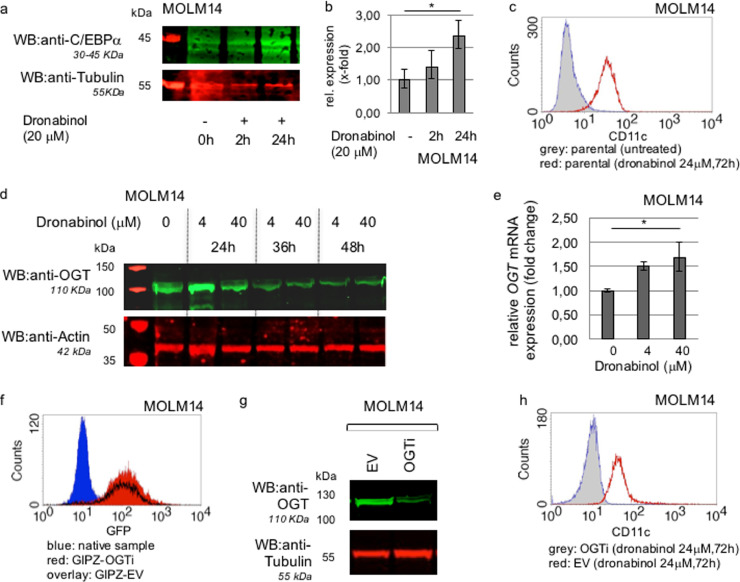
Fig. 10.
In vitro modeling of release of the differentiation blockage upon treatment with dronabinol. (a) Representative western immunoblot for C/EBPalpha expression in response to dronabinol in MOLM14 leukemia cells. Probing for Tubulin serves as a loading control. First lane: protein ladder. (b) qRT-PCR approach to measure C/EBPalpha mRNA expression levels in response to dronabinol. * significance at p < 0.05 (t-test). (c) Representative flow cytometry assay to determine relative expression of CD11c in MOLM14 cells as assessed by flow cytometry in response to dronabinol. (d) Representative western immunoblot for OGT expression in response to dronabinol. Probing for Actin serves as loading control. First lane: protein ladder. (e) qRT-PCR of MOLM14 cells treated with dronabinol for 24 h. Relative fold change of OGT expression levels are shown. * confidence interval 95%. GAPDH serves as housekeeping gene. (f) OGT-interference in MOLM14 cells: Flow cytometry assay to determine transduction efficiency as assessed by green fluorescence protein (GFP) reactivity. GIPZ-OGTi (lentiviral plasmid OGT shRNA vector), EV (empty vector control). (g) Representative western immunoblot demonstrating successful suppression of OGT translation. EV (empty vector control). Probing for tubulin serves as loading control. (h) Representative analyses of relative expression of CD11c as assessed by flow cytometry in response to dronabinol. OGT-interferenced (OGTi) MOLM14 cells vs. empty vector control cell strains.