ABSTRACT
Background
Metabolomics has proven useful for detecting objective biomarkers of diet that may help to improve dietary measurement. Studies to date, however, have focused on a relatively narrow set of lipid classes.
Objective
The aim of this study was to uncover candidate dietary biomarkers by identifying serum metabolites correlated with self-reported diet, particularly metabolites in underinvestigated lipid classes, e.g. triglycerides and plasmalogens.
Methods
We assessed dietary questionnaire data and serum metabolite correlations from 491 male and female participants aged 55–75 y in an exploratory cross-sectional study within the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO). Self-reported intake was categorized into 50 foods, food groups, beverages, and supplements. We examined 522 identified metabolites using 2 metabolomics platforms (Broad Institute and Massachusetts General Hospital). Correlations were identified using partial Pearson's correlations adjusted for age, sex, BMI, smoking status, study site, and total energy intake [Bonferroni-corrected level of 0.05/(50 × 522) = 1.9 × 10−6]. We assessed prediction of dietary intake by multiple-metabolite linear models with the use of 10-fold crossvalidation least absolute shrinkage and selection operator (LASSO) regression.
Results
Eighteen foods, beverages, and supplements were correlated with ≥1 serum metabolite at the Bonferroni-corrected significance threshold, for a total of 102 correlations. Of these, only 5 have been reported previously, to our knowledge. Our strongest correlations were between citrus and proline betaine (r = 0.55), supplements and pantothenic acid (r = 0.46), and fish and C40:9 phosphatidylcholine (PC) (r = 0.35). The multivariate analysis similarly found reasonably large correlations between metabolite profiles and citrus (r = 0.59), supplements (r = 0.57), and fish (r = 0.44).
Conclusions
Our study of PLCO participants identified many novel food-metabolite associations and replicated 5 previous associations. These candidate biomarkers of diet may help to complement measures of self-reported diet in nutritional epidemiology studies, though further validation work is still needed.
Keywords: metabolites, dietary questionnaire, biomarkers, food, metabolomics
Introduction
Many epidemiological studies rely on self-reported diet to assess habitual food and nutrient intake. Unfortunately, self-reported diet is subject to random and systematic errors (1–3) that can cause observed associations between diet and disease to be attenuated or even biased (4). In addition, studies of self-reported diet rarely examine how diet affects endogenous metabolism, yet such effects may be important for understanding the mechanisms underlying diet-disease relations.
Nutritional metabolomics is a developing field that has proven useful for detecting objective biomarkers of dietary exposure. Over the past several years, nutritional metabolomics studies have identified hundreds of correlations between self-reported diet and serum metabolite concentrations (5–9), with findings catalogued in nutritional biomarker databases (10–12). These candidate dietary biomarkers include both exogenous metabolites that act as specific indicators of consumption of certain foods (e.g. proline betaine as a marker for citrus intake) and endogenous metabolites that reflect food effects upon host metabolism (e.g. sex steroid hormones affected by alcohol intake), in addition to biomarkers of nutritional status. Such biomarkers can be exploited in nutritional epidemiology studies to improve dietary measurement (9).
A limitation of the current studies assessing metabolites in comparison to self-reported diet is the restricted coverage of some lipid classes. Previous studies have had strong representation of acylcarnitines and lysophospholipids (13). Analyses evaluating additional lipid classes, such as triacylglycerols and plasmalogens, could uncover new relations and help to identify new candidate biomarkers.
In the present study, we evaluated cross-sectional correlations between diet and concentrations of metabolites from 2 metabolomics platforms. Previous studies using these platforms have evaluated specific dietary interventions in relation to metabolite concentrations (14, 15) but not associations for individual foods, beverages, and vitamin supplements. In addition, the 2 platforms used in the present study contain a higher proportion of lipids (69%) than analyzed in prior studies (13), and they particularly emphasize a broad range of diacylglycerols and triacylglycerols of varying carbon lengths and saturation (16). Our objective was to expand upon existing dietary biomarker studies by identifying novel candidate nutritional metabolites of habitual diet.
Methods
Study design and population
We analyzed FFQ and serum metabolite data collected from the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial (PLCO). The PLCO was a multicenter trial that randomly assigned >150,000 US men and women to a screening or control arm between 1993 and 2001. Eligible participants included those aged 55–74 y at baseline with no previous history of prostate, lung, colorectal, or ovarian cancer. All participants provided consent and completed a self-administered questionnaire on demographic and lifestyle characteristics. In the screening arm, participants completed a dietary questionnaire (DQx) and provided nonfasting blood samples at defined times during the study. The PLCO was approved by the Institutional Review Boards of the US National Cancer Institute and the 10 screening centers.
We used serum metabolomics data from a nested case-control study of kidney cancer among 534 individuals. We excluded participants missing quantitative responses to ≥8 food-frequency questions or who were in the highest or lowest 1% of caloric intake (n = 43) so that we could adjust for caloric intake, resulting in a final sample size of 491 individuals. The study included 246 participants who were diagnosed with kidney cancer and 245 controls with no history of cancer who were matched to cases by age, sex, recruitment site, menopausal status (for women), and season and year of blood draw. Cases were followed for a median of 7.4 y (IQR: 4.3–9.7) from the time of blood collection to the date of diagnosis. All participants were cancer-free at the time of blood collection.
Dietary assessment
The DQx measured the typical frequency of intake during the 12 mo before baseline for 137 foods, including alcohol, and typical portion sizes for 77 items. The frequency of intake and serving size of each food item were converted into grams per day of the food and categorized into 50 food groups based on the USDA My Plate classifications adapted to prior nutritional metabolomics analyses (9). The DQx ascertained vitamin C, D, and E and calcium supplement intakes (pills per day) at baseline as well as recent multivitamin use (yes/no).
Metabolite assessment
Nonfasting serum samples were collected at the baseline visit. Blood samples were stored at −70°C and analyzed in 2014–2015. Metabolites were assessed using the metabolomics platforms of the Broad Institute and Massachusetts General Hospital. Methodologies for both platforms have been described previously (17, 18), and details of the methodologies can be found in the Supplemental Methods. The workflows for these 2 labs include; chromatography and positive ion mode MS detection (C8)-pos, hydrophilic interaction liquid chromatography (HILIC)-pos, and triple quadrupole with multiple reaction monitoring (MRM). A flowchart to describe these workflows can be found in Supplemental Figure 1.
In the present study, we measured 531 identified, named metabolites. Duplicate metabolites between the 2 platforms were deleted (n = 9), leaving 522 identified metabolites for analysis. We have provided a complete list of these metabolites, compounds, methods, and representative identification used to detect each in Supplemental Table 1. Metabolites were log-transformed (natural log), values below the detection threshold were set to the lowest observed value, and the distribution was centered. Across the 522 metabolites, the median intraclass correlation coefficient (ICC) was 0.95 (IQR: 0.91–0.99), indicating a high level of technical reliability (calculated by using 80 replicates from 10 study participants and 10 pooled samples).
Statistical analysis
We estimated correlations between 522 identified, named metabolites and 50 foods, beverages, and vitamin supplements by assessing partial Pearson's correlations adjusted for age, sex, BMI, smoking status (never, former, current), study site, and total energy intake (kcal/d). The threshold for statistical significance was Bonferroni-adjusted [a = 0.05/(50 × 522) = 1.9 × 10−6]. Demographic and lifestyle characteristics and dietary intakes for cases and controls were compared using 2-sided statistical tests (the chi-square test for categorical variables and Wilcoxon rank-sum test for continuous variables). In sensitivity analyses, we examined diet-metabolite correlations among controls and cases separately and evaluated whether correlations differed using the Wald test for homogeneity. Analyses were conducted in SAS 9.4 (SAS Institute).
Some of the metabolites associated with diet were intercorrelated as part of a metabolic pathway. To account for—and to help illustrate—these interdependencies, we used Gaussian graphical modeling (GGM) to assess the conditional correlations of diet-related metabolites and to display their relation as a metabolic network. Conditional correlations between metabolites (r ≥0.40) were visualized by linking them together by a line to represent direct relations. Prior tests of GGMs against known metabolic pathways have found that GGMs perform well at recapitulating these pathways and that they outperform alternative approaches (e.g. heat maps) at identifying metabolic interrelations (19). GGM was conducted in RStudio version 1.1.453 (RStudio Inc.) with visualization using Cytoscape 3.7.1 (The Cytoscape Consortium) (20).
We assessed the prediction of dietary intake by multiple-metabolite linear models with the use of 10-fold crossvalidation least absolute shrinkage and selection operator (LASSO) regression. The LASSO model imposed a penalty on the β-coefficients to reduce any potential overfitting that may occur. Ten iterations of predictive models were generated with the use of 80% data and tested in the remaining 20%. Final estimates averaged correlations between observed and predicted dietary intake levels. Crossvalidation LASSO was done in RStudio version 1.1.453 (RStudio Inc.) (cv.glmnet from the glmnet package). To adjust for age, sex, BMI, smoking status, study site, and daily caloric intake, we residually adjusted metabolites for these factors before including them in the LASSO analysis.
Results
Baseline demographic characteristics are shown in Table 1. Of the 491 participants, 66% were male. The mean age was 63 y and the sample was predominately white. Most participants were never (48%) or former (43%) smokers. For most characteristics, kidney cancer cases and controls did not have statistically significant differences. However, there was a significant difference in BMI (P = 0.0004), with a mean ± SD BMI (of 28 ± 5 kg/m2 for cases as compared with 27 ± 4 in controls). Self-reported dietary intake, estimated from the DQx, is shown in Table 2. There were no statistically significant differences in self-reported dietary intake between cases and controls; we therefore present results for diet-metabolite correlations only for the overall sample.
TABLE 1.
Participant characteristics in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial1,2
| Characteristic | Value |
|---|---|
| Sex, n(%) | |
| Men | 326 (66) |
| Women | 165 (34) |
| Age, y | 63 ± 5 |
| Race, n(%) | |
| White | 448 (91) |
| Smoking status, n(%) | |
| Current | 47 (9) |
| Former | 209 (43) |
| Never | 235 (48) |
| Total energy intake, kcal/d | 2171 ± 806 |
| BMI, kg/m2 | 28 ± 5 |
n = 491.
Values are mean ± SDs or n(%).
TABLE 2.
Self-reported usual dietary consumption in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial1, 2, 3
| Category and dietary group | Value |
|---|---|
| Fruit, g/d | |
| Citrus: oranges, orange juice, grapefruit | 113 (35–228) |
| Berries: strawberries | 5 (2–10) |
| Apples, pears | 30 (13–65) |
| Watermelon | 7 (2–14) |
| Juice | 117 (27–220) |
| Other: plums, bananas, apricots, peaches, prunes, raisins, grapes, pineapples | 76 (78–123) |
| Vegetables, g/d | |
| Cruciferous: broccoli, cabbage, Brussels sprouts, cauliflower, turnip greens, mustard greens, collards, kale, Swiss chard | 27 (15–49) |
| Leafy greens: lettuce, spinach | 29 (14–48) |
| Corn | 9 (4–17) |
| Tomatoes: canned, fresh, sauce, ketchup | 55 (33–88) |
| Yellow/orange vegetables: carrots, squash | 15 (8–29) |
| Sweet potatoes | 2 (1–4) |
| White potatoes | 63 (24–82) |
| Total potatoes: white, sweet, fried | 75 (43–108) |
| Other: celery, green beans, beets, peppers, cucumbers | 61 (37–95) |
| Meat/fish, g/d | |
| Red meat | 38 (21–67) |
| Processed meat: cold cuts, hot dogs, bacon, sausage | 14 (6–28) |
| Chicken | 16 (8–34) |
| Fish (excluding shellfish) | 18 (8–33) |
| Shellfish | 1 (0–2) |
| Snack foods, g/d | |
| Sweets: candy, chocolate, cookies, donuts, pies | 22 (11–45) |
| Ice cream | 8 (2–19) |
| French fries | 7 (2–16) |
| Chips | 4 (1–14) |
| Dairy, g/d | |
| Milk | 195 (79–400) |
| Cheese | 12 (5–23) |
| Yogurt | 11 (2–44) |
| Condiments: sour cream, sweet cream | 1 (0–2) |
| Grains, g/d | |
| High fiber: dark bread, high-fiber cereal, good-fiber cereal, brown rice | 33 (13–56) |
| Low fiber: biscuit, corn bread, white bread, hot cereal, fortified cereal, other cereal, crackers, pancakes, waffles, white rice, spaghetti, other grains | 129 (86–219) |
| Other foods, g/d | |
| Tofu | 0 (0–1) |
| Legumes: beans, peas | 20 (9–37) |
| Eggs | 11 (3–22) |
| Oil | 13 (8–19) |
| Butter | 3 (1–7) |
| Margarine | 2 (0–5) |
| Salad dressing | 3 (1–10) |
| Peanuts | 3 (1–9) |
| Added sugars | 4 (1–8) |
| Gravy | 2 (0–7) |
| Beverages, g/d | |
| Coffee | 843 (150–1517) |
| Tea | 22 (3–258) |
| Sugar-sweetened beverages: soda, fruit punch | 23 (5–148) |
| Beer | 4 (0–64) |
| Wine | 1 (0–12) |
| Liquor | 1 (0–4) |
| Total alcohol | 12 (1–179) |
| Reported multivitamin use, n(%) | 237 (44) |
n = 491.
FFQ measurement of 12-mo intake.
Values are medians (IQR).
Eighteen foods, beverages, and supplements were correlated with ≥1 serum metabolite at the Bonferroni-corrected threshold for significance and, in total, 102 different correlations met the threshold of significance (Table 3). The strongest correlation overall was between citrus fruit intake and concentrations of proline betaine (r = 0.55). Intake of yellow vegetables and high-fiber grains were inversely associated with cotinine (r = −0.23 and r = −0.25, respectively). The consumption of sweets was correlated with C36:1 phosphatidylcholine (PC) plasmalogen (r = 0.25) and ice-cream intake was associated with concentrations of many different lipids, with the strongest association for C36:1 phosphatidylethanolamine (PE) plasmalogen (r = 0.28). French fry intake was associated with concentrations of 1,2,4-trimethylbenzene (r = 0.24), and eggs and gravy consumption were both correlated with concentrations of C38:4 PC plasmalogen (r = 0.23 and 0.25, respectively). Oil consumption was correlated with C14:0 cholesterol ester (CE) (r = −0.26). Meat and fish consumption were correlated with many lipids, with the strongest relations as follows: red meat with C34:5 PC plasmalogen (r = 0.34), processed meat with C36:2 PE plasmalogen (r = 0.25), chicken with C36:5 PE plasmalogen (r = 0.26), and fish with C40:9 PC (r = 0.35). Tea intake was correlated with N-acetylornithine-2 (putative) (r = 0.39), beer with C24:0 sphingomyelin (SM) (r = 0.22), and total alcohol with C36:1 phosphatidylserine (PS) plasmalogen (r = 0.21). Supplement use was highly correlated with pantothenic acid (r = 0.46).
TABLE 3.
Top metabolites associated with dietary intake of foods in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial1,2
| Foods and metabolites | Super class | Correlation (r) | P |
|---|---|---|---|
| Citrus: oranges, orange juice, grapefruit | |||
| Proline betaine | Organic acids and derivatives | 0.55 | 0 |
| Histamine | Organonitrogen compounds | −0.33 | 8.30 × 10−4 |
| Arginine | Organic acids and derivatives | −0.32 | 9.92 × 10−3 |
| Acetylcholine | Organonitrogen compounds | 0.23 | 3.02 × 10−7 |
| Carnosine | Organic acids and derivatives | −0.23 | 3.70 × 10−7 |
| Glyoxylic acid | Lipids and lipid-like molecules | 0.22 | 6.19 × 10−7 |
| Juice | |||
| Proline betaine | Organic acids and derivatives | 0.43 | 2.16 × 10−3 |
| Arginine | Organic acids and derivatives | −0.26 | 9.68 × 10−9 |
| Acetylcholine | Organonitrogen compounds | 0.24 | 1.63 × 10−7 |
| Histamine | Organonitrogen compounds | −0.22 | 1.34 × 10−6 |
| Yellow/orange vegetables: carrots, squash | |||
| Cotinine | Organoheterocyclic compounds | −0.23 | 4.18 × 10−7 |
| Sweets: candy, chocolate, cookies, donuts, pies | |||
| C36:1 PC plasmalogen | Lipids and lipid-like molecules | 0.25 | 2.16 × 10−8 |
| Ice cream | |||
| C36:1 PE plasmalogen | Lipids and lipid-like molecules | 0.28 | 4.11 × 10−10 |
| C56:8 TAG | Lipids and lipid-like molecules | −0.26 | 5.10 × 10−9 |
| C56:7 TAG | Lipids and lipid-like molecules | −0.26 | 1.05 × 10−8 |
| C56:9 TAG | Lipids and lipid-like molecules | −0.26 | 1.22 × 10−8 |
| C38:3 PE plasmalogen | Lipids and lipid-like molecules | 0.24 | 5.58 × 10−8 |
| C58:10 TAG | Lipids and lipid-like molecules | −0.24 | 5.75 × 10−8 |
| C58:9 TAG + NH4 | Lipids and lipid-like molecules | −0.24 | 8.11 × 10−8 |
| C58:10 TAG + NH4 | Lipids and lipid-like molecules | −0.24 | 1.74 × 10−7 |
| C58:9 TAG | Lipids and lipid-like molecules | −0.23 | 2.00 × 10−7 |
| C36:1 PC plasmalogen | Lipids and lipid-like molecules | 0.23 | 2.00 × 10−7 |
| C56:8 TAG + NH4 | Lipids and lipid-like molecules | −0.23 | 3.60 × 10−7 |
| C36:0 PE | Lipids and lipid-like molecules | 0.22 | 6.54 × 10−7 |
| C56:9 TAG + NH4 | Lipids and lipid-like molecules | −0.22 | 9.49 × 10−7 |
| C58:11 TAG | Lipids and lipid-like molecules | −0.22 | 9.84 × 10−7 |
| C56:7 TAG + NH4 | Lipids and lipid-like molecules | −0.22 | 1.01 × 10−6 |
| C54:7 TAG | Lipids and lipid-like molecules | −0.22 | 1.02 × 10−6 |
| C9 carnitine | Lipids and lipid-like molecules | 0.22 | 1.28 × 10−6 |
| C18 carnitine | Lipids and lipid-like molecules | 0.22 | 1.60 × 10−6 |
| C38:4 PC plasmalogen | Lipids and lipid-like molecules | 0.22 | 1.67 × 10−6 |
| French fries | |||
| 1,2,4-trimethylbenzene | Benzenoids | 0.24 | 9.20 × 10−8 |
| C34:1 PC plasmalogen-B | Lipids and lipid-like molecules | 0.22 | 1.17 × 10−6 |
| High-fiber grains: dark bread, high-fiber cereal, brown rice | |||
| Cotinine | Organoheterocyclic compounds | −0.25 | 4.00 × 10−8 |
| Eggs | |||
| C38:4 PC plasmalogen | Lipids and lipid-like molecules | 0.23 | 2.09 × 10−7 |
| C26 carnitine | Lipids and lipid-like molecules | 0.22 | 5.90 × 10−7 |
| Oil | |||
| C14:0 CE | Lipids and lipid-like molecules | −0.25 | 1.57 × 10−8 |
| C16:1 CE | Lipids and lipid-like molecules | −0.24 | 6.87 × 10−8 |
| C14:0 CE + NH4 | Lipids and lipid-like molecules | −0.23 | 3.83 × 10−7 |
| C16:1 CE + NH4 | Lipids and lipid-like molecules | −0.22 | 1.22 × 10−6 |
| Gravy | |||
| C38:4 PC plasmalogen | Lipids and lipid-like molecules | 0.25 | 1.57 × 10−8 |
| C36:1 PE plasmalogen | Lipids and lipid-like molecules | 0.25 | 2.59 × 10−8 |
| C36:1 PC plasmalogen | Lipids and lipid-like molecules | 0.24 | 1.05 × 10−7 |
| C38:2 PE | Lipids and lipid-like molecules | 0.23 | 2.16 × 10−7 |
| C38:3 PE plasmalogen | Lipids and lipid-like molecules | 0.22 | 6.50 × 10−7 |
| C36:2 PC plasmalogen | Lipids and lipid-like molecules | 0.22 | 1.76 × 10−6 |
| C36:3 PE plasmalogen | Lipids and lipid-like molecules | 0.22 | 1.78 × 10−6 |
| Red meat | |||
| C34:5 PC plasmalogen | Lipids and lipid-like molecules | 0.34 | 2.13 × 10−14 |
| C38:3 PE plasmalogen | Lipids and lipid-like molecules | 0.32 | 1.04 × 10−12 |
| C36:2 PE plasmalogen | Lipids and lipid-like molecules | 0.30 | 1.17 × 10−11 |
| C36:3 PE plasmalogen | Lipids and lipid-like molecules | 0.27 | 1.61 × 10−9 |
| C34:2 PE plasmalogen | Lipids and lipid-like molecules | 0.27 | 2.55 × 10−9 |
| C38:5 PE plasmalogen | Lipids and lipid-like molecules | 0.26 | 3.74 × 10−9 |
| C36:1 PE plasmalogen | Lipids and lipid-like molecules | 0.26 | 4.90 × 10−9 |
| C38:4 PC plasmalogen | Lipids and lipid-like molecules | 0.25 | 4.03 × 10−8 |
| C36:4 PE plasmalogen | Lipids and lipid-like molecules | 0.22 | 6.27 × 10−7 |
| C36:5 PC plasmalogen | Lipids and lipid-like molecules | 0.22 | 9.63 × 10−7 |
| C34:3 PE plasmalogen | Lipids and lipid-like molecules | 0.22 | 1.02 × 10−6 |
| Processed meat: cold cuts, hot dogs, bacon, sausage | Lipids and lipid-like molecules | ||
| C36:2 PE plasmalogen | Lipids and lipid-like molecules | 0.25 | 2.10 × 10−8 |
| C38:3 PE plasmalogen | Lipids and lipid-like molecules | 0.25 | 4.79 × 10−8 |
| C38:4 PC plasmalogen | Lipids and lipid-like molecules | 0.24 | 5.85 × 10−8 |
| C36:3 PE plasmalogen | Lipids and lipid-like molecules | 0.24 | 9.37 × 10−8 |
| C36:1 PC plasmalogen | Lipids and lipid-like molecules | 0.23 | 1.81 × 10−7 |
| C36:4 PE plasmalogen | Lipids and lipid-like molecules | 0.23 | 2.14 × 10−7 |
| C34:2 PE plasmalogen | Lipids and lipid-like molecules | 0.22 | 8.57 × 10−7 |
| Chicken | |||
| C36:5 PE plasmalogen | Lipids and lipid-like molecules | 0.26 | 4.12 × 10−9 |
| Ectoine | 0.26 | 9.81 × 10−9 | |
| 1-methylhistidine | Organic acids and derivatives | 0.25 | 1.40 × 10−8 |
| C38:7 PC plasmalogen | Lipids and lipid-like molecules | 0.23 | 4.45 × 10−7 |
| 3-methylhistamine | Organonitrogen compounds | 0.22 | 7.11 × 10−7 |
| Fish (excluding shellfish) | |||
| C40:9 PC | Lipids and lipid-like molecules | 0.35 | 1.57 × 10−15 |
| C38:7 PC plasmalogen | Lipids and lipid-like molecules | 0.33 | 4.58 × 10−14 |
| C38:7 PE plasmalogen | Lipids and lipid-like molecules | 0.32 | 2.36 × 10−13 |
| C22:6 CE + NH4 | Lipids and lipid-like molecules | 0.32 | 3.42 × 10−13 |
| C60:12 TAG | Lipids and lipid-like molecules | 0.32 | 3.97 × 10−13 |
| C40:6 PC | Lipids and lipid-like molecules | 0.31 | 6.21 × 10−12 |
| C58:9 TAG + NH4 | Lipids and lipid-like molecules | 0.30 | 1.61 × 10−11 |
| C58:8 TAG | Lipids and lipid-like molecules | 0.29 | 5.56 × 10−11 |
| C58:9 TAG | Lipids and lipid-like molecules | 0.29 | 5.71 × 10−11 |
| C22:6 LPC | Lipids and lipid-like molecules | 0.29 | 1.47 × 10−10 |
| C36:5 PC plasmalogen-A | Lipids and lipid-like molecules | 0.28 | 2.80 × 10−10 |
| C56:8 TAG | Lipids and lipid-like molecules | 0.28 | 3.80 × 10−10 |
| C40:7 PE plasmalogen | Lipids and lipid-like molecules | 0.28 | 4.57 × 10−10 |
| C22:6 LPE | Lipids and lipid-like molecules | 0.28 | 5.68 × 10−10 |
| C38:6 PE | Lipids and lipid-like molecules | 0.28 | 5.81 × 10−10 |
| C58:8 TAG + NH4 | Lipids and lipid-like molecules | 0.27 | 1.19 × 10−9 |
| C58:10 TAG | Lipids and lipid-like molecules | 0.26 | 4.75 × 10−9 |
| C42:11 PE plasmalogen | Lipids and lipid-like molecules | 0.26 | 6.48 × 10−9 |
| C56:9 TAG | Lipids and lipid-like molecules | 0.25 | 2.06 × 10−8 |
| C40:10 PC | Lipids and lipid-like molecules | 0.25 | 3.07 × 10−8 |
| C60:12 TAG + NH4 | Lipids and lipid-like molecules | 0.25 | 3.24 × 10−8 |
| C58:10 TAG + NH4 | Lipids and lipid-like molecules | 0.25 | 3.52 × 10−8 |
| C58:11 TAG | Lipids and lipid-like molecules | 0.25 | 4.17 × 10−8 |
| C56:8 TAG + NH4 | Lipids and lipid-like molecules | 0.24 | 9.00 × 10−8 |
| C56:9 TAG + NH4 | Lipids and lipid-like molecules | 0.24 | 1.19 × 10−7 |
| C56:7 TAG + NH4 | Lipids and lipid-like molecules | 0.23 | 4.89 × 10−7 |
| Tea | |||
| N-acetylornithine-2 (putative) | 0.39 | 1.45 × 10−18 | |
| Beer | |||
| C24:0 SM | Lipids and lipid-like molecules | 0.22 | 1.55 × 10−6 |
| Total alcohol | |||
| C36:1 PS plasmalogen | Lipids and lipid-like molecules | 0.21 | 1.89 × 10−6 |
| Supplements | |||
| Pantothenic acid | Amino acids, peptides, and analogs | 0.46 | 3.1 × 10−27 |
| Thiamin | Organoheterocyclic compounds | 0.31 | 5.4 × 10−12 |
| 5-methyltetrahydrofolate | Organoheterocyclic compounds | 0.27 | 1.83 × 10−9 |
n = 491.
Abbreviations: CE, cholesterol ester; LPC, lysophosphatidylcholine; NH4, ammonium; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PS, phosphatidylserine; SM, sphingomyelin; TAG, triacylglycerol.
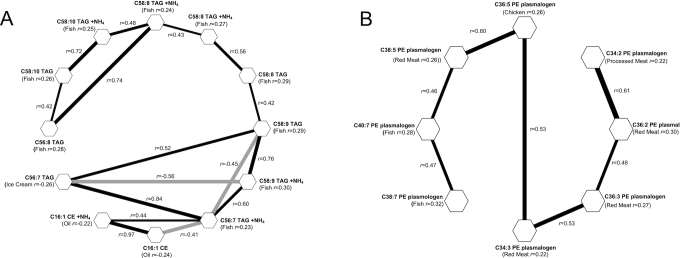
The association networks for pairs of diet-related metabolites with conditional correlations represented by connected lines are shown in Figure 1A and B (r ≥0.40). Most diet-related metabolites did not constitute a part of any metabolite cluster of appreciable size (>2 metabolites). However, there were 2 clusters, of 12 and 8 metabolites, consisting predominately of lipids that tracked with intake of oil, ice cream, fish, red meat, chicken, and processed meat.
FIGURE 1.
A–B. Gaussian graphical models of 2 clusters of diet-related metabolites measured in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Metabolites are depicted as hexagons, and pairs with an absolute value of conditional correlation >0.4 are connected by a line. Black lines represent positive conditional correlations. Gray lines represent inverse conditional correlations. Clusters with <8 metabolites not shown.
Figure 2 displays the correlations and 95% CIs between self-reported dietary intake and predicted intake based on 10-fold crossvalidated LASSO regression. These correlations, which broadly reflect the strength of multiple-metabolite prediction of diet based on statistical modeling (i.e. prediction based on a training data set and testing with the use of a testing data set), appear to reiterate the findings of the univariate analysis. Like in the analysis of individual metabolites, citrus intake (r = 0.59), multivitamin use (r = 0.57), and juice consumption (r = 0.53) had the strongest associations. Correlations were generally higher in the LASSO than in the univariate analysis, reflecting the ability of the LASSO to integrate information from multiple metabolites. However, the differences were generally modest, i.e. 0.01–0.11 larger than the correlation from the univariate model. A full list of the metabolites contributing to the multi-metabolite LASSO models and their β-coefficients can be found in Supplemental Table 2.
FIGURE 2.
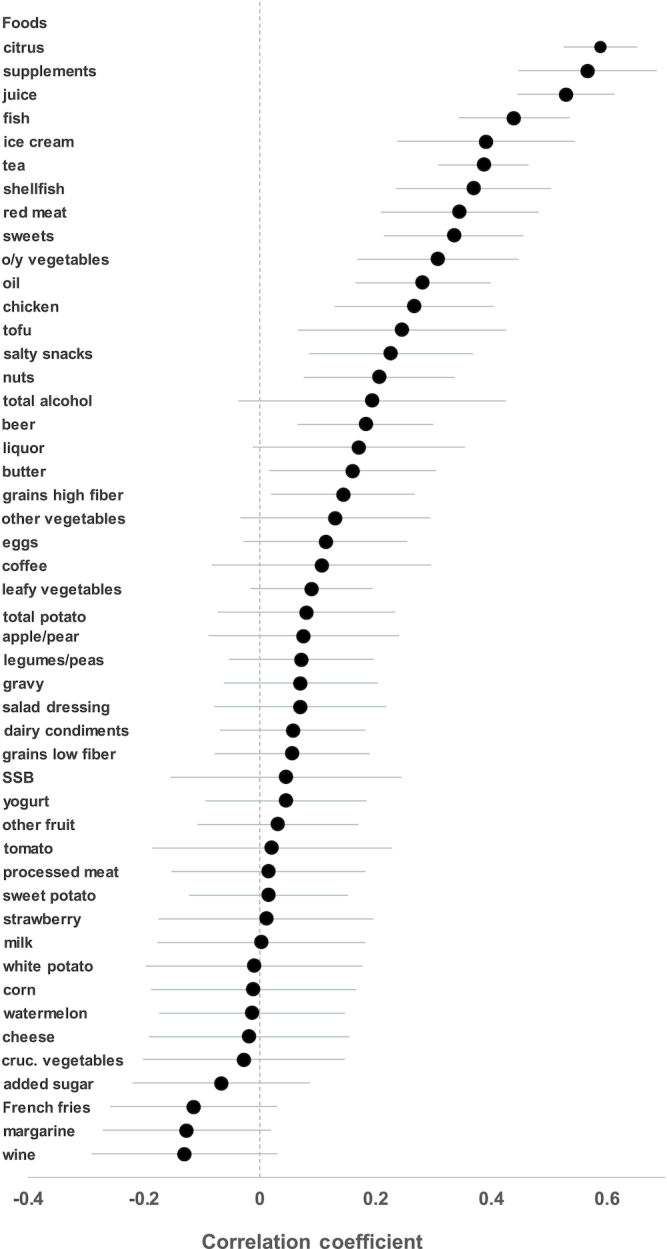
Correlations and 95% CIs between self-reported dietary intake and predicted intake based on metabolites and 10-fold crossvalidated LASSO regression in the Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial. Cruc. vegetables, cruciferous vegetables; LASSO, least absolute shrinkage and selection operator; o/y vegetables, orange/yellow vegetables; SSB, sugar sweetened beverage.
In sensitivity analyses, we identified 39 correlations (out of 26,100 diet-metabolite correlations examined) that differed by future case status (P < 1.9 × 10−6). Virtually all involved qualitative heterogeneity, meaning the correlations were in opposite directions for cases versus controls. The most heterogeneous of these associations was between total alcohol consumption and xanthine, where the correlation was −0.17 in cases and 0.23 in controls.
One metabolite associated (inversely) with citrus intake was carnosine, a metabolite that has previously been linked with beef intake (21). To determine whether this association was driven by a tendency for meat consumers to eat less citrus, we evaluated whether the self-reported intake of citrus was inversely correlated with self-reported meat intake. We found a weak, inverse correlation between citrus and meat consumption (r = −0.06), though it was not statistically significant (P = 0.18).
Discussion
In a large cross-sectional study, we were able to identify 18 foods, food groups, beverages, and supplements associated with ≥1 metabolite, and identified a total of 102 diet-metabolite correlations. Of these, only 5 to our knowledge have been previously reported in population and feeding studies (proline betaine and citrus, 1-methylhistidine and chicken, and pantothenic acid, thiamin, 5-methyltetrahydrofolate, and supplements) (6, 9, 12, 22). Our findings add to the growing list of candidate dietary biomarkers to be validated and potentially used in nutritional epidemiology studies.
To date, these 2 metabolomics platforms have been used in 2 prior dietary studies (14, 15), both of which focused on specific investigator-assigned diets. In contrast to these studies, the present study covers a broad span of different aspects of diet, which likely explains the large number of novel findings. As compared with other platforms (13), these platforms emphasize lipids, such as triacylglycerols and plasmalogens, which provided us with a different set of metabolites.
Several lines of evidence support the plausibility of the metabolites we identified as biomarkers of intake. Citrus fruits and juice are known to contain proline betaine, otherwise known as stachydrine, and the positive correlation we observed was similar in magnitude to what has been previously reported (6, 9). Citrus intake was also inversely associated with carnosine, predominately found in animal tissue (21), and arginine, which can be animal or plant derived (12); possibly because individuals who ate more citrus tended to consume less red meat, though this finding was not statistically significant. The inverse associations between the intake of yellow vegetables and high-fiber grains with cotinine are likely due to imperfect adjustment for smoking status, as smokers tend to consume fewer vegetables (23). The consumption of sweets and ice cream were positively correlated with phosphatidylethanolamines, whose acid moieties are derived from animal fats and cocoa butter (12), ingredients typically present in these foods. French fries were correlated with 1,2,4-trimethylbenzene, an environmental toxin previously found in fried beef liver, margarine, and butter (24). French fries are predominately cooked in oil, which may be the source of this metabolite. We observed an inverse relation between dietary oils (primarily unsaturated oils) and cholesterol esters (components of the cholesterol molecule), which may reflect the benefits of unsaturated fats on plasma cholesterol (25). The intake of eggs was positively associated with C38:4 PC plasmalogen, a phosphatidylcholine, and eggs are known to be rich sources of choline (26). Gravy consumption was positively associated with lipids that have animal fat-derived acid moieties (12), and gravy often contains animal products or is coconsumed with meat. Red meat and processed meat were associated with several phosphatidylethanolamines and phosphatidylcholines, all of which contain ≥1 acid moiety derived from animal tissues (12). Chicken was correlated with C36:5 PE plasmalogen and C38:7 PC plasmalogen, both of which contain acid moieties derived from animal fats (12). Chicken was also correlated with 1-methylhistidine, a metabolite that has previously been associated with meat intake (27), and chicken specifically (22). Fish was strongly correlated with C40:9 PC and C38:7 PC plasmalogen. Based on the bond content of these lipids, we can infer that both contain a moiety of DHA, which is derived from fish oils (12). Fish was also correlated with C38:7 PE plasmalogen, which contains acid moieties found in animal tissues and marine lipids (12). The metabolites we found to be associated with supplements are components of vitamins, for example, pantothenic acid is another name for vitamin B5, and thiamin is synonymous for vitamin B1 (12).
We observed that food intake was more often associated with ether lipids, rather than with ester lipids. Ether lipids are typically less abundant than ester lipids. Though speculative, the greater number of associations for ether lipids could occur if they are more specific markers, which would also contribute to their lower abundance.
In addition to identifying biomarkers of intake that, if validated (28), could be used in future studies, some findings suggest specific noteworthy effects of certain foods. For example, researchers have hypothesized for at least a decade that citrus fruits may increase circulating concentrations of histamine, thereby exacerbating seasonal and other allergies, and have therefore advocated to avoid their intake during allergic episodes (29). We found, however, that citrus and juice (primarily from citrus) were negatively correlated with histamine, thus contradicting this hypothesis. Mechanistically, this inverse association could be due to the ability of ascorbic acid to accelerate histamine degradation (30). Furthermore, alcohol, meats, poultry, and fish were each associated with several lipids, including phosphatidylethanolamines, cholesterol esters, and triacylglycerols, which may explain their links to cardiovascular disease (31, 32).
In our analysis, we did identify some diet-metabolite correlations that appeared to vary by case status. However, since every one of these instances involved qualitative rather than quantitative heterogeneity, these findings do not appear to be particularly biologically plausible and should be treated with caution.
The strengths of our study include a large sample size, many detected metabolites, and an extensive dietary questionnaire. Our study also has several limitations. Our results were based on a self-reported survey, which likely attenuated some correlations. We believe our results would be stronger with measures of dietary intake less susceptible to random errors. Participants were also not required to fast, however, prior studies did not show obvious differences by fasting status (8). Metabolomics in general has its own limitations, including missed peaks and integrated noise peaks (33), which we cannot rule out as a limitation of our own data. The amount of serum being analyzed limits which metabolites can be measured and the platforms we used are not optimized for particular metabolites. Since the level of certainty used to identify lipids was less definitive, a representative Human Metabolome Database (HMDB) identifier was used for some metabolites. Though some literature is available on these metabolites, we are limited in our ability to clarify whether many of our associations are direct biomarkers of intake or rather reflect endogenous effects of eating these foods. Future feeding studies could disentangle these possibilities by measuring concentrations of metabolites both before and after consumption to determine whether the metabolite is present even if the food had not been consumed. Lastly, our sample was largely white; thus, generalizability to other groups is unknown.
In conclusion, our findings build upon previous nutritional metabolomics studies by providing novel specific lipid-food relations and replicating previous associations. These candidate biomarkers, once validated, could potentially be used to complement self-reported measures of diet in nutritional epidemiology studies.
Supplementary Material
Acknowledgments
The authors’ responsibilities were as follows—SCM and LL: designed the research; KMMa: conducted the research; SCM, MCP, JNS, REG, CBC, and NDF: provided essential materials; KMMa and SCM: analyzed data; KMMa, KMMc, and SCM: wrote the manuscript; KMMa and SCM: had primary responsibility for final content; all authors have provided important intellectual input and have read and approved the final manuscript. This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov).
Notes
This work was supported by the Intramural Research Program of the National Cancer Institute, NIH, and Department of Health and Human Services.
Author disclosures: The authors report no conflicts of interest.
Supplemental Methods, Supplemental Tables 1 and 2, and Supplemental Figure 1 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn.
Abbreviations used: DQx, dietary questionnaire; GGM, Gaussian graphical modeling; LASSO, least absolute shrinkage and selection operator; PLCO, Prostate, Lung, Colorectal, and Ovarian Cancer Screening Trial.
References
- 1. Bingham SA, Luben R, Welch A, Wareham N, Khaw KT, Day N. Are imprecise methods obscuring a relation between fat and breast cancer?. Lancet. 2003;362(9379):212–4. [DOI] [PubMed] [Google Scholar]
- 2. Johansson L, Solvoll K, Bjorneboe GE, Drevon CA. Under- and overreporting of energy intake related to weight status and lifestyle in a nationwide sample. Am J Clin Nutr. 1998;68(2):266–74. [DOI] [PubMed] [Google Scholar]
- 3. Neuhouser ML, Tinker L, Shaw PA, Schoeller D, Bingham SA, Horn LV, Beresford SA, Caan B, Thomson C, Satterfield S et al.. Use of recovery biomarkers to calibrate nutrient consumption self-reports in the Women's Health Initiative. Am J Epidemiol. 2008;167(10):1247–59. [DOI] [PubMed] [Google Scholar]
- 4. Kaaks RJ. Biochemical markers as additional measurements in studies of the accuracy of dietary questionnaire measurements: conceptual issues. Am J Clin Nutr. 1997;65(4(Suppl):1232S–9S. [DOI] [PubMed] [Google Scholar]
- 5. Guertin KA, Loftfield E, Boca SM, Sampson JN, Moore SC, Xiao Q, Huang WY, Xiong X, Freedman ND, Cross AJ et al.. Serum biomarkers of habitual coffee consumption may provide insight into the mechanism underlying the association between coffee consumption and colorectal cancer. Am J Clin Nutr. 2015;101(5):1000–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Guertin KA, Moore SC, Sampson JN, Huang WY, Xiao Q, Stolzenberg-Solomon RZ, Sinha R, Cross AJ. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am J Clin Nutr. 2014;100(1):208–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Playdon MC, Moore SC, Derkach A, Reedy J, Subar AF, Sampson JN, Albanes D, Gu F, Kontto J, Lassale C et al.. Identifying biomarkers of dietary patterns by using metabolomics. Am J Clin Nutr. 2017;105(2):450–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Playdon MC, Sampson JN, Cross AJ, Sinha R, Guertin KA, Moy KA, Rothman N, Irwin ML, Mayne ST, Stolzenberg-Solomon R et al.. Comparing metabolite profiles of habitual diet in serum and urine. Am J Clin Nutr. 2016;104(3):776–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Playdon MC, Ziegler RG, Sampson JN, Stolzenberg-Solomon R, Thompson HJ, Irwin ML, Mayne ST, Hoover RN, Moore SC. Nutritional metabolomics and breast cancer risk in a prospective study. Am J Clin Nutr. 2017;106(2):637–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Neveu V, Moussy A, Rouaix H, Wedekind R, Pon A, Knox C, Wishart DS, Scalbert A. Exposome-explorer: a manually-curated database on biomarkers of exposure to dietary and environmental factors. Nucleic Acids Res. 2017;45(D1):D979–D84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Neveu V, Perez-Jimenez J, Vos F, Crespy V, du Chaffaut L, Mennen L, Knox C, Eisner R, Cruz J, Wishart D et al.. Phenol-explorer: an online comprehensive database on polyphenol contents in foods. Database (Oxford). 2010;2010:bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, Sajed T, Johnson D, Li C, Karu N et al.. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608–D17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yu B, Zanetti KA, Temprosa M, Albanes D, Appel N, Barrera CB, Ben-Shlomo Y, Boerwinkle E, Casas JP, Clish C et al.. The Consortium of Metabolomics Studies (COMETS): metabolomics in 47 prospective cohort studies. Am J Epidemiol. 2019;188(6):991–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Esko T, Hirschhorn JN, Hsu Y-HH, Feldman HA, Ebbeling CB, Ludwig DS, Clish CB, Deik AA. Metabolomic profiles as reliable biomarkers of dietary composition. Am J Clin Nutr. 2017;105(3):547–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Toledo E, Wang DD, Ruiz-Canela M, Clish CB, Razquin C, Zheng Y, Guasch-Ferré M, Hruby A, Corella D, Gómez-Gracia E et al.. Plasma lipidomic profiles and cardiovascular events in a randomized intervention trial with the Mediterranean diet. Am J Clin Nutr. 2017;106(4):973–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rhee EP, Cheng S, Larson MG, Walford GA, Lewis GD, McCabe E, Yang E, Farrell L, Fox CS, O'Donnell CJ et al.. Lipid profiling identifies a triacylglycerol signature of insulin resistance and improves diabetes prediction in humans. J Clin Invest. 2011;121(4):1402–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Paynter NP, Balasubramanian R, Giulianini F, Wang DD, Tinker LF, Gopal S, Deik AA, Bullock K, Pierce KA, Scott J et al.. Metabolic predictors of incident coronary heart disease in women. Circulation. 2018;137(8):841–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kimberly WT, O'Sullivan JF, Nath AK, Keyes M, Shi X, Larson MG, Yang Q, Long MT, Vasan R, Peterson RT et al.. Metabolite profiling identifies anandamide as a biomarker of nonalcoholic steatohepatitis. JCI Insight. 2017;2(9):92989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Krumsiek J, Suhre K, Illig T, Adamski J, Theis FJ. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst Biol. 2011;5(1):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park YJ, Volpe SL, Decker EA. Quantitation of carnosine in humans plasma after dietary consumption of beef. J Agric Food Chem. 2005;53(12):4736–9. [DOI] [PubMed] [Google Scholar]
- 22. Steele BF, Hubbard RW, Block WD. Excretion of histidine and histidine derivatives by human subjects ingesting protein from different sources. J Nutr. 1965;85(4):419–25. [DOI] [PubMed] [Google Scholar]
- 23. Birkett NJ. Intake of fruits and vegetables in smokers. Public Health Nutr. 1999;2(2):217–22. [DOI] [PubMed] [Google Scholar]
- 24. Heikes DL, Jensen SR, Fleming-Jones ME. Purge and trap extraction with GC-MS determination of volatile organic compounds in table-ready foods. J Agric Food Chem. 1995;43(11):2869–75. [Google Scholar]
- 25. Sacks FM, Lichtenstein AH, Wu JHY, Appel LJ, Creager MA, Kris-Etherton PM, Miller M, Rimm EB, Rudel LL, Robinson JG et al.. Dietary fats and cardiovascular disease: a presidential advisory from the American Heart Association. Circulation. 2017;136(3):e1–e23. [DOI] [PubMed] [Google Scholar]
- 26. Zeisel SH, da Costa K-A. Choline: an essential nutrient for public health. Nutr Rev. 2009;67(11):615–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cross AJ, Major JM, Rothman N, Sinha R. Urinary 1-methylhistidine and 3-methylhistidine, meat intake, and colorectal adenoma risk. Eur J Cancer Prev. 2014;23(5):385–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dragsted LO, Gao Q, Scalbert A, Vergères G, Kolehmainen M, Manach C, Brennan L, Afman LA, Wishart DS, Andres Lacueva C et al.. Validation of biomarkers of food intake–critical assessment of candidate biomarkers. Genes Nutr. 2018;13:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maintz L, Novak N. Histamine and histamine intolerance. Am J Clin Nutr. 2007;85(5):1185–96. [DOI] [PubMed] [Google Scholar]
- 30. Johnston CS. The antihistamine action of ascorbic acid. In: Harris JR. Subcellular biochemistry: ascorbic acid: biochemistry and biomedical cell biology. Boston (MA): Springer US; 1996. pp. 189–213. [DOI] [PubMed] [Google Scholar]
- 31. Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U, Menni C, Moayyeri A, Santer P, Rungger G et al.. Lipidomics profiling and risk of cardiovascular disease in the Prospective Population-Based Bruneck Study. Circulation. 2014;129(18):1821–31. [DOI] [PubMed] [Google Scholar]
- 32. Jaremek M, Yu Z, Mangino M, Mittelstrass K, Prehn C, Singmann P, Xu T, Dahmen N, Weinberger KM, Suhre K et al.. Alcohol-induced metabolomic differences in humans. Transl Psychiatry. 2013;3(7):e276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Scalbert A, Brennan L, Fiehn O, Hankemeier T, Kristal BS, van Ommen B, Pujos-Guillot E, Verheij E, Wishart D, Wopereis S. Mass-spectrometry-based metabolomics: limitations and recommendations for future progress with particular focus on nutrition research. Metabolomics. 2009;5(4):435–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.