Abstract
Background
Many studies have investigated the association between matrix metalloproteinase polymorphisms and lung cancer susceptibility. However, the results are still controversial. To clarify these associations, we conducted a meta-analysis.
Methods
A systematic search of studies was conducted in PubMed, Embase, and China National Knowledge Infrastructure. Overall and subgroup analysis stratified by ethnicity was conducted. OR with 95% CI was used to assess the strength of the association. Furthermore, false-positive report probability (FPRP) tests were also performed for associations obtained in this meta-analysis.
Results
Twenty-four studies, including 10,099 cases and 9,395 controls, were analyzed. Nine polymorphisms were reported. For MMP1 -1607 1G/2G and MMP7 -181 A/G, increased lung cancer risk was found in Asians. For MMP2 -1306 C/T and MMP2 -735 C/T, decreased lung cancer risk was found in both “diverse populations” and Asians. For MMP9 -1562, C/T decreased lung cancer risk was found in both “diverse populations” and Caucasians. For MMP13 -77A/G, the A/G genotype decreased lung cancer risk in Asians. However, only associations between MMP1 -1607 1G/2G, MMP2 -1306 C/T, MMP2 -735 C/T, and MMP7 -181 A/G and lung cancer risk were considered noteworthy according to FPRP tests. There was no association between MMP3 -1171 5A/6A, MMP9 R279Q, and MMP12 -82A/G and lung cancer risk.
Conclusions
Our meta-analysis suggested that MMP1 -1607 1G/2G and MMP7 -181 A/G were risk factors for lung cancer, while MMP2 -1306 C/T, MMP2 -735 C/T, MMP9 -1562 C/T, and MMP13 -77A/G might be protective factors. However, results for MMP9 -1562 C/T and MMP13 -77A/G should be interpreted with caution due to the probability of false-positive reports.
Keywords: Matrix metalloproteinases, polymorphisms, lung cancer, meta-analysis
Introduction
Lung cancer is one of the leading causes of cancer-related death globally and is still a huge health threat to human beings (1,2). Although tobacco smoking is the major risk factor for lung cancer, lung cancer develops in less than 20% throughout life smokers, indicating that other factors such as genetic susceptibility may also contribute to lung carcinogenesis (3,4). The matrix metalloproteases (MMPs) family, including at least 26 human MMPs belong to a larger family of proteases named metzincin superfamily. They are zinc-dependent endopeptidases that collectively degrade all extracellular matrix (ECM) components (5,6). According to the main substrates, MMPs are traditionally classified as collagenases (e.g., MMP-1, MMP-8 and MMP-13), gelatinases (e.g., MMP-2 and MMP-9), stromelysins (e.g., MMP-3, MMP-10 and MMP-11), matrilysins (e.g., MMP-7 and MMP-26) and macrophage metalloelastase (e.g., MMP-12) (7). Egeblad et al. demonstrated that these MMPs influenced tumor cell behavior and played an important role in several steps of cancer development, including immune surveillance, angiogenesis, and regulation of cell growth and apoptosis (8).
MMP-1 and MMP-13, belong to the collagenase, is related to the ability of neoplastic cells to cross the basal membrane of both the vascular endothelium and the epithelium (9). Studies have reported that MMP-1 might contribute to tumor growth and spread by altering the cellular microenvironment to favor tumor formation (8,10), and overexpression of MMP-13 is related to poor prognosis and more aggressive tumors (11,12). MMP-2 and MMP-9, members of gelatinases, can degrade the major basal membrane component-type IV collagen and, therefore, are involved in cancer invasion and metastasis (13). MMP-7, the member of the matrilysins, can degrade proteoglycans, elastin, type IV collagen, and fibronectin (14). Also, MMP-7 has the so-called “sheddase function” that cleave non-matrix substrates from the cell surface, such as pro-tumor necrosis factor from thecadherin (15). Several studies have proved that MMP-7 has a statistically significant positive correlation with invasive tumor potential and contributes to early tumor development (16-18). MMP-12, known as macrophage metalloelastase, exhibits the same ability as MMP-7 to degrade elastin. The roles of MMP-12 in cancers are still controversial. However, overexpression of MMP-12 is reported to be positively associated with not only tumor invasion and progression but also the poor outcome of patients in multiple cancers, including lung cancer (19-21). MMP-3 belongs to the stromelysins, is known to induce the synthesis of other MMPs (9).
Last decades, a surge of studies investigating the association between genetic polymorphisms and lung cancer risk was published. Polymorphisms in MMP genes were also considered to be related to lung cancer risk. However, the results remained ambiguous and controversial because the relatively small sample size of a single study was underpowered to detect the effect of these polymorphisms. Several meta-analyses have been conducted to assess the association between MMP polymorphisms and lung cancer risk (22-24). Nevertheless, the latest one was published four years ago, and the data were updated. Therefore, we conducted this meta-analysis based on 24 case-control studies and aimed to better assess the association between MMP polymorphisms and lung cancer risk to date.
Methods
Identification of eligible studies
Two independent investigators conducted a systematic search strategy. Firstly, we searched Pubmed, EMBASE, and China National Knowledge Infrastructure (CNKI) with the terms: “lung cancer or lung carcinoma” and “MMP or matrix metallopeptidase” on or before Sept 30, 2019. Secondly, after the title and abstract manually screened, all references cited in relevant studies were also reviewed to identify other studies.
Inclusion and exclusion criteria
Studies included in this meta-analysis must meet the following inclusion criteria: (I) case-control study about the association between MMP polymorphisms and lung cancer risk; (II) genotype and allele data were available; (III) all studies must conform to Hardy-Weinberg equilibrium (HWE) in the control group. Exclusion criteria: (I) duplication of publications; (II) studies that were not about MMP polymorphisms and the etiology of lung cancer; (III) no sufficient data to calculate the odds ratios (ORs) and 95% confidence intervals (CIs). If more than one study using the same case series was published, only the study with the largest sample size was included.
Data extraction
Two investigators (Li and Liu) extracted data from eligible studies independently and then followed by data exchange and cross-check. Any disagreement was settled by rechecking the original data and further discussion together. The following contents were collected: first author, publication year, country of origin, genes and polymorphisms, source of control (hospital-based or population-based), ethnicity, genotyping methods, number of cases and controls, and genotype distributions in cases and controls.
Quality assessment
We assessed the quality of included studies using the Newcastle-Ottawa Scale (NOS) (25). The NOS rating system was based on three aspects of the case-control study: selection, comparability, and exposure. For each of the three aspects, four, one, and three parameters were assigned, respectively. Scores were ranged from 0 to 9, and studies were of high quality if scores ≥7. Two investigators assessed the quality of the studies through consultations to reach consensus.
Statistics analysis
We evaluated HWE for each study in control groups by the chi-square goodness-of-fit test, and P<0.05 was considered a significant departure from HWE. ORs assessed the strength of association between MMP polymorphisms and lung cancer risk with 95% CIs. The pooled ORs were performed for five genetic models: allelic model (x versus X), heterozygote model (Xx versus XX), homozygote model (xx versus XX), dominant model (xx + Xx versus XX) and recessive model (xx versus Xx + XX), x represented the minor allele and X represented the major allele. Z-test determined the statistical significance level with a P value of less than 0.05. Heterogeneity was evaluated by both Q statistic and the I2 statistic. A P value of less than 0.1 and I2 greater than 50% was a significant inconsistency (26). The random effects model was used if there was significant inconsistency; otherwise, the fixed-effects model was used (27). For each genetic comparison, subgroup analysis stratified by ethnicity was conducted. Sensitivity analysis was also performed by omitting each study in each turn to evaluate the effect of each study on the combined ORs. Potential publication bias was checked by Egger’s test (28) and Begg’s funnel plots (29). The P value of Egger’s test less than 0.05, and an asymmetric plot was considered a significant publication bias. All statistical analyses were performed using Stata version 12.0 (StataCorp LP, College Station, TX, USA).
False-positive report probability (FPRP) tests
We performed FPRP tests for all the significant associations obtained in this meta-analysis. FPRP was determined by three parameters: the observed P value, the prior probability, and the statistical power of the test. The approach developed by Wacholder et al. was used (30). He advocated presetting the FPRP noteworthiness value at 0.2, and the prior probability of 0.01 and power OR 1.5 were used in our study. FPRP values were calculated by the excel spreadsheet offered by Wachoder et al.
Results
Characteristics of studies
A total of 4,469 articles were retrieved from PubMed, Embase, and CNKI databases due to less stringent terms we used. The literature selection process was shown in Figure 1. Full-text assessment was conducted for 39 articles and, as a result, 15 articles were excluded, among which 4 had no usable data (31-34), 3 used the same case series as another study (35-37), 6 deviated from HWE (38-43), and 2 were not about MMP polymorphisms and the etiology of lung cancer (44,45). Finally, 24 eligible case-control studies were included in our meta-analysis (21,46-68). Five studies presented genotype distributions of more than one polymorphisms separately; thus, each of them was treated as separate studies (21,50,51,57,58). Nine polymorphisms (MMP1 -1607 1G/2G, MMP2 -1306 C/T and -735 C/T, MMP3 -1171 5A/6A, MMP7 -181 A/G, MMP9 -1562 C/T and R279Q, MMP12 -82A/G, and MMP13 -77A/G) were reported in the 24 included studies containing a total of 10,099 cases and 9,395 controls. For most of the polymorphisms, studies were conducted in “diverse populations”, while studies about MMP7 -181 A/G and MMP9 R279Q focused on Asians. Different genotyping methods were utilized, including polymerase chain reaction-restriction fragment length polymorphism, TaqMan, and polymerase chain reaction-denaturing high-performance liquid chromatography. The genotype distributions in the controls of all studies were consistent with HWE. More details about the characteristics of these studies were shown in Table 1. Genotype counts, and the P value of HWE were shown in Table 2.
Figure 1.
Selection of studies.
Table 1. Characteristics of studies included in the meta-analysis.
| First author | Year | Country | Ethnicity | Source of control | Genotyping method | Cases/controls | Genes | Polymorphisms | NOS, total scores |
|---|---|---|---|---|---|---|---|---|---|
| Bayramoglu | 2009 | Turkey | Asian | HB | PCR-RFLP | 200/100 | MMP9 | –1562 C/T | 6 |
| Bayramoglu | 2011 | Turkey | Asian | HB | PCR-RFLP | 200/100 | MMP2 | –1306 C/T | 6 |
| Fakhoury | 2012 | Lebanon | Caucasian | PB | PCR-RFLP | 41/51 | MMP1 | –1607 1G/2G | 8 |
| Fang | 2005 | China | Asian | PB | PCR-RFLP | 243/350 | MMP3 | –1171 5A/6A | 8 |
| Gonzalez-Arriaga | 2012 | Spain | Caucasian | HB | PCR-RFLP | 716/534 | MMP3, MMP9 | –1171 5A/6A, –1562 C/T |
6 |
| Gonzalez-Arriaga | 2008 | Spain | Caucasian | HB | PCR-RFLP | 501/510 | MMP1, MMP13 | –1607 1G/2G, –77 A/G |
7 |
| Hart | 2011 | Norway | Caucasian | PB | TaqMan | 436/434 | MMP1 | –1607 1G/2G | 9 |
| Hu | 2005 | China | Asian | PB | PCR-RFLP | 744/747 | MMP9 | R279Q | 8 |
| Jia | 2009 | China | Asian | PB | PCR-RFLP | 370/436 | MMP2 | –735 C/T | 8 |
| Liu | 2011 | China | Asian | PB | PCR-RFLP | 825/825 | MMP1 | –1607 1G/2G | 9 |
| Lu | 2017 | China | Asian | PB | PCR-RFLP | 260/219 | MMP7 | –181 A/G | 8 |
| Peng | 2010 | China | Asian | PB | PCR-RFLP | 420/419 | MMP12, MMP13 | –82 A/G, –77 A/G | 8 |
| Rollin | 2007 | France | Caucasian | HB | PCR-RFLP | 90/90 | MMP2, MMP9 | –1306 C/T, –735 C/T, –1562 C/T | 7 |
| Sanli | 2013 | Turkey | Asian | PB | PCR-RFLP | 132/80 | MMP7 | –181 A/G | 7 |
| Song | 2007 | China | Asian | PB | PCR-RFLP | 163/148 | MMP2 | –1306 C/T | 8 |
| Su | 2006 | USA | Caucasian | PB | TaqMan | 2,014/1,323 | MMP1, MMP3, MMP12 | –1607 1G/2G, –1171 5A/6A, –82 A/G | 7 |
| Wang | 2013 | China | Asian | PB | PCR-RFLP | 300/300 | MMP12 | –82 A/G | 8 |
| Wang | 2013 | China | Asian | PB | PCR-RFLP | 300/300 | MMP13 | –77 A/G | 8 |
| Yu | 2002 | China | Asian | PB | PCR-DHPLC | 781/852 | MMP2 | –1306 C/T | 8 |
| Zhang | 2005 | China | Asian | PB | PCR-RFLP | 243/350 | MMP7 | –181 A/G | 8 |
| Zhang | 2006 | China | Asian | PB | PCR-RFLP | 150/200 | MMP1 | –1607 1G/2G | 8 |
| Zhao | 2007 | China | Asian | PB | PCR-RFLP | 50/50 | MMP9 | R279Q | 8 |
| Zhou | 2005 | China | Asian | PB | PCR-RFLP | 770/777 | MMP2 | –735 C/T | 8 |
| Zhang | 2005 | China | Asian | PB | PCR-RFLP | 150/200 | MMP9 | –1562 C/T | 8 |
HB, hospital-based; PB, population-based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism; PCR-DHPLC, polymerase chain reaction-denaturing high-performance liquid chromatography.
Table 2. Genotype counts of the analyzed polymorphisms included in the meta-analysis.
| Study | Ethnicity | Cases/controls | Cases | Controls | PHWE | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1G/1G | 1G/2G | 2G/2G | 1G/1G | 1G/2G | 2G/2G | |||||
| MMP1 -1607 1G/2G | ||||||||||
| Su | Caucasian | 2,014/1,323 | 541 | 1,015 | 458 | 367 | 642 | 314 | 0.310 | |
| Gonzalez-Arriaga | Caucasian | 501/510 | 128 | 248 | 125 | 119 | 259 | 132 | 0.712 | |
| Hart | Caucasian | 436/434 | 115 | 207 | 114 | 132 | 198 | 104 | 0.081 | |
| Liu | Asian | 825/825 | 74 | 323 | 428 | 100 | 367 | 358 | 0.691 | |
| Fakhoury | Caucasian | 41/51 | 5 | 17 | 19 | 7 | 16 | 28 | 0.081 | |
| Zhang | Asian | 150/200 | 32 | 70 | 48 | 60 | 98 | 42 | 0.865 | |
| MMP2 -1306 C/T | CC | CT | TT | CC | CT | TT | ||||
| Yu | Asian | 781/852 | 644 | 127 | 10 | 585 | 248 | 19 | 0.220 | |
| Rollin | Caucasian | 90/90 | 60 | 28 | 2 | 60 | 29 | 1 | 0.217 | |
| Bayramoglu | Asian | 200/100 | 123 | 73 | 4 | 65 | 32 | 3 | 0.692 | |
| Song | Asian | 163/148 | 129 | 32 | 2 | 100 | 44 | 4 | 0.747 | |
| MMP2 -735 C/T | CC | CT | TT | CC | CT | TT | ||||
| Zhou | Asian | 770/777 | 506 | 230 | 34 | 425 | 313 | 39 | 0.052 | |
| Rollin | Caucasian | 89/90 | 69 | 18 | 2 | 67 | 21 | 2 | 0.816 | |
| Jia | Asian | 370/436 | 260 | 96 | 14 | 292 | 123 | 21 | 0.092 | |
| MMP3 -1171 5A/6A | 5A/5A | 5A/6A | 6A/6A | 5A/5A | 5A/6A | 6A/6A | ||||
| Fang | Asian | 243/350 | 7 | 73 | 163 | 8 | 105 | 237 | 0.358 | |
| Su | Caucasian | 2,014/1,323 | 485 | 1,012 | 517 | 325 | 648 | 350 | 0.466 | |
| Gonzalez-Arriaga | Caucasian | 716/534 | 164 | 367 | 185 | 119 | 276 | 139 | 0.417 | |
| MMP7 -181 A/G | A/A | A/G | G/G | A/A | A/G | G/G | ||||
| Zhang | Asian | 243/350 | 200 | 40 | 3 | 316 | 33 | 1 | 0.888 | |
| Sanli | Asian | 132/80 | 120 | 10 | 2 | 79 | 1 | 0 | 0.955 | |
| Lu | Asian | 260/219 | 182 | 78 | 0 | 176 | 43 | 0 | 0.107 | |
| MMP9 -1562 C/T | CC | CT | TT | CC | CT | TT | ||||
| Zhang | Asian | 150/200 | 83 | 60 | 7 | 155 | 42 | 3 | 0.936 | |
| Rollin | Caucasian | 90/90 | 68 | 22 | 0 | 64 | 21 | 5 | 0.085 | |
| Bayramoglu | Asian | 200/100 | 150 | 48 | 2 | 67 | 30 | 3 | 0.871 | |
| Gonzalez-Arriaga | Caucasian | 762/649 | 581 | 174 | 7 | 483 | 148 | 18 | 0.110 | |
| MMP9 R279Q | RR | RQ | RR | RQ | ||||||
| Hu | Asian | 744/747 | 357 | 323 | 64 | 343 | 323 | 81 | 0.704 | |
| Zhao | Asian | 50/50 | 24 | 20 | 6 | 14 | 28 | 8 | 0.335 | |
| MMP12 -82A/G | A/A | A/G | G/G | A/A | A/G | G/G | ||||
| Su | Caucasian | 2,014/1,323 | 1,535 | 449 | 30 | 1,008 | 289 | 26 | 0.324 | |
| Wang | Asian | 300/300 | 290 | 10 | 0 | 287 | 13 | 0 | 0.701 | |
| Peng | Asian | 420/419 | 404 | 16 | 0 | 399 | 20 | 0 | 0.617 | |
| MMP13 -77A/G | A/A | A/G | G/G | A/A | A/G | G/G | ||||
| Gonzalez-Arriaga | Caucasian | 501/506 | 248 | 208 | 45 | 267 | 197 | 42 | 0.508 | |
| Wang | Asian | 300/300 | 85 | 132 | 83 | 55 | 156 | 89 | 0.354 | |
| Peng | Asian | 420/419 | 105 | 207 | 108 | 91 | 227 | 101 | 0.085 | |
PHWE, P value of Hardy-Weinberg Equilibrium.
MMP1 -1607 1G/2G and lung cancer susceptibility
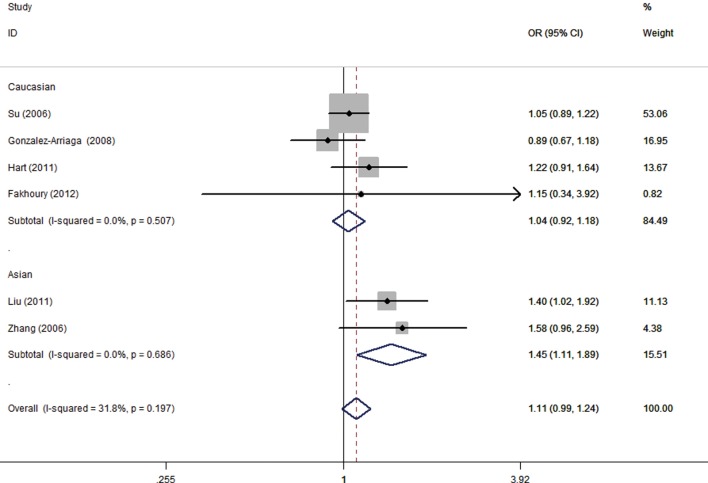
Six studies involving 3,967 cases and 3,343 controls were pooled. The random effects model was used in the allelic model, homozygote model, and recessive model. A fixed-effects model was used in the other two genetic models. Overall, no significant association was identified in any of the genetic models. Next, subgroup analysis stratified by ethnicity was conducted and increased lung cancer risk was found in Asians (allelic model: OR =1.34, 95% CI: 1.18–1.53, P<0.001; homozygote model: OR =1.73, 95% CI: 1.29–2.31, P<0.001; dominant model: OR =1.45, 95% CI: 1.11–1.89, P=0.006, Figure 2; recessive model: OR =1.45, 95% CI: 1.21–1.74, P<0.001, Table 3). No significant association was found in Caucasians.
Figure 2.
The association between MMP1 -1607 1G/2G and lung cancer risk (dominant model: OR =1.11, 95% CI: 0.99–1.24, P=0.077; P=0.197 for heterogeneity; fixed-effects model).
Table 3. Summary of pooled ORs in the meta-analysis and FPRP.
| Ethnicity | N † | Allelic model | Heterozygote model | Homozygote model | Dominant model | Recessive model | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | I2 (%) | FPRP‡ | OR | I2 (%) | FPRP | OR | I2 (%) | FPRP | OR | I2 (%) | FPRP | OR | I2 (%) | FPRP | ||||||
| MMP1 -1607 1G/2G | ||||||||||||||||||||
| Asian | 2 | 1.34 (1.18–1.53) | 0.0 | 0.002 | 1.23 (0.93–1.63) | 0.0 | – | 1.73 (1.29–2.31) | 0.0 | 0.107 | 1.45 (1.11–1.89) | 0.0 | 0.498 | 1.45 (1.21–1.74) | 0.0 | 0.010 | ||||
| Caucasian | 4 | 1.00 (0.93–1.09) | 0.0 | – | 1.06 (0.93–1.21) | 0.0 | – | 1.01 (0.87–1.18) | 0.0 | – | 1.04 (0.92–1.18) | 0.0 | – | 0.97 (0.85–1.10) | 0.0 | – | ||||
| Total | 6 | 1.12 (0.97–1.29) | 70.0 | – | 1.09 (0.97–1.23) | 0.0 | – | 1.22 (0.95–1.58) | 61.6 | – | 1.11 (0.99–1.24) | 31.8 | – | 1.13 (0.92–1.39) | 67.2 | – | ||||
| MMP2 -1306 C/T | ||||||||||||||||||||
| Asian | 2 | 0.53 (0.43–0.64) | 0.0 | 0.000 | 0.48 (0.39–0.60) | 0.0 | 0.000 | 0.46 (0.23–0.93) | 0.0 | 0.953 | 0.48 (0.39–0.59) | 0.0 | 0.000 | 0.55 (0.27–1.10) | 0.0 | – | ||||
| Caucasian | 1 | 1.04 (0.60–1.79) | – | – | 0.97 (0.51–1.81) | – | – | 2.00 (0.18–22.65) | – | – | 1.00 (0.54–1.86) | – | – | 2.02 (0.18–22.71) | – | – | ||||
| Total | 3 | 0.64 (0.44–0.93) | 64.1 | 0.821 | 0.58 (0.39–0.87) | 56.6 | 0.770 | 0.52 (0.27–1.02) | 0.0 | – | 0.59 (0.39–0.89) | 61.2 | 0.808 | 0.61 (0.31–1.18) | 0.0 | – | ||||
| MMP2 -735 C/T | ||||||||||||||||||||
| Asian | 2 | 0.76 (0.66–0.87) | 31.1 | 0.007 | 0.72 (0.51–1.01) | 69.4 | – | 0.74 (0.50–1.09) | 0.0 | – | 0.72 (0.53–0.97) | 64.1 | 0.814 | 0.84 (0.57–1.24) | 0.0 | – | ||||
| Caucasian | 1 | 0.87 (0.47–1.62) | – | – | 0.83 (0.41–1.70) | – | – | 0.97 (0.13–7.09) | – | – | 0.84 (0.42–1.68) | – | – | 1.01 (0.14–7.34) | – | – | ||||
| Total | 3 | 0.76 (0.66–0.88) | 0.0 | 0.024 | 0.73 (0.56–0.95) | 43.1 | 0.717 | 0.75 (0.51–1.10) | 0.0 | – | 0.73 (0.58–0.92) | 34.9 | 0.493 | 0.85 (0.58–1.24) | 0.0 | – | ||||
| MMP3 -1171 5A/6A | ||||||||||||||||||||
| Asian | 1 | 0.96 (0.71–1.30) | – | – | 0.79 (0.28–2.29) | – | – | 0.79 (0.28–2.21) | – | – | 0.79 (0.28–2.20) | – | – | 0.97 (0.69–1.38) | – | – | ||||
| Caucasian | 2 | 0.99 (0.91–1.08) | 0.0 | – | 1.02 (0.88–1.19) | 0.0 | – | 0.98 (0.83–1.16) | 0.0 | – | 1.01 (0.88–1.16) | 0.0 | – | 0.97 (0.85–1.11) | 0.0 | – | ||||
| Total | 3 | 0.99 (0.91–1.07) | 0.0 | – | 1.02 (0.88–1.18) | 0.0 | – | 0.98 (0.83–1.15) | 0.0 | – | 1.01 (0.88–1.15) | 0.0 | – | 0.97 (0.86–1.10) | 0.0 | – | ||||
| MMP7 -181 A/G | ||||||||||||||||||||
| Asian | 3 | 1.89 (1.41–2.54) | 29.7 | 0.037 | 1.92 (1.40–2.63) | 0.0 | 0.072 | 4.11 (0.67–25.80) | 0.0 | – | 1.98 (1.44–2.70) | 2.0 | 0.038 | 3.81 (0.61–23.86) | 0.0 | – | ||||
| Total | 3 | 1.89 (1.41–2.54) | 29.7 | – | 1.92 (1.40–2.63) | 0.0 | – | 4.11 (0.67–25.80) | 0.0 | – | 1.98 (1.44–2.70) | 2.0 | – | 3.81 (0.61–23.86) | 0.0 | – | ||||
| MMP9 -1562 C/T | ||||||||||||||||||||
| Asian | 1 | 0.68 (0.43–1.08) | – | – | 0.72 (0.42–1.23) | – | – | 0.30 (0.05–1.82) | – | – | 0.68 (0.40–1.15) | – | – | 0.33 (0.05–1.99) | – | – | ||||
| Caucasian | 2 | 0.83 (0.67–1.02) | 0.0 | – | 0.98 (0.77–1.24) | 0.0 | – | 0.27 (0.12–0.62) | 0.0 | 0.924 | 0.89 (0.71–1.12) | 0.0 | – | 0.27 (0.12–0.62) | 0.0 | 0.924 | ||||
| Total | 3 | 0.80 (0.67–0.97) | 0.0 | 0.704 | 0.93 (0.75–1.16) | 0.0 | – | 0.28 (0.13–0.59) | 0.0 | 0.878 | 0.86 (0.69–1.05) | 0.0 | – | 0.28 (0.13–0.59) | 0.0 | 0.878 | ||||
| MMP9 R279Q | ||||||||||||||||||||
| Asian | 2 | 0.88 (0.76–1.02) | 45.5 | – | 0.71 (0.32–1.56) | 69.8 | – | 0.73 (0.52–1.03) | 0.0 | – | 0.69 (0.33–1.45) | 68.9 | – | 0.78 (0.55–1.07) | 0.0 | – | ||||
| Total | 2 | 0.88 (0.76–1.02) | 45.5 | – | 0.71 (0.32–1.56) | 69.8 | – | 0.73 (0.52–1.03) | 0.0 | – | 0.69 (0.33–1.45) | 68.9 | – | 0.78 (0.55–1.07) | 0.0 | – | ||||
| MMP12 -82A/G | – | |||||||||||||||||||
| Asian | 2 | 0.78 (0.47–1.32) | 0.0 | – | 0.78 (0.46–1.32) | 0.0 | – | – | – | – | 0.78 (0.46–1.32) | 0.0 | – | – | – | – | ||||
| Caucasian | 1 | 0.98 (0.84–1.13) | – | – | 1.02 (0.86–1.21) | – | – | 0.76 (0.45–1.29) | – | – | 1.00 (0.85–1.18) | – | – | 0.75 (0.44–1.28) | – | – | ||||
| Total | 3 | 0.96 (0.83–1.11) | 0.0 | – | 0.99 (0.85–1.17) | 0.0 | – | 0.76 (0.45–1.29) | – | – | 0.98 (0.84–1.14) | 0.0 | – | 0.75 (0.44–1.28) | – | – | ||||
| MMP13 -77A/G | ||||||||||||||||||||
| Asian | 2 | 0.88 (0.72–1.08) | 46.7 | – | 0.67 (0.47–0.96) | 45.3 | 0.849 | 0.76 (0.50–1.16) | 49.5 | – | 0.70 (0.48–1.01) | 55.3 | – | 1.01 (0.80–1.27) | 0.0 | – | ||||
| Caucasian | 1 | 1.10 (0.91–1.34) | – | – | 1.14 (0.88–1.47) | – | – | 1.15 (0.73–1.82) | – | – | 1.14 (0.89–1.46) | – | – | 1.09 (0.70–1.69) | – | – | ||||
| Total | 3 | 0.95 (0.79–1.14) | 59.4 | – | 0.81 (0.54–1.22) | 78.3 | – | 0.87 (0.61–1.24) | 51.1 | – | 0.83 (0.56–1.23) | 78.3 | – | 1.02 (0.83–1.26) | 0.0 | – | ||||
†, N number of included studies; ‡, FPRP false-positive report probabilities (power OR =1.5, prior probability =0.01).
MMP2 -1306 C/T and lung cancer susceptibility
Three studies involving 1,034 cases and 1,090 controls were pooled after one study (47) excluded according to the sensitivity analysis (Table S1). Random effects model was used in allelic model, heterozygote model, and dominant model. Fixed-effects model was used in the other two genetic models. Decreased lung cancer risk was found in “diverse populations” (allelic model: OR =0.64, 95% CI: 0.44–0.93, P=0.020; heterozygote model: OR =0.58, 95% CI: 0.39–0.87, P=0.007; dominant model: OR =0.59, 95% CI: 0.39–0.89, P=0.011,Table 3) and Asians (allelic model: OR =0.53, 95% CI: 0.43–0.64, P<0.001; homozygote model: OR =0.48, 95% CI: 0.39–0.60, P<0.001; heterozygote model: OR =0.46, 95% CI: 0.23–0.93, P=0.032; dominant model: OR =0.48, 95% CI: 0.39–0.59, P<0.001, Table 3). No significant association was found in Caucasians.
Table S1. Positive results from the sensitivity analysis.
| Study omitted | Allelic model | Heterozygote model | Homozygote model | Dominant model | Recessive model | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | OR (95% CI) | P | I2 (%) | |||||
| MMP1 -1607 1G/2G | |||||||||||||||||||
| Su | 1.17 (1.00–1.38) | 0.050 | 36.5 | ||||||||||||||||
| Gonzalez-Arriaga | 1.15 (1.02–1.30) | 0.023 | 14.3 | ||||||||||||||||
| Hart | 1.09 (0.96–1.23) | 0.171 | 41.4 | ||||||||||||||||
| Liu | 1.07 (0.95–1.20) | 0.272 | 18.3 | ||||||||||||||||
| Fakhoury | 1.11 (0.99–1.24) | 0.079 | 45.4 | ||||||||||||||||
| Zhang | 1.08 (0.97–1.22) | 0.167 | 23.1 | ||||||||||||||||
| Combined | 1.11 (0.99–1.24) | 0.077 | 31.8 | ||||||||||||||||
| MMP2 -1306 C/T | |||||||||||||||||||
| Yu | 0.86 (0.58–1.29) | 0.479 | 54.0 | 0.87 (0.54–1.38) | 0.546 | 53.3 | 0.69 (0.25–1.87) | 0.467 | 0.0 | 0.86 (0.53–1.37) | 0.517 | 56.3 | |||||||
| Rollin | 0.67 (0.43–1.05) | 0.082 | 78.4 | 0.66 (0.38–1.16) | 0.153 | 81.6 | 0.49 (0.26–0.94) | 0.030 | 0.0 | 0.65 (0.38–1.12) | 0.118 | 81.0 | |||||||
| Bayramoglu | 0.64 (0.44–0.93) | 0.020 | 64.1 | 0.58 (0.39–0.86) | 0.007 | 56.6 | 0.52 (0.27–1.02) | 0.055 | 0.0 | 0.59 (0.39–0.89) | 0.011 | 61.2 | |||||||
| Song | 0.81 (0.46–1.41) | 0.452 | 84.4 | 0.79 (0.40–1.55) | 0.486 | 85.2 | 0.57 (0.30–1.10) | 0.096 | 0.0 | 0.79 (0.40–1.53) | 0.479 | 85.4 | |||||||
| Combined | 0.74 (0.49–1.11) | 0.142 | 76.8 | 0.72 (0.44–1.16) | 0.176 | 77.8 | 0.55 (0.30–1.00) | 0.051 | 0.0 | 0.71 (0.44–1.14) | 0.160 | 78.2 | |||||||
| MMP2 -735 C/T | |||||||||||||||||||
| Zhou | 0.86 (0.68–1.10) | 0.228 | 0.0 | 0.87 (0.65–1.16) | 0.340 | 0.0 | 0.86 (0.65–1.13) | 0.266 | 0.0 | ||||||||||
| Rollin | 0.76 (0.66–0.87) | 0.000 | 31.1 | 0.72 (0.51–1.01) | 0.061 | 69.4 | 0.72 (0.53–0.97) | 0.032 | 64.1 | ||||||||||
| Jia | 0.72 (0.61–0.85) | 0.000 | 0.0 | 0.63 (0.52–0.78) | 0.000 | 0.0 | 0.65 (0.53–0.79) | 0.000 | 0.0 | ||||||||||
| Combined | 0.76 (0.66–0.88) | 0.000 | 0.0 | 0.73 (0.56–0.95) | 0.019 | 43.1 | 0.73 (0.58–0.92) | 0.007 | 34.9 | ||||||||||
| MMP9 -1562 C/T | |||||||||||||||||||
| Zhang | 0.80 (0.66–0.97) | 0.021 | 0.0 | 0.29 (0.14–0.62) | 0.002 | 0.0 | 0.30 (0.14–0.64) | 0.002 | 0.0 | ||||||||||
| Rollin | 1.12 (0.56–2.23) | 0.758 | 91.4 | 0.75 (0.13–4.29) | 0.746 | 80.8 | 0.69 (0.15–3.20) | 0.641 | 75.2 | ||||||||||
| Bayramoglu | 1.12 (0.54–2.34) | 0.758 | 91.0 | 0.61 (0.07–5.02) | 0.644 | 82.7 | 0.56 (0.08–3.74) | 0.552 | 78.6 | ||||||||||
| Gonzalez-Arriaga | 1.05 (0.43–2.56) | 0.923 | 90.5 | 0.60 (0.06–6.44) | 0.673 | 77.7 | 0.58 (0.07–4.79) | 0.612 | 72.0 | ||||||||||
| Combined | 1.00 (0.57–1.75) | 0.988 | 87.9 | 0.53 (0.11–2.53) | 0.426 | 75.1 | 0.51 (0.13–2.07) | 0.348 | 68.8 | ||||||||||
| MMP9 R279Q | |||||||||||||||||||
| Hu | 0.42 (0.18–0.97) | 0.041 | – | ||||||||||||||||
| Zhao | 0.92 (0.75–1.13) | 0.424 | – | ||||||||||||||||
| Combined | 0.69 (0.33–1.45) | 0.330 | 68.9 | ||||||||||||||||
| MMP13 -77A/G | |||||||||||||||||||
| Gonzalez-Arriaga | 0.67 (0.47–0.96) | 0.029 | 45.3 | ||||||||||||||||
| Wang | 0.96 (0.68–1.37) | 0.838 | 64.1 | ||||||||||||||||
| Peng | 0.80 (0.39–1.64) | 0.548 | 88.5 | ||||||||||||||||
| Combined | 0.81 (0.54–1.22) | 0.312 | 78.3 | ||||||||||||||||
MMP2 -735 C/T and lung cancer susceptibility
Three studies involving 1,229 cases and 1,303 controls were pooled. Although no obvious heterogeneity was found in the overall analysis, significant heterogeneity was identified in subgroup analysis under the heterozygote model and dominant model. So, the random-effects model was used in these two genetic models. As a result, decreased lung cancer risk was found (allelic model: OR =0.76, 95% CI: 0.66–0.87, P<0.001; heterozygote model: OR =0.73, 95% CI: 0.56–0.95, P=0.019; dominant model: OR =0.73, 95% CI: 0.58–0.92, P= 0.007, Table 3). When subgroup analysis stratified by ethnicity was conducted, this association was lost in Caucasians.
MMP3 -1171 5A/6A and lung cancer susceptibility
Three studies involving 2,973 cases and 2,207 controls were pooled. No significant heterogeneity was identified, so a fixed-effects model was used. There was no association between MMP3 -1171 5A/6A polymorphism and lung cancer risk in both overall and subgroup analysis (Table 3).
MMP7 -181 A/G and lung cancer susceptibility
Three studies performed in Asians involving 635 cases and 649 controls were pooled. No significant heterogeneity was identified, so the fixed-effects model was used. As a result, increased lung cancer risk was found (allelic model: OR =1.89, 95% CI: 1.41–2.54, P<0.001; heterozygote model: OR =1.92, 95% CI: 1.40–2.63, P<0.001; dominant model: OR =1.98, 95% CI: 1.44–2.70, P<0.001, Table 3).
MMP9 -1562 C/T and lung cancer susceptibility
Three studies involving 1,052 cases and 839 controls were pooled after one study (68) excluded according to the sensitivity analysis (Table S1). No significant heterogeneity was identified, so the fixed-effects model was used. Decreased lung cancer risk was found in overall analysis (allelic model: OR =0.80, 95% CI: 0.67–0.97, P=0.021; homozygote model: OR =0.28, 95% CI: 0.13–0.59, P=0.001; recessive model: OR =0.28, 95% CI: 0.13–0.59, P=0.001, Table 3). The same association was also found in Caucasians (homozygote model: OR =0.27, 95% CI: 0.12–0.62, P=0.002; recessive model: OR =0.27, 95% CI: 0.12–0.62, P=0.002, Table 3).
MMP9 R279Q and lung cancer susceptibility
Two studies performed in Asians involving 794 cases and 797 controls were pooled. The random-effects model was used in the heterozygote model and the dominant model. A fixed-effects model was used in the other three genetic models. There was no association between MMP9 R279Q polymorphism and lung cancer risk (Table 3).
MMP12 -82A/G and lung cancer susceptibility
Three studies involving 2,734 cases and 2,042 controls were pooled. No significant heterogeneity was identified, so the fixed-effects model was used. There was no association between MMP12 -82A/G polymorphism and lung cancer risk in both overall and subgroup analysis. It was worth noting that there was no GG genotype in two studies (57,61), therefore pooled ORs under the homozygote model and recessive model were not available (Table 3).
MMP13 -77A/G and lung cancer susceptibility
Three studies involving 1,221 cases and 1,225 controls were pooled. The random-effects model was used in all genetic models except for the recessive model. No significant association was found in the overall analysis, but A/G genotype decreased the risk of lung cancer in Asians (OR =0.67, 95% CI: 0.47–0.96, P=0.029, Table 3).
Sensitivity analysis and publication bias
Sensitivity analyses were performed to assess the stability of results by removing each study once in every polymorphism and genetic model. The corresponding results were materially altered after a single study was excluded at a time for 6 polymorphisms (Table S1). Positive results for three polymorphisms were notable. For MMP2 -1306 C/T and MMP2 -735 C/T, excluding Ayşegül et al. (47) study and Zhou et al. (67) study respectively could reverse the results under the allelic model, heterozygote model, and dominant model. For MMP9 -1562 C/T, the exclusion of Zhang et al. (68) study could reverse the results under the allelic model, homozygote model, and recessive model.
Interestingly, heterogeneity between studies was decreased after these three studies were excluded, indicating that these studies might contribute as a source of heterogeneity. Ayşegül et al. (47) study and Zhang et al. (68) study were finally excluded from our meta-analysis but not Zhou et al. (67) study because the other two studies investigating MMP2 -735 C/T were relatively small. No publication bias for the association between MMP1 -1607 1G/2G and lung cancer susceptibility was identified by Begg’s test (P=0.452) or Egger’s test (P=0.376) under the dominant model. Symmetrical funnel plots were also obtained in all the genetic models (Figure 3). We did not evaluate publication bias of the rest polymorphisms owing to the limited study number.
Figure 3.
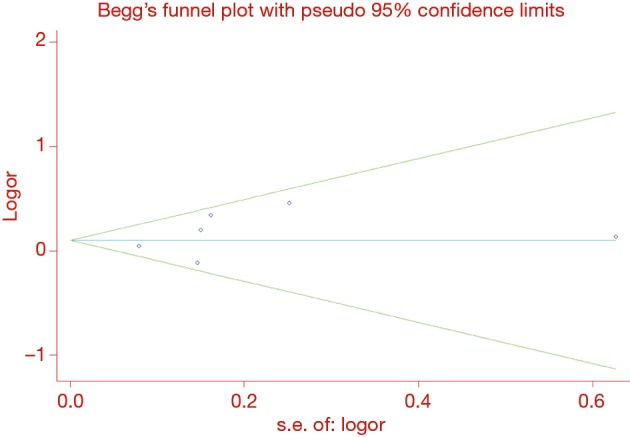
Begg’s funnel plot for the association between MMP1 -1607 1G/2G and lung cancer risk (dominant model: Z=0.75, P=0.452).
FPRP tests
We performed FPRP tests for all the significant associations obtained in this meta-analysis. Eleven associations involved 4 SNPs (MMP1 -1607 1G/2G, MMP2 -1306 C/T, MMP2 -735 C/T, and MMP7 -181 A/G) were considered to be noteworthy (FPRP value less than 0.2), indicating a true association, Table 3.
Discussion
Twenty-four eligible case-control studies, including 10,099 cases and 9,395 controls about nine polymorphisms in MMP genes, were analyzed. We found that MMP1 -1607 1G/2G and MMP7 -181 A/G increased lung cancer risk and MMP2 -1306 C/T, MMP2 -735 C/T, MMP9 -1562 C/T, and MMP13 -77A/G might confer protection against lung cancer. No association was found between MMP3 -1171 5A/6A, MMP9 R279Q, and MMP12 -82A/G and lung cancer risk. However, when FPRP tests were performed, only associations between MMP1 -1607 1G/2G, MMP2 -1306 C/T, MMP2 -735 C/T, and MMP7 -181 A/G and lung cancer risk were considered noteworthy.
MMPs, degrading basal membranes, and ECM, as we know, was involved in many critical physiological and pathological processes of lung cancer and inflammation (5,6,58,63). Nowadays, a large number of studies have reported that the expression of MMPs plays a critical role in tumor development, invasion, and poor prognosis of multiple cancers, including lung cancer (8,46,58,63). Polymorphisms in MMP genes were widely studied. Functional analyses are indicating alterations in the gene expression due to the modulatory effect of these polymorphisms on transcriptional activity (69-73). It could be presumed that MMP polymorphisms contributed to the development of lung cancer, and this conclusion might be biologically plausible.
MMP1 -1607 1G to 2G substitution might lead to significantly higher transcriptional activity because the 2G allele created an E26 (ETS) transcription factor binding site and increased transcription capacity (70). The study had reported that the Ets site was a notable correlation with the expression of MMP-1 (74). Enhancing Ets activity up-regulated the expression of MMP-1, and a reduction in Ets activity led to suppression (75,76). Two studies in Asians supported that individuals carrying the 2G/2G genotype had a higher risk of developing lung cancer than 1G/1G genotype (55,65). Our meta-analysis also observed that 2G/2G genotype carriers had a 1.73-fold increased risk of developing lung cancer compared with 1G/1G genotype carriers in Asians.
Nevertheless, no association was found in Caucasians. Besides, no heterogeneity between studies was found after subgroup analysis stratified by ethnicity was conducted. So, the loss of association in Caucasians could be explained by different frequencies of the polymorphism between different ethnicities.
MMP2 -1306 C/T and -735 C/T were in linkage disequilibrium (77). These two genotypes, within a haplotype, had a strong interaction and were correlated with lung cancer risk. Earlier studies demonstrated that CC genotype had higher promoter activity compared with the TT genotype, thus leading to the overexpression of MMP-2 (72,73). Although the differences resulting from polymorphisms were subtle, a long-time overexpression of MMP-2 might also increase the risk of lung cancer. Zhou et al. (67) reported that individuals with -1306 C/C or -735 C/C genotype were more likely to develop lung cancer, and the risk was even higher in smokers. However, Rollin et al. (58) observed opposite results due to different ethnicities and limited sample size. Our meta-analysis revealed that the T allele was a protective factor for lung cancer. That was exactly consistent with Zhou et al. (67) study. However, we did not investigate genetic-environment interaction effects.
The study has shown that MMP7 -181 G allele has a 2- to 3-fold higher promoter activity than that of the 181 A allele (78). Higher promoter activity induced an elevated level of MMP-7 mRNA and subsequently led to overexpression of MMP-7. “Sheddase function” of MMP-7 protein and the ability to increase activation of other MMPs worked together might predispose to malignant transformation (78,79). Although, to the best of our knowledge, all of the association studies investigating MMP7 -181 A/G were conducted in Asians, inconsistent results were still reported by previous studies. The reason might be listed as follows limited sample size and low frequency of the G allele. Our meta-analysis pooled data from 3 studies, suggesting that G allele increased lung cancer risk in Asians. However, further studies that focus on Caucasians are called for.
MMP9 -1562 C/T, the presence of the T allele was found to be involved in the decrease of the capacity of a putative transcription repressor protein with a subsequent increase in gene expression (71). MMP13 -77A/G, reduced the transcriptional activity of the MMP13 gene due to the modification of a PEA3 binding site (80). These two polymorphisms were also found significantly associated with lung cancer susceptibility. However, as Wacholder et al. reminded us, we should “protect ourselves from overinterpreting statistically significant findings that are not likely to signify a true association” (30). FPRP tests were indicating the probability of false-positive reports on these two polymorphisms. So, results for MMP9 -1562 C/T and MMP13 -77A/G should be interpreted with caution, and more studies were needed to replicate these findings.
The association between polymorphisms in MMP genes and lung cancer susceptibility has been investigated by several meta-analyses (22-24). The latest one was conducted by Li et al. (23) in 2015 and, in agreement with our meta-analysis, they demonstrated that significantly increased and reduced lung cancer risk were found in Asians for MMP1 -1607 1G/2G and MMP2 -1306 C/T, -735 C/T respectively. However, opposite to our result, they found a significantly increased risk for MMP9 -1562 C/T while a decreased risk was identified in ours. They yielded this result based on only one study (68), which was excluded in ours according to the sensitivity analysis. Our finding was consistent with another meta-analysis performed by Hu et al. (22). Compared with Li’s work, we excluded one letter (31) but identified more eligible studies (49,53,56,59,61,64,66). We also found two significant associations that were not observed in Li’s study—MMP13 -77A/G decreased lung cancer risk and MMP7 -181 A/G increased lung cancer risk. Furthermore, data for three new polymorphisms (MMP7 -181 A/G, MMP9 R279Q, and MMP12 -82A/G), which have never been investigated by previous meta-analyses, were also analyzed.
Our meta-analysis has several strengths. First, less stringent terms were used in the literature search, and no limitation was made. Thus, selection bias was well controlled. Additionally, compared with the prior meta-analyses, more studies were included, and three new polymorphisms were also explored. Finally, we performed FPRP tests to confirm if the obtained associations were noteworthy or not. However, some limitations should also be addressed. Firstly, for several polymorphisms, the number of included studies limited further analysis. Secondly, significant heterogeneity was detected. Although we used the random effects model to calculate the pooled ORs, the precision of the outcome would be affected. Finally, the lack of more detailed individual data preventing a more precise evaluation with adjusted ORs.
In conclusion, our results suggested that MMP1 -1607 1G/2G and MMP7 -181 A/G were risk factors for lung cancer in Asians, while MMP2 -1306 C/T, MMP2 -735 C/T, MMP9 -1562 C/T, and MMP13 -77A/G might be protective factors. There was no association between MMP3 -1171 5A/6A, MMP9 R279Q, and MMP12 -82A/G and lung cancer risk. However, results for MMP9 -1562 C/T and MMP13 -77A/G should be interpreted with caution. Well-designed studies with larger sample sizes and more ethnic groups are required to confirm the association identified in our meta-analysis.
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Ettinger DS, Wood DE, Aisner DL, et al. Non–Small Cell Lung Cancer, Version 5.2017, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw 2017;15:504-35. 10.6004/jnccn.2017.0050 [DOI] [PubMed] [Google Scholar]
- 2.Shankar A, Saini D, Dubey A, et al. Feasibility of lung cancer screening in developing countries: challenges, opportunities and way forward. Transl Lung Cancer Res 2019;8:S106-21. 10.21037/tlcr.2019.03.03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tseng TS, Gross T, Celestin MD, et al. Knowledge and attitudes towards low dose computed tomography lung cancer screening and smoking among African Americans—a mixed method study. Transl Cancer Res 2019;8:S431-42. 10.21037/tcr.2019.04.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gkolfinopoulos S, Mountzios G. Beyond EGFR and ALK: targeting rare mutations in advanced non-small cell lung cancer. Ann Transl Med 2018;6:142. 10.21037/atm.2018.04.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galateau-Salle FB, Salle G, Luna RE, et al. Matrix metalloproteinases and tissue inhibitors of metalloproteinases in bronchial squamous preinvasive lesions. Hum Pathol 2000;31:296-305. 10.1016/S0046-8177(00)80242-7 [DOI] [PubMed] [Google Scholar]
- 6.Frezzetti D, De Luca A, Normanno N. Extracellular matrix proteins as circulating biomarkers for the diagnosis of non-small cell lung cancer patients. J Thorac Dis 2019;11:S1252-6. 10.21037/jtd.2019.02.46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Biancheri P, Sabatino AD, Corazza GR, et al. Proteases and the gut barrier. Cell Tissue Res 2013;351:269-80. 10.1007/s00441-012-1390-z [DOI] [PubMed] [Google Scholar]
- 8.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2002;2:161-74. 10.1038/nrc745 [DOI] [PubMed] [Google Scholar]
- 9.Brinckerhoff CE, Rutter JL, Benbow U. Interstitial Collagenases as Markers of Tumor Progression. Clin Cancer Res 2000;6:4823-30. [PubMed] [Google Scholar]
- 10.Vincenti MP, White LA, Schroen DJ, et al. Regulating expression of the gene for matrix metalloproteinase-1 (collagenase): mechanisms that control enzyme activity, transcription, and mRNA stability. Crit Rev Eukaryot Gene Expr 1996;6:391-411. 10.1615/CritRevEukarGeneExpr.v6.i4.40 [DOI] [PubMed] [Google Scholar]
- 11.Gupta GP, Nguyen DX, Chiang AC, et al. Mediators of vascular remodelling co-opted for sequential steps in lung metastasis. Nature 2007;446:765-70. 10.1038/nature05760 [DOI] [PubMed] [Google Scholar]
- 12.Hsu CP, Shen GH, Ko JL. Matrix metalloproteinase-13 expression is associated with bone marrow microinvolvement and prognosis in non-small cell lung cancer. Lung Cancer 2006;52:349-57. 10.1016/j.lungcan.2006.01.011 [DOI] [PubMed] [Google Scholar]
- 13.Björklund M, Koivunen E. Gelatinase-mediated migration and invasion of cancer cells. Biochim Biophys Acta 2005;1755:37-69. [DOI] [PubMed]
- 14.Quantin B, Murphy G, Breathnach R. Pump-1 cDNA codes for a protein with characteristics like those of classical collagenase family members. Biochemistry 1989;28:5327-34. 10.1021/bi00439a004 [DOI] [PubMed] [Google Scholar]
- 15.Haro H, Crawford HC, Fingleton B, et al. Matrix metalloproteinase-7-dependent release of tumor necrosis factor-alpha in a model of herniated disc resorption. J Clin Invest 2000;105:143. 10.1172/JCI7091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ajisaka H, Yonemura Y, Miwa K. Correlation of lymph node metastases and expression of matrix metalloproteinase-7 in patients with gastric cancer. Hepatogastroenterology 2004;51:900-5. [PubMed] [Google Scholar]
- 17.Kioi M, Yamamoto K, Higashi S, et al. Matrilysin (MMP-7) induces homotypic adhesion of human colon cancer cells and enhances their metastatic potential in nude mouse model. Oncogene 2003;22:8662-70. 10.1038/sj.onc.1207181 [DOI] [PubMed] [Google Scholar]
- 18.Leeman MF, Curran S, Murray GI. New insights into the roles of matrix metalloproteinases in colorectal cancer development and progression. J Pathol 2003;201:528-34. 10.1002/path.1466 [DOI] [PubMed] [Google Scholar]
- 19.Cho NH, Hong KP, Hong SH, et al. MMP expression profiling in recurred stage IB lung cancer. Oncogene 2004;23:845-51. 10.1038/sj.onc.1207140 [DOI] [PubMed] [Google Scholar]
- 20.Hofmann HS, Hansen G, Richter G, et al. Matrix metalloproteinase-12 expression correlates with local recurrence and metastatic disease in non-small cell lung cancer patients. Clin Cancer Res 2005;11:1086-92. [PubMed] [Google Scholar]
- 21.Su L, Zhou W, Asomaning K, et al. Genotypes and haplotypes of matrix metalloproteinase 1, 3 and 12 genes and the risk of lung cancer. Carcinogenesis 2006;27:1024-9. 10.1093/carcin/bgi283 [DOI] [PubMed] [Google Scholar]
- 22.Hu C, Wang J, Xu Y, et al. Current evidence on the relationship between five polymorphisms in the matrix metalloproteinases (MMP) gene and lung cancer risk: a meta-analysis. Gene 2013;517:65-71. 10.1016/j.gene.2012.12.085 [DOI] [PubMed] [Google Scholar]
- 23.Li H, Liang X, Qin X, et al. Association of matrix metalloproteinase family gene polymorphisms with lung cancer risk: logistic regression and generalized odds of published data. Sci Rep 2015;5:10056. 10.1038/srep10056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei Z, Liu R, Chen J, et al. Meta Analysis of Association between Polymorphisms in Promoter Region of MMPs gene and Risk of Lung Cancer. Chin J Lung Cancer 2009;12. [DOI] [PubMed] [Google Scholar]
- 25.Wells G. The Newcastle-Ottawa Scale (NOS) for assessing the quality of non randomised studies in meta-analyses. Available online: http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp, 2001.
- 26.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med 2002;21:1539-58. 10.1002/sim.1186 [DOI] [PubMed] [Google Scholar]
- 27.DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled Clin Trials 1986;7:177-88. 10.1016/0197-2456(86)90046-2 [DOI] [PubMed] [Google Scholar]
- 28.Egger M, Smith GD, Schneider M, et al. Bias in meta-analysis detected by a simple, graphical test. BMJ 1997;315:629-34. 10.1136/bmj.315.7109.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics 1994:1088-101. 10.2307/2533446 [DOI] [PubMed] [Google Scholar]
- 30.Wacholder S, Chanock S, Garcia-Closas M, et al. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. J Natl Cancer Inst 2004;96:434-42. 10.1093/jnci/djh075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biondi ML, Turri O, Leviti S, et al. MMP1 and MMP3 polymorphisms in promoter regions and cancer. Clin Chem 2000;46:2023-4. 10.1093/clinchem/46.12.2023 [DOI] [PubMed] [Google Scholar]
- 32.Sun T, Gao Y, Tan W, et al. Haplotypes in Matrix Metalloproteinase Gene Cluster on Chromosome 11q22 Contribute to the Risk of Lung Cancer Development and Progression. Clin Cancer Res 2006;12:7009-17. 10.1158/1078-0432.CCR-06-0464 [DOI] [PubMed] [Google Scholar]
- 33.Sauter W, Rosenberger A, Beckmann L, et al. Matrix Metalloproteinase 1 (MMP1) Is Associated with Early-Onset Lung Cancer. Cancer Epidemiol Biomarkers Prev 2008;17:1127-35. 10.1158/1055-9965.EPI-07-2840 [DOI] [PubMed] [Google Scholar]
- 34.Chen XL. Correlation of single nucleotide polymorphism in the matrix metalloproteinase-1 gene promoter(-1607)1G/ 2G with risk of lung cancer. J Shanxi Med Univ 2007;38:777-9. [Google Scholar]
- 35.Fang SM, Jin X, Li Y, et al. Correlation of matrix metalloproteinase-3 polymorphism to genetic susceptibility and lymph node metastasis of non-small cell lung cancer. Ai Zheng 2005;24:305-10. [PubMed] [Google Scholar]
- 36.Su L, Zhou W, Park S, et al. Matrix metalloproteinase-1 promoter polymorphism and lung cancer risk. Cancer Epidemiol. Biomarkers Prev 2005;14:567-70. 10.1158/1055-9965.EPI-04-0482 [DOI] [PubMed] [Google Scholar]
- 37.Wang B. Correlation of the Matrix Metalloproteinases Polymorphisms to Non-Small Cell Lung Cancer. [Article in Chinese]. Wanfang Master Thesis Database. (2010) Available online: http://d.wanfangdata.com.cn/Thesis_Y1800003.aspx
- 38.Zhu Y, Spitz MR, Lei L, et al. A Single Nucleotide Polymorphism in the Matrix Metalloproteinase-1 Promoter Enhances Lung Cancer Susceptibility. Cancer Res 2001;61:7825-9. [PubMed] [Google Scholar]
- 39.Chen GL, Shen TC, Chang WS, et al. The contribution of MMP-7 promoter polymorphisms to Taiwan lung cancer susceptibility. Anticancer Res 2018;38:5671-7. 10.21873/anticanres.12903 [DOI] [PubMed] [Google Scholar]
- 40.Wang Y, Fang S, Wei L, et al. No association between the C-1562T polymorphism in the promoter of matrix metalloproteinase-9 gene and non-small cell lung carcinoma. Lung Cancer 2005;49:155-61. 10.1016/j.lungcan.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 41.Shen TC, Chang WS, Tsai CW, et al. The contribution of matrix metalloproteinase-1 promoter genotypes in Taiwan lung cancer risk. Anticancer Res 2018;38:253-7. [DOI] [PubMed] [Google Scholar]
- 42.Li W, Jia MX, Wang JH, et al. Association of MMP9-1562C/T and MMP13-77A/G Polymorphisms with Non-Small Cell Lung Cancer in Southern Chinese Population. Biomolecules 2019;9:107. 10.3390/biom9030107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei WQ, Liang XQ, Wen XP. The Association of MMP-1-1607(1G-2G) Single Nucleotide Polymorphism with the Susceptibility to Non-small Cell Lung Carcinoma in Guizhou Han Nationality. J Guiyang Med Coll 2007;32:267-9. [Google Scholar]
- 44.Schabath MB, Delclos GL, Martynowicz MM, et al. Opposing Effects of Emphysema, Hay Fever, and Select Genetic Variants on Lung Cancer Risk. Am J Epidemiol 2005;161:412-22. 10.1093/aje/kwi063 [DOI] [PubMed] [Google Scholar]
- 45.Lai CY, Chang WS, Hsieh YH, et al. Association of Tissue Inhibitor of Metalloproteinase-1 Genotypes with Lung Cancer Risk in Taiwan. Anticancer Res 2016;36:155-60. [PubMed] [Google Scholar]
- 46.Bayramoglu A, Gunes HV, Metintas M, et al. The association of MMP-9 enzyme activity, MMP-9 C1562T polymorphism, and MMP-2 and-9 and TIMP-1,-2,-3, and-4 gene expression in lung cancer. Genet Test Mol Biomarkers 2009;13:671-8. 10.1089/gtmb.2009.0053 [DOI] [PubMed] [Google Scholar]
- 47.Ayşegül B, Veysi GH, Muzaffer M, et al. Is a single nucleotide polymorphism a risk factor for lung cancer in the matrix metalloproteinase-2 promoter? Mol Biol Rep 2011;38:1469-74. 10.1007/s11033-010-0253-5 [DOI] [PubMed] [Google Scholar]
- 48.Fakhoury H, Noureddine S, Chmaisse HN, et al. MMP1-1607 (1G> 2G) polymorphism and the risk of lung cancer in Lebanon. Ann Thorac Med 2012;7:130. 10.4103/1817-1737.98844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang S, Jin X, Wang R, et al. Polymorphisms in the MMP1 and MMP3 promoter and non-small cell lung carcinoma in North China. Carcinogenesis 2005;26:481-6. 10.1093/carcin/bgh327 [DOI] [PubMed] [Google Scholar]
- 50.González-Arriaga P, Pascual T, García-Alvarez A, et al. Genetic polymorphisms in MMP 2, 9 and 3 genes modify lung cancer risk and survival. BMC Cancer 2012;12:121. 10.1186/1471-2407-12-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.González-Arriaga P, López-Cima MF, Fernández-Somoano A, et al. Polymorphism+ 17 C/G in matrix metalloprotease MMP8 decreases lung cancer risk. BMC Cancer 2008;8:378. 10.1186/1471-2407-8-378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hart K, Landvik NE, Lind H, et al. A combination of functional polymorphisms in the CASP8, MMP1, IL10 and SEPS1 genes affects risk of non-small cell lung cancer. Lung Cancer 2011;71:123-9. 10.1016/j.lungcan.2010.04.016 [DOI] [PubMed] [Google Scholar]
- 53.Hu Z, Xiang H, Lu D, et al. Functional Polymorphisms of Matrix Metalloproteinase-9 Are Associated with Risk of Occurrence and Metastasis of Lung Cancer. Clin Cancer Res 2005;11:5433-9. 10.1158/1078-0432.CCR-05-0311 [DOI] [PubMed] [Google Scholar]
- 54.Jia SX. Association of single nucleotide polymorphisms in the promoter of MMP-2 and TIMP-2 genes with Lung cancer.[Article in Chinese]. Wanfang Master Thesis Database. (2010) Available online: http://d.wanfangdata.com.cn/Thesis_Y1636837.aspx
- 55.Liu L, Wu J, Wu C, et al. A functional polymorphism (− 1607 1G→ 2G) in the matrix metalloproteinase-1 promoter is associated with development and progression of lung cancer. Cancer 2011;117:5172-81. 10.1002/cncr.26154 [DOI] [PubMed] [Google Scholar]
- 56.Lu LJ. Studies on Correlation between Genetic Polymorphisms in Matrix Metalloproteinase Genes and the Risk of Lung Cancer. [Article in Chinese]. Wanfang Master Thesis Database. (2019) Available online: http://d.wanfangdata.com.cn/Thesis_D01363925.aspx
- 57.Peng JC. Association of single nucleotide polymorphisms in the MMP-12 and MMP-13 genes with Lung cancer.[Article in Chinese]. Wanfang Master Thesis Database. (2010) Available online: http://d.wanfangdata.com.cn/Thesis_Y1779924.aspx
- 58.Rollin J, Régina S, Vourc’h P, et al. Influence of MMP-2 and MMP-9 promoter polymorphisms on gene expression and clinical outcome of non-small cell lung cancer. Lung Cancer 2007;56:273-80. 10.1016/j.lungcan.2006.11.021 [DOI] [PubMed] [Google Scholar]
- 59.Sanli M, Akar E, Pehlivan S, et al. The relationship of metalloproteinase gene polymorphisms and lung cancer. J Surg Res 2013;183:517-23. 10.1016/j.jss.2013.01.045 [DOI] [PubMed] [Google Scholar]
- 60.Song XY, Li L, Zhang L, et al. Association Polymorphisms in the Matrix Metalloproteinases-2 (MMP-2) Gene with Non-small Cell Lung Cancer. Sichuan J Cancer Control 2007;20:257-8. [Google Scholar]
- 61.Wang W, Zeng H, Wang R. Association between the matrix metalloproteinase-12 gene polymorphism and susceptibility of non-small cell lung cancer in northern China. Med J Nat Defending Forces Northwest China 2013;34:301-3. [Google Scholar]
- 62.Wang W, Wang ZC, Liu XY, et al. Assosiation between matrix metalloproteinase 13 gene polymorphism and susceptibility to non-small cell lung cancer. Chin J Mod Med 2013;(1):35-9.
- 63.Yu C, Pan K, Xing D, et al. Correlation between a single nucleotide polymorphism in the matrix metalloproteinase-2 promoter and risk of lung cancer. Cancer Res 2002;62:6430-3. [PubMed] [Google Scholar]
- 64.Zhang J, Jin X, Fang S, et al. The functional polymorphism in the matrix metalloproteinase-7 promoter increases susceptibility to esophageal squamous cell carcinoma, gastric cardiac adenocarcinoma and non-small cell lung carcinoma. Carcinogenesis 2005;26:1748-53. 10.1093/carcin/bgi144 [DOI] [PubMed] [Google Scholar]
- 65.Zhang WQ, Lin H, Zhou YA, et al. Association of MMP1-1607(1G/2G)single nucleotide polymorphism with susceptibility to lung cancer in Northwestern Chinese population of Han nationality. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2006;23:313-5. [PubMed] [Google Scholar]
- 66.Zhao YH. Functional polymorphisms of matrix metalloproteinase-9 and P53 are associated with risk of occurrence of non-small-cell lung cancer in Chinese population [Article in Chinese]. CNKI Master Thesis Database. (2007) Available online: http://kns.cnki.net/KCMS/detail/detail/FileName=2007091361.aspx
- 67.Zhou Y, Yu C, Miao X, et al. Functional haplotypes in the promoter of matrix metalloproteinase-2 and lung cancer susceptibility. Carcinogenesis 2005;26:1117-21. 10.1093/carcin/bgi057 [DOI] [PubMed] [Google Scholar]
- 68.Zhang W. A study of the relationship between lung cancer sensibility and gene polymorphisms of MMP-1,-9 [Article in Chinese]. CNKI Master Thesis Database. (2005) Available online: http://d.wanfangdata.com.cn/Thesis_Y726994.aspx
- 69.Stetler-Stevenson WG. The role of matrix metalloproteinases in tumor invasion, metastasis, and angiogenesis. Surg Oncol Clin N Am 2001;10:383-92. 10.1016/S1055-3207(18)30071-1 [DOI] [PubMed] [Google Scholar]
- 70.Rutter JL, Mitchell TI, Butticè G, et al. A single nucleotide polymorphism in the matrix metalloproteinase-1 promoter creates an Ets binding site and augments transcription. Cancer Res 1998;58:5321-5. [PubMed] [Google Scholar]
- 71.Zhang B, Ye S, Herrmann SM, et al. Functional polymorphism in the regulatory region of gelatinase B gene in relation to severity of coronary atherosclerosis. Circulation 1999;99:1788-94. 10.1161/01.CIR.99.14.1788 [DOI] [PubMed] [Google Scholar]
- 72.Nelson AR, Fingleton B, Rothenberg ML, et al. Matrix metalloproteinases: biologic activity and clinical implications. J Clin Oncol 2000;18:1135-49. 10.1200/JCO.2000.18.5.1135 [DOI] [PubMed] [Google Scholar]
- 73.Price SJ, Greaves DR, Watkins H. Identification of Novel, Functional Genetic Variants in the Human Matrix Metalloproteinase-2 Gene role of Sp1 in allele-specific transcriptional regulation. J Biol Chem 2001;276:7549-58. 10.1074/jbc.M010242200 [DOI] [PubMed] [Google Scholar]
- 74.Westermarck J, Seth A, K?H?Ri VM. Differential regulation of interstitial collagenase (MMP-1) gene expression by ETS transcription factors. Oncogene, 1997;14:2651-60. 10.1038/sj.onc.1201111 [DOI] [PubMed] [Google Scholar]
- 75.Tower GB, Coon CC, Benbow U, et al. Erk 1/2 differentially regulates the expression from the 1G/2G single nucleotide polymorphism in the MMP-1 promoter in melanoma cells. Biochim Biophys Acta 2002;1586:265-74. 10.1016/S0925-4439(01)00105-3 [DOI] [PubMed] [Google Scholar]
- 76.Park YH, Jung HH, Ahn JS, et al. Ets-1 upregulates HER2-induced MMP-1 expression in breast cancer cells. Biochemical Biophy Res Commun 2008;377:0-394. [DOI] [PubMed]
- 77.Yu C, Zhou Y, Miao X, et al. Functional haplotypes in the promoter of matrix metalloproteinase-2 predict risk of the occurrence and metastasis of esophageal cancer. Cancer Res 2004;64:7622-8. 10.1158/0008-5472.CAN-04-1521 [DOI] [PubMed] [Google Scholar]
- 78.Jormsjö S, Whatling C, Walter DH, et al. Allele-Specific Regulation of Matrix Metalloproteinase-7 Promoter Activity Is Associated With Coronary Artery Luminal Dimensions Among Hypercholesterolemic Patients. Arterioscler Thromb Vasc Biol 2001;21:1834. 10.1161/hq1101.098229 [DOI] [PubMed] [Google Scholar]
- 79.Barillé S, Bataille R, Rapp MJ, et al. Production of Metalloproteinase-7 (Matrilysin) by Human Myeloma Cells and Its Potential Involvement in Metalloproteinase-2 Activation. J Immunol 1999;163:5723-8. [PubMed] [Google Scholar]
- 80.Yoon S, Kuivaniemi H, Gatalica Z, et al. MMP13 promoter polymorphism is associated with atherosclerosis in the abdominal aorta of young black males. Matrix Biol 2002;21:487-98. 10.1016/S0945-053X(02)00053-7 [DOI] [PubMed] [Google Scholar]