Summary
Many human pathogens use Type III, Type IV and Type VI secretion systems to deliver effectors into their target cells. The contribution of these secretion systems to microbial virulence was the main focus of a Workshop organized by the International University of Andalusia in Spain. The meeting addressed structure-function, substrate recruitment and translocation processes, which differ widely on the different secretion machineries, as well as the nature of the translocated effectors and their roles in subverting the host cell. An excellent panel of worldwide speakers presented the state of the art of the field, highlighting the involvement of bacterial secretion in human disease and discussing mechanistic aspects of bacterial pathogenicity, which can provide the bases for the development of novel anti-virulence strategies.
Keywords: Bacterial secretion, Type III Secretion System, Type IV Secretion System, Type VI Secretion System, machinery assembly, protein translocation, effector protein, toxin, microbial virulence, host subversion, pathogenicity, antibacterial
Bacterial injection systems: all roads lead to Rome
Bacterial secretion systems are trans-envelope multi-protein assemblies devoted to the transport of specific macromolecules to the outside. These nanostructures connect bacteria with the outer world, playing a pivotal role in essential bacterial life processes such as finding nutrients, sharing genetic information, or communicating with other organisms in the context of pathogenic or symbiotic interactions (Green et al., 2016). Among the many different families of secretion machineries, the Type III, Type IV and Type VI protein secretion systems (T3/T4/T6SS) share the ability to inject their substrates directly into human cells (Galan et al., 2018), playing an important role in pathogenesis. The transported substrates, collectively known as effectors, have the capacity to modulate host cellular processes for the benefit of the bacterial pathogens that encode them. The role of translocated effectors in virulence is well established for an increasing number of human pathogens such as Escherichia coli, Salmonella enterica, Helicobacter pylori, Legionella pneumophila, Pseudomonas aeruginosa, or Brucella melitensis. These three secretion machineries are also found within phytopathogens whose effectors target plant cells. T4SS also play a role in horizontal gene transfer, one of the main causes for spreading antibiotic resistance genes within bacterial populations. This mechanism contributes to the generation of multi-resistant bacteria, which pose a major threat to global health (Aslam et al., 2018). T6SS are mainly recognized as antibacterial injection machineries, able to deliver toxic effectors in prey bacteria (Coulthurst, 2019).
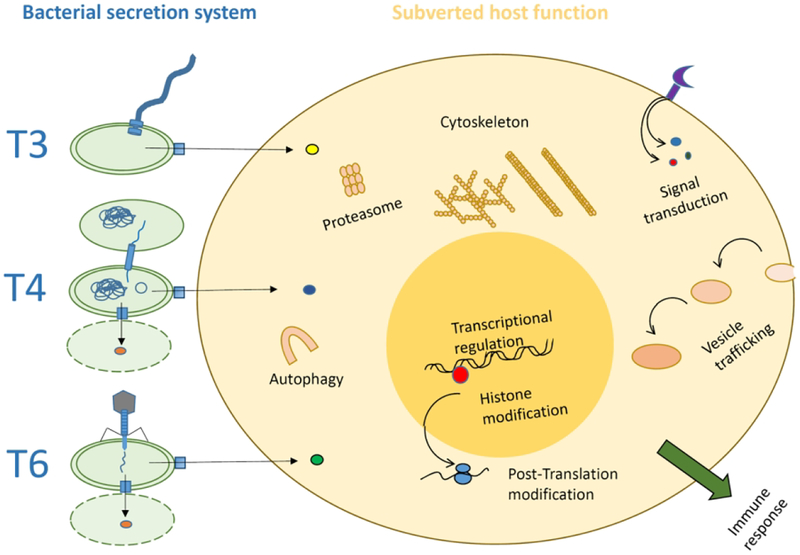
Remarkably, the evolutionary origin of these protein secretion systems differs significantly. T3SS are evolutionary related to the flagellar apparatus (Bhattacharya et al., 2019); T4SS were originally conceived as conjugative bridges for DNA transfer (Christie, 2001), and T6SS are evolutionary related to contractile phage tails (Leiman et al., 2009). Thus, evolution has shaped multiprotein complexes with the capacity to inject bacterially-encoded proteins into another cell multiple times. In spite of their different evolutionary origins, these secretion machines converged mechanistically into the ability to transport macromolecules across phospholipids bilayers, resulting in the delivery of proteins synthesized by one bacterium into another cell target, prokaryotic or eukaryotic. In the injection systems involved in pathogenicity, there is also a functional convergence in the effectors delivered by the different machines, which have the capacity to target similar cellular processes, such as signal transduction pathways, vesicular trafficking, or the immune response (Personnic et al., 2016, Ratner et al., 2017, Pisano et al., 2018, Tsai et al., 2019) (Figure 1).
Figure 1.
Schematic representation of T3, T4 and T6SS showing their relation to flagellum, conjugative DNA transfer and phage structures, and their common targets in the host cell. Bacteria (in green) are shown hosting the secretion system and related machineries: flagella for T3SS, conjugative DNA transfer complexes for T4SS, and phage tails for T6SS. Their bacterial targets are also shown: recipient bacteria for DNA transfer (T4SS), or bacteria killed (dotted lines) by T6SS-injected toxins. The three secretion systems can translocate effectors to the human cell, shown to the right, together with the most common subverted cellular functions.
The number of effector proteins transported by a given machine varies significantly and can reach hundreds in some exceptional cases. As these effectors have the capacity to modulate a vast range of cell physiology processes, their study are not only providing insight into mechanisms of pathogenicity but also illuminating fundamental principles of cell biology. Furthermore, the molecular knowledge of these secretion machineries may pave the way to the design of new anti-infective drugs. Thus, the search for inhibitors of the secretion machinery assembly or functioning/operation or the effector themselves is an active field of research (Fasciano et al., 2019, Graf et al., 2019). In addition, secretion systems can be converted into intracellular delivery machines which could also have therapeutic applications (Walker et al., 2017).
Our knowledge on the architecture and function of these different secretion machines has improved significantly over the last few years. Work during the 90’s led to the realization that T3SS had the capacity to deliver bacterially encoded proteins into host cells, and in particular, virulence factors into human cells. The discovery of the needle complex and associated structures led to the concept of the injectisome. Combined with the description of a plethora of effector proteins and their biochemical activities, these studies led to the emergence of novel paradigms in bacterial pathogenesis, host pathogen interactions, protein secretion, and molecular machines (Galan et al., 2014). Furthermore, because of their high degree of conservation across many important bacterial pathogens independently of their phylogeny and the commonality of secretion mechanisms, T3SS emerge as a potential target for the development of novel antimicrobial strategies (Lara-Tejero et al., 2019). Overall, the study of T3SS has serve as useful paradigm for research on other protein secretion systems.
T4SSs stand out for their plasticity, since they can translocate both protein and DNA molecules, and the destiny can be their secretion into the milieu or the delivery into another cell, being either prokaryotic or eukaryotic (Li et al., 2019). This versatility allows T4SS to play a key role in many essential aspects of bacterial life. These functions can be grossly classified in two: (i) contribution to the horizontal gene pool, both in Gram-negative and positive bacteria, and (ii) modulation of eukaryotic host cells. DNA transport is a particularity of T4SSs, which are an integral part of bacterial conjugation machineries. A T4SS subfamily adapted to interact with eukaryotic cells mediates secretion or direct cell-to-cell transfer of virulence factors to modulate host cells (Hayek et al., 2019). Members of this family contribute to the virulence of significant human pathogens, such as Helicobacter pylori, Legionella pneumophila, or Brucella melitensis. T4SS can also mediate interkingdom DNA transfer, as exemplified by the prototypical T4SS VirB of Agrobacterium tumefaciens, responsible for the transfer and integration of bacterial DNA into the plant cell genome (Li et al., 2018).
Finally, T6SSs were described only a decade ago, but in spite of this, their structure and mechanism have been deciphered in much detail quickly (Cherrak et al., 2019). The T6SS functions as a dynamic contractile phage tail-like structure anchored in the bacterial cell envelope to deliver effectors directly into the target cell. The T6SS primary role is to target competitor bacteria in the environment or in the context of host infection (Hood et al., 2010, Coulthurst, 2019); however, interestingly some T6SS effectors are also recognized as virulence factors that target eukaryotic cell components (Hachani et al., 2016). T6SSs are present among pathogens such as P. aeruginosa or Acinetobacter, classified by the WHO as antibiotic-resistant “priority pathogens”.
The role of bacterial secretion in disease is an especially relevant research field where to promote cooperation: the socio-economic importance of the issue raises interests among scientists from different backgrounds, such as medicine, biochemistry, structural, cellular and molecular biology, genetics, or microbiology, working on a variety of organisms including microbes, plants, animals, and humans. A detailed understanding of bacterial secretion systems requires a multidisciplinary approach that addresses all structural, biochemical, microbiological, cellular and medical aspects: from the elucidation of the molecular structure of the protein components to the cell biology studies to elucidate the activities of the translocated effector proteins in eukaryotic host cells. The ultimate goal of the Workshop “Contribution of bacterial injection systems to human disease”, which took place at the International University of Andalusia (UNIA, Spain) in November 2018, was to promote cross-talk between scientists working in different secretion systems at different organization levels, pursuing as goals a holistic comprehension of bacterial pathogenesis, and discussing possible common strategies for anti-pathogen therapies.
Knowledge on the mechanism of secretion is essential to understand pathogenicity and to design therapies that block the secretion process. The first session of the workshop addressed the assembly of the multiprotein transmembrane complex, the 3D structure of the individual protein components, and structure-function analysis of the nanomachines. A second session dealt with the nature and recruitment of substrates, and the active translocation process of both protein and DNA molecules. The third session focused on the subversion of the human cell by translocated effectors, including interaction of the delivered substrates with host elements and modulation of the host response.
Structure and function of T3, T4 and T6 Secretion Systems
Structural biology provides an essential knowledge on which we base to great extent our research for understanding multiprotein complexes. The size of these complexes makes electron microscopy the ideal technology to elucidate their three-dimensional architecture. In the last years, technologies requiring particle purification have been complemented by other approaches, which allow the elucidation of complex structures in situ. In this new context, cryo-electron tomography (cryo-ET) has proven an excellent tool, and new exciting results argue for the importance of the biological context on the elucidation of three dimensional structures. María Lara-Tejero (Yale University, New Haven, USA) presented the in situ structure of the Salmonella T3SS obtained by cryo-ET. Comparison with the cryo-electron microscopy (cryo-EM) structure of the isolated needle complexes revealed significant conformational differences resulting from the docking of the sorting platform in the needle complex, providing the symmetry adaptation required for the assembly of the entire secretion within the bacterial envelope (Hu et al., 2017). Expanding on the T3SS structure, Samuel Wagner (University of Tübingen, Germany) presented work in collaboration with Susan Lea (Oxford) on the T3SS core export apparatus components SctR, SctS, and SctT, (Wagner et al., 2010). A cryo-EM structure of the flagellar SctRST homologs FliPQR showed that these proteins assemble into a unique helical complex with a central pore, that is closed in the structure of the isolated complex, but that is thought to open when located in the membrane. Despite their prediction as membrane proteins, the helical SctRST complex resides within the bacterial inner membrane during assembly, but it locates to a periplasmic supramembrane position in the assembled injectisome. The details of its function in gating of and secretion through the T3SS injectisome awaits further investigation.
There were a number of reports providing new, data on the structure of T4SS. Until recently, our knowledge came from single particle negative staining electron microscopy (nsEM) and reconstruction of the full transmembrane complex of the conjugative plasmid R388 (Low et al., 2014), and the nsEM structure of the core complex of the Cag T4SS (Frick-Cheng et al., 2016) involved in H pylori pathogenicity. Although different in size and shape, both T4SS showed an outer membrane complex (OMC) with a 14-fold symmetry. The atomic structure of the OMC of the VirB T4SS from Xanthomonas citrii, which mediates bacterial killing, confirmed this 14-fold symmetry, as reported Tiago R. Costa from Gabriel Waksman lab (Imperial College, London, UK) (Sgro et al., 2018). Intriguingly, the fine structure of two different T4SS, solved in situ by cryo-ET, shed different results. Craig R Roy (Yale University, New Haven, USA) presented a refined structure of the Dot-Icm T4SS from L. pneumophila (Chetrit et al., 2018), previously reported at lower resolution (Ghosal et al., 2017); the Dot-Icm T4SS is the prototype of so-called “Type B” T4SS, due to their partial homology to the canonical “Type A” T4SS. Peter J. Christie (U Texas, Houston, USA) presented the architecture of the T4SS of the conjugative plasmid F (Hu et al., 2019a), which shows little homology to that of R388. In both cases, a surprising 13-fold symmetry in the OMC was evident.
The stoichiometry of the inner membrane / cytoplasmic complexes (IMCC) received much focus. The reconstruction by Low et al (2014) proposed an inner membrane complex formed by two side-by-side hexameric barrels of the cytoplasmic ATPase VirB4; more recently, a five-barrel cytoplasmic structure was proposed for the Cag T4SS (Chang et al., 2018), although the level of resolution left open the question of the structure/s of these subcomplexes. The new fine structures of the Dot-Icm and F plasmid allowed detailed observation of the IMCC. Craig Roy reported a DotB-DotO hexameric complex, involved in substrate recruitment, which creates a cytoplasmic channel whose assembly opens up the T4SS, thus directing the translocation of substrates through the T4SS (Chetrit et al., 2018). A very similar structure is revealed in the case of the system associated with the conjugative plasmid F, with a 13-fold symmetry OMC linked to a 6-fold symmetry IMCC (Hu et al., 2019a). In addition, Bo Hu from University of Houston reported a refined cryo-ET structure of the Cag T4SS confirming the previously reported 14-fold symmetry at the OMC, but showing a clear 6-fold symmetry in the cytoplasmic complex (Hu et al., 2019b).
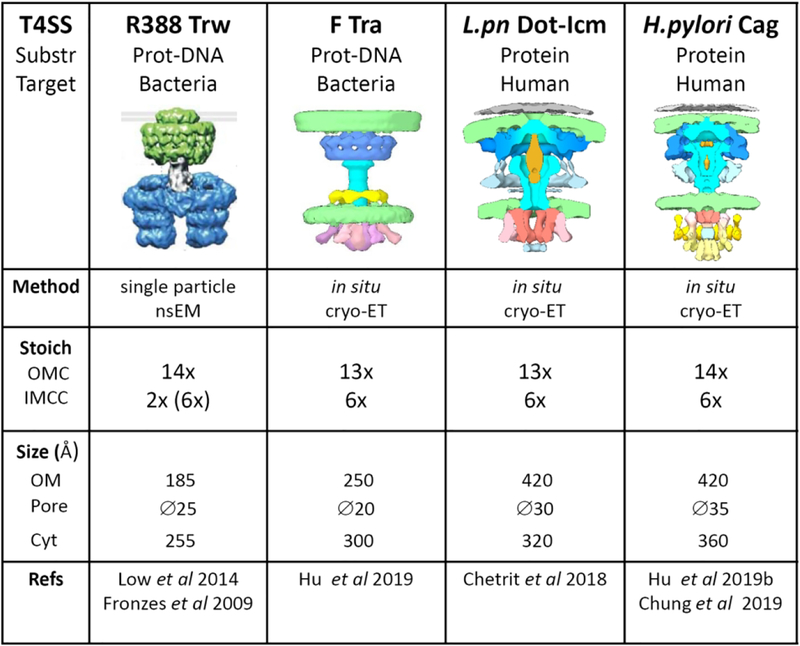
Figure 2 shows a comparison of the features of the different T4SS structures elucidated so far. Apart from the valuable insight into each particular T4SS, the set of results opens up the question as to whether there are significant structural variations among the T4SS, maybe adapted to their biological roles, and/or the variations are due to the differences in the technology used to solve the structures. The fact that both the 14-fold and 13-fold symmetry OMC were solved in different systems from independent labs argues for the co-existence of these alternative oligomeric structures in the T4SS world. The newly solved structures provide strong evidence for a 6-fold symmetry IMCC to which the 14-fold or 13-fol symmetric OMC has to engage. The apparent stoichiometric mismatches among subcomplexes could be in fact an important flexibility factor key to T4SS function. In this respect, it is interesting to note that the recently reported near-atomic resolution structure of the Cag T4SS (Chung et al., 2019) shows that the 14-fold symmetric outer membrane core complex connects to a newly described 17-fold symmetric periplasmic ring complex (PRC). This new structure illustrates the structural diversity among T4SS.
Figure 2.
Comparison of the 3D structure of T4SS from plasmid R388 (Low et al., 2014), plasmid F (Hu et al., 2019a), L pneumophila Dot-Icm (Chetrit et al., 2018), and H. pylori Cag (Hu et al., 2019b). Below each T4SS, the nature of the translocated substrate (Protein or protein-DNA complexes) and the target cell (bacteria or human) is indicated. The R388 T4SS image is from (Galan et al., 2018). The images for the F-Tra, Dot-Icm and Cag T4SS were kindly provided by Bo Hu (University of Texas, USA). The lanes below show the method for 3D structure elucidation, the stoichiometry of the subcomplexes, and the size of the structure (in Armstrongs) at the outer membrane (OM) side (the diameter of the pore is shown below) or the cytoplasmic (Cyt) side.
Another significant difference concerns the dimensions of the OMC. While the IMCC has a similar diameter in all the reported T4 structures (around 25–30 nm wide), the OMC is about 20 nm wide in the case of the R388, F, and Xanthomonas T4SS, while the OMC of Cag and Dot-Icm T4SS double this size. The obvious difference between these two types of T4SS is the target cell (prokaryotic or eukaryotic), raising the possibility that the extended OMC in the case of the H. pylori and L. pneumophila T4SS is designed to target the human cell.
Structural studies not only shed light on the architecture of mature, functional secretion machines. They can also illustrate the biogenesis pathway of the transmembrane complexes. Peter Christie reported various structures of the F T4SS, which may represent assembly intermediates or alternative biological states of the T4SS, such as F pili associated with distinct basal platforms. Bo Hu showed the Cag T4SS structures from mutants lacking the cytoplasmic ATPases, which do not affect the formation of the outer membrane complex, but suggest a pathway for the assembly of the inner membrane complex (Hu et al., 2019b). Joseph Vogel (Washington University, St Louis, USA) combined cryo-EM with immunofluorescence microscopy to address the assembly process of the L. pneumophila Dot-Icm T4SS, revealing that early-stage assembly process begins with the targeting of Dot/Icm components to the bacterial poles. Interestingly, polar targeting is mediated by two T4 components, DotU and IcmF, which have homologues in T6SS, raising the possibility that these elements may have been recruited by a T6SS (Ghosal et al., 2019).
Eric Cascales (CNRS, Marseille, France) reported the state of the art on T6SS structure and assembly. T6SS includes a contractile sheath that covers a hollow tube topped with a membrane-puncturing spike. The sheath can contract and inject the arrow loaded with effectors into the prey cell. The cryo-EM structure of the wedge complex of the enteroaggregative Escherichia coli T6SS and how the TssK wedge subunit anchors the baseplate to the trans-membrane complex were briefly presented at the workshop (Cherrak et al., 2018). The group has also dissected T6SS biogenesis using an original in vivo technique based on APEX2-dependent biotinylation to determine proximity partners of TssA. TssA, by stabilizing the baseplate and coordinating the polymerization of the tail plays a central role in the assembly of two T6SS subcomplexes. The results revealed a new partner, TagA, which holds the distal extremity of the sheath at the opposite site of the bacterial membrane (Santin et al., 2018). In fact, these authors have recently reported that cell width determines the length of the T6SS tail, since T6SS sheath polymerization is arrested upon contact with the opposite membrane, likely by the TagA stopper (Santin et al., 2019).
The intriguing relationships between T4SS and T6SS were also raised at the Workshop. It was already known that the conjugative pilus triggers T6SS activity in the recipient cell (Ho et al., 2013), so T6SS-containing bacteria are unlike recipients of conjugative plasmids. Mario Feldman (Washington University School of Medecine at St Louis, USA) built further into this relationship. Coexistence of T4 and T6SS in donor bacteria is functionally contradictory, since the former is intended to transfer DNA to recipient bacteria, while the latter kills potential recipients. In Acinetobacter baumanii, where most strains harbour a constitutively expressed T6SS, the solution is simple: large conjugative plasmids repress co-existing T6SS in order to allow their spread and the conjugation of other plasmids through their T4SS (Weber et al., 2015). In fact, they show that this repression is essential for conjugation to occur (Di Venanzio et al., 2019). The molecular components involved in these interactions remain to be determined.
Translocated substrates
The workshop devoted its attention also to different aspects of the substrates translocated by T3, T4 and T6SS, from their evolutionary origin to their recruitment and translocation by the secretion machineries, their detection and action in the target cell, and even their possible biotechnological applications. The varied nature of the substrates was represented in different talks, including the nucleoprotein complexes transferred by T4SS, the toxins targeted to other bacteria through T6SS, or the protein effectors injected into human cells by T3SS.
Christoph Dehio (University of Basel, Switzerland) addressed the evolution of effectors, using the example of the Bartonella VirB/D4 T4SS substrates, collectively known as Beps. Their group reported that Beps evolved from a single ancestral effector that emerged by fusion of a bacterial toxin-antitoxin module and a T4 secretion signal (Harms et al., 2017). This ancestral Bep became the primordial interkingdom effector when VirB/VirD4-T4SS was co-opted for host interaction. By parallel evolution, specific sets of Bep repertoires have been adapted to three different Bartonella lineages, in a remarkable example of host adaptation (Wagner et al., 2019a).
Substrate recruitment and translocation are especially intertwined in the case of T6SS. Luke Allsopp, from Alain Filloux lab (Imperial College London, UK), presented the fine characterization of the secretion of a DNAse toxin by the P. aeruginosa T6SS into competitor bacteria (Pissaridou et al., 2018). This evolved toxin, whose effector domain is fused to a structural component of the T6 puncturing device, was shown to fit exquisitely in the tip of the T6SS spike thanks to interactions by its PAAR domain. In addition, they showed that immunity proteins are highly specific for their cognate toxins, allowing interstrain competition. T6 effector genes can also be localized at the vicinity of genes encoding the phage tail like structure. Like this, Sophie Bleves (Aix-Marseille University, France) discovered a novel antibacterial phospholipase of P. aeruginosa whose secretion requires an adaptor protein for the targeting to the T6 spike and an immunity protein to protect from sister cell attack (Berni et al., 2019). The existence of toxin-antitoxin pairs was the also the basis for the search for new effectors associated with the P. aeruginosa H1-T6SS by David Albesa (CIC-Biogune, Spain) in collaboration with Alain Filloux and Julian Parkhill. They developed a methodology named TraDIS (Transposon directed insertion-site sequencing) and mutagenized in parallel strains with active/inactive T6SS. By comparing the number of transposon insertions in each gene, they identified putative immunity proteins (which are essential genes), allowing the search for adjacent toxins. Following this approach they discovered Tse8, an antibacterial toxin with an original mechanism of action, the inhibition of protein synthesis, and its cognate immunity protein Tsi8 (Nolan et al., 2019).
Access of the effectors to their host target requires different strategies depending on the pathogen. In the case of the Helicobacter pylori Cag T4SS, the effector oncoprotein CagA is secreted at the surface of the gastric cell and then translocated into the target cell. Steffen Backert (Friedrich-Alexander University Erlangen-Nuremberg, Germany) presented a novel basolateral T4 secretion model for CagA. While the gastric pathogen is exposed only to the apical surface of epithelial cells, the T4 needle-like pilus binds basolateral integrins to translocate CagA. The access to these basolateral receptors is allowed by the secretion of the HtrA serine protease that cleaves cell-to-cell junction factors as well as extracellular matrix proteins, disrupting the epithelial barrier and allowing transmigration of the pathogen (Backert et al., 2018). Exported HtrA proteases represent highly attractive targets for antibacterial treatment by inhibiting their proteolytic activity or for its use in vaccine development. In addition to integrins, H. pylori exploits at the basolateral surface the carcinoembryonic antigen-related cell adhesion molecules (CEACAM) receptors via the outer membrane adhesin HopQ (Moonens et al., 2018). This binding is required for CagA delivery into gastric epithelial cells. CEACAMs are also exploited by pathogenic bacteria such as Neisseria gonorrhoeae, N. meningitidis, and E. coli during mucosal colonization.
Bacterial secretion systems can target specifically a diversity of host cells and translocate very different cargos, making them ideal systems to customize as substrate delivery tools. Luis Angel Fernández (CNB, Spain) utilizes the ability of T3SS to translocate proteins into human cells for biotechnological purposes. His group has designed a non-pathogenic E. coli strain which produces a functional T3SS from enteropathogenic E. coli, that injects the desired proteins into human cells (Ruano-Gallego et al., 2015). They also showed that E. coli can be targeted to specific cell types, and in particular tumor cells, by expressing synthetic adhesins consisting of nanobodies (Pinero-Lambea et al., 2015). By joining both approaches, he presented encouraging preliminary results of the engineered E. coli strain injecting customized substrates specifically to tumor cells. Matxalen Llosa (Universidad de Cantabria, Spain) aims to customize T4SS to deliver protein-DNA complexes for genomic editing purposes. T4SS involved in effector translocation can also mediate DNA delivery into human cells, when combined with a plasmid conjugation machinery (Llosa et al., 2012). The translocated DNA is covalently attached to the conjugative relaxase. They showed that certain relaxases promote random integration of the incoming DNA into the recipient human genome (Gonzalez-Prieto et al., 2017). Through protein engineering, relaxase chimeric proteins can acquire new functions, including the ability to integrate the attached DNA (Agundez et al., 2018). Preliminary results suggest that a site-specific nuclease fused to the relaxase can be translocated through the T4SS in order to attain site-specificity in the integration of T4SS-delivered DNA.
Francisco Ramos-Morales (Universidad de Sevilla, Spain) presented a different example of possible biotechnological/biomedical exploitation of the secreted substrates, using Salmonella T3SS effectors as carriers in a live vaccine against P. aeruginosa. A fusion between the S. enterica effector SseJ and the P.aeruginosa antigen PcrV was translocated by Salmonella into host cells in vitro, eliciting the generation of specific antibodies in mice, which showed enhanced survival (Aguilera-Herce et al., 2019).
Effector-mediated subversion of host cells
Although the T3, T4 and T6SS are evolutionary and structurally quite diverse, many of the effector proteins that they deliver target common core eukaryotic functions. Consequently, the study of these effectors share common experimental approaches. One of the main limitations to study effector function in the host cell is their visualization after translocation. Amy E. Palmer (University of Colorado, Boulder, USA) reviewed the techniques they have developed to monitor T3SS effector injection in human cells, which they also used to visualize Listeria protein secretion in infection, and could ideally extend to imaging of T4 and T6 effectors. Effectors tagged with a fluorescent peptide allow following the secretion process from the intracellular bacteria by the loss of fluorescence. Another approach consists on an adaptation of the split-GFP assay, fusing the FP11 GFP domain to the effector and expressing the rest of the GFP protein (FP1–10) in the host cell (Young et al., 2017, Batan et al., 2018).
Ulla Bonas (Martin Luther University Halle-Wittenberg, Halle, Germany) discussed how bacterial plant pathogens subvert their host by T3SS-mediated effector translocation. Interestingly, effectors from human and plant pathogens share common host targets. The repertoire of T3 effectors of Xanthomonas campestris pv. vesicatoria comprises more than 35 proteins, among them the AvrBs3 transcription factor, cell death inducers, plant immunity suppressors (Adlung et al., 2017), or the XopL ubiquitine ligase. The characterization of the latter reveals and unprecedented role of microtubules in stromule extension and dynamics in Nicotiana benthamiana (Erickson et al., 2018).
Christoph Dehio addressed the functional versatility of the Bartonella VirB/D4 T4SS Bep substrates. The effectors are composed of only three basic domain types that are highly versatile in function, allowing them to adapt their original function to host cell subversion, by post-translation modification of target proteins, e.g. AMPylation by FIC domains, interfering with signaling pathways mimicking eukaryotic protein pY motifs, etc (Wagner et al., 2019b). Another fine example of host cell subversion by a T4SS effector was reported by Maria Lucas (University of Cantabria, Spain), who showed that the effector RavN of the L. pneumophila Dot/Icm T4SS mimics host cell E3 ligases to exploit the ubiquitylation pathway (Lin et al., 2018). Based on the structural information obtained from this effector, they propose and validate other four effectors with E3 ligase activity, underscoring the importance of manipulation of the host ubiquitylation pathway for Legionella infection.
Since the main function of T6SS is to target other bacteria, there are few examples of T6 anti-eukaryotic effectors (Hachani et al., 2016). Some of them provide unique cases of trans-kingdom toxins, which allow the search for antibacterial compounds that may limit bacterial competition for the niche colonization and further host subversion. While toxins are usually directed against eukaryotic cells (like AB toxins) or against rival bacteria (like bacteriocins), three T6 phospholipases of P. aeruginosa were previously shown to affect both, prokaryotic and eukaryotic target cells (Jiang et al., 2014, Jiang et al., 2016). These T6 effectors disrupt membrane integrity of their prey bacteria and promote internalization of P. aeruginosa into epithelial cells and autophagy. Sophie Bleves reported a fourth trans-kingdom effector of a novel type, called VgrG2b, which acts as an antibacterial metallopeptidase implicated in bacterial competition (Berni et al., 2019). VgrG2b was previously shown to facilitate the uptake of P. aeruginosa into non-phagocytic cells by recruiting microtubules through an interaction with the gamma-tubulin ring complex, the microtubule nucleating centre (Sana et al., 2015). As in the case of the Xanthomonas T3 effector XopL, the precise role on host microtubules remains to be evaluated in future studies. This represents a novel internalization strategy, since most pathogens manipulate the actin cytoskeleton to enter host cells. One such example was described by Miguel Valvano (University of Belfast, UK), who presented a T6 effector that targets the host cytoskeleton to survive in macrophages: the protease TecA of Burkholderia cenocepacia (Aubert et al., 2016) inactivates Rho GTPases by deamidation, thus inducing actin disruption and caspase-1 inflammation through activation of the Pyrin inflammasome. Furthermore, his lab addressed the role of T2, T4 and T6SS in the interactions between B. cenocepacia and macrophages by constructing mutant strains lacking each secretion system. They appear to increase pathogenicity in macrophages, while limiting the spread of infection.
Finally, three talks addressed the complex interplay between enteropathogens and the host immune response. Jorge E. Galán (Yale University, New Haven, USA) provided the demonstration that three well-characterized T3 effectors of Salmonella Typhimurium, SopE, SopE2 and SopB, trigger intestinal inflammation bypassing innate immune receptors. Instead, they do so through the activation of Cdc42 and Pak1. These effectors conserve signalling pathways that operate downstream from canonical innate immune receptors and involve Tak1 and TRAF6 (Sun et al., 2018). These findings illustrate the unique balance that emerges from the host/pathogen co-evolution, in that pathogen-initiated responses that help its replication are also important to prevent pathogen spread to deeper tissues. Furthermore, the mechanisms describe here could help develop anti-pathogen therapeutic strategies by targeting specific host-signaling pathways. Elisabeth L. Hartland (Monash University, Clayton, Australia) reported on the suppression of host innate immunity pathways in enteropathogenic E. coli and Shigella by T3SS effectors. EspL defines a family of cysteine protease effectors which directly target RHIM-dependent inflammatory and necroptotic signalling pathways (Pearson et al., 2017). Using the proteomic approach ProtoMap (Fuhrman-Luck et al., 2017), they have defined the host interacting partners of the Shigella EspL homologues, and found that they target components of the type I interferon signalling pathway. This suggests that Shigella activity blocks the host type I interferon response and that type I interferon induced proteins mediate host defense against Shigella infection. Feng Shao (National Institute of Biological Sciences, Beijing, China) reported the interaction of the T3 effector IpaH9.8 of Shigella flexneri with the host immune response. IpaH9.8 suppresses host defense through ubiquitination and degradation of guanylate-binding proteins (Li et al., 2017). Lack of IpaH9.8 or its binding to GBPs provoked translocation of GBPs such as hGBP1 and mGBP2 to intracellular S. flexneri, where they inhibited bacterial replication, highlighting the functional importance of GBPs in antibacterial defenses. The recently reported structure of the IpaH9.8:GBP interaction reveals clues on how IpaH proteins discriminate among different GBP targets through its LRR domain (Ji et al., 2019). This same group recently reported that the Salmonella SopF effector specifically blocks xenophagy by blocking V-ATPase recruitment of ATG16L1 onto bacteria-containing vacuole, without affecting canonical autophagy (Xu et al., 2019). This is just the last example of how the study of bacterial subversion of the human cell serves to elucidate basic aspects of cell biology.
Conclusions and prospects
The Workshop gathered an excellent panel of experts addressing different aspects of the biology of T3, T4 and T6SS and their involvement in bacterial virulence. The data presented reflected the exciting scientific moment in the field, and underscored the fact that these evolutionary distant nanomachines converge functionally in the host human cell, where they elicit pathogenic effects. This situation calls for a joint action among experts in these fields to find common anti-virulence strategies. In this context, the workshop constituted an excellent forum for merging scientists from different backgrounds, which undoubtedly leads to fruitful synergies. The authors hope similar meetings continue to take place in the future.
In the workshop, the reports on new 3D structures of the different secretion machineries highlighted the importance of solving structures that are relevant in vivo. The differences observed in the structures of the T3SS when it is embedded in the membrane, or the different IMCC observed in T4SS structures solved in situ compared to single particle reconstruction by cryo-EM (Fig 2), emphasizes the importance of the physical context in biological structures. It is evident that structural biology of secretion systems is moving towards the in situ characterization of the nanomachines (Oikonomou et al., 2019). Still, cryo-ET needs to reach a better level of resolution to provide full answers to the structure-function relations.
Elucidation of the structures is essential to understand the secretion mechanism, and from a practical point of view, to identify potential targets for blocking secretion, and thus, microbial virulence. Such anti-virulence molecules would be an alternative to classical bacteria-killing antibiotics. In addition, bacterial secretion systems offer numerous possibilities to develop biotechnological and biomedical applications, and some of them were illustrated here, such as customization of DNA- and protein-delivery systems which access in vivo specific target cells for the purpose of human cell modification, or even the construction of live vaccines based on secreted effectors.
Another highlight from the workshop is the increasing evidence in support of convergent strategies for host cell subversion, regardless of the type of secretion system involved in the translocation of the effectors. Different effectors, translocated by different nanomachines, often end up interfering with the same host functions, key for infection, survival and propagation of pathogens in the host cell, as illustrated in Fig 1. Typical examples are the manipulation of host cytoskeleton, subversion of host immune response, interference with host signaling pathways, or alteration of vesicular trafficking. This convergence implies that the identification of a novel host target for a TxSS effector can pave the way to find the intracellular target of another, even if they are not transported by the same machinery. Another important consequence is the possibility of finding common effector motifs that could be targeted for broad-spectrum anti-virulence strategies, which could block the action of effectors secreted by different systems. Last, but not least, the study of host cell subversion through bacterial secreted effectors keeps providing us with basic understanding of the human cell physiology.
Acknowledgements
We wish to thank the UNIA staff, and in particular Joaquin Torreblanca, for organization of the Workshop. We are grateful to the speakers for presenting unpublished data and allowing their citation. We are especially indebted to Bo Hu for kindly providing the Tra, Dot and Cag images shown in Fig 2.
Work in SB lab is supported by recurrent funding from the CNRS and Aix-Marseille University and by a grant from the Excellence Initiative of Aix-Marseille University-A*Midex, a French “Investissements d’Avenir” program (“Emergence & Innovation” AM-AAP-EI-17-139-170301-10.31-BLEVES-HLS). Work in JEG lab is supported by National Institutes of Health grant AI030492 to JEG. Work in ML lab is funded by grants BIO2017-87190-R from the Spanish Ministerio de Ciencia e Innovación, and IDEASLLO17 from the Fundación Científica Asociación Española Contra el Cáncer (AECC) to ML. The authors have no conflicts of interest to declare.
Abbreviations:
- cryo-EM
cryo-electron microscopy
- cryo-ET
cryo-electron tomography
- IMCC
inner membrane – cytoplasmic complex
- nsEM
negative staining electron microscopy
- OMC
outer membrane complex
- T3/T4/T6SS
Type III / Type IV / Type VI secretion system(s)
Footnotes
Conflict of Interests
The authors declare no competing financial interests.
References
- Adlung N and Bonas U (2017). Dissecting virulence function from recognition: cell death suppression in Nicotiana benthamiana by XopQ/HopQ1-family effectors relies on EDS1-dependent immunity. Plant J 91, 430–442. [DOI] [PubMed] [Google Scholar]
- Aguilera-Herce J, Garcia-Quintanilla M, Romero-Flores R, McConnell MJ and Ramos-Morales F (2019). A Live Salmonella Vaccine Delivering PcrV through the Type III Secretion System Protects against Pseudomonas aeruginosa. mSphere 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agundez L, Zarate-Perez F, Meier AF, Bardelli M, Llosa M, Escalante CR, et al. (2018). Exchange of functional domains between a bacterial conjugative relaxase and the integrase of the human adeno-associated virus. PLoS One 13, e0200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam B, Wang W, Arshad MI, Khurshid M, Muzammil S, Rasool MH, et al. (2018). Antibiotic resistance: a rundown of a global crisis. Infect Drug Resist 11, 1645–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert DF, Xu H, Yang J, Shi X, Gao W, Li L, et al. (2016). A Burkholderia Type VI Effector Deamidates Rho GTPases to Activate the Pyrin Inflammasome and Trigger Inflammation. Cell Host Microbe 19, 664–674. [DOI] [PubMed] [Google Scholar]
- Backert S, Bernegger S, Skorko-Glonek J and Wessler S (2018). Extracellular HtrA serine proteases: An emerging new strategy in bacterial pathogenesis. Cell Microbiol 20, e12845. [DOI] [PubMed] [Google Scholar]
- Batan D, Braselmann E, Minson M, Nguyen DMT, Cossart P and Palmer AE (2018). A Multicolor Split-Fluorescent Protein Approach to Visualize Listeria Protein Secretion in Infection. Biophys J 115, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berni B, Soscia C, Djermoun S, Ize B and Bleves S (2019). A Type VI Secretion System Trans-Kingdom Effector Is Required for the Delivery of a Novel Antibacterial Toxin in Pseudomonas aeruginosa. Front Microbiol 10, 1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya S, Baidya AK, Pal RR, Mamou G, Gatt YE, Margalit H, et al. (2019). A Ubiquitous Platform for Bacterial Nanotube Biogenesis. Cell Rep 27, 334–342 e310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthurst S (2019). The Type VI secretion system: a versatile bacterial weapon. Microbiology 165, 503–515. [DOI] [PubMed] [Google Scholar]
- Chang YW, Shaffer CL, Rettberg LA, Ghosal D and Jensen GJ (2018). In Vivo Structures of the Helicobacter pylori cag Type IV Secretion System. Cell Rep 23, 673–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrak Y, Flaugnatti N, Durand E, Journet L and Cascales E (2019). Structure and Activity of the Type VI Secretion System. Microbiol Spectr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrak Y, Rapisarda C, Pellarin R, Bouvier G, Bardiaux B, Allain F, et al. (2018). Biogenesis and structure of a type VI secretion baseplate. Nat Microbiol 3, 1404–1416. [DOI] [PubMed] [Google Scholar]
- Chetrit D, Hu B, Christie PJ, Roy CR and Liu J (2018). A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat Microbiol 3, 678–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ (2001). Type IV secretion: intercellular transfer of macromolecules by systems ancestrally related to conjugation machines. Mol Microbiol 40, 294–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung JM, Sheedlo MJ, Campbell AM, Sawhney N, Frick-Cheng AE, Lacy DB, et al. (2019). Structure of the Helicobacter pylori Cag type IV secretion system. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Venanzio G, Moon KH, Weber BS, Lopez J, Ly PM, Potter RF, et al. (2019). Multidrug-resistant plasmids repress chromosomally encoded T6SS to enable their dissemination. Proc Natl Acad Sci U S A 116, 1378–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson JL, Adlung N, Lampe C, Bonas U and Schattat MH (2018). The Xanthomonas effector XopL uncovers the role of microtubules in stromule extension and dynamics in Nicotiana benthamiana. Plant J 93, 856–870. [DOI] [PubMed] [Google Scholar]
- Fasciano AC, Shaban L and Mecsas J (2019). Promises and Challenges of the Type Three Secretion System Injectisome as an Antivirulence Target. EcoSal Plus 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick-Cheng AE, Pyburn TM, Voss BJ, McDonald WH, Ohi MD and Cover TL (2016). Molecular and Structural Analysis of the Helicobacter pylori cag Type IV Secretion System Core Complex. MBio 7, e02001–02015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuhrman-Luck RA, Silva LM, Hastie ML, Gorman JJ and Clements JA (2017). Determining Protease Substrates Within a Complex Protein Background Using the PROtein TOpography and Migration Analysis Platform (PROTOMAP). Methods Mol Biol 1574, 145–170. [DOI] [PubMed] [Google Scholar]
- Galan JE, Lara-Tejero M, Marlovits TC and Wagner S (2014). Bacterial type III secretion systems: specialized nanomachines for protein delivery into target cells. Annual review of microbiology 68, 415–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE and Waksman G (2018). Protein-Injection Machines in Bacteria. Cell 172, 1306–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D, Chang YW, Jeong KC, Vogel JP and Jensen GJ (2017). In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep 18, 726–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosal D, Jeong KC, Chang YW, Gyore J, Teng L, Gardner A, et al. (2019). Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat Microbiol 4, 1173–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Prieto C, Gabriel R, Dehio C, Schmidt M and Llosa M (2017). The Conjugative Relaxase TrwC Promotes Integration of Foreign DNA in the Human Genome. Appl Environ Microbiol 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf FE, Palm M, Warringer J and Farewell A (2019). Inhibiting conjugation as a tool in the fight against antibiotic resistance. Drug Dev Res 80, 19–23. [DOI] [PubMed] [Google Scholar]
- Green ER and Mecsas J (2016). Bacterial Secretion Systems: An Overview. Microbiol Spectr 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachani A, Wood TE and Filloux A (2016). Type VI secretion and anti-host effectors. Curr Opin Microbiol 29, 81–93. [DOI] [PubMed] [Google Scholar]
- Harms A, Liesch M, Korner J, Quebatte M, Engel P and Dehio C (2017). A bacterial toxin-antitoxin module is the origin of inter-bacterial and inter-kingdom effectors of Bartonella. PLoS Genet 13, e1007077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayek I, Berens C and Luhrmann A (2019). Modulation of host cell metabolism by T4SS-encoding intracellular pathogens. Curr Opin Microbiol 47, 59–65. [DOI] [PubMed] [Google Scholar]
- Ho BT, Basler M and Mekalanos JJ (2013). Type 6 secretion system-mediated immunity to type 4 secretion system-mediated gene transfer. Science 342, 250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood RD, Singh P, Hsu F, Guvener T, Carl MA, Trinidad RR, et al. (2010). A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Khara P and Christie PJ (2019a). Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli. Proc Natl Acad Sci U S A 116, 14222–14227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Khara P, Song L, Lin AS, Frick-Cheng AE, Harvey ML, et al. (2019b). In Situ Molecular Architecture of the Helicobacter pylori Cag Type IV Secretion System. MBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Lara-Tejero M, Kong Q, Galan JE and Liu J (2017). In Situ Molecular Architecture of the Salmonella Type III Secretion Machine. Cell 168, 1065–1074 e1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji C, Du S, Li P, Zhu Q, Yang X, Long C, et al. (2019). Structural mechanism for guanylate-binding proteins (GBPs) targeting by the Shigella E3 ligase IpaH9.8. PLoS Pathog 15, e1007876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Wang X, Wang B, Chen L, Zhao Z, Waterfield NR, et al. (2016). The Pseudomonas aeruginosa Type VI Secretion PGAP1-like Effector Induces Host Autophagy by Activating Endoplasmic Reticulum Stress. Cell Rep 16, 1502–1509. [DOI] [PubMed] [Google Scholar]
- Jiang F, Waterfield NR, Yang J, Yang G and Jin Q (2014). A Pseudomonas aeruginosa type VI secretion phospholipase D effector targets both prokaryotic and eukaryotic cells. Cell Host Microbe 15, 600–610. [DOI] [PubMed] [Google Scholar]
- Lara-Tejero M and Galan JE (2019). The Injectisome, a Complex Nanomachine for Protein Injection into Mammalian Cells. EcoSal Plus 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiman PG, Basler M, Ramagopal UA, Bonanno JB, Sauder JM, Pukatzki S, et al. (2009). Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc Natl Acad Sci U S A 106, 4154–4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Jiang W, Yu Q, Liu W, Zhou P, Li J, et al. (2017). Ubiquitination and degradation of GBPs by a Shigella effector to suppress host defence. Nature 551, 378–383. [DOI] [PubMed] [Google Scholar]
- Li YG and Christie PJ (2018). The Agrobacterium VirB/VirD4 T4SS: Mechanism and Architecture Defined Through In Vivo Mutagenesis and Chimeric Systems. Curr Top Microbiol Immunol 418, 233–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YG, Hu B and Christie PJ (2019). Biological and Structural Diversity of Type IV Secretion Systems. Microbiol Spectr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH, Lucas M, Evans TR, Abascal-Palacios G, Doms AG, Beauchene NA, et al. (2018). RavN is a member of a previously unrecognized group of Legionella pneumophila E3 ubiquitin ligases. PLoS Pathog 14, e1006897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low HH, Gubellini F, Rivera-Calzada A, Braun N, Connery S, Dujeancourt A, et al. (2014). Structure of a type IV secretion system. Nature 508, 550–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M, Schroder G and Dehio C (2012). New perspectives into bacterial DNA transfer to human cells. Trends in microbiology 20, 355–359. [DOI] [PubMed] [Google Scholar]
- Moonens K, Hamway Y, Neddermann M, Reschke M, Tegtmeyer N, Kruse T, et al. (2018). Helicobacter pylori adhesin HopQ disrupts trans dimerization in human CEACAMs. EMBO J 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan LM, Cain AK, Manoli E, Sainz-Polo MA, Dougan G, Mavridou DAE, et al. (2019). Discovery of a Pseudomonas aeruginosa type VI secretion system toxin targeting bacterial protein synthesis using a global genomics approach. [Preprint] Available from : 10.1101/733030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou CM and Jensen GJ (2019). Electron Cryotomography of Bacterial Secretion Systems. Microbiol Spectr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson JS, Giogha C, Muhlen S, Nachbur U, Pham CL, Zhang Y, et al. (2017). EspL is a bacterial cysteine protease effector that cleaves RHIM proteins to block necroptosis and inflammation. Nat Microbiol 2, 16258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personnic N, Barlocher K, Finsel I and Hilbi H (2016). Subversion of Retrograde Trafficking by Translocated Pathogen Effectors. Trends Microbiol 24, 450–462. [DOI] [PubMed] [Google Scholar]
- Pinero-Lambea C, Bodelon G, Fernandez-Perianez R, Cuesta AM, Alvarez-Vallina L and Fernandez LA (2015). Programming controlled adhesion of E. coli to target surfaces, cells, and tumors with synthetic adhesins. ACS Synth Biol 4, 463–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisano A, Albano F, Vecchio E, Renna M, Scala G, Quinto I and Fiume G (2018). Revisiting Bacterial Ubiquitin Ligase Effectors: Weapons for Host Exploitation. Int J Mol Sci 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pissaridou P, Allsopp LP, Wettstadt S, Howard SA, Mavridou DAI and Filloux A (2018). The Pseudomonas aeruginosa T6SS-VgrG1b spike is topped by a PAAR protein eliciting DNA damage to bacterial competitors. Proc Natl Acad Sci U S A 115, 12519–12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratner D, Orning MP and Lien E (2017). Bacterial secretion systems and regulation of inflammasome activation. J Leukoc Biol 101, 165–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano-Gallego D, Alvarez B and Fernandez LA (2015). Engineering the Controlled Assembly of Filamentous Injectisomes in E. coli K-12 for Protein Translocation into Mammalian Cells. ACS Synth Biol 4, 1030–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sana TG, Baumann C, Merdes A, Soscia C, Rattei T, Hachani A, et al. (2015). Internalization of Pseudomonas aeruginosa Strain PAO1 into Epithelial Cells Is Promoted by Interaction of a T6SS Effector with the Microtubule Network. MBio 6, e00712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santin YG, Doan T, Journet L and Cascales E (2019). Cell Width Dictates Type VI Secretion Tail Length. Curr Biol 29, 3707–3713 e3703. [DOI] [PubMed] [Google Scholar]
- Santin YG, Doan T, Lebrun R, Espinosa L, Journet L and Cascales E (2018). In vivo TssA proximity labelling during type VI secretion biogenesis reveals TagA as a protein that stops and holds the sheath. Nat Microbiol 3, 1304–1313. [DOI] [PubMed] [Google Scholar]
- Sgro GG, Costa TRD, Cenens W, Souza DP, Cassago A, Coutinho de Oliveira L, et al. (2018). Cryo-EM structure of the bacteria-killing type IV secretion system core complex from Xanthomonas citri. Nat Microbiol 3, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Kamanova J, Lara-Tejero M and Galan JE (2018). Salmonella stimulates pro-inflammatory signalling through p21-activated kinases bypassing innate immune receptors. Nat Microbiol 3, 1122–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai AY, English BC and Tsolis RM (2019). Hostile Takeover: Hijacking of Endoplasmic Reticulum Function by T4SS and T3SS Effectors Creates a Niche for Intracellular Pathogens. Microbiol Spectr 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A and Dehio C (2019a). Role of distinct type-IV-secretion systems and secreted effector sets in host adaptation by pathogenic Bartonella species. Cell Microbiol 21, e13004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A, Tittes C and Dehio C (2019b). Versatility of the BID Domain: Conserved Function as Type-IV-Secretion-Signal and Secondarily Evolved Effector Functions Within Bartonella-Infected Host Cells. Front Microbiol 10, 921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner S, Konigsmaier L, Lara-Tejero M, Lefebre M, Marlovits TC and Galan JE (2010). Organization and coordinated assembly of the type III secretion export apparatus. Proc Natl Acad Sci U S A 107, 17745–17750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Stan GV and Polizzi KM (2017). Intracellular delivery of biologic therapeutics by bacterial secretion systems. Expert reviews in molecular medicine 19, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber BS, Ly PM, Irwin JN, Pukatzki S and Feldman MF (2015). A multidrug resistance plasmid contains the molecular switch for type VI secretion in Acinetobacter baumannii. Proc Natl Acad Sci U S A 112, 9442–9447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Zhou P, Cheng S, Lu Q, Nowak K, Hopp AK, et al. (2019). A Bacterial Effector Reveals the V-ATPase-ATG16L1 Axis that Initiates Xenophagy. Cell 178, 552–566 e520. [DOI] [PubMed] [Google Scholar]
- Young AM, Minson M, McQuate SE and Palmer AE (2017). Optimized Fluorescence Complementation Platform for Visualizing Salmonella Effector Proteins Reveals Distinctly Different Intracellular Niches in Different Cell Types. ACS Infect Dis 3, 575–584. [DOI] [PMC free article] [PubMed] [Google Scholar]