Abstract
Higher probability of the development of Crohn’s disease (CD) and ulcerative colitis (UC) as a possible consequence of the north-south gradient has been recently suggested. Living far north or south of the equator is manifested in fluctuation of vitamin D (vitD) levels depending on the season in both healthy and affected individuals. In the present study we investigate the possible link between the seasonal serum vitD level to the microbial composition of the lower gut of Inflammatory Bowel disease (IBD) patients using 16S rRNA sequencing. Decrease of serum vitD level in winter/spring season in a cohort of 35 UC patients and 39 CD patients was confirmed. Low gut microbiota composition of patients with IBD correlated with the serum level of 25(OH)D that directly coupled to seasonal variability of the sunshine in the central European countries. It is supposed to be related to increased abundance of Actinobacteria and Proteobacteria in UC and Actinobacteria, Fusobacteria, Firmicutes and Bacteroidetes in CD. In summer/autumn period, we observed a reduction in abundance of bacterial genera typical for inflammation like Eggerthella lenta, Fusobacterium spp., Bacteroides spp., Collinsella aerofaciens, Helicobacter spp., Rhodococcus spp., Faecalibacterium prausnitzii; and increased abundance of Pediococcus spp. and Clostridium spp. and of Escherichia/Shigella spp.
Subject terms: Metagenomics, Molecular medicine, Microbiome
Introduction
Inflammatory bowel disease (IBD) is a modern life-style disease with a worldwide prevalence. IBD involves mainly Crohn’s disease (CD) and ulcerative colitis (UC) the etiology of which differs, so the clinical determination of the disease is difficult. Chronic inflammation of the gastrointestinal tract defined as cycling of the acute inflammation phase and remission is typical for IBD, both CD and UC. While in UC the disease is specifically localized to the colon with different degree of continuous inflammation proximally from the rectum, the CD can manifest anywhere in the gut. To date, there is no clear evidence of a single factor causing the IBD. It is affected by genetic susceptibility, microbiome composition, immune response dysregulation as well as various environmental factors1.
The vitD defficiency is common among IBD patients2,3 and it has been speculated, that a low level of vitD might be one of the risk factors influencing the IBD4–6. The current epidemiological studies of IBD show and upward dynamics in countries distant from the equator7, strongly suggesting a correlation between IBD incidence and latitude. This observation is for instance supported by a study from New Zeland that has one of the highest rates of CD in the world4. Higher probability of the development of CD and UC8 and CD but not UC9 as a possible consequence of the latitude-dependent sunlight conditions has been supported by two more epidemiological studies, also reporting that the serum concentrations of vitD levels reached the lowest point after winter10,11. The seasonal variability of vitD level correlates with the seasonal pattern of IBD that peaks during the spring season12–14. On the other hand, living far north or south of the equator may cause a fluctuation in vitD levels depending on the season (the levels may go down during the winter months due to the lack of sufficient sunlight) also in healthy individuals15,16.
The positive effect of the vitD on human health is well known17,18, however, its effect on the gastrointestinal microbiome of IBD patients has not been properly elucidated19. The human microbiome represents a microecosystem, that is a consequence of mutualism of the host and the bacteria directly influence the health of an individual1. The disbalance of the microbiota can lead to dysbiosis of the gut that can mirror also in stool. Bacterial composition is said to play a key role in IBD development5,20. Some studies show quantitative reduction of certain bacteria in CD and UC patients compared to healthy individuals21–23, in others also qualitative differences in microbiome of the gut and fecal samples likely point to IBD24–26. On the contrary some studies reporting no changes in analysed microbiota have been also published27. In an open-label study Bashir et al.28 pointed to the positive effect of vitD supplementation on the microbiome composition in the upper part of the gastrointestinal tract (gastric corpus, antrum, duodenum), but not in terminal ileum, appendiceal orifice, ascending and sigmoid colon and in stools of healthy individuals.
Furthermore, the active metabolite of vitamin D3 exerts its regulatory function by binding to the vitD receptor (VDR). Regarding IBD pathogenesis, VDR expression at mRNA and protein levels is significantly decreased in IBD patients29. Of the 618 reported variants, the most common SNPs studied regarding various inflammatory based diseases are FokI, ApaI, BsmI and TaqI RFLP polymorphisms the association of that with UC or CD varies between populations. In order to avoid the possibility of the vitD level alterations linked to VDR genetic variability of Slovak population, the association between the most common polymorphisms of VDR and serum vitD level was determined30. Since there is still a discussion whether lack of vitD could trigger the microbial changes in IBD a need for a study focused on the bacterial composition characterization in connection with the serum vitD 25(OH)D status and season has emerged. Our group therefore decided to determine the impact of the vitD levels on the bacterial community of adult IBD patients.
In this paper, a metagenomic analysis of ileum and/or colon pinch biopsies from CD (47) and UC (40) patients is presented. To our knowledge, this is the first such study undertaken including samples from both inflamed as well as noninflamed mucosa and stool. The data from the shotgun next-generation sequencing of almost full-length 16S rRNA gene (V1-V6) were used for microbiome alpha-diversity determination and association of vitD and bacterial composition changes, answering the question whether the vitD levels can be related to the microbial composition and consequently the gut homeostasis in IBD patients.
Material and methods
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the local ethical committee of the Bratislava Self - Governing region, Slovak republic and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
The authors confirm that all methods and experimental protocols were performed in accordance with the relevant guidelines and regulations and were approved by the of the committee of the Ministry of Health of the Slovak republic.
Informed consent
Individuals were recruited between August 2012 and August 2015 by the IBD center of the Department of Internal Medicine, Division of Gastroenterology and Hepatology, University Hospital Bratislava, Slovak Republic. Informed consent was obtained from all individual participants included in the study. Additional informed consent was obtained from all individual participants for whom identifying information is included in this article.
Patients and sample collection
All patients, who met the criteria for Caucasian, aged more than 18 and diagnosed UC or CD, were implicated. The heterogenic group of recruited patients involved both genders (male, female), smokers and non-smokers, obese and lean, young and older people. Each patient was characterized by its demographic and clinical status regarding gender, age, duration of the disease, anatomic alternations (fistula, stenosis), surgery, distribution of the disease according to Montreal classification31, activity of the disease32, serum vitD level and vitD supplementation. In total, 87 patients were included in the study, 40 with an established diagnosis of UC and 47 with CD. Disease activity of individuals was assessed by Mayo score (0–3) for UC and CDAI index (0–352) for CD. 15 patients with active UC (Mayo score 2–3), 25 patients with inactive UC or in remission (Mayo score 0–1) were recruited together with 12 patients with active CD (CDAI > 150) and 35 patients with inactive CD (CDAI ≤ 150). Purposely selected patients for this study represent different clinically significant categories of IBD (Table 1).
Table 1.
Characteristics of the cohort of patients recruited in the study.
| Characteristics | UC (n = 40) | CD (n = 47) | |
|---|---|---|---|
| gender | female | 17 | 13 |
| male | 23 | 34 | |
| age (median ± STDEV) | 41 ± 13 (23–69) | 33 ± 12,4 (20–68) | |
| smoking | smoker | 8 | 9 |
| non-smoker | 32 | 38 | |
| vit.D supplementation | supplemented | 5 | 8 |
| non-supplemented | 35 | 39 | |
| CDAI | ≤150 | 35 | |
| >150 | 12 | ||
| MAYO | 0–1 | 25 | |
| 2–3 | 15 |
Sigmoidoscopy and/or colonoscopy using PENTAX 3885LK conventional white-light colonoscopes were performed in a group of patients. Biopsy and fecal samples were taken from patients undergoing routine examination. 3–5 days before examination the patient was instructed to minimalize the intake of heavily digestive food encompassing seeds, stones etc. The day before colonoscopy laxative agens (Fortrans) was applied. The biopsy sample collection was performed by sigmoidoscopy of mucosa from large intestine (CD, UC) as well as from terminal ileum of the small intestine (CD). Mucosal samples taken from macroscopically inflamed and non-inflamed areas of the gut using standard gape forceps were immediately frozen at −70 °C until further total DNA isolation. The stool samples were collected at home in the evening before the visit of the clinician and stored at −20 °C. The biopsy samples and feces were used for total DNA isolation.
The control group comprised 155 cadaveric organ donors registered in the biobank of National Transplant Organization, Bratislava.
VitD concentration assessment
Patients were recruited during two seasons of the year, summer/autumn period with high sunlight exposure, from August to October, included and winter-spring period with low sunlight exposure, from February to April, included. These periods were selected according to sunshine conditions in Central Europe; the periods met the criteria of the highest and the lowest expected vitD levels in the serum. Serum vitD, 25(OH)D2 and 25(OH)D3, levels were determined by high-performance liquid chromatography (HPLC, Agilent 1200) under specified conditions (UV detection at 264 nm, flow rate 1 ml/min, temperature 40 °C) for 10 min11.
Tissue and stool sample processing for next-generation library preparation
Total DNA isolation from a single pinch of the mucosa was carried out, after treatment with glass beads and enzymatic lysis, by Qiagen Blood and Tissue kit (Hilden, Germany) according to the manufacturer’s protocol. Briefly, mucosal sample was treated with 140 μl of lysing buffer (20 mM Tris-HCl, 2 mM EDTA, 1.2% Triton-X-100 in distilled water), 30 μl of mutanolysin (600 U), 20 µl of lysozyme and 10 µl of RNase (10 mg/ml). For sample homogenization glass beads (425–600 µm, Sigma) were added. Incubation at 37 °C for 40 min with regular brief vortexing every 10 min was followed by incubation with proteinase K and 200 µl of AL buffer at 56 °C for 1 h and final incubation at 70 °C for 30 min. After addition of 200 µl of absolute ethanol (stored at −20 °C), DNA was purified with QIAamp columns and eluted into 100 µl of ultra-clean millipore water.
For total DNA isolation from fecal samples, stored at −70 °C until use, QIAamp DNA Stool Mini Kit (Qiagen, Germany), was used. Briefly, 180–220 mg of frozen stool samples were homogenized after 1 min of vortexing in 1.4 ml of ASL lysis buffer. After heating to 90 °C for 5 min, vortexing for 15 s and centrifugation for 1 min at 20.817 g, 1.2 ml of supernatants were treated with InhibitEx tablet, vortexed for 1 min and incubated at room temperature for 1 min. Samples were then centrifuged at 20.817 g for 3 min. 200 µl of supernatants were incubated with 15 µl of proteinase K and 200 µl of AL lysis buffer at 70 °C for 10 min. DNA was precipitated with absolute ethanol (stored at −20 °C) and purified with QIAamp column according to manufacturer’s protocol. Concentration and purity of acquired DNA samples were assessed spectrophotometrically by NanoDrop ND-1000 (ThermoFisher Scientific, USA) and Qubit® 2.0 Fluorometer (ThermoFisher Scientific, Waltham, MA USA). Isolates were stored at −70 °C in the DNA bank.
PCR amplification of the 16 S rDNA region (V1–V6) was carried out in a volume of 20 μl using HotStartTaq Plus Master Mix (Qiagen, Germany), 0.5 μM forward primer 27 F (AGAGTTTGATCMTGGCTCAG) and 0.5 μM reverse primer 1062 R (ACAGCCATGCAGCACCT) and 30 ng of template DNA. PCR was performed using the following cycle conditions: an initial denaturation at 95 °C for 5 min, 35 cycles (denaturation at 94 °C for 1 min, annealing at 54 °C for 30 s, extension at 72 °C for 1 min 30 s) and final extension at 72 °C for 10 min. The cleanup of PCR products was carried out by ExoSAP-IT (Affymetrix, Santa Clara, CA) treatment. Final amplicon concentrations were determined using the Qubit Fluorometer (ThermoFisher Scientific, Waltham, MA USA) with Qubit dsDNA HS (High Sensitivity) Assay Kit (Life Technologies, Carlsbad, CA, USA).
Samples were diluted to 2.5 ng/μl in 20 μl volume to meet the requirements in initial step of library preparation. For the sample processing Nextera DNA library kit (Illumina, San Diego, CA, USA) was utilized and the procedure was carried out according to standard library protocol. Briefly, preamplified DNA samples were fragmented by transpozon to app. 300 bp sequences that were purified by DNA Clean & Concentrator™−5 columns (Zymo research, Irvine, CA, USA) and further amplified for 5 cycles by PCR using indexed forward and reverse primers. Size selection of the DNA fragments was carried out by magnetic beads Agencourt® AMPure® XP Reagent beads (Beckman Coulter, Brea, CA, USA). Each final amplicon library was checked for its quality and quantity using Agilent 2200 TapeStation (Agilent Technologies, USA) and Qubit Fluorometer. Amplicon libraries were pooled in equimolar concentration of 4 nM for sequencing run. For sequencing analysis MiSeq Sequencing kit v3 (Illumina, San Diego, CA, USA) was used. Paired end sequencing was carried out with 192 bp read length setting.
High-throughput data analysis
Data preprocessing
Adapters and low-quality ends of sequenced reads were removed using Trimmomatic33, based on quality control statistics generated by FastQC34. After trimming, only fragments with sufficient length of both reads (>35 bp) were kept for further analysis. Sequences were mapped to the human genome, version hg19 (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/bigZips/), using Bowtie235 with default parameters. Concordantly mapped reads were excluded from the further analysis to eliminate contamination from human tissue. We used prealigned set of 5,181 sequences (http://www.mothur.org/wiki/Silva_reference_files) from Silva database v11936 to identify and remove chimeric artefacts. Analysis were performed with tools Mothur37 and UCHIME38. Chimeric-free sequences were labeled with taxonomy from the full set of the Silva database (153,307 sequences) using Metaxa2 classifier39. Overall taxonomic composition of each sample was visualized in form of multi-level pie chart generated by Krona40.
Statistical analysis of metagenomic data
First, only samples matching filtering criteria were kept (for example, samples with the Crohn disease from Stool). Then, the linear discriminant analysis effect size (LEfSe)41 tool was used to identify taxons, which are populated differently according to presence of a certain target condition (for example abundance of vitD). In other words, if a taxon is populated significantly higher (or lower) in high vitD environment than in a low vitD environment, it will be identified and marked. The marked taxons were visualized in separate figures and in a cladogram42. This analysis was performed for various filtering criteria and target conditions.
ANOSIM is a permutation-based test of the null hypothesis that within-group distances are not significantly smaller than between-group distances43. Univariate Mann-Whitney and multivariate Anosim tests were used to assess significance of difference between taxonomic abundances under various conditions. Statistical analysis and visualization were performed using Python and R scripts. Nonparametric measure of rank correlation (Spearman’s rank correlation) was used to determine the statistical dependence between the ranking of two variables.
Results
Biological material
Seasonal sample collection of biopsy and stool samples was performed during the two-year period. During the low sunlight period, 42 biopsy samples from patients with UC and 101 samples from patients with CD were obtained. From the high sunlight period 56 biopsy samples from UC and 65 samples from CD patients were available. Based on actual endoscopic findings and the the origin of the biopsy, samples were divided into groups. Samples from CD patients were assessed: 1. inflamed sigma (14), 2. non-inflamed sigma (44), 3. Inflamed terminal ileum (26), 4. Non-inflamed terminal ileum (37). Since UC is exclusively a chronic inflammation of the colon, two groups of samples could be determined: 1. Inflamed sigma (14), 2. Non-inflamed sigma (44). Samples from 8 CD and 5 UC patients supplemented with vitD were excluded from the analysis based on serum vitD level. Stool samples were obtained from 40 UC and 45 CD patients. Additional information on characteristics of patients recruited in the study is included in Table 1.
VitD and the gut microbiome composition in IBD
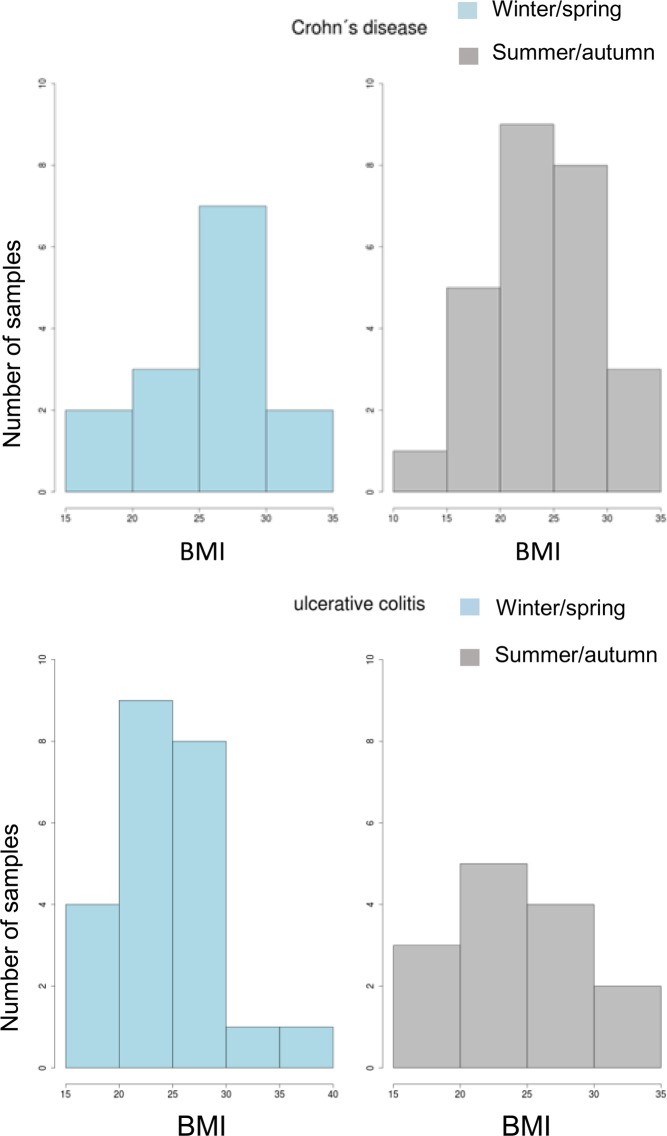
For the microbiome analysis each patient meeting the criteria was included in the study without categorization regarding gender, age or BMI. However, to exclude the possible influence of the BMI status of patients on the microbial composition of the gut in further analysis, we have tested the probability of the significance of distinction of BMI between cohorts of UC and CD patients in winter/spring and summer/autumn. No significant differences could be detected between IBD patients during any season (CD, p = 0.36; UC, p = 0.97) (Fig. 1).
Figure 1.
BMI of the cohort of IBD patients involved in the study.
Ulcerative colitis
Correlation of serum vitamin D level with the season
To determine the seasonal variation of serum vitD level, samples were classified into two seasons: winter/spring and summer/autumn. The serum vitD concentration varied among the seasons, winter/spring (in average 25.05 ng/ml) and summer/autumn period (in average 37.26 ng/ml). Without further categorization of samples according to other variables (gender, smoking, age) during winter/spring season the serum vitD level was significantly lower than in summer/autumn season (p = 7.163e-06) (Fig. 2).
Figure 2.
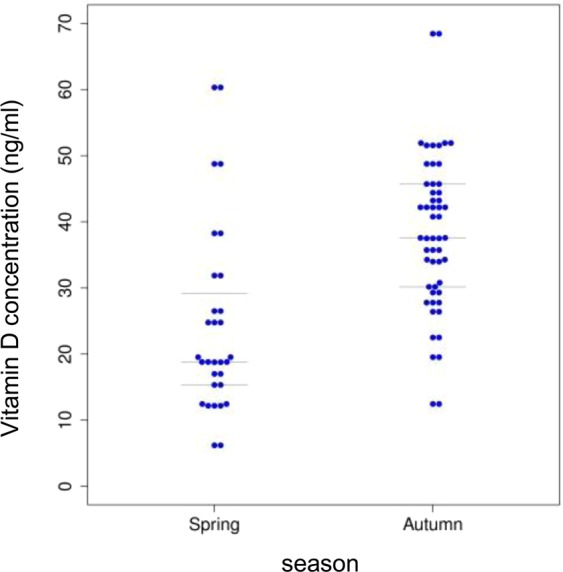
Serum vitD level determined in the cohort of UC patients in relation to the season (winter/spring, summer/autumn).
VitD level correlation to microbiome of patients with Ulcerative colitis
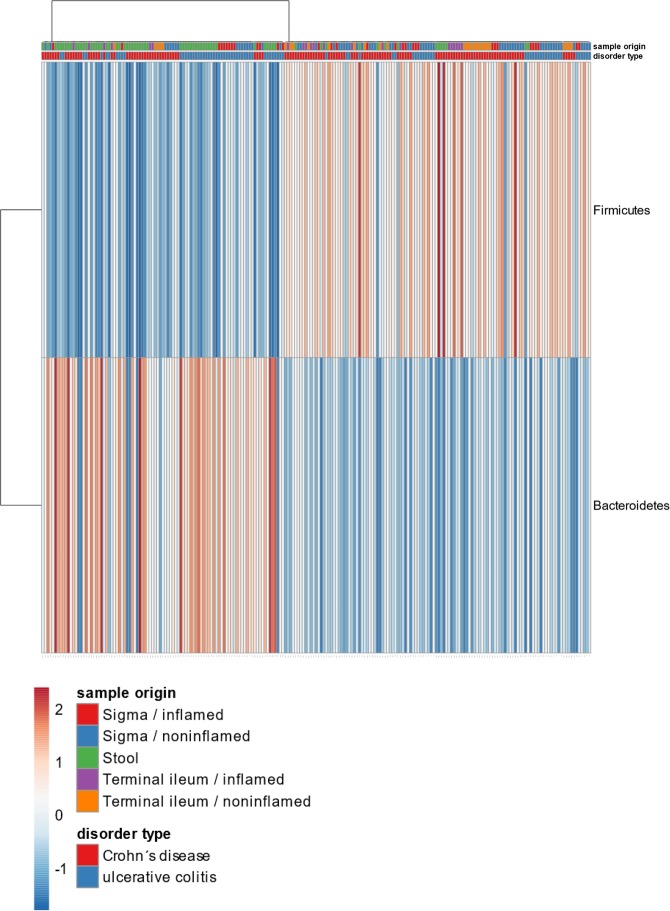
Regarding the microbiome the dominating phylum of the sigmoid mucosal samples either noninflamed (NI) or inflamed (I) was Firmicutes 49,9% (53,9%/43,4%). Bacteroidetes represented 34,3% (35,3%/32,7%) and Proteobacteria app. 12% (7,5%/19,3%). Actinobacteria were the fourth most abundant phylum 3,6% (3,1%/4,4%). The microbiome of stool samples corresponded with mucosal samples in the four most abundant phyla, although the relative amount of Firmicutes was lower (23,2%). Here Bacteroidetes was the most abundant phylum (63,1%) and Proteobacteria (12,3%) and Actinobacteria (0,5%) formed the least abundant proportion of the analyzed microbiome (Fig. 3). The observed difference in the proportion of Firmicutes and Bacteroidetes in mucosal samples and stool was investigated and the diversity of samples was visualized by heatmap (Fig. 4). In most stool samples Bacteroidetes were more abundant than Firmicutes.
Figure 3.
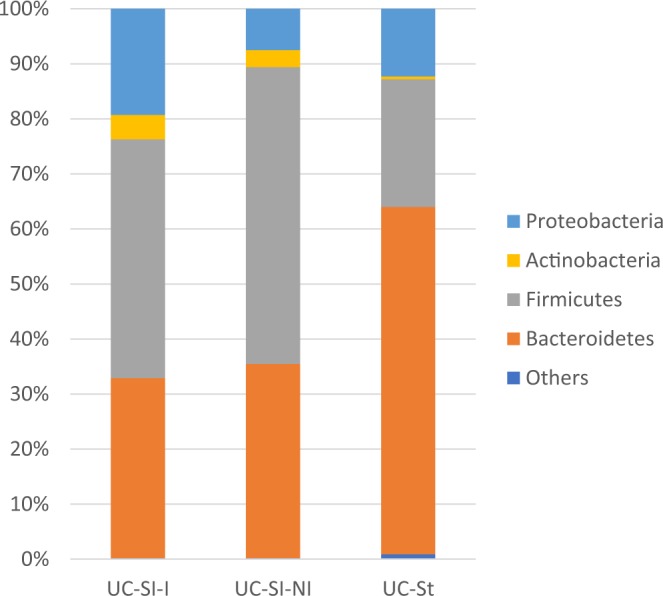
The proportional representation of the most abundant bacterial phyla in human lower gut (sigma) and stool within the cohort of patients with ulcerative colitis involved in the study. UC – ulcerative colitis, Si – sigma, St – stool, I – inflamed, NI –noninflamed.
Figure 4.
Heatmap visualization of Firmicutes and Bacteroidetes proportion in mucosal and fecal samples. Almost all stool samples (Crohn’s disease, ulcerative colitis) are dominated by Bacteroidetes, while mucosal samples by Firmicutes.
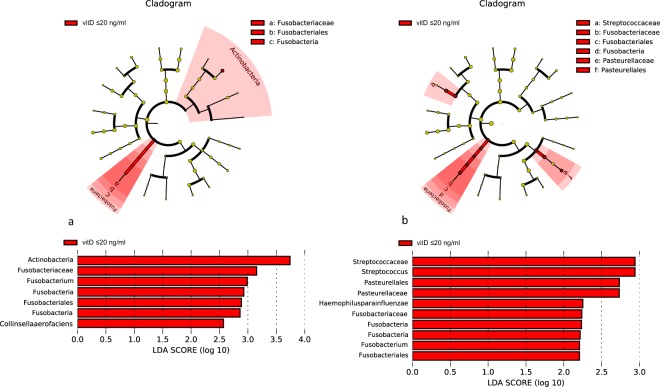
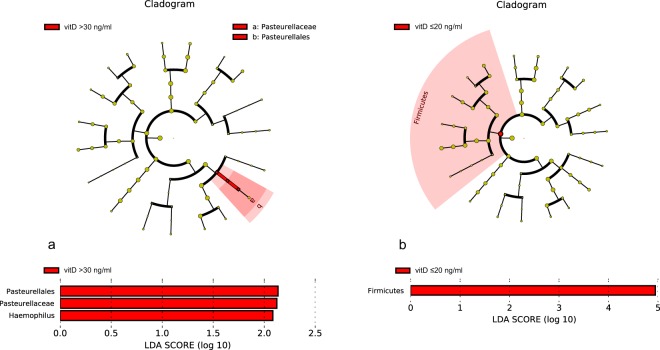
Regarding the serum vitD concentration, group A ≤ 20 ng/ml (low), group B 20–30 ng/ml (medium) and group C ≥ 30 ng/ml (high) serum vitD concentration, the microbiome of sigma (both inflamed and noninflamed) and stool was compared. In the cohort of UC patients with low vitD level (≤20 ng/ml) increase of Fusobacteria, Streptococcaceae and Pasteurellaceae represented by Haemophillus parainfluenzae was detected (Fig. 5b) in inflamed sigma samples. In noninflamed tissue increase of Actinobacteria followed by Fusobacteria, especially Fusobacterium spp. but also of Collinsella aerofaciens was detected, too (Fig. 5a). Microbiome analysis of stool samples haven’t revealed any significant changes.
Figure 5.
Lefse visualization of significantly altered microbiome taxa of sigmoid tissue of UC patients with low level of serum vitD (≤20 ng/ml; group A). Noninflamed (a) and inflamed (b) samples are shown. The level of significance ≤0.05 was applied.
Exposure to sun has been reported as a factor directly influencing the level of vitD and consequently possibly also the gut microbiome. To assess the influence of the seasonal serum vitD level, mucosal tissue and stool samples collected in winter/spring and summer/autumn period were examined for microbiome composition.
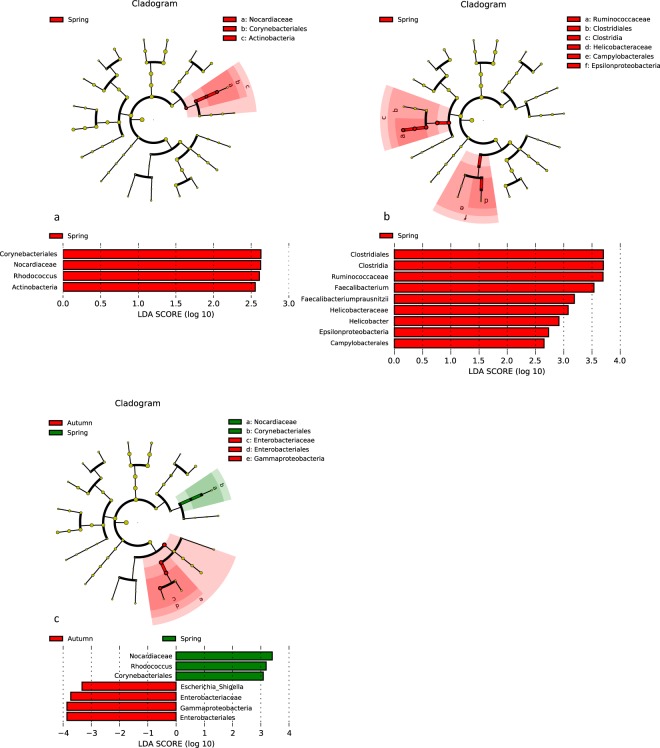
During the winter/spring period higher level of Proteobacteria, esp. Campylobacteralles and Helicobacteraceae in inflamed sigma (Fig. 6b); but more increased Actinobacteria, especially Corynebacteriales, Nocardiaceae and Rhodococcus were assessed in noninflamed sigma (Fig. 6a). The significant changes in the fecal microbiome correlated with the status of the healthy, noninflamed tissue of the patient during the same period. The sequencing of the bacteria from stool samples collected during summer/autumn period showed almost 4-fold increase of Gammaproteobacteria represented by Escherichia/Shigella (Fig. 6c).
Figure 6.
Lefse visualization of significantly altered microbiome taxa of sigmoid tissue of UC patients in winter/spring period in comparison to summer/autumn period. Noninflamed (a) and inflamed (b) sigma, stool (c) samples are shown. The level of significance ≤0.05 was applied.
To check the microbial composition of the inflamed and the noninflamed analyzed tissue, comparison of two sets of samples regarding their inflammation status and the collection period was carried out. Interestingly, during the winter/spring season, no significant differences in the beta diversity of inflamed and noninflamed samples were detected. However, healthy noninflamed samples from the summer/autumn period showed higher abundance of the representatives of the phylum Firmicutes (data not shown).
Crohn’s disease
Correlation of serum vitamin D level with the season
The seasonal variability of the serum vitD level in blood of CD patients was assessed and significant correlation of the two-season period (winter/spring, summer/autumn) was found. The serum vitD level varied among the seasons, winter/spring (20.23 ng/ml) and summer/autumn period (34.72 ng/ml) significantly (p ≤ 0.05). Without further categorization of samples according to other variables (gender, smoking, age) during winter/spring season the serum vitD level was significantly lower than in summer/autumn season (p = 7.837e-10) (Fig. 7).
Figure 7.
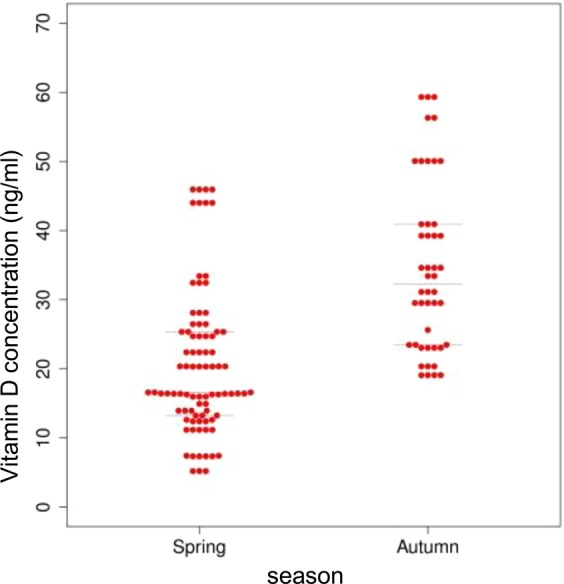
Serum vitD level estimated in the cohort CD patients in relation to the season (winter/spring, summer/autumn).
Analysis of the vitD correlation to microbiome of patients with Crohn’ disease
The microbiome analysis of mucosal tissue of the GIT (sigma, terminal ileum) revealed the same profile of main bacterial phyla in both noninflamed and inflamed samples. Presence of the most abundant phylum Firmicutes in sigma 52,6% (54,4%/47,2%) and terminal ileum 54,6% (53,1%/57,0%), followed by Bacteroidetes in sigma 29,1% (28,4%/31,4%); terminal ileum 26,5% (26,7%/25,3%), Proteobacteria in sigma 12,2% (11,7%/13,8%); terminal ileum 11,9% (12,9%/10,3%) and Actinobacteria in sigma 3,8% (3,4%/4,8%); terminal ileum 5,5% (5,9%/4,9%). A remarkable difference could be seen between the most abundant phyla of mucosal tissue and stool samples; here Bacteroidetes was the most abundant taxon (39,7%), followed by Firmicutes (24,8%) and Proteobacteria (27,1%) with app. 1:1 ratio. Actinobacteria formed only 0,7% of the total amount of analyzed bacteria (Fig. 8).
Figure 8.
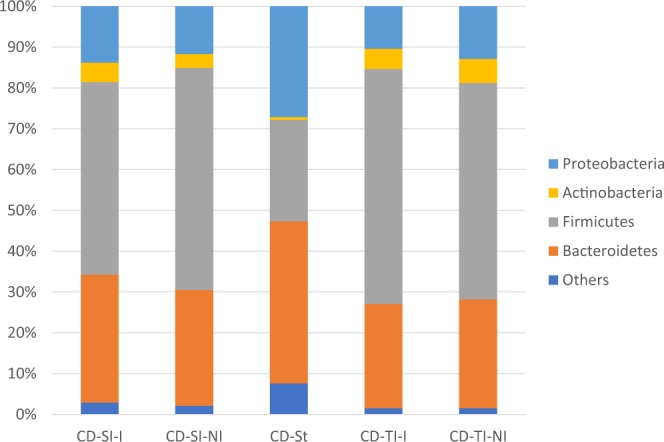
The proportional representation of the most abundant bacterial phyla in human lower gut (sigma, terminal ileum and stool) within the cohort of patients with Crohn’s disease involved in the study. CD – Crohn’s disease, Si – sigma, TI – terminal ileum, St – stool, I – inflamed, NI –noninflamed.
A search for significant change in microbiome possibly correlating with the level of serum vitD was observed as well as by UC patients. The same categorization criteria were applied (Hlavaty et al., 2014). The complex microbiome analysis of terminal ileum revealed no significant changes in any inflammation state. However, in the group of patients with low vitD level an increase of Firmicutes in the inflamed sigma was observed. On the contrary, high level of vitD significantly correlated with increase of Haemophilus spp. in stool (group C) (Fig. 9).
Figure 9.
Lefse visualization of significantly altered microbiome taxa of stool (a) and inflamed sigmoid tissue (b) of patients with Crohn’s disease in relation to vitD level. The level of significance ≤0.05 was applied.
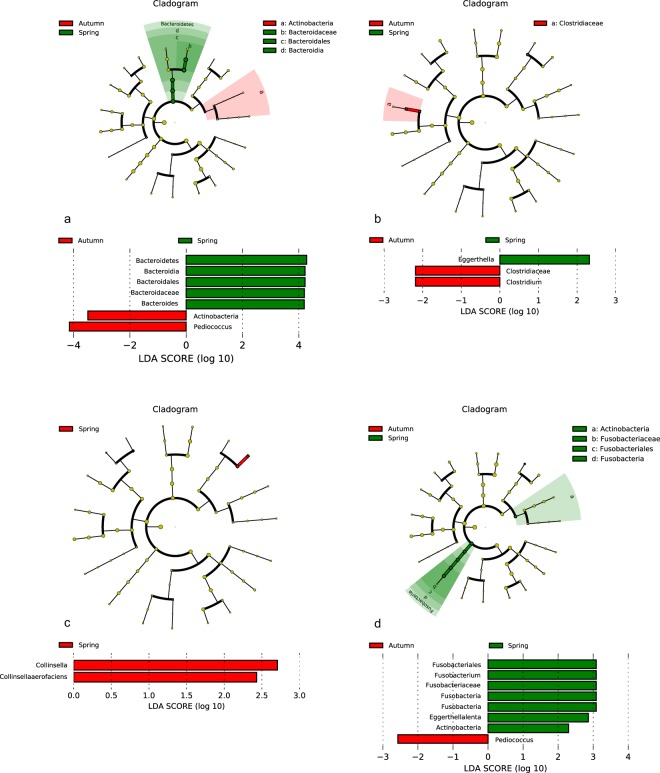
Regarding the exposition of individuals to sun the correlation of the seasonal vitD level and the bacterial composition could be observed. In the summer/autumn period significant decrease of Actinobacteria (especially Eggerthella lenta) in sigma (inflamed and noninflamed) and of Fusobacteria (Fusobacterium) (noninflamed sigma) was detected. Furthermore, higher abundance of Clostridium spp. (inflamed sigmoid tissue) and Pediococcus spp. (noninflamed sigma) was detected in lower gut region. On the contrary in stool samples increase of Actinobacteria and of Pediococcus spp. (Firmicutes) accompanied by decrease of Bacteroides spp. has been detected. A seasonal decrease in abundance of Collinsella aerofaciens in the noninflamed terminal ileum was observed, too (Fig. 10).
Figure 10.
Lefse visualization of significantly altered microbiome taxa of stool (a) and mucosal samples of lower gut regarding season. (a) stool sample; (b) inflamed sigma, (c) noninflamed terminal ileum, (d) noninflamed sigma of patients with Crohn’s disease. The level of significance ≤0.05 was applied.
Discussion
Inflammatory bowel disease comprises a group of heterogenous and complex disorders with chronic inflammation. The most common and the most studied are the Crohn’s disease and ulcerative colitis that are known also for their microbial disbalance in the gastrointestinal tract. The positive effect of a therapy using vitD have been proved in several studies. However, little is known about the season-dependent vitD fluctuation in Central European region that can have an impact on the lower gut bacterial composition. In this study we detected the association between the level of serum vitD (25(OH)D2 and 25(OH)D3) in the blood of patients with IBD excluding the possible interventions of the most common SNPs in the VDR receptor and the microbiome alternation of the gastrointestinal tract (GIT). Another important observation was the significant change of seasonal serum vitD level in IBD patients during winter/spring and summer/autumn period that could be related to the bacterial composition of the GIT in IBD patients. In addition, the microbiome composition was followed also in stool were distinct changes of bacterial taxa regarding altered vitD level were found.
Most of the cells in the healthy human body are bacteria of that the dominating phyla are in favor of Bacteroidetes, Firmicutes44 and Actinobacteria45. However, intestinal inflammation can disturb this balanced community and a reduction of strict anaerobes is accompanied with increase of Proteobacteria, mainly Enterobacteriaceae family24. IBD patients involved in our study possessed the characteristics of typical microbial composition in which the Firmicutes and Bacteroidetes are dominating taxa, followed by Proteobacteria and Actinobacteria both in mucosa as well as fecal samples. Whether stool can be used as a reliable marker for gut dysbiosis is still not clear enough. Although the Firmicutes to Bacteroidetes (F/B) ratio in mucosal samples compared to stool was not significantly altered, vitD level significantly selectively influenced its alfa diversity. Despite several papers showed differences in the composition of the human intestinal and fecal microbiome44,46,47, there are studies using the fecal microbiota as a benchmark for the changes of the upper or lower gut microbiome48.
Since some studies point to the obesity as an important factor affecting microbiome composition27, we investigated its potential influence on our study. Balanced BMI among IBD patients during the two-year period did not indicate any significant shift in the F/B ratio, therefore the obesity could be excluded as a variable with significant impact on our results.
In our study we could confirmed the phenomenon of the seasonality of the vitD level in blood of IBD patients, both CD and UC. During winter/spring season the concentration of serum vitD in UC and CD patients was lower than in summer/autumn period; furthermore, the lack of vitD in CD patients was more severe. Compared to multiethnic study of49 Chatu et al. in which Caucasians examined from January to June, possessed 41 nmol/L (app. 16 ng/ml) of vitD considered as vitD insufficiency, in the cohort of Slovak Caucasians examined during winter/spring period still higher level of vitD was investigated (CD −20.23 ng/ml, UC 25.05 ng/ml). So far there is one more study of Caucasian IBD population involved in larger cohort of patients50 regarding influence of vitD however the vitD level of exclusively Caucasians was not estimated.
The wide positive effect of vitD on human health (cancer, heart disease, diabetes, osteoporosis…) has been extensively investigated and a couple of studies focused on IBD as well. In previous studies positive influence of vitD on UC patients was detected5,51, but on the contrary no correlation between the vitD level and the disease activity could be found52,53. Our work assessed significant changes in the lower gut microbiome of both UC and CD patients in relation to the seasonal change of sunlight. The key observation in CD cohort is the drop in the level of Actinobacteria that is in line with the study on multiple sclerosis54. This effect was associated with both, inflamed as well as noninflamed lower gut tissue. Since increased level of Actinobacteria in CD patients was established to be associated with the disease47,55, obtained results point to the improvement of the gut microbiome. Interesting is the counter status observed within fecal microbiome what indicates that at this stage stool is not suitable mirror of the diseased mucosa status.
In the light of previous study which also reported increased abundance of Fusobacteriaceae and decreased level of Bacteoidales56, the opposite trend observed in our study (decrease of Fusobacteria and increase of Bacteroides spp.) suggests improvement in the microbiome homeostasis. There are also studies reporting increased24 or not significantly altered48,57 level of Bacteroides in biopsy samples, however, the discrepancy could be explained by various factors: age, diet, disease activity etc.
Patients suffering from UC are commonly vitD deficient58,59. According to the results of previous studies47,55 increased level of Actinobacteria is typical for UC. This observation correlates with the increase of Rhodococcus spp., Helicobacter pylori and Campylobacteralles, that are associated with the dysbiosis in UC25 and is a characteristic feature of winter/spring period. The current findings regarding Escherichia/Shigella group are controversial; according to Pascal et al.60 it is almost not detected in UC, however certain phylotypes harboring pathogenicity factors ompA, afae and USP belong to more prevalent pathogens in UC25. The effect of vitD on the penetration of virulent E. coli strains was proved in an in vitro study on Caco cells and although it was sufficient to increase clinical and histological parameters of the inflammation, by itself could not sufficiently induce adherent-invasive Escherichia coli (AIEC) strain LF82 induced gut injury61. In our study no significant changes of Escherichia spp. abundance in the sigmoid part of the colon could be observed as well as in healthy volunteers supplemented by vitD29, and but 4-fold increase of Escherichia/Shigella genera in fecal samples in the summer/autumn season was detected. This observation is in accordance with the study of 62 in that supplementation of UC patients with vitD lead to no overall fecal microbiome differences, but significant increase of Enterobacteriaceae. It is indicated that differences in microbiome composition and individual taxa abundances are pronounced in fecal and mucosal samples63. So far higher abundance of Escherichia/Shigella in stool has been associated rather with a diet rich in animal proteins and saturated fats64, that represents a suitable environment for Escherichia spp.65 or together with increased Fusobacterium as a marker for CD-type IBD66. To our knowledge there has been no study describing such an increase of Escherichia in fecal samples associated with seasonal increase of vitD.
The effect of the exposure to the sunlight was estimated by significant decrease of abundance of Actinobacteria also in CD although in UC only the noninflamed tissue was associated with this change. Furthermore, decrease in Proteobacteria, that were proved to be elevated in IBD48,67 and more abundant in inflamed UC that inflamed CD68 was detected, too. Here also the opposite effect of increase of Proteobacteria in stool could be observed. More detailed investigation revealed Helicobacter spp. as the only representative of Epsilonproteobacteria decreased in tissue samples69, while higher abundance of Escherichia/Shigella spp. (Gammaproteobacteria) was detected in stool. Interesting observation regarding decrease in abundance of Clostridiales, especially Faecalibacterium prausnitzii and Ruminococcaceae was associated with the inflamed biopsy of UC patients what correlates with previous study62. Reduction of Faecalibacterium is set to be primarily associated with ileal CD70 and decrease of Firmicutes with lower level of vitD58. In the study of Machiels et al.21 with 127 UC patients, also inverted association with the disease activity was detected. However, according to71 Prideaux et al. ethnicity could be a key factor that might play a role in the Faecalibacterium dysbiosis. The influence of the inflammatory status of the tissue in the terms of microbiome changes remains not fully elucidated. Some significant changes in the microbiome composition were assigned to the noninflamed, others to the inflamed part of the intestine. However, there are contradictory findings reporting no significant differences between the status of inflammation within the disease68 as well as significant differences in the composition of the mucosal tissue24. This could indicate that there must be other factor that selectively influences the microbiome composition, that may influence the vitD accessibility in pathologically altered parts of the intestine.
In our study, lower levels of vitD in winter/spring season are rather associated with more balanced microbiome composition in IBD, higher abundance of Faecalibacterium prausnitzii in an intestine and lower level of Escherichia/Schigella in stool of UC patients. Although in CD higher vitD level was associated with lower proportion of Actinobacteria and pathogens like Eggerthella lenta and Fusobacterium spp., the microbiome composition in winter/spring season was more favorable for lower proportion of Clostridium spp., higher proportion of Clostrida (Firmicutes) in mucosa and increased level of Bacteroidetes in stool.
Investigation of the relationship between the serum vitD level and the microbiome composition of IBD patients revealed increased proportion of Pasteurellales in fecal samples of patients with hypervitaminosis (vitD over 30 ng/ml) and higher abundance of Haemophillus spp. Interestingly, lower level of Firmicutes was detected in mucosal samples from inflamed sigma in the cohort of patients with normal vitD level.
In contrast to the previous statements that for patients with CD decrease in Firmicutes to Bacteroidetes ratio is typical72, in our study, the decreased serum vitD level was associated with more favorable microbiome of the gut of IBD patients.
The Human Microbiome Project showed that besides the fact that there is a set of common microorganisms present in human gut, there are also significant intrapopulation and interindividual differences. According to Conlon and Bird64 one of the main factors contributing to shifts in the composition and ratios of bacterial taxa could be a diet. Another study correlates high fat diet with increase of Firmicutes73. However, the results of individual studies are not consistent. The discrepancies might be explained by sample source (biopsy or stool), sampling location (inflammatory or noninflammatory sites), disease activity (active or quiescent), medication, diet, age, smoking, and methods used to analyse the microbiota74. Since also our study was focused on the effect of the sunshine period on the microbiome status of UC and CD patients, only patients who visited the IBD center during the selected period (August–October, February–April) were screened. During this period 220 IBD patients (79 UC and 141 CD) visited the IBD center. For 116 IBD patients (141 CD patients and 79 UC patients) the serum vitD level in both seasons was eligible. Since CD as well as UC are very heterogeneous diseases it was very difficult to obtain homogenous groups of patients, even more sets of samples. Many variables considering gender, age, smoking, therapy, supplementation, medication, IBD related surgeries, duration of the disease, disease location and disease behavior have been considered. Finally, 87 IBD patients (47 CD patients and 40 UC patients) were involved, of that samples from 8 CD and 5 UC patients supplemented with vitD were excluded. These are the limits of our study; however, we believe that precise cohort selection can help to clarify and improve the obtained results and emphasize their value.
Nevertheless, the Slovak population of Caucasian origin suffering from IBD, similarly to other geographically distinct populations, show decrease in the level of serum vitD. However, its effect on the gut microbiota composition is more prominent at the genus level differences, rather than at their overall abundancies.
Acknowledgements
This work was supported by projects APVV–0672-11, APVV-17-0099 and Comenius University Science Park in Bratislava (ITMS: 26240220086).
Author contributions
K.S. performed the high-throughput sequencing, M.S. conducted the laboratory work, T.H., H.D. and S.S. designed the research, J.B. bioinformatically processed the sequencing data, J.G. performed statistical analysis, B.G. performed DNA isolation, T.H. and Z.Z. performed colonoscopy and A.K. provided vitamin D level measurements, T.S., J.T. and D.K. helped in the analysis of the data sets. K.S. wrote the article. All authors discussed and commented on the final version of the manuscript.
Data availability
Results of all analyses are included in this published article. The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sun J, Chang EB. Exploring gut microbes in human health and disease: Pushing the envelope. Genes Dis. 2014;1:132–139. doi: 10.1016/j.gendis.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilman J, Shanahan F, Cashman KD. Determinants of vitamin D status in adult Crohn’s disease patients, with particular emphasis on supplemental vitamin D use. Eur. J. Clin. Nutr. 2006;60:889–896. doi: 10.1038/sj.ejcn.1602395. [DOI] [PubMed] [Google Scholar]
- 3.Blanck S, Aberra F. Vitamin D deficiency is associated with ulcerative colitis disease activity. Dig. Dis. Sci. 2013;58:1698–1702. doi: 10.1007/s10620-012-2531-7. [DOI] [PubMed] [Google Scholar]
- 4.Carvalho AYOM, et al. The role of vitamin D level and related single nucleotide polymorphisms in Crohn’s disease. Nutrients. 2013;5:3898–3909. doi: 10.3390/nu5103898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ananthakrishnan AN. Environmental Risk Factors for Inflammatory Bowel Disease. Gastroenterol. Hepatol. 2013;9:367–374. [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao, J., Wang, Y., Gu, Q., Du, Z. & Chen, W. The association between serum vitamin D and in fl ammatory bowel disease. Medicine (Baltimore). 98, 10.1097/MD.0000000000015233 (2019). [DOI] [PMC free article] [PubMed]
- 7.Molodecky NA, et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42. doi: 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 8.Jussila A, et al. High and increasing prevalence of inflammatory bowel disease in Finland with a clear North-South difference. J. Crohn’s Colitis. 2013;7:e256–e262. doi: 10.1016/j.crohns.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Jantchou P, et al. High residential sun exposure is associated with a low risk of incident Crohn’s disease in the prospective E3N cohort. Inflamm. Bowel Dis. 2014;20:75–81. doi: 10.1097/01.MIB.0000436275.12131.4f. [DOI] [PubMed] [Google Scholar]
- 10.Bours PHA, Wielders JPM, Vermeijden JR, Van De Wiel A. Seasonal variation of serum 25-hydroxyvitamin D levels in adult patients with inflammatory bowel disease. Osteoporos. Int. 2011;22:2857–2867. doi: 10.1007/s00198-010-1484-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hlavaty T, et al. Higher vitamin D serum concentration increases health related quality of life in patients with inflammatory bowel diseases. World J. Gastroenterol. 2014;20:15787–15796. doi: 10.3748/wjg.v20.i42.15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aratari A, et al. Seasonal variations in onset of symptoms in crohn’s disease. Dig. Liver Dis. 2006;38:319–23. doi: 10.1016/j.dld.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Lewis J, Aberra F, Lichtenstein G. Seasonal variation in flares of inflammatory bowel disease. Gastroenterology. 2004;126:665–73. doi: 10.1053/j.gastro.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 14.Tysk, C. & Järnerot, G. Seasonal Variation in Exacerbations of Ulcerative Colitis. Scand. J. Gastroenterol. 95–96, 10.3109/00365529309096052 (1993). [DOI] [PubMed]
- 15.Gozdzik A, et al. Serum 25-Hydroxyvitamin D Concentrations Fluctuate Seasonally in Young Adults of Diverse Ancestry Living in Toronto. J. Nutr. 2010;140:2213–2220. doi: 10.3945/jn.110.126284. [DOI] [PubMed] [Google Scholar]
- 16.Holick MF. Vitamin D: A millenium perspective. J. Cell. Biochem. 2003;88:296–307. doi: 10.1002/jcb.10338. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen, O. H., Rejnmark, L. & Moss, C. Role of Vitamin D in the Natural History of Inflammatory Bowel Disease 742–752, 10.1093/ecco-jcc/jjy025 (2018). [DOI] [PubMed]
- 18.Schardey, J. et al. Vitamin D Inhibits Pro-Inflammatory T Cell Function in Patients With Inflammatory Bowel Disease. 1–12, 10.1093/ecco-jcc/jjz090 (2019). [DOI] [PubMed]
- 19.Gominak SCMD. Vitamin D deficiency changes the intestinal microbiome reducing B vitamin production in the gut. The resulting lack of pantothenic acid adversely affects the immune system, producing a “pro-inflammatory” state associated with atherosclerosis and autoimmunity. Med. Hypotheses. 2016;94:103–107. doi: 10.1016/j.mehy.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 20.Ryz, N. R. et al. Dietary vitamin D3 deficiency alters intestinal mucosal defense and increases susceptibility to Citrobacter rodentium -induced colitis. 730–742, 10.1152/ajpgi.00006.2015 (2019). [DOI] [PMC free article] [PubMed]
- 21.Machiels K, et al. A decrease of the butyrate-producing species roseburia hominis and faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut. 2014;63:1275–1283. doi: 10.1136/gutjnl-2013-304833. [DOI] [PubMed] [Google Scholar]
- 22.Swidsinski A, Loening-Baucke V, Vaneechoutte M, Doerffel Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 2008;14:147–161. doi: 10.1002/ibd.20330. [DOI] [PubMed] [Google Scholar]
- 23.Sanderson J, et al. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011;11:7. doi: 10.1186/1471-2180-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sasaki M, Klapproth J-MA. The Role of Bacteria in the Pathogenesis of Ulcerative Colitis. J. Signal Transduct. 2012;2012:1–6. doi: 10.1155/2012/704953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martín-de-Carpi J, et al. Increasing incidence of pediatric inflammatory bowel disease in Spain (1996–2009): The SPIRIT registry. Inflamm. Bowel Dis. 2013;19:73–80. doi: 10.1002/ibd.22980. [DOI] [PubMed] [Google Scholar]
- 26.Walters W, Xu Z, Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;17:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jess T, et al. Enteric Salmonella or Campylobacter infections and the risk of inflammatory bowel disease. Gut. 2011;60:318–324. doi: 10.1136/gut.2010.223396. [DOI] [PubMed] [Google Scholar]
- 28.Bashir M, et al. Effects of high doses of vitamin D3 on mucosa-associated gut microbiome vary between regions of the human gastrointestinal tract. Eur. J. Nutr. 2016;55:1479–1489. doi: 10.1007/s00394-015-0966-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada K, et al. Vitamin D receptor expression is associated with colon cancer in ulcerative colitis. Oncol. Rep. 2009;22:1021–1025. doi: 10.3892/or_00000530. [DOI] [PubMed] [Google Scholar]
- 30.Stuchlíková, M. et al. The relationship between selected VDR gene polymorphisms and susceptibility to inflammatory bowel disease in Slovak population. Biologia (Bratisl), 10.2478/s11756-019-00212-3 (2019).
- 31.Dignass A, et al. Second European evidence-based Consensus on the diagnosis and management of ulcerative colitis Part 1: Definitions and diagnosis. Rev. Gastroenterol. Mex. 2014;79:263–289. doi: 10.1016/j.rgmx.2014.10.001. [DOI] [PubMed] [Google Scholar]
- 32.D’Haens G, et al. A Review of Activity Indices and Efficacy End Points for Clinical Trials of Medical Therapy in Adults With Ulcerative Colitis. Gastroenterology. 2007;132:763–786. doi: 10.1053/j.gastro.2006.12.038. [DOI] [PubMed] [Google Scholar]
- 33.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andrews, S. FastQC: a quality control tool for high throughput sequence data. 175–176 (2010).
- 35.Langmead B, Slazberg SL. Fast gapped-read alignmnet with Bowtie 2. Nat. Methods. 2013;9:357–359. doi: 10.1038/nmeth. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quast C, et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bengtsson-Palme J, et al. metaxa2: Improved identification and taxonomic classification of small and large subunit rRNA in metagenomic data. Mol. Ecol. Resour. 2015;15:1403–1414. doi: 10.1111/1755-0998.12399. [DOI] [PubMed] [Google Scholar]
- 40.Ondov BD, Bergman NH, Phillippy AM. Interactive metagenomic visualization in a Web browser. BMC Bioinformatics. 2011;12:385. doi: 10.1186/1471-2105-12-385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paulson JN, Stine OC, Bravo HC, Pop M. Robust methods for differential abundance analysis in marker gene surveys. Nat. Methods. 2014;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clarke KR, et al. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993;18:117–143. doi: 10.1111/j.1442-9993.1993.tb00438.x. [DOI] [Google Scholar]
- 44.Eckburg PB, et al. Diversity of the Human Intestinal Microbial Flora. Science (80-.) 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Simreń M, et al. Intestinal microbiota in functional bowel disorders: A Rome foundation report. Gut. 2013;62:159–176. doi: 10.1136/gutjnl-2012-302167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lepage P, et al. Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 2005;11:473–80. doi: 10.1097/01.MIB.0000159662.62651.06. [DOI] [PubMed] [Google Scholar]
- 47.Morgan XC, et al. Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 2012;13:R79. doi: 10.1186/gb-2012-13-9-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang W, et al. Increased proportions of Bifidobacterium and the Lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J. Clin. Microbiol. 2014;52:398–406. doi: 10.1128/JCM.01500-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chatu S, et al. Factors associated with vitamin D deficiency in a multicultural inflammatory bowel disease cohort. Frontline Gastroenterol. 2013;4:51–56. doi: 10.1136/flgastro-2012-100231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fu YTN, Chatur N, Cheong-Lee C, Salh B. Hypovitaminosis D in adults with inflammatory bowel disease: Potential role of ethnicity. Dig. Dis. Sci. 2012;57:2144–2148. doi: 10.1007/s10620-012-2130-7. [DOI] [PubMed] [Google Scholar]
- 51.Tan B, et al. Vitamin D levels and bone metabolism in Chinese adult patients with inflammatory bowel disease. J. Dig. Dis. 2014;15:116–123. doi: 10.1111/1751-2980.12118. [DOI] [PubMed] [Google Scholar]
- 52.Ulitsky A, et al. Vitamin D deficiency in patients with inflammatory bowel disease: association with disease activity and quality of life. JPEN J Parenter Enter. Nutr. 2011;35:308–16. doi: 10.1177/0148607110381267. [DOI] [PubMed] [Google Scholar]
- 53.Dumitrescu G, Mihai C, Dranga M, Prelipcean CC. Serum 25-hydroxyvitamin D concentration and inflammatory bowel disease characteristics in Romania. World J. Gastroenterol. 2014;20:2392–2396. doi: 10.3748/wjg.v20.i9.2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mielcarz, D. W. & Kasper, L. H. The Gut Microbiome in Multiple Sclerosis. Curr. Treat. Options Neurol. 17, 10.1007/s11940-015-0344-7 (2015). [DOI] [PubMed]
- 55.Gophna U, Sommerfeld K, Gophna S, Doolittle WF, Veldhuyzen Van Zanten SJO. Differences between tissue-associated intestinal microfloras of patients with Crohn’s disease and ulcerative colitis. J. Clin. Microbiol. 2006;44:4136–4141. doi: 10.1128/JCM.01004-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gevers D, et al. The treatment-naïve microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Willing BP, et al. A Pyrosequencing Study in Twins Shows That Gastrointestinal Microbial Profiles Vary With Inflammatory Bowel Disease Phenotypes. Gastroenterology. 2010;139:1844–1854. doi: 10.1053/j.gastro.2010.08.049. [DOI] [PubMed] [Google Scholar]
- 58.Ooi, J., Yunfei, L., Rogers, C. & Cantorna, M. Vitamin D Regulates the Gut Microbiome and Protects Mice from Dextran Sodium. J. Nutr. Immunol. 1679–1686, 10.3945/jn.113.180794.strong (2013). [DOI] [PMC free article] [PubMed]
- 59.Proal A, Albert P, Marshall T. Autoimmune disease in the era of the metagenome. Autoimmun Rev. 2009;8:677–81. doi: 10.1016/j.autrev.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 60.Pascal V, et al. A microbial signature for Crohn’s disease. Gut. 2017;66:813–822. doi: 10.1136/gutjnl-2016-313235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Assa A, et al. Vitamin D deficiency predisposes to adherent-invasive escherichia coli-induced barrier dysfunction and experimental colonic injury. Inflamm. Bowel Dis. 2015;21:297–306. doi: 10.1097/MIB.0000000000000282. [DOI] [PubMed] [Google Scholar]
- 62.Garg, M., Hendy, P., Ding, N., Shaw, S. & Hold, G. The Effect of Vitamin D on Intestinal Inflammation and Faecal Microbiota in Patients with Ulcerative Colitis. 963–972, 10.1093/ecco-jcc/jjy052 (2018). [DOI] [PubMed]
- 63.Stearns JC, et al. Bacterial biogeography of the human digestive tract. Sci. Rep. 2011;1:1–9. doi: 10.1038/srep00170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Conlon MA, Bird AR. The impact of diet and lifestyle on gut microbiota and human health. Nutrients. 2015;7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pugin B, et al. A wide diversity of bacteria from the human gut produces and degrades biogenic amines. Microb. Ecol. Health Dis. 2017;28:1353881. doi: 10.1080/16512235.2017.1353881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Strauss J, et al. Invasive potential of gut mucosa-derived fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011;17:1971–1978. doi: 10.1002/ibd.21606. [DOI] [PubMed] [Google Scholar]
- 67.Kolho K, et al. Fecal Microbiota in Pediatric Inflammatory Bowel Disease and Its Relation to Inflammation. Am J Gastroenterol. 2015;110:921–30. doi: 10.1038/ajg.2015.149. [DOI] [PubMed] [Google Scholar]
- 68.Forbes JD, Van Domselaar G, Bernstein CN. Microbiome survey of the inflamed and noninflamed gut at different compartments within the gastrointestinal tract of inflammatory bowel disease patients. Inflamm. Bowel Dis. 2016;22:817–825. doi: 10.1097/MIB.0000000000000684. [DOI] [PubMed] [Google Scholar]
- 69.Hosoda, K. et al. Identification and characterization of a vitamin D3 decomposition product bactericidal against Helicobacter pylori. Sci. Rep. 5, 10.1038/srep08860 (2015). [DOI] [PMC free article] [PubMed]
- 70.Sokol H, Lay C, Seksik P, Tannock G. Analysis of bacterial bowel communities of IBD patients: what has it revealed? Inflamm Bowel Dis. 2008;14:858–67. doi: 10.1002/ibd.20392. [DOI] [PubMed] [Google Scholar]
- 71.Prideaux L, et al. Impact of ethnicity, geography, and disease on the microbiota in health and inflammatory bowel disease. Inflamm. Bowel Dis. 2013;19:2906–2918. doi: 10.1097/01.MIB.0000435759.05577.12. [DOI] [PubMed] [Google Scholar]
- 72.Kabeerdoss J, Jayakanthan P, Pugazhendhi S, Ramakrishna BS. Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J. Med. Res. 2015;142:23–32. doi: 10.4103/0971-5916.162091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Murphy A, Velazquez K, Herbert K. Influence of High-Fat-Diet on Gut Microbiota: A Driving Force for Chronic Disease Risk. Curr. Opin. Clin. Nutr. Metab. Care. 2015;18:515–20. doi: 10.1097/MCO.0000000000000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Matsuoka K, Kanai T. The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 2015;37:47–55. doi: 10.1007/s00281-014-0454-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Results of all analyses are included in this published article. The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.