ABSTRACT
Background
Although diet response prediction for cardiometabolic risk factors (CRFs) has been demonstrated using single genetic variants and main-effect genetic risk scores, little investigation has gone into the development of genome-wide diet response scores.
Objective
We sought to leverage the multistudy setup of the Women's Health Initiative cohort to generate and test genetic scores for the response of 6 CRFs (BMI, systolic blood pressure, LDL cholesterol, HDL cholesterol, triglycerides, and fasting glucose) to dietary fat.
Methods
A genome-wide interaction study was undertaken for each CRF in women (n ∼ 9000) not participating in the dietary modification (DM) trial, which focused on the reduction of dietary fat. Genetic scores based on these analyses were developed using a pruning-and-thresholding approach and tested for the prediction of 1-y CRF changes as well as long-term chronic disease development in DM trial participants (n ∼ 5000).
Results
Only 1 of these genetic scores, for LDL cholesterol, predicted changes in the associated CRF. This 1760-variant score explained 3.7% (95% CI: 0.09, 11.9) of the variance in 1-y LDL cholesterol changes in the intervention arm but was unassociated with changes in the control arm. In contrast, a main-effect genetic risk score for LDL cholesterol was not useful for predicting dietary fat response. Further investigation of this score with respect to downstream disease outcomes revealed suggestive differential associations across DM trial arms, especially with respect to coronary heart disease and stroke subtypes.
Conclusions
These results lay the foundation for the combination of many genome-wide gene-diet interactions for diet response prediction while highlighting the need for further research and larger samples in order to achieve robust biomarkers for use in personalized nutrition.
Keywords: gene-diet interactions, cardiometabolic, diet response, dietary fat, nutrigenetics
Introduction
Nutrigenetics approaches, in which genetic information is used to predict response to dietary inputs, are central to the emerging promise of personalized nutrition for cardiometabolic risk reduction. Interindividual differences in food preferences, metabolism, excretion, etc., affect our responses to diet, in a similar manner to the well-studied field of pharmacogenomics (1). Ideally, genotype-based nutrigenetic investigations would be conducted in large-scale dietary interventions. Two notable examples are the Prevención con Dieta Mediterránea (PREDIMED) and Preventing Overweight Using Novel Dietary Strategies (POUNDS Lost) trials, with findings including the interaction of a TCF7L2 variant with a Mediterranean diet pattern for glycemic traits (2) and the interaction of a PCSK9 variant with dietary carbohydrate for insulin resistance (3). However, such intervention-based studies can examine only a single dietary change (whether food, nutrient, or pattern) at a time, and are often limited to lower sample sizes (4).
To allow for more flexibility and greater sample sizes, gene-diet interactions (GDIs) are more commonly investigated in observational datasets. There is a rich literature of GDI discovery in the cardiometabolic realm. Typically, these focus on candidate genes/variants and cardiometabolic risk factors (CRFs) (5, 6), but some have looked at clinical outcomes [e.g., myocardial infarction (7)]. Other approaches use main-effect genetic risk scores, such as those for obesity interacting with sugar-sweetened beverage intake to influence anthropometric traits (8, 9).
Characterization of individuals based on single or small groups of single nucleotide polymorphisms (SNPs) likely neglects important signals elsewhere in the genome, especially for highly polygenic cardiometabolic traits. Thus, for effective personalized nutrition approaches to be realized, it is necessary to integrate many signals across the genome. A few investigations have explored GDIs genome-wide, such as for dairy and BMI (10) and for various dietary components and colorectal cancer (11). However, genome-wide interaction studies (GWIS) can be problematic due to the lower statistical power inherent in gene-environment interaction analyses (12). Furthermore, there is potential for confounding and reverse causation (i.e., cardiometabolic risk impacting dietary behavior) in statistical interactions from observational data. Given these limitations, it is not yet known whether collections of GDIs, discovered in observational datasets, can predict the effect of a dietary intervention on CRFs.
In order to provide proof-of-concept for the use of observational GDIs in developing comprehensive diet response genetic scores, we sought to develop a genome-wide, GDI-based dietary fat response score (FRS) for each of a series of CRFs. We performed GWIS for 6 CRFs and used these intermediate results to derive FRSs for each CRF. We tested the performance of these scores in the fat reduction-focused Women's Health Initiative (WHI) Dietary Modification trial, finding that an FRS for LDL cholesterol predicts 1-y LDL cholesterol changes and associates with incident coronary heart disease (CHD) and stroke subtypes over approximately 22 y of follow-up.
Methods
WHI dataset
The WHI study consists of a series of substudies: 3 clinical trials (related to cancer, cardiovascular disease, and osteoporosis) and an observational study (13). Over 160,000 participants were enrolled between 1993 and 1998, with the ability to enroll in ≤3 of the clinical trials simultaneously. For the purposes of this analysis, participants were categorized based only on whether or not they were enrolled in the dietary modification (DM) trial, which randomly assigned almost 50,000 women (not all of whom were genotyped) to a low-fat diet or a control diet with no recommended dietary changes, with primary outcomes being incidence of breast and colorectal cancers and heart disease (14). The study of these participants conformed to the ethical guidelines outlined in the Declaration of Helsinki, and this research was approved by the Tufts Health Sciences Institutional Review Board(protocol 12592).
Participants were comprehensively screened at baseline, including physical measurements, blood sample collection, and questionnaire administration, and only a subset of participants provided blood samples or returned questionnaires during later visits. The FFQ was designed specifically for the WHI study, emphasizing specific foods and preparation methods to maximize its sensitivity to changes in fat intake (15).
Phenotype data were accessed from the database of Genotypes and Phenotypes (dbGaP; accession: phs000746.v2.p3). Values shown in Table 1 only pertain to women whose genotypes were measured in 1 of a series of follow-up studies. For GDI analyses, systolic blood pressure (SBP), LDL cholesterol, and fasting glucose (FG) were adjusted for medication use: LDL cholesterol and FG values were divided by 0.75 for those on lipid-lowering and antidiabetic medication, respectively, and SBP values were increased by 15 mmHg for those on antihypertensive medication. This type of adjustment for medication use has precedent in gene-environment interaction analyses (16). CRFs were Winsorized at 5 SDs from the mean and those other than LDL cholesterol (BMI, SBP, HDL cholesterol, triglycerides (TG), and FG) were log-transformed prior to analysis. Longitudinal risk factor changes were calculated in DM trial participants as the difference between baseline and year 1. Adjudicated time-to-event data for chronic disease outcomes (CHD, myocardial infarction, ischemic stroke, hemorrhagic stroke, and noncardiovascular disease (CVD) death) were collected, whereas diabetes incidence was defined as the self-report of any: diabetes pills, insulin treatment, or general treatment of diabetes. Follow-up data was available for ∼22 y following enrollment. Phenotype data processing was performed using R version 3.4.3 [R Foundation] (17) and Python version 3.6.0 [Python Software Foundation].
TABLE 1.
Baseline characteristics of European-ancestry participants in the Women's Health Initiative study with available genetic data (n = 14,860 in total)
| DM trial | Non-DM trial | P value | |
|---|---|---|---|
| Sample size | 2165 (intervention); 3281 (control) | 9414 | |
| Age | 66 (60–70) | 68 (64–72) | <0.001 |
| Current smoking | 366 (7%) | 888 (9%) | <0.001 |
| Lipid-lowering medication | 583 (11%) | 1277 (14%) | <0.001 |
| Hypertension medication | 23,058 (38%) | 3416 (36%) | 0.07 |
| Diabetes medication | 269 (5%) | 498 (5%) | 0.37 |
| BMI, kg/m–2 | 28.9 (25.3–33.1) | 27.2 (24.0–31.2) | <0.001 |
| SBP, mm Hg | 128 (117–140) | 144 (133–156) | <0.001 |
| LDL cholesterol, mg/dL | 151 (128–175) | 161 (135–192) | <0.001 |
| HDL cholesterol, mg/dL | 49.8 (42–58) | 51 (44–60) | <0.001 |
| TG, mg/dL | 138 (100–195) | 128 (92–179.6) | < 0.001 |
| FG, mg/dL | 97 (90–108) | 97.5 (90–113) | 0.01 |
Continuous values shown as: median (IQR). P values are from Wilcoxon rank-sum test (continuous values) or chi-square test (discrete values). DM: dietary modification trial; FG, fasting glucose; Non-DM: all women not participating in the DM trial (enrolled in ≥1 of: Hormone Therapy Trial, Calcium and Vitamin D Trial, or Observational Study); SBP, systolic blood pressure; TG, triglycerides.
Genotype data and preprocessing
Imputed genotype data were retrieved from dbGaP (accession: phs000746.v2.p3) as a harmonized set of imputation outputs from a series of genotyping studies involving WHI participants. Prior to imputation, study-specific quality control steps had been undertaken on directly genotyped SNPs, with filters based on sample and call rate, Hardy–Weinberg equilibrium, and minor allele frequency (MAF). Phasing had been performed for autosomes using BEAGLE, followed by imputation using minimac [MaCH (Markov Chain Haplotyping algorithm) for the SHARe (SNP Health Association Resource)study subset]. After download from dbGaP, variants were converted from dose format using dose2plink (http://genepi.qimr.edu.au/staff/sarahMe/dose2plink.html), filtered for imputation r2 >0.3 and MAF >0.001, and annotated with reference SNP cluster IDs (rsIDs), loci, and allelic information using the 1000 Genomes Phase 3 download from dbSNP (download date: 13 April, 2018). Only variants passing the imputation quality threshold in all genotyping substudies were included in the final dosage dataset. Postimputation genotype data processing and score calculation were performed using PLINK 2.0, and clumping was performed using PLINK 1.9 (18).
GWIS
A GWIS was performed for each of the 6 CRFs. The genome-wide scan used an additive genotype model, adjusted for fixed effects including dietary fat (binary: percentage of kcals above or below the median), total kcals per day, age, 5 ancestry principal components, and genotyping substudy. Genotyping was performed in a series of ancillary studies in WHI, including Hip Fracture, GARNET (Genetics and Randomized Trials Network), WHIMS+ (Women's Health Initiative Memory Study), GECCO (Genetics and Epidemiology of Colorectal Cancer Consortium; initial or CytoSNP), and AS264/MOPMAP (Genetic Modification of Particulate Matter-Mediated Arrhythmogenesis in Populations). (Many participants were also genotyped as part of the SHARe effort, but those women were of African-American and Hispanic ancestry and thus were not included in the GWIS portion of this study.) The primary estimand of interest was the interaction term between dietary fat and minor allele count at the SNP of interest. Interaction analyses were carried out using PLINK 2.0 [www.cog-genomics.org/plink/2.0/] (18). Variants of interest were annotated to genes using Annovar (19).
Gene-environment interaction power calculations for single SNPs were performed using the Quanto tool (20). The following assumptions were made: additive model, variance explained by genotype alone = 0.5%, and binary environment with 50% prevalence and explaining 10% of the variance. (Note: there is no effect of MAF in this case given that variances explained are directly specified.)
Genetic responder score construction and evaluation
Given the lower power of gene-environment interaction detection, a subset of variants were prioritized for score derivation having nominal (P <0.05) marginal effects in large-scale meta-analyses. Summary statistics were retrieved from: Genetic Investigation of Anthropometric Traits (GIANT) for BMI (21); International Consortium for Blood Pressure for SBP (22); Global Lipid Genetics Consortium (GLGC) for LDL cholesterol, HDL cholesterol, and TG (23); and Meta-Analyses of Glucose- and Insulin-Related Traits Consortium (MAGIC) for fasting glucose (GLU) (24). After this main-effect filter, each FRS was constructed using summary statistics for the diet-SNP interaction terms from the associated GWIS. Interaction summary statistics were used as input to a pruning-and-thresholding (P&T) procedure (using the “—clump” function in PLINK 1.9), with a seed threshold of P = 0.05 and a linkage disequilibrium (LD) threshold of r2 = 0.5. The LD reference for the procedure was calculated from hard-called genotypes of the white DM trial participants. Genetic FRSs for each individual were then calculated as the weighted sum of allelic dosages for variants selected by the P&T procedure, with weights corresponding to the GWIS interaction term estimates. This type of diet interaction-based genetic score development has been described previously, for example in a Korean cohort with respect to body fat changes (25). A genetic risk score for LDL cholesterol (main-effect) was created using the GLGC LDL cholesterol meta-analysis summary statistics and the same P&T method and parameters as were used for the interaction analyses, resulting in a 26,467-SNP score. As an alternative to the P&T procedure, the LDpred method (which uses all variants without any main-effect filter) was used to calculate FRS weights for each CRF, incorporating a minor change to the code to allow for the slight genomic deflation observed for TG and GLU (26). LDpred was run using an LD radius of 500 variants, HapMap 3 variants only (962,057 variants in total), and causal variant fractions of 0.001, 0.01, and 0.1, and the resulting weights were then used to calculate dosage-based scores as done with the P&T method.
FRSs were used to test for discrimination of changes in CRFs over the first year of the DM trial. Risk factor changes were assessed using linear models in participants in the intervention arm, with and without adjustment for baseline CRF levels. The LDL cholesterol-specific FRS was then investigated further in a series of sensitivity analyses. First, P values were calculated for the interaction of the genetic score with trial arm (control compared with dietary modification). Second, principal components analysis was performed in DM intervention participants using 4 baseline metabolic biomarkers (total cholesterol, HDL cholesterol, TG, and FG) included in a prior clustering analysis for use in personalized nutrition stratification (27). Equivalent linear models to the original FRS assessment models were fit, with additional adjustment for the 4 resulting principal components.
The LDL cholesterol-fat response score (LDL-FRS) was further tested for the prediction of chronic disease development during follow-up across DM trial strata. Time-to-event for each CHD, myocardial infarction, ischemic stroke, hemorrhagic stroke, diabetes, and non-CVD death were used to fit age-adjusted Cox proportional hazards models, including a random effect term for genotyping substudy (cluster() term in the coxph function call). This frailty/random effects model has been recommended for optimizing power in multicenter time-to-event models (28). Estimated log-HRs were extracted from regressions conducted in the following strata: 1) DM trial intervention arm, 2) DM trial control arm, 3) DM trial intervention arm filtered for participants with 1-y fat reduction based on FFQ, and 4) DM trial control arm filtered for participants with 1-y fat increase based on FFQ.
Results
Dietary FRS development
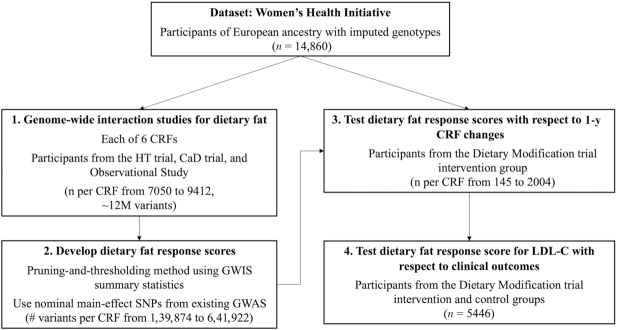
The study workflow is outlined in Figure 1. A series of GWIS were undertaken in cross-sectional data from the WHI. These GWIS incorporated only women who did not participate in the DM trial, using imputed genotypes along with baseline self-reported dietary intakes (from FFQs) and fasting blood biomarkers. Baseline characteristics of these women, along with those participating in the DM trial, are shown in Table 1. Although there were differences across groups in almost all characteristics, they were modest in size.
FIGURE 1.
Study workflow. First, a series of GWIS were conducted with dietary fat as the exposure for each of 6 cardiometabolic risk factors. Next, dietary fat response scores were developed using GWIS summary statistics and tested for the prediction of 1-y cardiometabolic risk factor changes in the dietary modification trial intervention (with sensitivity analyses including the control group). Finally, the LDL cholesterol score was tested for the prediction of differential effects on chronic disease development in the intervention and control groups over approximately 22 y of follow-up. CRF, cardiometabolic risk factor; GWAS, genome-wide association study; GWIS, genome-wide interaction study; SNP, single nucleotide polymorphism; HT, Hormone Therapy; CaD, calcium and vitamin D.
Preliminary power calculations were undertaken based on parameter assumptions including a modest SNP main effect (0.5%) under an additive model and a binary environment with 50% prevalence explaining 10% of the outcome phenotypic variance. The results showed that, at the sample sizes available for European ancestry non-DM participants (7050–9412 individuals for each CRF), this analysis was powered to detect only moderately large interaction effects (interaction variance explained greater than ∼0.5%) at genome-wide significance (Supplementary Table 1).
Dietary FRSs were generated for each CRF using results from the corresponding GWIS analysis (see Methods). Q–Q plots of the GWIS results showed that genomic inflation was fairly well-controlled (Supplementary Figure 1). For each CRF, the associated summary statistics (corresponding to the fat-genotype interaction term estimates) were filtered to include only those with nominal main-effect associations in large-scale published genome-wide association study (GWAS). This filter was informed by the power analysis above and chosen as a compromise between discovery and statistical power (alternative results using either a more stringent threshold or no filtering are shown in Supplementary Table 2). A P&T method was used to generate 6 FRS from these individual sets of summary statistics along with genotypes from the WHI DM participants as an LD reference. Using parameters of seed P value = 0.05 and LD r2 <0.5, 6 sets of score weights were generated, with relevant SNP set sizes ranging from 1536 (SBP) to 6042 (BMI). Scores were then calculated as the weighted sum of allele dosages across SNPs, normalized by the number of nonmissing SNPs per individual.
Dietary FRS assessment
As the scores were developed to predict a positive interaction with dietary fat intake, the expected direction of the FRS effect on risk factors in the present fat-reduction trial would be negative. Of the FRSs examined, only the LDL-FRS was predictive at P <0.05 of the associated CRF change in DM trial participants in the fat-reduction arm (passing a Bonferroni correction for the 6 CRFs tested in baseline-adjusted sensitivity models; results for all scores are shown in Table 2). For this score, the standardized effect size was −0.19 (corresponding to a 5.44 mg/dL greater decrease in LDL cholesterol per score SD; P = 0.020). We note that the sample size of European-ancestry DM trial participants with follow-up measurements was much smaller for biochemical variables (n ∼150) compared with BMI and SBP (n ∼2000). Using the score developed in European-ancestry individuals, score performance was then tested in a combined-ancestry group including black and Hispanic individuals, which almost doubled the sample size (Supplementary Table 3). Although some traits showed strong relations (e.g., SBP), the signs of many were in a counterintuitive direction, including a flip in sign for the previously strong LDL cholesterol relation, suggesting that these results may reflect the known difficulties in conducting transancestry polygenic prediction (due to differences in LD among other factors) rather than the intended biological differences. This observation was reinforced by the lack of association of the score with CRF changes in either blacks or Hispanics alone. Using the LDpred algorithm, which considers the full genome-wide set of SNPs, no predictive FRS effects were detected (Supplementary Table 4), possibly reflecting a lack of power to overcome the multiple testing burden without a nominal main-effect filter.
TABLE 2.
Responder score effects on 1-y CRF changes in DM trial participants
| Unadjusted | Baseline-adjusted1 | ||||||
|---|---|---|---|---|---|---|---|
| Risk factor | NGWIS2 | # SNPs in score | NDM3 | Std. effect4 | P value | Std. effect4 | P value |
| BMI | 9358 | 6042 | 1988 | 0.03 | 0.189 | 0.03 | 0.218 |
| SBP | 9412 | 1536 | 2004 | 0.03 | 0.125 | 0.04 | 0.041 |
| LDL-C | 7050 | 1760 | 145 | −0.19 | 0.020 | −0.21 | 0.005 |
| HDL-C | 7157 | 1731 | 150 | −0.08 | 0.320 | −0.08 | 0.351 |
| TG | 7158 | 1774 | 150 | −0.15 | 0.055 | −0.14 | 0.053 |
| FG | 7200 | 1924 | 281 | 0.01 | 0.853 | 0.02 | 0.689 |
Baseline-adjusted models are adjusted for the baseline value of the CRF being tested.
NGWIS = sample size available for the associated GWIS (non-DM participants).
NDM = sample size available for DM participants with 1-y follow-up measurements for the CRF in question.
Standard effect size represents the regression coefficient estimate in terms of CRF SD per responder score SD.
CRF, cardiometabolic risk factor; DM, dietary modification trial; FG, fasting glucose; GWIS, genome-wide interaction study; SBP, systolic blood pressure; SNP, single nucleotide polymorphism; TG, triglycerides.
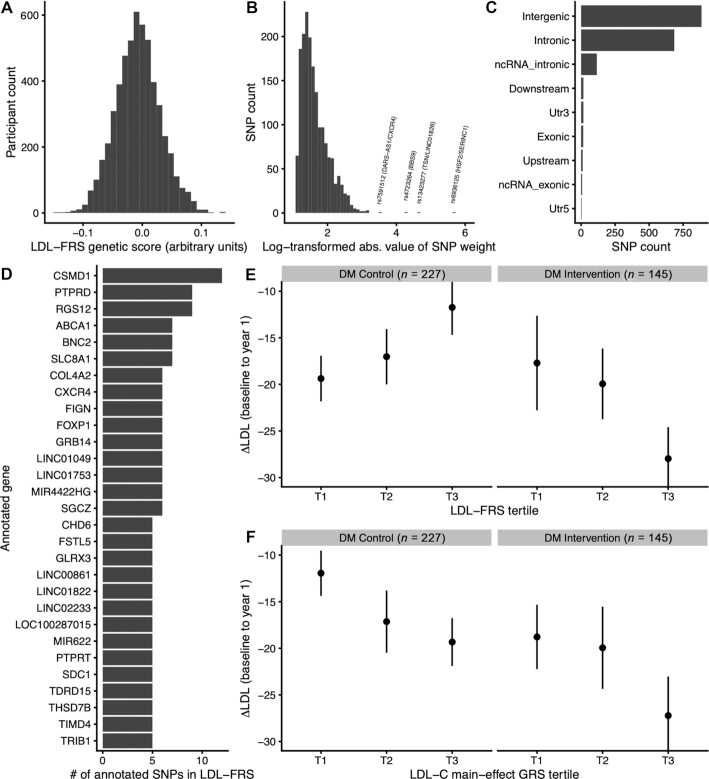
Based on its observed association in European-ancestry participants, the LDL-FRS was investigated further. Linear models showed that the LDL-FRS accounted for 3.7% (95% CI: 0.09, 11.9) of the variance in 1-y LDL cholesterol changes in the DM intervention arm. In baseline-adjusted models, this figure rose slightly to 4.3%, based on the change in R2compared with a baseline-only model. Further adjustment for 4 principal components of baseline metabolic markers (see Methods) did not materially affect this estimate (4.5%). For comparison, baseline LDL cholesterol alone accounted for 21.7% of this variance, although we note that this estimate is likely biased upward due to regression to the mean effects (arising from measurement error and stochastic biological fluctuations) (29). Additional baseline-adjusted models confirmed an interaction between the DM trial arm and the LDL-FRS (P = 0.002), supporting the specificity of this score for the fat-reduction arm, though the interaction directly with self-reported change in dietary fat was weaker (P = 0.11). The LDL-FRS also showed specificity for LDL cholesterol in that it did not predict changes in any other CRF (Supplementary Table 5). The 1760-component SNPs were annotated using Annovar (19), revealing a predominance of intergenic and intronic variants and a set of genes with high numbers of independent SNPs contributing to the score (Figure 2A–D). Top genes by number of contributing SNPs included CSMD1, PTPRD, and RGS12. Annotated genes and SNP weights for the LDL-FRS are available in Supplementary Table 6.
FIGURE 2.
LDL-FRS characterization. A) Distribution of LDL-FRS in Women's Health Initiative DM trial participants. B) Distribution of SNP weights constituting the LDL-FRS (shown as the natural log-transformed absolute values of the true weights). C) SNP counts in different loci types for LDL-FRS constituent SNPs. D) Genes are summarized by the number of annotated SNPs in the LDL-FRS (genes with ≥5 component SNPs are shown). E–F) 1-y changes in LDL cholesterol in DM trial participants as a function of genetic scores. Mean changes in LDL cholesterol (y-axis) are shown as a function of either LDL-FRS (E) or LDL-GRS (F) tertile (x-axis). Error bars represent SEs. DM, dietary modification trial; LDL-FRS, LDL cholesterol fat response genetic score; LDL-GRS, LDL cholesterol main-effect genetic risk score; SNP, single nucleotide polymorphism.
Differences in mean LDL cholesterol changes during the DM trial across genetic score strata are shown in Figure 2E, F. As suggested by the regression results, those in the control arm trended towards less substantial LDL cholesterol reductions in higher LDL-FRS strata, whereas those in the fat-reduction arm showed the opposite trend. Furthermore, isolation of individuals at the highest extreme of the score (top 10%) revealed an LDL cholesterol reduction of almost double that of the rest of the DM intervention group (−36.4 versus −20.3 mg/dL; 95% CI for the difference: [−1.0, 33.2]). For comparison to the FRS, a main-effect genetic risk score (GRS) for LDL cholesterol was developed using summary statistics from the GLGC meta-analysis (23) and an identical P&T procedure to that used for the FRS. As expected, this score was strongly predictive of baseline LDL cholesterol concentrations (P = 1.45 x 10-22). However, unlike the FRS, the GRS did not predict LDL cholesterol changes in the DM intervention group (P = 0.19; stratum-specific mean changes in Figure 2F).
LDL-FRS association with chronic disease outcomes
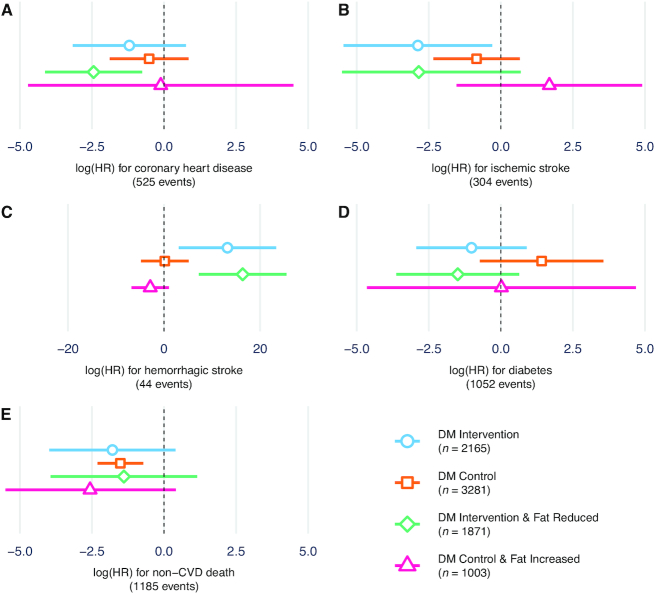
Next, the LDL-FRS was tested for relations with incident disease outcomes over ∼22 y of follow-up using Cox proportional hazards models (Figure 3). The mean time-to-event for incident CHD cases was 8.7 y (SD = 5.2), with similar values for other outcomes. In addition to intervention versus control arm, another set of “per protocol-like” strata was produced by additionally filtering for FFQ-based self-reported fat reduction (in the intervention group) or fat increase (in the control group). CHD qualitatively showed the expected interaction, i.e., a stronger inverse association between LDL-FRS and disease risk in the fat reduction group. Ischemic stroke showed a similar pattern, with a risk reduction only in the fat reduction group (P = 0.029). In contrast, hemorrhagic stroke, although having a low number of events (44 in total), showed a positive association only in the fat reduction group (P = 0.011). Results for diabetes qualitatively mirrored those for CHD and ischemic stroke, whereas those for non-CVD death did not vary across groups. These cross-arm differences were generally strengthened when comparing the per protocol-like strata, with a much stronger effect for CHD in the confirmed fat reduction stratum (P = 0.005). In DM trial arm interaction models (score × arm), none of the outcomes reached nominal statistical significance (P <0.05).
FIGURE 3.
LDL-FRS prediction of chronic disease development. Thex-axis shows log-HR estimates for the LDL-FRS from Cox proportional hazards regression for A) coronary heart disease, B) ischemic stroke, C) hemorrhagic stroke, D) diabetes, and E) non-CVD death. Separate estimates are shown for DM trial intervention arm, control arm, and the same strata filtered for FFQ-reported fat reduction or increase, respectively. Cox models are adjusted for age at baseline and include a random effect for Women's Health Initiative genotyping substudy. Error bars represent 95% CI estimates for the regression coefficients. CVD, cardiovascular disease; DM, dietary modification trial; LDL-FRS, LDL cholesterol-fat response genetic score.
Discussion
Diet response scores have shown some success in predicting the response of CRFs to nutritional interventions, but they are often based solely on main effects or single GDI SNPs. Here, we developed what to our knowledge is the first example of a diet response score based on a hypothesis-free genome scan for each of 6 risk factors, and showed preliminary evidence for the viability of an LDL cholesterol fat response score. The set of SNPs used for each score were limited to those showing nominal main effects in large-scale GWAS as a compromise between discovery and utilization of prior information, which was supported by the weaker results in sensitivity models incorporating either stronger (suggestive main-effect) or weaker (all SNPs, LDpred) variant filters (Supplementary Table 2).
Though FRS for 6 CRFs were developed and tested, only that for LDL cholesterol showed nominal significance in predicting 1-y changes in the corresponding CRF. Multiple factors could explain this lack of predictive performance, including residual confounding of the observed interactions and misclassification of the dietary exposure (despite the use of FFQs optimized for detection of dietary fat). Additionally, power calculations suggest that a cohort of this size may not be powered to detect gene-environment interactions with small effects.
CSMD1, PTPRD, and RGS12 stood out as genes containing the highest number of SNPs in the LDL-FRS (11, 9, and 9, respectively, after LD-pruning for r2 <0.5). CSMD1 variants are notably associated with LDL cholesterol response to statin treatment (30) as well as SBP response to a high-salt diet (31). CSMD1 has also shown epigenetic associations with LDL cholesterol (32) as well as response to modification of dietary fat composition (33). PTPRD variants modulate the response of type 2 diabetes patients to pioglitazone therapy (34) and show suggestive associations with eating behaviors (caloric intake at dinner) (35). RGS12 has been linked to LDL cholesterol in GWAS (36). Altogether, these genes have literature evidence for relations to dietary intake, response to cardiometabolic therapies, and LDL cholesterol, but have not been shown to directly modify the LDL cholesterol response to dietary fat proportions. We note that prioritizing main-effect SNPs for inclusion creates a bias towards identifying LDL cholesterol-related variants in the LDL-FRS.
A reasonable body of literature exists establishing GDIs for both dietary fat on CRFs (37, 38) and general dietary exposures on LDL cholesterol (39). Multiple studies have looked specifically at genetic variants modulating the LDL cholesterol response to dietary fat. For example, a caloric restriction intervention in type 2 diabetics was more effective in reducing LDL cholesterol in ApoE4 carriers (−15.6% versus −0.7%) (40). In the POUNDS Lost trial, carriers of specific alleles at APOA5and CETP variants saw 7.5 and 8.9 mg/dL greater LDL cholesterol decreases during a low-fat dietary intervention (41, 42). Our observed effect size of a 5.4 mg/dL decrease in LDL cholesterol is of a similar magnitude and emerged despite the multifactorial nature of the WHI dietary intervention. The observed variance explained of 3.7% for the LDL-FRS means that the score does not capture most of the interindividual variability in LDL cholesterol response to the WHI DM trial intervention. This explanatory power was modestly strengthened after adjustment for baseline LDL cholesterol as well as principal components reflecting baseline metabolic biomarker patterns. Based on prior observations of an inflection point in the impact of various genetic risk scores near the 90th percentile (43), we additionally evaluated the impact of LDL-FRS in the top 10%, finding almost double the LDL cholesterol reduction in these DM intervention participants.
The potential clinical utility of these findings can be evaluated in the context of a framework recently put forth for the scientific assessment of GDI (44). This genetic score was developed using a rigorous study design starting in an observational cohort and validating in a randomized trial, and relies on an “intermediate” interaction in which many nondietary factors are also expected to influence LDL cholesterol concentrations. Given that the biological plausibility is difficult to determine for a polygenic score and that the scientific validity of this FRS × diet interaction would be classified as “possible” to “probable,” additional validation of this or similar scores would be needed to render it clinically actionable.
Main-effect GRS have been used as genetic variables in order to improve statistical power to detect gene-environment interactions (45). Genetic risk is then modeled as a predisposition that is only triggered in certain environments (e.g., dietary behaviors). Here, we observed little association of a main-effect GRS for LDL cholesterol with greater LDL cholesterol reductions in the DM trial (P = 0.19). This trend runs counter to a prior observation of greater lifestyle intervention effectiveness for LDL cholesterol reduction in those with low genetic risk of hyperlipidemia (46), possibly due to differences between the DM trial and the personalized diet and lifestyle changes recommended in the intervention in question. Regardless, the meaningful increase in predictive power of the LDL-FRS compared with the main-effect GRS for LDL cholesterol indicates the value in using interaction-based genetic scores for personalized nutrition.
A useful diet response score should also predict downstream changes in chronic disease risk. Suggestive interactions for CHD, ischemic stroke, and diabetes were apparent across strata (Figure 3A, B, D), corresponding to a decreased risk in fat reduction participants (whose predicted LDL cholesterol drop would be larger). Hemorrhagic stroke showed the opposite trend, with a positive score-disease relation only in the fat reduction group, in line with existing evidence for the detrimental effects of low LDL cholesterol on hemorrhagic stroke risk (47). Non-CVD death showed no major associations, which could be expected due to the dominance of this category by cancer outcomes and the equivocal associations of cancer with lipids (48). We note that all disease outcome relations assessed here are subject to the major caveat that dietary evolution and decreased adherence likely developed over time in many subjects, diluting the utility of the randomization and 1-y changes used for stratification in Figure 3.
The present study had the advantage of developing a diet-focused genetic score in almost 10,000 women and testing in a dietary intervention trial using independent individuals from the same population. However, nominal main-effect SNPs were prioritized to improve statistical power given this moderate sample size, an approach that may fail to identify interactions with effect directions opposite to that of the main effect. Smaller fractions of alternate ancestries in this population also made the development of ancestry-specific response scores unrealistic. Additionally, the DM trial intervention included additional nonfat-related dietary recommendations and did not ultimately achieve its intended 20% fat reduction, making it an imperfect proxy for a pure fat reduction intervention. Finally, this study only examined women, despite the fact that CRF profiles and their genetic architectures vary across sexes (49).
In summary, we present a method for the development of diet response scores based on genome-wide, observational GDI study summary statistics. We provide proof-of-concept that a genetic score focused on LDL cholesterol may be useful for predicting changes in both CRFs and long-term disease risk during a dietary intervention. However, not all dietary FRSs derived here were informative, highlighting the continued need for increased sample sizes and improved diet measures for the discovery of sufficiently robust genetic interactions genome-wide. Our results provide a foundation for future investigations using new datasets and dietary variables to explore the genetic architecture of diet response.
Supplementary Material
ACKNOWLEDGEMENTS
The authors’ contributions were as follows—KW and JO: designed the research; KW: conducted the research and performed the statistical analysis; QL, SL, LP, PS, PJ, DD, and JO: advised the development of the analysis; KW: wrote the manuscript; QL, SL, LP, PS, PK, DD, and JO: provided substantive review of the manuscript; JO: had primary responsibility for final content; and all authors read and approved the manuscript. The authors report no conflicts of interest.
Notes
Supported by NHLBI T32 Nutrition and Cardiovascular Disease Predoctoral Training Program (T32HL069772-15) and by the United States Department of Agriculture, Agriculture Research Service (8050–51000‐098‐00D). The Women's Health Initiative (WHI) program is funded by the National Heart, Lung, and Blood Institute, NIH, US Department of Health and Human Services through contracts HHSN268201600018C, HHSN268201600001C, HHSN268201600002C, HHSN268201600003C, and HHSN268201600004C. This manuscript was prepared in collaboration with investigators of the WHI but has not been reviewed by the WHI, and does not necessarily reflect the opinions of the WHI investigators or the NHLBI. Funding support for WHI GARNET was provided through the NHGRI Genomics and Randomized Trials Network (GARNET) (grant number: U01 HG005152). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GARNET Coordinating Center (U01 HG005157). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Broad Institute of MIT (Massachusetts Institute of Technology) and Harvard, was provided by the NIH Genes, Environment and Health Initiative (GEI) (U01 HG004424). Funding for WHI SHARe genotyping was provided by NHLBI contract N02- HL-64278. The WHI Sight Exam and the Memory Study was funded in part by Wyeth Pharmaceuticals, Inc, St. Davids, PA.
SL is an editor of the American Journal of Clinical Nutrition and played no role in the journal's evaluation of the manuscript.
Supplemental Tables 1–6 and Supplemental Figure 1 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/ajcn/.
Data described in the manuscript and codebook are available by application through dbGaP (accession: phs000746.v2.p3). Analysis code can be found at https://github.com/kwesterman/diet-response.
Abbreviations used: CHD, coronary heart disease; CRF, cardiometabolic risk factor; CVD, cardiovascular disease; DM, dietary modification (trial); FG, fasting glucose; FRS, fat response score; GDI, gene-diet interaction; GLGC, Global Lipid Genetics Consortium; GRS, genetic risk score; GWAS, genome-wide association study; GWIS, genome-wide interaction study; LD, linkage disequilibrium; LDL-FRS, LDL cholesterol-fat response score; MAF, minor allele frequency; P&T, pruning-and-thresholding; SBP, systolic blood pressure; SNP, single nucleotide polymorphism; TG, triglycerides; WHI, Women's Health Initiative.
References
- 1. Ma Q, Lu AYH. Pharmacogenetics, pharmacogenomics, and individualized medicine. Pharmacol Rev. 2011;63:437–59. [DOI] [PubMed] [Google Scholar]
- 2. Corella D, Carrasco P, Sorli JV, Estruch R, Rico-Sanz J, Martinez-Gonzalez MA, Salas-Salvado J, Covas MI, Coltell O, Aros F et al.. Mediterranean diet reduces the adverse effect of the TCF7L2-rs7903146 polymorphism on cardiovascular risk factors and stroke incidence: a randomized controlled trial in a high-cardiovascular-risk population. Diabetes Care. 2013;36:3803–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Huang T, Huang J, Qi Q, Li Y, Bray GA, Rood J, Sacks FM, Qi L. PCSK7 genotype modifies effect of a weight-loss diet on 2-year changes of insulin resistance: the POUNDS LOST trial. Diabetes Care. 2015;38:439–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ordovas JM, Ferguson LR, Tai ES, Mathers JC. Personalised nutrition and health. BMJ. 2018;361:bmj.k2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corella D. APOA2, dietary fat, and body mass index. Arch Intern Med. 2009;169:1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cuda C, Badawi A, Karmali M, El-Sohemy A. Polymorphisms in Toll-like receptor 4 are associated with factors of the metabolic syndrome and modify the association between dietary saturated fat and fasting high-density lipoprotein cholesterol. Metabolism. 2011;60:1131–5. [DOI] [PubMed] [Google Scholar]
- 7. Cornelis MC, El-Sohemy A, Kabagambe EK, Campos H. Coffee, CYP1A2 genotype, and risk of myocardial infarction. JAMA. 2006;295:1135. [DOI] [PubMed] [Google Scholar]
- 8. Qi Q, Chu AY, Kang JH, Jensen MK, Curhan GC, Pasquale LR, Ridker PM, Hunter DJ, Willett WC, Rimm EB et al.. Sugar-sweetened beverages and genetic risk of obesity. N Engl J Med. 2012;367:1387–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Olsen NJ, Ängquist L, Larsen SC, Linneberg A, Skaaby T, Husemoen LLN, Toft U, Tjønneland A, Halkjær J, Hansen T et al.. Interactions between genetic variants associated with adiposity traits and soft drinks in relation to longitudinal changes in body weight and waist circumference. Am J Clin Nutr. 2016;104:816–26. [DOI] [PubMed] [Google Scholar]
- 10. Smith CE, Follis JL, Dashti HS, Tanaka T, Graff M, Fretts AM, Kilpeläinen TO, Wojczynski MK, Richardson K, Nalls MA et al.. Genome-wide interactions with dairy intake for body mass index in adults of European descent. Mol Nutr Food Res. 2018;62(3):1700347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Figueiredo JC, Hsu L, Hutter CM, Lin Y, Campbell PT, Baron JA, Berndt SI, Jiao S, Casey G, Fortini B et al.. Genome-wide diet-gene interaction analyses for risk of colorectal cancer. PLos Genet. 2014;10:e1004228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dempfle A, Scherag A, Hein R, Beckmann L, Chang-Claude J, Schäfer H. Gene-environment interactions for complex traits: definitions, methodological requirements and challenges. Eur J Hum Genet. 2008;16:1164–72. [DOI] [PubMed] [Google Scholar]
- 13. Anderson GL, Cummings SR, Freedman LS, Furberg C, Henderson MM, Johnson SR, Kuller LH, Manson JE, Oberman A, Prentice RL et al.. Design of the Women's Health Initiative clinical trial and observational study. Control Clin Trials. 1998;19:61–109. [DOI] [PubMed] [Google Scholar]
- 14. Ritenbaugh C, Patterson RE, Chlebowski RT, Caan B, Fels-Tinker L, Howard B, Ockene J. The Women's Health Initiative Dietary Modification Trial: overview and baseline characteristics of participants. Ann Epidemiol. 2003;13:S87–97. [DOI] [PubMed] [Google Scholar]
- 15. Patterson RE, Kristal AR, Tinker LF, Carter RA, Bolton MP, Agurs-Collins T. Measurement characteristics of the Women's Health Initiative food frequency questionnaire. Ann Epidemiol. 1999;9:178–87. [DOI] [PubMed] [Google Scholar]
- 16. Rao DC, Sung YJ, Winkler TW, Schwander K, Borecki I, Cupples LA, Gauderman WJ, Rice K, Munroe PB, Psaty BM. Multiancestry study of gene-lifestyle interactions for cardiovascular traits in 610 475 individuals from 124 cohorts. Circulation: Cardiovascular Genetics. 2017;10:e001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. R Core Team. R: A Language and Environment for Statistical Computing. [Internet] Vienna, Austria: R Foundation for Statistical Computing; 2017. Available from: https://www.r-project.org/. [Google Scholar]
- 18. Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaSci. 2015;4:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–84. [DOI] [PubMed] [Google Scholar]
- 21. Yengo L, Sidorenko J, Kemper KE, Zheng Z, Wood AR, Weedon MN, Frayling TM, Hirschhorn J, Yang J, Visscher PM. Meta-analysis of genome-wide association studies for height and body mass index in 700000 individuals of European ancestry. Hum Mol Genet. 2018;27:3641–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ehret GB, Munroe PB, Rice KM, Bochud M, Johnson AD, Chasman DI, Smith AV, Tobin MD, Verwoert GC, Hwang SJ et al.. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, Ganna A, Chen J, Buchkovich ML, Mora S et al.. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL et al.. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cha S, Kang J, Lee J-H, Kim J, Kim H, Yang Y, Park W-Y, Kim J. Impact of genetic variants on the individual potential for body fat loss. Nutrients. 2018;10:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vilhjálmsson BJ, Yang J, Finucane HK, Gusev A, Lindström S, Ripke S, Genovese G, Loh P-R, Bhatia G, Do R et al.. Modeling linkage disequilibrium increases accuracy of polygenic risk scores. Am J Hum Genet. 2015;97:576–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O'Donovan CB, Walsh MC, Nugent AP, McNulty B, Walton J, Flynn A, Gibney MJ, Gibney ER, Brennan L. Use of metabotyping for the delivery of personalised nutrition. Mol Nutr Food Res. 2015;59(3):377–85. [DOI] [PubMed] [Google Scholar]
- 28. Munda M, Legrand C. Adjusting for centre heterogeneity in multicentre clinical trials with a time-to-event outcome. Pharmaceut Statist. 2014;13:145–52. [DOI] [PubMed] [Google Scholar]
- 29. Barnett AG, van der Pols JC , Dobson AJ. Regression to the mean: what it is and how to deal with it. Int J Epidemiol. 2005;34:215–20. [DOI] [PubMed] [Google Scholar]
- 30. Thompson JF, Hyde CL, Wood LS, Paciga SA, Hinds DA, Cox DR, Hovingh GK, Kastelein JJ. Comprehensive whole-genome and candidate gene analysis for response to statin therapy in the Treating to New Targets (TNT) cohort. Circulation: Cardiovascular Genetics. 2009;2:173–81. [DOI] [PubMed] [Google Scholar]
- 31. Newton-Cheh C, Johnson T, Gateva V, Tobin MD, Bochud M, Coin L, Najjar SS, Zhao JH, Heath SC, Eyheramendy S et al.. Genome-wide association study identifies eight loci associated with blood pressure. Nat Genet. 2009;41:666–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bell JT, Tsai P-C, Yang T-P, Pidsley R, Nisbet J, Glass D, Mangino M, Zhai G, Zhang F, Valdes A et al.. Epigenome-wide scans identify differentially methylated regions for age and age-related phenotypes in a healthy ageing population. PLos Genet. 2012;8:e1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perfilyev A, Dahlman I, Gillberg L, Rosqvist F, Iggman D, Volkov P, Nilsson E, Risérus U, Ling C. Impact of polyunsaturated and saturated fat overfeeding on the DNA-methylation pattern in human adipose tissue: a randomized controlled trial. Am J Clin Nutr. 2017;105:991–1000. [DOI] [PubMed] [Google Scholar]
- 34. Pei Q, Huang Q, Yang G-P, Zhao Y-C, Yin J-Y, Song M, Zheng Y, Mo Z-H, Zhou H-H, Liu Z-Q. PPAR-γ2 and PTPRD gene polymorphisms influence type 2 diabetes patients’ response to pioglitazone in China. Acta Pharmacol Sin. 2013;34:255–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, Butte NF. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7:e51954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Spracklen CN, Chen P, Kim YJ, Wang X, Cai H, Li S, Long J, Wu Y, Wang YX, Takeuchi F et al.. Association analyses of East Asian individuals and trans-ancestry analyses with European individuals reveal new loci associated with cholesterol and triglyceride levels. Hum Mol Genet. 2017;26:1770–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Badawi A, Garcia-Bailo C, El-Sohemy A. A common polymorphism near the interleukin-6 gene modifies the association between dietary fat intake and insulin sensitivity. J Inflamm Res. 2012;5:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zheng Y, Huang T, Zhang X, Rood J, Bray GA, Sacks FM, Qi L. Dietary fat modifies the effects of FTO genotype on changes in insulin sensitivity. J Nutr. 2015;145:977–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ordovás JM, Robertson R, Cléirigh EN. Gene-gene and gene-environment interactions defining lipid-related traits. Curr Opin Lipidol. 2011;22:129–36. [DOI] [PubMed] [Google Scholar]
- 40. Saito M, Eto M, Nitta H, Kanda Y, Shigeto M, Nakayama K, Tawaramoto K, Kawasaki F, Kamei S, Kohara K et al.. Effect of apolipoprotein E4 allele on plasma LDL cholesterol response to diet therapy in type 2 diabetic patients. Diabetes Care. 2004;27:1276–80. [DOI] [PubMed] [Google Scholar]
- 41. Zhang X, Qi Q, Bray GA, Hu FB, Sacks FM, Qi L. APOA5 genotype modulates 2-y changes in lipid profile in response to weight-loss diet intervention: the POUNDS LOST Trial. Am J Clin Nutr. 2012;96:917–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu M, Ng SS, Bray GA, Ryan DH, Sacks FM, Ning G, Qi L. Dietary fat intake modifies the effect of a common variant in the LIPC gene on changes in serum lipid concentrations during a long-term weight-loss intervention trial. J Nutr. 2015;145:1289–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Khera AV, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, Natarajan P, Lander ES, Lubitz SA, Ellinor PT et al.. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet. 2018;50:1219–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grimaldi KA, van Ommen B, Ordovas JM, Parnell LD, Mathers JC, Bendik I, Brennan L, Celis-Morales C, Cirillo E, Daniel H et al.. Proposed guidelines to evaluate scientific validity and evidence for genotype-based dietary advice. Genes Nutr. 2017;12:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Aschard H. A perspective on interaction effects in genetic association studies. Genet Epidemiol. 2016;40:678–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zubair N, Conomos MP, Hood L, Omenn GS, Price ND, Spring BJ, Magis AT, Lovejoy JC. Genetic predisposition impacts clinical changes in a lifestyle coaching program. Sci Rep. 2019;9:6805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sun L, Clarke R, Bennett D, Guo Y, Walters RG, Hill M, Parish S, Millwood IY, Bian Z, Chen Y et al.. Causal associations of blood lipids with risk of ischemic stroke and intracerebral hemorrhage in Chinese adults. Nat Med. 2019;25:569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Koene RJ, Prizment AE, Blaes A, Konety SH. Shared risk factors in cardiovascular disease and cancer. Circulation. 2016;133:1104–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Knopp RH, Paramsothy P, Retzlaff BM, Fish B, Walden C, Dowdy A, Tsunehara C, Aikawa K, Cheung MC. Sex differences in lipoprotein metabolism and dietary response: basis in hormonal differences and implications for cardiovascular disease. Curr Cardiol Rep. 2006;8:452–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.