Abstract
Photobleaching limits extended imaging of fluorescent biological samples. Here, we developed DNA based “FluoroCubes” that are similar in size to the green fluorescent protein (GFP), have single-point attachment to proteins, have a ~54-fold higher photobleaching lifetime and emit ~43-fold more photons than single organic dyes. We demonstrate that DNA FluoroCubes provide outstanding tools for single-molecule imaging, allowing the tracking of single motor proteins for >800 steps with nanometer precision.
Introduction
Imaging proteins and macromolecular complexes at the single-molecule level is a powerful method to study distribution, stoichiometry, dynamics and precise motion of molecular machines1. To achieve high spatiotemporal resolution, proteins of interest are often labeled with fluorescent probes1. An ideal probe is photostable (long half-life), (continuously) bright, small, and can be monovalently attached to biological molecules. While organic dyes2,3 and fluorescent proteins4 fulfill the latter two criteria, they often suffer from photobleaching, which leads to a low signal. Alternative probes such as quantum dots2,5 and other fluorescent nanoparticles6 are very bright and extremely photostable, but frequently exhibit large fluctuations in intensity (blinking). The relatively large (~15 nm) size of these probes (Supplementary Fig. 1 a) can perturb protein function and complicates multicolor labeling of proteins2,3,5. Large probes often lack control over surface chemistry, which can lead to multiple proteins attaching to the same fluorescent probe7.
Here, we developed small (~6 nm) DNA-based FluoroCubes that have single-point attachment, exhibit continuous emission, emit up to ~43-fold more photons than single organic dyes, and have an up to ~54-fold longer half-life than single organic dyes. We show that the photostability of the DNA FluoroCubes varies with the type of organic dye attached to the DNA. We also provide some preliminary insights into the mechanism(s) for the increased photostability of DNA FluoroCubes. Attaching these DNA FluoroCubes to the microtubule-based motor protein kinesin, we were able to track its movement for more than 800 steps with nanometer precision.
Results
Design and assembly of six dye DNA FluoroCubes
Previous work established that placing organic dyes within 2 nm results in quenching7, while dye spacing of >5 nm results in a linear intensity increase with the number of dyes8. We were interested in the properties of probes separated by 2 to 5 nm.
To set the position of dyes with nanometer precision, we took advantage of tools from DNA nanotechnology8–10. Using the single-stranded tiles (SST) approach11,12, we designed a DNA FluoroCube composed of four 16 base-pair (bp) long double-stranded DNA helices labeled with six of the same organic dyes such as ATTO 488, ATTO 565, ATTO 647N, Cy3N (sulfonated Cy3), Cy3, or Cy5 (Fig. 1 a, b, Supplementary Fig. 1 b) separated by distances ranging from ~2 to ~6 nm. We reserved one position for the placement of a functional tag such as HALO-ligand, benzylguanine (for SNAP tag), thiol, biotin, or amine to label proteins at specific locations (Fig. 1 b).
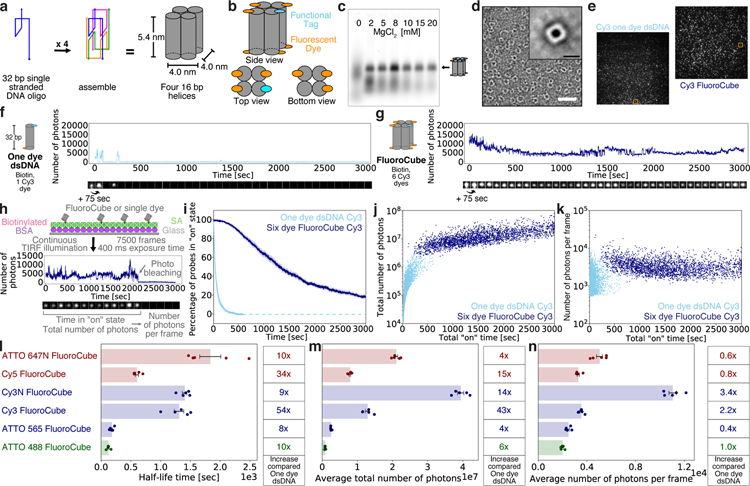
Figure 1 |. Design, assembly, and photophysical properties of DNA FluoroCubes.
(a) Design and shape of DNA FluoroCubes. Four 32 bp long single-stranded DNAs (ssDNA) are connected using crossovers resulting in four connected 16 bp long double-stranded DNAs (dsDNA) with a size of approximately 5.4 x 4.0 x 4.0 nm. A detailed map of oligonucleotide routing and the bases is depicted in Supplementary Figure 1 b. (b) Each of the 5’ and 3’ ends of the DNA can be functionalized. For the DNA FluoroCube design we used six fluorophores and one functional tag such as HALO-ligand, benzylguanine for SNAP tag, thiol, biotin, or amine. (c) 3.0% agarose gel of DNA FluoroCubes after thermal annealing. The four ssDNA strands are annealed with different MgCl2 concentrations. Quantification of assembly yield is shown in Supplementary Figure 1 c. Note, that the different brightness of FluoroCube bands is mainly due to variation of material loaded into the gel. Here the ssDNAs were modified with six ATTO 647N dyes and one biotin. (d) Negative stain electron microscopy image of DNA FluoroCubes. Insert shows class average of 983 particles. More images are shown in Supplementary Figure 1 e and 2. Here the FluoroCube was labeled with six ATTO 647N dyes and one biotin. White scale bar: 30 nm. Black scale bar: 6 nm. Class averaging was performed once. (e) Example TIRF image of a biotin functionalized six Cy3 dye FluoroCube and a 32 bp long double-stranded DNA (dsDNA) with one Cy3 dye. Orange boxes show molecules whose intensity traces are shown in f (one dye dsDNA) and g (FluoroCube). (f, g) Example intensity trace of (f) Cy3 one dye dsDNA and (g) six Cy3 dye FluoroCube with one biotin. More intensity traces with different fluorophores are shown in Supplementary Figure 4. (h) Experimental setup for quantification of photophysical properties of six dye FluoroCubes and one dye dsDNA. Biotinylated samples are attached to a cover slip (Glass) with biotinylated BSA, and streptavidin (SA). Then movies with 7,500 frames total (or until all probes photobleached) and 400 ms exposure each are recorded under continuous laser illumination. Intensity traces of single-molecules are analysed for time in “on” state (half-life), total number of photons, and number of photons per frame. A detailed description of the analysis can be found in Online Methods. (i) Photostability of six Cy3 dye FluoroCubes and Cy3 one dye dsDNA. The survival rate was quantified by counting the percentage of probes in the “on” state at any given time from 0 to 3,000 seconds. Opaque color is the standard error of the mean of five or four repeats (six dye FluoroCubes or one dye dsDNA, respectively) with more than 500 molecules each. Once all probes photobleached data analysis was terminated. This is indicated by the dashed line. (j) Total number of photons of six Cy3 dye FluoroCubes and Cy3 one dye dsDNA as a function of the total “on” time at the single-molecule level (pooled from all experiments). (k) Average number of photons per frame of six Cy3 dye FluoroCubes and Cy3 one dye dsDNA as a function of the total “on” time at the single-molecule level (pooled from all experiments). (l-n) Bar plot of the (l) half-life time, (m) average of the total number of photons, and (n) average number of photons per frame of six dye DNA FluoroCubes with different fluorophores. The error bars show the standard error of the mean of five repeats with more than 100 molecules each. Dots show the values of individual experiments. The tables on the right show fold increase of six dye DNA FluoroCubes compared to one dye dsDNA. Single-molecule distributions are shown in Supplementary Figure 7. Note, that the average of the total number of photons will be slightly higher for some six dye FluoroCubes than shown here because not all probes bleached within 3,000 seconds. (i-n) Each experiment was repeated five or four times with freshly assembled six dye FluoroCubes or one dye dsDNA, respectively. For every experiment we prepared new microscope slides. Exact numbers (also of the sample size) are given in Supplementary Table 1 and 2. “Cy3” stands for the non-sulfonated version of Cy3 whereas “Cy3N” stands for the sulfonated version of Cy3. “n.m.” is not measured.
Since small DNA nanostructures are difficult to assemble because of high electrostatic repulsion between the negatively-charged DNA strands13, we optimized the folding yield and determined the structural integrity of the DNA FluoroCubes after thermal annealing. Using agarose gel electrophoresis, we measured a folding yield of over 60% (Fig. 1 c, Supplementary Fig. 1 c) and found that only folding reactions that contained all four oligos assembled into FluroroCubes (Supplementary Fig. 1 d). Negative stain transmission electron microscopy (TEM) showed that FluoroCubes assembled into the desired shape, with slight variations (Fig. 1 d, Supplementary Fig. 1 e, 2). In summary, the protocol for the assembly of DNA FluoroCubes is easy and optimized for high yield (Supplementary Protocol).
Photophysical properties of six dye DNA FluoroCubes
Next, we examined the fluorescent properties of DNA FluoroCubes prepared with the commonly used fluorescent probe Cy3. We compared the six Cy3 dye FluoroCube to a single Cy3 dye attached to double-stranded DNA (dsDNA). Surface-immobilized samples were imaged by total internal reflection fluorescence (TIRF) microscopy (Fig. 1 e–h). To quantify photostability, we determined the time when 50% of all probes photobleached (half-life) and the total number of photons emitted. To quantify brightness, we measured the average number of photons per frame.
A single Cy3 dye conjugated to dsDNA or protein (streptavidin) (Supplementary Fig. 3) displayed a constant intensity followed by one-step photobleaching (Fig. 1 f, Supplementary Fig. 4). However, the six Cy3 dye FluoroCube was significantly more photostable than a single Cy3 dye bound to dsDNA (up to ~54-fold increase in half-life) (Fig. 1 i, I), and sometimes even increased in brightness during the time of acquisition (Supplementary Fig. 4). For instance, after 99% of the single Cy3 dyes bleached, more than 80% of the six Cy3 dye FluoroCubes were still in the “on” state. The total number of photons per six Cy3 dye FluoroCube was ~43-fold higher than for a single Cy3 dye bound to dsDNA (Fig. 1 j, m; 1.29 ± 0.06 x 107 photons for the six Cy3 dye FluoroCube versus 3.1 ± 0.5 x 105 photons for a single Cy3 dye bound to dsDNA, the latter being consistent with previous reports14). The six Cy3 dye FluoroCubes also blinked less than the single Cy3 dye bound to dsDNA (Supplementary Fig. 5 i). However, even though there are 6-fold more dyes, the brightness of a six Cy3 dye FluoroCubes was similar to a single Cy3 dye bound to dsDNA (Fig. 1 k, n). With increasing light exposure, the photon output of six Cy3 dye FluoroCubes behaved nonlinearly, as they plateaued in brightness at higher excitation powers (Supplementary Fig. 6 b). In summary, six Cy3 dye FluoroCubes emit an average of ~43-fold more photons than single Cy3 dyes but are similar in overall brightness.
We next tested the behavior of other dyes coupled to FluoroCubes and found variable photophysical properties (Fig. 1 l–n, Supplementary Fig. 5–7, Supplementary Movies 1–6, Supplementary Table 1). The six Cy5 dye, six ATTO 647N dye, and six Cy3N dye FluoroCube also demonstrated a significant increase in photostability (34-fold, 10-fold, and 9-fold respectively) (Fig. 1 l, Supplementary Fig. 7 a) and total number of photons (15-fold, 4-fold, and 14-fold respectively) (Fig. 1 m). The six ATTO 488 dye and six ATTO 565 dye FluoroCubes were less photostable in absolute terms, but still showed a 10-fold and 8-fold increase relative to single dyes attached to dsDNA (Fig. 1 l). Intermediate photostability was observed for the six dye Janelia Fluorophores JF549 and JF64615 (Supplementary Fig. 8). As with the six Cy3 dye FluoroCube, all of the FluoroCubes were similar in brightness to their respective single dyes (Fig. 1 n). The various six dye FluoroCubes performed better with oxygen scavengers16 and triplet quenchers17 (Supplementary Fig. 6 d–f). However, even without these aids all six dye FluoroCubes outperformed single dyes on dsDNA with oxygen scavengers16 and triplet quenchers17. Taken together, the performance of the six dye FluoroCubes is dye dependent, but all are significantly more photostable than commonly used single organic dyes (Fig. 1 l–n) and some emit up to ~43-fold more photons than a single dye.
To further characterize our new fluorescent probes, we examined additional properties of the DNA FluoroCubes. Comparing the absorbance, excitation, and emission spectra of various six dye FluoroCubes and their respective single dyes, we found a dye charge-dependent, large increase in absorbance in a “shoulder” blue shifted from the main peak (Supplementary Fig. 9). This “shoulder” absorbance is often observed for face-to-face dimers of dyes18 and may result from the flexibility of FluoroCubes in solution and the relatively long linker (up to 1.5 nm) between dye and oligo. An alternative explanation for the observed shoulder absorbance is that face-to-face dimers form between dyes on different FluoroCubes. To test whether our six dye FluoroCubes are truly monomeric, we performed dynamic light scattering (DLS) and found that the measured diameters of different FluoroCubes agree well with their predicted diameters (Supplementary Fig. 10). Moreover, the diameter distributions of the FluoroCubes are symmetric (no shoulders visible) indicating that the FluoroCubes are monodisperse in size. Additional experiments such as the measurement of the number of photobleaching steps (Supplementary Fig. 11) and our previous negative stain imaging (Supplementary Fig. 2, Supplementary Table 1) confirmed the DLS data. Thus, we concluded that six dye FluoroCubes are predominantly monomeric and that the shoulder absorbance is likely caused by dyes on the same FluoroCube. Interestingly, the shoulder absorbance for six dye FluoroCubes mainly represents non-fluorescent dyes (except for the six Cy3 dye FluoroCube) as shown by comparing excitation and absorption spectra (Supplementary Fig. 9, 12). Lastly, we measured fluorescence lifetime and fluorescence anisotropy (Supplementary Fig. 13), but did not find any change that might be correlated with increased photostability. Taken together, six dye FluoroCubes are predominantly monomeric and present a dye charge-dependent “shoulder” in absorbance that is blue shifted from the main peak.
Comparison of six dye DNA FluoroCubes and quantum dots
Next, we compared the behavior of the six ATTO 647N dye FluoroCubes to quantum dots which are known for their photostability. Quantum dots (655 Qdot Nanocrystals) emitted about four-fold more photons than six ATTO 647N dye FluoroCubes, were about four-fold brighter than six ATTO 647N dye FluoroCubes, and have a similar half-life time as six ATTO 647N dye FluoroCubes (Supplementary Fig. 14 c, d). However, quantum dots blink significantly more than DNA FluoroCubes (Supplementary Fig. 14 b). In addition, quantum dots tend to enter dark states in which they do not emit photons for a couple of seconds (Supplementary Fig. 14 a), making it difficult to track quantum dots continuously. Moreover, quantum dots are significantly larger in size (10–20 nm) than the six dye FluoroCubes2,3,5 (Supplementary Fig. 1 a) and typically not monovalent. In conclusion, six dye FluoroCubes display much more stable and uniform fluorescence than quantum dots.
Insights into potential mechanism(s) of increased photostability of six dye DNA FluoroCubes
To begin to understand the mechanism(s) behind the increase in photostability of the six dye FluoroCubes, we investigated how the DNA scaffold might influence dye photophysics. A single dye on the same FluoroCube scaffold (Single Dye Cube) (Supplementary Fig. 15 a) displayed a slight increase in photostability and emitted between ~2 and ~10-fold more photons than corresponding one dye dsDNA (Supplementary Fig. 15). Permuting through all six positions on the cube with a single Cy3N dye (Supplementary Fig. 16 a), we noticed a higher photostability (~3-fold longer half-life time) when the dye is attached to the 5’ end compared to the 3’ end of the same oligo (Supplementary Fig. 16). Together, these data show that the local environment of the DNA (Single Dye Cube vs. one dye dsDNA) and the dye position (attachment linkage) can influence photostability.
To further investigate the effect of the geometry of dye placement on photostability, we compared the photostability of the six Cy3 dye FluoroCubes to the same number of Cy3 dyes on an 8-fold (by volume) larger structure (the previously described Compact Cube13), which places the dyes between ~6 and ~10 nm from one another (Supplementary Fig. 17 a). The Compact Cube with one Cy3 dye behaved very similar to a Single Cy3 Dye Cube (Supplementary Fig. 17), while the Compact Cube with six Cy3 dyes had a slight increase in photostability but far less than the six Cy3 dye FluoroCube. Thus, increasing the distances between the dyes from ~2–6 nm (FluoroCube) to ~6–10 nm (Compact Cube) decreases photostability, indicating the importance of distance between dyes for the photostability effect. We speculate that both direct dye interactions as well as resonance energy transfer between individual dyes in a FluoroCube play roles in this phenomenon (Supplementary Fig. 18), although additional work is required to fully understand the photophysical mechanisms underlying six dye DNA FluoroCube photostability.
Tracking steps of a single kinesin over more than 6 µm
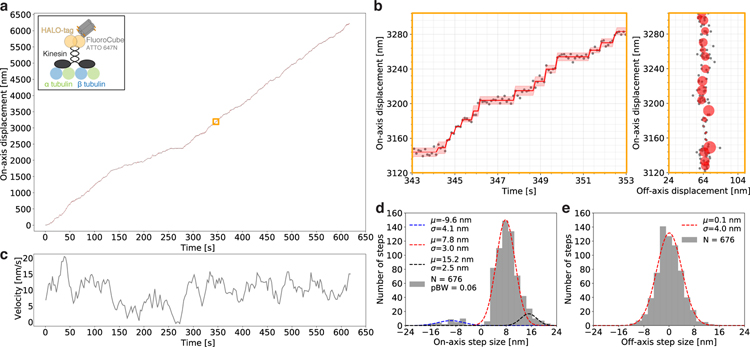
We next tested whether FluoroCubes can be attached to proteins and used for prolonged readouts of activity. For this purpose, we labeled an ultra processive kinesin KIF1A19 with a six ATTO 647N dye FluoroCube using a C-terminal HALO-tag (Fig. 2 a) and imaged it moving along axonemes. To ensure that a FluoroCube labeled kinesin behaves similar to a single dye labeled kinesin (single TMR HALO-dye), we compared velocity and processivity and found almost identical values indicating that FluoroCubes do not interfere with protein function in this assay (Supplementary Fig. 19). Labeling kinesin with a single six dye FluoroCube enabled us to record more than 800 steps of an individual motor with nanometer precision (Fig. 2 a, b, Supplementary Movie 7). The trace in Figure 2 revealed an on-axis step size of 7.8 nm with almost no off-axis stepping (Fig. 2 d, e) which is in good agreement with previous reports19,20 (discussion in Supplementary Fig. 20). Moreover, using six ATTO 647N dye FluoroCubes allowed us to collect more than 6,000 data points of an individual motor compared to approximately 200 data points that can be collected at a similar resolution with a single organic dye21,22. By recording very long traces, we could detect occasional pausing and velocity fluctuations within the trace of an individual kinesin (Fig. 2 c). Previous work demonstrated that different kinesins can have different velocities23, but our prolonged observations showed that even individual kinesins undergo considerable velocity fluctuations over time.
Figure 2 |. Tracking steps of a single kinesin over more than 6 µm.
(a) Raw stepping data with position versus time of one kinesin (grey dots) over 6 µm with detected steps (red line) along an axoneme. The opaque red line shows the standard deviation for each step. The insert shows the experimental setup for which a kinesin is labeled with one six dye ATTO 647N FluoroCube with a HALO-tag (for details see Online Methods). The orange box is enlarged in b. The movement of this particular kinesin is shown in Supplementary Movie 7. (b) Left: Zoom-in on the raw stepping data with position versus time of a single kinesin as shown in a. Right: Same trace as on the left but in XY space. The grey dots are raw data and the red circles show the fitted position for which the radius corresponds to the standard deviation. (c) Velocity over time for the stepping trace of a single kinesin as shown in a. The grey line shows a moving average of velocity binned into 15.6 sec (for details see Online Methods). (d) Histogram of the on-axis step size distribution of the data shown in a. The data was split into positive and negative steps and fit with Gaussians. For the negative steps, a single Gaussian was used (blue) whereas for the positive steps two Gaussians were used (red, black). pBW is the fraction of backward steps. (e) Histogram of the off-axis step size distribution of the data shown in a fitted with a single Gaussian. (d, e) We detected 821 steps but only used 676 steps for further quantification because we only counted steps for which the step itself and the previous as well as following step had a standard deviation of less than 4 nm. (a-e) All data shown here is from a single kinesin stepping trace (sample size n =1). However, we analyzed and quantified additional stepping traces of more motors and found very similar results (Supplementary Fig. 20).
Discussion
We developed small (~6-nm), ultra-photostable fluorescent probes. We have shown that six dye DNA FluoroCubes can emit up to 43-fold more photons and have an up to ~54-fold longer half-life than single organic dyes, making them ideal for long-term imaging. Comparing six dye FluoroCubes to single organic dyes, we found that the increase in photostability is dye specific. Currently, the six ATTO 647N dye and six Cy3N dye FluoroCubes demonstrate the most desirable properties, since they have the longest half-life and emit the most photons per probe (Fig. 1). Even the poorest performing six dye FluoroCube (ATTO 488) has a 10-fold longer half-life and emits 6-fold more photons than a single organic dye. Quantum dots are fluorescence emitters with comparable long half-lives. However, quantum dots exhibit significantly more blinking than FluoroCubes, and their larger size and multivalent attachment2,3,5 makes them more challenging to use as a non-invasive probe for protein activity. Applying the six ATTO 647N dye FluoroCube to single-molecule imaging, we were able to track the movement of the motor protein kinesin with nanometer precision over more than 800 steps (Fig. 2) without any evidence of perturbation by the probe.
We investigated potential mechanism(s) that contribute to increased photostability of the six dye DNA FluoroCubes and found that the local environment of the DNA and the dye position (attachment linkage) influence photostability. We also found that increased photostability disappears when the spacing between dyes increases from ~2–6 nm (FluoroCubes) to ~6–10 nm (Compact Cubes13) in a similar DNA-based scaffold. Based on these observations, we speculate that resonance energy transfer between individual dyes in a six dye DNA FluoroCube contributes to the increased photostability. However, a more detailed understanding of the mechanism(s) for the increased photostability awaits further studies. It is also likely that further optimizations could be made to improve photostability. Based on our current work, we suggest that improvements could be achieved by using other dyes such as self-healing fluorophores14, by adding DNA intercalating dyes24, by increasing the number of fluorophores on the FluoroCube8, by changing the spacing between dyes (either by changing the cube size7,13 or by alternating the dye linker length), or by changing the DNA sequence close to the fluorophores25.
DNA FluoroCubes are easily prepared from commercially available reagents and can be attached to all commonly used protein tags, making them simple to use for in vitro studies. We anticipate that DNA FluoroCubes will become the reagent of choice for in vitro and extracellular single-molecule imaging experiments. Beyond single-molecule studies, the long photobleaching-lifetime and high number of total photons of DNA FluoroCubes could prove useful in numerous other fluorescence imaging applications, including FISH26, MERFISH27, DNA-PAINT28, or immunofluorescence for research and medical diagnosis.
Online Methods
Flow-cell preparation.
Flow-cells were assembled as previously described29. Briefly, we made custom three-cell flow chambers using laser-cut double-sided adhesive sheets (Soles2dance, 9474–08x12 – 3M 9474LE 300LSE), glass slides (Thermo Fisher Scientific, 12–550-123), and 170 μm thick coverslips (Zeiss, 474030–9000-000). The coverslips were cleaned in a 5% v/v solution of Hellmanex III (Sigma, Z805939–1EA) at 50° C overnight and washed extensively with Milli-Q water afterwards.
Assembly and analysis of DNA FluoroCubes, Single Dye Cubes, double-stranded DNA, and Compact Cubes.
For each six dye FluoroCube and Single Dye Cube four 32 bp long oligonucleotide strands are required, each modified either with dyes or a functional tag such as biotin or HALO-ligand30 (Supplementary Fig. 1b, Supplementary Table 3, 4). Two oligos were used for the double-stranded DNA with one dye and 28 oligos were used for the Compact Cubes13 (Supplementary Table 3, 4). For each of the four samples oligos were mixed to a final concentration of 10 μM (if not stated otherwise) in folding buffer (5 mM Tris pH 8.5, 1 mM EDTA and 40 mM MgCl2 (if not stated otherwise)) and annealed by denaturation at 85° C for 5 min followed by cooling from 80° C to 65° C with a decrease of 1° C per 5 min. Afterwards the samples were further cooled from 65° C to 25° C with a decrease of 1° C per 20 min and then held at 4° C. Folding products were analyzed by 3.0% agarose gel electrophoresis (if not stated otherwise) in TBE (45 mM Tris-borate and 1 mM EDTA) with 12 mM MgCl2 at 70 V for 2.5 hours on ice and purified by extraction and centrifugation in Freeze ‘N Squeeze columns (BioRad Sciences, 732–6165). The gels were scanned using a Typhoon 9400 scanner (GE Healthcare). A step-by-step protocol on FluoroCube assembly can be found in the Supplementary Protocol and on protocols.io (dx.doi.org/10.17504/protocols.io.8k2huye)31.
Agarose gel-based yield estimation was carried out using ImageJ32. The percentage of FluoroCubes that ran as a monomeric band was estimated as the background-subtracted integrated intensity value divided by the background-subtracted integrated intensity value enclosing the material from the well, down to the bottom of the leading band (single oligos).
Negative stain electron microscopy data collection and processing.
For negative-stain EM, unpurified, but folded FluoroCubes at 300 nM (diluted in FluoroCube Buffer: 20 mM Tris pH 8.0, 1 mM EDTA, 20 mM Mg-Ac, and 50 mM NaCl) were incubated on freshly glow discharged carbon coated 400 mesh copper grids for 1 min and blotted off. Immediately after blotting, a 0.75% uranyl formate solution was applied for staining and blotted off without incubation. This staining was repeated four times and followed by a last incubation for which the stain was incubated for 45 sec before blotting. Samples were allowed to air dry before imaging. Data were acquired at UCSF, on a Tecnai T12 microscope operating at 120 kV, using a 4k×4k CCD camera (UltraScan 4000, Gatan) and a pixel size of 1.7 Å/pixel. For the class average in Figure 1 d, 1,743 Particles were picked and boxed using EMAN 2.2133. Then a 2D classification was performed to remove junk and noisy particles, leading to 983 particles selected.
Mono-Q clean-up of DNA FluoroCubes.
Thermally annealed DNA FluoroCubes were purified using anion exchange chromatography with a GE Source 15Q 4.6/100 PE column (Supplementary Fig. 21). DNA FluoroCubes were bound to the column in Buffer A (20 mM Tris pH 8.0, 1 mM EDTA, 10 mM Mg-Ac, and 10% Glycerol) and afterwards the ionic strength was increased linearly by adding Buffer B (20 mM Tris pH 8.0, 2 M K-Ac, 1 mM EDTA, 10 mM Mg-Ac, and 10% Glycerol) to 100% over 80 min.
Labeling of oligonucleotides with Janelia Fluorophores.
We mixed 5’ and 3’ amino modified oligos (Supplementary Table 3) at a final concentration of 500 µM (in water) with NHS ester modified Janelia Fluorophores JF549 or JF64615 at a final concentration of 5 mM (in DMSO) in 15 mM HEPES pH 8.5 buffer. These solutions were incubated for 4 h at room temperature. We then removed excess dye by four subsequent spins of the solution over Micro Bio-Spin 6 Columns (Bio-Rad) at 700 g for 2 min. The final oligo concentration was determined with a UV spectrophotometer. Afterwards six dye FluoroCubes were assembled as described above.
Preparation of flow-cells with DNA FluoroCubes, Single Dye Cubes, double-stranded DNA, Compact Cubes, biotinylated dyes and quantum dots.
The preparation of flow cells is identical for 6-dye Cubes (FluoroCubes), 1-dye Cubes (Single Dye Cubes), the double-stranded DNA with one dye, biotinylated dyes, and the one and six dye Compact Cubes13. In either case, samples were folded with biotin as the functional tag (except the single, biotinylated dye) and we used unpurified, but folded samples. We first added 10 µl of 5 mg/ml Biotin-BSA (Thermo Scientific, 29130) in BRB80 to the flow-cell and incubated for 2 min. Afterwards, we added another 10 µl of 5 mg/ml Biotin-BSA in BRB80 and incubated for 2 min. Then we washed with 20 µl of FluoroCube Buffer (20 mM Tris pH 8.0, 1 mM EDTA, 20 mM Mg-Ac, and 50 mM NaCl) with 2 mg/ml β-casein (Sigma, C6905), 0.4 mg/ml κ-casein (Sigma, C0406). This was followed by addition of 10 µl of 0.5 mg/ml Streptavidin (Vector Laboratories, SA-5000) in PBS (pH 7.4) and a 2 min incubation. We then washed with 20 µl of FluoroCube Buffer with 2 mg/ml β-casein, and 0.4 mg/ml κ-casein. Next, we either added DNA based samples or the single, biotinylated dye in FluoroCube Buffer or Quantum dots (Qdot™ 655 Streptavidin Conjugate, ThermoFisher Scientific, Q10121MP) and incubated for 5 min. Finally, we washed with 30 µl of FluoroCube Buffer with 2 mg/ml β-casein, and 0.4 mg/ml κ-casein. We then added the PCA/PCD/Trolox oxygen scavenging system16,17 in FluoroCube Buffer with 2 mg/ml β-casein, and 0.4 mg/ml κ-casein for the DNA based samples and or the single, biotinylated dye. For the Quantum dots we added the PCA/PCD oxygen scavenging system16 and 1% β-mercaptoethanol in FluoroCube Buffer with 2 mg/ml β-casein, and 0.4 mg/ml κ-casein.
We note that the concentration of the PCA/PCD/Trolox oxygen scavenging system16,17 is critical and small deviations had large effects on the performance of all samples used in our experiments. We used the following concentrations in all our experiments: 2.5 mM of protocatechuic acid (PCA) (Sigma: 37580) at pH 9.0, 5 units of protocatechuate-3,4-dioxygenase (PCD) (Oriental yeast company Americas Inc.: 46852004), and 1 mM Trolox (Sigma: 238813) at pH 9.5.
Measurements of fluorescence anisotropy, fluorescence lifetime, as well as absorption, excitation and emission spectra.
We determined fluorescence anisotropy, fluorescence lifetime, as well as excitation and emission spectra using an ISS K2 multifrequency fluorometer in bulk measurements. All experiments were performed at room temperature (21–23° C). Instrument settings for the excitation and emission spectra are listed in Supplementary Table 5, settings for the fluorescence anisotropy measurements are listed in Supplementary Table 6, and settings for the fluorescence lifetime measurements are listed in Supplementary Table 7. For the absorption spectra we used an Eppendorf Spectrophotometer (UV-Vis BioSpectrometer) and measured the absorbance from 240 nm to 800 nm. For all samples we used a concentration of 500 nM. In addition, we normalized the absorption spectra based on the 260 nm absorbance.
Dynamic light scattering (DLS) of DNA FluoroCubes and Compact Cubes.
Dynamic light scattering measurements were performed using a Zetasizer ZS90 (Malvern Panalytical) at a wavelength of 633 nm. 60 µl of unpurified FluoroCubes or Compact Cubes were measured at 25°C at a concentration of 5 μM in FluoroCube Buffer (20 mM Tris pH 8.0, 1 mM EDTA, 20 mM Mg-Ac, and 50 mM NaCl).
Kinesin cloning, purification and labeling.
The kinesin construct was cloned and purified as previously described19 except that the GFP was replaced with a HALO-tag30.
The plasmid was transfected and expressed in Agilent BL21(DE3) cells. Cells were grown in LB at 37° C until they reached 1.0 OD600 and the expression was induced by addition of 0.2 mM IPTG. Then the cells were incubated overnight at 20° C. Cells were pelleted and harvested in lysis buffer (25 mM Pipes (pH 6.8), 2 mM MgCl2, 250 mM NaCl, 20 mM imidazole, 1 mM BME, 0.1 mM ATP, and 0.4 mM PMSF), and lysed in the EmulsiFlex homogenizer (Avestin). After a spin in a Sorvall SS-34 rotor for 30 min at 30,000 x g, the supernatant was loaded onto a Ni-NTA resin (QIAGEN, 30210) and washed with additional lysis buffer. We then took 500 μl of beads slur in lysis buffer supplemented with 10 mM MgCl2 and added either six dye ATTO 647N FluoroCubes with a HALO-tag30 to 5 μM final or 5 μM final of HALO-tag TMR dye (Promega). These mixtures were incubated on a Nutator for 3 h at 4 ° C. Afterwards we washed with additional lysis buffer supplemented with 10 mM MgCl2. Then the protein of both labeling reactions was eluted by adding 300 mM of imidazole to the lysis buffer supplemented with 10 mM MgCl2. Subsequently the samples were purified by gel filtration over a S200 10/300GL column from GE Healthcare. Gel filtration buffer was composed of 25 mM Pipes (pH 6.8), 10 mM MgCl2, 200 mM NaCl, 1 mM EGTA, 1 mM DTT, and 10% sucrose. Finally the samples were flash frozen and stored at −80° C.
Preparation of flow-cells with kinesin.
Single-molecule assays with kinesin in flow-cells were prepared as previously described19,21. We first added 10 µl of Alexa 488 labeled axonemes in BRB80 (80 mM Pipes (pH 6.8), 1 mM MgCl2, 1 mM EGTA) and incubated for 5 min. Then, we washed with 60 µl of BRB80 with 1.0 mg/ml κ-casein (Sigma, C0406) supplemented with 5 mM MgCl2. For the comparison between six dye ATTO 647N FluoroCubes labeled kinesin and HALO-tag TMR dye labeled kinesin, we added 10 µl of labeled motor in BRB80 with additional 5 mM MgCl2, 1.0 mg/ml κ-casein, 1 mM ATP, and the PCA/PCD/Trolox oxygen scavenging system16,17. For the high resolution stepping data acquisition, we added 10 µl of six dye ATTO 647N FluoroCubes labeled kinesin in BRB80 with additional 5 mM MgCl2, 1.0 mg/ml κ-casein, 1.5 µM ATP, an ATP regeneration system (1 mM phosphoenolpyruvate (Sigma, 860077), ~0.01 U pyruvate kinase (Sigma, P0294), ~0.02 U lactate dehydrogenase (Sigma, P0294)), and the PCA/PCD/Trolox oxygen scavenging system16,17.
Microscope setup.
All data collections were carried out at room temperature (∼23° C) using a total internal reflection fluorescence (TIRF) inverted microscope (Nikon Eclipse Ti microscope) equipped with a 100× (1.45 NA) oil objective (Nikon, Plan Apo ƛ). We used an Andor iXon 512x512 pixel EM camera, DU-897E and a pixel size of 159 nm. We used two stepping motor actuators (Sigma Koki, SGSP-25ACTR-B0) mounted on a KS stage (KS, Model KS-N) and a custom-built cover to reduce noise from air and temperature fluctuations. A reflection based autofocus unit (FocusStat4) was custom adapted to our microscope (Focal Point Inc.). For the data collection we used a 488 nm laser (Coherent Sapphire 488 LP, 150 mW), a 561 nm laser (Coherent Sapphire 561 LP, 150 mW), and a 640 nm laser (Coherent CUBE 640–100C, 100 mW). A TIRF cube containing excitation filter (Chroma, zet405/491/561/638x), dichroic mirror (zt405/488/561/638rpc), and emission filter (Chroma, zet405/491/561/647m) was mounted in the upper turret of the microscope. The lower turret contained an ET450/50m (Chroma) filter for the 488 nm laser, an ET600/50m (Chroma) filter for the 561 nm laser, and an ET700/75m (Chroma) filter for the 640 nm laser.
Single-molecule TIRF data collection and analysis of DNA FluoroCubes, Single Dye Cubes, double-stranded DNA, biotinylated dyes, Compact Cubes and quantum dots.
The TIRF data of surface immobilized six dye DNA FluoroCubes, Single Dye Cubes, double-stranded DNA with one dye, biotinylated dyes, quantum dots or one and six dye Compact Cubes13 was acquired under continuous laser illumination with an intensity (irradiance) of 120 W/cm2 (488 nm laser), 120 W/cm2 (561 nm laser), and 160 W/cm2 (640 nm laser) and an exposure of 400 ms if not specified otherwise. The acquisition length varied based on the experiment and how fast the respective probe bleached. Thus, before conducting an experiment we pre-established the acquisition length. Typically we either recoded 7,500 frames (3,000 sec), 2,000 frames (800 sec), 1,500 frames (600 sec), or 300 frames (120 sec). We used the camera in conventional CCD mode (i.e., no EM gain). All datasets were acquired with a ‘16 bit, conventional, 3 MHz’ setting and a preamp gain of 5x. All experiments were performed at room temperature (21–23° C). The acquisition software was μManager34 2.0 and data was analyzed in ImageJ32. Single molecules were located and traced using the Spot Intensity Analysis plugin in ImageJ32 (https://imagej.net/Spot_Intensity_Analysis) with the following settings: Time interval of 0.4 sec (except for data in Supplementary Figure 6 for which the exposure time was used as listed in Supplementary Table 8), Electron per ADU of 1.84, Spot radius of 3, Noise tolerance of 100 (except for the Cy3 one dye dsDNA and the single, biotinylated Cy3 dye for which we used a Noise tolerance of 50), and a Median background estimation. The number of frames to check is shown in Supplementary Table 9 since it varies for each sample depending on how fast they bleach. Afterwards the data were further analyzed and plotted with a custom written python script and only localized particles with >500 photons were counted as in the “on” state.
Single-molecule TIRF data collection and analysis of kinesin stepping.
For the comparison between six dye ATTO 647N FluoroCubes labeled kinesin and HALO-tag TMR dye labeled kinesin, the motors were continuously illuminated with an effective exposure time of 0.103 s with a 640 nm laser (160 W/cm2) or a 561 nm laser (120 W/cm2), respectively.
For the high resolution stepping data acquisition, kinesins labeled with six dye ATTO 647N FluoroCubes with a HALO-tag30 were continuously illuminated with a 640 nm laser (160 W/cm2) with an effective exposure time of 0.103 s. We used the camera in conventional CCD mode (i.e., no EM gain). All datasets were acquired with a ‘16 bit, conventional, 3 MHz’ setting and a preamp gain of 5x. All experiments were performed at room temperature (21–23° C). The acquisition software was μManager34 2.0. All emitters were fitted and localized using μManager’s34 “Localization Microscopy” plug-in as previously described29. Parameters for analysis are shown in Supplementary Table 10. Tracks of individual motors were extracted using the μManagers34 “Localization Microscopy” plug-in. We set the minimum frame number to 1000, the maximum number of missing frames to 100, the maximum distance between frames to 100 nm and the total minimum distances of the full track to 500 nm. Then tracks of individual motors were rotated using a principal component analysis (PCA) implemented in python. Afterwards we used a custom Matlab (Matlab R2016b) script to identify individual steps using Chung--Kennedy edge--detecting algorithm as previously described35 and further analyzed the data in a custom written python script. Only steps for which the step itself and the previous as well as following step had a standard deviation of less than 4 nm were considered for further quantification. The velocity over time for the stepping trace of a single kinesin was analyzed with a moving average for which we first binned the data into 2.6 sec bins and then grouped six of these 2.6 sec bins into 15.6 sec bins. We used these 15.6 sec bins to calculate the average velocity at any given point, but used the 2.6 sec intervals to move along the time axis.
Figure preparation.
Figures and graphs were created using ImageJ32 (light microscopy data), Affinity designer (version 1.6.1, Serif (Europe) Ltd) and Python (version 2.7, Python Software Foundation).
Statistics.
For each result the inherent uncertainty due to random or systematic errors and their validation are discussed in the relevant sections of the manuscript. Details about the sample size, number of independent calculations, and the determination of error bars in plots are included in the figures and figure captions.
Data availability
Example raw datasets of DNA FluoroCubes with six dyes, Single Dye Cubes, one dye double-stranded DNA constructs, single, biotinylated dyes, and Compact Cube used to determine the photophysical properties are hosted on Zenodo: https://doi.org/10.5281/zenodo.356102436. All other data files are available from the authors upon request.
Code availability
μManager acquisition and analysis software is available partly under the Berkeley Software Distribution (BSD) license, partly under the GNU Lesser General Public License (LGPL) and development is hosted on GitHub at https://github.com/nicost/micro-manager. The latest version for MacOS and Windows can be downloaded here: https://micro-manager.org/wiki/Download%20Micro-Manager_Latest%20Release (version 2.0 gamma). The custom written Python code used to determine the total number of photons, the average number of photons per frame, and the half-life (photostability) for all fluorescent samples used in this study is hosted on Zenodo: https://doi.org/10.5281/zenodo.362974637. All other code is available from the authors upon request.
Supplementary Material
Acknowledgments
We are grateful to Jongmin Sung (University of California, San Francisco) for critical discussions of the manuscript. We thank Dyche Mullins (University of California, San Francisco) for teaching us how to use the ISS K2 multifrequency fluorometer. We are thankful to Young-wook Jun and Yuetao Zhao (University of California, San Francisco) for teaching us how to use the Malvern Zetasizer ZS90. We thank Luke Lavis (Janelia Research Campus) for the suggestion of comparing sulfonated versus non-sulfonated dyes and for providing the JF549 and JF646 dyes. We are thankful to Shawn Douglas (University of California, San Francisco) and Andrew G. York (Calico Labs) for feedback on the manuscript after pre-printing. Andrew Carter (MRC Laboratory of Molecular Biology) and Elizabeth Villa (University of California, San Diego) supplied the Matlab script for step detection of kinesin. The authors gratefully acknowledge funding from the National Institutes of Health: R01GM097312, 1R35GM118106 (R.D.V., S.N.), and the Howard Hughes Medical Institute (R.D.V. and N.S.).
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Joo C, Balci H, Ishitsuka Y, Buranachai C & Ha T Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem 77, 51–76 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Resch-Genger U, Grabolle M, Cavaliere-Jaricot S, Nitschke R & Nann T Quantum dots versus organic dyes as fluorescent labels. Nat. Methods 5, 763–775 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Zheng Q et al. Ultra-stable organic fluorophores for single-molecule research. Chem. Soc. Rev 43, 1044–1056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shaner NC, Steinbach PA & Tsien RY A guide to choosing fluorescent proteins. Nat. Methods 2, 905–909 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Wichner SM et al. Covalent Protein Labeling and Improved Single-Molecule Optical Properties of Aqueous CdSe/CdS Quantum Dots. ACS Nano 11, 6773–6781 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfbeis OS An overview of nanoparticles commonly used in fluorescent bioimaging. Chem. Soc. Rev 44, 4743–4768 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Schröder T, Scheible MB, Steiner F, Vogelsang J & Tinnefeld P Interchromophoric Interactions Determine the Maximum Brightness Density in DNA Origami Structures. Nano Lett 19, 1275–1281 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Woehrstein JB et al. Sub–100-nm metafluorophores with digitally tunable optical properties self-assembled from DNA. Science Advances 3, e1602128 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rothemund PWK Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–302 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Douglas SM et al. Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res 37, 5001–5006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ke Y, Ong LL, Shih WM & Yin P Three-dimensional structures self-assembled from DNA bricks. Science 338, 1177–1183 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wei B, Dai M & Yin P Complex shapes self-assembled from single-stranded DNA tiles. Nature 485, 623–626 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scheible MB et al. A Compact DNA Cube with Side Length 10 nm. Small 11, 5200–5205 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zheng Q et al. Electronic tuning of self-healing fluorophores for live-cell and single-molecule imaging. Chem. Sci 8, 755–762 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm JB et al. A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 12, 244–50, 3 p following 250 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aitken CE, Marshall RA & Puglisi JD An oxygen scavenging system for improvement of dye stability in single-molecule fluorescence experiments. Biophys. J 94, 1826–1835 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rasnik I, McKinney SA & Ha T Nonblinking and long-lasting single-molecule fluorescence imaging. Nat. Methods 3, 891–893 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Nicoli F et al. Proximity-Induced H-Aggregation of Cyanine Dyes on DNA-Duplexes. J. Phys. Chem. A 120, 9941–9947 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tomishige M, Klopfenstein DR & Vale RD Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science 297, 2263–2267 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Okada Y, Higuchi H & Hirokawa N Processivity of the single-headed kinesin KIF1A through biased binding to tubulin. Nature 424, 574–577 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Yildiz A, Tomishige M, Vale RD & Selvin PR Kinesin walks hand-over-hand. Science 303, 676–678 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Stepp WL, Merck G, Mueller-Planitz F & Ökten Z Kinesin-2 motors adapt their stepping behavior for processive transport on axonemes and microtubules. EMBO Rep 18, 1947–1956 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reddy BJN et al. Heterogeneity in kinesin function. Traffic 18, 658–671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ozhalici-Unal H & Armitage BA Fluorescent DNA nanotags based on a self-assembled DNA tetrahedron. ACS Nano 3, 425–433 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Kretschy N, Sack M & Somoza MM Sequence-Dependent Fluorescence of Cy3- and Cy5-Labeled Double-Stranded DNA. Bioconjug. Chem 27, 840–848 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Femino AM, Fay FS, Fogarty K & Singer RH Visualization of single RNA transcripts in situ. Science 280, 585–590 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Chen KH, Boettiger AN, Moffitt JR, Wang S & Zhuang X RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells. Science 348, aaa6090 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jungmann R et al. Multiplexed 3D cellular super-resolution imaging with DNA-PAINT and Exchange-PAINT. Nat. Methods 11, 313–318 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
References for Online Methods
- 29.Niekamp S et al. Nanometer-accuracy distance measurements between fluorophores at the single-molecule level. Proc. Natl. Acad. Sci. U. S. A (2019) doi: 10.1073/pnas.1815826116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Los GV et al. HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol 3, 373–382 (2008). [DOI] [PubMed] [Google Scholar]
- 31.Niekamp S, Stuurman N & Ronald D Folding of FluoroCubes v1 (protocols.io.8k2huye). doi: 10.17504/protocols.io.8k2huye. [DOI] [Google Scholar]
- 32.Schneider CA, Rasband WS & Eliceiri KW NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tang G et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol 157, 38–46 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Edelstein A, Amodaj N, Hoover K, Vale R & Stuurman N Computer control of microscopes using µManager. Curr. Protoc. Mol. Biol Chapter 14, Unit14.20 (2010). [DOI] [PMC free article] [PubMed]
- 35.Qiu W et al. Dynein achieves processive motion using both stochastic and coordinated stepping. Nat. Struct. Mol. Biol 19, 193–200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niekamp S, Stuurman N & Vale RDA 6-nm ultra-photostable DNA FluoroCube for fluorescence imaging. (2019) doi: 10.5281/ZENODO.3561024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niekamp S, Stuurman N & Vale RD Python code for ‘A 6-nm ultra-photostable DNA FluoroCube for fluorescence imaging’. (2020). doi: 10.5281/zenodo.3629746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Example raw datasets of DNA FluoroCubes with six dyes, Single Dye Cubes, one dye double-stranded DNA constructs, single, biotinylated dyes, and Compact Cube used to determine the photophysical properties are hosted on Zenodo: https://doi.org/10.5281/zenodo.356102436. All other data files are available from the authors upon request.