, Wautier MP, Cassinat B, Chomienne C, Colin Y, Wautier JL, Le Van Kim C and El Nemer W (2013) JAK2V617F activates Lu/BCAM‐mediated red cell adhesion in polycythemia vera through an EpoR‐independent Rap1/Akt pathway. Blood 121, 658–665. [DOI] [PubMed] [Google Scholar]
- Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22, 1775–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F et al (2012) Landscape of transcription in human cells. Nature 489, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewniok C, Wienrich BG, Schon M, Ulrich J, Zen Q, Telen MJ, Hartig RJ, Wieland I, Gollnick H and Schon MP (2004) Molecular interactions of B‐CAM (basal‐cell adhesion molecule) and laminin in epithelial skin cancer. Arch Dermatol Res 296, 59–66. [DOI] [PubMed] [Google Scholar]
- El Nemer W, Gane P, Colin Y, Bony V, Rahuel C, Galacteros F, Cartron JP and Le Van Kim C (1998) The Lutheran blood group glycoproteins, the erythroid receptors for laminin, are adhesion molecules. J Biol Chem 273, 16686–16693. [DOI] [PubMed] [Google Scholar]
- Gauthier E, Rahuel C, Wautier MP, El Nemer W, Gane P, Wautier JL, Cartron JP, Colin Y and Le Van Kim C (2005) Protein kinase A‐dependent phosphorylation of Lutheran/basal cell adhesion molecule glycoprotein regulates cell adhesion to laminin alpha5. J Biol Chem 280, 30055–30062. [DOI] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL et al (2010) Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464, 1071–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutschner T and Diederichs S (2012) The hallmarks of cancer: a long non‐coding RNA point of view. RNA Biol 9, 703–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines PC, Zen Q, Burney SN, Shea DA, Ataga KI, Orringer EP, Telen MJ and Parise LV (2003) Novel epinephrine and cyclic AMP‐mediated activation of BCAM/Lu‐dependent sickle (SS) RBC adhesion. Blood 101, 3281–3287. [DOI] [PubMed] [Google Scholar]
- Jiao S, Wang H, Shi Z, Dong A, Zhang W, Song X, He F, Wang Y, Zhang Z, Wang W et al (2014) A peptide mimicking VGLL4 function acts as a YAP antagonist therapy against gastric cancer. Cancer Cell 25, 166–180. [DOI] [PubMed] [Google Scholar]
- Kikkawa Y and Miner JH (2005) Review: Lutheran/B‐CAM: a laminin receptor on red blood cells and in various tissues. Connect Tissue Res 46, 193–199. [DOI] [PubMed] [Google Scholar]
- Kikkawa Y, Ogawa T, Sudo R, Yamada Y, Katagiri F, Hozumi K, Nomizu M and Miner JH (2013) The lutheran/basal cell adhesion molecule promotes tumor cell migration by modulating integrin‐mediated cell attachment to laminin‐511 protein. J Biol Chem 288, 30990–31001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BK, Kim I and Yoon SK (2015) Identification of miR‐199a‐5p target genes in the skin keratinocyte and their expression in cutaneous squamous cell carcinoma. J Dermatol Sci 79, 137–147. [DOI] [PubMed] [Google Scholar]
- Kong R, Zhang EB, Yin DD, You LH, Xu TP, Chen WM, Xia R, Wan L, Sun M, Wang ZX et al (2015) Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer 14, 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W and Anders S (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Bajic VB and Zhang Z (2013) On the classification of long non‐coding RNAs. RNA Biol 10, 925–933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manhong D, Pinglang W, Boyd AD, Georgi K, Brian A, Jones EG, Bunney WE, Myers RM, Speed TP and Huda A (2005) Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res 33, e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons SF, Mallinson G, Holmes CH, Houlihan JM, Simpson KL, Mawby WJ, Spurr NK, Warne D, Barclay AN and Anstee DJ (1995) The Lutheran blood group glycoprotein, another member of the immunoglobulin superfamily, is widely expressed in human tissues and is developmentally regulated in human liver. Proc Natl Acad Sci USA 92, 5496–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL and Reik W (2009) Evolution and functions of long noncoding RNAs. Cell 136, 629–641. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W and Smyth GK (2015) limma powers differential expression analyses for RNA‐sequencing and microarray studies. Nucleic Acids Res 43, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Z, Wu Y, Yang J, Yang D and Fang X (2017) Progress in the treatment of advanced gastric cancer. Tumour Biol 39 10.1177/1010428317714626 [DOI] [PubMed] [Google Scholar]
- Spitale RC, Tsai MC and Chang HY (2011) RNA templating the epigenome: long noncoding RNAs as molecular scaffolds. Epigenetics 6, 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spizzo R, Almeida MI, Colombatti A and Calin GA (2012) Long non‐coding RNAs and cancer: a new frontier of translational research? Oncogene 31, 4577–4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M, Jin FY, Xia R, Kong R, Li JH, Xu TP, Liu YW, Zhang EB, Liu XH and De W (2014) Decreased expression of long noncoding RNA GAS5 indicates a poor prognosis and promotes cell proliferation in gastric cancer. BMC Cancer 14, 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Yang Y, Xu C and Guo J (2017) Regulatory mechanisms of long noncoding RNAs on gene expression in cancers. Cancer Genet 216–217, 105–110. [DOI] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA et al (2010) The nuclear‐retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39, 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I and Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154, 26–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KC and Chang HY (2011) Molecular mechanisms of long noncoding RNAs. Mol Cell 43, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H and Spector DL (2009) Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23, 1494–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie SS, Jin J, Xu X, Zhuo W and Zhou TH (2016) Emerging roles of non‐coding RNAs in gastric cancer: pathogenesis and clinical implications. World J Gastroenterol 22, 1213–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing R, Li W, Cui J, Zhang J, Kang B, Wang Y, Wang Z, Liu S and Lu Y (2012) Gastrokine 1 induces senescence through p16/Rb pathway activation in gastric cancer cells. Gut 61, 43–52. [DOI] [PubMed] [Google Scholar]
- Xu TP, Huang MD, Xia R, Liu XX, Sun M, Yin L, Chen WM, Han L, Zhang EB, Kong R et al (2014) Decreased expression of the long non‐coding RNA FENDRR is associated with poor prognosis in gastric cancer and FENDRR regulates gastric cancer cell metastasis by affecting fibronectin1 expression. J Hematol Oncol 7, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Xue X, Zheng L, Bi J, Zhou Y, Zhi K, Gu Y and Fang G (2014) Long non‐coding RNA GHET1 promotes gastric carcinoma cell proliferation by increasing c‐Myc mRNA stability. FEBS J 281, 802–813. [DOI] [PubMed] [Google Scholar]
- Zhang EB, Kong R, Yin DD, You LH, Sun M, Han L, Xu TP, Xia R, Yang JS, De W et al (2014) Long noncoding RNA ANRIL indicates a poor prognosis of gastric cancer and promotes tumor growth by epigenetically silencing of miR‐99a/miR‐449a. Oncotarget 5, 2276–2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo W, Liu Y, Li S, Guo D, Sun Q, Jin J, Rao X, Li M, Sun M, Jiang M et al (2019) Long non‐coding RNA GMAN, upregulated in gastric cancer tissues, is associated with metastasis in patients and promotes translation of Ephrin A1 by competitively binding GMAN‐AS. Gastroenterology 156, 676–691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. The expression of BCAM and BAN in gastric cancer cell lines.
Fig. S2. The localization of BAN in gastric cancer cells.
Fig. S3. The effects of BAN knockdown on the half‐life (t1/2) of BCAM protein.
Data S1 . Supplementary Methods.


