ABSTRACT
Background
Human milk peptides released by gastrointestinal proteases have been identified with bioactivities that can benefit the infant but must first reach their respective sites of activity. Peptides in the stool either survived to or were released inside the intestinal tract, and thus had the opportunity to exert bioactivity there. However, it is unknown whether any milk peptides, bioactive or not, can survive in the stool of infants.
Objective
The aim of this study was primarily to identify milk peptides in infant stool samples and secondarily test the hypotheses that the milk peptide profiles of stools are different between preterm infants at different days of life and between preterm and term infants.
Methods
Infant stool samples were collected from 16 preterm infants (<34 weeks gestational age) at 8 or 9 and 21 or 22 days of life (DOL), and from 10 term infants (>34 weeks gestational age) at 8 or 9 DOL. Milk peptides were isolated from the stool samples and identified using tandem MS. The peptide counts and abundances were compared between infant groups.
Results
In total, 118 exclusively milk-derived peptides from the caseins and α-lactalbumin were present in the stool samples, including some peptides with known or potential bioactivity. The remaining 8014 identified peptides could be derived either from milk or endogenous proteins. Although many individual milk peptides were significantly different between preterm infants at 8/9 and 21/22 DOL and between preterm and term infants, total peptide abundance and count were similar for all 3 groups.
Conclusions
This is the first study to confirm the survival of milk peptides in the stool of infants. Some of the peptides had potential bioactivities that could influence infant gut development. These results are important to understand the physiological relevance of human milk peptides to the infant.
Keywords: peptides, human milk, stool, infant, bioactive
Introduction
Over the course of digestion, human milk proteins are broken down by the action of milk, gastric, intestinal, and bacterial proteases. These proteases release amino acids that the infant can absorb and utilize for protein synthesis and growth, but the first compounds cleaved from proteins are larger peptides. Many milk peptides have been identified with bioactivities that may be useful to the developing infant by killing pathogenic microbes (1, 2), stimulating the growth of commensal bacteria (3, 4), stimulating the immune system (5), stimulating mucin secretion (6), and extending gastrointestinal (GI) transit time (7). In many instances, the released peptides have distinct and more potent bioactivity than their intact parent proteins (8, 9). However, most bioactive peptides have been identified from in vitro digested milk and milk products. Only 1 study has identified novel bioactive peptides from undigested human milk (10), whereas others have confirmed the in vivo release of bioactive peptides in human milk and infant gastric digesta through homology searches (11, 12). As milk and gastric digesta represent only the beginning of protein digestion, little is known about how bioactive peptides are released and survive digestion throughout the entire GI tract.
For bioactive milk peptides to be relevant to the developing infant, they must first be released from their parent protein by digestion and then survive until they reach their site of action. For many bioactivities, this site is the upper intestinal tract and colon, where they can either be absorbed into the bloodstream to act systemically, or can act locally on bacteria, immune cells, and intestinal epithelial cells. The different proteolytic environments of the GI tract alter the peptide profile at each site by cleaving new peptides and breaking down those already released (11, 13). Milk proteins are exposed to different proteases with different cleavage site specificities at each site of digestion. In human milk within the mammary gland, proteases such as plasmin, thrombin, elastase, and kallikrein initiate the breakdown of milk proteins into peptides (14–16). In the stomach, pepsin is secreted, and cathepsin D from milk is activated by the high acidity (17). Protein digestion continues in the intestinal tract with the addition of pancreatic proteases like trypsin and chymotrypsin and brush border exopeptidases. In the colon, microbes can contribute to further proteolysis. Indeed, fecal proteolysis experiments on pancreatectomized and healthy subjects show that the gut microbiota contributes to digestive proteolysis (18). Any peptides that are present in the stool must therefore either have been released in the colon or survived proteolysis long enough to make it to the colon. These peptides would have had the potential to exert site-specific bioactivities in the colon and upper intestinal tract relevant to the infant's health, such as antimicrobial or immunomodulatory activity.
There have been few studies on the survival of milk proteins in stool. Human whey proteins such as lysozyme, lactoferrin, and IgA have been identified in both preterm and term infant stool through immunoblotting (19–21) and proteomics (22). Intact casein proteins have not been identified in the stool. Although lactoferrin and many other milk proteins can be produced endogenously by humans, human lactoferrin and its fragments were identified only in the stool of infants fed human milk and not bovine milk, suggesting that no endogenous lactoferrin is secreted by the infant (23). No studies have yet been performed on the survival of milk protein-derived peptides.
The primary objective of this study was to determine whether any human milk peptides survive GI digestion to reach the stool of infants. Secondary objectives were to compare the milk peptides in the stools of preterm infants between 8 or 9 days of life (8/9 DOL) and 21 or 22 (21/22) DOL, and between preterm and term infants at 8/9 DOL. Peptidomics analysis was used to identify milk peptides from the stool of term and preterm infants. The data was analyzed for similarities and differences in the peptide profiles of stools from the infants and to identify the presence of bioactive peptides.
Methods
Materials
Ammonium bicarbonate and HPLC-grade acetonitrile were obtained from Thermo Fisher Scientific, trifluoroacetic acid and HPLC-grade formic acid were obtained from EMD Millipore, HPLC-grade ethanol, iodoacetamide, and trichloroacetic acid were obtained from Sigma-Aldrich, and dithiothreitol was obtained from Promega.
Participants and enrollment
This study was approved by Institutional Review Boards at Legacy Health Systems and Oregon State University. Infant subjects were enrolled at Randall Children's Hospital and were grouped as either preterm (preterm infants of <34 weeks gestational age) or term (late-preterm and full-term infants of >34 weeks gestational age). As this study was primarily exploratory as to the presence of milk peptides in stool, the sample size of the groups was not calculated. Clinical data for each infant was collected upon enrollment and at each feeding and are listed in Table 1. Eligibility criteria for enrollment included having an indwelling naso/orogastric feeding tube, tolerating <60 min bolus feeding and ≥4 mL feed volumes, and mothers that could produce a volume of milk for 1 full day of feeding. Exclusion criteria included life-threatening diagnoses, GI anomalies or major GI surgeries, genitourinary anomalies, and any significant metabolic or endocrine diseases.
TABLE 1.
Characteristics for infants included in this study
| Infant | Maturity | 8/9 DOL | 21/22 DOL | Postmenstrual age, wk | Birth weight, g |
|---|---|---|---|---|---|
| 1 | Preterm | X | X | 30.3 | 1205 |
| 2 | Preterm | X | X | 30.3 | 1105 |
| 3 | Preterm | X | X | 31.7 | 2070 |
| 4 | Preterm | X | — | 26.6 | 695 |
| 5 | Preterm | X | — | 27.9 | 1165 |
| 6 | Preterm | — | X | 31.4 | 1843 |
| 7 | Preterm | X | — | 31.9 | 1920 |
| 8 | Preterm | — | X | 26.4 | 1140 |
| 9 | Preterm | X | — | 26.4 | 880 |
| 10 | Preterm | — | X | 27.7 | 1080 |
| 11 | Preterm | — | X | 27.7 | 1050 |
| 12 | Preterm | X | X | 26 | 900 |
| 13 | Preterm | X | X | 26 | 900 |
| 14 | Preterm | X | X | 31 | 1340 |
| 15 | Preterm | X | — | 31 | 1220 |
| 16 | Preterm | X | X | 32 | 1245 |
| 17 | Term | X | — | 38.7 | 1930 |
| 18 | Term | X | — | 34.7 | 1625 |
| 19 | Term | X | — | 35.9 | 3657 |
| 20 | Term | X | — | 39.6 | 2455 |
| 21 | Term | X | — | 34 | 2105 |
| 22 | Term | X | — | 35.7 | 2785 |
| 23 | Term | X | — | 37.3 | 3040 |
| 24 | Term | X | — | 34.4 | 2280 |
| 25 | Term | X | — | 34.4 | 2135 |
| 26 | Term | X | — | 34 | 2570 |
Stool was collected from preterm infants once during a 2-d period at 8/9 DOL and/or 21/22 DOL and from term infants at 8/9 DOL. As all enrolled term infants were discharged from the hospital prior to reaching 21 DOL, no stool was able to be collected for the 21/22 DOL time point for term infants. Additionally, several preterm infants did not stool over the course of one or the other of the 2-d periods and only had 1 stool sample collected. Feedings were prepared at Randall Children's Hospital using aseptic techniques. Frozen human milk was thawed at 37°C and delivered to the infant through the naso/orogastric feeding tube over a time period of 30–60 min. Nurses attempted to collect all stool produced over 48 h by the infant after their first feeding on days 8 and 21. In total, 16 preterm infants and 10 term infants were included in this study. After stool was collected, it was immediately frozen at −80°C and transported to Oregon State University on dry ice for sample analysis.
Sample preparation
Initial stool preparation
Stool samples were thawed on ice, and a small portion (∼500 mg) was collected, weighed, and dissolved in water to a concentration of 10% m/v. The samples were thoroughly agitated with a vortex mixer and sonicated for 10 s at 60 amps to ensure homogenization of the mixture. The samples were centrifuged at 4000 × g for 10 min at 4°C to precipitate remaining large solids, and the supernatant was centrifuged at 12,000 × g for 20 min at 4°C to remove cellular matter and lipids. The infranatant was pipetted from below the lipid layer and stored at −80°C until analysis.
Protein and peptide concentration determination
The combined protein and peptide concentrations and peptide isolate concentrations of the stool samples were determined in duplicate with the Pierce™ Quantitative Colorimetric Peptide Assay (Thermo Fisher Scientific) based on the reduction of Cu2+ to Cu1+ by peptide bonds. Two aliquots of 40 μL were removed from the stool infranatants. The first aliquot was analyzed for combined protein and peptide following the protocol for the kit. The concentration of only the peptide (peptide isolate) was determined in the second aliquot after ethanol precipitation of intact proteins. The samples were mixed with 160 μL of ice-cold ethanol and incubated for 2 h at −20°C. Samples were centrifuged at 12,000 × g for 30 min at 4°C and the pellet was discarded. The supernatant was lyophilized, and the peptides were reconstituted in 40 μL of water for concentration determination.
Total peptide extraction
Peptides were extracted from 100 μL of the infranatant as described in our previous peptidomic publication, with some modifications (16). To prevent milk peptides from potentially being precipitated with intact proteins, any disulfide bonds between the peptides and proteins were reduced and alkylated. The samples were mixed with 100 μL of 200 mM ammonium bicarbonate. Dithiothreitol was added to the samples to a final concentration of 40 mM, and the samples were incubated at 56°C for 45 min. Iodoacetamide was added to a final concentration of 100 mM and the samples were incubated at room temperature in the dark for 1 h. Intact proteins were precipitated as described previously (16). The peptides in the supernatant were treated by C18 reverse-phase extraction as described previously (16). After elution from the C18 column, the peptides were lyophilized and rehydrated in 100 μL of nanopure water prior to MS analysis.
LCMS
Peptides were analyzed with MS as described in our previous publication (12), with some modifications as follows. The LC phase was condensed so that the peptides were eluted from the ultra-performance liquid chromatography column over a period of 60 min. The separation gradient was 3–10% solvent B over 3 min, 10–30% solvent B over 42 min, 30–90% solvent B over 3 min, held at 90% solvent B for 4 min, 90–3% solvent B over 1 min, and held at 3% solvent B for 7 min. A 30-min column wash was included between sample separations. The mass spectra were collected and analyzed with the following altered parameters. Peptides were ionized with an electrospray voltage of 2300 V. Scanned masses were between 375 and 1500 m/z and with a charge state of 2–8. Precursor ions were fragmented with higher-energy collisional dissociation with a collision energy of 35%. Peptides were detected from the raw files using Thermo Proteome Discoverer 2.2.0.388 [Thermo Fisher Scientific]. Dynamic peptide modifications allowed were phosphorylation of serine and threonine, oxidation of methionine, and carbamidomethylation of cysteine.
Data analysis
Stool samples were analyzed for the combined protein and peptide concentration, peptide isolate concentration, peptide abundance, and peptide count. Peptide abundance is unitless and represents the summed ion intensities from the mass spectra, whereas peptide count is the number of unique peptide sequences. Milk peptides from proteins of similar function were sorted for analysis into the following groups: Igs, antiproteases, proteases, nutrient-binding proteins, caseins, mucins, and other milk proteins. The protein composition of the groups are defined in Supplemental Table 1.
Statistical comparisons were performed with RStudio 1.2.1335 (RStudio). Stool peptides were grouped into preterm infants at 8/9 DOL, preterm infants at 21/22 DOL, and term infants at 8/9 DOL. Paired t-tests were used to compare stool peptide isolate concentrations and stool protein and peptide concentrations within each group. ANOVA followed by the posthoc Tukey–Kramer test was used to compare peptide abundance, count, and concentration between preterm 8/9, 21/22, and term 8/9 groups. Significance was determined by a P value of <0.05. Data are presented as mean ± SE.
Milk peptides were analyzed for sequence homology with bioactive peptides in the Milk Bioactive Peptide Database (MBPDB) (24). The search type was “Sequence” with a similarity threshold of 80%. PepEx (http://mbpdb.nws.oregonstate.edu/pepex/) was used to map the identified milk peptides to their location in the parent protein sequence.
Results
Milk peptide profile of infant stool
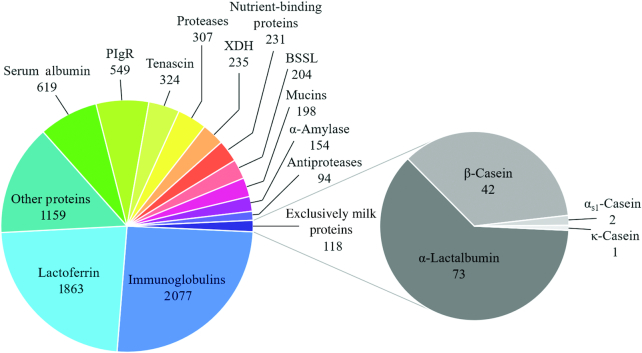
The overall peptide profile of all stool samples was composed of 8132 peptides divided amongst 169 unique proteins previously identified in human milk. Although the majority of the peptides are derived from proteins that could be from either milk or endogenous sources (hereafter referred to as “potential milk peptide”), 118 peptides were derived from proteins that are exclusive to breast milk: 73 peptides from α-lactalbumin, 42 from β-casein, 2 from αs1-casein, and 1 from κ-casein. Of the remaining peptides that are potentially derived from milk, lactoferrin was the single largest contributor with 1863 peptides (Figure 1). There were 2077 combined peptides from all Ig proteins, and the largest Ig protein contributors were Ig heavy constant α (IGHA) 1 and 2, which encode the constant segment of the heavy chain of IgA. IGHA1 had 360 peptides and IGHA2 had 197; an additional 408 were indistinguishable between IGHA1 and IGHA2. The heavy chains of IgM and IgG contributed 166 and 118 peptides, respectively. The full list of proteins and how many peptides were identified from each is included as Supplemental Table 2.
FIGURE 1.
Pie chart of the breakdown of milk peptides identified in the infant stool samples. Numbers underneath the pie section labels represent the number of unique peptides identified from all infant stool samples, n = 33. BSSL, bile salt-stimulated lipase; PIgR, polymeric Ig receptor; XDH, xanthine dehydrogenase.
The mean concentrations of combined proteins and peptides in the stool samples were 28.2 ± 4.3 μg/mg for preterm infants at 8/9 DOL, 20.6 ± 2.6 μg/mg for preterm infants at 21/22 DOL, and 17.8 ± 2.7 μg/mg for term infants at 8/9 DOL (Supplemental Figure 1). The mean concentrations of peptide were less than half of the combined proteins and peptides, at 9.6 ± 2.2 μg/mg, 7.7 ± 1.1 μg/mg, and 7.6 ± 1.4 μg/mg for preterm 8/9, preterm 21/22, and term 8/9 infant stools, respectively. All infant groups had significantly different stool peptide and protein concentrations. There were no significant differences between preterm 8/9, 21/22, and term 8/9 infant stools for peptide concentration or combined protein and peptide concentration.
Bioactive milk peptides in the infant stool
To predict the bioactivity of identified milk peptides, the peptides were compared for sequence homology with known bioactive peptides in the MBPDB (24). There were 26 peptides that matched with ≥80% sequence homology with the database, but as some query peptides matched with multiple known peptides, only 19 of the matches were unique peptides (Table 2). All homologous peptides derived from lactoferrin and β-casein, which is unsurprising as the majority of bioactive peptides in the MBPDB are from these 2 proteins. Two peptides, VVPYPQR and DLENLHLPLPL, matched with peptides derived from bovine milk β-casein; the rest matched with peptides from human milk proteins. Antimicrobial activity was the most prevalent function with 12 homologous peptides, followed by DNA synthesis-stimulatory with 4, antioxidant with 2, antihypertensive with 2, and opioid with 1. Only 1 query peptide, RETIESLSSSEESITEYK from β-casein, was 100% homologous with a known bioactive peptide. This peptide was identified to stimulate DNA synthesis and cell proliferation of BALB/c3T3 mice fibroblasts (25) and was previously found in milk and gastric samples (11, 12).
TABLE 2.
Milk peptides from stool that matched with ≥80% sequence homology with a known bioactive milk peptide from the MBPDB
| Known peptide | Query peptide | Protein | % Sequence homology | Function | No. of infants |
|---|---|---|---|---|---|
| AVPYPQR1 | VVPYPQR | β-casein | 85.7 | Antihypertensive | 31 |
| AVPYPQR1 | VVPYPQR | β-casein | 85.7 | Antimicrobial | 31 |
| AVPYPQR1 | VVPYPQR | β-casein | 85.7 | Antioxidant | 31 |
| ENLHLPLPLL1 | DLENLHLPLPL2 | β-casein | 81.8 | Antihypertensive | 13 |
| LENLHLPLP | DLENLHLPLPL2 | β-casein | 81.8 | Antihypertensive | 13 |
| LLNQELLLNPTHQIYPV | NQELLLNPTHQIYPV2 | β-casein | 88.2 | Antimicrobial | 6 |
| LLNQELLLNPTHQIYPV | QALLLNQELLLNPTHQIYP2 | β-casein | 85 | Antimicrobial | 25 |
| QELLLNPTHQIYPVTQPLAPVHNPISV | NQELLLNPTHQIYPVTQPLAPVHNPISV2 | β-casein | 96.4 | Antimicrobial | 29 |
| QELLLNPTHQIYPVTQPLAPVHNPISV | LLNPTHQIYPVTQPLAPVHNPISV2 | β-casein | 88.9 | Antimicrobial | 21 |
| QELLLNPTHQIYPVTQPLAPVHNPISV | LLLNQELLLNPTHQIYPVTQPLAPVHNPISV2 | β-casein | 87.1 | Antimicrobial | 24 |
| QVVPYPQ | QVVPYPQR | β-casein | 87.5 | Antioxidant | 16 |
| QVVPYPQ | VVPYPQR | β-casein | 85.7 | Antioxidant | 31 |
| RETIESLSSSEESITEYK | RETIESLSSSEESITEYK2 | β-casein | 100 | Stim. DNA synthesis | 30 |
| RETIESLSSSEESITEYK | ETIESLSSSEESITEYK2 | β-casein | 94.4 | Stim. DNA synthesis | 33 |
| RETIESLSSSEESITEYK | RETIESLSSSEESITEYKQK2 | β-casein | 90 | Stim. DNA synthesis | 20 |
| RETIESLSSSEESITEYK | RETIESLSSSEESIT2 | β-casein | 83.3 | Stim. DNA synthesis | 16 |
| VENLHLPLPLL1 | DLENLHLPLPL2 | β-casein | 81.8 | Antihypertensive | 13 |
| EATKCFQWQRNMRKVR | SQPEATKCFQWQRNMR | Lactoferrin | 81.3 | Antimicrobial | 7 |
| FFSASCVPGADKGQFPNLCRLCAGTGENKCA | FFSASCVPGADKGQFPNLCRLCAGTGENK | Lactoferrin | 93.6 | Antimicrobial | 6 |
| KYLGPQY | KYLGPQYV | Lactoferrin | 87.5 | Opioid | 23 |
| PEATKCFQWQRNMRKVR | SQPEATKCFQWQRNMR | Lactoferrin | 82.4 | Antimicrobial | 7 |
| QPEATKCFQWQRNMRKVR | AVSQPEATKCFQWQRNMR | Lactoferrin | 83.3 | Antimicrobial | 17 |
| QPEATKCFQWQRNMRKVR | SQPEATKCFQWQRNMR | Lactoferrin | 83.3 | Antimicrobial | 7 |
| TKCFQWQRN | ATKCFQWQR | Lactoferrin | 88.9 | Antimicrobial | 21 |
| TKCFQWQRN | TKCFQWQR | Lactoferrin | 88.9 | Antimicrobial | 23 |
| TKCFQWQRN | EATKCFQWQR | Lactoferrin | 80 | Antimicrobial | 15 |
Known peptide was derived from a bovine milk protein.
Peptide has been previously identified in human milk or infant gastric samples.
MBPDB, milk bioactive peptide database; Stim. DNA synthesis, stimulates cell DNA synthesis.
Twelve homologous peptides were identified in ≥50% of the infants’ stools, and 4 in ≥80% of the infants. RETIESLSSSEESIT, with potential DNA synthesis-stimulating activity, was the only homologous peptide identified in every infant stool. Each infant had a mean of 11.4 ± 3.6 homologous peptides in their stool samples (between 4 and 17 homologous peptides per infant). Every peptide was present in ≥1 infant stool from each of the 3 groups (preterm at 8/9 DOL, preterm at 21/22 DOL, and term at 8/9 DOL).
Comparison of peptides from preterm and term infant stools
The total abundance and count of the peptides were highly similar among stools from preterm infants at 8/9 DOL, preterm infants at 21/22 DOL, and term infants at 8/9 DOL (Figure 2). There were no significant differences in peptide abundance or count between preterm 8/9, 21/22, and term 8/9 infants.
FIGURE 2.
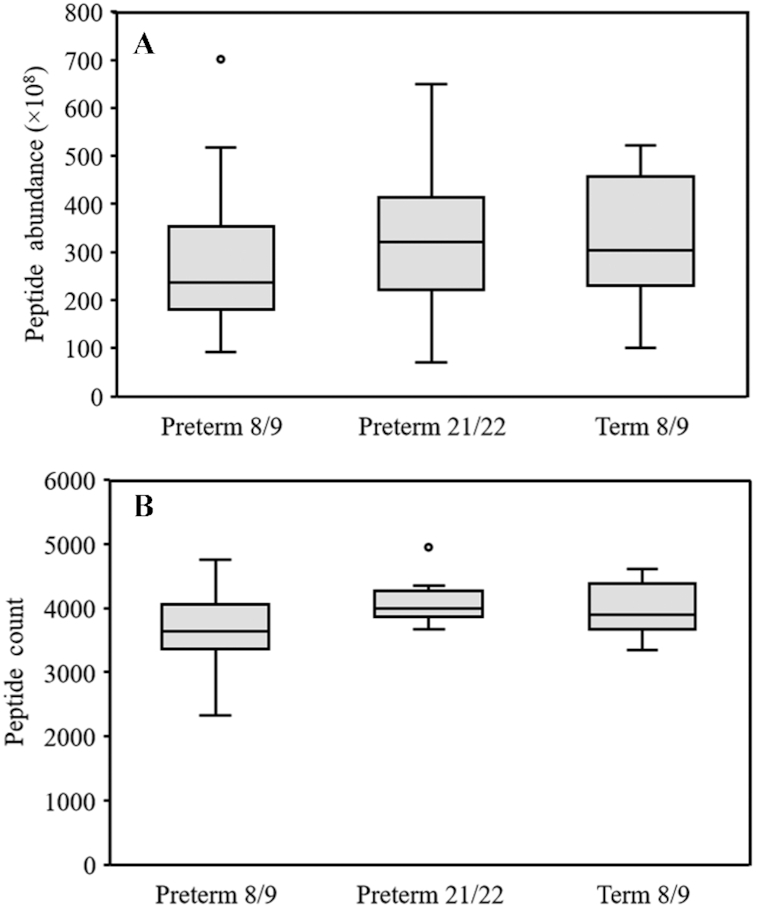
Boxplots of the abundance (A) and count (B) of total potential milk peptides from the stool of preterm infants at 8/9 DOL and 21/22 DOL and term infants at 8/9 DOL. Preterm 8/9 n = 12, preterm 21/22 n = 11, and term n = 10. DOL, days of life.
The 3 infant groups remained similar when peptides from individual potential milk proteins or groups of related milk proteins were analyzed for abundance (Table 3) and count (Table 4). No significant differences were found for peptide abundance in individual proteins, only for peptide count. Preterm 8/9 infant stool peptide counts were significantly lower than preterm 21/22 for lactoferrin, polymeric Ig receptor (PIgR), xanthine dehydrogenase, and mucins, and were significantly lower than term 8/9 for PIgR. The peptides from individual milk proteins and potential milk proteins were thus highly similar in stools among the 3 infant groups for both count and abundance.
TABLE 3.
Comparison of the mean abundance of peptides from select milk proteins and potentially milk proteins between preterm infants at 8/9 DOL, preterm infants at 21/22 DOL, and term infants at 8/9 DOL1
| Preterm 8/92 | Preterm 21/222 | Term2 | |
|---|---|---|---|
| n = 12 | n = 11 | n = 10 | |
| Immunoglobulins | 7.13 × 109 ± 1.08 × 109 | 6.78 × 109 ± 1.12 × 109 | 7 × 109 ± 9.55 × 108 |
| Lactoferrin | 6.15 × 109 ± 1.33 × 109 | 7.7 × 109 ± 1.23 × 109 | 7.16 × 109 ± 1.03 × 109 |
| PIgR | 1.9 × 109 ± 3.92 × 108 | 2.35 × 109 ± 3.05 × 108 | 2.45 × 109 ± 3.74 × 108 |
| Serum albumin | 1.82 × 109 ± 2.8 × 108 | 2.08 × 109 ± 2.37 × 108 | 2.18 × 109 ± 3.28 × 108 |
| Antiproteases | 1.08 × 109 ± 2.28 × 108 | 1.18 × 109 ± 1.62 × 108 | 1.31 × 109 ± 2.46 × 108 |
| Proteases | 1.11 × 109 ± 3.24 × 108 | 1.27 × 109 ± 2.34 × 108 | 9.54 × 108 ± 1.69 × 108 |
| Tenascin | 9.05 × 108 ± 1.38 × 108 | 1.09 × 109 ± 1.39 × 108 | 1.12 × 109 ± 1.83 × 108 |
| Nutrient-binding proteins | 1.06 × 109 ± 2.2 × 108 | 1.02 × 109 ± 1.71 × 108 | 1 × 109 ± 1.72 × 108 |
| Xanthine dehydrogenase | 7.58 × 108 ± 1.71 × 108 | 9.78 × 108 ± 1.38 × 108 | 1.12 × 109 ± 2.13 × 108 |
| α-Amylase | 7.04 × 108 ± 1.77 × 108 | 7.45 × 108 ± 1.46 × 108 | 9.68 × 108 ± 1.82 × 108 |
| Bile salt-stimulated lipase | 6.02 × 108 ± 1.57 × 108 | 7.53 × 108 ± 1.71 × 108 | 6.01 × 108 ± 1.57 × 108 |
| Mucins | 4.15 × 108 ± 1.03 × 108 | 6.94 × 108 ± 8.66 × 107 | 7.43 × 108 ± 1.21 × 108 |
| α-Lactalbumin | 3 × 108 ± 7.45 × 107 | 4.05 × 108 ± 7.94 × 107 | 5.42 × 108 ± 1.31 × 108 |
| Caseins | 3.01 × 108 ± 9.19 × 107 | 4.74 × 108 ± 1.26 × 108 | 3.56 × 108 ± 7.6 × 107 |
Data are represented as mean ± SE.
Abundance units are the summed ion intensities for each peptide from the mass spectra.
DOL, days of life; PIgR, polymeric Ig receptor.
TABLE 4.
Comparison of the mean count of peptides from select milk proteins and potentially milk proteins between preterm infants at 8/9 DOL, preterm infants at 21/22 DOL, and term infants at 8/9 DOL1
| Preterm 8/92 | Preterm 21/222 | Term2 | |
|---|---|---|---|
| n = 12 | n = 11 | n = 10 | |
| Immunoglobulins | 916 ± 45 | 987 ± 29.6 | 972 ± 30.3 |
| Lactoferrin | 789 ± 58.5 | 945 ± 26.6* | 916 ± 38.6 |
| Serum albumin | 269 ± 14.7 | 309 ± 10 | 299 ± 8.7 |
| PIgR | 256 ± 16 | 302 ± 8.1* | 300 ± 9.4* |
| Proteases | 136 ± 7.9 | 159 ± 5 | 146 ± 6.4 |
| Tenascin | 136 ± 8.9 | 159 ± 5.6 | 144 ± 6.8 |
| Xanthine dehydrogenase | 103 ± 8 | 128 ± 5.6* | 125 ± 4.1 |
| Nutrient-binding proteins | 111 ± 5.7 | 115 ± 3.9 | 120 ± 4.4 |
| Mucins | 89.1 ± 7.7 | 111 ± 4.1* | 106 ± 4.9 |
| Bile salt-stimulated lipase | 87.2 ± 5.1 | 90.3 ± 4 | 89 ± 3.19 |
| α-Amylase | 70.2 ± 3.6 | 78.7 ± 3.5 | 74.5 ± 3.5 |
| Antiproteases | 59.3 ± 2.6 | 60 ± 2.2 | 58.5 ± 1.8 |
| α-Lactalbumin | 28.3 ± 3 | 32.9 ± 1.7 | 34.6 ± 1.7 |
| Caseins | 21.2 ± 2.7 | 26.5 ± 1.5 | 26.6 ± 1.4 |
Data are represented as mean ± SE.
Indicates the mean is significantly different from the mean for preterm 8/9 in the same row, P < 0.05.
Count units are the number of unique peptide sequences.
DOL, days of life; PIgR, polymeric Ig receptor.
The peptide profiles of each group were highly conserved, with peptides common to all groups accounting for the majority of the peptide abundance and count. One hundred and twenty-five peptides were identified in 100% of the samples in each group and accounted for 18.8% of the mean abundance of peptides in the samples, 550 peptides were identified in ≥80% of the samples in each group and accounted for 32.5% of the mean abundance, 1716 peptides were identified in ≥50% of the samples in each group and accounted for 33.5% of the mean abundance, and 5742 peptides were identified in <50% of the samples in ≥1 group and accounted for only 15.2% of the mean abundance. On an individual protein level, peptides present in ≥50% of the samples in each group made up the majority of the mean peptide abundance for all proteins except bile salt-stimulated lipase and the caseins (Figure 3). These results suggest that although each group had different peptide profiles, those peptides that were the same between the groups were present at much higher relative amounts than peptides that were different.
FIGURE 3.

Stacked column charts of the abundance (A) and count (B) of peptides from individual milk proteins and protein groups. Stacking is based on the percentage of infants in each group that the peptides were identified in. Data are represented as means, n = 33. BSSL, bile salt-stimulated lipase; PIgR, polymeric Ig receptor; XDH, xanthine dehydrogenase.
Only once individual peptides were analyzed did many of the differences between groups begin to appear. Both preterm 21/22 and term 8/9 infants had many more individual stool peptides that were in significantly higher abundance (>5-fold increase) than preterm 8/9 (Figure 4A, B). This pattern was conserved for both lactoferrin peptides (Figure 4C, D) and combined α-lactalbumin and casein peptides (Figure 4E, F). Three homologous peptides were also significantly different among the groups. DLENLHLPLPL from β-casein, with potential antihypertensive activity, was significantly higher in preterm 21/22 and term 8/9 infant stools compared with preterm 8/9, and LLNPTHQIYPVTQPLAPVHNPISV from β-casein, with potential antimicrobial activity, was significantly higher in term than in preterm 8/9. Only ATKCFQWQR from lactoferrin, with potential antimicrobial activity, was significantly higher in the preterm 8/9 infant stools than in term 8/9.
FIGURE 4.
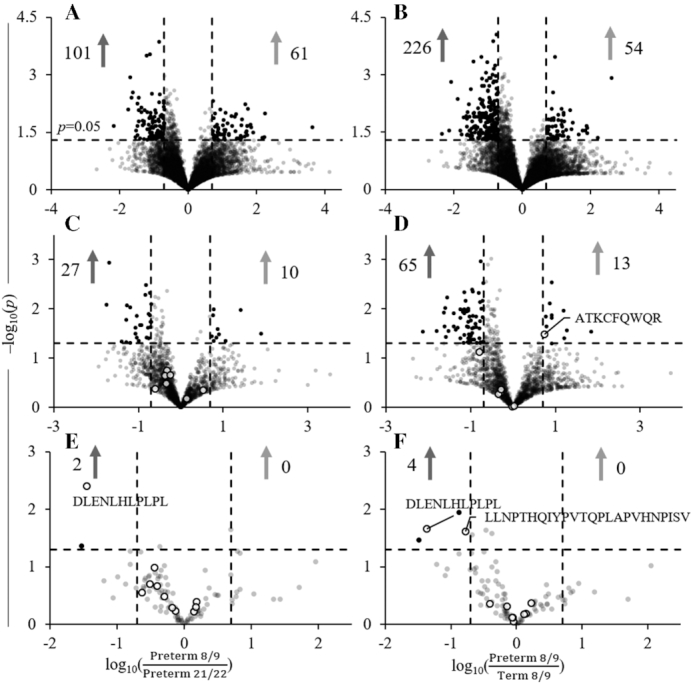
Volcano plots of total peptide abundance for preterm 8/9 and preterm 21/22 (A) and preterm 8/9 and term 8/9 (B), lactoferrin peptide abundance for preterm 8/9 and preterm 21/22 (C), and preterm 8/9 and term 8/9 (D), and combined α-lactalbumin and casein peptide abundance for preterm 8/9 and preterm 21/22 (E), and preterm 8/9 and term 8/9 (F). Filled dots represent peptides that were both significantly different (P < 0.05) and present in 1 of the groups at 5-fold increased abundance. For lactoferrin and α-lactalbumin/casein, white dots with black borders represent peptides with ≥80% sequence homology to a known bioactive peptide from the MBPDB. Light gray arrows represent the number of peptides increased in the numerator, and dark gray arrows represent the number of peptides increased in the denominator. Preterm 8/9 n = 12, preterm 21/22 n = 11, and term n = 10. MBPDB, milk bioactive peptide database.
Preterm 8/9, 21/22, and term 8/9 infant stools had similar counts and abundances of peptides, but there was considerable variation in the peptide profiles of each individual infant's stool. Compared with the mean peptide abundance in stools across all infants, the stools of infants 3, 16, 19, and 20 had higher peptide abundances for nearly all major proteins and protein groups, whereas stools of infants 2, 12, 13, 21, and 26 had much lower peptide abundances (Figure 5A). There were no clear patterns of increase or decrease in peptide abundance in preterm infant stools from 8/9 to 21/22 DOL. Some infants, such as infant 2, had a decreased stool peptide abundance between measurements, whereas the abundance in stools of others, such as infants 12 and 13, was increased. The percentage composition of each infant's stool peptide profile also varied between and within groups (Figure 5B). Each group exhibited variation in the relative abundance of peptides from the measured milk proteins and potential milk proteins. Ig and lactoferrin peptides dominated the profiles in stools from infants 4 and 13 at 8/9 DOL compared with the other infants, and stools from infants 9 at 8/9 DOL, 12 at 21/22 DOL, and 23 had increased percentages of lower abundant peptides such as α-lactalbumin, caseins, α-amylase, and nutrient-binding proteins.
FIGURE 5.
Variation in the peptide profiles of each infant. (A) Heatmap of the fold-change in peptide abundance from a specific protein or group of proteins for each infant compared with the mean. (B) Percent contribution of each protein or group of proteins to the total peptide profile of each infant. BSSL, bile salt-stimulated lipase; PIgR, polymeric Ig receptor; XDH, xanthine dehydrogenase.
Mapping the stool peptides to their parent protein
Peptide abundances were mapped to the parent protein sequence for lactoferrin, α-lactalbumin, and β-casein (Supplemental Figure 2). Peptides came from nearly identical regions of the proteins for all 3 groups. There were clearly defined regions of peptides at α-lactalbumin f(42–64) and f(71–93), and β-casein f(1–24) and f(161–211). However, lactoferrin peptides were more spread out across the lactoferrin sequence. There were distinct peaks of peptide abundance in lactoferrin, but there was a nearly continuous abundance of peptides across the entire sequence. For peptide count, the pattern of release was similar to that of abundance but with more similar peptide counts across the sequences of the proteins for the 3 groups (Supplemental Figure 3). For both count and abundance, stools from term 8/9 infants had slightly more peptides at each of the sites in the protein except for some regions in lactoferrin and the N-terminus of β-casein, where stools from preterm infants at 21/22 DOL had the most.
Discussion
This study is the first to profile the milk peptidome of infant stool. Previous investigations have profiled the peptidomes of human milk and infant gastric samples (11, 12, 26). Whereas those samples represent the beginning of protein digestion in the infant, stool represents the endpoint. Any milk proteins and peptides remaining in the stool are unavailable to the infant for nutrition but may be important bioactive factors in milk by influencing the gut environment through interactions with intestinal epithelial cells, immune cells, or the microbiota. The identification of remaining milk peptides in stool, some with known bioactivity or potential for bioactivity, represents another step along the path to determining whether the release and survival of milk peptides is coordinated to provide physiological benefit to the infant.
The peptide profile of stools differs significantly from the peptide profiles previously identified in undigested human milk and preterm gastric samples. The peptides in undigested human milk are primarily sourced from the caseins, with some from whey proteins such as osteopontin, PIgR, and butyrophilin subfamily 1 member a1 (10, 16). Almost no lactoferrin or α-lactalbumin peptides are in human milk, suggesting these proteins are relatively undigested. In the infant stomach, casein peptides still dominate the profile, but lactoferrin and α-lactalbumin begin to release small amounts of peptides at select regions in their sequences (11, 12, 26). The peptides in stool are completely different from those in milk and gastric digesta, with a predominance of lactoferrin peptides from across the entire sequence and little to no casein peptides. As the casein proteins are rapidly digested in the stomach and intestine (27), it is likely they are almost entirely broken down and absorbed as amino acids, di-, and tripeptides before they reach the stool, with only the N- and C-termini of β-casein remaining in significant quantities. On the other hand, although lactoferrin is cleaved at many more sites along its sequence as it progresses through the GI tract, the released peptides persist in the stool. Lactoferrin, especially iron-saturated lactoferrin, is highly resistant to pepsin, trypsin, and chymotrypsin proteolysis (28, 29), so the presence of its peptides in stool provides evidence that the peptides may retain some of this resistance.
Many of the peptides detected in this study could be derived from either milk proteins or endogenous proteins. However, there was evidence that at least the peptides from lactoferrin, the caseins, and α-lactalbumin were from milk. β-casein, αs1-casein, κ-casein, and α-lactalbumin are only secreted into milk, so any peptides from these proteins must have derived from the ingested milk. Lactoferrin is produced endogenously by mucosal linings and neutrophils in the intestine as part of the innate immune system (30), but a previous study comparing human lactoferrin in the stool of both human milk-fed and bovine milk-fed infants discovered no human lactoferrin from the bovine milk-fed infants (23). Therefore, the majority of the lactoferrin peptides identified in this study were also likely from the milk, rather than from the infants’ endogenous production. IgA, IgM, and IgG, bile salt-stimulated lipase, and PIgR are found in large quantities in breast milk, but these are also produced endogenously so it remains unclear whether their peptides derive from milk or from the infant (31–33).
There were very few differences in the stool peptide profiles between the preterm infants at 8/9 DOL, 21/22 DOL, and the term infants at 8/9 DOL. The majority of the peptide abundance in stool was composed of peptides that were conserved in both preterm and term infants. Similar results were reported for peptides in human milk, where a minority of shared peptides accounted for the majority of milk peptide abundance (11, 16). The overall peptide content and the protein-specific peptides were similar between groups, although there was a large degree of variation in the stool peptide profiles of individual infants within groups. Only at the individual peptide level did many of the differences become apparent. The results show that although similar numbers of milk peptides and potential milk peptides survived in the stool, the specific peptides differed by infant age and birth maturity. However, as no sample size was calculated for this study, the significance of the comparisons between infant groups are considered exploratory in nature. Due to the high amount of variation between the peptides in the infant stools, a larger sample size may be required to more accurately determine the differences or similarities in peptides between infants.
Previous peptidomics studies comparing term and preterm milk peptides were performed on only human milk (34, 35). Milk peptide abundances are higher in preterm milk during early lactation ≤41 d, but then decrease to match term levels. The infants in this study were all aged 8–22 d, so would have been at the stage of life when preterm infants are consuming larger quantities of milk peptides than term infants. However, as the milk and potential milk peptide abundances in term and preterm infant stool were similar, something may have occurred during digestion to equalize the peptide concentrations. Compared with term infants, preterm infants have lower gastric proteolysis (14), and some studies report higher gastric pH (36), which could inhibit further milk protein digestion and peptide release in the stomach. In the duodenum, preterm infants produce less trypsin than term infants, which could also impact milk peptide release (37), but little else is known about infant intestinal digestion. Thus, it could be that preterm infants start with a higher peptide abundance in milk (34, 35) but are unable to cleave milk peptides as efficiently as term infants, leading to a similar milk peptide content in stool. Studies that clarify the digestive differences in the stomach and intestines of preterm and term infants are necessary for a full understanding of milk peptide release inside the infants.
Compared with the peptides identified in stool, many more homologous peptides have been identified in human milk and infant gastric samples. Previous searches with the MBPDB identified between 37 and 55 ≥80% homologous peptides in undigested human milk samples with 2 being identical to known bioactive peptides (11, 12, 16). Infant gastric samples contained 85 and 123 homologous human milk peptides, with ≤8 being identical to known bioactive peptides (11, 12). None of the homologous lactoferrin peptides identified in the present study have previously been identified, but 9 of the 12 homologous β-casein peptides were identified in both milk and gastric samples (11, 12). Only VVPYPQR (antihypertensive, antimicrobial, antioxidant), QVVPYPQR (antioxidant), and QALLLNQELLLNPTHQIYPV (antimicrobial) from β-casein are homologous peptides that are, for the first time, identified in stool but not milk or gastric samples, although peptides that differ by the addition or subtraction of 1 residue were previously identified (11, 12).
Although many peptides homologous to known bioactive peptides are released at the beginning of digestion, the present data show that most are further degraded until no longer bioactive by the time they reach the stool. However, some peptides with possible bioactivities relevant to infant health do persist. Many of the query peptides matched bioactive peptides with antimicrobial activity against bacteria and fungi that can infect the infant gut, such as Escherichia coli, Staphylococcus aureus, Klebsiella pneumoniae, and Candida albicans (38, 39). Antioxidant milk peptides scavenge reactive oxygen species and prevent lipid hydroxylation to reduce oxidative stress (40). The only peptide that matched 100% with a known bioactive peptide had DNA synthesis-stimulating properties for mouse fibroblasts (25). Human milk naturally contains peptide growth factors that stimulate intestinal development (41), so it could be that this DNA synthesis-stimulating peptide and others like it contribute to the trophic effects of milk. As stool represents the endpoint of digestion, it is unknown if other bioactive peptides are present inside the small intestine and gut that are broken down to amino acids before excretion. The majority of the peptides in the stool, as in milk and gastric digesta, did not match a known bioactive peptide, but it may be that some of these peptides possess a bioactivity that is currently undetermined. Furthermore, it is unknown whether the identified peptides are present in high enough quantities to have an appreciable effect on the infant. Further studies are needed to answer these remaining questions if the full relevance of milk peptides to infant health is to be determined.
This study is the first to identify that milk peptides of both whey and casein origin survive GI digestion to reach the stool of infants. Several of these peptides have sequences homologous to bioactive milk peptides with antimicrobial and DNA-stimulating activity. These peptides are present in the gut of the infant, where they have the opportunity to influence intestinal cells and the microbiota. The majority of the peptides were not homologous to a known bioactive milk peptide, but they should be investigated for bioactivity due to their confirmed presence in the infant gut. This study contributes to the developing map of milk peptide release across infant digestion and can provide directions for future studies that will determine the relevance of milk peptides to infant health.
Supplementary Material
Acknowledgments
We acknowledge the nurses and physicians of Randall Children's Hospital for facilitating the collection of samples, the Oregon State University Mass Spectrometry Facility for use of their instrumentation, and Yanming Di for statistical assistance. The authors’ responsibilities were as follows—DCD and RLB: designed the research; RLB, RKH, AMM, EAM, and RLM: conducted the research; RLB: analyzed data and performed statistical analysis; RLB, RKH, AMM, EAM, RLM and DCD: wrote the manuscript; DCD: had primary responsibility for final content; and all authors have read and approved the final manuscript.
Notes
Supported by NIH K99/R00 Pathway to Independence Career Award (R00HD079561) (DCD), NIH Oregon Clinical and Translational Research Institute TL1 Program (5TL1TR002371-03) (RLB), USDA National Institute of Food and Agriculture (DCD), the Gerber Foundation (DCD), and NIH Shared Instrument Grant (1S10OD020111-01).
Author disclosures: The authors report no conflicts of interest.
Supplemental Tables 1 and 2 and Supplemental Figures 1–3 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn.
Abbreviations used: DOL, days of life; GI, gastrointestinal; IGHA, Ig heavy constant α; MBPDB, milk bioactive peptide database; PIgR, polymeric Ig receptor.
References
- 1. Lupetti A, Paulusma-Annema A, Welling MM, Senesi S, van Dissel JT, Nibbering PH. Candidacidal activities of human lactoferrin peptides derived from the N-terminus. Antimicrob Agents Chemother. 2000;44(12):3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Håversen L, Kondori N, Baltzer L, Hanson LA, Dolphin GT, Dunér K, Mattsby-Baltzer I. Structure-microbicidal activity relationship of synthetic fragments derived from the antibacterial alpha-helix of human lactoferrin. Antimicrob Agents Chemother. 2010;54(1):418–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liepke C, Adermann K, Raida M, Mägert HJ, Forssmann WG, Zucht HD. Human milk provides peptides highly stimulating the growth of bifidobacteria. Eur J Biochem. 2002;269(2):712–8. [DOI] [PubMed] [Google Scholar]
- 4. Oda H, Wakabayashi H, Yamauchi K, Sato T, Xiao J-Z, Abe F, Iwatsuki K. Isolation of a bifidogenic peptide from the pepsin hydrolysate of bovine lactoferrin. Appl Environ Microbiol. 2013;79(6):1843–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Parker F, Migliore-Samour D, Floch F, Zerial A, Werner GH, Jollès J, Casaretto M, Zahn H, Jollès P. Immunostimulating hexapeptide from human casein: amino acid sequence, synthesis and biological properties. Eur J Biochem. 1984;145(3):677–82. [DOI] [PubMed] [Google Scholar]
- 6. Martínez-Maqueda D, Miralles B, De Pascual-Teresa S, Reverón I, Muñoz R, Recio I. Food-derived peptides stimulate mucin secretion and gene expression in intestinal cells. J Agric Food Chem. 2012;60(35):8600–5. [DOI] [PubMed] [Google Scholar]
- 7. Aslam H, Ruusunen A, Berk M, Loughman A, Rivera L, Pasco JA, Jacka FN. Unravelled facets of milk derived opioid peptides: a focus on gut physiology, fractures and obesity. Int J Food Sci Nutr. 2019:1–14. [DOI] [PubMed] [Google Scholar]
- 8. Sinha M, Kaushik S, Kaur P, Sharma S, Singh TP. Antimicrobial lactoferrin peptides: the hidden players in the protective function of a multifunctional protein. Int J Pept. 2013;2013:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mukhopadhya A, Noronha N, Bahar B, Ryan MT, Murray BA, Kelly PM, O'Loughlin IB, O'Doherty JV, Sweeney T. Anti-inflammatory effects of a casein hydrolysate and its peptide-enriched fractions on TNFα-challenged Caco-2 cells and LPS-challenged porcine colonic explants. Food Sci Nutr. 2014;2(6):712–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dallas DC, Guerrero A, Khaldi N, Castillo PA, Martin WF, Smilowitz JT, Bevins CL, Barile D, German JB, Lebrilla CB. Extensive in vivo human milk peptidomics reveals specific proteolysis yielding protective antimicrobial peptides. J Proteome Res. 2013;12(5):2295–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nielsen SD, Beverly RL, Underwood MA, Dallas DC. Release of functional peptides from mother's milk and fortifier proteins in the premature infant stomach. PLoS One. 2018;13(11):e0208204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beverly RL, Underwood MA, Dallas DC. Peptidomics analysis of milk protein-derived peptides released over time in the preterm infant stomach. J Proteome Res. 2019;18(3):912–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao D, Li L, Le TT, Larsen LB, Xu D, Jiao W, Sheng B, Li B, Zhang X. Digestibility of glycated milk proteins and the peptidomics of their in vitro digests. J Sci Food Agric. 2019;99(6):3069–77. [DOI] [PubMed] [Google Scholar]
- 14. Demers-Mathieu V, Nielsen SD, Underwood MA, Borghese R, Dallas DC. Analysis of milk from mothers who delivered prematurely reveals few changes in proteases and protease inhibitors across gestational age at birth and infant postnatal age. J Nutr. 2017;146(6):1152–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Heegaard CW, Larsen LB, Rasmussen LK, Højberg KE, Petersen TE, Andreasen PA. Plasminogen activation system in human milk. J Pediatr Gastroenterol Nutr. 1997;25(2):159–66. [DOI] [PubMed] [Google Scholar]
- 16. Nielsen SD, Beverly RL, Dallas DC. Milk proteins are predigested within the human mammary gland. J Mammary Gland Biol Neoplasia. 2017;22(4):251–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Demers-Mathieu V, Nielsen SD, Underwood MA, Borghese R, Dallas DC. Changes in proteases, antiproteases and bioactive proteins from mother's breast milk to the premature infant stomach. J Pediatr Gastroenterol Nutr. 2018;66(2):318–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Macfarlane GT, Allison C, Gibson SAW, Cummings JH. Contribution of the microflora to proteolysis in the human large intestine. J Appl Bacteriol. 1988;64(1):37–46. [DOI] [PubMed] [Google Scholar]
- 19. Davidson LA, Lönnerdal BO. Persistence of human milk proteins in the breast-fed infant. Acta Paediatr. 1987;76(5):733–40. [DOI] [PubMed] [Google Scholar]
- 20. Haneberg B, Finne P. Lysozymes in feces from infants and children. Acta Paediatr. 1974;63(4):588–94. [DOI] [PubMed] [Google Scholar]
- 21. Carbonare SB, Silva MLM, Palmeira P, Carneiro-Sampaio MMS. Human colostrum IgA antibodies reacting to enteropathogenic Escherichia coli antigens and their persistence in the faeces of a breastfed infant. J Diarrhoeal Dis Res. 1997;15(2):53–8. [PubMed] [Google Scholar]
- 22. Young JC, Pan C, Adams RM, Brooks B, Banfield JF, Morowitz MJ, Hettich RL. Metaproteomics reveals functional shifts in microbial and human proteins during a preterm infant gut colonization case. Proteomics. 2015;15(20):3463–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goldman AS, Garza C, Schanler RJ, Goldblum RM. Molecular forms of lactoferrin in stool and urine from infants fed human milk. Pediatr Res. 1990;27(3):252–5. [DOI] [PubMed] [Google Scholar]
- 24. Nielsen SD, Beverly RL, Qu Y, Dallas DC. Milk bioactive peptide database: a comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 2017;232:673–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Azuma N, Nagaune S-I, Ishino Y, Mori H, Kaminogawa S, Yamauchi K. DNA-synthesis stimulating peptides from human β-casein. Agric Biol Chem. 1989;53(10):2631–4. [Google Scholar]
- 26. Dallas DC, Guerrero A, Khaldi N, Borghese R, Bhandari A, Underwood MA, Lebrilla CB, German JB, Barile D. A peptidomic analysis of human milk digestion in the infant stomach reveals protein-specific degradation patterns. J Nutr. 2014;144(6):815–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Inglingstad RA, Devold TG, Eriksen EK, Holm H, Jacobsen M, Liland KH, Rukke EO, Vegarud GE. Comparison of the digestion of caseins and whey proteins in equine, bovine, caprine and human milks by human gastrointestinal enzymes. Dairy Sci Technol. 2010;90(5):549–63. [Google Scholar]
- 28. Line WF, Sly DA, Bezkorovainy A. Limited cleavage of human lactoferrin with pepsin. Int J Biochem. 1976;7(5):203–6. [Google Scholar]
- 29. Brines RD, Brock JH. The effect of trypsin and chymotrypsin on the in vitro antimicrobial and iron-binding properties of lactoferrin in human milk and bovine colostrum: unusual resistance of human apolactoferrin to proteolytic digestion. Biochim Biophys Acta, Gen Subj. 1983;759(3):229–35. [DOI] [PubMed] [Google Scholar]
- 30. Siqueiros-Cendón T, Arévalo-Gallegos S, Iglesias-Figueroa BF, García-Montoya IA, Salazar-Martínez J, Rascón-Cruz Q. Immunomodulatory effects of lactoferrin. Acta Pharmacol Sin. 2014;35(5):557–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Goldsmith SJ, Dickson JS, Barnhart HM, Toledo RT, Eiten-Miller RR. IgA, IgG, IgM and lactoferrin contents of human milk during early lactation and the effect of processing and storage. J Food Prot. 1983;46(1):4–7. [DOI] [PubMed] [Google Scholar]
- 32. Hernell O, Bläckberg L. Human milk bile salt-stimulated lipase: functional and molecular aspects. J Pediatr. 1994;125(5, Part 2):S56–61. [DOI] [PubMed] [Google Scholar]
- 33. Turula H, Wobus CE. The role of the polymeric immunoglobulin receptor and secretory immunoglobulins during mucosal infection and immunity. Viruses. 2018;10(5):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Dallas DC, Smink CJ, Robinson RC, Tian T, Guerrero A, Parker EA, Smilowitz JT, Hettinga KA, Underwood MA, Lebrilla CB et al.. Endogenous human milk peptide release is greater after preterm birth than term birth. J Nutr. 2015;145(3):425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Dingess KA, de Waard M, Boeren S, Vervoort J, Lambers TT, van Goudoever JB, Hettinga K. Human milk peptides differentiate between the preterm and term infant and across varying lactational stages. Food Funct. 2017;8(10):3769–82. [DOI] [PubMed] [Google Scholar]
- 36. Miclat NN, Hodgkinson R, Marx GF. Neonatal gastric pH. Anesth Analg. 1978;57(1):98–101. [DOI] [PubMed] [Google Scholar]
- 37. Borgström B, Lindquist B, Lundh G. Enzyme concentration and absorption of protein and glucose in duodenum of premature infants. AMA Am J Dis Child. 1960;99(3):338–43. [PubMed] [Google Scholar]
- 38. Viejo-Díaz M, Andrés MT, Fierro JF. Different anti-candida activities of two human lactoferrin-derived peptides, lfpep and kaliocin-1. Antimicrob Agents Chemother. 2005;49(7):2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moriarty LC, Joannou CL, van den Berg JJM, Gorinsky B, Evans RW. Factors contributing to the potency of antimicrobial cationic peptides from the N-terminal region of human lactoferrin. FEMS Microbiol Lett. 2004;239(2):295–9. [DOI] [PubMed] [Google Scholar]
- 40. Tsopmo A, Romanowski A, Banda L, Lavoie JC, Jenssen H, Friel JK. Novel anti-oxidative peptides from enzymatic digestion of human milk. Food Chem. 2011;126(3):1138–43. [Google Scholar]
- 41. Donovan SM, Odle J. Growth factors in milk as mediators of infant development. Ann Rev Nutr. 1994;14(1):147–67. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.