Graphical Abstract
The development of a multicellular organism starts with the division of a single cell, the zygote. The descendants of the zygote divide and gradually differentiate and form lineages that comprise all the tissues in the organism. The division process resembles a tree, where each cell as a node is connected to its two daughter cells in one direction and to its parent cell in the opposite direction. Reconstructing these lineage relationships for an organism will guide us in uncovering the developmental programs and help us characterize their defects.
From the 19th century on, embryologists have attempted to map lineages using a variety of methods, including direct observation of growing embryos under the microscope, grafting cells and tissues, or creating chimeric animals.1 One of the most common methods is the clonal labeling of cells: marking progenitor cells at a given developmental stage with a detectable label that is inherited by their progeny, throughout the development and possibly in the adult stage. In this fashion, the lineages of the progenitors can be traced, and their fate may be delineated. For instance, a common labeling method is to inject progenitors with dyes that will be passed on to their progeny; the primary limitation here is the small number of different dyes that can be used, thus limiting the number of different lineages that can be distinguished simultaneously.
Inspired by modern phylogenetic analysis, developmental labeling is the storing of lineage information in the form of genetic labels within the genome of dividing cells, such that detailed lineage relationships between cells can be inferred by comparing their labels. This is further than clonal analysis, as it can capture developmental dynamics and cell divisions over a stretch of time rather than a snapshot of the embryo’s state. One way to implement this strategy is to use the DNA targeting power of CRISPR to induce continuous and stochastic mutations in targeted regions of the genome of cells during development and thereby marking cells with cumulative mutational signatures that reflect their developmental history. These mutations, which are usually in the form of short indels, can be recovered by high-throughput DNA or RNA sequencing. In recent years, this idea was implemented in cell cultures, Caenorhabditis elegans, and zebrafish with very promising results.2 During the past year, however, the idea has been extended to mice in studies by Kalhor et al.3 and Chan et al.,4 which focused on the lineages that form up to day 8.5 post fertilization (E8.5).
Lineage tracing in mice is particularly challenging because of its developmental complexity in the uterus and challenges associated with its genetic manipulation. The first study approached the problem by creating a transgenic mouse line (MARC1) that expresses 60 loci of homing CRISPR guide RNAs (hgRNAs), which start accumulating mutations upon activation. These mutations served as developmental barcodes to reconstruct the lineage relationships between embryonic and adult tissues at the cell-population level.
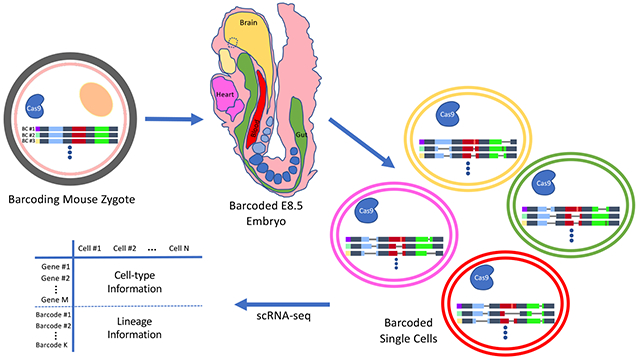
More recently, Chan et al.4 described a significant advance in the field by performing simultaneous lineage tracing and single-cell transcriptional profiling in mice. They fertilized eggs by injection of sperm that also carry Cas9 genes, a cassette of guide RNAs, and a cassette of target sites for the guide RNAs. The target cassettes start accumulating mutations over time upon integration into as many as 15 stochastic locations on the genome and thus can serve as developmental barcodes, verified by the ability to recapitulate the relationships between different embryonic tissues at the population level only based on the similarity between their mutational profiles.
The target cassettes are designed to be transcribed by the RNA polymerase II machinery; hence, they can be captured along with mRNA transcripts by single-cell mRNA sequencing (scRNA-seq) methods. Chan et al.4 performed scRNA-seq on E8.5 barcoded embryos and obtained gene expression profiles and lineage information for up to 13 000 and 8000 single cells, respectively. The reconstructed lineage trees overlaid with cell type information could map the relationships between several embryonic and extra-embryonic tissues. They also observed that, in general, cells with a closer lineage distance show higher similarity in their transcriptional state. Importantly, they confirmed and quantified the contribution of extra-embryonic lineages to embryonic tissues5 by detecting a subpopulation of embryonic endodermal cells with lineage barcodes closer to that of the extra-embryonic cells.
The CRISPR-based lineage tracing in model organisms has opened up opportunities for detailed analysis of dynamic developmental processes. To increase the depth of the data, the sensitivity of single-cell capturing of the recording elements has to be improved. Recording lineage information while preserving the spatial data of single cells can be the next important step in the field. Another important avenue is using inducible or tissue-specific promoters for driving Cas9 that can focus the recording capability of the system on a specific lineage including diseased tissues.
ACKNOWLEDGMENTS
This work is supported by NIH grants MH103910 and HG005550 and the Intelligence Advanced Research Projects Activity (IARPA) via the Department of Interior/Interior Business Center (DoI/IBC) contract number D16PC00008.
The authors declare the following competing financial interest(s): Dr. Church’s Advisory Roles, Tech Transfer, and Funding Sources can be found here: http://v.ht/PHNc.
REFERENCES
- (1).Kretzschmar K, and Watt FM (2012) Lineage Tracing. Cell 148 (1), 33–45. [DOI] [PubMed] [Google Scholar]
- (2).McKenna A, and Gagnon JA (2019) Recording development with single cell dynamic lineage tracing. Development 146, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Kalhor R, Kalhor K, Mejia L, Leeper K, Graveline A, Mali P, and Church GM (2018) Developmental barcoding of whole mouse via homing CRISPR Science 361, 6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, Khodaverdian A, Yosef N, Meissner A, and Weissman JS (2019) Molecular recording of mammalian embryogenesis. Nature 570, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Kwon GS, Viotti M, and Hadjantonakis A-K (2008) The Endoderm of the Mouse Embryo Arises by Dynamic Widespread Intercalation of Embryonic and Extraembryonic Lineages. Dev. Cell 15 (4), 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]


