Abstract
Background
Recently, long non-coding RNAs (lncRNAs) are important populations of non-coding RNAs with defined key roles in normal breast development as well as breast tumorigenesis. Triple-negative breast cancer (TNBC) is a particular breast cancer subtype with poor prognosis because of highly invasive and no specific drug treatment yet. Breast cancer stems cells (BCSCs) cause a high risk of invasion, metastasis and drug resistance for breast cancer patients.
Methods
Two microarrays of BCSCs and no-BCSCs were isolated from mammosphere-3D-cultured MCF-7 cells, differentially expressed lncRNAs (DELs) were screened out by Gene Expression Omnibus (GEO). Gene ontology enrichment analysis and the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were also performed to analyze DELs features. Using the STRING database to analyze DELs interaction network module to further screen the hub lncRNAs related to tumor stemness and make functional annotations. The expressions of hub DELs were validated using data from The Cancer Genome Atlas database. In addition, the expression analysis and survival analysis were conducted using GEO was utilized to analyze DELs in TNBC using GEPIA database.
Results
A total of 143 aberrantly expressed lncRNAs in BCSCs were identified, and 25 lncRNAs were downregulated and 118 lncRNAs were upregulated compared to non-BCSCs. Up- and downregulated top 3 lncRNAs were selected and verified by RT-PCR. Notably, GO enrichment analysis and KEGG pathway analysis indicated that RNA transport, spliceosome, oxidative phosphorylation, NOD-like receptor signaling pathway, PI3K-Akt signaling pathway, and metabolic pathways may serve important roles in BCSCs. Additionally, the function loss assay indicated that ZGRF1 positively upregulated phenotype and biological functions of BCSCs in vitro. Collectively, our work establishes the lncRNAs signature in BCSCs and these findings assess us with evidence to explore further functionalities of lncRNAs in BCSCs and provide a novel therapeutic strategy for breast cancer.
Conclusion
Our work establishes the lncRNAs signature in BCSCs and these findings assess us with evidence to explore further functionalities of lncRNAs in BCSCs and provide a novel therapeutic strategy for breast cancer. ZGRF1 expression is upregulated in TNBC patients and has a poor prognosis, which can be promising biomarkers.
Keywords: lncRNAs, breast cancer stem cells, ZGRF1, triple-negative breast cancer
Introduction
Breast cancer (BC) is the most commonly diagnosed cancer and the leading cause of cancer death in females worldwide,1 while in China BC is currently the most common cancer in women and the fifth leading cause of cancer-related death.2 In the past few decades, breast cancer mortality has been declining, mainly due to the early detection and application of targeted therapy.3,4 The invasion and metastasis of BC are the main causes of BC-related deaths.
Breast cancer stems cells (BCSCs) are the tumor-derived cells with self-renewal and unlimited proliferation potential, which cause a high risk of disease invasion and metastasis. Therefore, eliminating BCSCs may provide a therapeutic avenue for overcoming chemoresistance, metastasis, and relapse in BC patients. Al-Hajj M and his colleagues provided the first evidence for the existence of BCSCs, and they revealed that only the minority of BC cells with a CD44+/CD24−/low/ESA+ phenotype could form new tumors in NOD/SCID mice.5 Besides, BCSCs also presented with features of enhanced “side population” (SP) cells that efflux the Hoechst dye via ABCG2, high aldehyde dehydrogenase 1 (ALDH1) expression, and the CD44+/CD24− phenotype. Furthermore, BCSCs possess the property to form mammospheres, which are thought to be enriched mammary stem cells.6–8
Of note, BCSCs cause a high risk of invasion, metastasis and drug resistance for breast cancer patients. Therefore, there is an urgent need to discover new molecules to directly target BCSCs for preventing breast cancer progression and recurrence. Long non-coding RNAs (lncRNAs) were an important population of non-coding RNAs with defined key roles in normal development as well as tumorigenesis. Deregulations of several lncRNAs, such as HOTAIR, XIST, MALAT, H19, etc. have been detected in breast cancer samples and cell lines.9 In addition, the association between lncRNAs signature and BC patients’ survival has been assessed in various studies.10,11 Although recent findings have indicated that lncRNAs have important roles in regulating tumorigenicity and stemness of BC, little is known about the total lncRNAs profiles and mechanisms by which lncRNAs control BCSCs maintenance. Here, the expression patterns of lncRNAs in BCSCs, as well as their biological functions and significance in prognosis are discussed.
Materials and Methods
Cell Lines, Culture Conditions
Human breast cancer cell lines MCF-7 were obtained from ATCC and grown in RPMI 1640 medium (HyClone) supplemented with 1% penicillin-streptomycin, 10% fetal bovine serum at 37°C with 5% CO2 (2D culture). The sphere (cancer stem cell) medium was similar to stem-cell culture medium of human samples with serum-free suspension in RPMI medium supplemented with antibiotic-antimycotic (1%, Thermo Fisher Scientific), insulin (5 mg/mL, Sigma), EGF (10 ng/mL, Invitrogen), FGF (20 ng/mL, Invitrogen), B27 (10 μL/mL, Invitrogen) and heparin (4 μL/mL, Invitrogen). Cells were cultured in 6-well low adhesion plates (HyClone) for 14 days (3D culture). Each group of cells was observed by an inverted microscope (HyClone). Photographs were taken at 20× and 60× magnifications, separately.
Western Immunoblotting
Total cell proteins were extracted with RIPA Lysis and Extraction Buffer (Thermo Scientific) supplemented with 1% protease inhibitor cocktail, 1mM PMSF, 10mM NaF, 1mM Na3VO4 (Sigma). Cell lysates were subjected to 10% SDS-PAGE, transferred to nitrocellulose membrane and probed with primary antibodies, followed by HRP-linked secondary antibodies (Sigma-Aldrich and Abcam) and detected using Super Signal chemiluminescence reagent (34095, Thermo Scientific). The antibodies used for immunoblotting were rabbit anti-CD24 (ab202073, Abcam, 1:1000), rabbit anti-CD44 (ab157107, Abcam 1:1000), rabbit anti-ALDHA1 (ab52492, Abcam 1:1000), rabbit anti-SOX2 (ab93689, Abcam 1:1000), and mouse anti-β-Actin (A1978, Sigma-Aldrich, 1:4000).
Acquisition and Analysis of Microarray Data
Procedures of microarray analysis were as follows: 1) the sample was divided into two groups, the 2D culture group (n=3) and the 3D culture group (n=3). 2) Three qualified RNA samples were amplified and transcribed into fluorescent RNAs for each group. 3) After labeling and purifying, 1ug of RNAs were used for hybridization on microarrays (GeneChip Human Exon 1.0 ST Array). 4) Microarrays were washed, fixed, and scanned using an Agilent DNA Microarray Scanner. The annotation of lncRNAs was accessed directly from additional files of the microarrays or query from the LNCipedia database (http://www.lncipedia.org, version 5.0). If a lncRNA has an official gene symbol according to Hugo Gene Nomenclature Committee (HGNC), that symbol was used as the name of lncRNA. Otherwise, the accession number or gene ID was used. We then screen out significant differentially expressed lncRNAs for further analysis with the criteria of |lgFC| ≥2.5 and P-value ≤0.05. In order to reduce the difference screening error caused by small samples, we compare these two groups with the T-test modified by random variance model (RVM), calculating the significance level between the differential genes (p < 0.05 were considered to have statistical significance for DELs). The software Morpheus (https://software.broadinstitute.org/morpheus/) was used to draw heatmap.
Functional Enrichment Analysis of Differentially Expressed lncRNAs
The Database for Annotation, Visualization, and Integrated Discovery (DAVID; v.6.8) provides a synthetic set of functional annotation tools for users to reveal the biological significance among a huge scale of genes.12,13 Gene Ontology (GO) (http://www.geneontology.org/) function enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp/kegg/) pathway enrichment analysis were performed using DAVID online tool.14,15 GO functional categories include molecular function (MF), biological process (BP), and cellular component (CC). In this study, GO functions assembled by differentially expressed lncRNAs (DELs) in the severe group, as well as KEGG pathways enriched by the co-regulated DELs were investigated. P <0.05 and enriched gene numbers (count) ≥2 were considered as cut-off values.
STRING Network Analysis
We constructed STRING networks for the up- and downregulated DELs, separately. A cytoscape app that can be used to construct STRING networks (https://string-db.org/) was performed a network-based analysis of our data to extract the hub genes.16 In this network, each node represents a gene. An edge between two nodes represents an interaction between two genes in the network. The edge weights corresponding to the Pearson correlation coefficient between the two genes. The STRING network was then subjected to network clustering analysis with the Markov clustering algorithm and the most highly connected network modules were extracted from the network, in which the hub gene in the module was referred to the genes with the largest interaction.17,18
GEPIA Analysis
To further study the expression and prognosis of DELs with TNBC, the cancer genome atlas and the genotype-tissue expression database (GEPIA) (http://www.cbioportal.org/) were performed to analysis the expressional and prognostic patterns of DELs in the triple-negative breast cancer (TNBC) and non-triple-negative breast cancer (non-TNBC).19,20
RNA Isolation and Real-Time Polymerase Chain Reaction (PCR)
Total RNAs were prepared from cells using Trizol reagent (Invitrogen, CA, USA) and cleaned up using the RNeasy Mini Kit (Qiagen, Hilden, Germany). Samples with optical density (OD) 260/280 ratio between 1.8 and 2.1 and OD260/230 ratio >1.8 were considered as qualified. The cDNAs were synthesized with the PrimerScript RT reagent Kit (Takara, Otsu, Shiga, Japan). The real-time PCR reactions were performed according to the protocols of the SYBR Premix Ex Taq kit (Takara) and using StepOnePlus Real-Time PCR System (Applied Biosystems, USA). The fluorescence data of the detected genes were normalized to the expression of β-actin using the 2-ΔΔCT method. The primers utilized in our investigation are listed in Table 1.
Table 1.
The lncRNA Primer for q-PCR Validation
| Up-lncRNA | Down-lncRNA | ||
|---|---|---|---|
| Primer | Sequence | Primer | Sequence |
| 5′-GATGCTTGTGAACTAATTGAAAGGT-3′ | 5′-GACCCTCGGCCTGAGAAC-3′ | ||
| TMA16 | 3′-CCCTGCCTGTCCCTGATAC-5′ | LOC105378960 | 3′-GGAAGGGACTGGGTGAAAG-5′ |
| 5′-TATGTGACAGAGTGGGGCCTA-3′ | 5′-GGAAGCAGGCAGGATACTTG-3′ | ||
| GOLPH3L | 3′-AGGCCTCCATATGGGTGAAG-5′ | MIR6727 | 3′-CACTGCCACTACCTAATCAGCA-5′ |
| 5′-TTTCTGCTTTGTTTGGGACA-3′ | 5′-AAGGACAGAAACACGGCGAT-3′ | ||
| ETFA | 3′-CCTGTTATGATTCAGCTTCCATT-5′ | MARCH5 | 3′-GTCTGAAGGGATGCACGGAT-5′ |
Transient Transfection of siRNA
Control and ZGRF1-specific siRNAs were synthesized by Shanghai GenePharma Co., Ltd (Shanghai, China). Transient transfections of ZGRF1 siRNA were used for transfection into MCF-7 cells according to the manufacturer’s protocols.
The siRNA sequences:
ZGRF1-homo-sence1 CCUUGAGAUUCCAGACAUUTT; ZGRF1-homo-anti-sence 1 AAUGUCUGGAAUCUCAAGGTT.
ZGRF1-homo- sence 2 GGAGCAGAUUGAGUUACAUTT; ZGRF1-homo- anti-sence2 AUGUAACUCAAUCUGCUCCTT.
Cells were collected 48 h after transfection. Total cell protein was extracted with RIPA Lysis and Extraction Buffer (Thermo Scientific), and then total RNA was extracted using the Trizol (Invitrogen).
Sphere Formation
Cells plated at a density of 50 cells per well with serum-free suspension in RPMI medium supplemented with antibiotic-antimycotic (1%, Thermo Fisher Scientific), insulin (5 mg/mL, Sigma), EGF (10 ng/mL, Invitrogen), FGF (20 ng/mL, Invitrogen), B27 (10 μL/mL, Invitrogen) and heparin (4 μL/mL, Invitrogen). After 2 weeks of cultivation, these cells were observed by an inverted microscope. Photographs were taken at 20× and 60× magnifications separately. The relative numbers of spheres were counted.
Colony Formation Assay
Cells were suspended in 0.3% agarose (Sigma-Aldrich) and placed at a density of 50 cells per well. After 2 weeks of cultivation, the cells were fixed and stained with 10% formalin and 0.1% crystal violet. The relative numbers of colonies were counted.
Flow Cytometry Analysis
Antibodies for the human antigens CD24 (PE 1:100, eBioscience) and CD44 (APC 1:100, eBioscience) were used for detecting the proportion of breast cancer stem cells (CD44+/CD24−). Antibodies are validated according to the manufacturer’s website. The side scatter width versus side scatter region (SSC-W versus SSC-A) and the forward scatter width versus forward scatter height (FSC-W versus FSC-H) were used to eliminate dead cells and cell clumps.
Results
Suspension Culture Promotes Cancer Stemness
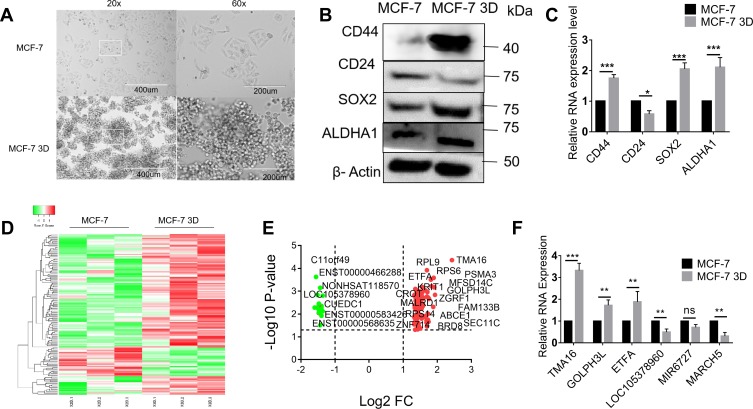
MCF-7 breast cell lines were subjected to the adherent culture (2D group) and the suspension culture (3D group). Cells of each group were observed by an inverted microscope after 14 days’ culture. The results showed that cells of the 2D group grew in an adherent pattern, neatly arranged, with clear cell boundary and spindle-shaped. In the 3D group, cells grew in suspension and globular aggregated (Figure 1A). Generally, CD44 high CD24 low subgroup of BC cells was recognized to be BCSCs.21 Compared to cells of 2D culture groups, cells of 3D culture groups enriched breast cancer stem cells easily and efficiently, of which CD44+/CD24−, SOX2, as well as ALDHA1 had a higher expression at the protein level and mRNA level, respectively, by Western immunoblotting and q-PCR (Figure 1B and C).
Figure 1.
Suspension culture promotes cancer stemness and dysregulated lncRNAs in BCSCs: (A) MCF-7 breast cell lines were subjected to 2D and 3D culture groups of each cell were observed by an inverted microscope. (B) Western blotting for detecting the expression of stem marker CD44, CD24, ALDHA1, SOX2. (C) q-PCR detected the expression of stem marker CD44, CD24, ALDHA1, SOX2. (D, E) Volcano map and heatmap for 143 DELs. (F) qRT-PCR validation differential expression of top 3 Downregulated (LOC105378960, MIR6727, MARCH5) and upregulated (TMA16, GOLPH3L, ETFA) hub gene Two-tailed Student’s t test was used for statistical analysis. *P < 0.05, **P < 0.01, and ***P < 0.001 and error bars represented standard deviation from three times of independent experiments.
Dysregulated lncRNAs in BCSCs
A total of 143 DELs (FC ≥ 2.50 or ≤0.50) comprising 25 downregulated and 118 upregulated genes were obtained from human lncRNA microarray. Volcano map and heatmap for 143 DELs (Figure 1D–E), among which there are top 20 most significantly up- and downregulated DELs listed in Table 2. To validate the results of the array, down- and upregulated top 3 hub genes were selected for confirmation of microarray results using qRT-PCR (Figure 1F). TMA16, GOLPH3L, ETFA were upregulated, LOC105378960, MIR6727 and MARCH5 were downregulated. The results were consistent with the microarray.
Table 2.
Top 20 Most Significantly Upregulated and Downregulated DELs
| Up-lncRNA | Down-lncRNA | ||
|---|---|---|---|
| LncRNA | Fold change | LncRNA | Fold change |
| TMA16 | 5.39 | LOC105378960 | 0.39 |
| GOLPH3L | 3.82 | MIR6727 | 0.39 |
| ETFA | 3.76 | 5-Mar | 0.38 |
| RPS6 | 3.72 | LOC101928402 | 0.38 |
| FAM133B | 3.68 | LOC105370802 | 0.38 |
| MFSD14C | 3.56 | CXCR4 | 0.37 |
| PSMA3 | 3.51 | TUBA1B | 0.36 |
| ZGRF1 | 3.38 | SPRED3 | 0.36 |
| MRPL42 | 3.35 | C11orf49 | 0.34 |
| CD44 | 3.32 | CUEDC1 | 0.33 |
| ABCE1 | 3.32 | n410150:3108009 | 0.39 |
| TBC1D15 | 3.31 | n372417:3338127 | 0.39 |
| CMTR2 | 3.27 | ENST00000496353 | 0.39 |
| RPL9 | 3.24 | n411152:2894191 | 0.39 |
| KRIT1 | 3.24 | n345209:3482792 | 0.38 |
| SEC11C | 3.24 | n376618:3900407 | 0.38 |
| BRD8 | 3.21 | ENST00000466288 | 0.37 |
| THEM4 | 3.21 | n325109:2604194 | 0.36 |
| NCL | 3.19 | ENST00000332858 | 0.36 |
| PRKCI | 3.15 | ENST00000553466 | 0.36 |
Module-Based Functional Characterization of DELs with Co-Expression Network Analysis
To explore potential functions of the 143 DELs, we used STRING to construct a co-expression network based on their normalized expression data of the 143 DELs. Finally, 6 cancer stem cell-associated modules that contained 60 hub-lncRNAs were selected for downstream analysis. Details of the 6 modules are listed in Table 3.
Table 3.
Overview of 6 Breast Cancer Stem Cell-Associated Modules
| Module | PCGs Counts | Onco-lncRNAs Counts | Knowm-Function lncRNA | Module Cancer P value | Functional Category |
|---|---|---|---|---|---|
| Red | 10 | 11 | CCT8,RPL9,RPS14,DNAJC2,ABCE1,SERBP1,NCL,RPS6,SHOC2,SEC11C,TEX10 | 2.87E-07 | RNA transport |
| Cyan | 12 | 7 | CMTR2,DMTF1,ZRANB2,RNPS1,THOC2 | 3.06E-14 | Spliceosome |
| Orchid | 6 | 2 | MRPL42,ATP5A1 | 5.26E-08 | Oxidative phosphorylation |
| Violet | 6 | 1 | CROT | 0.00000285 | NOD-like receptor signaling pathway |
| Purple | 4 | 2 | CXCR4,PRKCI | 0.00014 | PI3K-Akt signaling pathway |
| Other | 43 | 23 | L3HYPDH,GOLPH3L,RBM47,API5,TMEM116,INHBA-AS1,TTC17 | 2.22E-05 | Metabolic pathways |
Functional Characterization of Hub Genes
To further interpretation of the function of 143 DELs, KEGG and GO annotations were performed in DAVID.22
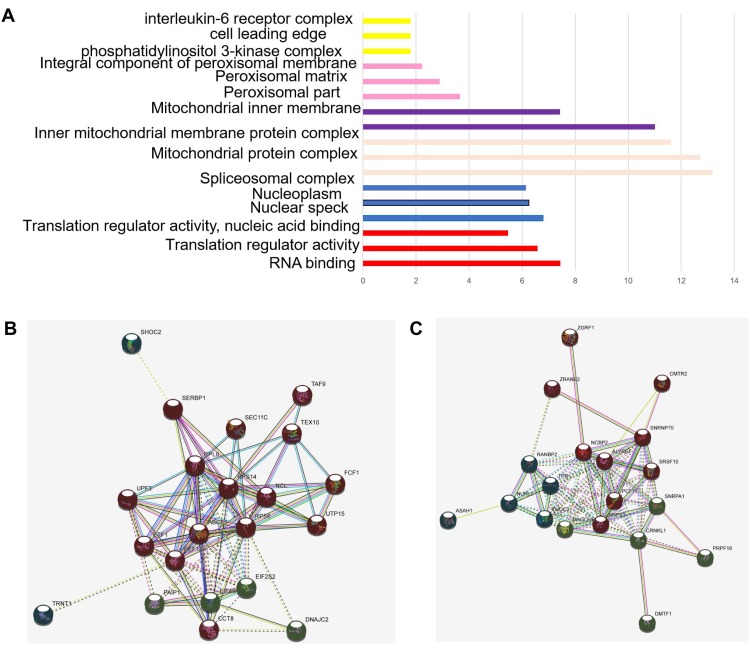
For BP, the DELs were mainly involved in biological process such as RNA binding translation regulator activity, translation regulator activity, nucleic acid binding, nuclear speck nucleoplasm spliceosomal complex, mitochondrial protein complex, inner mitochondria, integral component of peroxisomal membrane phosphatidylinositol 3-kinase complex, cell leading edge, interleukin-6 receptor complex and delta DNA polymerase complex ciliary neurotrophic factor receptor complex (Figure 2C). Regarding KEGG, the top six enriched pathways were RNA transport, spliceosome, oxidative phosphorylation, NOD-like receptor signaling pathway, PI3K-Akt signaling pathway, and metabolic pathways (Table 4).
Figure 2.
Network and expression levels of the red and cyan modules: (A, B) in which only edges with weight (w) above a threshold of 0.4 are displayed. Line color indicated the type of interaction evidence. (C) Bar plot of representative GO biological process (BP) terms of 6 modules. The y-axis depicted names of BP terms, and the x-axis depicted −log10 (P-value). Bar color denoted the module color. The red dotted line denoted a p-value of 0.05.
Table 4.
Significant KEGG Pathways (P≤0.05, Counts ≥5) Associated with 6 Breast Cancer Stem Cell-Associated Modules
| Module | Entry | Name | Count | P-value |
|---|---|---|---|---|
| hsa03013 | RNA transport | 6 | 2.87E-07 | |
| hsa03008 | Ribosome biogenesis in eukaryotes | 3 | 0.00072 | |
| Red | hsa03010 | Ribosome | 3 | 0.0022 |
| hsa03015 | mRNA surveillance pathway | 2 | 0.019 | |
| hsa04150 | mTOR signaling pathway | 2 | 0.04 | |
| hsa03040 | Spliceosome | 9 | 3.06E-14 | |
| Cyan | hsa03013 | RNA transport | 8 | 8.24E-12 |
| hsa03015 | mRNA surveillance pathway | 5 | 8.41E-08 | |
| hsa00190 | Oxidative phosphorylation | 7 | 6.50E-14 | |
| hsa04714 | Thermogenesis | 7 | 1.43E-12 | |
| hsa05012 | Parkinson’s disease | 6 | 1.71E-11 | |
| hsa05010 | Alzheimer’s disease | 6 | 0.0074 | |
| Orchid | hsa05016 | Huntington’s disease | 6 | 6.20E-11 |
| hsa04932 | Non-alcoholic fatty liver disease (NAFLD) | 5 | 2.85E-09 | |
| hsa01100 | Metabolic pathways | 7 | 5.26E-08 | |
| hsa04723 | Retrograde endocannabinoid signaling | 3 | 3.37E-05 | |
| hsa04621 | NOD-like receptor signaling pathway | 4 | 2.85E-06 | |
| hsa05164 | Influenza A | 4 | 2.85E-06 | |
| hsa05162 | Measles | 3 | 7.54E-05 | |
| hsa05152 | Tuberculosis | 3 | 0.00012 | |
| Violet | hsa04060 | Cytokine-cytokine receptor interaction | 3 | 0.00034 |
| hsa04623 | Cytosolic DNA-sensing pathway | 2 | 0.00091 | |
| hsa05140 | Leishmaniasis | 2 | 0.00091 | |
| hsa05321 | Inflammatory bowel disease (IBD) | 2 | 0.00091 | |
| hsa05132 | Salmonella infection | 2 | 0.0011 | |
| hsa04620 | Toll-like receptor signaling pathway | 2 | 0.0014 | |
| hsa05142 | Chagas disease (American trypanosomiasis) | 2 | 0.0014 | |
| hsa05145 | Toxoplasmosis | 2 | 0.0014 | |
| hsa04380 | Osteoclast differentiation | 2 | 0.0016 | |
| hsa04650 | Natural killer cell mediated cytotoxicity | 2 | 0.0016 | |
| hsa04217 | Necroptosis | 2 | 0.0022 | |
| hsa04630 | Jak-STAT signaling pathway | 2 | 0.0022 | |
| hsa05167 | Kaposi’s sarcoma-associated herpesvirus infection | 2 | 0.0026 | |
| hsa05168 | Herpes simplex infection | 2 | 0.0026 | |
| hsa05200 | Pathways in cancer | 2 | 0.0179 | |
| hsa04151 | PI3K-Akt signaling pathway | 4 | 0.00014 | |
| hsa04015 | Rap1 signaling pathway | 3 | 0.00019 | |
| Purple | hsa04062 | Chemokine signaling pathway | 3 | 0.00019 |
| hsa04360 | Axon guidance | 3 | 0.00019 | |
| hsa04611 | Platelet activation | 3 | 0.00019 | |
| hsa04630 | Jak-STAT signaling pathway | 3 | 0.00019 | |
| hsa01100 | Metabolic pathways | 15 | 2.22E-05 | |
| hsa03430 | Mismatch repair | 3 | 0.0021 | |
| hsa04950 | Maturity onset diabetes of the young | 3 | 0.0021 | |
| hsa03030 | DNA replication | 3 | 0.0028 | |
| other | hsa03410 | Base excision repair | 3 | 0.0028 |
| hsa00510 | N-Glycan biosynthesis | 3 | 0.0047 | |
| hsa03420 | Nucleotide excision repair | 3 | 0.0047 | |
| hsa05200 | Pathways in cancer | 7 | 0.0047 | |
| hsa04210 | Apoptosis | 4 | 0.0049 |
Co-Expression Network and Hub Genes
The co-expression analysis (STRING) showed that there were 60 DELs and 754 protein-coding genes (PCGs) whose expressions were significantly correlated. A co-expression network of these DELs was constructed based on their correlation coefficients. The 20 top hub-genes were selected out and visualized. The red and cyan modules of significantly enriched terms were shown in (Figure 2A and B).
ZGRF1 Expression Is Upregulated in TNBC Patients and Has a Poor Prognosis
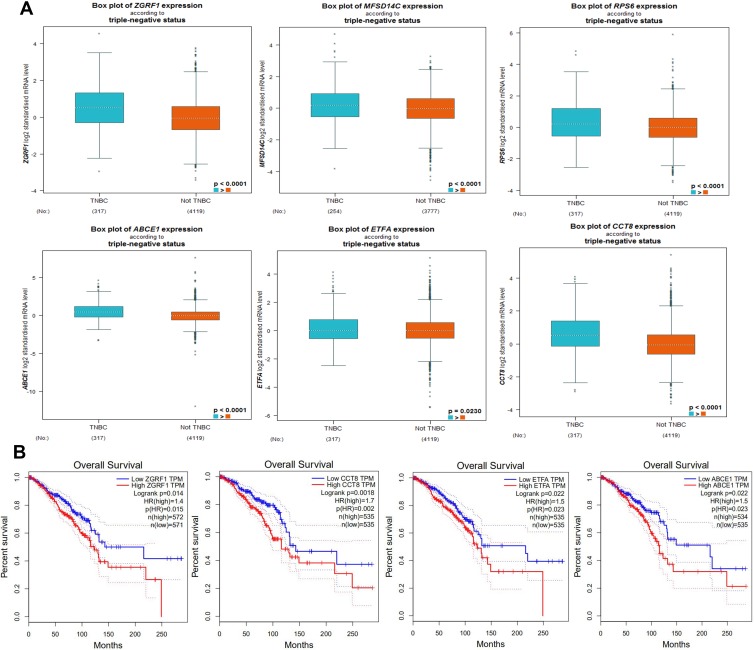
Due to the claudin-low subtype of breast cancer, especially TNBC, was enriched with an EMT gene signature and stem cell-like features.23,24 To further verify the expression in breast cancer tissue with high stem cells, we selected TNBC (n=317) as high BCSCs and non-TNBC (n=4119) patient samples as low BCSCs in the GEPIA database for further analyzed the expression level of 20 top hub-genes, we found that ZGRF1, ETFA, RPL9, MFSD14C, ABCE1, and CCT8 showed higher expression level than those in non-TNBC patient samples (Figure 3A). To explore the prognostic values of the significantly hub-genes, we analyzed the associations of the expression level of them in the GEPIA database with the survival of TNBC patients. ZGRF1, ETFA, ABCE1, CCT8 were found to associate with overall survival (OS) of TNBC (P < 0.05, Figure 3B). What is more, patients with high expression of ZGRF1 had more beneficial for OS (HR = 1.4) than TNBC patients (Figure 3B).
Figure 3.
Expression and prognostic value of hub genes in TNBC and not-TNBC: (A) the expression of ZGRF1, ETFA, RPL9, MFSD14C, ABCE1 and CCT8 between TNBC (n=317) and not TNBC patient samples (n=4119) in GEPIA database. (B) the prognostic value of the ZGRF1, ETFA, ABCE1 and CCT8 were found to associate with OS of TNBC (P < 0.05 n=572) (GEPIA).
ZGRF1 Promoted Cancer Stemness
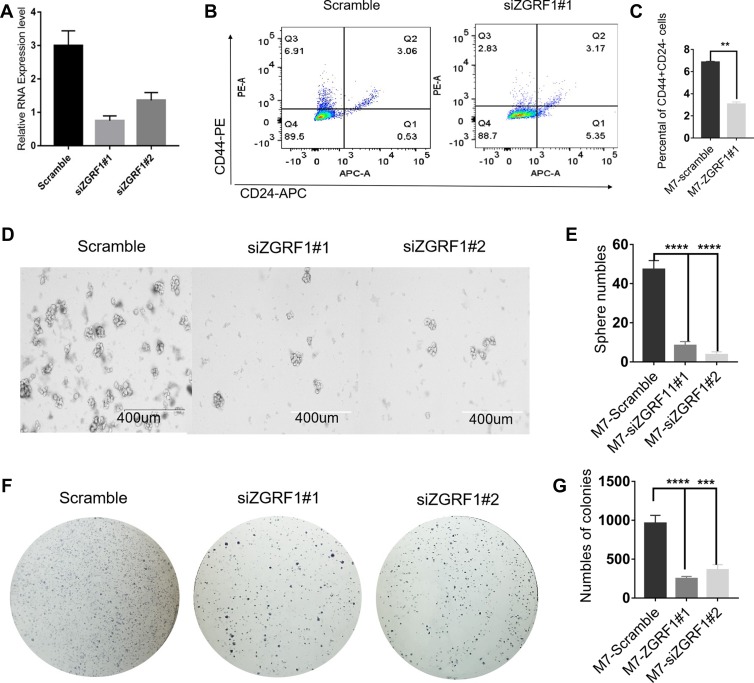
To further explore the biological functions of ZGRF1 in BCSCs regulation, we used two siRNA to interfere ZGRF1. And qRT-PCR analysis revealed the efficiency of the siRNA at 48 h after transfection was 85% and 75%, respectively (Figure 4A). ZGRF1 knockdown decreases the number of cells with CD44+/CD24− phenotype (Figure 4B and C).ZGRF1 knockdown decreased sphere formation efficiency, including the size and numbers of spheres (Figure 4D and E), colony formation capacity and numbers (Figure 4F and G) revealed that ZGRF1 promoted cancer cell stemness.
Figure 4.
ZGRF1 promoted cancer stemness: (A) Real-time PCR analysis revealed interference efficiency of the siRNA against ZGRF1 at 48 h after transfection was 85% and 75%, respectively. (B, C) Flow cytometry analyses of the ratios with CD44+/CD24− cells in the indicated cells with or without ZGRF1 knockdown were performed. (D, E) Tumor spheres of representative images of the indicated cells with or without ZGRF1 Knockdown; Scale bar = 400 μm. Histograms showed the mean numbers and diameters of spheres cultured. (F, G) Colony formation assay (n = 3). **P < 0.01, ***P < 0.001, and ****P < 0.0001 and error bars represent standard deviation from three times of independent experiments.
Statistical Analysis
All data were statistically analyzed with GraphPad Prism 6.0 software. Two-tailed t test was utilized to analyze the difference between the two groups. Pearson’s test was applied to determine the correlation between clinicopathological parameters and protein expression. Data were presented as mean ± SD or SEM. Differences at P < 0.05 were considered statistically significant.
Discussion
Tumors contain a functional subpopulation of cells that exhibit stem cell properties. These cells, named cancer stem cells (CSCs), play significant roles in the initiation and progression of cancer. LncRNAs are defined as a large class of transcripts longer than 200 nucleotides, with no function of encoding proteins, which act at the transcriptional, post-transcriptional and translational level. Previous studies indicate that lncRNAs may be involved in various biological processes such as DNA damage repair, metabolism, cell growth, survival, differentiation, and signaling. Accumulating evidence indicate that lncRNAs are key regulators of the cancer stem cell (CSC) subpopulation, including breast cancer.25 Thereby contributing to cancer progression. Our research in the function and mechanism of lncRNAs in the initiation, maintenance and regulation of BCSCs derived from MCF-7 cell line vie 3D mammosphere culture in order to search for new targets for cancer treatment through lncRNAs to selectively eliminate BCSC and ultimately improve clinical outcomes of breast cancer patients.
In our previous study, the mammosphere formation efficacy (MFE) certified that there were cancer stem cells in MCF-7 which was 11.7 per 1000 cells and suggested that mammospheres enriched a part of BCSCs.26 In this study, MCF-7 cells were cultured in suspension to generate mammospheres. The expression of stem cell-related genes of CD44, CD24, SOX2, and ALDHA1 was detected by quantitative real-time PCR (qRT-PCR) (Figure 1C) and western bolt (Figure 1B). We investigated the expression profile of lncRNAs in BCSCs with the 3D culture mammospheres (secondary mammospheres) from MCF-7 and MCF-7 2D culture (non-BCSCs) via microarray analysis (Figure 1A). To the best of our knowledge, we are the first to report the lncRNA signatures in BCSCs. In the present study, we obtained a total 143 variations of the lncRNA between the BCSCs and non-BCSCs (Figure 1D). Importantly, among the 143 lncRNAs identified, 118 lncRNAs were upregulated and 25 were downregulated (Figure 1F). In addition, Up-and-down regulated top 3 lcnRNAs (TMA16, GOLPH3L, ETFA, LOC105378960, MIR6727, and MARCH5) were selected and verified with qRT-PCR (Figure 1G). These findings suggested that lncRNAs were really differentially expressed in BCSCs, and they might be related to CSC initiation, differentiation and regulation.
Previous studies showed that lncRNAs were important in the genesis and progression of cancer. Various lncRNAs, such as ROR, HOTAIR, H19, UCA1, and ARSR were involved in cancer stemness. These lncRNAs could regulate the expression of CSC-related transcriptional factors, such as SOX2, OCT4, and NANOG, in colorectal, prostate, bladder, breast, liver, and other cancer types.27 For example, it was reported that the lncRNA FEZF1-AS1 promoted breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis.28 While LincK contributed to breast tumorigenesis by promoting proliferation and epithelial-to-mesenchymal transition.29 TNBC is a malignant subtype of breast cancer with the absence of targeted therapy. In recent years, lncRNAs have been implicated in multiple biological functions in TNBC. LncRNA NEAT1 conferred the oncogenic role in TNBC through modulating chemoresistance and cancer stemness.30 Furthermore, these data identified lncRNA NRAD1 as a downstream effector of ALDH1A3, and a target for TNBCs and CSCs, functioning in cell survival and regulation of gene expression.31 In summary, our study, at least partially, corroborates previously published data and validates the important roles of lncRNAs in breast cancer stemness and tumorigenesis, providing a novel insight in the treatment strategy for breast cancer.
Currently, due to dramatic advances in bioinformatics and sequencing technologies, increasing numbers of lncRNAs have been found. Recent studies have reported that lncRNAs play critical roles in the regulation of BC invasion and metastasis through a variety of molecule mechanisms, such as participating in epithelial-to-mesenchymal transition, strengthening cancer stem cell generation, serving as competing endogenous lncRNAs, influencing multiple signaling pathways, as well as regulating expressions of invasion-metastasis-related factors, including cell adhesion molecules, extracellular matrix, and matrix metalloproteinases. In the current study, GO and KEGG pathway analyses were consulted to predict the potential roles of the lncRNAs that were identified in the present study. GO analysis showed that most frequently enriched functions of lncRNAs were involved in a membrane protein complex, regulation of peroxisomal matrix, RNA binding, and nuclear speck. These results indicated that the lncRNAs identified here might be involved in RNA transportation, oxidative phosphorylation, PI3K-Akt and metabolic pathways to some extent (Figure 2C). Additionally, the KEGG pathway analysis revealed that the thermogenesis, oxidative phosphorylation, PI3K-Akt pathway and metabolic signaling pathway were among the top predicted pathways (Table 3). These pathways were demonstrated in sustaining the stemness of breast cancer cells and took part in regulating the behaviors of CSCs in previous studies.32–34 Therefore, our data had provided a panoramic view of the pathways underpinning the contribution of lncRNAs to BCSC stemness and regulation, which may lay the foundation for the development of new strategies to prevent BC invasion and metastasis.
The potential functionalities of lncRNAs in BCSCs remain unexplored. Generally, the functions of lncRNAs were predicted through their counterpart mRNAs. Previous reports showed that lncRNAs LINC00511 contributed to breast cancer tumorigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis.35 The other function patterns for lncRNAs are mediating signal transduction. For example, LINC01638 lncRNA activated MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in TNBC.36 A recent study also reported that lncRNA NEAT1 conferred an oncogenic role in TNBC through modulating chemoresistance and cancer stemness.30 To explore the identity and putative function of selected lncRNAs, we analyzed the most significantly differentially expressed lncRNA related genes from KEGG pathway analysis. We did a correlation analysis of 20 top hub genes in the red and cyan module (Figure 2A and B). TNBC is a malignant subtype of BC with the absence of targeted therapy, resulting in poor prognosis in patients. We know that the current cancer therapies fail to repress tumor recurrence and metastasis in TNBC because they fail to target cells that possess epithelial–mesenchymal (E-M) plasticity and acquire cancer stem cell (CSC) properties.23,37,38 To examine hub lncRNAs difference expression level between TNBC and non-TNBC samples. GEPIA database was used to analyze (http://www.cbioportal.org/). Among 40 top hub genes, 6 were highly expressed in the samples of TNBC than in non-TNBC (Figure 3A). In order to explore the prognostic values of the significantly lncRNAs, we analyzed the associations of the expression level of 40 top hub-genes in the co-expression network with the survival statistics of breast cancer patients. Four lncRNA-related genes were found associating with overall survival (OS) of TNBC patients (P < 0.05, Figure 3B). Patients with an elevated level of ZGRF1 tend to have a poor overall survival (HR = 1.4 and P=0.015). The encoded protein contains GRF zinc finger (zf-GRF) and transmembrane domains. GRF zinc fingers are found in a number of DNA-binding proteins. Alternative splicing results in multiple transcript variants encoding different isoforms. Previous genome-wide association studies have identified that ZGRF1 was associated with obesity39 and childhood apraxia of speech.40 Therefore, based on these findings, we hypothesized that ZGRF1 might exert stimulating effects on BCSCs. Here we testified that BCSCs expressed high levels of ZGRF1, and ectopic overexpression of ZGRF1 significantly promoted breast cancer cell clonogenicity, migration, and mammosphere-forming ability. Conversely, silencing of ZGRF1 repressed these BCSCs properties. In concordance, knockdown of ZGRF1 markedly inhibits tumor growth and suppresses tumorigenesis. To confirm our hypothesis, we designed siRNAs to knock down the expression of ZGRF1 in MCF-7 and explored its effects on the stemness of BCSCs. Based on the BCSC concept, breast cancer is derived from a small fraction of breast cancer cells with the CD44+CD24- phenotype.41 Therefore, we tested the impact of ZGRF1 on the BCSC phenotype. The results showed that the number of CD44+CD24- cells were decreased substantially after the loss of ZGRF1 (Figure 4A–C). Sphere formation (Figure 4D and E) and colony formation (Figure 4F and G) profile showed that MFE levels were decreased when the expression of ZGRF1 was knocked down. These results indicated that ZGRF1 was a potential modulator of the stemness of BCSCs. Our data highlighted the roles of ZGRF1 in breast cancer stemness, suggesting that it might be considered targets of therapeutic intervention in breast cancer patients, especially for those with TNBC. Further work is needed to bring the promise of regulating their activities to clinical use.
In summary, our findings revealed the lncRNAs profile in BCSCs by using a microarray assay. Of these lncRNAs, loss of function assays for ZGRF1 was demonstrated that it might be a novel functional driver of stemness of BCSCs and a potential prognostic marker in breast cancer. These findings provide evidence that lncRNAs may possess considerable potential as a therapeutic target for BCSCs.
Funding Statement
This work was funded by the National Natural Science Foundation of China (No. 81172522 and No. 81301858), the Suzhou science and technology project (No. SYS201508 and SYS201308) and the Jiangsu Natural Science Foundation Project (BK20181186).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.v68.6 [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Sun K, Zheng R, et al. Cancer incidence and mortality in China, 2014. Chin J Cancer Res. 2018;30(1):1–12. doi: 10.21147/j.issn.1000-9604.2018.01.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marcu M. Population and social conditions. Eurostat; 2014. April 18 Available from: http://epp.eurostat.ec.europa.eu. Accessed February5, 2020.
- 4.The World Bank. Population (total); April 18, 2014. Available from: http://data.worldbank.org/. Accessed February5, 2020.
- 5.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh SK, Clarke ID, Terasaki M, et al. Identification of a cancer stem cells in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- 7.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponti D, Costa A, Zaffaroni N, et al. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65(13):5506–5511. doi: 10.1158/0008-5472.CAN-05-0626 [DOI] [PubMed] [Google Scholar]
- 9.Soudyab M, Iranpour M, Ghafouri-Fard S. The role of long non-coding RNAs in breast cancer. Arch Iran Med. 2016;19(7):508–517 [PubMed] [Google Scholar]
- 10.Liu YR, Jiang YZ, Xu XE, Hu X, Yu KD, Shao ZM. Comprehensive transcriptome profiling reveals multigene signatures in triple-negative breast cancer. Clin Cancer Res. 2016;22(7):1653–1662. doi: 10.1158/1078-0432.CCR-15-1555 [DOI] [PubMed] [Google Scholar]
- 11.Liu Z, Mi M, Li X, Zheng X, Wu G, Zhang L. lncRNA OSTN-AS1 may represent a novel immune-related prognostic marker for triple-negative breast cancer based on integrated analysis of a ceRNA network. Front Genet. 2019;10:850. doi: 10.3389/fgene.2019.00850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484. doi: 10.1126/science.1142109 [DOI] [PubMed] [Google Scholar]
- 13.Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563. [DOI] [PubMed] [Google Scholar]
- 14.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases, and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Franceschini A, Szklarczyk D, Frankild S, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41(Database issue):D808–15. doi: 10.1093/nar/gks1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498‑2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. PIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Honeth G, Bendahl PO, Ringnér M, et al. The CD44+/CD24-phenotype is enriched in basal-like breast tumors. Breast Cancer Res. 2008;10(3). doi: 10.1186/bcr2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu G, Feng X, Stein L. A human functional protein interaction network and its application to cancer data analysis. Genome Biol. 2010;11(5):R53. doi: 10.1186/gb-2010-11-5-r53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheele CLGJ, Hannezo E, Muraro MJ, et al. Identity and dynamics of mammary stem cells during branching morphogenesis. Nature. 2017;542(7641):313–317. doi: 10.1038/nature21046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prat A, Parker JS, Karginova O, et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12(5):R68. doi: 10.1186/bcr2635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hennessy BT, AM Gonzalez-Angulo, Stemke-Hale K, et al. Characterization of a naturally occurring breast cancer subset enriched in epithelial-to-mesenchymal-transition and stem cell characteristics. CancerRes. 2009;69:4116–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klinge CM. Non-coding RNAs in breast cancer: intracellular and intercellular communication. Noncoding RNA. 2018;4(4):E40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang F, Song C, Ma Y, Tang L, Xu Y, Wang H. Effect of fibroblasts on breast cancer cell mammosphere formation and regulation of stem cell-related gene expression. Int J Mol Med. 2011;28(3):365–371. doi: 10.3892/ijmm.2011.700 [DOI] [PubMed] [Google Scholar]
- 27.Chen S, Zhu J, Wang F, et al. LncRNAs and their role in cancer stem cells. Oncotarget. 2017;8(66):110685–110692. doi: 10.18632/oncotarget.22161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Z, Sun L, Zhang Y, Lu G, Li Y, Wei Z. Long non-coding RNA FEZF1-AS1 promotes breast cancer stemness and tumorigenesis via targeting miR-30a/Nanog axis.J. Cell Physiol. 2018;233(11):8630–8638. doi: 10.1002/jcp.26611 [DOI] [PubMed] [Google Scholar]
- 29.Li J, Hao Y, Mao W, et al. LincK contributes to breast tumorigenesis by promoting proliferation and epithelial-to-mesenchymal transition. J Hematol Oncol. 2019;12(1):19. doi: 10.1186/s13045-019-0707-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.VY S, Chen J, Cheuk IW, et al. Long non-coding RNA NEAT1 confers oncogenic role in triple-negative breast cancer through modulating chemoresistance and cancer stemness. Cell Death Dis. 2019;10(4):270. doi: 10.1038/s41419-019-1513-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vidovic D, Huynh TT, Konda P, et al. ALDH1A3-regulated long non-coding RNA NRAD1 is a potential novel target for triple-negative breast tumors and cancer stem cells. Cell Death Differ. 2019. doi: 10.1038/s41418-019-0362-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee KM, Giltnane JM, Balko JM, et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 2017;26(4):633–647. doi: 10.1016/j.cmet.2017.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Almozyan S, Colak D, Mansour F, et al. PD-L1 promotes OCT4 and nanog expression in breast cancer stem cells by sustaining PI3K/AKT pathway activation. Int J Cancer. 2017;141(7):1402–1412. doi: 10.1002/ijc.v141.7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou S, He Y, Yang S, et al. The regulatory roles of lncRNAs in the process of breast cancer invasion and metastasis. Biosci Rep. 2018;38(5):BSR20180772. doi: 10.1042/BSR20180772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu G, Li Y, Ma Y, et al. Long noncoding RNA LINC00511 contributes to breast cancer tumourigenesis and stemness by inducing the miR-185-3p/E2F1/Nanog axis. J Exp Clin Cancer Res. 2018;37(1):289. doi: 10.1186/s13046-018-0945-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo L, Tang H, Ling L, et al. LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene. 2018;37(47):6166–6179. doi: 10.1038/s41388-018-0396-8 [DOI] [PubMed] [Google Scholar]
- 37.Junk D-J, Cipriano R, Bryson B-L, Gilmore H-L, Jackson MW. Tumor microenvironmental signaling elicits epithelial-mesenchymal plasticity through cooperation with transforming genetic events. Neoplasia. 2013;15:1100–1109. doi: 10.1593/neo.131114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Doherty M-R, Smigiel J-M, Junk D-J, Jackson M-W. Cancer stem cell plasticity drives therapeutic resistance. Cancers (Basel). 2016;8:1–13. doi: 10.3390/cancers8010008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao C, Wang N, Guo X, et al. A comprehensive analysis of common and rare variants to identify adiposity loci in hispanic americans: the IRAS family study (IRASFS). PLoS One. 2015;10(11):e0134649. doi: 10.1371/journal.pone.0134649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peter B, Wijsman EM, Nato AQ Jr, et al. Genetic candidate variants in two multigenerational families with childhood apraxia of speech. PLoS One. 2016;11(4):e0153864. doi: 10.1371/journal.pone.0153864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Marcu M. Population and social conditions. Eurostat; 2014. April 18 Available from: http://epp.eurostat.ec.europa.eu. Accessed February5, 2020.