Introduction
Current technological level made possible the quick identification and culture of the SARS-CoV-2, an emerging coronavirus that caused until March 3, 2020, 90,870 confirmed cases of infections, with outside China a still limited spread (10,556 cases) and low mortality (166 deaths) (https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200303-sitrep-43-covid-19.pdf?sfvrsn=2c21c09c_2). However, it is necessary to quickly diagnose this virus in order to manage people’s fears and SARS-CoV-2-infected individuals and to quickly sequence its genome to possibly detect mutations. It is also crucial to quickly cultivate the viral strain, which will allow to test its sensitivity to a large panel of molecules with potential antiviral activity, including those that are known to interfere with the replication process of RNA viruses and, among others, chloroquine, hydroxychloroquine [1–3], and teicoplanin [4–7], as well as other molecules that were not yet evaluated but that may be efficient on RNA viruses, such as azithromycin [8]. The Hospital University Institute (IHU) Méditerranée Infection, Southeastern France, was built in order to be able to coordinate all possible efforts at the level of health care but also of diagnosis (https://www.mediterranee-infection.com/). As proof of concept, we report the first case of SARS-CoV-2 we diagnosed at our institution.
Results and discussion
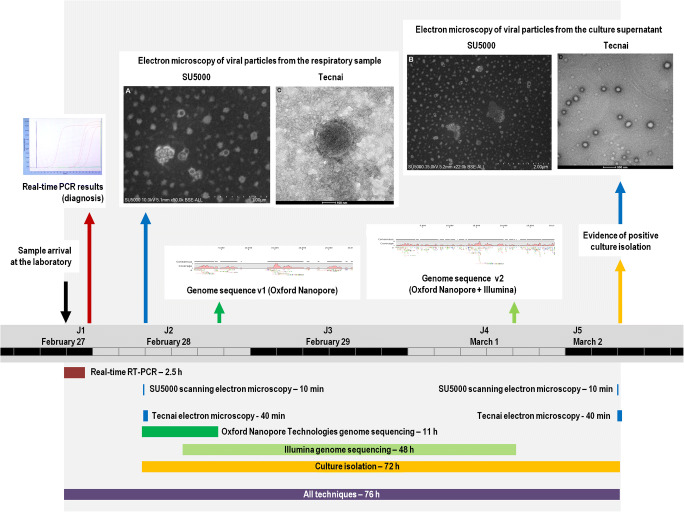
The respiratory sample was collected for diagnosis of SARS-CoV-2 on February 27, 2020. This nasopharyngeal swab sample arrived at 8:30 pm in our clinical microbiology and virology laboratory at IHU Méditerranée Infection, which has been performing the routine diagnosis of SARS-CoV-2, 24 h a day, 7 days a week since the end of January 2020 for all suspected cases of Covid-19 (the SARS-CoV-2-associated disease). We have tested more than 4084 respiratory samples since January 2020 and until February 19, 2020 [9]. We carried out SARS-CoV-2 RNA detection by two first-line real-time reverse transcription (RT)-PCR assays [9], one of which (SpikeP_ps80) was developed in-house as soon as the first SARS-CoV-2 genome (GenBank Accession no. MN908947) was released by Chinese teams (on January 10, 2020). Results of PCR testing of the nasopharyngeal swab sample were provided to clinicians at 11:30 pm (Fig. 1). Both PCR tests were positive, with a Ct value of 13 with the “gene E” system [10]. On February 28 at 8:30 am, we decided to examine the viral particles by both scanning and transmission electron microscopy. We directly used the clinical sample without time-consuming and laborious preparation, and directly screened the sample in 10 min, using a SU5000 scanning electron microscope (SEM) (Hitachi High-Tech Corporation, Tokyo, Japan). The clinical specimen was fixed using glutaraldehyde and deposited on carbon grids. The SU5000 SEM has a capability of observing specimens in both low and high vacuum pressure with a short time for evacuation after specimen loading. Micrographs were generated at × 35,000 magnification. Images were compatible in size and shape with those of coronaviruses. We also screened the sample on a Tecnai G20 electron microscope (FEI, Germany). Micrographs generated in approximately 40 min by this electron microscope at × 29,000 magnification also showed particles compatible in shape and size with coronavirus virions.
Fig. 1.
Timeline of tests performed and results availability
Furthermore, in parallel to electron microscopy observation, we launched on February 28 at 8:30 am the process for the next-generation sequencing of the viral genome by using Oxford Nanopore Technology (ONT). The viral RNA previously extracted from 200 μL of nasopharyngeal swab fluid using the EZ1 Virus Mini Kit v2.0 (Qiagen, Courtaboeuf, France) was reverse transcribed with SuperScript IV (ThermoFisher Scientific, Waltham, MA, USA), and cDNA second strand synthesis was performed with Klenow Fragment DNA polymerase (New England Biolabs, Beverly, MA, USA). Then, the generated DNA was purified with Agencourt AMPure XP beads (Beckman Coulter, Villepinte, France) before ONT library preparation. At 7:00 pm the same day, after a run of 4 h on the GridION X5 instrument (Oxford Nanopore Technologies Ltd., Oxford, UK), we were able to obtain using the CLC Genomics tool (https://digitalinsights.qiagen.com/), by mapping 42 reads with an average length of 1433 nucleotides on a set of coronavirus genomes from the Virus Pathogen Database and Analysis Resource [11], a 18,611-nucleotide-long consensus sequence corresponding to approximately 62% of the viral genome (EMBL Accession no. ERS4366776). This sequence, by similarity search using the BLASTn tool [12] against the NCBI nucleotide sequence database (GenBank), found as best hit the genome of a SARS-CoV-2 (GenBank Accession no. MT106053.1). Additionally, we had launched at 2:00 pm the same day another next-generation sequencing using Illumina technology with the Illumina Nextera XT paired-end strategy on a MiSeq instrument (Illumina Inc., San Diego, CA, USA), as previously described [13]. A genome consensus sequence was further generated with the CLC Genomics software using reads obtained by both Oxford Nanopore and Illumina technologies. A total of 18,585 reads could be mapped on the SARS-CoV-2 genome GenBank Accession no. MN908947, allowing to obtain a 25,799-nucleotide-long consensus genomic sequence (ca. 86% of the full-length genome).
Finally, on February 28 at 8:30 am, we also launched the culture isolation of the viral strain. Thus, 500 μL of the liquid collected from the nasopharyngeal swab was passed through a 0.22-μm pore-sized centrifugal filter (Merck Millipore, Darmstadt, Germany), then was inoculated in wells of 96-well culture microplates, of which 4 wells contained Vero E6 cells (ATCC CRL-1586) and 4 wells contained LLC-MK2 cells (ATCC CCL-7) in Minimum Essential Medium culture medium with 4% fetal calf serum and 1% glutamine. After centrifugation at 4,000 g, microplates were incubated at 37 °C. On Monday morning (March 2) at 8:00 am, moderate cytopathic effects consisting of clusters of rounding cells were detected for Vero E6 cells. A sample of 50 μL of the co-culture supernatant showed on the SU5000 SEM the presence of particles that were compatible with the size and morphology of a coronavirus, which was then confirmed by the observation on the Tecnai G20 EM. After subculture and definitive identification by PCR and sequencing, this isolate was named Covid-19-IHUMI1.
This observation is the proof of concept that from the clinical sample, it was possible to have the result of the PCR in 3 h, to have the genome in 11 h, and to recover using culture in 72 h the viral strain whose availability makes it possible to study drugs with recognized activity including those (hydroxychloroquine and chloroquine) with reported clinical activity, as communicated by Chinese teams [14]. Thus, it shows that we can, in specialized centers, extremely rapidly respond to the emergence of any viral strain pathogenic for humans.
Acknowledgments
We sincerely thank Takashi Irie, Kyoko Imai, Shigeki Matsubara, Taku Sakazume, Toshihide Agemura, Yusuke Ominami, and the Hitachi team of Japan (Hitachi High-Tech Corporation, Toranomon Hills Business Tower, 1-17-1 Toranomon, Minato-ku, Tokyo 105-6409, Japan) for the collaborative study conducted together with IHU Méditerranée Infection, and for the installation of a SU5000 microscope at IHU Méditerranée Infection. We are also grateful to Priscilla Jardot, Vincent Bossi, Anthony Fontanini, and Marion Le Bideau for their technical help.
Author contributions
Conceived and designed the experiments: DR. Contributed materials/analysis tools: PC, JCL, JPB, JB, BLS, and DR. Analyzed the data: PC, JB, BLS, and DR. Wrote the manuscript: PC, JB, BLS, and DR. Reviewed and approved the final version of the manuscript: all authors.
Funding information
This work was supported by the French Government under the “Investments for the Future” program managed by the National Agency for Research (ANR), Méditerranée-Infection 10-IAHU-03, and was also supported by the Région Provence Alpes Côte d’Azur and European funding FEDER PRIMMI (Fonds Européen de Développement Régional - Plateformes de Recherche et d’Innovation Mutualisées Méditerranée Infection). This work received partial support by Hitachi High-Tech Corporation.
Compliance with ethical standards
Conflict of interest
Authors would like to declare that Didier Raoult is a consultant for Hitachi High-Tech Corporation. Funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; and preparation, review, or approval of the manuscript.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rolain JM, Colson P, Raoult D. Recycling of chloroquine and its hydroxyl analogue to face bacterial, fungal and viral infections in the 21st century. Int J Antimicrob Agents. 2007;30:297–308. doi: 10.1016/j.ijantimicag.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Colson P, Rolain JM, Raoult D (2020) Chloroquine for the 2019 novel coronavirus SARS-CoV-2. Int J Antimicrob Agents 55:105923 [DOI] [PMC free article] [PubMed]
- 3.Colson P, Rolain JM, Lagier JC, Brouqui P, Raoult D (2020) Chloroquine and hydroxychloroquine as available weapons to fight COVID-19. Int J Antimicrob Agents 105932. 10.1016/j.ijantimicag.2020.105932 [DOI] [PMC free article] [PubMed]
- 4.Wang Y, Cui R, Li G, Gao S, Yuan S, et al. Teicoplanin inhibits Ebola pseudovirus infection in cell culture. Antivir Res. 2016;125:1–7. doi: 10.1016/j.antiviral.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balzarini J, Keyaerts E, Vijgen L, Egberink H, De Clercq E, et al. Inhibition of feline (FIPV) and human (SARS) coronavirus by semisynthetic derivatives of glycopeptide antibiotics. Antivir Res. 2006;72:20–33. doi: 10.1016/j.antiviral.2006.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Colson P, Raoult D. Fighting viruses with antibiotics: an overlooked path. Int J Antimicrob Agents. 2016;48:349–352. doi: 10.1016/j.ijantimicag.2016.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baron SA, Devaux CA, Colson P, Raoult D, Rolain JM (2020) Teicoplanin: an alternative drug for the treatment of new coronavirus COVID-2019? Int J Antimicrob Agents 13:105944. 10.1016/j.ijantimicag.2020.105944 [DOI] [PMC free article] [PubMed]
- 8.Bosseboeuf E, Maite A, Nhan T, de Pina JJ, Rolain JM, et al. Azithromycin inhibits the replication of Zika virus. J Antivir Antiretrov. 2018;10:6–11. doi: 10.4172/1948-5964.1000173. [DOI] [Google Scholar]
- 9.Colson P, La Scola B, Esteves-Vieira V, Ninove L, Zandotti C, et al. Plenty of coronaviruses but no SARS-CoV-2. Eurosurveillance. 2020;25:2000171. doi: 10.2807/1560-7917.ES.2020.25.8.2000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:10–7917. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pickett BE, Sadat EL, Zhang Y, Noronha JM, Squires RB, et al. ViPR: an open bioinformatics database and analysis resource for virology research. Nucleic Acids Res. 2012;40:D593–D598. doi: 10.1093/nar/gkr859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 13.Reteno DG, Benamar S, Khalil JB, Andreani J, Armstrong N, et al. Faustovirus, an asfarvirus-related new lineage of giant viruses infecting amoebae. J Virol. 2015;89:6585–6594. doi: 10.1128/JVI.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gao J, Tian Z, Yang X (2020) Breakthrough: chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci Trends 14(1):72–73 [DOI] [PubMed]