Abstract
The mitochondrion is an extremely important organelle that performs various functions in the cell: e.g. energy production, regulation of respiration processes and maintenance of calcium homeostasis. Disruption of the biogenesis and functioning of this organelle can lead to cell damage and cell death. Mitochondrial dysfunction has been shown to possibly be involved in the pathogenesis of Parkinson's disease. However, the role of genes associated with mitochondrial biogenesis in the early stages of disease remains poorly understood. The objective of the present study was to analyze changes in the expression of activator (Nrf1, Ppargc1a, Prkn, and Kif1b) and repressor (Zfp746 and Mybbp1a) genes of mitochondrial biogenesis in the early stages of the development of neurodegeneration in an MPTP-induced model of presymptomatic and early symptomatic stages of PD. Statistically significant changes in expression at the mRNA level were detected for all studied genes. There was mainly a decrease in the expression of activator genes (Nrf1, Ppargc1a, Prkn, and Kif1b) at all stages of neurodegeneration, which seemed to be associated with impaired mitochondrial biogenesis and the development of neurodegeneration processes. A predominant decrease in the expression was detected for the Zfp746 and Mybbp1a repressor genes of mitochondrial biogenesis. However, in this case, it was associated with the emergence of compensatory mechanisms during the development of Parkinson's disease. The largest number of statistically significant changes was detected for the Nrf1 activator gene and the Mybbp1a repressor gene. Apparently, these two genes play the most important role in this disease.
Keywords: Neurodegeneration, Mitochondrial biogenesis, Parkinson's disease, MPTP-models, mRNA
1. Introduction
Mitochondria are important organelles that perform various functions in the cell: e.g. energy production, regulation of respiration processes and maintenance of calcium homeostasis [1]. Disturbance of the biogenesis and functioning of this organelle can lead to cell damage and cell death. Mitochondrial dysfunction can be associated mainly with disturbances in various complexes of the respiratory electron transfer chain, especially in complex I. These disturbances lead to a reduction in the production of adenosine triphosphate (ATP) and the formation of reactive oxygen species (ROS). In addition, the levels of ROS increase rapidly. As a result, oxidation and subsequent disruption of the normal functioning of proteins, lipids, and DNA occur [2]. Moreover, mitochondrial dysfunction can lead to a fusion/division imbalance of these organelles [3] and to the late activation of apoptosis [[4], [5], [6]].
Various works have shown that mitochondrial dysfunction may be associated with the development of the pathogenesis of several diseases, including Parkinson's disease (PD) [[7], [8], [9], [10], [11]]. Analysis of gene functions in monogenic forms of PD indicates that nine of them (PINK1, PRKN, SNCA, LRRK2, PARK7, ATP13A2, FBXO7, VPS35 and CHCHD2) are somehow related to the functioning of mitochondria [[12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25]]. The results of several meta-analyzes of whole-transcriptome data obtained in studies of patients with PD revealed a change in the expression of a large number of genes associated with the functioning of mitochondria, which also confirms the important role of this process in the pathogenesis of the disease [26].
Interestingly, there are very few studies of the expression of individual genes associated with mitochondrial functioning, especially in the earliest stages of the development of neurodegeneration in PD; most of them were based on cell culture experiments [17,[27], [28], [29]].
Because it is impossible to study the endogenous processes occurring in the brains of patients during the earliest stages of the development of the pathological process in PD, it remains unknown how the expression profile of genes involved in mitochondrial biogenesis changes in the earliest stages of the disease. One of the main approaches to the investigation of these stages of the disease is the development of different PD models that reproduce the early stages of neurodegeneration.
Previously, we conducted a whole-transcriptome study of such models [30,31]. In that study, it was shown that mitochondrial dysfunction is involved in the development of neurodegeneration at its earliest stages. However, it remains unclear which genes of mitochondrial biogenesis are involved in the pathogenesis of PD at the earliest stages. In this regard, an expression analysis of genes involved in the regulation of mitochondrial biogenesis was performed at the early stages of the development of the pathological process in PD in brain tissues and peripheral blood of mice with MPTP-induced models of early stages of PD [32,33].
2. Material and methods
2.1. Parkinson's disease models
We used models of the presymptomatic stage of PD with decapitation of mice 6 h after (6 h-PSS), the presymptomatic stage of PD with decapitation of mice 24 h after (24 h-PSS), the advanced presymptomatic (AdvPSS) and early symptomatic (ESS) stages of PD. Models were created as previously described [32,33].
In brief, male mice C57BL/6 at the age 8–12 weeks weighing 22–26 g were used in this study. The animals were maintained at 21–23 °C in a light-dark cycle having free access to food and tap water. Animals (n = 80) were divided into 4 control groups (n = 10 each) and 4 experimental groups 6h-PSS group (n = 10), 24h-PSS group (n = 10), AdvPSS group (n = 10) and ESS group (n = 10). Four groups of the experiments were made with the s.c. injections of MPTP hydrochloride (Sigma, St. Louis, MO, USA). The control animals received saline only. In the first group, MPTP was injected twice with 2 h intervals between the injections at the individual dose of 12 mg/kg. In another groups, MPTP was injected four times at the individual dose of 12 mg/kg with a 2-h interval between the injections (30 animals in the experiment and 30 animals in the control). In the first group (AdvPSS group), 2 weeks after the MPTP administration the animals were decapitated. In another groups, the animals were decapitated after 6 h (6h-PSS group), 24 h (24h-PSS group) and 2 weeks after the last MPTP administration (ESS group).
The brain was removed from the skull and cut along the midsagittal plane. Frontal cortex, dorsal striatum, and substantia nigra were dissected under a dissecting microscope with an ocular micrometer (Nikon SMZ660, Nikon, Melville, NY, USA). Also, from all animals samples of peripheral blood were obtained. The tissue pieces were weighed, frozen in liquid nitrogen and kept at —70 °C until RNA isolation.
Experimental work with laboratory animals was carried out in accordance with “The Guide for the Care of Animals” [34] and was approved by the ethical committee of the Institute of Development Biology. N.K. Koltsov RAS.
2.2. RNA isolation
During isolating RNA, tissue samples from each animal were homogenized using TRI Reagent (MRC, USA) [35]. Further phase separation was performed with addition of chloroform to the brain tissue homogenate with TRI Reagent according to the manufacturer's recommendations. After phase separation, the aqueous phase containing RNA was taken and then ethanol was introduced in a 1: 1 ratio. Further, the isolation of total RNA was performed using RNAeasy Mini Kit (Qiagen, Germany) according to the manufacturer's recommendations. RNA concentration was measured using a Quant-iT RNA BR Assay Kit and a Qubit 3.0 fluorimeter (Invitrogen, USA).
RNA quality was assessed using the Experion RNA HighSens Analysis Kit and the Experion instrument (Bio-Rad, USA).
2.3. Expression analysis of individual candidate genes
Gene expression analysis was performed using reverse transcription and qPCR with TaqMan probes. The reverse transcription reaction was performed on a T3 Thermocycler amplifier (T3 Thermoblock, Biometra, Germany) using the RevertAid ™ H Minus Reverse Transcriptase kit (Thermo Fisher Scientific, USA) according to the manufacturer's recommendations. A mixture of Random Hexamer Primer (Thermo Fisher Scientific, USA) and Oligo (dT)18 primer (“Thermo Fisher Scientific”, USA) was used in a ratio of 3: 2, respectively.
The sequences of the primers and probes for expression analysis of the candidate genes were designed using Beacon designer 7.0 software (Premier Biosoft, USA) and the nucleotide sequences of the Nrf1, Ppargc1a, Zfp746, Prkn, Mybbp1a and Kif1b genes, and the housekeeping genes Bcat2 and Psmd7. The sequences are based on mouse genome (Mus musculus (assembly GRCm38.p6), [36] https://www.ncbi.nlm.nih.gov/genome/?term=Mus+Musculus, NCBI). The sequences of gene-specific primers and probes are presented in Table 1.
Table 1.
Nucleotide sequences of gene-specific primers and probes.
| Gene | Nucleotide sequence |
|---|---|
| Mybbp1a | Probe: 5’-VIC-CGCCTAATCACTGGGCTTGG-BHQ2–3’ Forward primer: 5’-TCGGAGATGAAATATGCC-3’ Reverse primer: 5’-ATGGGATGTCTTCAAAAGA-3’ |
| NM_016776.2a | |
| Ppargc1a | Probe: 5’-VIC-TAACTCCTCCCACAACTCCTCCTC-BHQ2–3’ Forward primer: 5’-CGAACCTTAAGTGTGGAA-3’ Reverse primer: 5’-AGCCTTGAAAGGGTTATC-3’ |
| NM_008904.2 | |
| Prkn | Probe: 5’-VIC-CACGATGCTCAACTTGGCTACTC-3’ Forward primer: 5’-GTCACAAGACTCAACGATC-3’ Reverse primer: 5’-TGCTCTTCTCCAAGGATC-3’ |
| NM_016694.3 | |
| Nrf1 | Probe: 5’-VIC-AGCATGATCCTGGAAGACCTCG-BHQ2–3’ Forward primer: 5’-CAGCACCTTTGGAGAATG-3’ Reverse primer: 5’-GGCAGCTCTGAATTAACC-3’ |
| NM_001164226.1 | |
| Zfp746 | Probe: 5’-VIC-ACTGGTCTCCCTGGACTATGCC-BHQ2–3’ Forward primer: 5’-GAGCTCTACAAGCATGTG-3’ Reverse primer: 5’-TTGCCTTGTTCAATCTGG-3’ |
| NM_001163475.1 | |
| Kif1b | Probe: 5’-VIC-CAAGGAGTCCAAGTGCATCATTCAG-BHQ2–3’ Forward primer: 5’-CCTTCAATTCTCGAGAGA-3’ Reverse primer: 5’-CCAGTAGGAGTAGTCAAA-3’ |
| NM_207682.2 | |
| Bcat2 NM_001243053.1 | Probe: 5’-FAM-CGGATACACTCCAACAGCTCCTGCTTGT-BHQ1–3’ Forward primer: 5’-TCAACATGGACAGGATGCTACG-3’ Reverse primer: 5’-CCAGTCTTTGTCTACTTCAATGAGC-3’ |
| Psmd7 NM_010817.2 | Probe: 5’-FAM-AGTCCTAGGTCCTTTGGCTTCACGTCGA-BHQ1–3’ Forward primer: 5’-CTGCACAAGAATGATATCGCCATC-3’ Reverse primer: 5’-CTCCACTGAGATGTAGGCTTCG-3’ |
FAM and VIC – fluorescent dyes; BHQ1 and BHQ2 – fluorescence quenchers.
Numbers in the database GenBank (Accession numbers).
For real-time PCR, cDNA obtained in the reverse transcription reaction was used as a template. Before being added to the reaction mix, cDNA was diluted in an aqueous solution of tRNA from Escherichia coli (100 ng/μl) [37] to concentration of 0.02 ng/μl. Real-time PCR was performed using the StepOnePlus™ System (Applied Biosystems, USA). The composition of 30 μl of the reaction mixture consisted of: 5 μl of cDNA (0.02 ng/μl); 3 μl of PCR buffer (× 10) (Synthol, Russia); 3 μl 25 mM MgCl2; 10 PM primers (Evrogen, Russia); 2.5 pM probe (DNA synthesis, Russia); 200 μM of each dNTP; 1 enzyme unit Taq DNA Polymerase (Synthol, Russia). Thermal cycling was performed as follows: 50 °C, 60 s; 95 °C, 600 s; further 40 cycles of 95 °C, 20 s; and 61 °C, 50 s. Each sample was analyzed three times to correct for differences in sample quality and the efficiency of the reverse transcription reaction.
2.4. Statistical and bioinformatic analyzes
Nucleotide sequences of gene-specific primers and probes were designed using the Biosoft International Beacon designer 7.0 program (Palo Alto, USA) and the corresponding sequences from NCBI database. Checking the specificity of primers and probes was performed using the resource Primer3 and BLAST (Madden 2003) (https://www.ncbi.nlm.nih.gov/tools/primer-blast/), as well as the base IDT OligoAnalyzer 3.1 (http://eu.idtdna.com/calc/analyzer).
The relative levels of the transcripts in the test groups were calculated as R = 2–ΔΔCt [38]. Statistical data processing was performed using the “Statistica for Windows 8.0” software package (StatSoft, Inc. (2007), STATISTICA (version 8.0. Www.statsoft.com) and MS Excel 2013 software (Microsoft). Data were analyzed using the nonparametric Mann–Whitney U test. Gene interaction networks were made using the Pathway Studio® 12.0 (“Elsevier”) program [39,40].
3. Results
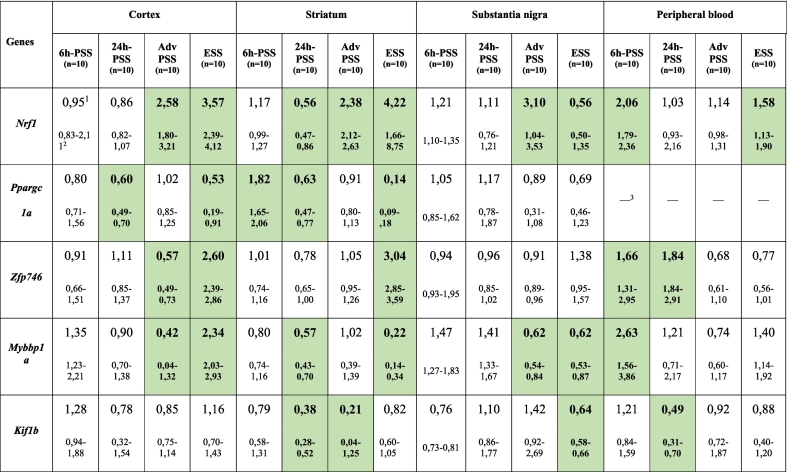
In the present study, an expression analysis of the Nrf1, Ppargc1a, Zfp746, Mybbp1a, Prkn and Kif1b genes was performed in samples from the cortex, striatum, and substantia nigra, and in the peripheral blood from mice with MPTP-induced models of 6h-PSS, 24h-PSS, AdvPSS and ESS of PD. These genes were selected based on their possible role in the pathogenesis of PD. The results of the expression analysis of candidate genes are presented in Table 2.
Table 2.
Relative mRNA levels of the genes studied in brain and peripheral blood of mice with MPTP-induced models of early stages of PD.
1Median.
225–75- quartiles. The levels of the mRNA of the genes studied in the control group were taken as 1. The data with p < .05 is bold and highlighted.
3The mRNA levels in these samples are below the detection level of the method used in this study. N = 10 for each group (control and experimental).
A much lower number of significantly changed genes was found in animal tissues with 6h-PSS and 24h-PSS models than in those from models of more advanced stages of PD. In fact, only one gene, Prkn, increased its expression in the substantia nigra with 6h-PSS model. Moreover, no changes in the relative mRNA levels of genes were shown in the substantia nigra 24h after MPTP administration. There was a decrease in the mRNA levels of the Prkn and Ppargc1a genes in the frontal cortex with models 6h-PSS and 24h-PSS respectively. However, six significantly different changes were found in the striatum with models 6h-PSS and 24h-PSS of PD: e.g., an increase in the mRNA levels of the Ppargc1a and Prkn genes in mice with 6h-PSS model, and a decrease in the expression of Nrf1, Ppargc1a, Mybbp1a, and Kif1b genes in mice with 24h-PSS model.
Changes in mRNA levels in the tissues with the AdvPSS model were detected in a larger number of the studied samples compared with models of early presymptomatic stages. As shown in Table 2, there was an increase in the relative mRNA levels of the Nrf1 gene in all studied brain tissues, and a decrease in the relative mRNA levels of the Mybbp1a and Prkn genes in the substantia nigra, of the Kif1b and Prkn genes in the striatum, and of the Zfp746 and Mybbp1a genes in the frontal cortex.
A statistically significant decrease in the mRNA levels of the Nrf1, Mybbp1a, Kif1b, and Prkn genes was found in the substantia nigra with model ESS of PD. A significant change was detected for four genes (Nrf1, Ppargc1a, Zfp746, and Mybbp1a) in the frontal cortex. In addition, these changes were co-directed in these tissues for three genes (Nrf1, Ppargc1a, and Zfp746).
The largest number of statistically significant changes (in 9 out of the 16 studied samples) was detected for the Nrf1 gene. Most changes corresponded to increasing of mRNA levels, whereas decreasing in the expression was found only in two cases (in the striatum with the 24h-PSS model and in the substantia nigra with the ESS model of PD). The Mybbp1a gene was significantly changed in 7 samples, with an increase in its mRNA levels in 2 out of 7 samples (in the frontal cortex with the ESS model and in the peripheral blood with the 6h-PSS model).
4. Discussion
At present time, there is no doubt that impaired functioning of mitochondria can play an important role in the development of the pathological process in PD. This is indicated by the presence of mutations in the genes of the familial form of PD (PINK1, PRKN, SNCA, LRRK2, PARK7, ATP13A2, FBXO7, VPS35, and CHCHD2), which are encoded by the nuclear genome [[12], [13], [14], [15], [16], [17], [18], [19], [20], [21], [22], [23], [24], [25]]. At the same time, almost nothing is known about the role of other nuclear genes associated with mitochondrial biogenesis in the development of PD. In this regard, the aim of this work was to study changes in the expression of a number of genes encoded by the nuclear genome and involved in the regulation of mitochondrial biogenesis in the early stages of the development of the pathological process in PD in the brain tissues and peripheral blood of mice with MPTP-induced models of early stages of PD [32,33].
In the present study, we used MPTP-induced models of the presymptomatic stages of PD (with decapitation of mice 6 h after MPTP administration (6h-PSS) and decapitation of mice 24 h after MPTP administration(24h-PSS)) and of the advanced presymptomatic (AdvPSS) and early symptomatic (ESS) stages of PD (with decapitation 2 weeks after MPTP administration). Administration of MPTP allows modeling the death of DAergic neurons in the substantia nigra, the deficiency of DA in the striatum, and the manifestation of the classic signs of PD [41,42]. PD models based on injections of MPTP are one of the most adequate experimental models of this disease. An important advantage of the use of MPTP is that it selectively penetrates in DAergic neurons because of its high affinity for the DA transporter DAT and selectively inhibits complex I of the mitochondrial electron transport chain, causing oxidative stress and impaired calcium homeostasis. These events lead to the degeneration of DAergic neurons by necrosis or apoptosis [[43], [44], [45]].
It should be noted that one of the disadvantages of the models we use is that they are acute, and the development of symptoms of PD occurs quite quickly. At the same time, the advantage of these models is that they are simple to implement and require a small amount of time to create them. The different dosage of MPTP toxin allows us to simulate different stages and quickly conduct an initial assessment of the involvement of certain genes in the development of the disease.
In this work, the analysis of the tissues of the frontal cortex, striatum, substantia nigra, and peripheral blood of mice with MPTP-induced models of PD was carried out. The striatum and substantia nigra are directly involved in the pathogenesis of PD in the very early stages. The frontal cortex is not the primary target in the MPTP model of PD – but its involvement in the pathological process in humans is observed in the late stages of the disease. Blood was examined to further examine the candidate genes being studied as transcriptional peripheral markers of this disease.
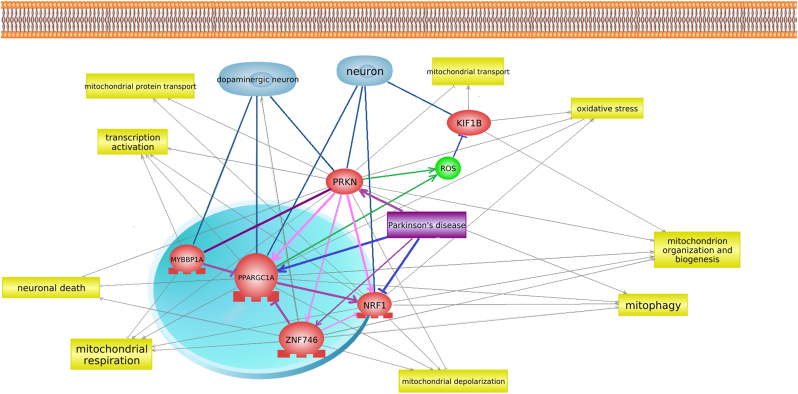
Genes under study were selected using a literature data analysis and the bioinformatics resource Pathway Studio. The functional interactions between these participants in mitochondrial biogenesis are shown in the network of interactions presented in Fig. 1. The main functions of the studied genes are presented in Table 3.
Fig. 1.
Network for the genes included in mitochondrial functioning processes. The network was built using Pathway Studio. A yellow rectangle indicates cellular process, red ellipse indicates studied genes, purple rectangle indicates disease (PD), light blue indicates cell (neuron). Arrows indicates interaction between gene and object: grey – regulation, burgundy – Genetic change, dark blue - quantitative change, green - Promoter Binding, pink – Expression, aquamarine - Cell expression.
Table 3.
The main functions of the Nrf1, Ppargc1a, Zfp746, Prkn, Mybbp1a, and Kif1b genes in molecular-cell processes.
| Gene | Function in the cell | Participation in the functioning of mitochondria | References |
|---|---|---|---|
| Nrf1 | Transcription factor of key genes involved in metabolic and cell development processes. | Participates in the control of nuclear genes necessary for transcription and replication of mitochondrial DNA. | [46,47] |
| Ppargc1a | Transcriptional activator of genes involved in various metabolic pathways. | It is one of the main transcription factors for mitochondrial biogenesis genes | [48,49] |
| Zfp746 | Transcriptional repressor. It plays an important role in the death of neurons. | It is a transcriptional repressor of Ppargc1a | [50,51] |
| Mybbp1a | Transcriptional corepressor. | Transcriptional repressor of myitochondrial biogenesis genes | [52,53] |
| Kif1b | A protein that actively interacts with cell cytoskeleton proteins. | Activator of the antitrade transport of mitochondria | [[54], [55], [56]] |
| Prkn | Ubiquitin E3 ligase, which catalyzes the ubiquitination of a large number of substrates in the cell. | It participates in the degradation of malfunctioning mitochondria using mitophagy. | [[14], [15], [16], [17]]. |
These genes belonged to both the activator (Nrf1, Pprargc1a, Prkn, Kif1b) [[14], [15], [16], [17],[57], [58], [59], [60], [61]] and repressor (Zfp746 и Mybbp1a) [27,62] groups of genes implicated in mitochondrial biogenesis. It should be noted that the genes we selected are encoded by the nuclear genome. Possible changes in the expression of these genes in response to mitochondrial dysfunction can play an important role in the development of neurodegeneration.
As shown in Table 2, there were a greater number of statistically significant changes in the tissues of the brain and peripheral blood of mice with models of AdvPSS and ESS of PD compared with those from models of decapitation performed 6 h and 24 h after the last injection of the toxin. At advanced stages of development of the pathological process in PD, we observed a more-active involvement of changes at the transcriptome level. These results correlate with our previous data which showed that the number of differentially expressed genes increased with the progression of the disease [31].
In general, we observed both a significant decrease and a significant increase in the expression levels of the studied genes in all the studied samples (Table 2). Such changes affect both activators and repressors of mitochondrial biogenesis. Taken together, these results may indicate that the mouse brain tissues studied here possess their own unique scenario of variability in expression levels at different stages of the pathological process.
There is an only a statistically significant decrease in expression of activator genes of mitochondrial biogenesis in the early stages of the developing pathological processes (6h-PSS and 24h-PSS models of PD). This may indicate a disruption in the normal functioning of mitochondria, which may be associated with the development of neurodegeneration. At the same time, the absence of significant changes for the Zfp746 and Mybbp1a genes indicates that the repressors of mitochondrial biogenesis are not involved in the development of the pathological process at these stages.
In general, there was a predominant decrease in the expression of activator genes of mitochondrial biogenesis at all studied stages of the developing neurodegenerative process. Thus, a significant decrease in the mRNA levels of the Ppargc1a, Nrf1, Prkn, and Kif1b genes was detected in 13 cases. In contrast, these genes increased their expression in only 8 cases. This suggests the activator genes are primarily associated with impaired mitochondrial biogenesis and are involved in the processes of neurodegeneration. A significant increase in mRNA levels was observed in the 3 cases studied and a significant decrease in expression was detected in 6 cases, when considering changes in the expression profiles of the repressor genes Zfp746 and Mybbp1a. Such results may conversely be associated with compensatory mechanisms; e.g., the cell tries to restore and maintain mitochondrial biogenesis by suppressing the expression of mitochondrial repressors.
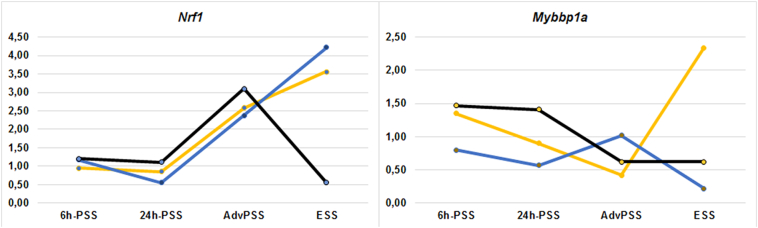
The most conspicuous findings regarding levels of expression were identified for 2 genes - the Nrf1 and Mybbp1a genes (Fig. 2). Statistically significant changes were detected in 9 out of the 16 investigated samples for the Nrf1 gene. NRF1 is a transcription factor that regulates directly the function of a large number of mitochondrial genes, both by itself and through the upregulation by PGC-1α [63,64]. In turn, PGC-1α is a transcriptional activator of mitochondrial biogenesis genes, including NRF1. Shin et al. showed that PRKN knock out leads to a decrease in the expression levels of NRF1 and PPARGC1A in cell culture with a mitochondrial dysfunction model [17].
Fig. 2.
Relative mRNA levels of Nrf1 and Mybbp1a in cortex, striatum and substantia nigra of mice with MPTP-induced models of PD. The expression level studied in the control was taken as 1.
In our study, a statistically significant decrease in the mRNA level of the Nrf1 genes was detected in the striatum of mice with the 24h-PSS model of PD. We observed an increase in Nrf1 expression at more-advanced modeling stages in all studied mouse brain tissues, with the exception of the substantia nigra with the ESS model of PD. These results suggest that in the early stages of neurodegeneration the NRF1 is extensively involved in the compensatory mechanisms, which contribute to the activation of the expression of mitochondrial genes. The significant decrease in the mRNA level of the Nrf1 gene detected in the substantia nigra with the ESS model of PD also confirmed our hypothesis that the cell in the substantia nigra does not have the resources that are necessary to maintain compensatory mechanisms at this stage in the development of the pathological process.
A statistically significant increase in the expression of the Nrf1 gene in the cortex in mice with models of AdvPSS and ESS of PD suggests that this part of the brain can be involved in the development of the pathological process in PD even at the model of presymptomatic stages of the disease. In addition, we found a statistically significant increase in the mRNA level of the Nrf1 gene in the peripheral blood of mice with the 6 h-PSS and ESS models of PD. These data indicate that changes in the expression of the Nrf1 gene can occur in the peripheral blood. NRF1 gene can be considered as a potential biomarker of the development of the pathological process in PD.
Among the repressor genes of mitochondrial biogenesis, Mybbp1a exhibited significant changes in expression in 7 out of the 16 studied samples. The MYBBP1A protein is a transcriptional repressor of rRNA genes and several other genes, including PPARGC1A [[65], [66]]. Currently, its role in the pathogenesis of PD is poorly understood. In our work, we observed a predominant decrease in the mRNA levels of this gene in the striatum and substantia nigra from earlier stages (6 h-PSS) to later stages (ESS). This allows us to suggest that MYBBP1A is also involved in the compensatory mechanisms that reduce the probability of the occurrence of oxidative stress and slow down neurodegenerative processes. In the cortex, in the early stages, a decrease in the expression of this Mybbp1a is also observed, however, mice with the ESS model of PD shows a significant increase, this may indicate the preservation of the ratio of activators and repressors of mitochondrial biogenesis due to the compensatory increase in Nrf1 gene expression. The preservation of the activator/repressor ratio is observed at the ESS stage in the substantia nigra too - but it is achieved due to, on the contrary, a simultaneous decrease in the expression of the Nrf1 and Mybbp1a genes.
It should be noted that the level of expression of the Mybbp1a gene in peripheral blood changes only at the earliest stage (6 h-PSS) and this gene cannot be considered as a candidate expression biomarker of PD.
Here, we studied the changes in the expression of activators (Nrf1, Pprargc1a, Prkn, and Kif1b) and repressors (Zfp746 and Mybbp1a) of mitochondrial biogenesis in the brain and peripheral blood of mice with MPTP-induced models of PD. Statistically significant changes in expression at the mRNA level were detected for all studied genes, while an increase in the number of statistically significant changes in the expression of the studied genes was observed from the early modeled stages to the late stage of PD. This may indicate the gradual involvement of mitochondrial biogenesis genes in the development of pathological processes in PD.
In addition, there was a predominant decrease in activator genes of mitochondrial biogenesis in the studied samples. Apparently, this was because these genes are associated with impaired mitochondrial biogenesis and the development of neurodegenerative processes. A predominant decrease in expression was also detected for repressor genes; however, in this case, this was attributable to the inclusion of compensatory mechanisms during the development of neurodegeneration in mouse brain with models of PD.
The most pronounced changes in expression levels identified for the Nrf1 activator gene and for the Mybbp1a repressor gene. Apparently, these two genes play the most important role in the processes of mitochondrial biogenesis in this disease.
Declartion of competing interest
The authors declare that they have no conflicts of interest regarding the publication of this paper.
CRediT authorship contribution statement
M.M. Rudenok:Validation, Investigation, Data curation, Writing - original draft, Writing - review & editing, Visualization.A.Kh. Alieva:Software, Formal analysis, Investigation, Data curation, Visualization, Funding acquisition.J.S. Starovatykh:Validation, Investigation.M.S. Nesterov:Methodology, Validation, Formal analysis.V.A. Stanishevskaya:Validation, Investigation.A.A. Kolacheva:Methodology, Validation.M.V. Ugryumov:Methodology, Resources.P.A. Slominsky:Conceptualization, Writing - review & editing, Supervision, Funding acquisition.M.I. Shadrina:Conceptualization, Writing - review & editing, Supervision, Project administration, Funding acquisition.
Acknowledgements
This work was carried out on the basis of the center for collective use of IMG RAS “Center of Cell and Gene Technologies”. The work was supported by the financing of research work of IMG RAS (State budget financing, Russian Federation, Moscow, no.AAA-A19-119031390032-2).
Contributor Information
M.M. Rudenok, Email: margaritamrudenok@gmail.com.
P.A. Slominsky, Email: slomin@img.ras.ru.
M.I. Shadrina, Email: shadrina@img.ras.ru.
References
- 1.Galluzzi L., Kepp O., Trojel-Hansen C., Kroemer G. Mitochondrial control of cellular life, stress, and death. Circ. Res. 2012;111:1198–1207. doi: 10.1161/CIRCRESAHA.112.268946. [DOI] [PubMed] [Google Scholar]
- 2.Perier C., Vila M. Mitochondrial biology and Parkinson's disease. Cold Spring Harb. Perspect. Med. 2012;2 doi: 10.1101/cshperspect.a009332. a009332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta. 2012;1817:1833–1838. doi: 10.1016/j.bbabio.2012.02.033. [DOI] [PubMed] [Google Scholar]
- 4.Indran I.R., Tufo G., Pervaiz S., Brenner C. Recent advances in apoptosis, mitochondria and drug resistance in cancer cells. Biochim. Biophys. Acta. 2011;1807:735–745. doi: 10.1016/j.bbabio.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 5.Perier C., Tieu K., Guegan C., Caspersen C., Jackson-Lewis V., Carelli V., Martinuzzi A., Hirano M., Przedborski S., Vila M. Complex I deficiency primes Bax-dependent neuronal apoptosis through mitochondrial oxidative damage. Proc. Natl. Acad. Sci. U. S. A. 2005;102:19126–19131. doi: 10.1073/pnas.0508215102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maharjan S., Oku M., Tsuda M., Hoseki J., Sakai Y. Mitochondrial impairment triggers cytosolic oxidative stress and cell death following proteasome inhibition. Sci. Rep. 2014;4 doi: 10.1038/srep05896. 5896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verstraeten A., Theuns J., Van Broeckhoven C. Progress in unraveling the genetic etiology of Parkinson disease in a genomic era. Trends Genet. 2015;31:140–149. doi: 10.1016/j.tig.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 8.Mahalingam S., Levy L.M. Genetics of Huntington disease. Am. J. Neuroradiol. 2014;35:1070–1072. doi: 10.3174/ajnr.A3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tabrizi S.J., Cleeter M.W., Xuereb J., Taanman J.W., Cooper J.M., Schapira A.H. Biochemical abnormalities and excitotoxicity in Huntington's disease brain. Ann. Neurol. 1999;45:25–32. doi: 10.1002/1531-8249(199901)45:1<25::aid-art6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 10.Swomley A.M., Forster S., Keeney J.T., Triplett J., Zhang Z., Sultana R., Butterfield D.A. Abeta, oxidative stress in Alzheimer disease: evidence based on proteomics studies. Biochim. Biophys. Acta. 2014;1842:1248–1257. doi: 10.1016/j.bbadis.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cottrell D.A., Borthwick G.M., Johnson M.A., Ince P.G., Turnbull D.M. The role of cytochrome c oxidase deficient hippocampal neurones in Alzheimer's disease. Neuropathol. Appl. Neurobiol. 2002;28:390–396. doi: 10.1046/j.1365-2990.2002.00414.x. [DOI] [PubMed] [Google Scholar]
- 12.Schiesling C., Kieper N., Seidel K., Kruger R. Review: Familial Parkinson's disease – genetics, clinical phenotype and neuropathology in relation to the common sporadic form of the disease. Neuropathol. Appl. Neurobiol. 2008;34:255–271. doi: 10.1111/j.1365-2990.2008.00952.x. [DOI] [PubMed] [Google Scholar]
- 13.Youle R.J., Narendra D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011;12:9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park J.S., Davis R.L., Sue C.M. Mitochondrial dysfunction in parkinson's disease: new mechanistic insights and therapeutic perspectives. Curr. Neurol. Neurosci. Rep. 2018;18 doi: 10.1007/s11910-018-0829-3. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pickrell A.M., Youle R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron. 2015;85:257–273. doi: 10.1016/j.neuron.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scarffe L.A., Stevens D.A., Dawson V.L., Dawson T.M. Parkin and PINK1: much more than mitophagy. Trends Neurosci. 2014;37:315–324. doi: 10.1016/j.tins.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin J.H., Ko H.S., Kang H., Lee Y., Lee Y.I., Pletinkova O., Troconso J.C., Dawson V.L., Dawson T.M. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson's disease. Cell. 2011;144:689–702. doi: 10.1016/j.cell.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith W.W., Pei Z., Jiang H., Dawson V.L., Dawson T.M., Ross C.A. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat. Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 19.Ryan B.J., Hoek S., Fon E.A., Wade-Martins R. Mitochondrial dysfunction and mitophagy in Parkinson's: from familial to sporadic disease. Trends Biochem. Sci. 2015;40:200–210. doi: 10.1016/j.tibs.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 20.Mandemakers W., Morais V.A., Strooper D. A cell biological perspective on mitochondrial dysfunction in Parkinson disease and other neurodegenerative diseases. J. Cell Sci. 2007;120:1707–1716. doi: 10.1242/jcs.03443. [DOI] [PubMed] [Google Scholar]
- 21.Guardia-Laguarta C., Area-Gomez E., Rub C., Liu Y., Magrane J., Becker D., Voos W., Schon E.A., Przedborski S. alpha-Synuclein is localized to mitochondria-associated ER membranes. J. Neurosci. 2014;34:249–259. doi: 10.1523/JNEUROSCI.2507-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park J.S., Blair N.F., Sue C.M. The role of ATP13A2 in Parkinson's disease: clinical phenotypes and molecular mechanisms. Mov. Disord. 2015;30:770–779. doi: 10.1002/mds.26243. [DOI] [PubMed] [Google Scholar]
- 23.Fonzo A.D., Dekker M.C.J., Montagna P., Baruzzi A., Yonova E.H., Guedes L.C., Szczerbinska A., Zhao T., Dubbel-Hulsman L.O.M., Wouters C.H., de Graaff E., Oyen W.J.G., Simons E.J., Breedveld G.J., Oostra B.A., Horstink M.W., Bonifati V. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology. 2009;72:240–245. doi: 10.1212/01.wnl.0000338144.10967.2b. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Z.D., Xie S.P., Sathiyamoorthy S., Saw W.T., Sing T.Y., Ng S.H., Chua H.P., Tang A.M., Shaffra F., Li Z., Wang H., Ho P.G., Lai M.K., Angeles D.C., Lim T.M., Tan E.K. F-box protein 7 mutations promote protein aggregation in mitochondria and inhibit mitophagy. Hum. Mol. Genet. 2015;24:6314–6330. doi: 10.1093/hmg/ddv340. [DOI] [PubMed] [Google Scholar]
- 25.Tang F.L., Liu W., Hu J.X., Erion J.R., Ye J., Mei L., Xiong W.C. VPS35 deficiency or mutation causes dopaminergic neuronal loss by impairing mitochondrial fusion and function. Cell Rep. 2015;12:1631–1643. doi: 10.1016/j.celrep.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Borrageiro G., Haylett W., Seedat S., Kuivaniemi H., Bardien S. A review of genome-wide transcriptomics studies in Parkinson's disease. Eur. J. Neurosci. 2018;47:1–16. doi: 10.1111/ejn.13760. [DOI] [PubMed] [Google Scholar]
- 27.Castillo-Quan J.I. Parkin' control: regulation of PGC-1alpha through PARIS in Parkinson's disease. Dis. Model. Mech. 2011;4:427–429. doi: 10.1242/dmm.008227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bendikov-Bar I., Rapaport D., Larisch S., Horowitz M. Parkin-mediated ubiquitination of mutant glucocerebrosidase leads to competition with its substrates PARIS and ARTS. Orphanet J. Rare Dis. 2014;9 doi: 10.1186/1750-1172-9-86. 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye Q., Chen C., Si E., Cai Y., Wang J., Huang W., Li D., Wang Y., Chen X. Mitochondrial effects of PGC-1alpha silencing in MPP(+) treated human SH-SY5Y neuroblastoma cells. Front. Mol. Neurosci. 2017;10 doi: 10.3389/fnmol.2017.00164. 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alieva A.K., Filatova E.V., Kolacheva A.A., Rudenok M.M., Slominsky P.A., Ugrumov M.V., Shadrina M.I. Transcriptome profile changes in mice with MPTP-induced early stages of Parkinson's disease. Mol. Neurobiol. 2017;54:6775–6784. doi: 10.1007/s12035-016-0190-y. [DOI] [PubMed] [Google Scholar]
- 31.Alieva A.K., Zyrin V.S., Rudenok M.M., Kolacheva A.A., Shulskaya M.V., Ugryumov M.V., Slominsky P.A., Shadrina M.I. Whole-transcriptome analysis of mouse models with MPTP-induced early stages of Parkinson's disease reveals stage-specific response of transcriptome and a possible role of myelin-linked genes in neurodegeneration. Mol. Neurobiol. 2018;55:7229–7241. doi: 10.1007/s12035-018-0907-1. [DOI] [PubMed] [Google Scholar]
- 32.Ugrumov M.V., Khaindrava V.G., Kozina E.A., Kucheryanu V.G., Bocharov E.V., Kryzhanovsky G.N., Kudrin V.S., Narkevich V.B., Klodt P.M., Rayevsky K.S., Pronina T.S. Modeling of presymptomatic and symptomatic stages of parkinsonism in mice. Neuroscience. 2011;181:175–188. doi: 10.1016/j.neuroscience.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 33.Kolacheva A.A., Kozina E.A., Volina E.V., Ugryumov M.V. Time course of degeneration of dopaminergic neurons and respective compensatory processes in the nigrostriatal system in mice. Dokl. Biol. Sci. 2014;456:160–164. doi: 10.1134/S0012496614030041. [DOI] [PubMed] [Google Scholar]
- 34.The Guide for the Care and Use of Laboratory Animals. 8th ed. National Academies Press; Washington (DC): 2011. [PubMed] [Google Scholar]
- 35.I Molecular Research Center . 2017. Tri Reagent® - RNA/DNA/Protein Isolation Reagent. [Google Scholar]
- 36.Mus musculus (assembly GRCm38.p6), https://www.ncbi.nlm.nih.gov/genome/?term=Mus+Musculus, NCBI.
- 37.Suslov O., Steindler D.A. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gni176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. (San Diego, Calif.) [DOI] [PubMed] [Google Scholar]
- 39.Hettne K.M., Thompson M., van Haagen H.H., van der Horst E., Kaliyaperumal R., Mina E., Tatum Z., Laros J.F., van Mulligen E.M., Schuemie M., Aten E., Li T.S., Bruskiewich R., Good B.M., Su A.I., Kors J.A., den Dunnen J., van Ommen G.J., Roos M., t Hoen P.A., Mons B., Schultes E.A. The implicitome: a resource for rationalizing gene-disease associations. PLoS One. 2016;11 doi: 10.1371/journal.pone.0149621. e0149621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Au W.L., Adams J.R., Troiano A.R., Stoessl A.J. Parkinson’s disease: in vivo assessment of disease progression using positron emission tomography brain research. Mol. Brain Res. 2005;134:24–33. doi: 10.1016/j.molbrainres.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 41.German D.C., Nelson E.L., Liang C.L., Speciale S.G., Sinton C.M., Sonsalla P.K. The neurotoxin MPTP causes degeneration of specific nucleus A8, A9 and A10 dopaminergic neurons in the mouse. Neurodegeneration. 1996;5:299–312. doi: 10.1006/neur.1996.0041. [DOI] [PubMed] [Google Scholar]
- 42.Meredith G.E., Totterdell S., Potashkin J.A., Surmeier D.J. Parkinsonism Relat. Disord. 2008;14(Suppl. 2):S112–S115. doi: 10.1016/j.parkreldis.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gerlach M., Riederer P. Animal models of Parkinson’s disease: an empirical comparison with the phenomenology of the disease in man. J. Neural Transm. 1996;103:987–1041. doi: 10.1007/BF01291788. [DOI] [PubMed] [Google Scholar]
- 44.Langston J.W., Ballard P., Tetrud J.W., Irwin I. Chronic Parkinsonism in humans due to a product of meperidine-analog synthesis. Science. 1983;219:979–980. doi: 10.1126/science.6823561. [DOI] [PubMed] [Google Scholar]
- 45.Przedborski S., Jackson-Lewis V., Djaldetti R., Liberatore G., Vila M., Vukosavic S., Almer G. The parkinsonian toxin MPTP: action and mechanism. Restor. Neurol. Neurosci. 2000;16:135–142. [PubMed] [Google Scholar]
- 46.Lee D., Ryu K.Y. Effect of cellular ubiquitin levels on the regulation of oxidative stress response and proteasome function via Nrf1. Biochem. Biophys. Res. Commun. 2017;485:234–240. doi: 10.1016/j.bbrc.2017.02.105. [DOI] [PubMed] [Google Scholar]
- 47.Kiyama T., Chen C.K., Wang S.W., Pan P., Ju Z., Wang J., Takada S., Klein W.H., Mao C.A. Essential roles of mitochondrial biogenesis regulator Nrf1 in retinal development and homeostasis. Mol. Neurodegener. 2018;13 doi: 10.1186/s13024-018-0287-z. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scarpulla R.C. Metabolic control of mitochondrial biogenesis through the PGC-1 family regulatory network. Biochim. Biophys. Acta. 2011;1813:1269–1278. doi: 10.1016/j.bbamcr.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Theilen N.T., Kunkel G.H., Tyagi S.C. The role of exercise and TFAM in preventing skeletal muscle atrophy. J. Cell. Physiol. 2017;232:2348–2358. doi: 10.1002/jcp.25737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevens D.A., Lee Y., Kang H.C., Lee B.D., Lee Y.I., Bower A., Jiang H., Kang S.U., Andrabi S.A., Dawson V.L., Shin J.H., Dawson T.M. Parkin loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc. Natl. Acad. Sci. U. S. A. 2015;112:11696–11701. doi: 10.1073/pnas.1500624112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee Y., Stevens D.A., Kang S.U., Jiang H., Lee Y.I., Ko H.S., Scarffe L.A., Umanah G.E., Kang H., Ham S., Kam T.I., Allen K., Brahmachari S., Kim J.W., Neifert S., Yun S.P., Fiesel F.C., Springer W., Dawson V.L., Shin J.H., Dawson T.M. PINK1 primes parkin-mediated ubiquitination of PARIS in dopaminergic neuronal survival. Cell Rep. 2017;18:918–932. doi: 10.1016/j.celrep.2016.12.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.George B., Horn D., Bayo P., Zaoui K., Flechtenmacher C., Grabe N., Plinkert P., Krizhanovsky V., Hess J. Regulation and function of Myb-binding protein 1A (MYBBP1A) in cellular senescence and pathogenesis of head and neck cancer. Cancer Lett. 2015;358:191–199. doi: 10.1016/j.canlet.2014.12.042. [DOI] [PubMed] [Google Scholar]
- 53.Fan M., Rhee J., St-Pierre J., Handschin C., Puigserver P., Lin J., Jäeger S., Erdjument-Bromage H., Tempst P., Spiegelman B.M. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1alpha: modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pareyson D., Saveri P., Sagnelli A., Piscosquito G. Mitochondrial dynamics and inherited peripheral nerve diseases. Neurosci. Lett. 2015;596:66–77. doi: 10.1016/j.neulet.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Gentil B.J., Cooper L. Molecular basis of axonal dysfunction and traffic impairments in CMT. Brain Res. Bull. 2012;88:444–453. doi: 10.1016/j.brainresbull.2012.05.003. [DOI] [PubMed] [Google Scholar]
- 56.Hirokawa N., Takemura R. Biochemical and molecular characterization of diseases linked to motor proteins. Trends Biochem. Sci. 2003;28:558–565. doi: 10.1016/j.tibs.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 57.Cam H., Balciunaite E., Blais A., Spektor A., Scarpulla R.C., Young R., Kluger Y., Dynlacht B.D. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 58.Adam T., Opie L.H., Essop M.F. AMPK activation represses the human gene promoter of the cardiac isoform of acetyl-CoA carboxylase: role of nuclear respiratory factor-1. Biochem. Biophys. Res. Commun. 2010;398:495–499. doi: 10.1016/j.bbrc.2010.06.106. [DOI] [PubMed] [Google Scholar]
- 59.Wareski P., Vaarmann A., Choubey V., Safiulina D., Liiv J., Kuum M., Kaasik A. PGC-1{alpha} and PGC-1{beta} regulate mitochondrial density in neurons. J. Biol. Chem. 2009;284:21379–21385. doi: 10.1074/jbc.M109.018911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lin M.Y., Sheng Z.H. Regulation of mitochondrial transport in neurons. Exp. Cell Res. 2015;334:35–44. doi: 10.1016/j.yexcr.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao C., Takita J., Tanaka Y., Setou M., Nakagawa T., Takeda S., Yang H.W., Terada S., Nakata T., Takei Y., Saito M., Tsuji S., Hayashi Y., Hirokawa N. Charcot-Marie-Tooth disease type 2A caused by mutation in a microtubule motor KIF1Bbeta. Cell. 2001;105:587–597. doi: 10.1016/s0092-8674(01)00363-4. [DOI] [PubMed] [Google Scholar]
- 62.Hochstatter J., Holzel M., Rohrmoser M., Schermelleh L., Leonhardt H., Keough R., Gonda T.J., Imhof A., Eick D., Langst G., Nemeth A. Myb-binding protein 1a (Mybbp1a) regulates levels and processing of pre-ribosomal RNA. J. Biol. Chem. 2012;287:24365–24377. doi: 10.1074/jbc.M111.303719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang G.M., Deng M.T., Lei Z.H., Wan Y.J., Nie H.T., Wang Z.Y., Fan Y.X., Wang F., Zhang Y.L. Effects of NRF1 on steroidogenesis and apoptosis in goat luteinized granulosa cells. Reproduction. 2017;154:111–122. doi: 10.1530/REP-16-0583. [DOI] [PubMed] [Google Scholar]
- 64.Scarpulla R.C. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 65.Kang H., Shin J.H. Repression of rRNA transcription by PARIS contributes to Parkinson's disease. Neurobiol. Dis. 2015;73:220–228. doi: 10.1016/j.nbd.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 66.Yoshihara E., Fujimoto S., Inagaki N., Okawa K., Masaki S., Yodoi J., Masutani H. Disruption of TBP-2 ameliorates insulin sensitivity and secretion without affecting obesity. Nat. Commun. 2010;1 doi: 10.1038/ncomms1127. 127. [DOI] [PMC free article] [PubMed] [Google Scholar]