Abstract
Objective
Decreased muscle mass is a major contributor to age-related morbidity, and strategies to improve muscle regeneration during ageing are urgently needed. Our aim was to identify the subset of relevant microRNAs (miRNAs) that partake in critical aspects of muscle cell differentiation, irrespective of computational predictions, genomic clustering or differential expression of the miRNAs.
Methods
miRNA biogenesis was deleted in primary myoblasts using a tamoxifen-inducible CreLox system and combined with an add-back miRNA library screen. RNA-seq experiments, cellular signalling events, and glycogen synthesis, along with miRNA inhibitors, were performed in human primary myoblasts. Muscle regeneration in young and aged mice was assessed using the cardiotoxin (CTX) model.
Results
We identified a hierarchical non-clustered miRNA network consisting of highly (miR-29a), moderately (let-7) and mildly active (miR-125b, miR-199a, miR-221) miRNAs that cooperate by directly targeting members of the focal adhesion complex. Through RNA-seq experiments comparing single versus combinatorial inhibition of the miRNAs, we uncovered a fundamental feature of this network, that miRNA activity inversely correlates to miRNA cooperativity. During myoblast differentiation, combinatorial inhibition of the five miRNAs increased activation of focal adhesion kinase (FAK), AKT, and p38 mitogen-activated protein kinase (MAPK), and improved myotube formation and insulin-dependent glycogen synthesis. Moreover, antagonizing the miRNA network in vivo following CTX-induced muscle regeneration enhanced muscle mass and myofiber formation in young and aged mice.
Conclusion
Our results provide novel insights into the dynamics of miRNA cooperativity and identify a miRNA network as therapeutic target for impaired focal adhesion signalling and muscle regeneration during ageing.
Keywords: microRNA network, Primary human muscle cells, Skeletal muscle regeneration, Focal adhesion signalling, Glycogen synthesis
Highlights
-
•
The microRNA pathway is connected to focal adhesion signalling during muscle cell differentiation through a small set of microRNAs.
-
•
microRNAs can form hierarchical networks independent of their genomic localization in which activity inversely correlates to target gene cooperativity.
-
•
Relieving the regulation of focal adhesion members by microRNA inhibition improves insulin signaling and glycogen synthesis in human muscle cells.
-
•
Targeting a microRNA network during muscle regeneration improves muscle formation in young and aged mice.
1. Introduction
Skeletal muscle comprises 40% of whole body mass and plays a critical role in the overall health of humans [1]. It not only ensures movement, posture, and stability, but also regulates heat production and whole body metabolism [2]. After 40 years of age, muscle mass starts to decline by approximately 8% per decade [3]. Loss of muscle mass during ageing, or sarcopenia, is a key contributor to adverse events in the elderly such as falls, disability and death, which are summarized under the term frailty [4]. Sarcopenia affects 5–13% of seniors aged 60–70 years and up to 50% of seniors aged more than 80 years [5]. It is therefore important to understand the mechanisms of muscle formation and to develop strategies that help to maintain muscle mass and function throughout life.
A vital requirement for skeletal muscle is its capacity to contract. To prevent muscle damage during contraction, the contractile apparatus is tightly connected to the extracellular matrix (ECM) through the costamere, a sarcolemmal lattice [6]. The costamere is comprised of the focal adhesion complex and the dystrophin glycoprotein complex that connect the sarcolemma to the extracellular matrix. Moreover, both protein complexes are interconnected and linked to the Z disc of the sarcomere. The periodicity of the costamere ensures an even distribution of force transduction from the muscle fiber to the ECM during contraction. Focal adhesion kinase (FAK, encoded by PTK2) is a non-receptor tyrosine kinase that acts as a mechanosensor at the core of the focal adhesion complex and receives signals from the ECM via integrins [7]. FAK integrates extracellular signals from the ECM and multiple growth factors to elicit the activation of anti-apoptotic pathways intracellularly. Unsurprisingly, FAK signaling is stimulated by exercise [8] as well as skeletal muscle hypertrophy [9]. However, FAK is also crucial for muscle stem cell differentiation as it is activated early in differentiating myoblasts [[10], [11], [12]] and can activate pathways that drive the myogenic differentiation program, such as phosphatidylinositol 3-kinase (PI3K) and p38 mitogen-activated protein kinase (MAPK) signaling [[13], [14], [15]].
MicroRNAs (miRNAs) are a family of short, non-coding RNA molecules that are potential therapeutic targets to improve skeletal muscle mass and function. miRNAs impact the protein output of a cell in a unique manner. Typically, one miRNA binds many of its target genes through the seed motif, 6 nucleotides at its 5′-end, which recognizes the miRNA response elements (MREs) in the 3′-untranslated regions (UTRs) of mRNAs [16]. miRNA binding leads to decapping of the mRNA, 5′-to-3′-decay and a subsequent decrease in protein output [17,18]. Although the effect of miRNAs on their target genes is typically modest, miRNAs have been assigned important functions in diverse biological aspects such as cancer [19], metabolism [20], genetic noise cancellation [21], biomarkers [22] and as therapeutic targets in a wide range of diseases [23]. Skeletal muscle formation has become an essential paradigm in understanding the role of miRNAs in the processes of development and differentiation. Over the last decade, miRNAs have emerged as powerful regulators of myogenesis [24], and aberrant expression of miRNAs underlie various skeletal muscle diseases [25]. Global loss of miRNAs by the deletion of essential components of the miRNA biosynthesis pathway, such as the RNAse Dicer, has underscored the role of miRNAs during the development of skeletal muscle. The deletion of dicer at embryonic days 10.5–12.5 (MyoDCre x Dicerfloxflox) [26] or in satellite cells in adult mice (Pax7CreER/+ x Dicerfloxflox) [27] severely decreased muscle fiber formation.
Cooperativity is emerging as an important determinant of miRNA function that can enhance target gene regulation. Co-regulation of target genes by a group of miRNAs can result in stronger effects than those achieved by individual miRNAs alone, depending on the distance of the MREs [28,29]. Accordingly, computational workflows that predict miRNA cooperativity at the target gene level indicate that miRNA cooperativity might be more frequent than previously anticipated and could affect thousands of human genes [30]. miRNA cooperativity is not only being discovered by target gene predictions, but also by the localization of miRNA clusters within the genome. miRNA clusters are polycistronic and form one long primary transcript (pri-miRNA) from which single miRNA precursors (pre-miRNAs) are cleaved [31]. MiRNAs that arise from clusters exhibit similar expression patterns, although their individual abundance can vary substantially [32,33]. Different miRNA clusters can contain miRNA paralogs, indicating a role of gene duplication in their evolution. When clusters are defined as miRNAs separated by a distance of less than 3000 nucleotides, an estimated 42% of human miRNAs appear in clusters of more than two, usually two to three, miRNAs [34]. In a different study, 146 human miRNAs were identified to form 51 clusters [35]. The true number of miRNA clusters might be even higher, since the abundance of miRNA sequences can correlate over a distance of up to 50000 nucleotides [36]. Large scale computational analysis predicted the presence of >400 miRNA clusters in the human genome [37]. Regardless of the exact numbers, miRNA clustering is higher in the genome than expected at random, suggesting that the co-expression of miRNAs and miRNA cooperativity provide an important evolutionary advantage.
miRNA cooperativity provides unique opportunities for enhanced target gene regulation and miRNA function, and its complex dynamics are slowly beginning to be unraveled. To uncover miRNA cooperativity in an unbiased and systematic fashion, irrespective of genomic clustering or the differential expression of miRNAs, we genetically deleted miRNA biosynthesis in primary muscle cells during differentiation. We identified a small set of miRNAs which targets the focal adhesion complex and which negatively influences the differentiation of myogenic progenitors in vitro and in vivo. We demonstrate that this hierarchical non-clustered network, consisting of miRNAs with varying degrees of activity, engage in target gene cooperativity. Finally, we provide evidence that targeting such a network could be used therapeutically to improve regeneration in aged muscle.
2. Results
2.1. Genetic deletion of DGCR8 in primary myoblasts and add-back of a small group of miRNAs significantly regulates the focal adhesion gene cluster
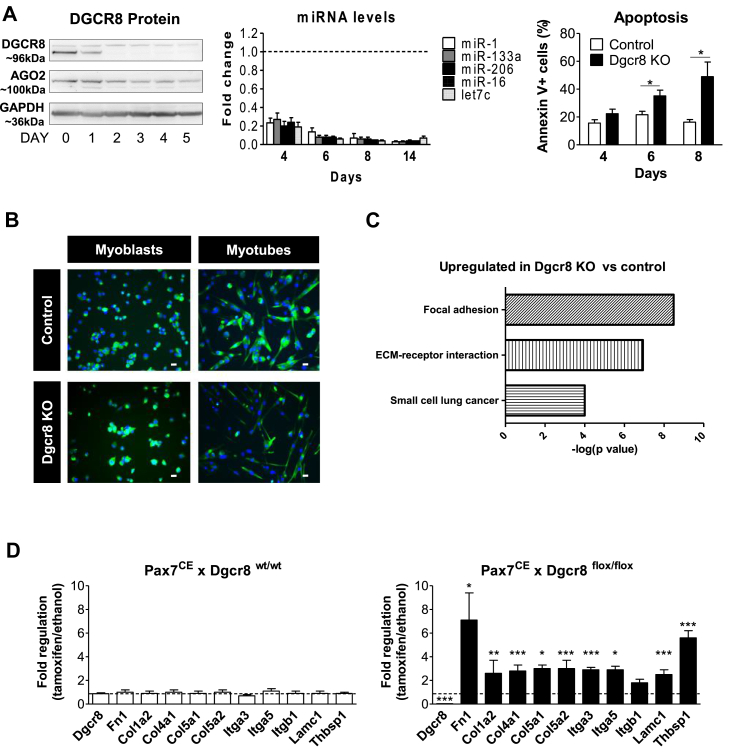
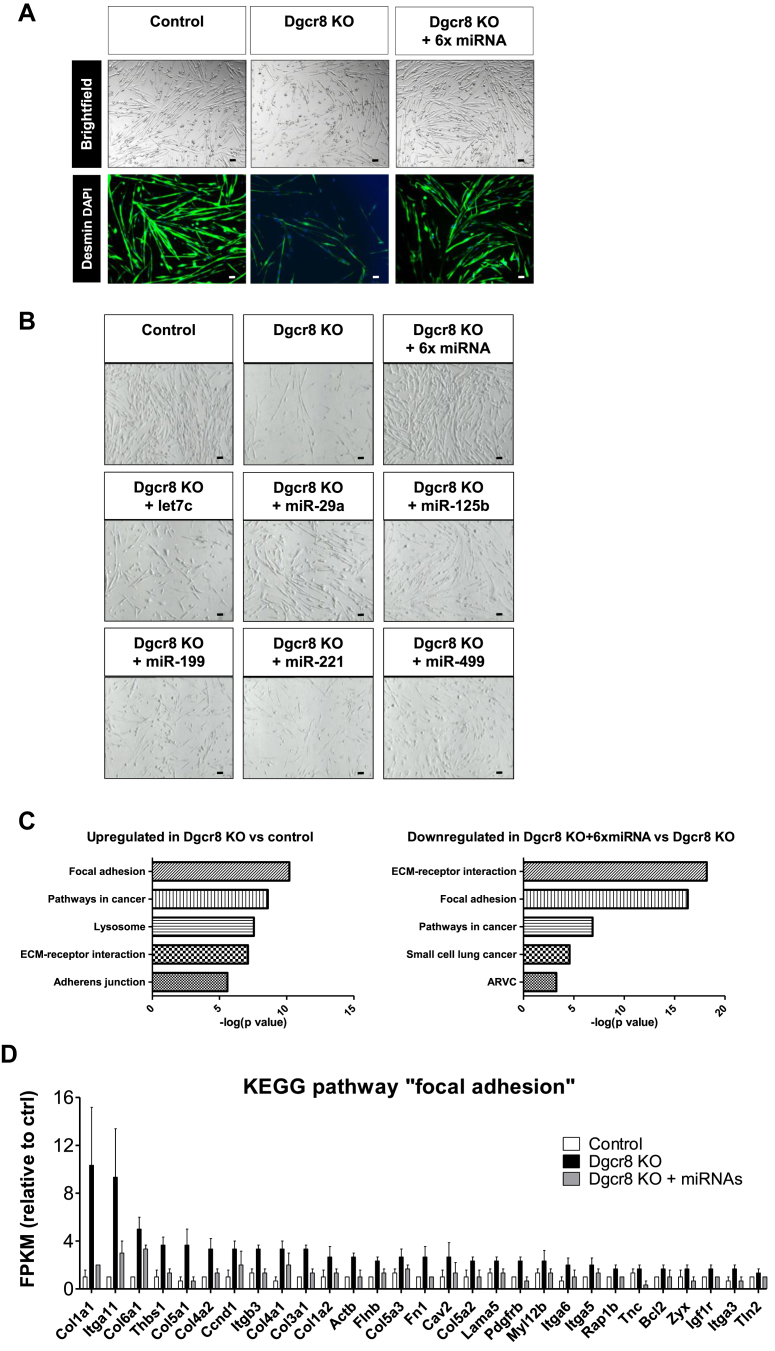
To identify signaling pathways interconnected to the miRNA pathway in skeletal muscle cells, we developed a genetic screening platform based on the depletion of myoblasts of all miRNAs and subsequently screening for gene regulation following miRNA add-back (Suppl.Fig.1). We chose to work with miRNA-depleted cells to avoid any potential redundancy of gene regulation by miRNA families and cooperativity of miRNAs. To achieve this aim, we generated the transgenic mouse line Pax7CE x Dgcr8flox/flox and isolated primary myoblasts from skeletal muscle based on positive selection for α7-integrin [33]. We chose to delete DGCR8 since this RNA-binding protein is necessary for pri-miRNA processing but not required for processing of exogenous RNA duplexes. The Pax7CE transgene contains a tamoxifen-inducible Cre recombinase driven by the endogenous Pax7 promoter and allows for activation of Cre expression in myogenic progenitor cells [38,39]. The deletion of Dgcr8 was induced by tamoxifen incubation and cells were harvested at different time points for analysis under proliferating conditions and during differentiation. Efficiency of tamoxifen-induced genomic recombination at exon 3 of the Dgcr8 allele was >95% (data not shown) and DGCR8 protein became undetectable 2 days after the tamoxifen treatment (Figure 1A). The expression of another important component of the miRNA pathway, argonaute 2 (AGO2), remained detectable throughout all time points tested (Figure 1A). Following the loss of DGCR8, miRNA levels gradually decreased and dropped to less than 10% after 6 days, at which point the onset of apoptosis was observed (Figure 1A). To avoid the influence of apoptosis, differentiation was subsequently induced at day 4 for 48 h. Although Dgcr8-depleted myoblasts were able to form multinucleated myotubes, they displayed a markedly thinner myotube morphology (Figure 1B). To identify pathways affected by the loss of miRNA biosynthesis, we determined the global gene expression profile four days after Cre induction in Pax7CE x Dgcr8flox/flox myoblasts using Agilent mRNA microarrays. At this time point, 458 transcripts were upregulated and 235 transcripts were downregulated >2-fold with a p-value <0.05. The most significant regulated KEGG pathway was “focal adhesion” with a p-value of 3.2E-9 (Figure 1C). This cluster contained 15 genes upregulated in the DGCR8 knockout cells, and we confirmed regulation of 10 of these genes using qRT-PCR (Figure 1D). The results of the complete KEGG pathway analysis is shown in Suppl. Table 1. Next we selected two genes as readouts for our miRNA screen, thrombospondin-1 (Thbs-1), as part of the KEGG pathway “focal adhesion”, and plasminogen activator inhibitor-1 (Pai-1), an inflammatory marker known to positively correlate with Thbs-1 [40]. Indeed, we reconfirmed a robust and time-dependent upregulation of both genes upon the deletion of Dgcr8 in myoblasts (Suppl.Fig.2). Additionally, we performed small RNA deep sequencing in wild type myoblasts to identify the highest expressed miRNAs in muscle cells. The top 85% of all miRNAs expressed in at least one myoblast culture were used to generate a library for our screen (Suppl. Fig. 3). miRNA seed families were represented by one member, e.g., let-7c for the let7 family, and three miRNA sequences were excluded since they were not conserved between mice and humans (mmu-miR-351–5p, mmu-miR-434–5p and mmu-miR-541–5p). Although not part of the top 85% of miRNAs in myoblasts, we included the muscle-specific miRNAs miR-1, miR-133, miR-206 and miR-499. Since we only studied miRNAs that are conserved between mice and humans, we omitted the prefix “mmu” or “hsa” before the miRNA name throughout this paper. miRNAs from the library were transfected into Dgcr8-depleted myoblasts, and the expression of Thbs-1 and Pai-1 was analyzed under both proliferating conditions (MB) and during differentiation (MT) (Suppl. Fig. 4A). First, we ranked the miRNAs based on downregulation of the reporter genes in two independent library transfections. We then selected those miRNAs that scored for downregulation of Thbs-1 or Pai-1 in the top 10 of all miRNAs independent of differentiation (in both MB and MT). Six miRNAs fulfilled these criteria as highlighted in Suppl. Fig. 4B. This set of six miRNAs was able to rescue the marked morphological alterations of Dcgr8 KO myotubes (Figure 2A). Importantly, adding back each of the six miRNAs alone was not sufficient to rescue cell morphology (Figure 2B). Finally, we assessed the ability of this group of miRNAs to rescue the loss of all miRNAs in muscle cells at the molecular level using RNA deep sequencing. Consistent with the microarray analysis on proliferating myoblasts, the KEGG pathway “focal adhesion” was again the most significantly upregulated gene cluster in DGCR8 KO cells during differentiation. Importantly, this upregulation was reversed by the add-back of the six miRNAs (Figure 2C,D). Together, these results demonstrate that the miRNA pathway is strongly linked to focal adhesion signaling in myoblasts through a very limited set of miRNAs.
Figure 1.
Genetic deletion of the miRNA pathway induces the gene cluster “focal adhesion” in proliferating primary myoblasts. A. Myoblast cultures (Pax7CExDgcr8flox/flox) were incubated for 48 h with tamoxifen to induce Cre recombinase (Dgcr8 KO) or with vehicle (Control). Deletion of Dgcr8 induced a time-dependent decrease of DGCR8 protein (western blotting), miRNA levels (qRT-PCR normalized for sno234, n = 4, control represented by the dashed line) and onset of apoptosis (flow cytometry for annexin V staining, n = 6–11). B. Primary myoblasts were treated as in A and differentiated into myotubes for 48 h four days after the beginning of the tamoxifen incubation (day 4). Desmin (green), nuclear DAPI ((blue), 20× magnification, scale bar = 50 μm. C. KEGG pathway analysis of RNA isolated from proliferating Dgcr8 KO and control myoblasts at day 4. D. Expression of Dcgr8 and members of the KEGG pathway “focal adhesion” in Pax7CExDgcr8wt/wt (control) and Pax7CExDgcr8flox/flox myoblasts measured using qRT-PCR at day 4 after tamoxifen incubation, n = 4. The dashed line represents incubation with vehicle. All results are shown as mean ± SEM. ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, student's t test.
Figure 2.
A combination of six miRNAs rescues myotube morphology and reverses induction of the focal adhesion gene cluster following DGCR8 deletion. Dgcr8 KO and control myoblasts were transfected with control mimics, the indicated individual six miRNA mimics or their combination (6× miRNA) two days after beginning of the tamoxifen incubation. Twenty-four hours after transfection, myoblast differentiation was induced for 48 h. Myotube morphology was analyzed using immunofluorescence for desmin (A) and brightfield microscopy (B), 10× magnification. Scale bar = 50 μm. C. KEGG pathway analysis of RNA isolated from control, Dgcr8 myotubes and Dgcr8 myotubes transfected with the combination of six miRNAs. D. RNA levels as determined by RNA deep sequencing in control and DGCR8 knockout cells with or without transfection of the six miRNA mimics, n = 3. Shown are all genes that are significantly upregulated in the knockout cells and significantly downregulated after transfection with the mimics (p value < 0.05 according to manufacturer's analysis).
2.2. miRNA cooperativity enhances differentiation and activates focal adhesion signaling in primary myoblasts from mice and humans
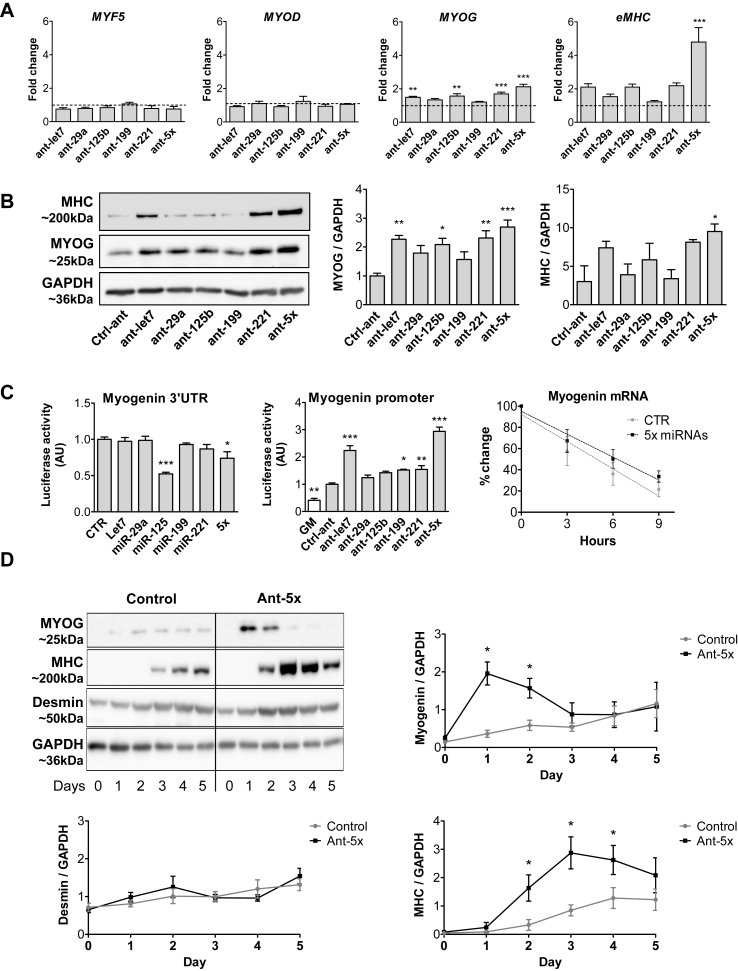
To further clarify the mechanisms and cooperativity of the identified set of miRNAs as well as its conservation between species, we turned to loss-of-function assays in wildtype skeletal muscle cells from mice and humans. Since miR-499 was not among the top 85% of the most abundant miRNAs in myoblasts (Suppl. Fig. 3), we continued with only five miRNAs with largely conserved expression levels in mouse and human primary myoblasts (Suppl. Fig. 5). Inhibition of these five miRNAs in human myoblasts at the onset of differentiation specifically upregulated myogenin among the myogenic regulatory factors (MRFs) and strongly induced embryonic myosin heavy chain (eMHC) at the mRNA and protein level (Figure 3A,B). Especially in the case of eMHC, this effect was most pronounced in the combination of all 5 antagomirs (Ant-5x) (Figure 3A,B). The same cooperative miRNA effect on MHC expression was observed during differentiation of primary myoblasts from mice (Suppl.Fig.6). We also transfected miRNA mimics into human myoblasts and analyzed the different stages of myogenic differentiation at the mRNA level (Suppl.Fig.7a). Interestingly, three out of the five miRNAs (let7, miR-199, miR-125) increased differentiation at the level of MyoD, myogenin, or myosin heavy chain, while miR-29a or miR-221 overexpression had no effect. The combinatorial overexpression overall caused no alterations in differentiation. We confirmed successful and marked overexpression for the single miRNAs as well as for their combination (Suppl. Fig. 7b). These results are consistent with our previous data obtained in the DGCR8 KO cells, where miRNA overexpression favored myotube formation and revealed a non-symmetrical effect of miRNA mimics and inhibitors during muscle cell differentiation. Our results are in line with published data of another miRNA network that acts during the differentiation of endothelial cells, where both gain and loss of function of the miRNAs displayed a non-symmetrical effect and affected differentiation in the same direction regardless of the manipulation [41]. We hypothesize that the complex equilibrium of a miRNA network during cellular differentiation requires controlled miRNA levels and that the marked overexpression of miRNAs cannot easily be compared with miRNA inhibitors under these conditions.
Figure 3.
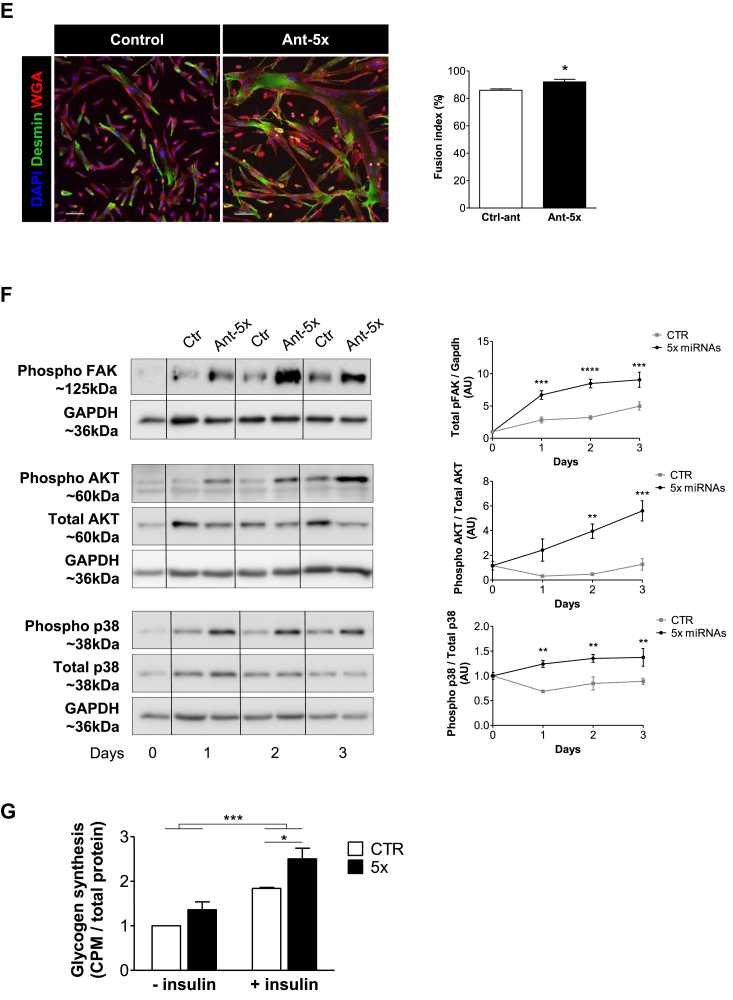
A combination of five miRNAs accelerates differentiation of human myoblasts and improves insulin sensitivity downstream of FAK signaling. Human primary myoblasts were transfected with equal concentrations of control antagomirs or antagomirs against the indicated miRNAs, either as single antagomirs or in combination (Ant-5x). Twenty-four hours after transfection, differentiation was induced for two days (A, B, C, E, G) or up to five days (D, F). A. Gene expression of myogenic regulatory factors and eMHC was analyzed by qRT-PCR and normalized for 18S RNA, n = 5. B. Protein expression of myogenin and eMHC was analyzed by western blot and normalized to GAPDH (n = 4–6). C. Luciferase vectors harboring either the myogenin 3′UTR (n = 4) or promoter region (n = 5) were transfected with miRNA mimics or antagomirs respectively. Myogenin mRNA from control and Ant-5x treated samples was measured by qRT-PCR at the indicated time points following Actinomycin D administration (n = 3). D. Time course of myogenin and eMHC protein expression during five days of differentiation, normalized to GAPDH (n = 4–6). E. Immunofluorescent analysis of myotube formation using anti-desmin and wheat germ agglutinin (WGA). Fusion index was calculated as percentage of nuclei present in cells containing at least two nuclei compared to all nuclei per well (scale bar 100um, n = 4). F. Phosphorylation of p38 MAPK, AKT and FAK during the first three days of differentiation, n = 4. G. Insulin-dependent glycogen synthesis, n = 3. ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, A, B, C: One way-ANOVA with Dunnett's multiple comparison test, D,E, F: student's t test. G: Two way-ANOVA. All results are shown as mean ± SEM.
To explore miRNA cooperativity at the level of the MYOG gene, we used the 3′UTR and the promoter region of myogenin in luciferase assays (Figure 3C). The myogenin 3′UTR was only regulated by miR-125 mimics, while the myogenin promoter responded to all single transfected antagomirs, albeit the strongest regulation was again observed for the Ant-5x combination. The rate of decay for myogenin mRNA did not differ between control and Ant-5x samples (Figure 3C). These data demonstrate that the set of miRNAs induces the transcription of myogenin upstream of its promoter activity, but does not affect myogenin mRNA degradation via binding to its 3′UTR. Time course experiments confirmed that Ant-5x accelerated myogenic differentiation, causing a surge in the expression of myogenin followed by MHC 24 h earlier compared to control human myotubes (Figure 3D), and resulting in more pronounced myotube formation with increased complexity and an increased fusion index (Figure 3E). Since depletion of the miRNA pathway significantly alters focal adhesion signaling in myoblasts, and given that FAK influences muscle differentiation [[10], [11], [12]], we evaluated the activation of FAK following the inhibition of the five miRNAs. Indeed, FAK phosphorylation was strongly induced during the differentiation of human myoblasts treated with Ant-5x as compared to control antagomir (Figure 3F). The phosphoinositide-3-kinase (PI3K)-Akt pathway and the p38 mitogen-activated protein kinase (MAPK) are downstream of FAK signaling [[13], [14], [15]] and are both necessary for the initiation of the myogenic program [42]. Accordingly, phosphorylation of both AKT and p38 MAPK was 2-fold higher in myotubes treated with the antagomir combination than control antagomir (Figure 3F). To investigate if enhanced phosphorylation of AKT also relates to improved muscle cell function, we measured insulin-dependent glycogen synthesis. Human myotubes transfected with Ant-5x had heightened insulin sensitivity and produced significantly more glycogen when challenged with insulin than control antagomir treated myotubes (Figure 3G). Together, these results support our initial observation that focal adhesion signaling couples the miRNA pathway to the activation of the myogenic lineage program. Lastly, we tested the difference between silencing the selected five miRNAs compared to silencing the previous six miRNAs including miR-499. Immunoblot analysis of muscle differentiation and intracellular signaling showed that adding miR-499 antagomirs to Ant-5x did not further enhance the activation of the myogenic lineage program (Suppl. Figure 8) supporting the view that it is indeed the set of five miRNAs that is critical for muscle cell differentiation.
2.3. Genome-wide analysis of miRNA cooperativity during human muscle cell differentiation reveals a hierarchical regulatory network of the focal adhesion complex
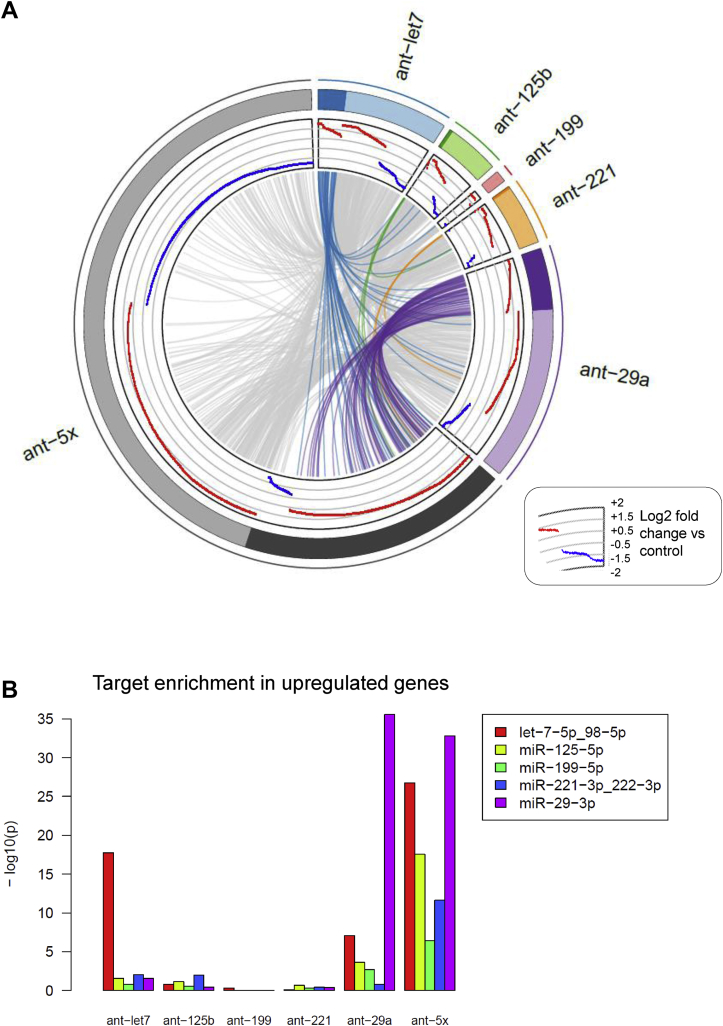
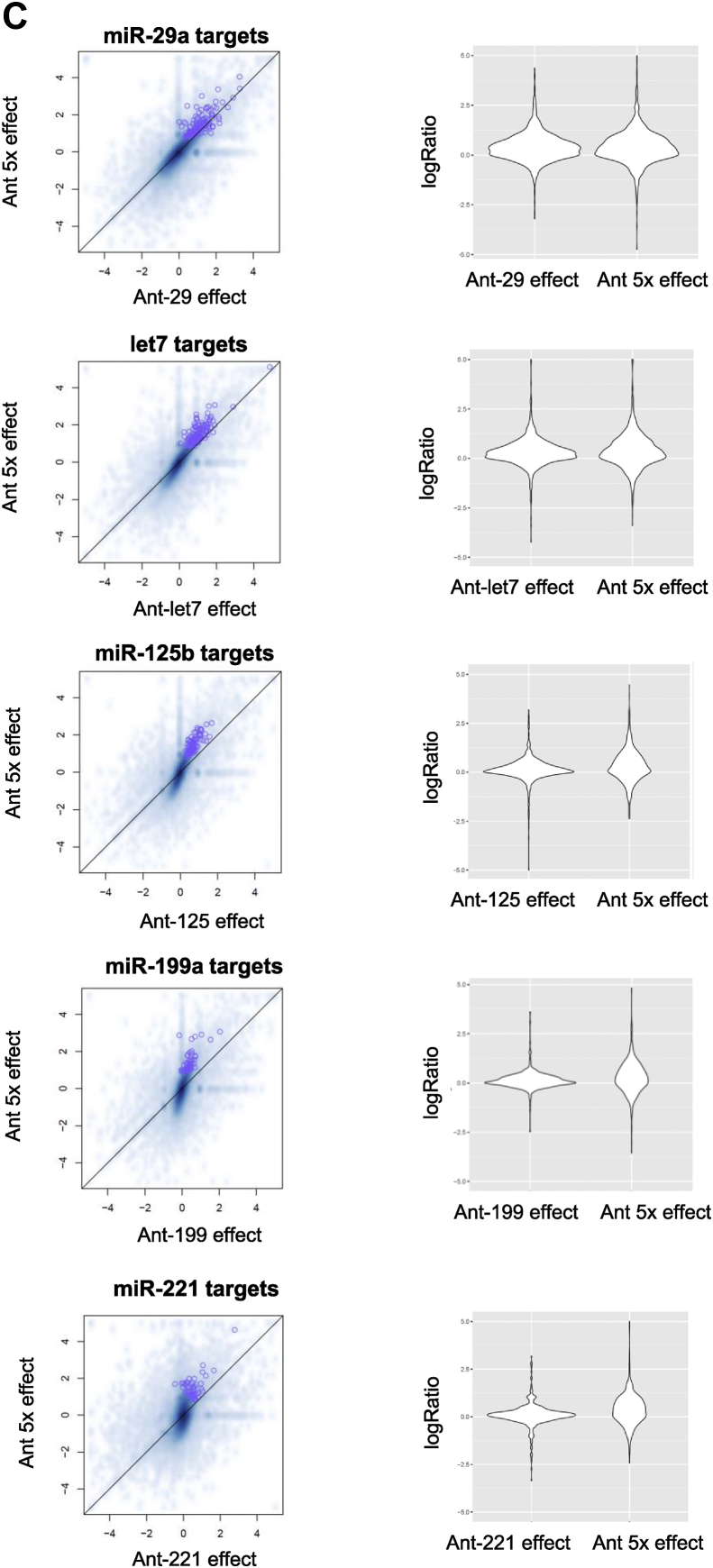
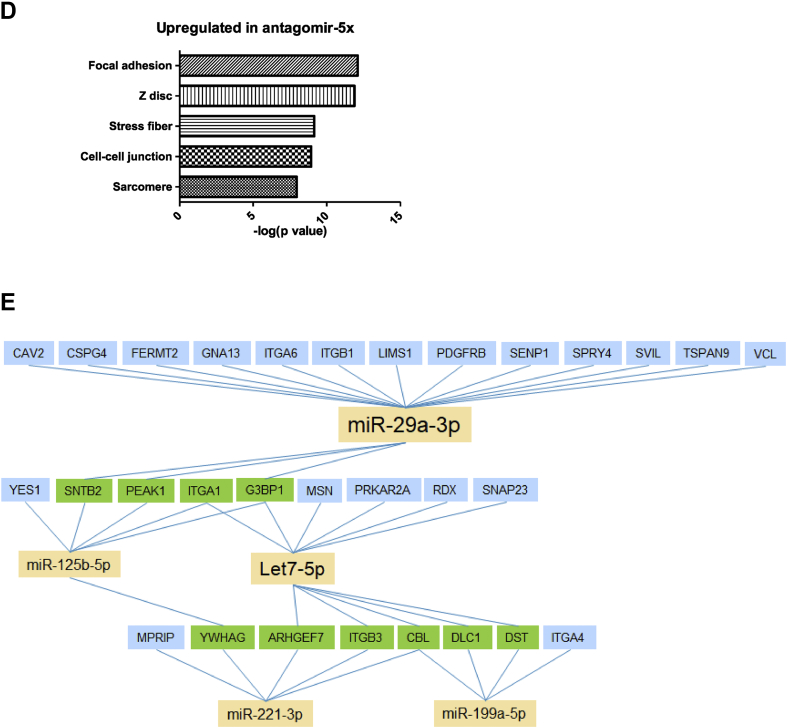
Our next aim was to investigate the level of cooperativity of the different miRNAs and compare the contributions of the miRNAs to muscle cell differentiation. Accordingly, we performed whole genome analysis using RNA-seq on differentiated human myoblasts treated with antagomirs either against the individual miRNAs or against all miRNAs (ant-5x). The circos plot of the significantly regulated genes for each antagomir and ant-5x provides an overview of the complex regulatory activity at the molecular level (Figure 4A). Strikingly, inhibition of all five miRNAs together regulated more genes than the inhibition of the individual miRNAs separately. This was observed for both the upregulated predicted target genes of the miRNAs as well as the non-target genes, indicating that gene regulation also occurred downstream of the predicted targets. Circos plot analysis also revealed the individual contributions of the miRNAs to the total number of regulated predicted target genes and non-target genes such that miRNA activity could be classified as highly active (miR-29a), moderately active (let-7) and mildly active (miR-125b, miR-199 and miR-221). The enrichment of predicted targets in the group of upregulated genes was only observed after single inhibition of miR-29a and let-7, the two most active miRNAs, but not after inhibition of the less active miR-125b, miR-199 or miR-221 (Figure 4B). However, the less active miRNAs strongly benefited from the combinatorial antagomir approach, since predicted targets of miR-125b, miR-199 and miR-221 were highly enriched in the ant-5x condition, while the enrichment of miR-29a targets rather decreased (Figure 4B). These results indicate that only moderate and mildly active miRNAs engaged in target cooperativity. Similar results were obtained when we compared the effect size of target gene regulation after single antagomir versus combinatorial antagomir inhibition. Although targets of miR-29a were similarly regulated in both conditions, there was a slight increase in the upregulation of let7 targets after ant-5x, while the strongest effect size was observed for the target genes of miR-125b, miR-199a and miR-221 in ant-5x compared to the single antagomir treatments (Figure 4C). To gain further insight into the cooperativity of the miRNAs, we analyzed the cumulative distribution fractions (CDF) of their predicted target genes compared to non-targets (Suppl. Fig. 9). Inhibition of miRNAs relieves repression of their predicted target genes compared to non-target genes, visualized as a right shift in CDF plots (Suppl. Fig. 9A, B). To provide a better overview of the results, we plotted the log2-fold differences of the average CDFs of targets and non-targets for each antagomir condition (Suppl. Fig. 9C), where increased CDF fold change reflects a larger average rightward shift of the group of predicted target genes compared to non-target genes and therefore a stronger inhibitory activity of the miRNA. Consistent with the observed enrichment of predicted miRNA targets shown in Figure 4B, miR-29a and let-7 had the largest CDF fold changes for their predicted target genes in antagomir-29a and antagomir-let7, respectively (Suppl. Fig. 9C). Differences for mildly active miRNAs were more subtle or not detected. Next, we asked whether any of the miRNAs would show cooperativity at the target gene level, which would manifest as an increase in the CDF fold difference in the ant-5x condition. Again, such evidence for target gene cooperativity was only observed for the mildly active miRNAs, miR-125b/miR-221, miR-125b/miR-199 and miR-221/miR-199 (Suppl. Fig. 9C). Lastly, we searched for enrichment of Gene Ontology (GO) terms within the ant-5x group to recapitulate our initial observation linking the miRNA pathway to the focal adhesion complex. Consistent with the results obtained for Dgcr8 KO cells and the add-back experiments, the GO term focal adhesion was the most significantly enriched gene cluster in ant-5x (Figure 4D). The focal adhesion gene cluster contained a total of 145 genes, of which 30 genes could be categorized as potential direct targets by comparing predicted target genes lists with significantly upregulated genes after single and combinatorial antagomir inhibition (Figure 4E). Interestingly, also in this analysis, the mildly active miRNAs are predominantly connected to genes that are targeted by more than one miRNA. Together, our results support the view that the focal adhesion pathway is targeted during muscle cell differentiation by a hierarchical network of miRNAs with different degrees of activity, where miRNA activity inversely correlates with miRNA cooperativity.
Figure 4.
Genome-wide identification of differentially expressed genes and miRNA target regulation after single or combinatorial miRNA inhibition in human myotubes. Human myoblasts were transfected with the indicated antagomirs and 24 h after transfection differentiation was induced for two days. RNA was isolated for RNA deep sequencing, n = 3. A. Circos plot of differentially expressed genes (p-value < 0.01 and the absolute log2 fold-change >0.5). Each sector of the plot represents a different antagomir or combination of the antagomirs (ant-5x). Genes with predicted binding sites for the respective miRNA are grouped under the darker color in each sector. Log2 fold-changes are presented in red (upregulation) or blue (downregulation). Genes differentially expressed in more than one condition are joined by grey lines or colored lines in case of predicted target genes. B. Enrichment for the predicted target genes (Targetscan) for the indicated miRNAs was analyzed in the group of upregulated genes for the different antagomir conditions using Fisher's Exact Test. C. Regulation of all predicted target genes after single antagomir treatment was plotted against their regulation after the combinatorial antagomir treatment (left panels). Full distribution of target gene regulation (log ratio) is shown using violin plots (right panels). D. Bar plot of GO terms enriched in the upregulated genes after combinatorial antagomir transfection. E. Network of miRNAs and their predicted target genes from the focal adhesion gene cluster. Blue boxes identify predicted target genes of one miRNA that are significantly upregulated after single and combinatorial antagomir inhibition. Green boxes indicate genes predicted to be targeted by at least two miRNAs that are significantly upregulated after the combinatorial antagomir inhibition.
2.4. Improved skeletal muscle regeneration in young and aged mice by combinatorial miRNA targeting
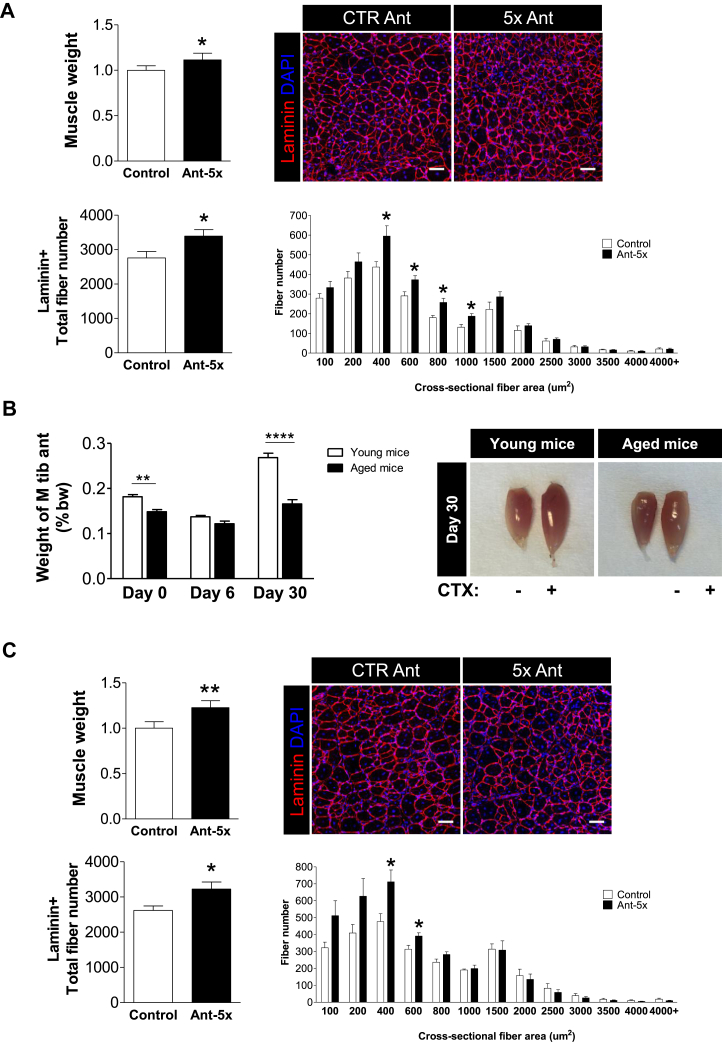
Finally, we asked whether the miRNA network that we identified also enhances muscle regeneration in vivo. To this end, we injected the combination of Ant-5x or control antagomirs i.m. following CTX-induced muscle regeneration (Figure 5). The inhibition of the miRNA network significantly enhanced muscle mass and fiber formation in young mice (Figure 5A). Since focal adhesion signaling is defective during muscle regeneration in aged mice [15], we sought to test whether the inhibition of the miRNA network could also improve muscle formation in this pathological condition. Indeed, muscle regeneration was largely inhibited in aged mice compared to young mice 30 days after CTX injection (Figure 5B). Importantly, i.m. injection of the miRNA inhibitors improved muscle weight and fiber formation in the aged mice. Together, these results reveal the utility of a miRNA network that regulates the focal adhesion complex to improve muscle formation in young and aged states (Figure 6).
Figure 5.
Combinatorial miRNA inhibition during skeletal muscle regeneration improves muscle weight and fiber number in young and aged mice in vivo. Three days following CTX injection in young mice (A) or aged mice (22 months) (C), TA muscles were injected with either control antagomir or a cocktail of the five antagomirs (total amount of 7.5ug per injection [32]). A,C. Muscle weight was measured and muscle cross sections were evaluated 9 days after antagomir injection using anti-laminin and DAPI immunofluorescence (scale bar 50um, n = 5). B. CTX was injected in TA muscles of young and aged mice and TA muscle weight was measured as percent of total body weight at the indicated time points. All results are shown as mean ± SEM, and evaluated with student's t test ∗: p < 0.05, ∗∗: p < 0.01, ∗∗∗: p < 0.001, n = 6–7.
Figure 6.
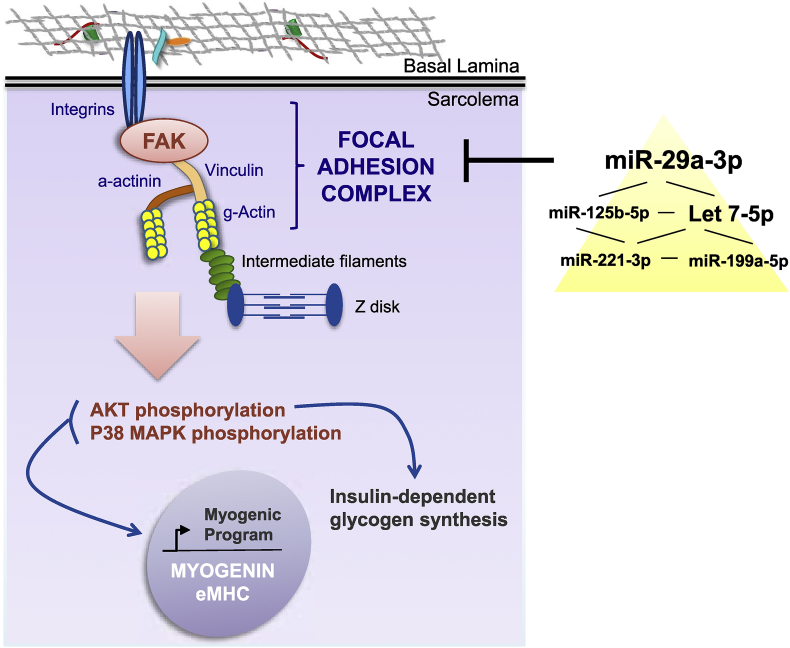
Regulation of the focal adhesion complex by a hierarchical network of miRNAs in myoblasts and its consequences for the myogenic program and insulin-dependent glycogen synthesis during muscle cell differentiation. The yellow triangle reflects the frequency of miRNA target gene cooperativity within the miRNA network.
3. Discussion
Our study provides an unbiased and genome-wide approach that uncovered a hierarchical miRNA network during muscle cell differentiation. The focal adhesion complex is central to the integration of multiple extracellular signals for muscle health, and we demonstrate that this complex is tightly connected to the miRNA pathway by a small set of miRNAs. The novelty of the miRNA network that we describe is its composition of non-clustered miRNAs, where miRNA activity is inversely proportional to target gene cooperativity. Importantly, combinatorial targeting of this miRNA network improved muscle regeneration in young and aged mice.
We propose that, within a miRNA network, miRNA activity inversely correlates to miRNA cooperativity. Predictions of miRNA cooperativity generally build upon the physical distance of miRNA binding sites on the 3′UTR of mRNA targets, with optimal downregulation when sites are separated by 13 to 35nt [29]. Our data suggest an additional criteria that affects the likelihood of a miRNA to be involved in the co-regulation of target genes and that is the activity of the miRNA. A highly active miRNA, such as miR-29a, exerts its effect on numerous targets that become upregulated upon inhibition of the miRNA, while other miRNAs, such as miR-221 and miR-199, appear to be less active because inhibition of both miRNAs is required to successfully upregulate target genes. We speculate that high activity might be a function of affinity between miRNA and target, rather than the expression of the miRNA itself since miR-29a is less abundant in myoblasts than miR-125b, miR-199 or let7 (Suppl. Fig 3, 5). Thus we reason that miRNA cooperativity hinges on the strength of the interaction between miRNA and target, such that the presence of a strongly engaged binding site will relieve the evolutionary pressure on the weak binding site of another miRNA on the same transcript, resulting in the loss of target cooperativity for the active miRNA. Such a scenario, high affinity binding of miRNA and target, would be reminiscent of the interactions between miRNA and competing endogenous (ce) RNA, where the highly abundant ceRNA needs to harbor high affinity sites to detract miRNAs [43]. On the other hand, two or more equally engaged binding sites would have similar evolutionary pressures and co-evolve, resulting in the synergistic regulation of the target by multiple miRNAs and miRNA target cooperativity. Further experiments are necessary to support our hypothesis.
The five miRNAs that we identified to exert cooperativity do not belong to a common miRNA cluster nor do they share a common seed motif, different from previous reports of miRNA cooperativity. For example, miRNA cooperativity within skeletal muscle was shown for miRNA isomers that contain identical seed regions. miR-133a1/miR-133a2 or miR-208b/miR-499, are expressed from different genetic locations, but share their target recognition motif. In these cases, only deletion of both miR-133a alleles (miR-133a1/a2 double knockout) resulted in adult onset myopathy [44], while only deletion of miR-208b and miR-499 together caused substantial loss of type 1 myofibers in the soleus muscle [45]. Such functional redundancy was also demonstrated in the model organism C. elegans for the miR-58 family members that share the same 6-mer seed motif [46]. Outside skeletal muscle, miRNA cooperativity also occurs between members of the same miRNA cluster that do not share the same target genes. Serial genetic deletions of the miRNA alleles from the miR-17-92 cluster containing four different seed families or the two miR-200 clusters containing two different seed families have demonstrated functional cooperativity across seed conservation [47,48]. Our findings emphasize that miRNA cooperativity can exist beyond miRNA clustering, and miRNA isomers and might be more frequent than previously appreciated. We show that predicted target regulation increases when miRNAs are collectively inhibited, similar to the functional redundancy described above during other types of miRNA cooperativity. A feature of miRNA cooperativity is that some miRNAs contribute a small set of target genes, yet the phenotype is most pronounced when all miRNAs are inhibited collectively [47]. In agreement, we show that the predicted target genes within the focal adhesion gene cluster are not equally distributed between the miRNAs, with the bulk of the targets being regulated by miR-29a. Notably, our data also confirms that miRNA cooperativity not only enhances the degree of regulation of gene expression, but also regulates a larger number of genes than observed for single miRNAs [47].
The role of individual miRNAs in skeletal muscle formation has been extensively studied, and numerous miRNAs have been shown to participate in this process [24]. An inhibitory role during muscle cell differentiation was assigned to four of the five miRNAs identified in our miRNA library screen. Single inhibition of miR-29a, let-7, miR-125b or miR-221 promotes muscle cell differentiation [33,[49], [50], [51], [52]], while the effects of inhibiting skeletal muscle miR-199a-5p are less clear [53]. Additionally, inhibition of let-7 by injection of miRNA inhibitors or transgenic expression of the let-7 inhibitor Lin28 increases muscle mass in mice [54,55]. This remarkable overlap provides further proof-of-concept for our study and underlines the potency of our assays to identify the few miRNAs from the total pool that partake in cooperativity to affect muscle cell differentiation. However, our study highlights the concept that these five miRNAs are integrated in a more complex regulatory network.
Our results suggest that the set of five miRNAs cooperate at the level of the focal adhesion complex to inhibit myoblast differentiation. During differentiation of human muscle cells, at least 20% of the genes from the focal adhesion gene cluster are potential direct targets of these miRNAs. This number includes only the evolutionary conserved binding sites and is likely to be even higher considering all possible non-conserved binding sites. FAK signaling is crucial for skeletal muscle regeneration and insulin-sensitivity and, consequently, inhibition of the set of miRNAs in our study closely mirrored its activation. Satellite cell-specific FAK knockout mice have a 4-fold decrease in regenerating small-diameter fibers 3 days after induction of muscle injury [12], and FAK is necessary for myotube formation [12,56]. In contrast, combinatorial miRNA inhibition in our study enhanced muscle mass and fiber size during muscle regeneration and improved myotube formation. FAK activates the PI3K and p38 MAPK pathways and improves insulin action in muscle cells, such as insulin-stimulated GLUT4 translocation and glycogen synthesis [[57], [58], [59], [60], [61]]. Ant-5x treatment in our study increased phosphorylation of p38 MAPK and AKT and enhanced glycogen synthesis in response to insulin. At the onset of differentiation, PI3K/AKT and MKK6/p38 MAPK pathways converge at the chromatin level to initiate transcription of the myogenic lineage program [42]. Consistent with our results, it has been shown in two separate studies that forced activation of p38 MAPK through a constitutive active mutant of MKK6 resulted in the accelerated differentiation of myoblasts and the early induction of both myogenin and MHC [62,63]. In addition, FAK translocation to the nucleus induced myogenin transcription by decreasing protein interactions on the myogenin gene promoter [64], in line with our findings using the myogenin promoter-driven reporter assays. Furthermore, inhibition of FAK during muscle cell differentiation prevented the induction of myogenin expression [12]. Therefore our results place the induction of the myogenic program by miRNA inhibition upstream of myogenin, driven by enhanced FAK signaling.
During ageing, myogenic progenitors (MPs) are depleted in the adult muscle stem cell niche and muscle regeneration is impaired [[65], [66], [67], [68]]. Decreased FAK signaling is associated with this regeneration defect since MPs from aged mice have lower FAK expression [15]. Furthermore, expression of the ECM member fibronectin is reduced in the aged stem cell niche of the skeletal muscle and integrin-mediated signaling through fibronectin, FAK and p38 MAPK is subsequently downregulated [15]. In humans, upregulation of ECM genes is part of the adaptation to exercise in middle-aged and old subjects [69,70], but older subjects failed to upregulate fibronectin expression upon exercise-induced muscle regeneration [71]. Our data reinforce the notion that regulation of focal adhesion signaling during ageing could be a valuable therapeutic approach.
In vivo, inhibition of the set of five miRNAs during skeletal muscle regeneration resulted in increased muscle mass and fiber number. Our protocol carefully timed the antagomir injections to day 3 after CTX injection, a time point where we expected to preferentially target muscle formation and myofiber differentiation, but not MP proliferation. In fact, perturbations in p38 MAPK signaling during the early proliferative phase of muscle regeneration can have deleterious effects. For example, in aged mice, increased FGF2 and p38 MAPK signaling in proliferating MPs are associated with impaired asymmetric division and a depletion of the quiescent stem cell pool [65,72]. Similarly, we have recently demonstrated that the genetic ablation of miR-29a in quiescent MPs decreases proliferation and subsequently muscle formation [33]. These results show that the timing of miRNA inhibition during muscle regeneration is likely critical to decide whether the predominant effect is on muscle cell proliferation or differentiation. Our approach would be most valuable in stem cell therapy where the outcome is to increase myofiber formation, but not MP proliferation.
Therapeutically, combinatorial targeting of miRNAs is advantageous since it reduces the concentration of pharmacological miRNA inhibitors and off-target effects. This has been demonstrated in human glioblastoma cells where inhibition of either miR-10b or miR-21 enhances apoptosis and reduces cell invasiveness. The amount of inhibitors needed to produce a therapeutic response was dramatically decreased when miR-10b and miR-21 inhibitors were combined compared to the use of the inhibitors alone [73]. Furthermore, we speculate that by using minimal inhibitor concentrations to downregulate a ubiquitous group of miRNAs, the inhibition will be sufficient to cause biological effects only in the tissue that concomitantly expresses the miRNAs, with minimal side effects to other organs. Prospective experiments will address whether targeting the 5× miRNA combination selectively affects the differentiation of skeletal muscle instead of the differentiation of other types of stem cells. In addition, it is an intriguing possibility that tissue-specific phenotypes can emerge when multiple miRNAs are targeted in combination. For example, contrary to the effects that we and others have observed for skeletal muscle stem cell differentiation as discussed above, miR-29 and miR-125b enhanced the differentiation of preadipocytes [74,75], while delivery of miR-125–5p, miR-199a-5p and miR-221 increased cardiomyocyte differentiation [76]. Furthermore, the 5× miRNA combination might be beneficially used as a therapeutic intervention in various pathological conditions. miRNA microarray analysis of human skeletal muscle from 10 different primary muscular disorders reported upregulation of four out of the five 5xAnt-miRNAs (let-7 in 5/10, miR-125a in 6/10, miR-199a in 8/10 and miR-221 in 10/10 primary muscular disorders) [25]. Strikingly, the most affected gene cluster in our study, the KEGG pathway “focal adhesion,” was also significantly regulated in seven out of nine primary muscular disorders [25]. The connection between the focal adhesion complex and the miRNA pathway might be more relevant for skeletal muscle diseases than previously appreciated.
In summary, we have discovered miRNA cooperativity that is independent of miRNA clustering and seed conservation and which regulates focal adhesion signaling during muscle stem cell differentiation. Our approach provides an alternative to purely computational methods to uncover miRNA cooperativity and introduces the notion that the activity of a miRNA could determine its capacity to engage in miRNA cooperativity. The discovery of the tight connection between focal adhesion and the miRNA pathway in muscle cells might lead to novel therapeutic protocols during ageing and other diseases where ECM-dependent signaling is altered.
4. Methods
4.1. Animals
C57BL6/6J mice were obtained from Harlan (Netherlands), DCGR8flox/flox mice were kindly provided by M. Stoffel (Zurich), Pax7tm2.1(cre/ERT2) mice were obtained from Jackson (US), and aged mice and control littermates (C57Bl/6JRj) were obtained from Janvier Labs (France). Animals were housed in a pathogen-free animal facility on an inverted 12-hour light cycle and fed ad libitum with a chow diet. For genotyping, genomic DNA was isolated from ear punches and analyzed using PCR. Primer sequences used were Dgcr8 Forward GACATCAATCTGAGTAGAGACAGG, Reverse CAGATGGTAACTAACCTGCCAACC, Pax7 Forward ACTAGGCTCCACTCTGTTCCTTC, and Reverse GCAGATGTAGGGACATTCCAGTG. Muscle regeneration in the TA muscle was induced by injection of cardiotoxin (CTX) as previously described [32]. All animal studies were approved by the ethics committee of the Canton of Zurich Veterinary Office.
4.2. Primary mouse and human myoblast cultures, apoptosis assay, western blotting and RNA decay
Procedures for isolation of primary muscle cells from human and mouse skeletal muscle were performed as previously described [33]. Briefly, skeletal muscle tissue was subjected to collagen digestion and myogenic progenitors (MPs) were isolated using flow cytometry. Human MP isolation was based on CD56 expression and absence of CD15, CD31 and CD45 staining, while mouse MP isolation was based on the presence of a7-integrin and the absence of Sca1, CD31 and CD45 staining. Cells were grown on collagen-coated plates in culture medium (1:1 v/v F10 nutrient mixture and DMEM (low glucose) containing 20% fetal bovine serum (FBS), 1% Penicillin/Streptomycin (P/S) and 5 ng/ml basic FGF (Invitrogen)). Tamoxifen was dissolved in 100% ethanol and diluted in growth media (final concentration 20 nM) for a 48-hour incubation period on primary myoblasts. Differentiation was initiated when myoblasts reached subconfluency by changing the media to DMEM containing 2% horse serum and 1% P/S. For apoptosis assays, myoblasts were trypsinized and resuspended in buffer (10 mM HEPES, 140 mM NaCl, 2.5 mM CaCl2, pH 7.4), followed by incubation with an Alexa Fluor® 647-conjugated Annexin V phosphoserine sensitive antibody (Biolegend, #422201) and fluorescence detection by flow cytometry. For western-blotting, cell lysates were separated by SDS-PAGE, transferred to PVDF membranes and proteins detected using primary antibodies for AGO2 (1:1000 Wako, 2D4), desmin (1:1000 Sigma, D1033), DGCR8 (1:1500 Protein tech, 10996-1-AP), GAPDH (1:2000 Protein tech, 10494), adult myosin heavy chain (MHC, 1:100 DSHB, MF-20), myogenin (1:1000 Santa Cruz, sc-12732), phospho p38 MAPK (1:1000 Cell Signaling 9211), p38 MAPK (1:1000 Cell Signaling 9212), phospho AKT (1:1000 Cell Signaling S473), AKT (1:1000 Cell Signaling 2920) and phospho FAK (1:1000 Cell Signaling 8556). For RNA decay, cells were differentiated for 3 days, treated with 10ug/ml Actinomycin D (Sigma) for the indicated time, then processed for qRT-PCR.
4.3. Immunofluorescence of primary myoblasts and skeletal muscle
Primary muscle cells were fixed with 4% paraformaldehyde at room temperature for 5 min and washed with PBS. Cells were subsequently blocked and permeabilized for 1 h (0.01% saponin, 1% BSA, 3% horse serum). Primary antibodies were incubated overnight at 4 °C and secondary antibodies were incubated for 1 h at room temperature (antibodies were used as indicated for western blotting except for anti-desmin [1:50, Abcam, ab8976]). Slides were mounted with DAPI (Vectashield) and visualized using Zeiss Axioplan2 imaging or Slide scanner Axio Scan Z1. The images were processed with Image J. Fusion index was calculated as the ratio of nuclei inside a myotube containing at least two nuclei compared to the number of all nuclei in the well. For skeletal muscle immunofluorescence, sections were processed as described in Mizbani et al. [32], incubated with anti-laminin antibody (1:500, Sigma L9393), imaged with a Leica CLSM SP8, and analyzed with Ilastic and ImageJ for fiber size and number.
4.4. Antagomir and miRNA mimic transfections
miRNA mimic and inhibitor transfection was performed as previously described [33]. In brief, primary myoblasts were transfected using the Lipofectamin RNAimax protocol (Invitrogen) and human miRNA mimics (30 nM, Mission, Sigma–Aldrich) or miRNA inhibitors (24 nM, antagomirs [77]) (Mission, Sigma–Aldrich). In case of the combination of multiple mimics or antagomirs, the total concentration per well was always kept constant at 30 nM or 24 nM, respectively (same as in the control mimic/antagomir transfections). Antagomirs against the let-7 family consisted of a 1:1 mixture of antagomirs directed against let7a-5p and let7c-5p.
4.5. RNA isolation, qRT-PCR, RNA microarrays and sequencing
Total RNA was isolated using Trizol Reagent (Invitrogen) according to the manufacturer's instruction. Total RNA was DNase treated and equal amounts of RNA were transcribed with random hexamer primers using SuperScript III reverse transcriptase (Invitrogen). qRT-PCRs for miRNA and mRNA levels were performed on an AB7500 FAST Real-time PCR system, using Taqman Universal PCR Master mix (Applied Biosystems) and FastStart Universal SYBR Green Master Mix (Roche), respectively. Primer sequences are available upon request. For miRNA qRT-PCR, the TaqMan miRNA assay (Applied Biosystems) was used. The transcript levels of mRNA were normalized to 18S ribosomal RNA and the levels of miRNAs were normalized to sno234. For Agilent mRNA microarrays of Dgcr8 KO mouse myoblasts and Illumina RNA deep sequencing of human myotubes, RNA was processed and analyzed at the Functional Genomics Center Zurich. For Illumina RNA deep sequencing of mouse myotubes and miRNA deep sequencing of mouse myoblasts, RNA was processed and analyzed at LC Sciences (Houston, TX, USA). The sequence results were obtained as FPKM (fragment per kilobase of exons per million reads) for each transcript. Predicted targets were downloaded from TargetScan v7.2.
4.6. Luciferase assays
The full length human myogenin 3′UTR was purchased from Genecopoeia (San Diego, US) in a Dual-Luciferase-Vector (HmiT102716) and transfected into C2C12 cells using lipofectamin 2000 (Life technologies, #11668–027) along with the miRNA mimics as described above. For myogenin promoter, 1300bp upstream of the transcription start site of the human promoter (primers: forward GCCGCCTCGGAGGAGATTTGGG; reverse CCCCCAAGCTCCAGCAGCCCCT) were cloned into the pGL3 vector and transfected along with pRLTK as control vector and the indicated antagomirs into mouse primary myoblasts. For both experiments, growth media was exchanged for differentiation media 24 h after transfection and cells were processed according to the protocol from Dual-Luciferase Reporter Assay (Promega, #1960) 72 h after transfection.
4.7. Glycogen synthesis
The assay was modified from Krützfeldt et al. [78]. Briefly, myoblasts were transfected with either control or Ant-5x as described above and differentiated for 3 days. Myotubes were then washed with HEPES-buffered saline (20 mmol/l HEPES, 140 mmol/l NaCl, 5 mmol/l KCl, 2.5 mmol/l MgSO4, 1 mmol/l CaCl2, 0.1% BSA, pH 7.4) and incubated in the same solution with insulin (1,000 nmol/l) for 1 h at 37 °C. d-glucose/D-[14C]glucose (5 mmol/l final concentration, 0.3 μCi/well) was added to the wells for an additional hour. Cells were then washed with ice-cold PBS and lysed in 30% (v/v) KOH. Small aliquots were removed for Bradford protein analysis. Samples were heated for 30 min at 95 °C, cooled on ice, and glycogen was precipitated with 1 ml 95% ethanol. After samples were centrifuged for 5 min at 9,000g, the resulting pellet was resuspended in water. The samples were precipitated one additional time, centrifuged and pellet resuspended in water. Radioactivity was determined by liquid scintillation counting. Results are expressed as counts per minute normalized to total protein content.
4.8. Statistical analyses
Student's t-tests were analyzed using excel software, classical one-way ANOVA with post tests and two-way ANOVA, as indicated in the figure legends, was analyzed using the GraphPad Prism software. P-values smaller than 0.05 were considered significant. Results are shown as means ± SEM.
Author contributions
E.L. wrote the manuscript, performed and analyzed all in vivo experiments and the myogenin reporter assays, and together with A.H. designed, performed and analyzed all experiments on human primary myoblasts. K.T. established primary cultures lacking DGCR8, performed and helped with design and analysis of the experiments on mouse primary myoblasts. S.M. performed the cumulative distribution analysis in human myotubes. H.R. analyzed the Ilumina RNA deep sequencing data on human myotubes. J.K. wrote the manuscript, supervised the study, and designed and analyzed all experiments. All authors discussed the results and commented on the manuscript.
Acknowledgements
This study was supported by the Swiss National Foundation (SNF) grant PP00P3_128474 to J.K., by the Clinical Research Priority Program “small RNAs” of the University of Zurich and an unrestricted grant by the Philhuman Foundation, Vaduz. We thank the Flow Cytometry Facility Zurich for their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2020.02.010.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Myers J., Prakash M., Froelicher V., Do D., Partington S., Atwood J.E. Exercise capacity and mortality among men referred for exercise testing. New England Journal of Medicine. 2002;346(11):793–801. doi: 10.1056/NEJMoa011858. [DOI] [PubMed] [Google Scholar]
- 2.Shulman G.I. Ectopic fat in insulin resistance, dyslipidemia, and cardiometabolic disease. New England Journal of Medicine. 2014;371(23):2237–2238. doi: 10.1056/NEJMc1412427. [DOI] [PubMed] [Google Scholar]
- 3.Rosenberg I.H. Sarcopenia: origins and clinical relevance. Journal of Nutrition. 1997;127(5 Suppl):990S–991S. doi: 10.1093/jn/127.5.990S. [DOI] [PubMed] [Google Scholar]
- 4.Baumgartner R.N., Koehler K.M., Gallagher D., Romero L., Heymsfield S.B., Ross R.R. Epidemiology of sarcopenia among the elderly in New Mexico. American Journal of Epidemiology. 1998;147(8):755–763. doi: 10.1093/oxfordjournals.aje.a009520. [DOI] [PubMed] [Google Scholar]
- 5.von Haehling S., Anker S.D. Cachexia as a major underestimated and unmet medical need: facts and numbers. J Cachexia Sarcopenia Muscle. 2010;1(1):1–5. doi: 10.1007/s13539-010-0002-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardo J.V., Siliciano J.D., Craig S.W. A vinculin-containing cortical lattice in skeletal muscle: transverse lattice elements ("costameres") mark sites of attachment between myofibrils and sarcolemma. Proceedings of the National Academy of Sciences of the U S A. 1983;80(4):1008–1012. doi: 10.1073/pnas.80.4.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graham Z.A., Gallagher P.M., Cardozo C.P. Focal adhesion kinase and its role in skeletal muscle. Journal of Muscle Research & Cell Motility. 2015;36(4–5):305–315. doi: 10.1007/s10974-015-9415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gehlert S., Suhr F., Gutsche K., Willkomm L., Kern J., Jacko D. High force development augments skeletal muscle signalling in resistance exercise modes equalized for time under tension. Pflügers Archiv. 2015;467(6):1343–1356. doi: 10.1007/s00424-014-1579-y. [DOI] [PubMed] [Google Scholar]
- 9.Fluck M., Carson J.A., Gordon S.E., Ziemiecki A., Booth F.W. Focal adhesion proteins FAK and paxillin increase in hypertrophied skeletal muscle. American Journal of Physiology. 1999;277(1):C152–C162. doi: 10.1152/ajpcell.1999.277.1.C152. [DOI] [PubMed] [Google Scholar]
- 10.Goel H.L., Dey C.S. Focal adhesion kinase tyrosine phosphorylation is associated with myogenesis and modulated by insulin. Cell Proliferation. 2002;35(3):131–142. doi: 10.1046/j.1365-2184.2002.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clemente C.F., Corat M.A., Saad S.T., Franchini K.G. Differentiation of C2C12 myoblasts is critically regulated by FAK signaling. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2005;289(3):R862–R870. doi: 10.1152/ajpregu.00348.2004. [DOI] [PubMed] [Google Scholar]
- 12.Quach N.L., Biressi S., Reichardt L.F., Keller C., Rando T.A. Focal adhesion kinase signaling regulates the expression of caveolin 3 and beta1 integrin, genes essential for normal myoblast fusion. Molecular Biology of the Cell. 2009;20(14):3422–3435. doi: 10.1091/mbc.E09-02-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen H.C., Appeddu P.A., Isoda H., Guan J.L. Phosphorylation of tyrosine 397 in focal adhesion kinase is required for binding phosphatidylinositol 3-kinase. Journal of Biological Chemistry. 1996;271(42):26329–26334. doi: 10.1074/jbc.271.42.26329. [DOI] [PubMed] [Google Scholar]
- 14.Han J.W., Lee H.J., Bae G.U., Kang J.S. Promyogenic function of integrin/FAK signaling is mediated by Cdo, Cdc42 and MyoD. Cellular Signalling. 2011;23(7):1162–1169. doi: 10.1016/j.cellsig.2011.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Lukjanenko L., Jung M.J., Hegde N., Perruisseau-Carrier C., Migliavacca E., Rozo M. Loss of fibronectin from the aged stem cell niche affects the regenerative capacity of skeletal muscle in mice. Nature Medicine. 2016;22(8):897–905. doi: 10.1038/nm.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huntzinger E., Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nature Reviews Genetics. 2011;12(2):99–110. doi: 10.1038/nrg2936. [DOI] [PubMed] [Google Scholar]
- 19.Svoronos A.A., Engelman D.M., Slack F.J. OncomiR or tumor suppressor? The duplicity of MicroRNAs in cancer. Cancer Research. 2016;76(13):3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krutzfeldt J., Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metabolism. 2006;4(1):9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Schmiedel J.M., Klemm S.L., Zheng Y., Sahay A., Bluthgen N., Marks D.S. Gene expression. MicroRNA control of protein expression noise. Science. 2015;348(6230):128–132. doi: 10.1126/science.aaa1738. [DOI] [PubMed] [Google Scholar]
- 22.Williams Z., Ben-Dov I.Z., Elias R., Mihailovic A., Brown M., Rosenwaks Z. Comprehensive profiling of circulating microRNA via small RNA sequencing of cDNA libraries reveals biomarker potential and limitations. Proceedings of the National Academy of Sciences of the U S A. 2013;110(11):4255–4260. doi: 10.1073/pnas.1214046110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krutzfeldt J. Strategies to use microRNAs as therapeutic targets. Best Practice & Research Clinical Endocrinology & Metabolism. 2016;30(5):551–561. doi: 10.1016/j.beem.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 24.Diniz G.P., Wang D.Z. Regulation of skeletal muscle by microRNAs. Comparative Physiology. 2016;6(3):1279–1294. doi: 10.1002/cphy.c150041. [DOI] [PubMed] [Google Scholar]
- 25.Eisenberg I., Eran A., Nishino I., Moggio M., Lamperti C., Amato A.A. Distinctive patterns of microRNA expression in primary muscular disorders. Proceedings of the National Academy of Sciences of the U S A. 2007;104(43):17016–17021. doi: 10.1073/pnas.0708115104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Rourke J.R., Georges S.A., Seay H.R., Tapscott S.J., McManus M.T., Goldhamer D.J. Essential role for Dicer during skeletal muscle development. Developmental Biology. 2007;311(2):359–368. doi: 10.1016/j.ydbio.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheung T.H., Quach N.L., Charville G.W., Liu L., Park L., Edalati A. Maintenance of muscle stem-cell quiescence by microRNA-489. Nature. 2012;482(7386):524–528. doi: 10.1038/nature10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grimson A., Farh K.K., Johnston W.K., Garrett-Engele P., Lim L.P., Bartel D.P. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Molecules and Cells. 2007;27(1):91–105. doi: 10.1016/j.molcel.2007.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saetrom P., Heale B.S., Snove O., Jr., Aagaard L., Alluin J., Rossi J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Research. 2007;35(7):2333–2342. doi: 10.1093/nar/gkm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmitz U., Lai X., Winter F., Wolkenhauer O., Vera J., Gupta S.K. Cooperative gene regulation by microRNA pairs and their identification using a computational workflow. Nucleic Acids Research. 2014;42(12):7539–7552. doi: 10.1093/nar/gku465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finnegan E.F., Pasquinelli A.E. MicroRNA biogenesis: regulating the regulators. Critical Reviews in Biochemistry and Molecular Biology. 2013;48(1):51–68. doi: 10.3109/10409238.2012.738643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizbani A., Luca E., Rushing E.J., Krutzfeldt J. MicroRNA deep sequencing in two adult stem cell populations identifies miR-501 as a novel regulator of myosin heavy chain during muscle regeneration. Development. 2016;143(22):4137–4148. doi: 10.1242/dev.136051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galimov A., Merry T.L., Luca E., Rushing E.J., Mizbani A., Turcekova K. MicroRNA-29a in adult muscle stem cells controls skeletal muscle regeneration during injury and exercise downstream of fibroblast growth factor-2. Stem Cells. 2016;34(3):768–780. doi: 10.1002/stem.2281. [DOI] [PubMed] [Google Scholar]
- 34.Altuvia Y., Landgraf P., Lithwick G., Elefant N., Pfeffer S., Aravin A. Clustering and conservation patterns of human microRNAs. Nucleic Acids Research. 2005;33(8):2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J., Wang F., Yang G.H., Wang F.L., Ma Y.N., Du Z.W. Human microRNA clusters: genomic organization and expression profile in leukemia cell lines. Biochemical and Biophysical Research Communications. 2006;349(1):59–68. doi: 10.1016/j.bbrc.2006.07.207. [DOI] [PubMed] [Google Scholar]
- 36.Baskerville S., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11(3):241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dambal S., Shah M., Mihelich B., Nonn L. The microRNA-183 cluster: the family that plays together stays together. Nucleic Acids Research. 2015;43(15):7173–7188. doi: 10.1093/nar/gkv703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lepper C., Conway S.J., Fan C.M. Adult satellite cells and embryonic muscle progenitors have distinct genetic requirements. Nature. 2009;460(7255):627–631. doi: 10.1038/nature08209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lepper C., Fan C.M. Inducible lineage tracing of Pax7-descendant cells reveals embryonic origin of adult satellite cells. Genesis. 2010;48(7):424–436. doi: 10.1002/dvg.20630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varma V., Yao-Borengasser A., Bodles A.M., Rasouli N., Phanavanh B., Nolen G.T. Thrombospondin-1 is an adipokine associated with obesity, adipose inflammation, and insulin resistance. Diabetes. 2008;57(2):432–439. doi: 10.2337/db07-0840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosano S., Cora D., Parab S., Zaffuto S., Isella C., Porporato R. A regulatory microRNA network controls endothelial cell phenotypic switch during sprouting angiogenesis. Elife. 2020;9 doi: 10.7554/eLife.48095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serra C., Palacios D., Mozzetta C., Forcales S.V., Morantte I., Ripani M. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Molecules and Cells. 2007;28(2):200–213. doi: 10.1016/j.molcel.2007.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bosson A.D., Zamudio J.R., Sharp P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Molecules and Cells. 2014;56(3):347–359. doi: 10.1016/j.molcel.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu N., Bezprozvannaya S., Shelton J.M., Frisard M.I., Hulver M.W., McMillan R.P. Mice lacking microRNA 133a develop dynamin 2-dependent centronuclear myopathy. Journal of Clinical Investigation. 2011;121(8):3258–3268. doi: 10.1172/JCI46267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van Rooij E., Quiat D., Johnson B.A., Sutherland L.B., Qi X., Richardson J.A. A family of microRNAs encoded by myosin genes governs myosin expression and muscle performance. Developmental Cell. 2009;17(5):662–673. doi: 10.1016/j.devcel.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subasic D., Brummer A., Wu Y., Pinto S.M., Imig J., Keller M. Cooperative target mRNA destabilization and translation inhibition by miR-58 microRNA family in C. elegans. Genome Research. 2015;25(11):1680–1691. doi: 10.1101/gr.183160.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han Y.C., Vidigal J.A., Mu P., Yao E., Singh I., Gonzalez A.J. An allelic series of miR-17 approximately 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nature Genetics. 2015;47(7):766–775. doi: 10.1038/ng.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belgardt B.F., Ahmed K., Spranger M., Latreille M., Denzler R., Kondratiuk N. The microRNA-200 family regulates pancreatic beta cell survival in type 2 diabetes. Nature Medicine. 2015;21(6):619–627. doi: 10.1038/nm.3862. [DOI] [PubMed] [Google Scholar]
- 49.Ge Y., Sun Y., Chen J. IGF-II is regulated by microRNA-125b in skeletal myogenesis. The Journal of Cell Biology. 2011;192(1):69–81. doi: 10.1083/jcb.201007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tan S.B., Li J., Chen X., Zhang W., Zhang D., Zhang C. Small molecule inhibitor of myogenic microRNAs leads to a discovery of miR-221/222-myoD-myomiRs regulatory pathway. Chemistry & Biology. 2014;21(10):1265–1270. doi: 10.1016/j.chembiol.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 51.Cardinali B., Castellani L., Fasanaro P., Basso A., Alema S., Martelli F. Microrna-221 and microrna-222 modulate differentiation and maturation of skeletal muscle cells. PloS One. 2009;4(10) doi: 10.1371/journal.pone.0007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Polesskaya A., Degerny C., Pinna G., Maury Y., Kratassiouk G., Mouly V. Genome-wide exploration of miRNA function in mammalian muscle cell differentiation. PloS One. 2013;8(8) doi: 10.1371/journal.pone.0071927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander M.S., Kawahara G., Motohashi N., Casar J.C., Eisenberg I., Myers J.A. MicroRNA-199a is induced in dystrophic muscle and affects WNT signaling, cell proliferation, and myogenic differentiation. Cell Death & Differentiation. 2013;20(9):1194–1208. doi: 10.1038/cdd.2013.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu H., Shyh-Chang N., Segre A.V., Shinoda G., Shah S.P., Einhorn W.S. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147(1):81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Frost R.J., Olson E.N. Control of glucose homeostasis and insulin sensitivity by the Let-7 family of microRNAs. Proceedings of the National Academy of Sciences of the U S A. 2011;108(52):21075–21080. doi: 10.1073/pnas.1118922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Quach N.L., Rando T.A. Focal adhesion kinase is essential for costamerogenesis in cultured skeletal muscle cells. Developmental Biology. 2006;293(1):38–52. doi: 10.1016/j.ydbio.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 57.Huang D., Khoe M., Ilic D., Bryer-Ash M. Reduced expression of focal adhesion kinase disrupts insulin action in skeletal muscle cells. Endocrinology. 2006;147(7):3333–3343. doi: 10.1210/en.2005-0382. [DOI] [PubMed] [Google Scholar]
- 58.Bisht B., Goel H.L., Dey C.S. Focal adhesion kinase regulates insulin resistance in skeletal muscle. Diabetologia. 2007;50(5):1058–1069. doi: 10.1007/s00125-007-0591-6. [DOI] [PubMed] [Google Scholar]
- 59.Bisht B., Dey C.S. Focal Adhesion Kinase contributes to insulin-induced actin reorganization into a mesh harboring Glucose transporter-4 in insulin resistant skeletal muscle cells. BMC Cell Biology. 2008;9:48. doi: 10.1186/1471-2121-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bisht B., Srinivasan K., Dey C.S. In vivo inhibition of focal adhesion kinase causes insulin resistance. Journal of Physiology. 2008;586(16):3825–3837. doi: 10.1113/jphysiol.2008.157107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lassiter D.G., Nylen C., Sjogren R.J.O., Chibalin A.V., Wallberg-Henriksson H., Naslund E. FAK tyrosine phosphorylation is regulated by AMPK and controls metabolism in human skeletal muscle. Diabetologia. 2018;61(2):424–432. doi: 10.1007/s00125-017-4451-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu Z., Woodring P.J., Bhakta K.S., Tamura K., Wen F., Feramisco J.R. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Molecular and Cellular Biology. 2000;20(11):3951–3964. doi: 10.1128/mcb.20.11.3951-3964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Suelves M., Lluis F., Ruiz V., Nebreda A.R., Munoz-Canoves P. Phosphorylation of MRF4 transactivation domain by p38 mediates repression of specific myogenic genes. The EMBO Journal. 2004;23(2):365–375. doi: 10.1038/sj.emboj.7600056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo S.W., Zhang C., Zhang B., Kim C.H., Qiu Y.Z., Du Q.S. Regulation of heterochromatin remodelling and myogenin expression during muscle differentiation by FAK interaction with MBD2. The EMBO Journal. 2009;28(17):2568–2582. doi: 10.1038/emboj.2009.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chakkalakal J.V., Jones K.M., Basson M.A., Brack A.S. The aged niche disrupts muscle stem cell quiescence. Nature. 2012;490(7420):355–360. doi: 10.1038/nature11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carlson M.E., Suetta C., Conboy M.J., Aagaard P., Mackey A., Kjaer M. Molecular aging and rejuvenation of human muscle stem cells. EMBO Molecular Medicine. 2009;1(8–9):381–391. doi: 10.1002/emmm.200900045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Watters J.M., Clancey S.M., Moulton S.B., Briere K.M., Zhu J.M. Impaired recovery of strength in older patients after major abdominal surgery. Annals of Surgery. 1993;218(3):380–390. doi: 10.1097/00000658-199309000-00017. discussion 390-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muller M., Tohtz S., Dewey M., Springer I., Perka C. Age-related appearance of muscle trauma in primary total hip arthroplasty and the benefit of a minimally invasive approach for patients older than 70 years. International Orthopaedics. 2011;35(2):165–171. doi: 10.1007/s00264-010-1166-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hjorth M., Norheim F., Meen A.J., Pourteymour S., Lee S., Holen T. The effect of acute and long-term physical activity on extracellular matrix and serglycin in human skeletal muscle. Physics Reports. 2015;3(8) doi: 10.14814/phy2.12473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hangelbroek R.W., Fazelzadeh P., Tieland M., Boekschoten M.V., Hooiveld G.J., van Duynhoven J.P. Expression of protocadherin gamma in skeletal muscle tissue is associated with age and muscle weakness. J Cachexia Sarcopenia Muscle. 2016;7(5):604–614. doi: 10.1002/jcsm.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sorensen J.R., Skousen C., Holland A., Williams K., Hyldahl R.D. Acute extracellular matrix, inflammatory and MAPK response to lengthening contractions in elderly human skeletal muscle. Experimental Gerontology. 2018;106:28–38. doi: 10.1016/j.exger.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 72.Bernet J.D., Doles J.D., Hall J.K., Kelly Tanaka K., Carter T.A., Olwin B.B. p38 MAPK signaling underlies a cell-autonomous loss of stem cell self-renewal in skeletal muscle of aged mice. Nature Medicine. 2014;20(3):265–271. doi: 10.1038/nm.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dong C.G., Wu W.K., Feng S.Y., Wang X.J., Shao J.F., Qiao J. Co-inhibition of microRNA-10b and microRNA-21 exerts synergistic inhibition on the proliferation and invasion of human glioma cells. International Journal of Oncology. 2012;41(3):1005–1012. doi: 10.3892/ijo.2012.1542. [DOI] [PubMed] [Google Scholar]
- 74.Ouyang D., Ye Y., Guo D., Yu X., Chen J., Qi J. MicroRNA-125b-5p inhibits proliferation and promotes adipogenic differentiation in 3T3-L1 preadipocytes. Acta Biochimica et Biophysica Sinica. 2015;47(5):355–361. doi: 10.1093/abbs/gmv024. [DOI] [PubMed] [Google Scholar]
- 75.Zhang X.M., Wang L.H., Su D.J., Zhu D., Li Q.M., Chi M.H. MicroRNA-29b promotes the adipogenic differentiation of human adipose tissue-derived stromal cells. Obesity. 2016;24(5):1097–1105. doi: 10.1002/oby.21467. [DOI] [PubMed] [Google Scholar]
- 76.Lee D.S., Chen J.H., Lundy D.J., Liu C.H., Hwang S.M., Pabon L. Defined MicroRNAs induce aspects of maturation in mouse and human embryonic-stem-cell-derived cardiomyocytes. Cell Reports. 2015;12(12):1960–1967. doi: 10.1016/j.celrep.2015.08.042. [DOI] [PubMed] [Google Scholar]
- 77.Krutzfeldt J., Rajewsky N., Braich R., Rajeev K.G., Tuschl T., Manoharan M. Silencing of microRNAs in vivo with 'antagomirs'. Nature. 2005;438(7068):685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 78.Krutzfeldt J., Kausch C., Volk A., Klein H.H., Rett K., Haring H.U. Insulin signaling and action in cultured skeletal muscle cells from lean healthy humans with high and low insulin sensitivity. Diabetes. 2000;49(6):992–998. doi: 10.2337/diabetes.49.6.992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.