Abstract
Insect effectors are mainly secreted by salivary glands, modulate plant physiology and favor the establishment and transmission of pathogens. Feeding is the principal vehicle of transmission of Candidatus Liberibacter asiaticus (Ca. Las) by the Asian citrus psyllid (ACP), Diaphorina citri. This study aimed to predict putative ACP effectors that may act on the Huanglongbing (HLB) pathosystem. Bioinformatics analysis led to the identification of 131 candidate effectors. Gene expression investigations were performed to select genes that were overexpressed in the ACP head and modulated by Ca. Las. To evaluate the actions of candidate effectors on D. citri feeding, six effectors were selected for gene silencing bioassays. Double-stranded RNAs (dsRNAs) of the target genes were delivered to D. citri adults via artificial diets for five days. RNAi silencing caused a reduction in the ACP lifespan and decreased the salivary sheath size and honeydew production. Moreover, after dsRNA delivery of the target genes using artificial diet, the feeding behaviors of the insects were evaluated on young leaves from citrus seedlings. These analyses proved that knockdown of D. citri effectors also interfered with ACP feeding abilities in planta, causing a decrease in honeydew production and reducing ACP survival. Electrical penetration graph (EPG) analysis confirmed the actions of the effectors on D. citri feeding behaviors. These results indicate that gene silencing of D. citri effectors may cause changes in D. citri feeding behaviors and could potentially be used for ACP control.
Subject terms: Entomology, RNAi
Introduction
Insect pests are one of the main factors that reduce agricultural plant productivity. Global losses caused by these animals reach 220 billion dollars annually1. In addition to the damage caused by their actions, problems caused by insects can be increased by the transmission of several plant pathogens2.
Asian citrus psyllid (ACP), Diaphorina citri Kuwayama (Hemiptera: Liviidae), stands out as the most important agricultural pest among citrus crops3. ACP is a vector for the bacteria Candidatus Liberibacter asiaticus (Ca. Las), the causal agent of Huanglongbing (HLB). This disease affects all commercial citrus varieties by promoting plant decline and reducing fruit quality4,5. In the last fifteen years, HLB has spread quickly across the American continent and the citrus production in the USA declined 58%4. In Brazil, 34 million citrus trees have been removed since the first detection of HLB in 20046.
So far, no efficient control for HLB has been found. Disease management consists of producing seedlings using Ca Las-free nursery stock, removal of infected trees and insecticide applications for vector control7,8. However, the extensive application of insecticides could select for ACP-resistant populations9–11.
Diaphorina citri acquire Ca. Las via a circulative-persistent manner during feeding12,13. The efficiency of Ca. Las transmission is affected by the duration of phloem ingestion by ACP14. Studies have been developed to understand the interactions of plants and ACP15–17. Nevertheless, this complex process remains unclear.
Additionally, salivary secretions have been described as important for insect-plant interactions18–21. These secretions may contain effector molecules that alter host plant physiology, activating or suppressing defense responses, consequently inhibiting or promoting plant infestation by the insects22,23. Insect effectors may also interfere with the transmission of microorganisms and plant-pathogen interactions as well24–26.
Many insect effectors have been described over the years as from saliva or salivary gland proteomic and transcriptomic data27–31. To unveil the actions of effector proteins on insect-hosts interactions, RNA interference (RNAi) has been applied. Knockdown of salivary effectors from Nephotettix cincticeps (Uhler, 1896) (Hemiptera: Cicadellidae), for example, reduced rice phloem ingestion by this insect18. Silencing of salivary mucin-like protein decreased the feeding performance and caused Nilaparvata lugens (Stål, 1854) (Hemiptera: Delphacidae) mortality through interactions with rice32. RNAi targeting whitefly, Bemisia tabaci (Genn.) (Hemiptera: Aleyrodidae), salivary laccase decreased the survival rates of this insect in tomato plants19.
Despite the existence of the ACP saliva proteome33, ACP effectors or salivary molecules that interact with host plants have not been identified until now34. In the present work, we predict secreted D. citri effectors using bioinformatics tools. Gene silencing through dsRNAs combined with an EPG technique was performed to verify the actions of the effectors on D. citri feeding behaviors.
Results
Candidate effector prediction
To generate a list of D. citri putative effectors, we used Myzus persicae Sulzer (Hemiptera: Aphididae) bioinformatics approach as a model for effector prediction25. With the intent of obtaining an accurate effectoromics dataset, proteins sequences used in these analyses were obtained from the following two distinct transcriptome and genomic sources: a transcriptome predicted from Arizona University studies35 and genomic sequences from the HLB consortium (citrusgreening.org).
Considering that insect effectors are secreted through the classical eukaryotic endoplasmic reticulum (ER)-Golgi pathway through salivary glands, the initial predicted D. citri secretome was composed of proteins presenting signal peptides and the absence of transmembrane domains. SignalP analysis allowed the identification of 2,099 proteins with signal peptides. Furthermore, screening using the TMHMM, Phorbius and GPI-SOM tools identified 670 proteins that contained transmembrane domains or GPI-anchor signals, and those proteins were removed from further analysis. Thereafter, the ACP secretome was composed of 1,443 proteins.
It has been reported that insect effectors are mainly species- or genus-specific36. Thus, we predicted conserved domains and identified characterized proteins through multiple alignments against public protein databases (Pfam and Uniprot). All proteins that showed ordinary domains were excluded. To select proteins that are only present in salivary glands, we also performed tBlast-X against the arthropod gut and salivary ESTs, removing proteins that may be secreted in the ACP gut. Thus, were identified 131 candidate effectors (Table S1) composed of 78% exclusively D. citri proteins and 22% showed similarity with arthropod saliva proteins (Table S2). Interestingly, many of the sequences showed intrinsically disordered regions (IDRs) (Table S1). These regions are abundant in phytopathogen effector proteins, interfere with protein secondary structure and allow protein folding in a stimulus-dependent manner37. The presence of enriched IDR proteins in the D. citri effectorome suggests a positive evolutionary selection of these proteins and potential functions through host interactions. Additionally, the subcellular localization of the predicted D. citri effectors suggests that the majority of the identified proteins act on the extracellular space (Table S1; Fig. S1). However, these proteins may also act on important host organelles such as the nucleus, chloroplast and mitochondrion (Table S1; Fig. S1).
Selection of effectors candidates of D. citri by RT-qPCR
After the bioinformatic predictions, the screened candidate effectors were analyzed for their gene expression. Twelve putative effectors were evaluated (Tables 1 and S3) to identify genes that were expressed in different life stages, upregulated in the ACP head and showed expression modulated by Ca. Las.
Table 1.
The twelve selected D. citri effectors for gene expression analysis.
| Effector | ID | Characterization | Size (aa) | Subcellular localization |
|---|---|---|---|---|
| DCEF08 | XP_008479196.1 | WD repeat-containing protein 92 | 191 | Extracellular space |
| DCEF10 | XP_008482426.1 | uncharacterized protein LOC103519088 | 228 | Mitochondrion/Chloroplast |
| DCEF11 | XP_026681229.1 | embryonal Fyn-associated substrate-like | 396 | Mitochondrion/Chloroplast |
| DCEF19 | DcWN_006647 | uncharacterized protein | 165 | Extracellular space |
| DCEF22 | DcWN_010436 | uncharacterized protein | 128 | Extracellular space/Nucleus |
| DCEF23 | DcWN_013243 | uncharacterized protein | 94 | Extracellular space/Nucleus |
| DCEF26 | DcWN_027964 | uncharacterized protein | 252 | Mitochondrion |
| DCEF27 | DcWN_028357 | uncharacterized protein | 259 | Mitochondrion/Chloroplast |
| DCEF28 | XP_026685929.1 | uncharacterized LOC113471181 | 567 | Nucleus |
| DCEF32 | XP_008477481.1 | uncharacterized protein LOC103514387 | 209 | Golgi apparatus |
| DCEF33 | XP_008475409.1 | uncharacterized protein LOC103512422 | 197 | Extracellular |
| DCEF35 | XP_008468032.1 | uncharacterized protein | 142 | Extracellular |
First, the mRNA abundance of the candidate effector genes was evaluated in two different ACP life stages (nymph and adult). For both analyzed life stages, similar mRNA patterns were observed for the most candidates of the effectors (Fig. S2). DCEF22, DCEF28 and DCEF33 showed elevated mRNA levels in the nymph stage compared with the adult phase (Fig. S2).
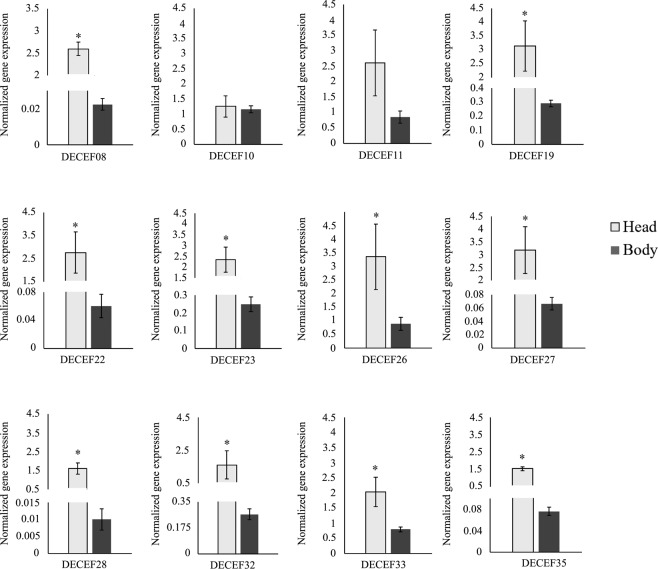
Our goal was to select effectors that are present in the D. citri salivary gland or could be secreted by saliva. To achieve this aim, we evaluated the gene expression of candidate effectors from different body parts (head and body). The mRNA abundance of ten of twelve candidate effector genes was higher in the head than in the ACP body (Fig. 1). Candidate effectors that were not majorly expressed in the ACP head (DCEF10 and DCEF11) were excluded from further analysis.
Figure 1.
Relative gene expression of the 12 D. citri candidate effectors in the head and body. These data consist of normalized target mRNA levels to the mRNA abundance of the housekeeping genes S13 and S20. The results are the mean of three biological replicates per set. *corresponds to statistically significant differences amongst the treatments (t-test, P-value < 0.05).
Furthermore, Ca. Las regulation of the gene expression of the ten candidate effectors was evaluated in nymphs and adults. Except for DCEF22, all the selected genes were significantly modulated by Ca. Las infection conditions in at least one of the analyzed life stages (Fig. 2). DCEF08 and DCEF23 were overexpressed in Ca. Las-infected insects compared with healthy insects in both the nymph and adult stages. The presence of Ca. Las contributes to increased DCEF19, DCEF26 and DCEF27 mRNA levels in ACP adults. DCEF33 was downregulated in both nymph and adult Ca. Las-infected ACPs. The DCEF32 and DCEF35 genes were downregulated in the Ca Las-infected adults. DCEF28 mRNA levels were reduced in Ca Las-infected nymphs compared with healthy nymphs (Fig. 2). Based on these results, the following six effectors were selected for the RNAi experiments: DCEF26, DCEF27, DCEF28, DCEF32, DCEF33 and DCEF35.
Figure 2.
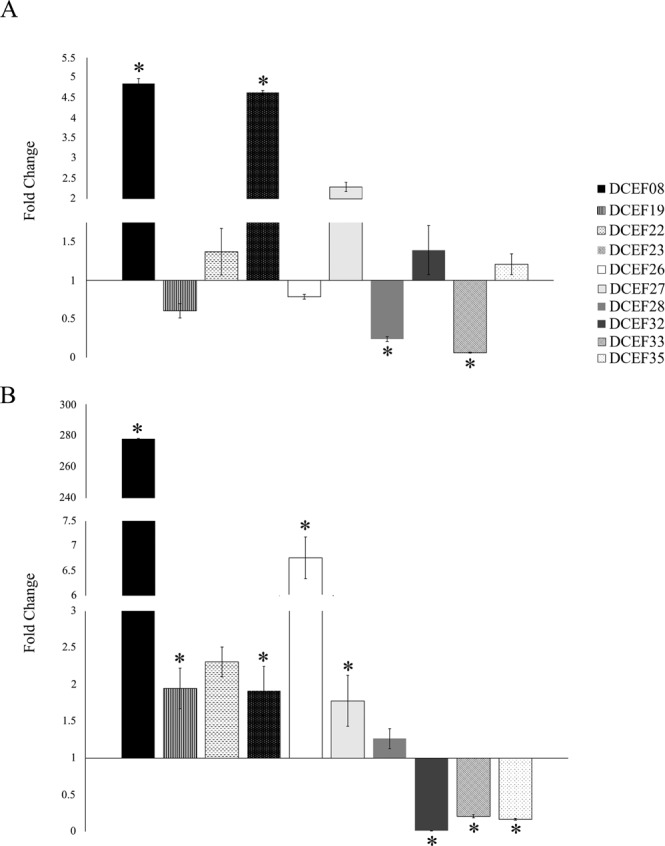
Relative gene expression of ten D. citri candidate effectors in nymph (A) and adult (B) Ca. Las-infected insects compared with Ca. Las-free nymph and adults. The results are the mean of three biological replicates per set. *corresponds to statistically significant differences amongst the healthy and Ca. Las-infected treatments (t-test, P-value < 0.05).
Delivery of dsRNA by artificial diet
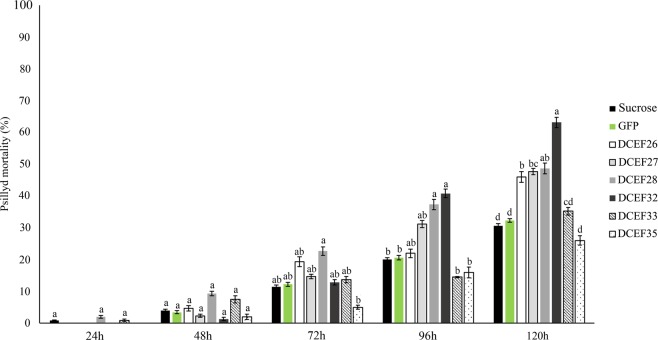
DsRNA was delivered to ACP adults using a 30% sucrose-artificial diet with 100 ng/µL of dsRNA. After five days of continuous dsRNA delivery, we observed 46, 47 and 61% mortality for the DCEF26, DCEF27, and DCEF32 gene-fed insects, respectively (Fig. 3). DCEF28 treatment caused significant mortality during the first 72 hours of the experiment, reaching 48% mortality after five days. DCEF33 and DCEF35 showed fewer expressive effects on ACP mortality, causing 35 and 26% mortality, respectively (Fig. 3).
Figure 3.
Mortality rates of D. citri fed an artificial diet containing 30% sucrose and 100 ng/µL of dsRNA for GFP or dsRNA for the effectors DCEF26, DCEF27, DCE28, DCEF32, DCEF33 or DCEF35. The mortality rates were evaluated daily during a 120-hour period. The data correspond to the average of three independent experiments. Different letters correspond to statistically significant differences amongst the treatments (Tukey test, P-value < 0.05).
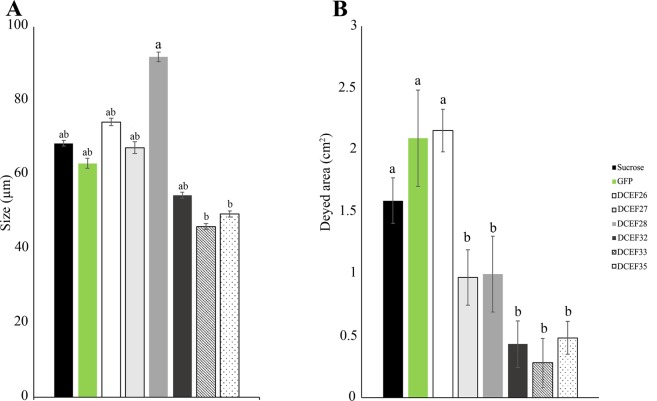
To verify the interference of silencing the effectors on ACP feeding, salivary sheath evaluation was performed through microscopy analysis. No structural differences in the salivary sheaths were observed between the treatments and control (Fig. S3). However, the salivary sheath sizes of dsDCEF32-, dsDCEF33- and dsDCEF35-treated insects were 20, 32 and 27% lower than the sucrose control, respectively. dsDCEF28-treated insects exhibited a 45% increase in salivary sheath size compared with the control (Fig. 4).
Figure 4.
(A) Measurement (µm) of the ACP salivary sheath after treatment with dsRNAs for GFP, DCEF26, DCEF27, DCEF28, DCEF32, DCEF33 or DCEF35. (B) Honeydew tinted areas after treatment with dsRNAs for GFP, DCEF26, DCEF27, DCEF28, DCEF32, DCEF33 or DCEF35. The data correspond to the average of three independent experiments. Different letters correspond to statistically significant differences amongst the treatments (Tukey test, P-value < 0.05).
Moreover, the effects of effector silencing on ACP ingestion abilities were evaluated by quantification of the excretions deposited on filter paper at the bottom of the cages. A significant reduction in honeydew was observed in five of six treatments (Fig. 4). The DCEF32, DCEF33 and DCEF35 treatments caused the strongest reduction in honeydew production, presenting tinted areas that were 72, 82 and 69% lower than the sucrose control, respectively. A 38 and 37% reduction in the tinted area was observed for the DCEF27 and DCEF28 treatments, respectively. No differences in honeydew production were observed between the DCEF26 treatment and control. These results corroborate the hypothesis that the silencing of selected candidate effectors may affect D. citri feed performance.
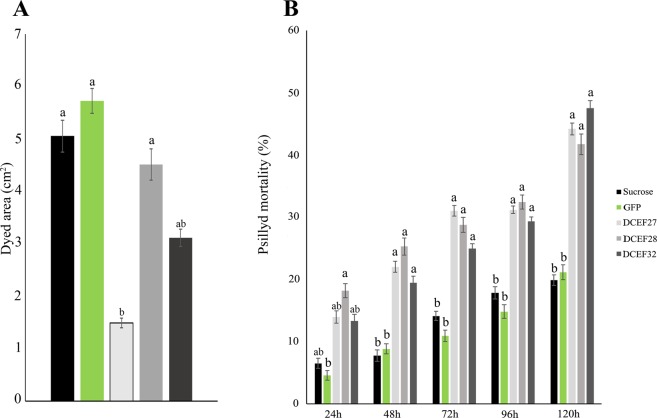
Compared with in vitro conditions, plant responses could alter insect feeding performance31. To confirm the silencing interference on ACP feeding in planta, we evaluated the ACP behaviors on citrus plants after delivery of dsRNAs. This analysis was performed using the following three candidate gene effectors: DCEF27, DCEF28 and DCEF32. ACP adults were treated with dsRNA via artificial diet for five days. Then, insects were transferred to ‘Rangpur lime’ seedling leaves for five days. Mortality was evaluated daily and the quantification of honeydew droplets and gene expression analysis were performed at the end of the assays. After the first 48 hours, all treatments showed elevated mortality compared with the sucrose and GFP controls (Fig. 5), presenting 44, 41 and 47% mortality for the DCEF27, DCEF28 and DCEF32 gene treatments after five days, respectively. The honeydew analysis demonstrated a reduction in excreta production by dsRNA-treated insects (Fig. 5). A decrease of 10 and 38% for the tinted area was observed for the DCEF28 and DCEF32 treatments, respectively, compared with sucrose. dsDCEF27-treated insects showed the strongest reduction in honeydew production, showing tinted areas that were 70% lower than the controls (Fig. 5). These results demonstrated that knockdown of D. citri effectors could also interfere with ACP feeding in planta.
Figure 5.
(A) Honeydew tinted area from DCEF27-, DCEF28-, and DCEF32- silenced ACPs after feeding with ‘Rangpur lime’ leaves. (B) Mortality rates of D. citri on Rangpur lime leaves after treatment with dsRNAs for the DCEF27, DCE28, or DCEF32 effectors. ACPs were treated with effector dsRNAs in vitro and then transferred to the citrus plants for feeding. The mortality rates were evaluated daily during a 120-hour period. The data correspond to the average of four independent experiments. Different letters correspond to statistically significant differences amongst the treatments (Tukey test, P-value < 0.05).
Evaluation of D. citri feeding behaviors
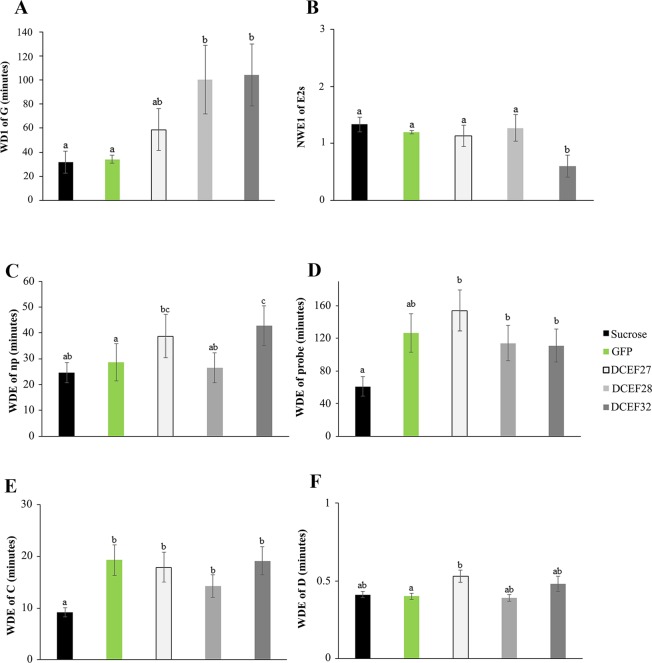
To confirm the changes to D. citri feeding behaviors caused by the silencing of candidate effectors, EPG analysis was applied to reveal the differences by the treatments. EPG recordings were performed on adult females after dsRNA delivery via an artificial diet for five days. The DCEF28 and DCEF32 treatments showed long feeding periods on xylem (waveform G) (Fig. 6A) compared with the controls (sucrose and GFP). Moreover, the proportion of individuals that produced waveform G (PPW) was lower for the treatment GFP than in the other treatments (Table 2).
Figure 6.
Comparison of ACP waveforms of feeding behaviors after in vitro delivery of dsRNAs for the DCEF27, DCEF28, or DCEF32 effectors and the sucrose and GFP dsRNA controls during an 8-h recording on the ‘Rangpur lime’ seedlings. The waveform duration per insect (WDI) of xylem (A). The number of waveform events per insect (NWEI) for E2s (B). The waveform duration (min) per event (WDE) for non-probe (np) (C), probe (D), waveform C (E), and waveform D (F). Different letters indicate statistically significant differences according to the Tukey test (for Gaussian variables) or Kruskal-Wallis test (for non-Gaussian variables) (P-value < 0.05).
Table 2.
Proportion of Diaphorina citri individuals that produced a specific waveform type (PPW) on ‘Rangpur lime’ seedlings during a 10-h recording.
| Waveform* | PPW | P | ||||||
|---|---|---|---|---|---|---|---|---|
| Sucrose | GFP | DCEF27 | DCEF28 | DCEF32 | X2 | df | ||
| G | 9/15 a | 6/15 b | 11/15 a | 13/15 a | 12/15 a | 9.44 | 4 | 0.049 |
| D | 15/15 a | 15/15 a | 13/15 a | 12/15 a | 11/15 a | 8.08 | 4 | 0.089 |
| E1 | 15/15 a | 15/15 a | 13/15 a | 12/15 a | 11/15 a | 8.08 | 4 | 0.089 |
| E2 | 15/15 a | 15/15 a | 12/15 a | 12/15 a | 8/15 b | 15.45 | 4 | 0.004 |
| E2 > 10 min | 15/15 a | 15/15 a | 12/15 a | 12/15 a | 7/15 b | 18.79 | 4 | <0.001 |
*Waveforms: (G) active intake of xylem sap, (D) first contact with phloem, (E1) salivation in the phloem sieve tubes, and (E2) phloem sap ingestion54. Proportions followed by the same letter in the same row do not differ significantly (P-value > 0.05) using chi-square (X2) test for pairwise comparisons.
Sustained phloem ingestion (E2 > 10 minutes) was significantly reduced for the DCEF32 treatment compared to the other treatments (Fig. 6B). Furthermore, the proportion of individuals that underwent ingestion in the phloem (E2 and E2s) was lower for the DCEF32 treatment when compared to the other treatments (Table 2).
The mean duration per event (WDE) of non-probing was significantly longer for the DCEF32 treatment than for the sucrose and GFP treatments (Fig. 6C). The WDE of C (stylet pathway) and probe were significantly shorter for the sucrose treatment control compared with the other treatments (Fig. 6D,E). The WDE of D was significantly longer for the DCEF27 treatment when compared with the GFP treatment (Fig. 6F).
Gene expression analysis
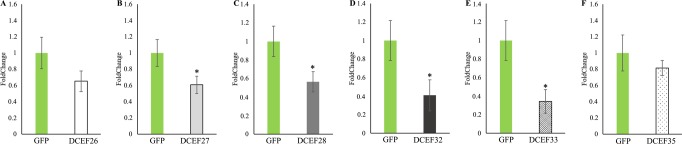
After five days of dsRNA delivery via an artificial diet, gene expression analysis showed a reduction in the mRNA levels of all the effector genes, confirming the knockdown of the candidate effectors (Fig. 7). The strongest mRNA reduction was observed for the DCEF33 and DCEF32 genes, which presented 63 and 59% inhibition in their mRNA levels, respectively. The DCEF26, DCEF27 and DCEF28 genes showed a reduction of 34, 39 and 43% of their mRNA levels, respectively, compared with the GFP control. The DCEF35 effector presented an 18% inhibition in gene expression (Fig. 7).
Figure 7.
Relative gene expression of the candidate effectors DCEF26 (A), DCEF27 (B), DCEF28 (C), DCEF32 (D), DCEF33 (E) and DCEF35 (F) compared with the GFP control. Gene expression evaluation occurred on ACP adults after feeding with an artificial diet containing dsRNAs for the effectors or dsRNAs for GFP for five days. The data correspond to the mean of three independent experiments. *corresponds to statistically significant differences amongst the treatments (t-test, P-value < 0.05).
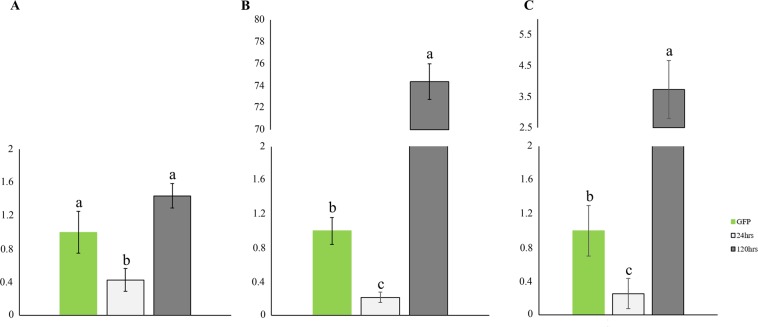
To confirm the maintenance of effector silencing in the in planta experiments, gene expression analysis was performed 24 and 120 hours after the transfer of the dsRNA-treated insects from an artificial diet to ‘Rangpur lime’ plants (Fig. 8). During the first 24 hours when the dsRNA-treated ACPs were fed plants, all the analyzed genes showed a reduction in their mRNA levels, proving that the D citri effectors still silenced (Fig. 8). However, the dsRNA-treated D. citri showed restoration of effector gene mRNA levels after 120 hours of feeding with citrus plants (Fig. 8).
Figure 8.
Relative gene expression of the candidate effectors DCEF27 (A), DCEF 28 (B), and DCEF32 (C) compared with GFP control observed 24 hours (light grey) and 120 hours (dark gray) after ACP adults were treated with dsRNAs for effectors and transferred for feeding on ‘Rangpur lime’ leaves. The data correspond to the average of four independent experiments. Different letters correspond to statistically significant differences amongst the treatments (Tukey test, P-value < 0.05).
Discussion
Candidate effectors prediction and validation
Salivary proteins are important for proper insect feeding and promoting lubrification, digestion and penetration38. These proteins also play important roles in plant-insect interactions34. For example, the potato aphid Macrosiphum euphorbiae Thoma (Hemiptera: Aphididae) presents an effector that interacts with two plant defense compounds to enhancing aphid fecundity39. The Hessian fly, Mayetiola destructor (Say 1817) (Diptera: Cecidomyiidae), phosphatase 2 C effector family elicits effector-triggered immunity in wheat by interacting with signal transduction pathways40. Furthermore, feeding is the principal vehicle of Ca. Las acquisition by ACP. Studies have demonstrated that long phloem ingestion by ACP nymph instars is responsible for the high acquisition efficiency of Ca. Las by immature D. citri instars14. Based on this, we proposed the identification of D. citri candidate effectors, looking for proteins important for interactions between ACP, citrus and Ca. Las.
Bioinformatics tools are the first line for the prediction of prokaryote or eukaryote effectors41,42. Independent of species, effector prediction is based on the following same criteria: amino acid composition and primary protein sequences, such as the presence of signal peptides, absence of anchor membrane domains, and secondary characteristics such as species-specific sequences, nuclear localization signals (NLSs), three-dimensional structure, positive selection, etc41. Thus, we use a Pea aphid bioinformatics pipeline25 as a model to predict ACP candidate effectors. Pea aphid effectors are some of the best characterized insect effectors28,43,44, and these tools allowed the identification of 131 ACP candidate effectors (Table S1). Saliva or salivary gland protein sequences are very often applied for the prediction of insect effectors45–47. However, in the absence of this kind of sequence source, the Pea aphid pipeline was successfully applied for mite effector prediction using full body genomics and transcriptomic data48, as was also applied in this work. These studies support the legitimacy of the results obtained here. Seeking to ensure that the predicted candidate effectors are manly secreted in ACP saliva, all proteins that presented similarity with arthropod gut proteins were excluded. We identified twenty-nine D. citri candidate effectors showing similarity with arthropod salivary proteins (Table S2), supporting the hypothesis that these effectors may be produced and secreted by D. citri salivary glands.
The major ACP candidate effectors are composed of D. citri-unique unknown function proteins (Table S1). Prediction of aphid effectors also showed a large number of unknown functional proteins28,29. Insect effectors are mostly species- or genus-specific36,43,49. An elevated number of unique uncharacterized proteins may indicate that the effectors are highly host-specific, resulting from strong selection pressures to adaptation between insects and their host plant28,29. Bioinformatics analysis revealed nuclear localization signals (NLSs) and chloroplast transit peptides (cTP) in several ACP effectors (Table S1; Fig. S1), showing that these putative effectors may act on important host organelles (Table S1; Fig. S1). The VPS52 effector protein from M. persicae is relocated to post-Golgi and pre-vacuolar compartments in Nicotiana benthamiana plants and interferes with aphid virulence50. The B. tabaci effector Bsp9 acts on the host cell cytoplasm, interacting with plant WRKY33 transcription factors to suppress defenses51.
D. citri effectors genes are expressed during different ACP life stages (Fig. S2). These candidate effectors genes are also overexpressed in D. citri head compared with their body, supporting the hypothesis that these proteins may be secreted by ACP saliva (Fig. 1). Over-expression in the insect head was also a criterion applied to discriminate effectors from aphid and mite species29,48. Moreover, gene expression analysis of Ca. Las-infected and Ca. Las-free nymphs and adults showed that the D. citri effectors genes are modulated in response to these bacteria (Fig. 2), which is indicative that these genes could interfere with the Ca. Las transmission process by D. citri.
Silencing of candidate effectors on D. citri feeding behavior
Effectors are strongly related to insect feed performance. Salivary compounds may elicit or suppress plant defenses, increasing or inhibiting insect feeding on its host plants52. Thereby, feeding performance and fecundity are the main phenotypes evaluated in insect effector studies. Silencing of angiotensin-converting enzymes (ACEs) genes from A. pisium interferes with phloem sap ingestion, enhancing the aphid mortality23. The knockdown of salivary endo-b-1,4-glucanase from N. lugens decreased its feeding performance in rice53. To ensure that the predicted D. citri effectors are feeding-related, we evaluated the ACP feeding performance after gene silencing.
Classical strategies for evaluating insect feeding include monitoring the salivary sheath and excreta production54,55. We observed that knockdown of ACP effectors in vitro assays promoted changes in the salivary sheath size and a strong reduction in honeydew production (Fig. 4), which indicates that the size of the salivary sheath may affect ACP ingestion.
The salivary sheath is one of two common salivary secretions produced by phytophagous hemipterans. Resulting from the solidification of insect gelling saliva, the salivary sheath is a solid structure that covers insect stylet providing orientation and stability during the piercing process and also protects the insect stylet from plant defenses56–58. The production of this structure is tightly related to appropriate insect feeding. Even so, excretion production is directly associated with insect ingestion59,60. Similar phenotypes were observed upon knockdown of salivary sheath proteins in the brown planthopper N. lugens, which also resulted in a reduction of honeydew amounts61.
However, these alterations did not directly affect mortality rates. The DCEF33 and DCEF35 treatments, which showed the strongest reduction in salivary sheath size and honeydew droplets, presented no significant mortality in the artificial diet experiments (Fig. 3). Similar patterns were observed when silencing the NlShp gene from N. lugens, which cause changes in insect feeding behaviors in vitro; however, knockdown of this gene did not cause significant insect mortality31.
Additionally, knockdown of ACP effectors also interferes with ACP feeding in planta. An expressive decrease in excreta production in dsDCEF27 and dsDCEF32-treated insects and elevated mortality were observed when these insects were fed the ‘Rangpur lime’ leaves (Fig. 5).
Interference of feeding performance caused by effector silencing was proven by evaluation of D. citri feeding behaviors through EPG analysis, which showed a significant reduction in phloem ingestion by the dsDCEF32-treated insects. Only 50% of the dsDCEF32-treated insects showed E2 and E2 > 10 min waveforms (Table 2). Moreover, the number (NWEI) of E2 > 10 min waveforms was reduced by 50% for the DCEF32 treatment compared with the other treatments (Fig. 6B). These results indicate that dsDCEF32-treated insects seldomly performed passive phloem ingestion. Since Ca. Las is a phloem-inhabiting bacteria62, these results allow the hypothesis that DCEF32 may be important for the Ca. Las transmission process. Luo and collaborators (2015) showed that ACP adults that displayed an E2 waveform (passive phloem-sap uptake from sieve elements) were subsequently detected as Ca. Las-positive, demonstrating these EPG waveforms are strongly associated with Ca. Las acquisition63. In contrast, the strong correlation between the duration of phloem salivation (E1 waveform) and Ca. Las inoculation was described by Wu and collaborators (2016). These studies demonstrated that phloem phases (E1 and E2) are a critical point for the transmission of Ca. Las by D. citri63–65.
An interesting observation was the increased frequency and time of waveform G, which represents xylem ingestion, for the DCEF28 and DCEF32 treatments (Fig. 6A). In general, the G waveform presents low frequency and duration during D. citri feeding behaviors. Previous analysis demonstrated that the G waveform occurs on approximately 30% EGP recordings for D. citri adults, with durations ranging from 9 to 18% of the recording time14,66,67. Surprisingly, our results showed a proportion of individuals that had G waveforms (PPW) greater than 70% for the dsRNA-effector treatments (Table 2). The dsDCEF28- and dsDCEF32-treated insects showed longer feeding periods on xylem vessels (Fig. 6A; Table S4). G waveform formation for phloem sap feeders is frequently associated with dehydration or osmotic balance68,69. During citrus-ACP interactions, an increase in the G waveform duration was reported when ACP feeds on CLas-infected plants14,64. D. citri that performed xylem ingestion more often and for long periods were also observed on mature leaves compared with young ones due to the more pronounced sclerenchymatous ring around phloematic vessels, which difficult for phloem ingestion67. In this case, we hypothesize that xylem ingestion occurs to compensate for inefficient phloem ingestion67.
Effect of silencing on candidate effectors gene expression
Knockdown of D. citri effectors was proved by the significant reduction of the mRNA levels of these genes after delivery of dsRNA via an artificial diet and 24 hours after transferring the dsRNA-treated insects to Rangpur plants (Figs. 7 and 8). Nevertheless, after 120 hours of constant feeding of dsRNA-treated insects with citrus plants, the mRNA levels of D. citri effector genes were restored (Fig. 8), demonstrating that the silencing effects of the analyzed genes are transient in the absence of a constant dsRNA source. The lack of an efficient silencing signal amplification system and the presence of nucleases that degrade dsRNAs may be factors that could explain the observed phenomena, though none of these observations have been previously described for ACP70. Several hemipterans species have been reported to have nucleases that quickly degrade dsRNAs in different body fluids71. Differences in nuclease activity in different tissues were also described as an important factor for determining dsRNA delivery strategies for some insects72–74.
Conclusion
Most of the RNAi studies applied to D. citri control are focused on identifying candidate genes that promote significant mortality or insecticide susceptibility as well as new efficient strategies for dsRNA delivery, however, no practical use of this technology against D. citri has emerged. Studies focused on D. citri-Citrus- Ca. Las are recent, and many gaps regarding the genetic and molecular aspects of these interactions need to be fulfilled. Thus, the increase of the knowledge about D. citri biology and its interactions with citrus and Ca. Las is essential to develop new and efficient control strategies for HLB. Despite of recent identification of the D. citri saliva proteome, ACP effectors or salivary molecules that interact with plant hosts have not been identified to date. This study predicted D. citri effectors using a classical bioinformatics pipeline. Gene expression analysis combined with RNAi and EPG techniques allowed us to prove that the predicted effectors are directly related to ACP feeding performance, which is similar to the majority of insect effectors identified to date. The DCEF32 gene stands out amongst the identified effectors due to its action on D. citri phloem ingestion, which is the main colonization site of Ca. Las in plants. Moreover, gene expression analysis showed that the DCEF32 gene is modulated in response to Ca. Las during both the nymph and adult stages. These results may indicate that the DCEF32 effector could participate in Ca. Las-D. citri interactions. However, how D. citri effectors interact with host plants and/or Ca. Las still needs to be investigated. Understanding the participation of D. citri effectors in the HLB pathosystem can elucidate the molecular mechanisms of Ca. Las transmission by D. citri and its interaction with citrus plants. This knowledge is essential to develop novel and efficient control strategies for HLB.
Materials and Methods
Sequence database
Transcripts sequences from Arizona University transcriptome studies35 were initially used as a database. Complementary version 0.9 of the transcriptome and version 2.0 of the D. citri genome available on Citrus greening Solutions website (citrusgreening.org) were added to the bioinformatics analysis.
Bioinformatics analysis
The bioinformatics for the identification of D. citri candidate effectors was based on a pipeline for the prediction of aphid effectors25. First, redundant sequences and proteins with 1000 > amino acids were removed from the analysis. Redundant sequences were those that presented a similarity > 95% and an e-value < 10−10. Secreted proteins were predicted using SignalP 4.075. Proteins that had transmembrane domains were predicted using the TMHMM and GPI-SOM software76,77 and removed from future analysis. InterPro, Uniprot and Prosite78,79 were used to identify and exclude ordinary proteins. Proteins that displayed similarity with arthropod gut proteins were removed through blastX against arthropod gut ESTs. To identify potential D. citri salivary proteins, blastX against arthropod salivary proteins was performed. Additionally, prediction of ACP candidate effector subcellular localization was performed using the NetNES 1.1 Server, Localizer and LocTree tools80–82.
Asian citrus psyllid colony
Mesh cages containing Murraya paniculata were used to rear healthy D. citri at 25 ± 2 °C under a 14:10 h (light:dark) photoperiod and 60 to 70% relative humidity (RH). CLas-infected ACP colonies were reared on Ca. Las-infected Pera sweet orange trees (symptomatic and PCR positive) were grown under greenhouse conditions for light, temperature and humidity. Both ACP populations were maintained at the biotechnology laboratory at the Centro de Citricultura ‘Sylvio Moreira’.
Nuclei acid isolation and cDNA synthesis
RNA extraction was performed using a Direct-zol RNA MiniPrep and DNAse I Set (Zymo Research, Irvine, CA, USA) according to the manufacturers’ instructions. A NanoDrop ND 8000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA) was used to measure the RNA integrity and quality. One microgram of extracted RNA was applied for cDNA synthesis using the GoScript reverse transcription system (Promega) according to the manufacturer’s protocol. After the reaction, the cDNA was diluted 1:25 for further analysis.
Double-strand RNA synthesis
The target sequences of ACP candidate effectors were amplified by RT-PCR using specific gene primers conjugated with 19 bases of the T7 RNA polymerase promoter (Table S3). The GFP sequence was amplified from the pIG1783f plasmid83. The PCR reaction was performed using 5 μL of cDNA, 25 μL of Gotaq colorless MasterMix (Promega), and 100 nM of each primer pair in a final volume of 50 μL. Thermal profiles were determined using 10 cycles of 94 °C for 30 s, 1 min at 10 °C above the optimal primer annealing temperature, and 72 °C for 1.3 min, followed by 35 cycles of 94 °C for 30 s, 1 min at the optimal primer annealing temperature, and 72 °C for 40 s.
DsRNAs were synthesized from purified PCR products using the MEGAscript RNAi kit (Ambion, catalog no. AM1626) according to the manufacturer’s protocol. The purified dsRNAs were quantified spectrophotometrically at 260/280 nm and the integrity was examined by agarose gel electrophoresis.
Delivery of dsRNA and bioassay in vitro
Ten-day-old adults were used for the RNAi experiments. The insects were maintained in plastic cages containing a sachet composed of 100 µL of an artificial diet (30% of sucrose and 0.1% green and 0.4% yellow food dyes) homogeneously distributed between two parafilm layers84 and filter paper on the bottom of the cage. Feeding assays were performed for a period of five days at room temperature under a 14:10 h (light: dark) photoperiod and 60 to 70% HR. Each treatment consisted of five biological replicates (10 insects per replicate). Insect mortality was evaluated daily. Three independent bioassays were performed.
After the feeding period, live insects were collected for further gene expression analysis. Salivary sheaths present on the parafilm membrane were dyed using 0.1% safranin, quantified and photographed with a light microscope using a 40x objective. The sizes of the salivary sheaths were measured using the ImageJ software85. Honeydew droplets present on the filter paper were dyed with 2% ninhydrin, counted and measured using the Quant software86. The remaining artificial diet was collected and examined by electrophoretic roll to observe the dsRNA stability (Fig. S4). Statistical analyses were performed using one-way ANOVA (P < 0.05) with the GraphPad Prism software87.
In vivo bioassay for silencing effect evaluation
After the in vitro dsRNA delivery described above, living insects were confined to ‘Rangpur lime’ seedling leaves using clip cages. The clip cages were made from 90-mm in diameter plastic Petri dishes. A hole (7 mm) was made on the lateral side of the Petri dishes to facilitate the accommodation of a leaf blade and petiole. The leaf was attached to the Petri lid using adhesive tape. The filter paper was placed in the bottom of the clip cage to collect the honeydew droplets. The plates were sealed using a cotton and PVC film. Mortality was evaluated for five days and the feeding abilities were analyzed based on the quantity of honeydew droplets present on filter paper as described before. The experiment was repeated four times using four biological replicates for each treatment (ten adults ACP per biological replicate). Statistical analyses were performed using one-way ANOVA (Tukey test) and the t-test (P < 0.05) using the GraphPad Prism software87.
Feeding behavior evaluation of D. citri by electron penetration graph (EPG)
After delivery of the dsRNA via the artificial diet as described above, we performed feeding behavior assays using an EPG technique, which allows for the real-time study of an insect’s stylet activities in plant tissues. Females ACPs were anesthetized for 5 seconds with CO2 and immobilized using a vacuum chamber under a dissecting microscope. Then, a gold wire (3 cm length, 20 µm in diameter; EPG Systems, Wageningen, The Netherlands) was attached to the ACP pronotum with a small droplet of water-based silver glue. The opposite end of the gold wire was glued to a thin copper wire (2 cm length), which was connected to the EPG probe. Another copper electrode (10 cm long, 2 mm wide) was inserted into the plant soil.
The insects were placed on the abaxial surface of a young ‘Rangpur lime’ leaf. The EPG waveforms were recorded using a Direct Current 8-channel EPG device model Giga-8d, with Stylet+ for Windows software (EPG Systems). The recordings were carried out in a room at 25 ± 1 °C inside a Faraday cage (for electrical noise isolation) for 8 hours. The waveforms recorded for D. citri feeding behaviors were characterized according to previous reports54. The output is given in an Excel workbook88 to calculate the treatment mean for each EPG variable. Fifteen individual recordings were performed for each treatment.
The EPG waveforms previously described for D. citri66 were identified as follows: waveform np (non-probing behavior); waveform C (which indicates movement of the stylets in the intercellular apoplastic space); waveform D (a short waveform always observed between waveforms C and E1); E1 waveform (indicates salivation into phloem sieve elements); waveform E2 (correlated with passive phloem-sap uptake from sieve elements); and waveform G (active intake of xylem sap).
The experimental design was entirely randomized. The EPG data were transformed when necessary with ln (x + 1) or √(x + 1) to reduce the heteroscedasticity and improve the normal distribution. All parameters were analyzed with Tukey’s test (P < 0.05). If the data did not follow a normal distribution according to the Shapiro-Wilk normality test, a nonparametric Kruskall-Wallis test (P < 0.05) was performed. The proportion of individuals that produced a specific waveform type (PPW) was compared among the different treatment groups using the chi-square test. All data were analyzed using the IBM Statistics SPSS 22.0 software89.
Gene expression analysis
To select candidate effectors by RT-qPCR, pools of 10 adults and 50 nymphs were employed for RNA extraction as described before. Gene expression analysis of different body parts was conducted using 10 heads and 10 bodies from ACP adults. The measurement of gene expression in insects silenced by dsRNA was performed using RNA extracted from a pool of four ACP adults per biological replicate from both the artificial diet and plant experiments.
RT-qPCR reactions were performed using 6.5 µL of GoTaq qPCR MasterMix (Promega), 120 nM of each gene-specific primer pair and 3 μL of the diluted cDNA in a final volume of 12.5 μL. The amplification cycles were performed on a 7500 Fast Real-Time PCR System device (Thermo Scientific, Waltham, MA, USA) using the following standard thermal profile: 95 °C for 20 s followed by 40 cycles of 95 °C for 3 s and 60 °C for 30 s. Five technical replicates were analyzed for each sample. The Cq values and the primer efficiency were estimated using the Miner software (http://miner.ewindup.info). The normalized gene expression analyses were performed with the 2ΔCq method and relative gene expression was estimated using the 2-ΔΔCq method using the ribosomal genes S20 and S13 as reference genes. For statistical analysis, one-way ANOVA (Tukey test) and the t-test (P < 0.05) were performed using the GraphPad Prism software87,90.
Supplementary information
Acknowledgements
This study was supported by INCT-Citros (FAPESP 2014/50880-0 and CNPq 465440/2014-2) and a PhD fellowship to I.S.P. (FAPESP 2015/13971-0) and Post-doc fellowship to D.M.G. (CNPq 103228/2018-7).
Author contributions
I.S.P. and M.A.M. conceived the study. I.S.P. and D.M.G. designed and executed the experiments. N.K.P.M., J.R.S.L. and I.S.P. performed the EPG experiments and analysis. All authors wrote and reviewed the manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Inaiara de Souza Pacheco and Diogo Manzano Galdeano.
Supplementary information
is available for this paper at 10.1038/s41598-020-62856-5.
References
- 1.FAO - News Article: Global body adopts new measures to stop the spread of plant pests. Available at: http://www.fao.org/news/story/en/item/1118322/icode/. (Accessed: 10th January 2019).
- 2.Emani, C. The biology of plant-insect interactions: a compendium for the plant biotechnologist. (Taylor & Francis Group, 2018).
- 3.Alves G, Diniz A, Parra J. Biology of the Huanglongbing Vector Diaphorina citri (Hemiptera: Liviidae) on Different Host Plants. J. Econ. Entomol. 2014;107:691–696. doi: 10.1603/EC13339. [DOI] [PubMed] [Google Scholar]
- 4.Munir, S. et al. Huanglongbing Control: Perhaps the End of the Beginning. Microbial Ecology 76, 192–204 (Springer US, 2018). [DOI] [PubMed]
- 5.da Graça JV, et al. Huanglongbing: An overview of a complex pathosystem ravaging the world’s citrus. J. Integr. Plant Biol. 2015;XXXX:n/a–n/a. doi: 10.1111/jipb.12437. [DOI] [PubMed] [Google Scholar]
- 6.Filho AB, et al. The importance of primary inoculum and area-wide disease management to crop health and food security. Food Secur. 2016;8:221–238. doi: 10.1007/s12571-015-0544-8. [DOI] [Google Scholar]
- 7.Craig AP, Cunniffe NJ, Parry M, Laranjeira FF, Gilligan CA. Grower and regulator conflict in management of the citrus disease Huanglongbing in Brazil: A modelling study. J. Appl. Ecol. 2018;55:1956–1965. doi: 10.1111/1365-2664.13122. [DOI] [Google Scholar]
- 8.Shimwela MM, et al. In-grove spatiotemporal spread of citrus huanglongbing and its psyllid vector in relation to weather. Phytopathology. 2019;109:418–427. doi: 10.1094/PHYTO-03-18-0089-R. [DOI] [PubMed] [Google Scholar]
- 9.Chen XD, Ashfaq M, Stelinski LL. Susceptibility of Asian citrus psyllid, Diaphorina citri (Hemiptera: Liviidae), to the insecticide afidopyropen: a new and potent modulator of insect transient receptor potential channels. Appl. Entomol. Zool. 2018;53:453–461. doi: 10.1007/s13355-018-0574-8. [DOI] [Google Scholar]
- 10.Tiwari S, Mann RS, Rogers ME, Stelinski LL. Insecticide resistance in field populations of Asian citrus psyllid in Florida. Pest Manag. Sci. 2011;67:1258–1268. doi: 10.1002/ps.2181. [DOI] [PubMed] [Google Scholar]
- 11.Pardo S, et al. Insecticide resistance of adults and nymphs of Asian citrus psyllid populations from Apatzingán Valley, Mexico. Pest Manag. Sci. 2018;74:135–140. doi: 10.1002/ps.4669. [DOI] [PubMed] [Google Scholar]
- 12.Canale MC, et al. Latency and Persistence of ‘ Candidatus Liberibacter asiaticus’ in Its Psyllid Vector, Diaphorina citri (Hemiptera: Liviidae) Phytopathology. 2017;107:264–272. doi: 10.1094/PHYTO-02-16-0088-R. [DOI] [PubMed] [Google Scholar]
- 13.Inoue H, et al. Enhanced proliferation and efficient transmission of Candidatus Liberibacter asiaticus by adult Diaphorina citri after acquisition feeding in the nymphal stage. Ann. Appl. Biol. 2009;155:29–36. doi: 10.1111/j.1744-7348.2009.00317.x. [DOI] [Google Scholar]
- 14.George J, Ammar ED, Hall DG, Shatters RG, Lapointe SL. Prolonged phloem ingestion by Diaphorina citri nymphs compared to adults is correlated with increased acquisition of citrus greening pathogen. Sci. Rep. 2018;8:10352. doi: 10.1038/s41598-018-28442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ebert TA, Backus EA, Shugart HJ, Rogers ME. Behavioral Plasticity in Probing by Diaphorina citri (Hemiptera, Liviidae): Ingestion from Phloem Versus Xylem is Influenced by Leaf Age and Surface. J. Insect Behav. 2018;31:119–137. doi: 10.1007/s10905-018-9666-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stockton DG, Martini X, Patt JM, Stelinski LL. The Influence of Learning on Host Plant Preference in a Significant Phytopathogen Vector, Diaphorina citri. PLoS One. 2016;11:e0149815. doi: 10.1371/journal.pone.0149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fletcher, J. et al. A Review of the Citrus Greening Research and Development Efforts Supported by the Citrus Research and Development Foundation. National Academies Press, 10.17226/25026 (2018).
- 18.Matsumoto Y, Hattori M. The green rice leafhopper, Nephotettix cincticeps (Hemiptera: Cicadellidae), salivary protein NcSP75 is a key effector for successful phloem ingestion. PLoS One. 2018;13:e0202492. doi: 10.1371/journal.pone.0202492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C-H, et al. Secretory laccase 1 in Bemisia tabaci MED is involved in whitefly-plant interaction. Sci. Rep. 2017;7:3623. doi: 10.1038/s41598-017-03765-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Naessens E, et al. A Secreted MIF Cytokine Enables Aphid Feeding and Represses Plant Immune Responses. Curr. Biol. 2015;25:1898–903. doi: 10.1016/j.cub.2015.05.047. [DOI] [PubMed] [Google Scholar]
- 21.Giron D, Huguet E, Stone GN, Body M. Insect-induced effects on plants and possible effectors used by galling and leaf-mining insects to manipulate their host-plant. J. Insect Physiol. 2016;84:70–89. doi: 10.1016/j.jinsphys.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 22.Mutti NS, et al. A protein from the salivary glands of the pea aphid, Acyrthosiphon pisum, is essential in feeding on a host plant. Proc. Natl. Acad. Sci. USA. 2008;105:9965–9969. doi: 10.1073/pnas.0708958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, et al. Angiotensin-converting enzymes modulate aphid–plant interactions. Sci. Rep. 2015;5:8885. doi: 10.1038/srep08885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinya T, et al. Modulation of plant defense responses to herbivores by simultaneous recognition of different herbivore-associated elicitors in rice. Sci. Rep. 2016;6:32537. doi: 10.1038/srep32537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bos, J. I. B. et al. A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet. 6 (2010). [DOI] [PMC free article] [PubMed]
- 26.Lee H-R, et al. Transient Expression of Whitefly Effectors in Nicotiana benthamiana Leaves Activates Systemic Immunity Against the Leaf Pathogen Pseudomonas syringae and Soil-Borne Pathogen Ralstonia solanacearum. Front. Ecol. Evol. 2018;6:90. doi: 10.3389/fevo.2018.00090. [DOI] [Google Scholar]
- 27.Rao W, et al. Secretome Analysis and In Planta Expression of Salivary Proteins Identify Candidate Effectors from the Brown Planthopper Nilaparvata lugens. Mol. Plant-Microbe Interact. 2019;32:227–239. doi: 10.1094/MPMI-05-18-0122-R. [DOI] [PubMed] [Google Scholar]
- 28.Carolan JC, et al. Predicted Effector Molecules in the Salivary Secretome of the Pea Aphid (Acyrthosiphon pisum): A Dual Transcriptomic/Proteomic Approach. J. Proteome Res. 2011;10:1505–1518. doi: 10.1021/pr100881q. [DOI] [PubMed] [Google Scholar]
- 29.Thorpe P, Cock PJAA, Bos J. Comparative transcriptomics and proteomics of three different aphid species identifies core and diverse effector sets. BMC Genomics. 2016;17:172. doi: 10.1186/s12864-016-2496-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Bel AJE, Will T. Functional Evaluation of Proteins in Watery and Gel Saliva of Aphids. Front. Plant Sci. 2016;7:1840. doi: 10.3389/fpls.2016.01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang HJ, et al. A salivary sheath protein essential for the interaction of the brown planthopper with rice plants. Insect Biochem. Mol. Biol. 2015;66:77–87. doi: 10.1016/j.ibmb.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 32.Shangguan X, et al. A mucin-like protein of planthopper is required for feeding and induces immunity response in plants. Plant Physiol. 2017;176:00755.2017. doi: 10.1104/pp.17.00755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu X, Killiny N. The secreted salivary proteome of Asian citrus psyllid Diaphorina citri. Physiol. Entomol. 2018;43:324–333. doi: 10.1111/phen.12263. [DOI] [Google Scholar]
- 34.Dalio RJD, et al. PAMPs, PRRs, effectors and R-genes associated with citrus–pathogen interactions. Ann. Bot. 2017;119:mcw238. doi: 10.1093/aob/mcw238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fisher TW, et al. Comparison of potato and asian citrus psyllid adult and nymph transcriptomes identified vector transcripts with potential involvement in circulative, propagative liberibacter transmission. Pathog. (Basel, Switzerland) 2014;3:875–907. doi: 10.3390/pathogens3040875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Boulain H, et al. Fast evolution and lineage-specific gene family expansions of aphid salivary effectors driven by interactions with host-plants. Genome Biol. Evol. 2018;10:1554–1572. doi: 10.1093/gbe/evy097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marín M, Uversky VN, Ott T. Intrinsic disorder in pathogen effectors: protein flexibility as an evolutionary hallmark in a molecular arms race. Plant Cell. 2013;25:3153–7. doi: 10.1105/tpc.113.116319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cherqui A, Tjallingii WF. Salivary proteins of aphids, a pilot study on identification, separation and immunolocalisation. J. Insect Physiol. 2000;46:1177–1186. doi: 10.1016/S0022-1910(00)00037-8. [DOI] [PubMed] [Google Scholar]
- 39.Kettles GJ, Kaloshian I. The Potato Aphid Salivary Effector Me47 Is a Glutathione-S-Transferase Involved in Modifying Plant Responses to Aphid Infestation. Front. Plant Sci. 2016;7:1–12. doi: 10.3389/fpls.2016.01142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao C, et al. Avirulence gene mapping in the Hessian fly (Mayetiola destructor) reveals a protein phosphatase 2C effector gene family. J. Insect Physiol. 2016;84:22–31. doi: 10.1016/j.jinsphys.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 41.Dalio RJD, Herlihy J, Oliveira TS, McDowell JM, Machado M. Effector Biology in Focus: A Primer for Computational Prediction and Functional Characterization. Mol. Plant-Microbe Interact. 2018;31:22–33. doi: 10.1094/MPMI-07-17-0174-FI. [DOI] [PubMed] [Google Scholar]
- 42.Varden FA, D la Concepcion JC, Maidment JH, Banfield MJ. Taking the stage: effectors in the spotlight. Curr. Opin. Plant Biol. 2017;38:25–33. doi: 10.1016/j.pbi.2017.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Pitino M, Hogenhout SA. Aphid Protein Effectors Promote Aphid Colonization in a Plant Species-Specific Manner. Mol. Plant-Microbe Interact. 2013;26:130–139. doi: 10.1094/MPMI-07-12-0172-FI. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, et al. Armet is an effector protein mediating aphid-plant interactions. FASEB J. 2015;29:2032–45. doi: 10.1096/fj.14-266023. [DOI] [PubMed] [Google Scholar]
- 45.Nicholson SJ, Hartson SD, Puterka GJ. Proteomic analysis of secreted saliva from Russian Wheat Aphid (Diuraphis noxia Kurd.) biotypes that differ in virulence to wheat. J. Proteomics. 2012;75:2252–2268. doi: 10.1016/j.jprot.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 46.Felton, G. W. et al. Herbivore Oral Secretions are the First Line of Protection Against Plant-Induced Defences. Annual Plant Reviews: Insect-Plant Interactions47, John Wiley & Sons, Ltd, (2014).
- 47.Johnson AJ, et al. Differential expression of candidate salivary effector proteins in field collections of Hessian fly, Mayetiola destructor. Insect Mol. Biol. 2015;24:191–202. doi: 10.1111/imb.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Villarroel CA, et al. Salivary proteins of spider mites suppress defenses in Nicotiana benthamiana and promote mite reproduction. Plant J. 2016;86:119–131. doi: 10.1111/tpj.13152. [DOI] [PubMed] [Google Scholar]
- 49.Vandermoten S, et al. Comparative analyses of salivary proteins from three aphid species. Insect Mol. Biol. 2014;23:67–77. doi: 10.1111/imb.12061. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez PA, Escudero-Martinez C, Bos JIBB. An Aphid Effector Targets Trafficking Protein VPS52 in a Host-Specific Manner to Promote Virulence. Plant Physiol. 2017;173:1892–1903. doi: 10.1104/pp.16.01458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang N, et al. A whitefly effector Bsp9 targets host immunity regulator WRKY33 to promote performance. Philos. Trans. R. Soc. B Biol. Sci. 2019;374:20180313. doi: 10.1098/rstb.2018.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mondal HA. Shaping the understanding of saliva-derived effectors towards aphid colony proliferation in host plant. J. Plant Biol. 2017;60:103–115. doi: 10.1007/s12374-016-0465-x. [DOI] [Google Scholar]
- 53.Ji R, et al. A salivary endo-β-1,4-glucanase acts as an effector that enables the brown planthopper to feed on rice. Plant Physiol. 2017;173:1920–1932. doi: 10.1104/pp.16.01493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hall DG, Shatters RG, Carpenter JE, Shapiro JP. Research Toward an Artificial Diet for Adult Asian Citrus Psyllid. Ann. Entomol. Soc. Am. 2010;103:611–617. doi: 10.1603/AN10004. [DOI] [Google Scholar]
- 55.Ammar E, Hall D. A new method for short-term rearing of citrus psyllids (Hemiptera: Pysllidae) and for collecting their honeydew excretions. Florida Entomol. 2011;94:340–342. doi: 10.1653/024.094.0229. [DOI] [Google Scholar]
- 56.Sharma A, et al. Salivary proteins of plant-feeding hemipteroids - implication in phytophagy. Bull. Entomol. Res. 2014;104:117–36. doi: 10.1017/S0007485313000618. [DOI] [PubMed] [Google Scholar]
- 57.Morgan JK, et al. Formation of Stylet Sheaths in āere (in air) from Eight Species of Phytophagous Hemipterans from Six Families (Suborders: Auchenorrhyncha and Sternorrhyncha) PLoS One. 2013;8:e62444. doi: 10.1371/journal.pone.0062444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Walling LL. Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol. 2008;146:859–866. doi: 10.1104/pp.107.113142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dugravot S, Backus EA, Reardon BJ, Miller TA. Correlations of cibarial muscle activities of Homalodisca spp. sharpshooters (Hemiptera: Cicadellidae) with EPG ingestion waveform and excretion. J. Insect Physiol. 2008;54:1467–1478. doi: 10.1016/j.jinsphys.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 60.Cid M, Fereres A. Characterization of the Probing and Feeding Behavior of Planococcus citri (Hemiptera: Pseudococcidae) on Grapevine. Ann. Entomol. Soc. Am. 2010;103:404–417. doi: 10.1603/AN09079. [DOI] [Google Scholar]
- 61.Ye W, et al. A salivary EF-hand calcium-binding protein of the brown planthopper Nilaparvata lugens functions as an effector for defense responses in rice. Sci. Rep. 2017;7:40498. doi: 10.1038/srep40498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Koh E-JJ, et al. Callose deposition in the phloem plasmodesmata and inhibition of phloem transport in citrus leaves infected with ‘Candidatus Liberibacter asiaticus’. Protoplasma. 2012;249:687–697. doi: 10.1007/s00709-011-0312-3. [DOI] [PubMed] [Google Scholar]
- 63.Luo X, et al. Feeding Behavior of Diaphorina citri (Hemiptera: Liviidae) and Its Acquisition of ‘ Candidatus Liberibacter Asiaticus’, on Huanglongbing-Infected Citrus reticulata Leaves of Several Maturity Stages. Florida Entomol. 2015;98:186–192. doi: 10.1653/024.098.0132. [DOI] [Google Scholar]
- 64.Cen Y, et al. Feeding behaviour of the Asiatic citrus psyllid, Diaphorina citri, on healthy and huanglongbing-infected citrus. Entomol. Exp. Appl. 2012;143:13–22. doi: 10.1111/j.1570-7458.2012.01222.x. [DOI] [Google Scholar]
- 65.Wu T, et al. Feeding behavior of Diaphorina citri and its transmission of ‘ Candidatus Liberibacter asiaticus’ to citrus. Entomol. Exp. Appl. 2016;161:104–111. doi: 10.1111/eea.12496. [DOI] [Google Scholar]
- 66.Bonani JP, et al. Characterization of electrical penetration graphs of the Asian citrus psyllid, Diaphorina citri, in sweet orange seedlings. Entomol. Exp. Appl. 2010;134:35–49. doi: 10.1111/j.1570-7458.2009.00937.x. [DOI] [Google Scholar]
- 67.George J, Ammar E-D, Hall DG, Lapointe SL. Sclerenchymatous ring as a barrier to phloem feeding by Asian citrus psyllid: Evidence from electrical penetration graph and visualization of stylet pathways. PLoS One. 2017;12:e0173520. doi: 10.1371/journal.pone.0173520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pompon J, Quiring D, Goyer C, Giordanengo P, Pelletier Y. A phloem-sap feeder mixes phloem and xylem sap to regulate osmotic potential. J. Insect Physiol. 2011;57:1317–1322. doi: 10.1016/j.jinsphys.2011.06.007. [DOI] [PubMed] [Google Scholar]
- 69.Spiller NJ, Koenders L, Tjallingii WF. Xylem ingestion by aphids - a strategy for maintaining water balance. Entomol. Exp. Appl. 1990;55:101–104. doi: 10.1111/j.1570-7458.1990.tb01352.x. [DOI] [Google Scholar]
- 70.Taning CNT, Andrade EC, Hunter WB, Christiaens O, Smagghe G. Asian Citrus Psyllid RNAi Pathway – RNAi evidence. Sci. Rep. 2016;6:38082. doi: 10.1038/srep38082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh IK, Singh S, Mogilicherla K, Shukla JN, Palli SR. Comparative analysis of double-stranded RNA degradation and processing in insects. Sci. Rep. 2017;7:17059. doi: 10.1038/s41598-017-17134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spit J, et al. Knockdown of nuclease activity in the gut enhances RNAi efficiency in the Colorado potato beetle, Leptinotarsa decemlineata, but not in the desert locust, Schistocerca gregaria. Insect Biochem. Mol. Biol. 2017;81:103–116. doi: 10.1016/j.ibmb.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 73.Song H, et al. A double-stranded RNA degrading enzyme reduces the efficiency of oral RNA interference in migratory locust. Insect Biochem. Mol. Biol. 2017;86:68–80. doi: 10.1016/j.ibmb.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 74.Prentice K, et al. RNAi-based gene silencing through dsRNA injection or ingestion against the African sweet potato weevil Cylas puncticollis (Coleoptera: Brentidae) Pest Manag. Sci. 2017;73:44–52. doi: 10.1002/ps.4337. [DOI] [PubMed] [Google Scholar]
- 75.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 76.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 77.Fankhauser N, Maser P. Identification of GPI anchor attachment signals by a Kohonen self-organizing map. Bioinformatics. 2005;21:1846–1852. doi: 10.1093/bioinformatics/bti299. [DOI] [PubMed] [Google Scholar]
- 78.Mitchell A, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2014;43:D213–21. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.The UniProt Consortium UniProt: a hub for protein information. Nucleic Acids Res. 2014;43:D204–212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.la Cour T, et al. Analysis and prediction of leucine-rich nuclear export signals. Protein Eng. Des. Sel. 2004;17:527–536. doi: 10.1093/protein/gzh062. [DOI] [PubMed] [Google Scholar]
- 81.Sperschneider J, et al. LOCALIZER: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci. Rep. 2017;7:44598. doi: 10.1038/srep44598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Goldberg T, et al. LocTree3 prediction of localization. Nucleic Acids Res. 2014;42:W350–W355. doi: 10.1093/nar/gku396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lange M, Oliveira-Garcia E, Deising HB, Peiter E. A modular plasmid system for protein co-localization and bimolecular fluorescence complementation in filamentous fungi. Curr. Genet. 2014;60:343–350. doi: 10.1007/s00294-014-0429-y. [DOI] [PubMed] [Google Scholar]
- 84.Galdeano DM, Breton MC, Lopes JRS, Falk BW, Machado MA. Oral delivery of double-stranded RNAs induces mortality in nymphs and adults of the Asian citrus psyllid, Diaphorina citri. PLoS One. 2017;12:e0171847. doi: 10.1371/journal.pone.0171847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vale FXR, Fernandes Filho EI, Liberato JR, Zambolim L. Quant - A software to quantify plant disease severity. Int. Work. Plant Dis. Epidemiol. 2001;8:160. [Google Scholar]
- 87.Swift ML. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Comput. Sci. 1997;37:411–412. doi: 10.1021/ci960402j. [DOI] [Google Scholar]
- 88.Sarria E, Cid M, Garzo E, Fereres A. Excel Workbook for automatic parameter calculation of EPG data. Comput. Electron. Agric. 2009;67:35–42. doi: 10.1016/j.compag.2009.02.006. [DOI] [Google Scholar]
- 89.Verma, J. P. Data analysis in management with SPSS software. Data Analysis in Management with SPSS Software (Springer India, 2013).
- 90.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:research0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.