Abstract
Albomycin δ2 is a sulfur-containing sideromycin natural product that shows potent antibacterial activity against clinically important pathogens. The l-serine-thioheptose dipeptide partial structure, known as SB-217452, has been found to be the active seryl-tRNA synthetase inhibitor component of albomycin δ2. Here it is demonstrated that AbmF catalyzes condensation between the 6′-amino-4′-thionucleoside with the d-ribo configuration and seryl-adenylate supplied via the serine adenylation activity of AbmK. Formation of the dipeptide is followed by C3′-epimerization to produce SB-217452 with the d-xylo configuration, which is catalyzed by the radical S-adenosyl-l-methionine enzyme AbmJ. Gene deletion suggests that AbmC is involved in peptide assembly linking SB-217452 with the siderophore moiety. This study establishes how the albomycin biosynthetic machinery generates its antimicrobial component SB-217452.
Keywords: albomycin, biosynthesis, amide bond formation, epimerization, radical S-adenosyl-l-methionine enzyme
Graphical Abstract
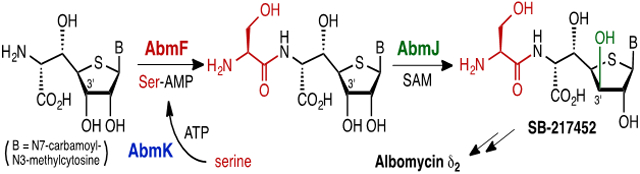
Biosynthesis of seryl-thioheptose core of albomycin δ2. In this study, AbmF is shown to catalyze condensation between a 6′-amino-4′-thionucleoside with the d-ribo configuration and seryl-adenylate provided by AbmK. This is followed by C3′-epimerization of the thiofuranose core catalyzed by the radical S-adenosyl-l-methionine enzyme AbmJ. This work highlights how the biosynthetic machinery is organized to generate albomycin without producing side products.
Albomycins (1-3), isolated from Strepromyces species, are sideromycins that comprises a thioheptose core (4) and a ferrichrome-type siderophore (5) conjugated via peptide linkages to an l-serine residue (6) (Scheme 1).1 These peptidyl thionucleosides exhibit potent antibacterial activities against a number of clinically important pathogens.2 It is believed that the high antimicrobial activity of the albomycins can be attributed in part to exploitation of the siderophore-dependent iron acquisition systems of the bacterial targets.3 When albomycin δ2 (1) is transported into the bacterial cell, it will be hydrolytically digested by the host peptidases to release the active antimicrobial agent SB-217452 (7), which inhibits the host seryl-tRNA synthetase (SerRS) thereby blocking protein synthesis.4 Thus, formation of the peptidyl linkages between 6′-amino-4′-thionucleoside (4), l-serine (6) and the siderophore 5 is a key biosynthetic maturation process crucial for the activity and potency of albomycins.
Scheme 1.
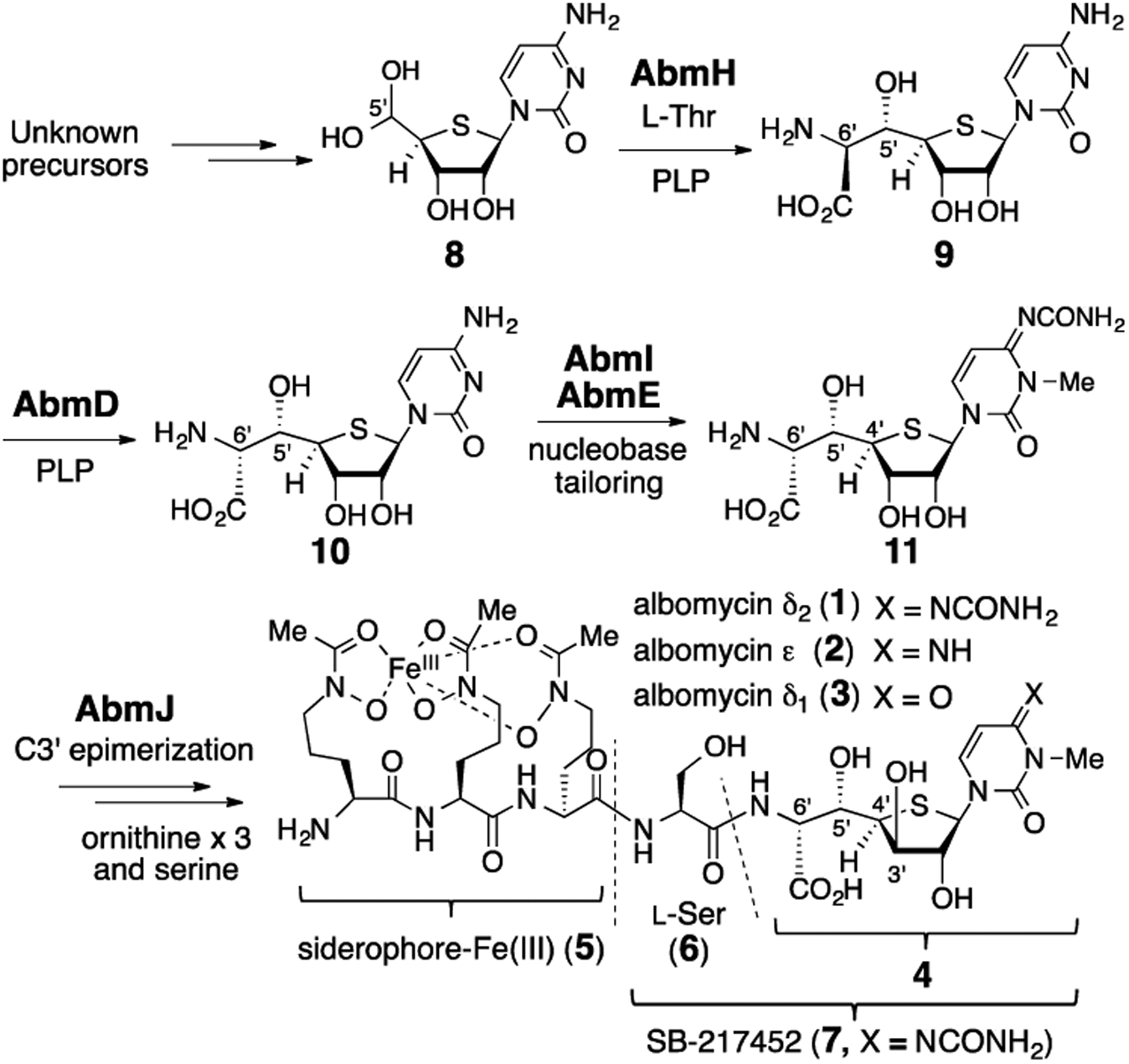
Proposed biosynthetic pathway of albomycins (1–3).
We have recently shown that the 6′-amino-4′-thioheptose moiety in 1 is constructed from thioribonucleoside 8 and l-threonine in a stereo-controlled manner in a reaction catalyzed by the pyridoxal 5′-phosphate (PLP)-dependent transaldolase AbmH.5 The resulting product 9 then undergoes inversion of stereoconfiguration at C6′ catalyzed by the PLP-dependent epimerase AbmD to give 10.5 The conversion of the d-ribo to the d-xylo configuration (i.e., epimerization at C3’) of the furanose ring is hypothesized to be catalyzed by the ambJ gene product, which is annotated as a radical S-adenosyl-l-methionine (SAM) enzyme, and this hypothesis is consistent with gene deletion experiments (Scheme 1).5 However, AbmJ-catalyzed C3′-epimerization remains to be tested, and the pathway leading to assembly of albomycins from 6′-amino-4′-thionucleoside (11 or 4), l-serine (6), and siderophore 5 is currently unknown.
To address these questions, the abm gene cluster was closely analyzed demonstrating the presence of a gene, abmQ, that is annotated to encode a modular nonribosomal peptide synthetase (NRPS) consistent with the peptide-based siderophore (5) in 1 (see Scheme 2).6 Sequence analysis suggested that AbmQ contains two condensation (C) domains leading to the hypothesis that AbmQ catalyzes construction of a tripeptide using three molecules of N5-acetyl-N5-hydroxy-l-ornithine (13) as building blocks.6 The building blocks (13) could in turn be biosynthesized from l-ornithine (12) in reactions catalyzed by AbmA (an N-acyltransferase) and AbmB (a flavin-dependent monooxygenase).6,7 A typical NRPS contains a C-terminal thioesterase (TE) domain to catalyze hydrolysis of the NRP thioester intermediate to release the peptide product from the phosphopantetheine group linked to the peptidyl carrier protein (PCP) domain.8 However, AbmQ lacks a C-terminal TE domain. This atypical domain organization implies that no free siderophore (5) is released from AbmQ and that the thioester intermediate 14 (see Scheme 2) may directly serve as the electrophile in the subsequent amide bond formation with either SB-217452 (7) or l-serine (6). It was further hypothesized that AbmC may participate in this amide bond formation, because it shows sequence similarity (albeit low with less than 20% overall identity) to ATP-dependent ligases and proteases that catalyze amide bond formation and hydrolysis.
Scheme 2.
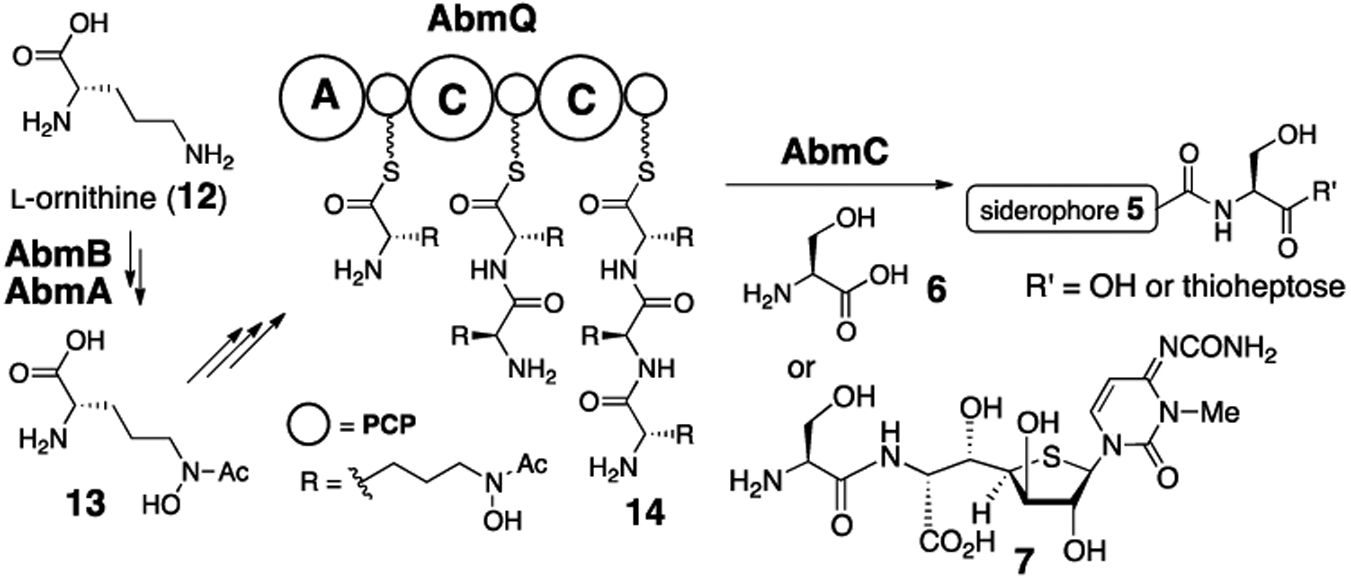
Proposed functions of AbmQ and AbmC.
To study the function of AbmC, the abmC gene in the albomycin-producing strain Streptomyces sp ATCC700974 was subjected to in-frame deletion (see Supporting Information).9 As expected, albomycin δ2 (1) production was completely abolished in the ΔabmC mutant strain (Figure 1). In contrast, LCMS analysis of the culture showed the production of SB-217452 (7, calcd m/z for C16H25N6O9S+ [M + H]+ 477.1398; obsd 477.1473, see Figure S1). Although co-production of albomycin δ2 (1) and SB-217452 (7) is always noted in the fermentation culture of the natural producers, whether SB-217452 (7) is a free biosynthetic intermediate or a degradation product of albomycin δ2 (1) has never been examined.6 The observation of SB-217452 (7) alone without albomycin δ2 (1) in the ΔabmC growth culture implied that SB-217452 (7) is a biosynthetic precursor to albomycin δ2 (1) and that the ligation of l-serine (6) with thioheptose 4 can precede ligation with the ferrichrome moiety (5), which was speculated to be catalyzed by AbmC. Furthermore, the carboxylic acid form of ferrichrome (5) was not detected in cultures of the ΔabmC strain even though it could be reproducibly detected in cultures of the wild type strain. This observation is consistent with the hypothesis that the ferrichrome moiety (see 14 in Scheme 2) is directly transferred from the C-terminal PCP domain of AbmQ to SB-217452 (7) without dissociation to generate the free carboxylic acid (5). This was also supported by the abmC gene complementation experiment using the ΔabmC mutant strain, in which the formation of albomycin δ2 (1) and ferrichrome (5) was restored (Figure 1).
Figure 1.
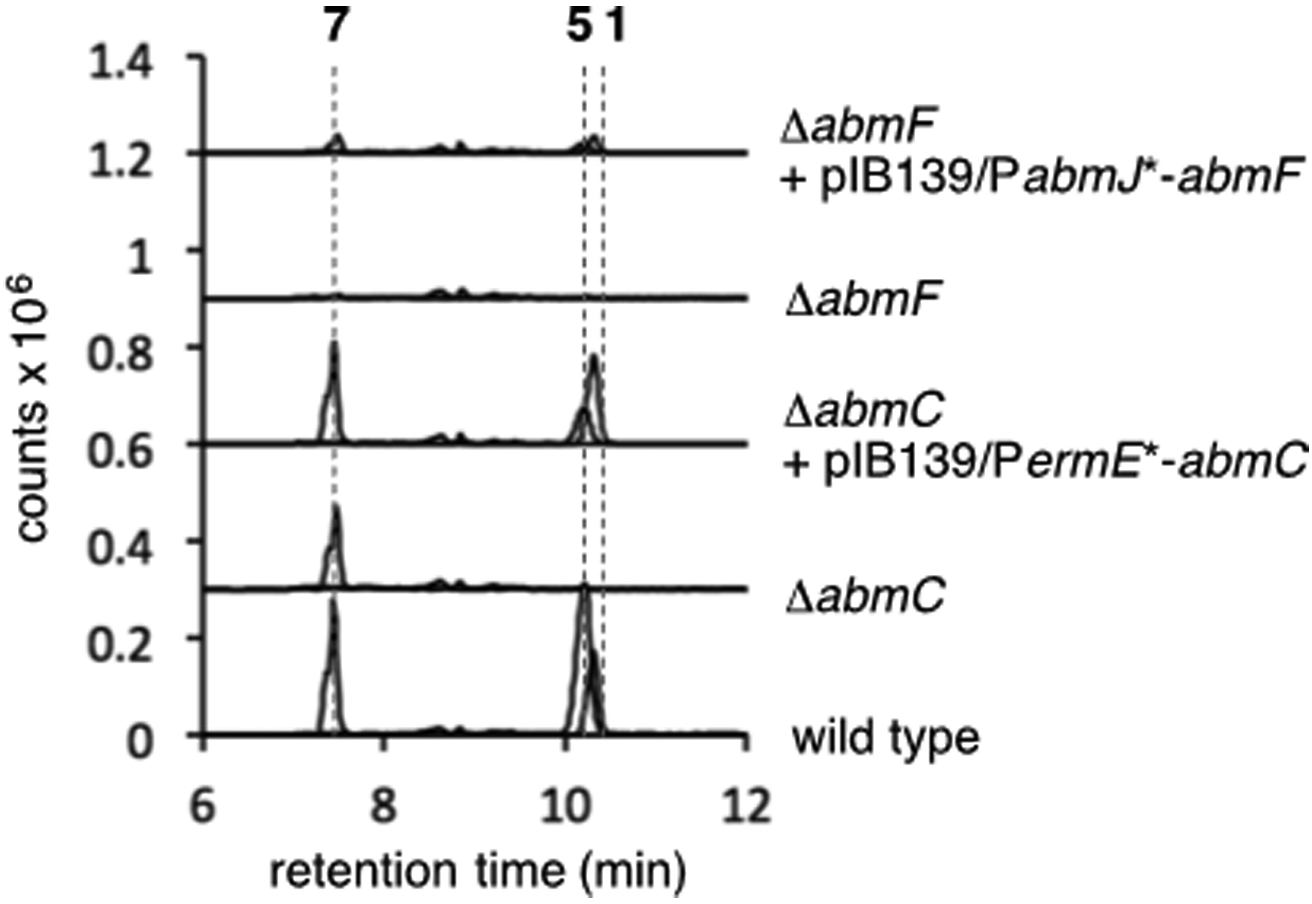
Gene deletion of abmC and abmF and complementation experiments. Three extracted ion chromatogram (EIC) traces (m/z = 1046.3, 588.2, and 477.1, corresponding to the [M + H]+ signals of 1, 5, and 7, respectively) are overlaid.
How the thioheptose moiety (e.g., 11) is converted to antibiotic SB-217452 (7) was next investigated. Based on BLAST analysis of the abm gene cluster, no obvious gene candidate for the peptide bond formation between l-serine (6) and the thioheptose (such as 11 or 4) could be identified. Nevertheless, structure-based computational analysis using I-TASSER10 and HHpred11 suggested that the gene product of abmF (AbmF) is similar to aminoacyl-tRNA synthetases (aaRS) despite its BLAST annotation as a hypothetical protein.6 Importantly, certain aaRS-like proteins are known to be capable of catalyzing non-ribosomal peptide synthesis.12 Hence, it was hypothesized that AbmF catalyzes peptide bond formation between the 6′-amino-4′-thionucleoside 11 (or 4) and l-serine to form 17 (or 7). This class of enzymes often utilize aminoacyl-tRNA as an aminoacyl group donor.12 The abmK gene in the abm gene cluster encodes a SerRS (AbmK) (Scheme 3A) that has been shown to be resistant to inhibition by SB-217452 (7).6,13 AbmK thus provides a functional seryl-tRNA (16) for ribosomal protein synthesis when the constitutive SerRS is inhibited by SB-217452 (7).6,13 Thus, AbmK may be involved in the biosynthesis of SB-217452 (7) by supplying seryl-tRNA (16) during the proposed AbmF-catalyzed condensation of 16 with 6′-amino-4′-thionucleoside (Scheme 3B). Alternatively, seryl-adenylate (15), which is an intermediate of AbmK-catalyzed seryl-tRNA biosynthesis, represents the seryl donor during the AbmF-catalyzed reaction (Scheme 3A and 3B). Finally, the 6′-amino-4′-thionucleoside substrate of AbmF may be 11 (3′S) and/or 4 (3′R) depending on the specificity of AbmF versus the putative C3′-epimerase AbmJ (Scheme 3B).
Scheme 3.
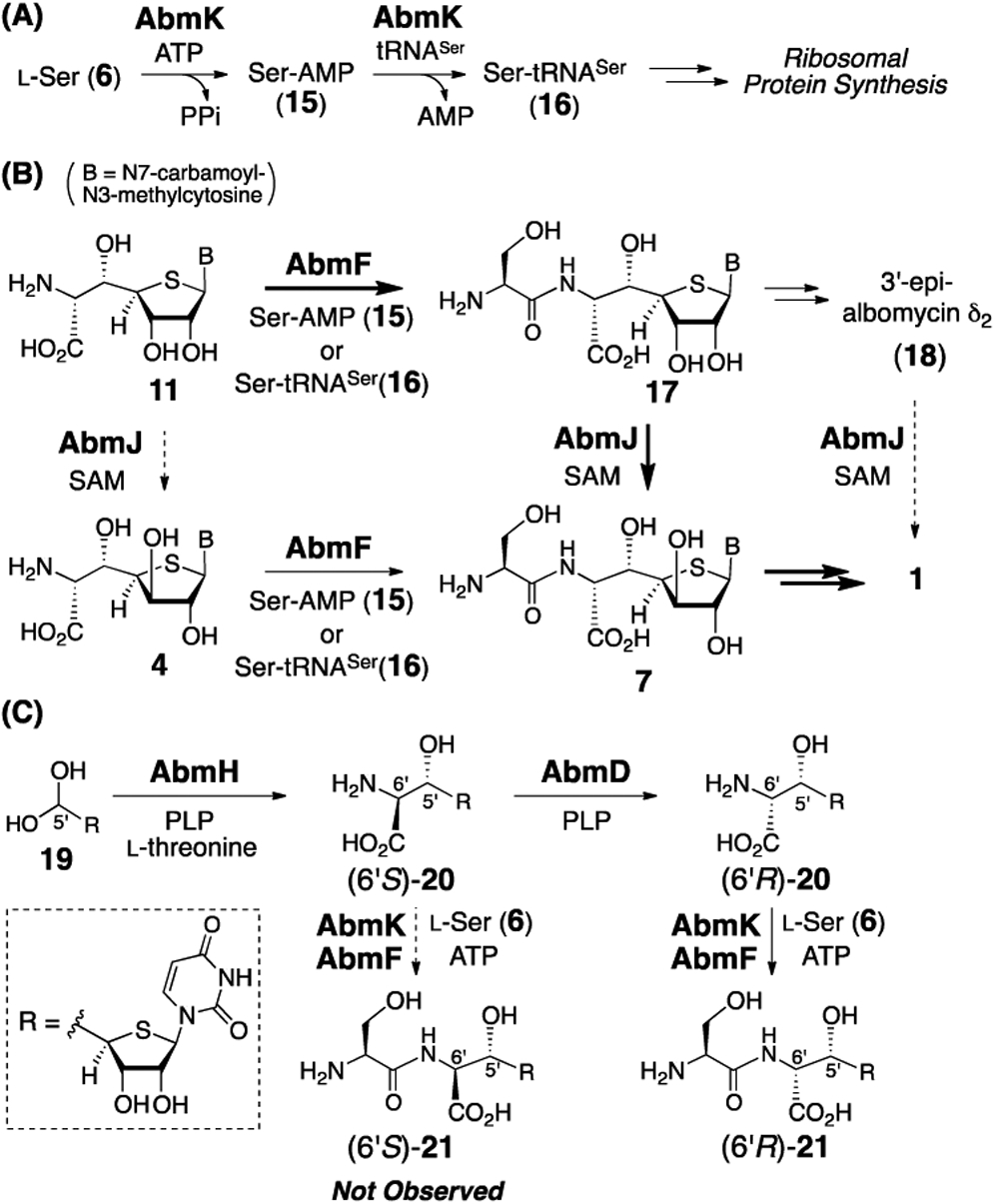
(A) Function of AbmK. (B) Proposed functions of AbmF and AbmJ. (C) AbmK/AbmF reactions of substrate analogues (6′S)-20 and (6′R)-20.
To probe the possible involvement of AbmF in the condensation between l-serine and the thionucleoside (11 or 4), deletion of the gene abmF in S. sp. ATCC700974 was carried out. As expected, disruption of abmF fully arrested the production of albomycin δ2 (1), SB-217452 (7), and free ferrichrome (5) as indicated by LCMS analysis (see Figure 1). Construction of these three metabolites was partially rescued when abmF was reintroduced into the ΔabmF strain. These observations indicated that abmF is for albomycin biosynthesis. To test whether AbmF catalyzes peptide bond formation between 11 or 4 and seryl-tRNA (16) or seryl-adenylate (15), both N-His6-tagged AbmF and N-His6-tagged AbmK13 were heterologously overexpressed and purified from E. coli (see Supporting Information). The putative substrate 11 was prepared by proteolysis of 3′-epi-albomycin δ2 (18) produced in the ΔabmJ strain (see Supporting Information).5 When 11 (0.20 mM) was incubated with AbmF (3.8 μM), AbmK (3.3 μM), l-serine (2.5 mM), ATP (2.5 mM), and E. coli total tRNA (5 μg/μL) in a buffer containing 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH 7.5, 50 mM), KCl (100 mM), and MgCl2 (10 mM), compound 11 was almost fully consumed with concomitant formation of 3′-epi-SB-217452 (17) as shown by ESI-MS analysis (calcd m/z for C16H23N6O9S+ [M – H]– 475.1253; found 475.1287, see Figure S7) and coelution with the standard 17 obtained from the ΔabmJ strain (Figure 2A). Control experiments in the absence of AbmF or ATP gave no significant formation of 17. Moreover, when tRNA was omitted, the production of 17 was still observed, which implied that AbmF could accept seryl-adenylate (15) as the seryl donor (Figure 2A). However, it remains an open possibility that seryl-tRNA can also serve as a substrate for AbmF. In the absence of AbmK, only trace amounts of 11 were converted to 17 during the incubation period (see Figure 2A, trace c, and Figure S7 in the Supporting Information). This observation is consistent with the hypothesis that AbmK is necessary for the AbmF-catalyzed coupling reaction. The formation of 17 without AbmK may be due to possible serine adenylation activity of AbmF, which is structurally similar to aminoacyl-tRNA synthetases and thus predicted to exhibit low levels of aminoacyl-adenylase activity.12c Incubation of 4 with AbmF under the above conditions also afforded SB-217452 (7) as a product (Figures S3 and S7).
Figure 2.
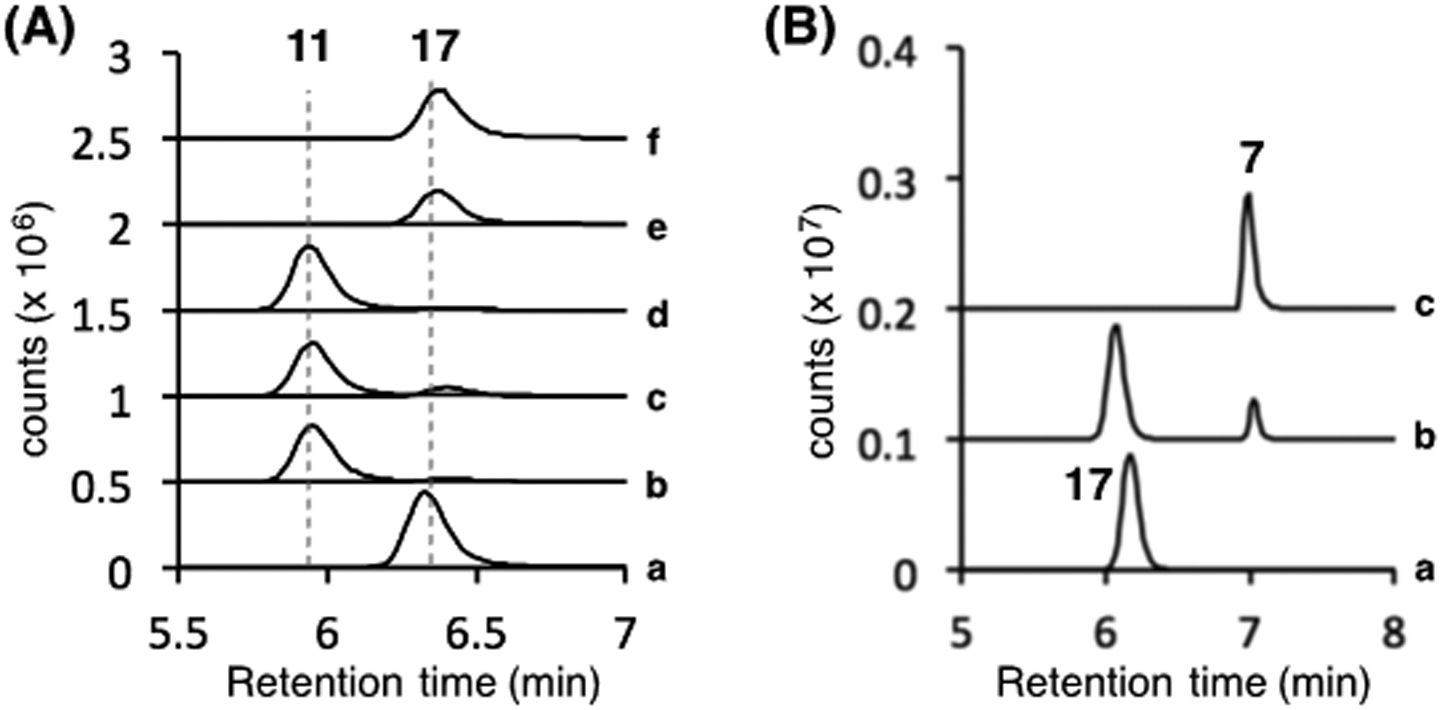
(A) LCMS analysis of in vitro AbmK/AbmF reaction of 11. Two EIC traces corresponding to [M – H]– signals from 11 (m/z = 388.1) and 17 (m/z = 475.1) are overlaid. a, Full reaction; b, without AbmF; c, without AbmK; d, without ATP; e, without tRNAs; f, standard sample of 17. (B) LCMS analysis of in vitro AbmJ reaction of 17. EIC traces corresponding to the [M + H]+ signal of 17 and 7 (m/z = 477.1) are shown. a, without AbmJ; b, with AbmJ; c, standard sample of 7.
These results indicate that AbmF accepts both C3′-epimers 11 and 4 of the 6′-amino-4′-thionucleoside substrate together with seryl-adenylate (15) to generate the serine-nucleoside conjugates 17 and 7, respectively. Recognition of the seryl-adenylate 15 by AbmF is notable because other aaRS-like ligases typically require an aminoacyl-tRNA as the activated amino acid donor.12 Furthermore, when the constitutive seryl-tRNA synthetase (SerRS1)13 encoded in S. sp. ATCC700974 was used instead of AbmK in the AbmF reaction of 4, the formation of SB-217452 (7) was significantly diminished likely due to product inhibition of SerRS1 by 7 (Figures S4 and S5). This implies that AbmK participates directly in the biosynthesis of SB-217452 in addition to being important for the resistance of the producing strain to inhibition of ribosomal peptide formation by SB-217452 as previously recognized.13
To gain insight into the specificity of AbmF, the substrate analogues (6′S)-20 and (6′R)-20, which differ from 11 in nucleobase structure (see Scheme 3C), were prepared and tested with the enzyme. It has been previously shown that incubation of 19 with AbmH and l-threonine produces (6′S)-20, which can be further converted to the C6′-epimerized product (6′R)-20 by AbmD.5 When (6′R)-20 was incubated with AbmF and AbmK in the presence of l-serine and ATP, (6′R)-20 was completely consumed and a product with a mass consistent with (6′R)-21 was detected by LCMS analysis (calcd m/z for C14H19N4O9S– [M – H]– 419.0878; found 419.0880, see Figures S6 and S7). On the other hand, (6′S)-20 obtained from AbmH reaction with 19 was not accepted by AbmF (Figure S6). These findings indicate that AbmF tolerates variations in the nucleobase (11 and 20) and the stereochemistry at C3′ (11 and 4) but is selective for a d-amino acid moiety as the nucleophile. Consequently, the PLP-dependent C6′-epimerase AbmD5 likely operates before AbmF in the biosynthetic pathway (Scheme 3C).
Furthermore, to investigate the aminoacyl-adenylate substrate tolerance of AbmF, four different aaRSs (threonyl-, tyrosyl-, cysteinyl-, and lysyl-tRNA synthetases, see Supporting Information for preparation) were heterologously expressed from E. coli and tested with AbmF and 4 in the absence of tRNAs; however formation of the corresponding dipeptide was not observed in all cases (Figure S8). These results demonstrated that AbmF is highly specific toward seryl-adenylate (15) and will not accept other aminoacyl-adenylates.
Although these results established the biosynthetic function of AbmF, the role played by the putative radical SAM enzyme AbmJ in catalyzing C3′-epimerization as well as the actual substrate for this reaction remained to be determined (Scheme 3B). To address this question, the abmJ gene was heterologously overexpressed in E. coli (see Supporting Information). Following purification, AbmJ was reconstituted with iron and sulfide according to established procedures (see Supporting Information).14 The reconstituted protein showed a broad absorbance band at 410 nm, which is indicative of the presence of one or more [4Fe-4S]2+ clusters bound to AbmJ (Figure S9).15 Subsequent iron quantification assays16 suggested that AbmJ binds either one or two [4Fe-4S] clusters per monomer consistent with the presence of 12 cysteine residues in its primary sequence (Table S1). In addition, the 410 nm absorption remains visible when the three cysteine residues associated with the radical SAM motif were all mutated to alanine (C12A/C16A/C19A) (Figure S9). Taken together, these results suggested that AbmJ binds at least two [4Fe-4S]2+ clusters.
When compound 11 was incubated with AbmJ under anaerobic, reducing conditions, no consumption of 11 was observed (Figure S12). While formation of a trace amount of 5′-deoxyadenosine (5′-dA), a typical byproduct in the catalysis of radical SAM enzymes,15 could be detected, it likely arose from uncoupled SAM cleavage which is a common phenomenon of radical SAM catalysis.15 In contrast, when the serine-thioheptose conjugate 17 (250 μM) was incubated with the reconstituted AbmJ (5 μM) in the presence of SAM (0.5 mM) and dithionite (2 mM), a new peak with the same mass as 17 (calcd m/z for C16H25N6O9S+ [M + H]+ 477.1398; found 477.1387, see Figure S11) was detected at a shifted retention time by LCMS analysis (Figures 2B). The identity of the product was confirmed to be SB-217452 (7) by coinjection with the standard sample. As expected, 5′-dA was also produced with 7 in an approximately 1:1 ratio (see Supporting Information). The production of 7 was only detected in the presence of SAM and a reductant such as dithionite (Figures S10). Furthermore, no epimerization activity of AbmJ was noted when 3′-epi-albomycin δ2 (18) was used in place of 17 (Figure S12). These in vitro assays indicate that AbmJ-catalyzed C3′-epimerization of the nucleoside core occurs after the AbmF-catalyzed ligation reaction but before conjugation with the siderophore moiety 5. Namely, albomycin biosynthesis likely proceeds via the pathway 11 → 17 → 7 → 1 (see Scheme 3B).
A possible reaction mechanism for the AbmJ catalyzed reaction is shown in Figure S13 of the Supporting Information. The 5′-deoxyadenosyl radical generated by reductive cleavage of SAM may abstract the C3′ hydrogen atom of 17. The resulting substrate radical may then be stereoselectively quenched by a H atom donor such as a Cys residue in the active site of AbmJ similar to other cases of radical SAM epimerases.17 However, further investigation is needed to determine the H atom donor and the role of the auxiliary iron sulfur cluster(s) in AbmJ.
In summary, this study has elucidated how the antibiotic core (i.e., SB-217452, 7) of the albomycins is generated from the previously recognized5 thioheptose nucleoside intermediate 11. Thus, AbmF is a unique ligase that catalyzes condensation between 6′-amino-4′-thioheptose 11 and seryl-adenylate. The latter, activated serine residue is a product generated in the reaction catalyzed by the SerRS AbmK, which not only confers resistance to albomycins on the producing strain but likely also participates directly in their biosynthesis. The role of the radical SAM enzyme AbmJ as a C3′-epimerase acting on the serine-thioheptose conjugated intermediate 17 to generate SB-217452 (7) has also been characterized in vitro. Finally, gene deletion experiments suggest that AbmC catalyzes the final reaction in this pathway during which SB-217452 (7) is conjugated with the siderophore moiety (5) to form 1. This study not only identifies three unique enzymes involved in the biosynthesis of albomycins but also highlights how the biosynthetic machinery is organized to efficiently generate the natural product without producing potential side products.
Supplementary Material
Acknowledgements
This work was supported by grants from the National Institutes of Health (GM035906 and 1 S10 OD021508-01 for NMR) and the Welch Foundation (F-1511).
Footnotes
Experimental Section
Experimental procedures and supplementary data are available in Supporting Information.
Supporting information for this article is given via a link at the end of the document
References
- [1].(a) Gause GF, Brit. Med. J 1955, 12, 1177–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Stapley EO, Ormond RE, Science 1957, 125, 587–589. [DOI] [PubMed] [Google Scholar]; (c) Benz G, Liebigs Ann Chem, 1984, 1399–1407. [Google Scholar]; (d) Benz G, Born L, Brieden M, Grosser R, Hurz J, Paulsen H, Sinnwell V, Weber B, Liebigs Ann Chem, 1984, 1408–1423. [Google Scholar]; (e) Paulsen H, Brieden M, Benz G, Liebigs Ann Chem, 1987, 565–575. [Google Scholar]
- [2].Lin Z, Xu X, Zhao S, Yang X, Guo J, Zhang Q, Jing C, Chen S, He Y, Nat. Commun 2018, 9, 3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].(a) Pramanik A, Braun V, J. Bacteriol 2006, 188, 3878–3886. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ferguson AD, Braun V, Fiedler HP, Coulton JW, Diederichs K, Welte W, Protein Sci. 2008, 9, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Clarke TE, Braun V, Winkelmann G, Tari LW, Vogel HJ, J. Biol. Chem 2002, 277, 13966–13972. [DOI] [PubMed] [Google Scholar]
- [4].(a) Stefanska AL, Fulston M, Houge-Frydrych CSV, Jones JJ, Warr SR, J. Antibiot 2000, 53, 1346–1353. [DOI] [PubMed] [Google Scholar]; (b) Saha A, Dutta S, Nandi N, J. Biomol. Struct. Dyn 2019. DOI: 10.1080/07391102.2019.1635912. [DOI] [PubMed] [Google Scholar]; (c) Cochrane RVK, Norquay AK, Vederas JC, Med. Chem. Commun 2016, 7, 1535–1545. [Google Scholar]
- [5].Ushimaru R, Liu H.-w., J. Am. Chem. Soc 2019, 141, 2211–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zeng Y, Kulkarni A, Yang Z, Patil PB, Zhou W, Chi X, Van Lanen S, Chen S, ACS Chem. Biol 2012, 7, 1565–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].(a) Schwecke T, Göttling K, Durek P, Dueñas I, Kaüfer NF, Zock-Emmenthal S, Staub E, Neuhof T, Dieckmann R, von Döhren H, ChemBioChem 2006, 7, 612–622. [DOI] [PubMed] [Google Scholar]; (b) Winterberg B, Uhlmann S, Linne U, Lessing F, Marahiel MA, Eichhorn H, Kahmann R, Schirawski J, Mol. Microbiol, 2010, 75, 1260–1271. [DOI] [PubMed] [Google Scholar]; (c) Bushley KE; Ripoll DR; Turgeon BG; (d) Welzel K, Eisfeld K, Antelo L, Anke T, Anke H. FEMS Microbiol. Lett 2005, 249, 157–163. [DOI] [PubMed] [Google Scholar]
- [8].(a) Fischbach MA, Walsh CT, Chem. Rev 2006, 106, 3468–3496. [DOI] [PubMed] [Google Scholar]; (b) Sieber SA, Chem. Rev 2005, 105, 715. [DOI] [PubMed] [Google Scholar]; (c) Süssmuth RD, Mainz A, Angew. Chem., Int. Ed 2017, 56, 3770–3821. [DOI] [PubMed] [Google Scholar]
- [9].Zeng et al. constructed a ΔabmC mutant strain; however, the metabolic profile of the strain is not reported in reference 6. [Google Scholar]
- [10].Roy A, Kucukural A, Zhang Y, Nat. Protoc 2010, 5, 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Sõding J, Biegert A, Lupas AN, Nucleic Acids Res. 2005, 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].(a) Moutiez M, Belin P, Gondry M, Chem. Rev 2017, 117, 5578–5618. [DOI] [PubMed] [Google Scholar]; (b) Aravind L, de Souza RF, Lyer LM, Biol. Direct 2010, 5, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Hu Z, Awakawa T, Ma Z, Abe I, Nat. Commun 2019, 10, 184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Zeng Y, Roy H, Patil PB, Ibba M, Chen S, Antimicrob. Agents Chemother 2009, 53, 4619–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ruszczycky MW, Choi S.-h., Liu H.-w., J. Am. Chem. Soc 2010, 132, 2359–2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].(a) Broderick JB, Duffus BR, Duschene KS, Shepard EM, Chem. Rev 2014, 114, 4229–4317. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE, Nucleic Acids Res. 2001, 29, 1097–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Frey PA, Hegeman AD, Ruzicka FJ, Crit. Rev. Biochem. Mol. Biol 2008, 43, 63–88. [DOI] [PubMed] [Google Scholar]; (d) Ruszczycky MW, Zhong A, Liu H.-w., Nat. Prod. Rep 2018, 35, 615–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].(a) Zhu W, Martins AM, Klinman JP, Methods Enzymol. 2018, 606, 389–420. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Fish WW, Methods Enzymol. 1988, 158, 357–364. [DOI] [PubMed] [Google Scholar]
- [17].(a) Morinaka BI, Verest M, Freeman MF, Gugger M, Piel J, Angew. Chem. Int. Ed 2017, 56, 762–766. [DOI] [PubMed] [Google Scholar]; (b) Kudo F, Hoshi S, Kawashima T, Kamachi T, Eguchi T, J. Am. Chem. Soc 2014, 136, 13909–13915. [DOI] [PubMed] [Google Scholar]; (c) Benjdia A, Gluillot A, Ruffié P, Leprince J, Berteau O, Nat. Chem 2017, 9, 698–707. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Parent A, Benjdia A, Gluillot A, Kubiak X, Balty C, Lefranc B, Leprince J, Berteau O, J. Am. Chem. Soc, 2018, 140, 2469–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


