ABSTRACT
Background
It is unclear whether the favorable effects of walnuts on the gut microbiota are attributable to the fatty acids, including α-linolenic acid (ALA), and/or the bioactive compounds and fiber.
Objective
This study examined between-diet gut bacterial differences in individuals at increased cardiovascular risk following diets that replace SFAs with walnuts or vegetable oils.
Methods
Forty-two adults at cardiovascular risk were included in a randomized, crossover, controlled-feeding trial that provided a 2-wk standard Western diet (SWD) run-in and three 6-wk isocaloric study diets: a diet containing whole walnuts (WD; 57–99 g/d walnuts; 2.7% ALA), a fatty acid–matched diet devoid of walnuts (walnut fatty acid–matched diet; WFMD; 2.6% ALA), and a diet replacing ALA with oleic acid without walnuts (oleic acid replaces ALA diet; ORAD; 0.4% ALA). Fecal samples were collected following the run-in and study diets to assess gut microbiota with 16S rRNA sequencing and Qiime2 for amplicon sequence variant picking.
Results
Subjects had elevated BMI (30 ± 1 kg/m2), blood pressure (121 ± 2/77 ± 1 mmHg), and LDL cholesterol (120 ± 5 mg/dL). Following the WD, Roseburia [relative abundance (RA) = 4.2%, linear discriminant analysis (LDA) = 4], Eubacterium eligensgroup (RA = 1.4%, LDA = 4), LachnospiraceaeUCG001 (RA = 1.2%, LDA = 3.2), Lachnospiraceae UCG004 (RA = 1.0%, LDA = 3), and Leuconostocaceae (RA = 0.03%, LDA = 2.8) were most abundant relative to taxa in the SWD (P ≤ 0.05 for all). The WD was also enriched in Gordonibacter relative to the WFMD. Roseburia (3.6%, LDA = 4) and Eubacterium eligensgroup (RA = 1.5%, LDA = 3.4) were abundant following the WFMD, and Clostridialesvadin BB60group (RA = 0.3%, LDA = 2) and gutmetagenome (RA = 0.2%, LDA = 2) were most abundant following the ORAD relative to the SWD (P ≤ 0.05 for all). Lachnospiraceae were inversely correlated with blood pressure and lipid/lipoprotein measurements following the WD.
Conclusions
The results indicate similar enrichment of Roseburia following the WD and WFMD, which could be explained by the fatty acid composition. Gordonibacter enrichment and the inverse association between Lachnospiraceae and cardiovascular risk factors following the WD suggest that the gut microbiota may contribute to the health benefits of walnut consumption in adults at cardiovascular risk. This trial was registered at clinicaltrials.gov as NCT02210767.
Keywords: butyrate, bioactive compounds, PUFAs, cardiovascular disease, α-linolenic acid
Introduction
Cardiovascular disease (CVD) is the leading cause of morbidity and mortality globally, and poor diet quality is the primary etiological risk factor underlying the development of CVD (1–3). Recent evidence suggests that the gut microbiota also plays an important role in cardiovascular risk (4–8). The gut microbiota is a complex ecosystem of organisms that are critical for human health. The microbes present in the lower gastrointestinal tract are involved in extraction and metabolism of nutrients not fully digested in the small intestine. Diet largely affects the composition and functionality of bacteria present in the large intestine (9).
There is evidence that the gut microbiota may be a mediator of the CVD benefits observed with walnut consumption. A study conducted by Baer et al. (10) demonstrated that the metabolizable energy from walnuts is overestimated by 21% by the predicted Atwater factors, suggesting that walnut-derived nutrients are accessible to the gut microbiota. Walnuts are uniquely rich in PUFAs, including α-linolenic acid (ALA), a plant-based omega-3 fatty acid that has cardiometabolic benefits (11, 12). However, walnuts also contain bioactive compounds, such as hydrolyzable tannins, and fiber that can be metabolized by gut bacteria and may confer additional benefits beyond the fatty acid profile of walnuts (13). Collectively, these walnut components may contribute to the cardiometabolic benefits of walnuts that have been demonstrated via different mechanisms of action.
The present study is a secondary analysis of a randomized, crossover, controlled-feeding trial that provided 3 study diets: a diet containing walnuts as an SFA replacement compared to 2 diets that replace SFA with a vegetable oil blend reflective of the fatty acid profile of walnuts and a vegetable oil blend 83% lower in ALA and proportionally higher in oleic acid (both without walnuts). This study examined between-diet differences in gut bacterial composition in individuals at risk for CVD. Exploratory analyses were also conducted to examine the relation between CVD risk factors and changes in bacterial abundance following each experimental diet. We hypothesized that following the diet containing walnuts, individuals would have a significantly different gut microbiota because of the bioactive content and whole food complex compared to the 2 other experimental diets and the baseline run-in diet, which was a standard Western diet (SWD).
Methods
Study design
Details of the study design and results of the CVD-related endpoints, including blood pressure (BP), vascular health, and lipids and lipoproteins, are reported elsewhere (14). Data from secondary analyses, including analysis of fecal bacteria diversity, enrichment, predictive functional analysis, and taxonomic co-occurrence, are reported here. Briefly, a randomized, crossover, controlled-feeding study with a 2-wk run-in on a SWD followed by three 6-wk diet periods (with compliance breaks between diet periods) on a walnut diet (WD), a walnut fatty acid-matched diet (WFMD), or an oleic acid replaces α-linolenic acid diet (ORAD), was conducted at The Pennsylvania State University. The fully controlled, weight-maintenance dietary feeding intervention provided similar macronutrient profiles (Table 1) in all 3 study diets (48% carbohydrate, 17% protein, 35% fat, 7% SFA). The run-in diet was designed to represent a SWD (50% carbohydrate, 17% protein, 33% fat, 12% SFA). The presence or absence of walnuts and the fatty acid profile of each of the diets differed: WD (contained 57–99 g/d walnuts; 2.7% ALA); WFMD (did not contain walnuts; 2.6% ALA); ORAD (did not contain walnuts; 0.4% ALA). Participants’ compliance was assessed with daily questionnaires for diet, medications, exercise, and general wellness. Participants were asked to refrain from medication use, including antibiotics, 48 h before sample collections. This trial is registered at clinicaltrials.gov as NCT02210767.
TABLE 1.
Nutrient profiles of the run-in, WD, WFMD, and ORAD tested in this study of adults at increased cardiovascular risk (n = 42)1
| Nutrient2 | SWD (run-in) | WD | WFMD | ORAD |
|---|---|---|---|---|
| Total fat, % | 34 | 35 | 35 | 35 |
| SFAs, % | 12 | 7 | 7 | 7 |
| MUFAs, % | 12 | 9 | 9 | 12 |
| PUFAs (ALA), % | 7 | 16 (3) | 16 (3) | 14 (0.4) |
| Carbohydrate, % | 50 | 48 | 48 | 48 |
| Protein, % | 16 | 17 | 17 | 17 |
| Fiber, g/d | 25 | 30 | 26 | 26 |
This table has been adapted from (14). % represents % total kcal based on a 2100 kcal/d diet. ALA, α-linolenic acid; ORAD, oleic acid replaces α-linolenic acid diet; SWD, standard Western diet; WD, walnut diet; WFMD, walnut fatty acid-matched diet.
All diets used the same 6-d cycle menu, developed with Food Processor SQL software, version 10.8 (ESHA Research).
Participants
Study participants were recruited from the State College, PA, area. Interested volunteers who were likely eligible after a telephone screen were scheduled for a clinic screening to confirm eligibility based on inclusion criteria. Eligible participants were men and women aged 30–65 y with overweight or obesity (BMI: 25.0–39.9 kg/m2) who had elevated BP (120–159/80–99 mmHg) and/or increased LDL cholesterol (128–177 mg/dL for men and 121–172 mg/dL for women). Written informed consent was obtained from all participants before enrollment in the study. All study samples were collected and procedures were conducted at The Pennsylvania State University Clinical Research Center. Differences between individuals included in the microbiota analysis and individuals not included in the analysis were computed in SAS (version 9.4; SAS Institute) with use of a Student's t test (α <0.05).
CVD risk factors
BP and vascular measures were assessed with the SphygmoCor XCEL system (AtCor Medical) at the end of each diet period and following the run-in diet. Fasting serum lipids [total cholesterol (TC), triglycerides] and lipoproteins (LDL cholesterol, HDL cholesterol, non-HDL cholesterol) were measured by a commercial laboratory (Quest Diagnostics) on 2 consecutive days. Additional details are published elsewhere (15).
Fecal sample collection
A ∼30 g fecal sample from a single defecation was collected from participants in the 72 h before completion of the run-in diet and each of the 3 diet periods with use of a collection kit (containing a stool collection hat, long-handled spoon, medical gloves, and Para-Pak clean vial; Meridian Bioscience, Inc.) provided by investigators (participants brought their fecal samples to clinic visit 1 or 2). Samples were stored in a freezer (−18°C) until the next day at which time they were given to research staff who stored the samples at −80°C at the Clinical Research Center until analysis.
DNA extraction and quantification
Nucleic acid extractions were performed on each sample with a Qiagen Powersoil DNA Isolation kit following the manufacturer's instructions (Qiagen Germantown) with lysing performed for 5 min on a Disruptor Genie cell disruptor (Scientific Industries). The resulting genomic DNA was eluted in 50 μL of 10 mM Tris. The Qubit 4.0 Fluorometer (Invitrogen) with the dsDNA High Sensitivity was used to quantify genomic DNA according to the assay.
PCR amplification
Illumina iTag PCR of the V4 region of the 16S gene was performed according to the Earth Microbiome Protocol 16S protocol at a total volume of 25 μL for each sample and contained final concentrations of 1X Ex Taq PCR buffer, 0.8 mM Ex Taq dNTPs, 0.625 U Ex Taq Polymerase (Takara Bio), 0.2 μM 515F forward barcoded primer, 0.2 μM Illumina 806R primer (16), and ∼10 ng of template DNA per reaction. PCR was performed under the following cycling conditions: 98°C for 3 min; 35 cycles of 98°C for 1 min, 55°C for 40 s, and 72°C for 1 min; 72°C for 10 min; then kept at 4°C. PCR products were visualized on a 2% Agarose E-gel (ThermoFisher Scientific) stained with ethidium bromide (17).
Library purification, verification, and sequencing
Equimolar concentrations of PCR amplicons were pooled together and were subsequently gel purified with the Qiagen Gel Purification Kit (Qiagen). Clean PCR products were quantified with the Qubit 4.0 Fluorometer (Life Technologies) and a 2100 Bioanalyzer DNA 1000 chip (Agilent Technologies) before they were shipped on dry ice to Laragen, Inc. (Culver City) for sequencing.
The library was then size verified with the Fragment Analyzer CE (Advanced Analytical Technologies Inc.) and quantified with the Qubit High Sensitivity dsDNA kit (Life Technologies). Finally, the library was denatured and diluted then spiked with 10% PhiX V3 library and loaded on an Illumina MiSeq V2 500 cycle kit for 250 base, paired-end reads.
Quality filtering and amplicon sequence variant picking
Qiime2 version 2018.8.0 was used to implement DADA2 (18) processing of sequence data. Initially, data were quality filtered to remove adapter and barcode sequences, trimmed to 240 base pairs on each read to remove portions of sequence that contained Q scores below 30, followed by discarding reads with expected errors >0.5%. Sequences were then dereplicated, checked for sequence variants, merged, and finally chimera checked by DADA2. A total of 3,008,191 sequences was obtained after quality filtering and processing. Representative sequences were then assigned with Qiime2 implementation of the naive bayes machine learning classifier (19) trained against the Silva 132 database (20). The resulting table and taxonomy artifacts were then exported as a biom table and text file, respectively, for subsequent analyses following addition of taxa data to the biom-formatted amplicon sequence variant (ASV) table.
α- and β-diversity analysis
α-Diversity plots were generated with a rarefied biom-formatted ASV table with all samples that had >10,000 sequences. Rarefaction was conducted on sequences across all samples to a maximum depth of 10,000 sequences. α-Diversities were then collated and plotted with observed species richness metric (α = 0.05).
Principal coordinates analysis plots and ANOSIM tests for significance were generated from a weighted UniFrac distance matrix that was produced from a cumulative sum scaling normalized biom-formatted ASV table within QIIME 1.9.0 (21).
Taxonomic comparisons
Taxonomy was assigned for ASVs against the Silva 132 database (as described above) and added into the biom-formatted ASV table. Relative abundance (RA) facet grid bar plots were generated within Rstudio through use of the Phyloseq package (22) to visualize the abundance of taxa across all samples, grouped by individual and divided by diet.
The RA of the most prominent phyla (Bacteroidetes, Firmicutes, and Proteobacteria) following each of the study diets (WD, WFMD, and ORAD) were compared. The RA of the Gordonibacter genus, a urolithin producer (23), following each of the study diets was also tested. The RA of Bacteroidetes, Firmicutes, and Proteobacteria by BMI status were evaluated between classifications (25.0–29.9: overweight, 30.0–34.9: obese, 35.0–39.9: morbid obesity) following the 3 study diets (diet by BMI interaction) and after pooling the study diets. Normality was assessed and variables not normally distributed were logarithmically transformed. Differences were tested with the mixed models procedure (PROC MIXED) in SAS (version 9.4; SAS Institute) at a predetermined α value of 0.05. Diet was considered a fixed effect and subject was considered a random effect. Tukey-Kramer corrected P values were used to correct for multiple comparisons.
RA of taxa were subject to linear discriminant analysis effect size (LEfSe version 1.0.8) (24) analysis to identify enriched taxa during pairwise comparisons of the diets. For each comparison, each participant's data for a respective diet were included. Linear discriminant analysis (LDA) scores of 0 were used to identify taxa that were significantly enriched with respect to the other diet in the comparison.
Co-occurrence Network analysis
Biom-formatted ASV tables were used to produce co-occurrence networks of ASVs present across 50% samples within a given diet with the Co-occurrence Network (CoNet) tool (25). Pearson and Spearman correlations were used to determine the relations (positive or negative) between the ASVs (α = 0.05) denoted by green edges (positive correlations) or red edges (negative correlations). Nodes are colored by phylum, and size is relative to the abundance of the taxa.
Predictive functional analysis with Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
Filtered sequences were used to predict the functional profile of the microbial communities within each sample via closed reference picking against Greengenes 13_5. Predicted KEGG Orthology genes were then grouped by functional category and results were then compared pairwise within LEfSe (LDA = 0) to determine enriched predicted functional pathways. In addition, individual KEGG Orthology terms were analyzed to determine individual genes that may be differentially present (LDA = 0.5).
Correlations with cardiovascular risk factors
Primary analyses from this study showed significant reductions in brachial and central mean arterial pressure (MAP), central diastolic BP, TC, LDL cholesterol, non-HDL cholesterol, and the ratio of TC to HDL cholesterol following the WD compared to baseline (14). In addition, TC, LDL cholesterol, non-HDL cholesterol, and HDL cholesterol were significantly reduced following the WFMD and ORAD compared to baseline. The bacteria that were significantly enriched (LEfSe plots) were correlated with the respective magnitudes of change for CVD risk factors that were significantly changed following each of the study diets. Statistical analyses were performed with SAS (version 9.4; SAS Institute). The correlation procedure (PROC CORR) was used with the Spearman correlation coefficient to assess associations between the magnitudes of change for CVD risk factors that were significantly changed from baseline with bacteria that were significantly enriched at a predetermined α value of 0.05.
Multivariate association with linear models (MaAsLin) (26) was also used to examine how the entire bacterial community, rather than only the significantly enriched bacteria, was associated with CVD risk factors. This exploratory analysis uses a novel multivariate algorithm. All of the bacteria present in the sample were mapped against the magnitudes of change for significantly changed CVD risk factors for each study diet and mean values for the same CVD risk factors were mapped against the run-in diet bacteria. A multivariate linear model associating metadata with each bacterium independently was boosted, and any metadata selected in at least 1% of these iterations was finally tested for significance (α <0.05) in a standard generalized linear model. Because this approach compared every bacteria present in the sample against each individual CVD risk factor, multiple comparisons were adjusted with a Bonferroni correction; multiple hypothesis tests over all bacteria and metadata were adjusted to produce a final Benjamini and Hochberg false discovery rate (Q-value). Statistical analyses were performed in R with the glm package.
Results
Participants
A detailed flow diagram of the phases of this trial is available elsewhere (14). Briefly, a total of 46 participants were enrolled in the study, 45 were randomly assigned, and 36 completed the study. Of the 45 participants who were randomly assigned, fecal samples were available from 42 participants. Thirty-four out of 42 participants completed all diet periods with available fecal samples. Thirty-five participants completed the WD, 36 completed the WFMD, and 38 completed the ORAD. There were no significant differences between participants included in the analysis compared to individuals not included (Table 2). Analysis was performed on all 42 participants, including participants who did not complete the trial but contributed partial data. Two participants were on an antibiotic regimen during the course of the study; both instances were ∼2.5 wk before endpoint fecal collections. A sensitivity analysis was conducted and removal of these participants did not affect the results. Participants included in the gut microbiota analysis had elevated BMI (30 ± 1), BP (121 ± 2/77 ± 1 mmHg), and LDL cholesterol (120 ± 5 mg/dL) (Table 2).
TABLE 2.
Baseline characteristics of enrolled participants1
| Characteristic2 | Fecal sample available | Fecal sample not available | P |
|---|---|---|---|
| n | 42 | 3 | — |
| Men, n | 23 | 2 | 0.76 |
| Age, y | 43 ± 2 (30–60) | 40 ± 7 (32–55) | 0.72 |
| BMI, kg/m2 | 30 ± 1 (24–42) | 30 ± 2 (27–33) | 0.89 |
| TG, mg/dL | 119 ± 9 (52–279) | 104 ± 15 (89–134) | 0.44 |
| TC, mg/dL | 191 ± 5 (120–260) | 170 ± 19 (132–194) | 0.39 |
| LDL cholesterol, mg/dL | 120 ± 5 (59–190) | 103 ± 14 (77–122) | 0.33 |
| HDL cholesterol, mg/dL | 47 ± 2 (29–76) | 46 ± 5 (38–55) | 0.93 |
| TC:HDL cholesterol, mg/dL | 4 ± 0.2 (2.4–7.6) | 4 ± 0.3 (3.4–4.3) | 0.16 |
| Non-HDL cholesterol, mg/dL | 144 ± 5 (75–212) | 124 ± 16 (94–149) | 0.34 |
| Glucose, mg/dL | 91 ± 1.1 (75–112) | 89 ± 7 (75–98) | 0.84 |
| Insulin, µIU/mL | 7 ± 0.6 (1.7–15) | 6 ± 1 (6–7) | 0.40 |
| bSBP, mmHg | 121 ± 1.7 (102–153) | 130 ± 3 (126–136) | 0.07 |
| bDBP, mmHg | 77 ± 1.2 (61–102) | 84 ± 5 (78–94) | 0.57 |
Data are presented as means ± SEMs (ranges). Baseline measurements were taken on the last 2 d of the run-in diet. Differences between individuals included in the microbiota analysis and individuals not included in the analysis were computed in SAS (version 9.4; SAS Institute) with use of a Student's t test (α <0.05). Baseline characteristics for total participants are available elsewhere (14). bDBP, brachial diastolic blood pressure; bSBP, brachial systolic blood pressure; TC, total cholesterol; TG, triglycerides.
Lipids, lipoproteins, and insulin were measured from serum and glucose was measured from plasma.
DNA extraction and quantification, PCR amplification, library purification, verification and sequencing, and quality filtering and ASV picking
16S rRNA gene PCR amplification of the V4 region was successful for all collected samples. High-quality sequence data were obtained for 144 out of 149 fecal samples. Overall, sequencing depth of DADA2 processed data ranged from 1955 to 95,549 sequences per sample. A total of 3,008,191 sequences were obtained after quality filtering, merging, and chimera checking. One-hundred and forty-four samples were able to be incorporated into community analyses and a cumulative sum scaling normalized ASV table, as sequencing depth exceeded 1000 sequences for each sample.
α- and β-diversity
α-Diversity was assessed with rarefaction curves (n = 101 subjects with >10,000 sequences), which show the average number of ASVs per study diet. Each diet had similar average observed species richness (Supplemental Figure 1). α-Diversity analyses did not reveal distinct differences in microbial community species richness between diets and ranged from 99 to 120 average ASVs (Supplemental Figure 2). A range of 18–246 total unique bacterial ASVs was observed within each sample when evaluating raw DADA2 processed ASVs.
The composition of the bacterial communities was similar across each diet. There was no distinct shaping or clustering observed between the samples by diet (ANOSIM P = 0.86). Similar to the above analyses, no distinct trends were evident in the bacterial community structure of the gut communities with respect to diet and the composition of the communities. Across all of the study diets, Bacteroidetes (average RA = 23%), Firmicutes (73%), and Proteobacteria (3%) were the most abundant bacteria phyla (Supplemental Figure 3) and similar abundances were observed in the run-in diet (Bacteroidetes = 24%, Firmicutes = 71%, Proteobacteria = 3%).
Taxonomic comparisons
LEfSe plot (LDA = 0) comparisons of diets to the SWD showed significantly enriched ASVs (Table 3; Supplemental Figure 4). There were 9 significantly enriched taxa following the WD relative to the SWD; Roseburia (RA = 4.2%, LDA = 4.2, P = 0.0008), Eubacterium eligensgroup (RA = 1.4%, LDA = 3.6, P = 0.05), Lachnospiraceae UCG001 (RA = 1.2%, LDA = 3.2, P = 0.03), and Lachnospiraceae UCG004 (RA = 1.0%, LDA = 3.1, P = 0.03) showed the greatest magnitude of enrichment. There were 4 enriched taxa following the WFMD; Roseburia (RA = 3.6%, LDA = 3.8, P = 0.02) and Eubacterium eligensgroup (RA = 1.5%, LDA = 3.4, P = 0.02) showed the greatest magnitude of enrichment relative to the SWD. Three taxa were enriched following the ORAD relative to the SWD; Clostridialesvadin BB60group (RA = 0.3%, LDA = 2.5, P = 0.04) was significantly enriched and was most abundant compared to the other significantly enriched ASVs. There were also significant differences between study diets. There were 3 enriched taxa following the WD relative to 8 enriched taxa following the ORAD; Roseburia (RA = 4.2%, LDA = 4.1, P = 0.02) was most abundant and significantly enriched following the WD relative to the ORAD. Ruminiclostridium (RA = 3.4%, LDA = 1.4, P = 0.04) and Clostridialesviadin BB60group (RA = 0.3% compared with 0.1%, LDA = 2.4, P = 0.03) were significantly enriched following the ORAD relative to the WD. There was 1 enriched taxa following the WD [Gordonibacter (RA = 0.04%, LDA = 3.2, P = 0.03)] relative to 2 enriched taxa following the WFMD [Faecalibacterium (RA = 4.4%, LDA = 4.3, P = 0.03) and Angelakisella (RA = 0.1%, LDA = 2.8, P = 0.04)]. There were no enriched taxa when comparing the WFMD to the ORAD diet.
TABLE 3.
Between-diet comparison of enriched bacteria in adults at increased cardiovascular risk with available fecal samples (n = 42)1
| Comparison2 | Diet | LDA score | P |
|---|---|---|---|
| WD vs. run-in | |||
| D_5__Roseburia | WD | 4.17 | 0.0008 |
| D_5__Eubacterium_eligensgroup | WD | 3.62 | 0.05 |
| D_5__Lachnospiraceae UCG_001 | WD | 3.19 | 0.03 |
| D_5__Lachnospiraceae UCG_004 | WD | 3.07 | 0.04 |
| D_4__Leuconostocaceae | WD | 2.8 | 0.05 |
| D_5__Ruminococcaceae UCG_003 | WD | 2.75 | 0.03 |
| D_5__Butyricicoccus | WD | 2.37 | 0.01 |
| WFMD vs. run-in | |||
| D_5__Roseburia | WFMD | 3.77 | 0.02 |
| D_5__Eubacterium_eligensgroup | WFMD | 3.44 | 0.02 |
| D_5__gutmetagenome | WFMD | 3.12 | 0.01 |
| D_5__Butyricicoccus | WFMD | 2.95 | 0.02 |
| D_4__Streptococcaceae | Run-in | 3.33 | 0.05 |
| D_5__Streptococcus | Run-in | 3.35 | 0.02 |
| ORAD vs. run-in | |||
| D_5__Streptococcus | Run-in | 2.83 | 0.03 |
| D_5__gutmetagenome | ORAD | 2.44 | 0.005 |
| D_4__Clostridiaesvadin BB60group | ORAD | 2.48 | 0.04 |
| WD vs. WFMD | |||
| D_5__Faecalibacterium | WFMD | 4.27 | 0.03 |
| D_5__Angelakisella | WFMD | 2.78 | 0.04 |
| D_5__Gordonibacter | WD | 3.22 | 0.03 |
| WD vs. ORAD | |||
| D_5__Roseburia | WD | 4.1 | 0.02 |
| D_5__ Defluviitaleaceae UCG_011 | WD | 1.63 | 0.01 |
| D_4__Defluviitaleaceae | WD | 1.62 | 0.01 |
| D_5__Ruminiclostridium1 | ORAD | 1.43 | 0.04 |
| D_4__Clostridialesvain BB60group | ORAD | 2.35 | 0.03 |
LEfSe plots for between-diet and study diet–run-in diet comparisons of enriched taxa. LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; ORAD, oleic acid replaces α-linolenic acid diet; WD, walnut diet; WFMD, walnut fatty acid matched diet.
D_5_ indicates that this bacterial species has been identified down to the genus taxonomic level and D_4_ indicates this bacteria has been identified down to the family level.
There were no significant differences in the relative abundance of Bacteroidetes (P = 0.7), Firmicutes (P = 0.7), or Proteobacteria (P = 0.5) among the WD, WFMD, or ORAD (Table 3). There were differences in relative abundance of Gordonibacter (P = 0.02) following the WD (RA = 0.0004), WFMD (RA = 0), and ORAD (RA = 0.00003). The RA of Gordonibacter was significantly greater following the WD compared to the WFMD (P = 0.04) and the ORAD (P = 0.04). After pooling the diets and categorizing by baseline BMI, there were no statistically significant differences between the RA of Bacteroidetes (P = 0.8), Firmicutes (P = 0.9), or Proteobacteria (P = 0.5) among individuals with overweight (n = 23), obesity (n = 9), and morbid obesity (n = 10) (Table 3; Supplemental Figure 5).
CoNet analysis
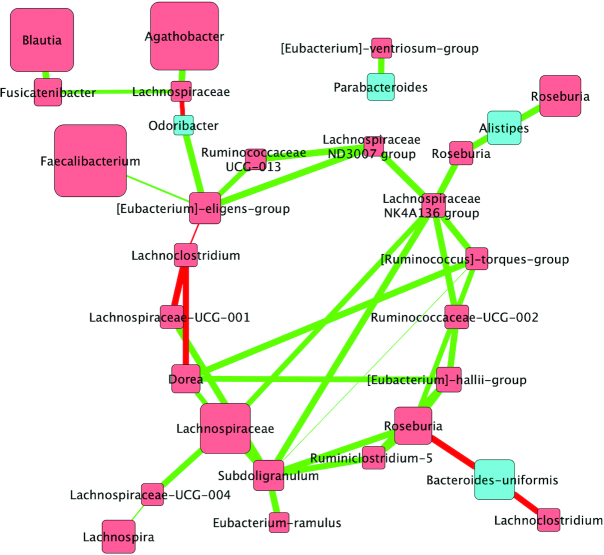
Following the 3 study diets (WD, WFMD, and ORAD) and the run-in, SWD, the ASV co-occurrence was quantitatively different (Figures 1–4). The WD network comprised 30 nodes; 26 were Firmicutes and 4 were Bacteroidetes. The WD was largely dominated by Firmicutes including: Agathobacter, Faecalibacterium, Blautia, Lachnospiraceae, and Roseburia and Bacteroidetes including: Alistipes, Bacteroides-uniformis, and Parabacteroides. There were 6 negative and 31 positive interactions between nodes. The nodes that showed the largest number of connections were Lachnospiraceae NK4A136 (6 interactions) and Subdoligranulum (6).
FIGURE 1.
Correlations between bacterial communities following the walnut diet in adults at increased cardiovascular risk (n = 42). Biom-formatted ASV tables were used to produce a CoNet of ASVs present across 50% samples within a given diet through use of the CoNet tool (25). Pearson and Spearman correlations were used to determine the relations (positive or negative) between the ASVs (α = 0.05) denoted by green edges (positive correlations) or red edges (negative correlations). Nodes are colored by phylum and size is relative to the abundance of the taxa analysis for the walnut diet. Nodes (colored squares) were colored by phylum (all members of the same phylum are identical colors) and labeled by the genus or lowest available assigned taxonomic level. Blue nodes represent Bacteroidetes and red nodes represent Firmicutes. In addition, node size is relative to the abundance of the taxa (larger squares indicate larger abundance, whereas smaller squares indicate a lower abundance). ASV, amplicon sequence variant; CoNet, Co-occurrence Network.
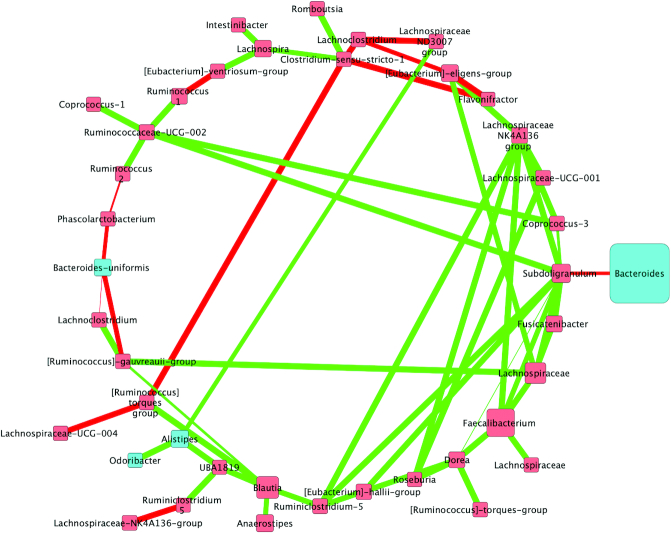
FIGURE 4.
Correlations between bacterial communities following the run-in diet in adults at increased cardiovascular risk (n = 42). Biom-formatted ASV tables were used to produce a CoNet of ASVs present across 50% samples within a given diet through use of the CoNet tool (25). Pearson and Spearman correlations were used to determine the relations (positive or negative) between the ASVs (α = 0.05) denoted by green edges (positive correlations) or red edges (negative correlations). Nodes are colored by phylum and size is relative to the abundance of the taxa analysis for the walnut diet. Nodes (colored squares) were colored by phylum (all members of the same phylum are identical colors) and labeled by the genus or lowest available assigned taxonomic level. Blue nodes represent Bacteroidetes and red nodes represent Firmicutes. In addition, node size is relative to the abundance of the taxa (larger squares indicate larger abundance, whereas smaller squares indicate a lower abundance). ASV, amplicon sequence variant; CoNet, Co-occurrence Network.
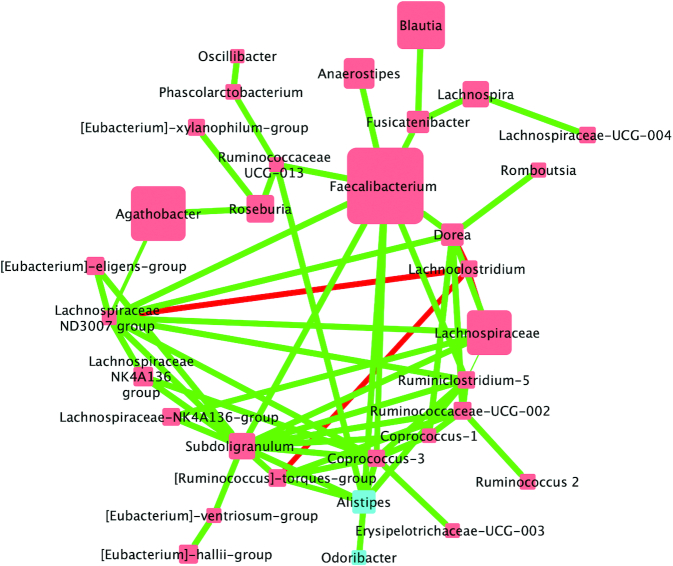
FIGURE 2.
Correlations between bacterial communities following the walnut fatty acid matched diet in adults at increased cardiovascular risk (n = 42). Biom-formatted ASV tables were used to produce a CoNet of ASVs present across 50% samples within a given diet through use of the CoNet tool (25). Pearson and Spearman correlations were used to determine the relations (positive or negative) between the ASVs (α = 0.05) denoted by green edges (positive correlations) or red edges (negative correlations). Nodes are colored by phylum and size is relative to the abundance of the taxa analysis for the walnut diet. Nodes (colored squares) were colored by phylum (all members of the same phylum are identical colors) and labeled by the genus or lowest available assigned taxonomic level. Blue nodes represent Bacteroidetes and red nodes represent Firmicutes. In addition, node size is relative to the abundance of the taxa (larger squares indicate larger abundance, whereas smaller squares indicate a lower abundance). ASV, amplicon sequence variant; CoNet, Co-occurrence Network.
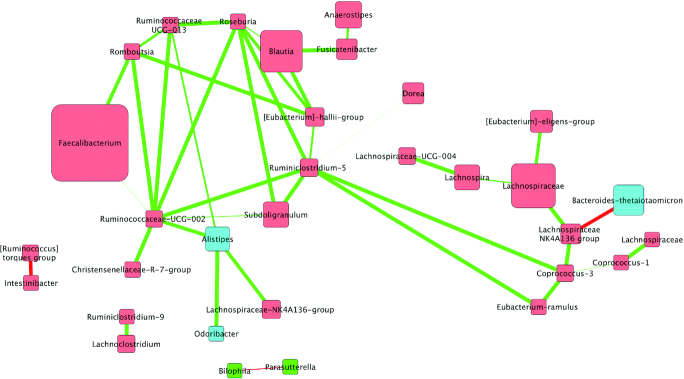
FIGURE 3.
Correlations between bacterial communities following the oleic acid replaces α-linolenic acid (ALA) diet in adults at increased cardiovascular risk (n = 42). Biom-formatted ASV tables were used to produce a CoNet of ASVs present across 50% samples within a given diet through use of the CoNet tool (25). Pearson and Spearman correlations were used to determine the relations (positive or negative) between the ASVs (α = 0.05) denoted by green edges (positive correlations) or red edges (negative correlations). Nodes are colored by phylum and size is relative to the abundance of the taxa analysis for the walnut diet. Nodes (colored squares) were colored by phylum (all members of the same phylum are identical colors) and labeled by the genus or lowest available assigned taxonomic level. Blue nodes represent Bacteroidetes, red nodes represent Firmicutes, and green nodes represent Proteobacteria. In addition, node size is relative to the abundance of the taxa (larger squares indicate larger abundance, whereas smaller squares indicate a lower abundance). ASV, amplicon sequence variant; CoNet, Co-occurrence Network.
The WFMD network contained 32 nodes; 27 Firmicutes, 3 Bacteroidetes, and 2 Proteobacteria. The WFMD network contained numerous Firmicutes including: Faecalibacterium, Lachnospiraceae, Blautia, Anaerostipes, and Subdoligranulum and was abundant in 3 Bacteroidetes: Alistipes, Bacteroides-thetalotaomicron, and Odoibacter and 2 Proteobacteria: Bilophila and Parasutterella. Three negative interactions were observed in the WFMD network and 34 positive interactions were among nodes. The nodes with the greatest number of connections were Ruminiclostridium (6 interactions), Ruminococcaceae UCG002 (6), and Roseburia (5).
The ORAD network was made up of 40 nodes; 36 Firmicutes and 4 Bacteroides. The ORAD did not have nodes as large as the WD, WFMD, or the SWD, which indicates it did not have a large relative abundance of any present taxa. The ORAD network contained Bacteroidetes, including Bacteroides, and Firmicutes, including Faecalibacterium. There were 13 negative and 44 positive interactions between nodes in the ORAD network. The nodes that had the greatest number of connections were Subdoligranulum (9 interactions) and Lachnospiraceae NK4A136 (6).
The SWD network contained 32 nodes; 30 Firmicutes and two Bacteroidetes. The network was dominated by Firmicutes: Faecalibacterium, Blautia, Agathobacter, Lachnospiraceae, and Anaerostipes. The SWD network was also rich in 2 Bacteroidetes: Alistipes and Odoribacter. The SWD network had 2 negative interactions and 58 positive among the nodes. The nodes with the greatest number of connections included Subdoligranulum (12 interactions), Faecalibacterium (9), and Lachnospiraceae (8).
Correlations with cardiovascular risk factors
There were significant correlations (Spearman) between the percentages of enriched bacteria following the WD with significantly changed CVD risk factors (Table 4). Eubacterium eligens was inversely associated with the change in brachial MAP (ρ = −0.50; P = 0.0009), central diastolic BP (ρ = −0.52; P = 0.0006), and central MAP (ρ = −0.47, P = 0.002). This indicates that a greater RA of Eubacterium eligens was associated with greater reductions in the aforementioned CVD risk factors. Greater RA of Lachnospiraceae was associated with greater reductions in brachial MAP (ρ = −0.37, P = 0.02), central diastolic BP (ρ = −0.32; P = 0.04), central MAP (ρ = −0.35; P = 0.02), TC (ρ = −0.35; P = 0.03), and non-HDL cholesterol (ρ = −0.37; P = 0.02). Leuconostocaceae was positively associated with the change in brachial MAP (ρ = 0.34; P = 0.03) and central MAP (ρ = 0.34; P = 0.03), which corresponds to a greater RA of Leuconostocaceae being associated with a greater increase in central MAP. There were no significant correlations between enriched bacteria following the WFMD or ORAD with CVD risk factors (Supplemental Table 3,Supplemental Table 4). There were also no significant correlations between Eubacterium eligens following the WFMD and brachial MAP, central MAP, or central diastolic BP.
TABLE 4.
Spearman correlations between significantly enriched taxa (n = 35) and change in (Δ) cardiovascular risk factors following the WD in adults at increased cardiovascular risk1
| Roseburia | Eubacterium_ eligensgroup | Lachnospiraceae UCG_001 | Lachnospiraceae UCG_004 | Leuconostocaceae | Ruminococcaceae | Butyricicoccus | Gordonibacter | Defluviitaleaceae UCG_011 | Defluviitaleaceae | |
|---|---|---|---|---|---|---|---|---|---|---|
| bMAP, Δ | –0.03 | –0.50** | –0.37* | –0.1 | 0.34* | –0.13 | –0.01 | 0.25 | 0.04 | 0.04 |
| cDBP, Δ | –0.12 | –0.52** | –0.32* | –0.17 | 0.31 | –0.11 | 0.03 | 0.27 | –0.11 | –0.11 |
| cMAP, Δ | –0.05 | –0.47** | –0.35* | –0.09 | 0.34* | –0.06 | –0.02 | 0.26 | –0.02 | –0.02 |
| TC, Δ | –0.23 | –0.26 | –0.06 | –0.35* | 0.24 | –0.16 | 0.02 | 0.16 | 0.17 | 0.17 |
| LDL cholesterol, Δ | –0.31 | –0.31 | –0.1 | –0.27 | 0.17 | –0.26 | –0.06 | 0.15 | 0.11 | 0.11 |
| Non–HDL cholesterol, Δ | –0.25 | –0.2 | –0.05 | –0.37* | 0.29 | –0.13 | 0.02 | 0.15 | 0.14 | 0.14 |
| TC:HDL cholesterol, Δ | –0.17 | –0.12 | 0.07 | –0.15 | 0.24 | 0.01 | 0.1 | 0.29 | –0.03 | –0.03 |
Statistical analyses were performed with SAS (version 9.4; SAS Institute). The correlation procedure (PROC CORR) was used with the Spearman correlation coefficient to assess association between the cardiovascular risk factors that were significantly changed with bacteria that were significantly enriched a predetermined α value of 0.05. bMAP, brachial mean arterial pressure; cMAP, central mean arterial pressure; cDBP, central diastolic blood pressure; TC, total cholesterol; WD, walnut diet. *indicates P < 0.05; **indicates P < 0.01.
The MaAsLin analysis showed when comparing every individual bacterium present in the sample to the magnitudes of change of CVD risk factors, there were significant associations between gut bacteria and CVD risk factors following the WFMD and ORAD; however, the false discovery rate (indicated by the Q-value) for the majority of the significant relations was 1, which indicates that there is 100% likelihood that these associations occurred by chance. There were no significant associations between the bacteria present following the WD and CVD risk factors. The run-in diet bacteria, Hungatella and Coprococcus were significantly associated with CVD risk factors including central MAP, C-reactive protein (CRP), and central systolic BP.
Phylogenetic Investigation of Communities by Reconstruction of Unobserved States
There were several pathways that were predicted to be enriched in pairwise comparisons (Table 5,Supplemental Figure 6). Genes related to bacterial invasion of epithelial cells were enriched following the SWD (LDA = 0.12, P = 0.049, 406 genes) compared to the WFMD (212 genes), and butirosin and neomycin biosynthesis genes were enriched following the SWD (LDA = 1.8, P = 0.04, 502,999 genes) compared to the WD (464,480 genes). Basal transcription factor genes were enriched following the WFMD (LDA = 1.5, P = 0.03, 7962 genes) relative to the WD (3268 genes). β-Alanine metabolism genes were enriched following the WD (LDA = 1.6, P = 0.03, 1,215,388 genes) compared to the WFMD (946,353 genes). There were no differential pathways between the SWD compared with ORAD, WD compared with ORAD, and WFMD compared with ORAD.
TABLE 5.
Predictive functional pathway comparisons between study diets and versus the run-in diet in adults at increased cardiovascular risk (n = 42)1
| Comparison | Diet | Gene count | LDA score | P |
|---|---|---|---|---|
| WD vs. run-in | ||||
| Butirosin and neomycin biosynthesis | Run-in | 502,999 | 1.77 | 0.041 |
| WFMD vs. run-in | ||||
| Bacterial invasion of epithelial cells | Run-in | 406 | 0.118 | 0.049 |
| WD vs. WFMD | ||||
| Basal transcription factors | WFMD | 7962 | 1.53 | 0.031 |
| β-Alanine metabolism | WD | 1,215,388 | 1.56 | 0.033 |
LEfSe plots were used to visualize PICRUSt predicted functional pathways of pairwise comparisons between diets. PICRUSt was performed on DADA2 processed sequences and clustered via closed reference clustering to Greengenes 13_5. Subsequently, predicted KO terms were then collapsed into KEGG pathways and compared pairwise between the diets. Four pathways were differentially abundant from 3 comparisons. KO, KEGG Orthology; LEfSe, linear discriminant analysis effect size; PICRUSt, Phylogenetic Investigation of Communities by Reconstruction of Unobserved States; WD, walnut diet; WFMD, walnut fatty acid-matched diet.
Discussion
To our knowledge, this is the first study to examine the effects of diets containing walnuts (and walnut-derived fiber) and vegetable oils compared to a diet with a greater % SFA (12% compared with 7%) on the human gut microbiota composition, predictive functional analysis, taxonomic co-occurrence, and associations with cardiovascular risk. Our findings, together with previous research on walnuts and the gut microbiota, assist in advancing our understanding of the role of the gut microbiota in disease risk and prevention, and how diet affects this. Relative to the current study, we have shown that a diet containing whole walnuts (including bioactive compounds), a diet with a matched fatty acid profile to the walnut-containing diet (devoid of whole walnuts and walnut bioactive compounds), and a diet matched for SFA but with oleic acid in place of ALA found in walnuts (devoid of walnuts and walnut bioactive compounds) differentially affected the gut microbiota. Our results demonstrate unique enrichment of bacteria, including butyrate producers such as Roseburia, following the WD and WFMD relative to the SWD. The bioactive compounds in walnuts may also play an important role in altering the gut environment. Walnuts contain ellagitannins that are metabolized by gut bacteria to form urolithins, which may provide cardiovascular benefits (27). Gordonibacter is a bacteria involved in ellagitannins metabolism, which was enriched following the WD relative to the WFMD. Further, this study showed possible microbiota-CVD interactions between enriched bacteria following the WD. Thus, consuming walnuts as a replacement for SFA elicits changes in gut bacteria that may have important health implications.
The present controlled-feeding study showed differentially enriched bacteria among all 3 study diets relative to the SWD run-in diet, which suggests that replacing SFA with unsaturated fatty acids in the presence or absence of walnuts affects the gut microbial environment. Both the WFMD and WD led to significant enrichment in Roseburia, Eubacteria eligens, and Butyricicoccus relative to the SWD. The enrichment of Roseburia following the WD is a similar finding to Holscher and colleagues (28), who reported that the RA of Roseburia was negatively associated with secondary bile acids. Certain gut bacteria can convert bile salts to secondary bile acids in the colon, which can damage the gut epithelial lining (29, 30). Dietary fat composition affects gut bacteria and can also affect bile acid metabolism and composition (31). SFAs in particular may promote the expansion of bacteria capable of converting bile salts into secondary bile acids (31). Our study demonstrates that replacing SFAs with walnuts or a walnut fatty acid profile match may beneficially affect the gut environment through increasing bacteria that are inversely associated with secondary bile acids.
Holscher et al. (28) also reported significant differences in β-diversity. Of note, our study and the study conducted by Holscher et al. (28) were controlled-feeding studies that provide a more robust assessment of diet-microbiota interactions because of the tightly controlled study design implemented. In contrast, Bamberger et al. (32) conducted a free living study and, consistent with our data and that of Holscher et al. (28), reported enrichment of Ruminococcaceae and Lachnospiraceae after walnut consumption compared to a walnut-free diet. Bamberger and colleagues (32) also reported significantly different β-diversity between diets. Although we did not see differences in β-diversity among diets, this may be because of our extremely similar treatment diets; diets only differed in the walnut bioactive and fiber profile or ALA content. Otherwise, our results are similar to the 2 previous studies (28, 32); however, those studies were conducted in healthy individuals and were not able to identify which component of walnuts may be inducing beneficial changes to the gut microbiota.
The significantly enriched bacteria and predictive functional analysis suggest walnut consumption and a diet that contains a comparable fatty acid profile promote a favorable environment in the gastrointestinal tract and may also be important in gut epithelial maintenance. Increased gut permeability or “leaky gut” is associated with a variety of diseases such as nonalcoholic fatty liver disease and obesity (33, 34). SCFAs, butyrate in particular, are a main energy source of enterocytes and are important for the maintenance of the intestinal epithelium (35, 36). Increased SCFA availability decreases intestinal permeability (37, 38). Although we did not observe significant enrichment in genes related to butyrate production or metabolism, Roseburia and Eubacterium were enriched following both the WD and WFMD and Leuconostocaceae was enriched following the WD; all of these bacteria are capable of SCFA production (39, 40). Ruminococcaceae was also enriched following the WD and may also be involved in intestinal epithelium maintenance as it is inversely correlated with intestinal permeability (41–43). Further, genes involved in bacterial invasion of epithelial cells were enriched following the Western-style, run-in diet relative to the WFMD. Unfortunately, when we examined the contributions of bacteria within the enriched Phylogenetic Investigation of Communities by Reconstruction of Unobserved States pathway (data not shown), the bacterial species had not been identified, but they possess genes related to the enriched pathway. Identification of SJTU_G_09_34, SHZO627, and aab28d05 could provide more information on the enrichment of invasion of epithelial cells. This pathway has been connected to increased gastrointestinal epithelial permeability (44) and suggests a Western-style diet that had a greater % SFA (compared to study diets), which participants followed previous to the study diets, may increase gut permeability. The MaAsLin analysis also showed a positive association between gut bacteria and systemic inflammation following the run-in diet, which could be linked to increased gut bacteria infiltration. Although we did not analyze gut-derived metabolites, the enrichment of SCFA-producing bacteria following the WD and WFMD suggests that the fatty acid composition, including ALA, may be protective against gut permeability.
The co-occurrence networks constructed to assess the bacterial community following each of the study diets illustrates potentially protective clustering following the WD and WFMD. Baldassano and Bassett (45) reported that healthy individuals had a high degree of bacterial modularity compared to individuals with inflammatory bowel disease, who had less distinctive community structures. The WD and WFMD networks had more modular bacterial clusters such as the Blautia, Agathobacter, Lachnospiraceae, Fusicatenibacter cluster in the WD co-occurrence network and the Lachnospiraceceae, Eubacterium eligens, Lachnospira cluster in the WFMD network. The ORAD network did not indicate any clustering and there was limited clustering in the SWD network. The interactions between bacterial nodes were primarily positive in the WD, WFMD, and SWD networks, with the greatest number of negative interactions following the ORAD. The large numbers of positive bacterial interactions correspond to a stable gut environment. A common trend in the co-occurrence network analyses is negative interactions among several bacteria that may be pathogenic with potentially beneficial bacteria, Lachnospiraceae indicating this bacterium may have a protective effect within the bacterial community. The observed positive interactions and modular bacterial clustering in the WD and WFMD networks could provide protection against pathogenic bacteria to maintain host health.
The enrichment of Gordonibacter following the WD relative to the WFMD (Supplemental Table 2) suggests that the walnuts may be altering the gut microenvironment to better metabolize the bioactive compounds present in walnuts. Gordonibacter is known to metabolize ellagitannins, which are uniquely high in walnuts, to urolithins, the primary form in which ellagitannins are absorbed into the body (23, 46). Urolithin A has been negatively correlated with CVD risk factors, such as plasma glucose. Further, when we examined the RA of Gordonibacter, the RA was significantly greater following the WD compared to both the WFMD and ORAD. The increase in ellagitannins-metabolizing bacteria after walnut consumption suggests that the gut microbiota may be involved in the underlying mechanisms associated with the cardiovascular benefits of walnut consumption.
In the absence of clearly defined underlying mechanisms for the association between the gut microbiota and cardiovascular risk, we performed correlation analyses between enriched bacteria and CVD risk factors examined in the primary analysis of this study. We demonstrated that members of the Lachnospiraceae family are inversely correlated with CVD risk factors following a diet containing walnuts. Increased Lachnospiraceae abundance was negatively correlated with changes in brachial MAP, central MAP, central diastolic BP, TC, and non-HDL cholesterol; greater abundance of Lachnospiraceae bacteria was associated with greater reductions in CVD risk factors. Increased abundance of Eubacterium eligens was also correlated with reductions in brachial MAP, central MAP, and central diastolic BP. Increased Leuconostocaceae was positively correlated with increases in brachial and central MAP. Although further research examining bacterial functionality and secondary metabolites is needed to provide more mechanistic insight, this exploratory analysis suggests that several bacteria may be involved in the underlying mechanisms for walnut consumption and reduced cardiovascular risk.
Previous reports of interactions between BMI and the gut microbiota (47) prompted us to perform an exploratory analysis on phylogenic taxonomy by BMI in the present study. Although we did not observe any differences between individuals with overweight, obesity, or morbid obesity in the prominent phyla, Firmicutes, Bacteroidetes, and Proteobacteria, there could be differences at lower taxonomic levels or functional differences that were not detected. These results are similar to a meta-analysis that examined whether α-diversity could predict BMI. The authors reported a relatively weak association between diversity and BMI that was confounded by large inter-personal variation (48). We did observe possible interactions between bacteria that may affect BMI. The CoNet of the WD showed Lachnospiraceae could be suppressing Lachnoclostridium, a bacteria that may be associated with increased BMI (49). The enrichment of Lachnospiraceae following the WD could play a role in weight status as it may be suppressing bacteria associated with increased BMIs. This warrants further exploration of interactions between weight status and gut bacteria given the increasing rates of obesity globally.
The strength of this study is the use of a controlled-feeding, randomized, controlled crossover trial with a run-in period to examine the differences in gut microbial composition and correlations with cardiovascular risk markers in individuals at risk for CVD following a diet containing walnuts as an SFA replacement. Study limitations include the lack of evaluation of secondary metabolites, such as SCFAs, and the use of Phylogenetic Investigation of Communities by Reconstruction of Unobserved States rather than a metatrancriptomics analysis to assess the functional capacity of gut bacteria. This study also did not assess the conversion of ALA to EPA which could affect the gut microbiota.
In conclusion, this study, which was designed to investigate which component(s) of walnuts underlie the beneficial cardiovascular effects, suggests that whole walnuts (fatty acids, fiber, and bioactive compounds), and the fatty acid profile may differentially affect the gut microbiota relative to a Western-style diet with a greater percentage of calories (or energy) from SFAs. Similarities between enrichment of SCFA-producing bacteria, including Roseburia and Eubacterium, following the WD and WFMD illustrate the effect that the high unsaturated fat content, including ALA, may have on gut bacteria. The unique enrichment of Gordonibacter following the WD suggests that walnut-derived bioactive compounds and fiber modulate gut microbiota. The associations between Lachnospiraceae and improved cardiovascular risk factors suggest that the gastrointestinal microbiota may contribute to the underlying mechanisms of the beneficial health effects of walnut consumption.
Supplementary Material
Acknowledgments
We thank the diet study manager, Marcella Smith, and research staff nurses, Cyndi Flanagan, Phyllis Martin, and Christa Oelhaf, for their assistance. The authors’ responsibilities were as follows—PMK-E and RL: designed the study; AMT: collected the data; CJM: analyzed fecal samples; AMT, KSP, and CJM: performed the statistical analyses; AMT, KSP, PMK-E, CJM, and RL: wrote the article; and all authors: read and approved the final manuscript.
Notes
This study was funded by The California Walnut Commission. Their staff were not involved with any aspects of conducting the study, data analyses, or interpretation of the results reported in this manuscript. This research was also supported by the Penn State Clinical and Translational Research Institute, Pennsylvania State University Clinical and Translational Science Award and NIH/National Center for Advancing Translational Sciences (grant no. UL1TR000127).
Author disclosures: The authors report no conflicts of interest.
Supplemental Tables 1–4 and Supplemental Figures 1–6 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn.
AMT and CJM share joint first authorship.
Abbreviations used: ALA, α-linolenic acid; ASV, amplicon sequence variant; BP, blood pressure; CoNet, Co-occurrence Network; CVD, cardiovascular disease; LDA, linear discriminant analysis; LEfSe, linear discriminant analysis effect size; MaAsLin, multivariate association with linear models; MAP, mean arterial pressure; ORAD, oleic acid replaces ALA diet; RA, relative abundance; SWD, standard Western diet; TC, total cholesterol; WD, walnut diet; WFMD, walnut fatty acid-matched diet.
References
- 1. Mozaffarian D, Wilson PWF, Kannel WB. Beyond established and novel risk factors. Circulation. 2008;117:3031–8. [DOI] [PubMed] [Google Scholar]
- 2. US Department of Agriculture and US Department of Health and Human Services. Scientific Report of the 2015 Dietary Guidelines Advisory Committee. Washington (DC); 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Afshin A, Sur PJ, Fay KA, Cornaby L, Ferrara G, Salama JS, Mullany EC, Abate KH, Abbafati C, Abebe Z et al.. Health effects of dietary risks in 195 countries, 1990–2017: a systematic analysis for the Global Burden of Disease Study 2017. Lancet. 2019;393(10184):1958–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tindall AM, Petersen KS, Kris-Etherton PM. Dietary patterns affect the gut microbiome—the link to risk of cardiometabolic diseases. J Nutr. 2018;148:1402–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Li J, Zhao F, Wang Y, Chen J, Tao J, Tian G, Wu S, Liu W, Cui Q, Geng B et al.. Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome. 2017;5:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brown JM, Hazen SL. Microbial modulation of cardiovascular disease. Nat Rev Microbiol. 2018;16:171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kelly TN, Bazzano LA, Ajami NJ, He H, Zhao J, Petrosino JF, Correa A, He J. Gut microbiome associates with lifetime cardiovascular disease risk profile among Bogalusa Heart Study participants. Circ Res. 2016;119:956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koeth RA, Wang Z, Levison BS, Buffa JA, Org E, Sheehy BT, Britt EB, Fu X, Wu Y, Li L et al.. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med. 2013;19:576–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA et al.. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baer DJ, Gebauer SK, Novotny JA. Walnuts consumed by healthy adults provide less available energy than predicted by the Atwater factors. J Nutr. 2016;146:9–13. [DOI] [PubMed] [Google Scholar]
- 11. Rajaram S. Health benefits of plant-derived α-linolenic acid. Am J Clin Nutr. 2014;100:443S–8S. [DOI] [PubMed] [Google Scholar]
- 12. West SG, Krick AL, Klein LC, Zhao G, Wojtowicz TF, McGuiness M, Bagshaw DM, Wagner P, Ceballos RM, Holub BJ et al.. Effects of diets high in walnuts and flax oil on hemodynamic responses to stress and vascular endothelial function. J Am Coll Nutr. 2010;29:595–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Selma MV, González-Sarrías A, Salas-Salvadó J, Andrés-Lacueva C, Alasalvar C, Örem A, Tomás-Barberán FA, Espín JC. The gut microbiota metabolism of pomegranate or walnut ellagitannins yields two urolithin-metabotypes that correlate with cardiometabolic risk biomarkers: comparison between normoweight, overweight-obesity and metabolic syndrome. Clin Nutr. 2018;37:897–905. [DOI] [PubMed] [Google Scholar]
- 14. Tindall AM, Petersen KS, Skulas‐Ray AC, Richter CK, Proctor DN, Kris‐Etherton PM. Replacing saturated fat with walnuts or vegetable oils improves central blood pressure and serum lipids in adults at risk for cardiovascular disease: a randomized controlled‐feeding trial. J Am Heart Assoc. 2019;8:e011512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tindall AM, Petersen KS, Skulas-Ray A, Richter C, Proctor DN, Kris-Etherton PM. Aging and chronic disease. Curr Dev Nutr. 2018;2. [Google Scholar]
- 16. Walters W, Hyde ER, Berg-Lyons D, Ackermann G, Humphrey G, Parada A, Gilbert JA, Jansson JK, Caporaso JG, Fuhrman JA et al.. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. mSystems. 2015;1(1):e00009–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Caporaso J, Lauber C. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci. 2011;108:(Suppl):4516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Callahan B, McMurdie P, Rose M, Han A, Johnson AJ, Holmes S. DADA2: high resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Bokulich NA, Kaehler BD, Rideout JR, Dillon M, Bolyen E, Knight R, Huttley GA, Caporaso JG. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Glo FO, Yarza P. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. McMurdie PJ, Holmes S. phyloseq: An R package for reproducible interactive analysis and graphics of Microbiome Census Data. PLoS One. 2013;8:e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Selma M V, Beltrán D, Luna MC, Romo-Vaquero M, García-Villalba R, Mira A, Espín JC, Tomás-Barberán FA. Isolation of human intestinal bacteria capable of producing the bioactive metabolite isourolithin A from ellagic acid. Front Microbiol. 2017;8:1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Faust K, Raes J. CoNet app: inference of biological association networks using Cytoscape. F1000Research. 2016;5:1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, Leleiko N, Snapper SB et al.. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Selma M V, Beltran D, Garcıa-Villalba R, Espin JC, Tomas-Barberan FA. Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species. Food Funct. 2014;5:1779–84. [DOI] [PubMed] [Google Scholar]
- 28. Holscher H, Guetterman H, Swanson K, An R, Matthan N, Lichtenstein A, Novotny J, Baer D. Walnut consumption alters the gastrointestinal microbiota, microbial-derived secondary bile acids, and health markers in healthy adults: a randomized controlled trial. J Nutr. 2018;148:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chiang JYL. Bile acid metabolism and signaling. Compr Physiol. 2013;3:1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ajouz H, Mukherji D, Shamseddine A. Secondary bile acids: an underrecognized cause of colon cancer. World J Surg Oncol. 2014;12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Devkota S, Wang Y, Musch MW, Leone V, Fehlner-Peach H, Nadimpalli A, Antonopoulos DA, Jabri B, Chang EB. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10-/- mice. Nature. 2012;487:104–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bamberger C, Rossmeier A, Lechner K, Wu L, Waldmann E, Fischer S, Stark RG, Altenhofer J, Henze K, Parhofer KG. A walnut-enriched diet affects gut microbiome in healthy Caucasian subjects: a randomized, controlled trial. Nutrients. 2018;10:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Damms-Machado A, Louis S, Schnitzer A, Volynets V, Rings A, Basrai M, Bischoff SC. Gut permeability is related to body weight, fatty liver disease, and insulin resistance in obese individuals undergoing weight reduction. Am J Clin Nutr. 2017;105:127–35. [DOI] [PubMed] [Google Scholar]
- 34. Soderborg TK, Friedman JE. Imbalance in gut microbes from babies born to obese mothers increases gut permeability and myeloid cell adaptations that provoke obesity and NAFLD. Microb Cell. 2019;6:102–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Scheppach W, Müller JG, Boxberger F, Dusel G, Richter F, Bartram HP, Christl SU, Dempfle CE, Kasper H. Histological changes in the colonic mucosa following irrigation with short-chain fatty acids. Eur J Gastroenterol Hepatol. 1997;9:163–8. [DOI] [PubMed] [Google Scholar]
- 36. Homann H-H, Kemen M, Fuessenich C, Senkal M, Zumtobel V. Reduction in diarrhea incidence by soluble fiber in patients receiving total or supplemental enteral nutrition. J Parenter Enter Nutr. 1994;18:486–90. [DOI] [PubMed] [Google Scholar]
- 37. Butzner JD, Parmar R, Bell CJ, Dalal V. Butyrate enema therapy stimulates mucosal repair in experimental colitis in the rat. Gut. 1996;38:568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan H, Ajuwon KM. Butyrate modifies intestinal barrier function in IPEC-J2 cells through a selective upregulation of tight junction proteins and activation of the Akt signaling pathway. PLoS One. 2017;12:e0179586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Meehan CJ, Beiko RG. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome Biol Evol. 2014;6:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nieminen TT, Säde E, Endo A, Johansson P, Björkroth J. The family leuconostocaceae. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson Feditors. The Prokaryotes: Firmicutes and Tenericutes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2014. pp. 215–40. [Google Scholar]
- 41. Mörkl S, Lackner S, Meinitzer A, Mangge H, Lehofer M, Halwachs B, Gorkiewicz G, Kashofer K, Painold A, Holl AK et al.. Gut microbiota, dietary intakes and intestinal permeability reflected by serum zonulin in women. Eur J Nutr. 2018;57:2985–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kang C, Wang B, Kaliannan K, Wang X, Lang H, Hui S, Huang L, Zhang Y, Zhou M, Chen M et al.. Gut microbiota mediates the protective effects of dietary capsaicin against chronic low-grade inflammation and associated obesity induced by high-fat diet. MBio. 2017;8:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leclercq S, Matamoros S, Cani PD, Neyrinck AM, Jamar F, Stärkel P, Windey K, Tremaroli V, Bäckhed F, Verbeke K et al.. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc Natl Acad Sci. 2014;111:E4485–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu LC-H, Wang J-T, Wei S-C, Ni Y-H. Host-microbial interactions and regulation of intestinal epithelial barrier function: from physiology to pathology. World J Gastrointest Pathophysiol. 2012;3:27–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Baldassano SN, Bassett DS.. Topological distortion and reorganized modular structure of gut microbial co-occurrence networks in inflammatory bowel disease. Sci Rep. 2016;6:26087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tulipani S, Urpi-Sarda M, García-Villalba R, Rabassa M, Loez-Uriarte P, Bullo N, Jaúregui O, Toma-Barbera F, Salas-Salvado J, Carlos Espín J et al.. Urolithins are the main urinary microbial-derived phenolic metabolites discriminating a moderate consumption of nuts in free-living subjects with diagnosed metabolic syndrome. J Agric Food Chem. 2012;60:8930–40. [DOI] [PubMed] [Google Scholar]
- 47. Haro C, Montes-Borrego M, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, Quintana-Navarro GM, Tinahones FJ, Landa BB et al.. Two healthy diets modulate gut microbial community improving insulin sensitivity in a human obese population. J Clin Endocrinol Metab. 2016;101:233–42. [DOI] [PubMed] [Google Scholar]
- 48. Sze MA, Schloss PD.. Looking for a signal in the noise: revisiting obesity and the microbiome. MBio. 2016;7:e01018–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Amadou T, Hosny M, La Scola B, Cassir N “Lachnoclostridium bouchesdurhonense,” a new bacterial species isolated from human gut microbiota. New Microbes New Infect. 2016;13:69–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.