Abstract
Background
Maturation of ultrasound myocardial tissue characterization may have far-reaching implications as a widely available alternative to cardiac magnetic resonance (CMR) for risk stratification in left ventricular (LV) remodeling.
Methods
We extracted 328 texture-based features of myocardium from still ultrasound images. After we explored the phenotypes of myocardial textures using unsupervised similarity networks, global LV remodeling parameters were predicted using supervised machine learning models. Separately, we also developed supervised models for predicting the presence of myocardial fibrosis using another cohort who underwent cardiac magnetic resonance (CMR). For the prediction, patients were divided into a training and test set (80:20).
Findings
Texture-based tissue feature extraction was feasible in 97% of total 534 patients. Interpatient similarity analysis delineated two patient groups based on the texture features: one group had more advanced LV remodeling parameters compared to the other group. Furthermore, this group was associated with a higher incidence of cardiac deaths (p = 0.001) and major adverse cardiac events (p < 0.001). The supervised models predicted reduced LV ejection fraction (<50%) and global longitudinal strain (<16%) with area under the receiver-operator-characteristics curves (ROC AUC) of 0.83 and 0.87 in the hold-out test set, respectively. Furthermore, the presence of myocardial fibrosis was predicted from only ultrasound myocardial texture with an ROC AUC of 0.84 (sensitivity 86.4% and specificity 83.3%) in the test set.
Interpretation
Ultrasound texture-based myocardial tissue characterization identified phenotypic features of LV remodeling from still ultrasound images. Further clinical validation may address critical barriers in the adoption of ultrasound techniques for myocardial tissue characterization.
Funding
None.
Keywords: Echocardiography, Machine learning, Radiomics, Clustering, Tissue characterization
Research in context.
Evidence before this study
We performed a literature search with PubMed on December 20, 2019 and found no study in which the radiomics-based texture analysis approach was employed using cardiac ultrasound images for assessing LV remodeling. Before this study, cardiac ultrasound was not clinically used for tissue characterization and cardiac MR was the leading imaging modality for the assessment of myocardial remodeling.
Added value of this study
This study demonstrated that by using a machine learning pipeline to integrate and process texture features, important information associated with myocardial remodeling can be derived from ultrasound images. Integrated texture features phenotypes showed discrimination of patients with impaired cardiac function from those without cardiac remodeling. Furthermore, machine learning models enabled the prediction of impaired systolic function and myocardial fibrosis.
Implications of all the available evidence
Given the portability, wide availability, and cost-effectiveness, tissue characterization using cardiac ultrasound can provide huge implications in clinical practice. Moreover, the application of this approach to other fields of medical research will accelerate them by using it as a noninvasive and repeatable evaluation of myocardial remodeling. Further studies are warranted to expand our approach to such a clinical and research world.
Alt-text: Unlabelled box
1. Introduction
A rise in cardiovascular risk factors, improved survival rate from ischemic heart disease, and population-ageing have contributed to the increasing global burden of heart failure [1]. An important step to prevent the progression of heart failure includes early detection of left ventricular (LV) remodeling – a process driven by architectural cellular and interstitial changes in the myocardium and identified clinically as global changes in LV size, geometry, and function [2]. Studies have shown that the degree of LV remodeling has a strong correlation with the impact of particular drugs or device therapies as well as with clinical outcomes [3].
Recent advancements in cardiac magnetic resonance (CMR) have revealed that myocardial tissue imaging characteristics alter under various cardiac conditions which reflect structural LV remodeling, including fibrosis, increased extracellular volume, and altered fibre orientation [4,5]. Cardiac ultrasound is not currently utilized clinically for myocardial tissue characterization although previous studies have reported that the intensity of the ultrasound backscatter is related to the physical properties of the myocardium and is influenced by tissue components (e.g. collagen, water, fat) [6,7]. Moreover, there has been limited information regarding the specific application of texture-based analysis for cardiac ultrasound imaging. The recent developments in image analysis and novel bioinformatics approaches have augmented methods that can extract information from the texture in a still image. The application of such texture-based image analysis has been increasingly utilized as a key function in various image-processing applications such as automated inspection, document processing, radiology image processing, and content-based image retrieval [8], [9], [10], [11]. Such techniques may also have direct relevance for cardiac ultrasound techniques like speckle tracking echocardiography where myocardial motion is analyzed using frame-by-frame tracking of natural acoustic markers (often referred in literature as “speckles”, “patterns”, or “fingerprints”) [12,13]. A functional unit (kernel) of speckles generated from ultrasound-tissue interactions (i.e., reflections, interference, and scattering) is unique, allowing software to track itself during the entire cardiac cycle [12]. Thus, an ultrasound texture of myocardium may carry unique and specific information of the indexed myocardium.
This study, therefore, presents the development and validation of a novel approach that combines the texture-based informatics of myocardium with machine learning techniques. We first extract texture-based tissue features from still ultrasound images and identify the association of texture feature-based patient phenotypes with LV remodeling. Subsequently, we illustrate the value of texture-based supervised machine learning models in predicting LV systolic dysfunction and the presence of myocardial fibrosis in a remodeled LV.
2. Materials and methods
2.1. Study participants
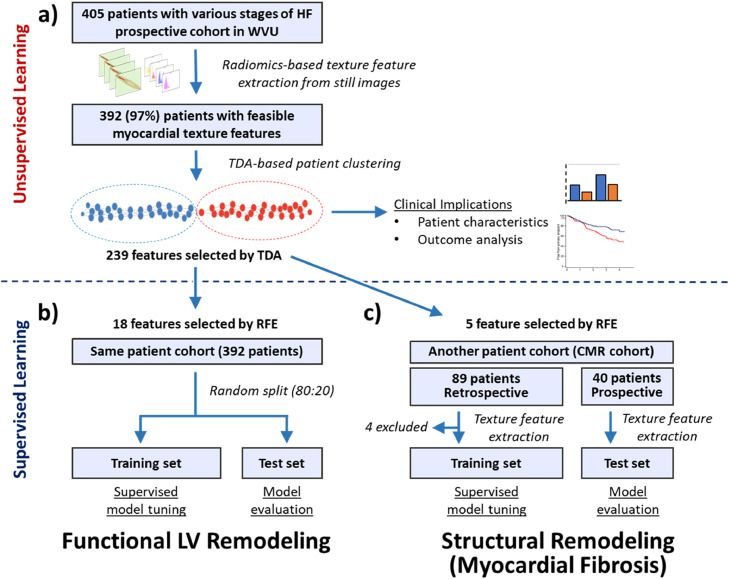
This study consisted of three parts. The detailed study design is presented in Fig. 1.
Fig. 1.
Study process. This study consisted of three parts as shown in panel a to c. In the first part (panel a), we used unsupervised clustering to explore patient phenotypes and their clinical implications in total 405 patients with various stages of HF. Next, supervised machine learning was used to predict the functional (impaired LVEF and GLS) LV remodeling using the texture features in the same patient cohort (panel b). Finally, in an independent cohort of patients who underwent CMR and echocardiography, we explored the usefulness of texture-based supervised models for predicting myocardial fibrosis (panel c). WVU, West Virginia University; HF, heart failure; ML, machine learning; TDA, topological data analysis, LV, left ventricular; RFE, recursive feature elimination; CMR, cardiac magnetic resonance.
2.1.1. Unsupervised phenotyping based on texture features (Fig. 1a)
We pooled 405 patients from three prospective studies conducted at West Virginia University between August 2017 and September 2018. Those studies used echocardiography as a reference standard of LV function and were evaluating [1] the value of a surface ECG algorithm to predict diastolic dysfunction (n = 196). This study included adult (>18 years old) subjects who underwent ECG and echocardiography on the same day [2]; a probe for estimating pulmonary artery pressure from chest wall (n = 145). This study included adults older than 18 years old, admitted to the hospital for HF who had an echocardiogram performed within 48 h of presentation, and [3] a software for the assessment of intracardiac flow (n = 64), which included consecutive adult patients referred for LV function assessment. The common exclusion criteria for all the three studies included [1] patients with inadequate echocardiographic views and [2] patients with chest deformities. Myocardial texture feature extraction was feasible in 392 patients. We performed an unsupervised machine learning using topological data analysis for aggregating patients with similar textural properties and compared the patient characteristics, cardiac function, and outcome between the phenogroups.
2.1.2. Supervised learning-based prediction of LV remodeling (Fig. 1b)
We used the 392 patient cohort as described above to develop supervised machine learning models for predicting functional markers of LV remodeling (impairment in LV ejection fraction [LVEF] and global longitudinal strain [GLS]), the cohort was randomly divided into a training (80%) and test (20%) set. Then, machine-learning models were trained in the training set (with cross-validation) and subsequently evaluated in the test set.
2.1.3. Supervised learning-based prediction of myocardial fibrosis (Fig. 1c)
To assess the value of texture features for predicting the presence of CMR delineated myocardial fibrosis, we retrospectively identified 89 patients who underwent clinically indicated CMR and cardiac ultrasound within 48 h between July 2017 and December 2018. Exclusion criteria were [1] patients with inadequate echocardiographic views [2], patients with chest deformities, and [3] patients who underwent CMR without gadolinium contrast. The retrospective cohort was used to train machine learning models with cross-validation and the developed model was tested in 40 prospective patients who were enrolled with the same inclusion/exclusion criteria.
2.2. Data collection
The New York Heart Association (NYHA) functional class and the heart failure stages defined by the American College of Cardiology and the American Heart Association were used to investigate clinical severity [14]. Major adverse cardiac event (MACE) was predefined as the composite of cardiac death, hospitalization due to myocardial infarction, acute coronary syndrome, heart failure, and arrhythmias and were tracked on an electronic chart and/or telephone interview. The Meta-Analysis Global Group in Chronic (MAGGIC) heart failure risk score was calculated as previously reported [15,16]. All enrolled patients underwent comprehensive 2-dimensional echocardiography using commercially available ultrasound equipment (Vivid-9/95, GE Healthcare; iE-33, Philips Healthcare; and LISENDO 880, Hitachi Healthcare) with 1 – 5 MHz phased array probes. Ultrasound images were stored in a DICOM format on the institute's local Picture Archiving and Communication System (PACS). Conventional echocardiographic parameters were analyzed per under the current guidelines [17]. LVEF was measured using 2D disk methods at end-diastole and end-systole. Speckle tracking strain analysis was performed offline using vendor-free software (ImageArena, TomTec Inc.) by observers who were blinded to other information, including the texture-based tissue features. The longitudinal strain was calculated using apical 4-, 2-, and long-axis views, and the averaged value was reported as the GLS [17]. CMR was performed using a 1.5 Tesla scanner (MAGNETOM Arena, Siemens Healthineers, Erlangen, Germany). Late gadolinium enhancement imaging was performed in all subjects in accordance with standard clinical protocols [4]. Late gadolinium enhancement was defined by hyperenhanced pixels with signal intensities of 5 standard deviations above the mean of normal myocardium. Patients were considered to have myocardial fibrosis in the studied segments if positive late gadolinium enhancement was seen in any of the anteroseptal and posterior wall myocardial segments (corresponding to the segments where ultrasound ROIs were placed for extracting texture features).
2.3. Quantitative texture-based tissue feature extraction
Texture-based tissue features of myocardium were extracted from still images of traditional parasternal long axis views using LIFEx software v4.5 (https://www.lifexsoft.org/) [18]. This technique of texture-based feature extraction has been popularized in radiology and referred to as ‘radiomics’ [10,11]. Using 2 still frames, an end-diastolic and an end-systolic frame, we placed circular regions of interest (ROIs) including 257 pixels (4 – 9 mm in diameter) per each, at the basal and mid-segments of the interventricular septum and the left ventricular posterior wall, respectively. The basal and mid-segments were defined as the level of the mitral valve leaflet tips and the papillary muscle. The ROI contents were first resampled in 64 discrete values using the formula:
Where R(x) is the resampled value of pixel x, I(x) is the intensity of pixel x in the original image, and max and min Intensity are the maximum and minimum intensities in the ROI, respectively. The software extracted 41 texture features, or radiomics features, from each ROI, including first-order statistics such as the maximum, minimum, standard deviation, and the mean value of intensity and histogram features, and second-order indices such as the gray-level co-occurrence matrix (GLCM) [19], gray-level run length matrix (GLRLM) [20], neighboring gray-level dependence matrices (NGLDM) [21], and gray-level zone length matrices (GLZLM) [22]. More details about the texture features are provided in the Supplemental Material.
2.4. Feature phenotyping using topological data analysis
A total of 328 texture features extracted during diastole and systole were included in the topological data analysis using Ayasdi Workbench v7.4 (Ayasdi Inc., Menlo Park, California). Topological data analysis is a novel mathematical and data analysis approach that establishes the topological and geometrical structure of the data to garner information and patterns from the features in a patient-patient similarity network [18,23]. The technical details are shown in the Supplemental Material. In a topological data analysis-based patient similarity network, patients with similar features (in this study, texture-based tissue features) form a node or a dot, and adjacent nodes, including similar patients, are connected with edges or lines. Accordingly, the relative distance between nodes (more precisely, the minimum number of edges between nodes) represents the similarity of features between the nodes. Thus, clusters or groups of patients with similar features can be identified based on the shape of the network. This notion of linking the shape to meaning using topological data analysis has been extensively validated and increasingly used in various areas of health sciences [23,24]. After we identified patient clusters using topological data analysis, the clinical significance of those clusters was studied by summarizing and comparing patient characteristics and outcomes.
2.5. Machine learning prediction of functional LV remodeling
As the number of texture features was large (328) and there were similar features with strong correlations, we used the differences in important features between patient clusters as highlighted topological data analysis for feature selection and pruned the list of features further with recursive feature elimination to build a supervised machine learning framework [25,26]. The final number of the features was determined by averaged Matthews correlation coefficient. Using a cloud-based automated machine learning platform (OptiML, BigML.com, Corvallis, Oregon, http://bigml.com), multiple algorithms, including decision trees, ensemble models, logistic regression models with least absolute shrinkage (LASSO) and selection operator, and deep neural networks, were trained to predict reduced LVEF (≤50%) and impaired GLS (<16% in absolute value). In the training sets, the platform performed automated model selection and hyperparameter tuning based on Bayesian parameter optimization using an optimization technique called sequential model-based algorithm configuration [27,28]. This process was performed with Monte-Carlo cross-validation. Finally, several models with the highest performance were selected and their prediction probabilities were averaged to create an ensemble model (fusion model), which were evaluated in the hold-out (not used in the training process) test set. Such techniques of making fusion models help combining diverse and independent models for reducing the generalization error.
2.6. Reproducibility of tissue texture
Top texture-based tissue features for predicting myocardial fibrosis were defined using the top importance gain of the 4 regions. We assessed variability related to the operator, device settings, and device vendors used in the study. We tested the interobserver variability of feature extraction in 2 blinded observers who independently analyzed 20 randomly selected patients and assessed the consistency of the texture features. To test the resistance of the texture features to device settings, we evaluated changes of texture features in different gain settings and image qualities. After adding 5 levels of gain (to I(x) + 20, I(x) + 40, I(x) + 60, I(x) + 80, I(x) + 100) and Gaussian additive noise (mean = 0, variance of 0.01, 0.02, 0.03, 0.04, and 0.05) to 10 images using MATLAB R2018a (The MathWorks, Natick, MA, USA), the texture features were extracted using exactly the same ROIs. Lastly, we verified the vendor dependency of the texture features by testing topological data analysis-based patient similarity networks generated using features extracted from 2 vendors (GE Healthcare and Hitachi Healthcare).
2.7. Statistical analysis
Data are presented as the median [1st and 3rd interquartile range] for continuous variables and as the frequency (%) for categorical variables. Group differences were evaluated using Mann–Whitney U tests for continuous variables and chi-square or Fisher's exact tests for categorical variables. Kaplan–Meier curve analysis, the log-rank test, and multivariable Cox proportional hazard models were used for survival analysis. The ROC curves of the machine learning models were drawn, and the best thresholds were identified based on the Youden index. Interobserver variability was evaluated using Pearson's r and interclass correlation coefficients. All statistical analyses were performed with R version 3.5.2 (The R Foundation for Statistical Computing, Vienna, Austria). A two-tailed p <0.05 indicated statistical significance.
2.8. Ethics and data sharing statement
The study protocol was approved by the institutional review board and written informed consents were obtained from all prospective patients. The study was compliant with the Declaration of Helsinki and the institutional review board approved the study protocol. Research data are not available for public access due to patient privacy concerns but can be obtained from the corresponding author on reasonable request approved by the institutional review boards of all participating institutions.
3. Results
3.1. Unsupervised phenotyping based on texture features
For the first part of the study (Fig. 1a), we successfully extracted 328 texture features from still-frame ultrasound images in 392 of the 405 (97%) subjects. Overall, the median age of the population was 58 [45 – 68] years, 55.9% were female, 26.3% had severe heart failure symptoms (NYHA class III or IV) and 32.9% had stage C or D heart failure [29].
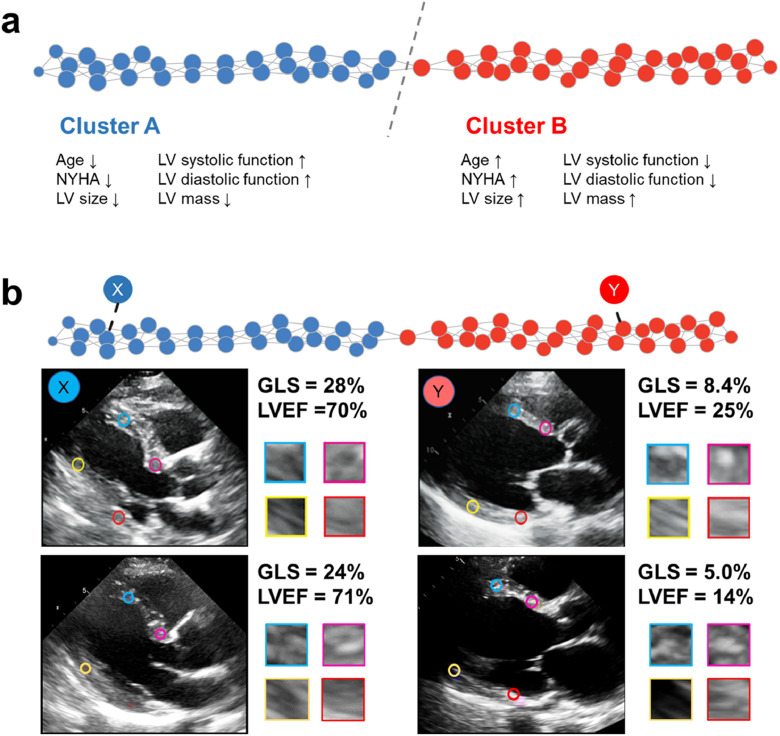
Using the extracted texture features, unsupervised topological data analysis identified a bar-shaped patient similarity network, where 2 clusters were connected by a single node (Fig. 2A). Clusters A and B included 196 and 210 patients, respectively, with 14 patients overlapping between the groups. Interestingly, these identified clusters had significantly different clinical and echocardiographic characteristics even though the clusters were created using only the texture features of still images. Group differences are summarized in Table 1. Compared with cluster A, cluster B was associated with greater age and more advanced heart failure. Furthermore, patients in cluster B had significant differences in LV remodeling: the intraventricular septum and LV mass index were greater, the LV dimensions and volumes were larger, the LVEF and LV GLS were reduced, the LV diastolic function represented by tissue Doppler e’ and E / e’ was impaired, and the left atrial volume was larger compared to those in cluster A (Table 1). The differences in texture features between the groups were summarized in Supplemental Table 1.
Fig. 2.
Patient similarity network based on myocardial texture features. Panel a: Extracted texture features were integrated using topological data analysis to create a patient similarity network. In the network, patients with similar features form a node, and adjacent nodes, including similar patients, are connected with edges. The network demonstrated the shape of a bar that was geometrically divided into two parts which had significantly different clinical and echocardiographic characteristics although the groups were created using only texture features. Panel b. Patients X, Y, Z, and W were identified in corresponding x, y, z, and w nodes, respectively. X and Y had a normal cardiac function and were found in cluster A, whereas Z and W were located in cluster B with significantly impaired cardiac function. GLS, global longitudinal strain; LVEF, left ventricular ejection fraction.
Table 1.
Patient characteristics.
| Factor | Overall | Cluster A | Cluster B | p value |
|---|---|---|---|---|
| Number of patients | 392 | 196 | 210 | |
| Age, years | 58 [45–68] | 54 [40–65] | 61 [50–71] | <0.001 |
| Female, n (%) | 219 (55.9) | 113 (57.7) | 113 (53.8) | 0.484 |
| BSA, m2 | 2.02 [1.8–2.22] | 2.03 [1.84–2.23] | 1.97 [1.77–2.20] | 0.041 |
| BMI, kg/m2 | 29.2 [25.7–36.0] | 29.9 [25.7–37.4] | 28.8 [25.7–33.8] | 0.113 |
| Coronary artery disease, n (%) | 148 (37.8) | 63 (32.1) | 90 (42.9) | 0.031 |
| Hypertension, n (%) | 272 (69.4) | 133 (67.9) | 148 (70.5) | 0.592 |
| Cerebral vessel disease, n (%) | 46 (11.8) | 21 (10.7) | 25 (12.0) | 0.755 |
| Diabetes mellitus, n (%) | 119 (30.4) | 50 (25.5) | 72 (34.3) | 0.065 |
| Atrial fibrillation, n (%) | 50 (12.8) | 12 (6.1) | 42 (20.0) | <0.001 |
| NYHA ≥III, n (%) | 103 (26.3) | 37 (18.9) | 73 (34.8) | <0.001 |
| HF stages C or D, n (%) | 129 (32.9) | 56 (28.6) | 78 (37.1) | 0.073 |
| Echocardiography | ||||
| IVSd, mm | 10 [9–12] | 10 [8–12] | 11 [9–13] | 0.005 |
| PWd, mm | 9 [8–11] | 9 [8–10] | 9 [8–11] | 0.068 |
| LVIDd, mm | 46 [42–51] | 46 [42–49] | 47 [42–52] | 0.036 |
| LVIDs, mm | 32 [28–37] | 31 [28–35] | 34 [29–40] | <0.001 |
| LVEDVi, mL/m2 | 52 [43–64] | 50 [41–60] | 56 [45–68] | <0.001 |
| LVESVi, mL/m2 | 21 [15–28] | 19 [15–26] | 22 [16–33] | <0.001 |
| LA volume index, mL/m2 | 25 [20–35] | 23 [18–32] | 28 [21–41] | <0.001 |
| LV mass index, mg/m2 | 76 [59–101] | 72 [56–90] | 82 [64–112] | <0.001 |
| E wave velocity, m/s | 0.84 [0.69–0.96] | 0.81 [0.69–0.94] | 0.87 [0.70–1.01] | 0.057 |
| A wave velocity, m/s | 0.70 [0.54–0.89] | 0.70 [0.53–0.89] | 0.70 [0.55–0.90] | 0.897 |
| E / A ratio | 1.14 [0.85–1.55] | 1.11 [0.85–1.51] | 1.16 [0.84–1.56] | 0.720 |
| e', cm/s | 8.1 [6.0–10.5] | 8.5 [7.0–10.7] | 7.6 [5.5–10.5] | 0.001 |
| E / e' | 9.3 [7.2–14.5] | 9.0 [7.2–12] | 10.1 [7.7–17.2] | 0.003 |
| LVEF, % | 60 [53–65] | 61 [55–65] | 59 [46–65] | 0.011 |
| GLS, absolute % | 19.3 [15.7–22.0] | 20.2 [18.1–23.5] | 17.3 [12.5–20.4] | <0.001 |
| LV hypertrophy, n (%) | 84 (21.4) | 23 (11.7) | 63 (30.0) | <0.001 |
| LV diastolic function, n (%) | <0.001 | |||
| normal | 188 (49.3) | 120 (61.9) | 74 (36.8) | |
| grade 1 | 29 (7.6) | 10 (5.2) | 20 (10.0) | |
| grade 2 | 63 (16.5) | 22 (11.3) | 45 (22.4) | |
| grade 3 | 34 (8.9) | 8 (4.1) | 27 (13.4) | |
| indeterminate | 61 (16.0) | 31 (16.0) | 32 (15.9) | |
| indeterminate grade | 6 (1.6) | 3 (1.5) | 3 (1.5) |
BSA, body surface area; BMI, body mass index; NYHA, New York Heart Association; HF, heart failure; IVSd, interventricular septum thickness in diastole; PWd, posterior wall thickness in diastole; LVIDd, left ventricular end-diastolic diameter; LVIDs, left ventricular end-systolic diameter; LVEDVi, left ventricular end-diastolic volume index; LVESVi, left ventricular end-systolic volume index; LA, left atrial; LVEF, left ventricular ejection fraction; GLS, global longitudinal strain.
Illustrative cases for each cluster are shown in Fig. 2B. Although functional evaluation of the myocardial textures in the images seems unfeasible with the human eye, this texture-based approach was able to identify important information for evaluating cardiac function from still routine ultrasound images and classified patients in a clinically meaningful way.
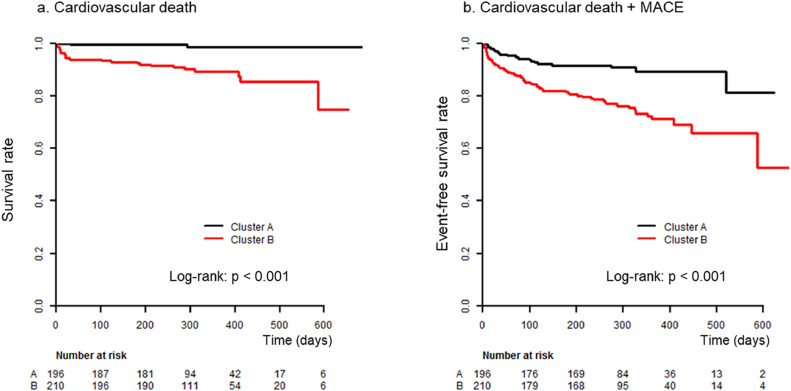
3.1.1. Comparison of clinical outcomes between clusters
We further compared the clinical prognosis of the two clusters. During the follow-up period of a median of 301 [268–323] days, 76 MACEs, including 26 cardiac deaths, were observed. Kaplan–Meier curves showed that patients in cluster B had a significantly higher incidence of cardiac death and MACE than those in cluster A (p < 0.001 by log-rank test, for both, Fig. 3). Cox proportional hazard models showed that even after adjusting for the MAGGIC score, a well-established risk score for patients with heart failure validated in various clinical settings [15,16], texture-based clustering was significantly associated with cardiac death (HR 6.23, 95% CI 1.46 – 26.5, p = 0.013 by Cox proportional hazard analysis). The association of texture-based clustering with MACE was significant in the univariate model (HR 2.85, 95% CI 1.69 – 4.78, p <0.001 by Cox proportional hazard analysis) and a model adjusted with other clinical factors (age, sex, body mass index, history of coronary heart disease, hypertension, and LVEF; HR 1.74, 95% CI 1.01 – 3.00, p = 0.047 by Cox proportional hazard analysis), and showed borderline significance in a model adjusted with the MAGGIC score (HR 1.68, 95% CI 0.99 – 2.87, p = 0.057 by Cox proportional hazard analysis). The results for the Cox models are summarized in Table 2 and HRs for all variables are listed in Supplementary Table 2.
Fig. 3.
Clinical outcomes between clusters. Kaplan–Meier curve analyses showed that cluster B had a significantly higher incidence of cardiovascular death (panel a) and of the composite of cardiovascular death and major adverse cardiac events (MACE; panel b) compared with cluster A.
Table 2.
Cox regression models.
| For cardiac death |
For MACE |
|||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p value | HR | 95% CI | p value | |
| Cluster B | 11.09 | 2.62–46.93 | 0.001 | 2.85 | 1.69–4.78 | <0.001 |
| Adjustment for MAGGIC score | 6.23 | 1.46–26.50 | 0.013 | 1.68 | 0.99–2.87 | 0.057 |
| Adjustment for clinical factors* | 1.74 | 1.01–3.00 | 0.047 | |||
The column names and the row names indicate the dependant and the independent variables, respectively. The full models are provided in the Supplementary materials.
MACE, major adverse cardiac event; HR, hazard ratio; CI, confidence interval; MAGGIC, Meta-Analysis Global Group in Chronic Heart Failure.
Clinical factors include age, sex, body mass index, history of coronary heart disease, hypertension, and left ventricular ejection fraction.
3.2. Supervised learning-based prediction of functional LV remodeling
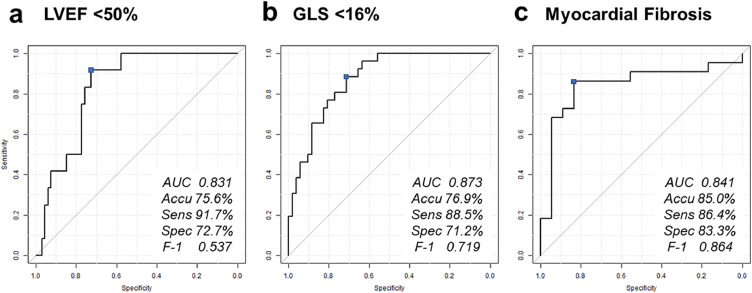
This part is summarized in the bottom left of Fig. 1. To explore the value of the texture-based tissue features to directly predict functional LV remodeling, the patient cohort was randomly divided into training (80%) and test set (20%), and supervised machine learning algorithms were trained in the training set using only the texture features extracted from the still images. Topological data analysis and subsequent recursive feature elimination were used for feature selection and 18 features per each were used to train prediction models for reduced LVEF (<50%) and GLS (<16%). Panel A and B in Fig. 4 show the ROC curves for the prediction of reduced LVEF and GLS obtained in the test set. The best model for predicting reduced LVEF was an ensemble of 10 models (2 boosted trees, 3 random decision forests, 2 LASSO regressions, 2 ridge regressions, and 1 neural network: the performance of each model in the cross-validation is summarized in Supplementary Table 3) and showed performance of ROC AUC 0.83, sensitivity 91.7%, and specificity 72.3%, whereas the one for impaired GLS was an ensemble of 10 different models (5 bootstrap decision forest, 2 ridge regressions, 2 LASSO regressions, and 1 neural network) and had ROC AUC of 0.87, sensitivity of 88.5%, and specificity of 71.2%.
Fig. 4.
Direct prediction of impaired cardiac function. Panels a to c show receiver operator characteristics curves for predicting reduced LVEF (<50%), impaired GLS (<16%), and the presence of myocardial fibrosis. Supervised machine learning models were successful in predicting these outcomes with the texture features extracted from still ultrasound images.
LVEF, left ventricular ejection fraction; GLS, global longitudinal strain; ROC AUC, area under the curve; Accu, accuracy; Sens, sensitivity; Spec, specificity; F-1, F1 score.
3.3. Supervised learning-based prediction of myocardial fibrosis
For investigating the value of the cardiac ultrasound texture-based features in predicting whether the patient has myocardial fibrosis detected by CMR, we studied 89 retrospectively identified patients who had undergone CMR and echocardiography within 48 h as the training set, and 40 independent prospective patients as the test set, as shown in Fig. 1b. Texture feature extraction was feasible in 85 (96%) and 40 (100%) patients, respectively. The clinical characteristics of the training and test set are summarized in Table 3. There were 48 (56.4%) and 22 (55.0%) patients who had myocardial fibrosis in at least one of the four ROIs where the texture features were extracted using the ultrasound images in the training and test set, respectively.
Table 3.
Patient characteristics of the patients with a magnetic resonance scan.
| Factor | Retrospective (training) | Prospective (test) | p value |
|---|---|---|---|
| Number of patients | 85 | 40 | |
| Age, years | 55 [41–66] | 56 [46–64] | 0.667 |
| Female, n (%) | 53 (62.4) | 16 (40.0) | 0.022 |
| BSA, m2 | 2.03 (0.35) | 2.03 (0.27) | 0.984 |
| BMI, kg/m2 | 55.37 (231.98) | 29.77 (6.23) | 0.488 |
| Coronary artery disease, n (%) | 38 (44.7) | 13 (33.3) | 0.246 |
| Hypertension, n (%) | 43 (50.6) | 27 (69.2) | 0.078 |
| Cerebral vessel disease, n (%) | 6 (7.1) | 3 (12.5) | 0.410 |
| Diabetes mellitus, n (%) | 21 (24.7) | 13 (33.3) | 0.387 |
| Atrial fibrillation, n (%) | 8 (9.4) | 4 (10.3) | >0.99 |
| NYHA ≥III, n (%) | 12 (14.1) | 16 (40.0) | 0.002 |
| HF stages C or D, n (%) | 17 (20.0) | 21 (52.5) | <0.001 |
| Echocardiography | |||
| IVSd, mm | 10 [8–12] | 10 [9–11] | 0.983 |
| PWTd, mm | 9 [8–11] | 10 [9–11] | 0.503 |
| LVIDd, mm | 48 [42–53] | 48 [45–57] | 0.165 |
| LVIDs, mm | 34 [29–42] | 35 [30–48] | 0.378 |
| LVEDVi, mL/m2 | 54 [40–67] | 57 [45–69] | 0.537 |
| LVESVi, mL/m2 | 23 [16–40] | 27 [20–45] | 0.168 |
| LA volume index, mL/m2 | 22 [18–33] | 32 [21–39] | 0.011 |
| LV mass index, mg/m2 | 79 [67–103] | 96 [70–132] | 0.057 |
| E wave velocity, m/s | 0.85 [0.62–1.00] | 0.86 [0.69–1.00] | 0.816 |
| A wave velocity, m/s | 0.60 [0.51–0.84] | 0.68 [0.49–0.83] | 0.980 |
| E / A ratio | 1.26 [0.84–1.77] | 1.16 [0.90–1.66] | 0.810 |
| e', cm/s | 9 [6–12] | 8 [6–11] | 0.317 |
| E / e' | 8.4 [6.6–11.3] | 10.4 [7.6–14.8] | 0.093 |
| LVEF, % | 55 [42–63] | 48 [34–56] | 0.025 |
| GLS, absolute % | NA | 12.8 [7.9–19.4] | NA |
| LV hypertrophy, n (%) | 0.136 | ||
| LV diastolic function, n (%) | 0.173 | ||
| Normal | 17 (20.0) | 3 (8.3) | |
| Grade 1 | 28 (32.9) | 11 (30.6) | |
| Grade 2 | 18 (21.2) | 15 (41.7) | |
| Grade 3 | 17 (20.0) | 5 (13.9) | |
| Indeterminate | 5 (5.9) | 2 (5.6) | |
| Indeterminate grade | 0 (0.0) | 0 (0.0) |
Abbreviations are the same as in Table 1.
Out of the extracted texture features, we performed feature selection and identified 5 best features to develop supervised machine learning models. The models were trained to predict whether the patient has myocardial fibrosis using cross-validation in the training set (Fig. 5). In the test set, the developed model (an ensemble of 2 LASSO regressions) predicted myocardial fibrosis with an ROC AUC of 0.84 (sensitivity 86.4%, and specificity 83.3%).
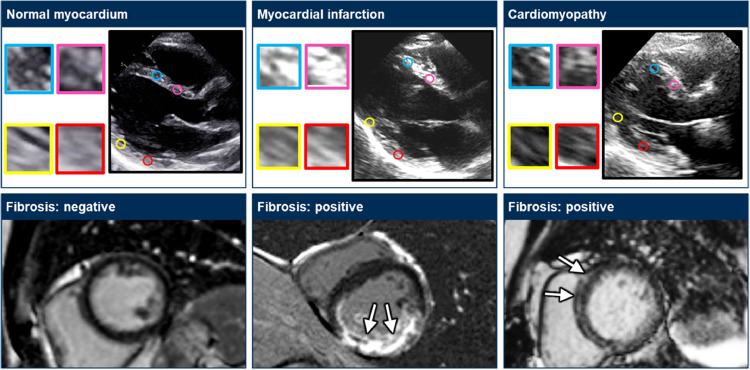
Fig. 5.
Prediction of myocardial fibrosis. The upper panels show still ultrasound images and the corresponding myocardial textures where the texture features were extracted. The lower panels show magnetic resonance images with late gadolinium enhancement.
3.4. Robustness of texture-based feature extraction
To confirm the stability of the texture features, we assessed variability related to the operator, image quality, and device vendors used in the study. Supplementary Table 4 summarizes the importance and the interobserver variability of top features. Briefly, most features had a good interobserver agreement with interclass correlation coefficient 0.74–0.96, except for correlation in GLCM (0.54). Supplementary Fig. 1 shows the variability of the features by the image quality. We artificially increased noise and gain on each image and tested the change of the features. As shown, each feature showed different behavior against increases in gain and noise. For example, gray-level non-uniformity of the GLRLM, the second most important feature for predicting functional LV remodeling, was resistant to an increase in gain, while it markedly increased with additional noise in the images. On the other hand, the high gray-level run emphasis of GLCM, one of the important features for predicting myocardial fibrosis, was relatively resistant to an increase in noise, whereas it dramatically increased with additional gains on the images.
To elucidate the value of the texture features among different vendors, patient similarity networks were created using data obtained from each of the two dominant vendors using topological data analysis. The created networks from both vendors formed similar loops, where most patients with reduced LV systolic function were segregated in a part of a loop (Supplementary Fig. 2), suggesting that the information content of the texture features were relatively invariant to the data source.
4. Discussion
In the present study, we illustrated that [1] texture-based analysis was feasible for most clinical cardiac ultrasound (97%) [2], unsupervised patient-similarity analysis revealed that a specific pattern of information from myocardial texture was associated with functional LV remodeling, advanced heart failure, and adverse clinical outcome, and [3] the texture features extracted from still cardiac ultrasound images could be used for developing supervised machine learning models that enable clinical prediction of functional and structural LV remodeling. Texture-based analysis has been recently used in radiology (also referred to as radiomics) to extract maximal information from standard-of-care images using high-throughput computing [10,11] Our work resembles the general principles of radiomics and specifically defines a computational pipeline where texture-based tissue features were extracted and used for building supervised machine learning models for individualized predictions. This approach may potentially address a long-described objective of cardiac ultrasound in providing myocardial tissue characterization in clinical practice.
It is well known that in typical cases, the pathological myocardial texture is visually distinguishable with ultrasound images. For example, scar lesions after myocardial infarction have high echo intensity and thin walls, and myocardium with infiltration of amyloid has a granular sparkling texture [30]. However, many previous attempts to characterize myocardial tissue using ultrasound images, such as integrated backscatter analysis, have resulted in suboptimal results because of variations in the quality and texture of the cardiac ultrasound images [31,32]. As a consequence, in current clinical practice, CMR imaging is preferred modality for myocardial tissue characterization using late gadolinium enhancement imaging and methods such as parametric and non-parametric T1, T2 and T2* imaging. However, due to its cost, accessibility, and contraindications, CMR is not available for every patient and in every place. Since cardiac ultrasound remains portable, low-cost, and the most common cardiac imaging procedure performed in clinical practice, implementation of tissue characterization with cardiac ultrasound may have a wider clinical impact. In our initial attempt, we illustrated that the use of cardiac ultrasound texture-based tissue features of myocardium was robust and concordant in several steps of analyses: i.e., cluster analysis with topological data analysis with clinical outcome prediction; supervised machine learning analysis for predicting impaired LV systolic function; and identification of the presence of myocardial fibrosis. We also showed that the segregation of high-risk myocardium was vendor-independent and that interobserver agreement was adequate for clinical application. These results reconfirmed that important information associated with myocardial remodeling that can be captured by CMR is also carried in ultrasound texture features and can be retrieved using a modern high-throughput computing pipeline, which possibly amended the signal to noise ratio and helped extraction of useful information from noisy ultrasound data. Although some features were sensitive to changes in gain or noise, the majority of the features were stable and resistant to the changes in image quality. More work would be needed for appropriate feature engineering and building models that are even more resistant to the changes in image quality.
Although we used radiomics-based texture analysis approach in this study, deep learning may be another choice of approaches with which images can be analyzed in an end-to-end pipeline. Both deep learning and radiomics have received considerable attention in recent years in radiology and the relative merits of both techniques remains an area of active investigation [33]. While some investigators have only recently compared the two approaches citing the advantage of deep learning approaches for radiological images [34], others have suggested that both approaches are complementary and can unite in the future to produce a single unified framework [34]. Such comparative studies have been performed mostly in radiology, in general, and the application of radiomics for cardiac imaging is still in its infancy. To the best of our knowledge, our study is the first to attempt the application of traditional radiomics approach to extract semantic and agnostic features from cardiac ultrasound images for predicting LV remodeling. The recent successful application of hand-crafted radiomics features in myocardial tissue characterization [35], [36], [37] further supports our choice of restricting the initial analysis to only using handcrafted radiomics approach. We agree that deep learning based radiomics may have several advantages including its generalization capability and its independence from the supervision of experts, however, the lack of reproducibility and interpretability, as well as over-fitting on small datasets like ours, poses substantial challenges in readily adapting deep networks for this study. We have added this information in the Supplementary Table 3
Our study results will open up new lines of investigations. Currently, our description of cardiac ultrasound texture-analysis addresses only texture-based myocardial properties. However, perhaps in the near future, the addition of newer tissue material properties such as myocardial stiffness obtained using ultrafast-ultrasound techniques could provide a similar multiparametric yield as described in CMR [38]. Furthermore, extracellular volume quantification and other myocardial characteristics seen in specific diseases such as cardiac amyloidosis and hypertrophic cardiomyopathy will merit important future considerations. Moreover, due to its noninvasive and repeatable nature, it may allow serial assessments of myocardial properties after clinical therapies.
The present study has several limitations that require future considerations. First, this study was a single-center study using ultrasound images acquired in only one laboratory. External validation in ultrasound images acquired in other centers is necessary before clinical application. Next, we used only parasternal long axis views because this view is the most standardized and feasible view in routine clinical cardiac ultrasound examination and provides good quality images in most cases. The usability of other views, such as apical four chamber views and parasternal short axis views, should also be tested. Moreover, in this study, as proof of the initial concept, we focused on the prediction of fibrosis within the four ROI applied in four-segments studied by cardiac ultrasound and CMR. Also, the association of texture features with quantitative measures of myocardial remodeling, such as extracellular volume, is of interest in future studies. The parasternal long-axis echocardiographic view provides images with the insonifying ultrasonic field primarily perpendicular to the predominant myocardial fibre orientation and, hence, reduces the confounding effects of tissue anisotropy on echocardiographic image quality. Extraction of the texture-based informatics for each myocardial segment and for all the views of the LV (from both apical and parasternal views) using novel segmentation and machine learning techniques could potentially improve the yield and applicability of the overall technique. Finally, although we showed that changes in myocardial texture were associated with impaired cardiac function, fibrosis and high-risk patient characteristics and adverse clinical outcomes, the direct comparison with histological features of LV remodeling would be desirable.
In conclusion, our new approach using a low-cost texture-based pipeline identifies a tissue texture of LV remodeling that is associated with clinical characteristics and outcomes in early and advanced stages of heart failure. Further clinical validation of this quantitative approach using advanced image processing and artificial intelligence may address critical barriers in the adoption of ultrasound techniques as a viable cost-effective alternative to CMR techniques for myocardial tissue characterization.
Declaration of Competing Interest
Nobuyuki Kagiyama is supported by a research grant from Hitachi Healthcare; Partho P. Sengupta is a consultant to Heart Sciences, Ultromics, and Kencor Health. The other authors have nothing to disclose
Acknowledgements/Funding
The authors thank the following individual for their excellent contributions:
Lan Hu, RN as a research coordinator from West Virginia University Heart and Vascular Institute; Naveena Yanamala, PhD (Carnegie Melon University) as a technical consultant regarding the machine learning pipeline; Pablo Gonzalez (BigML Inc.) for his support in optimizing the use of automated machine learning models for feature and model discovery.
This project was supported by a Research Fellowship Grant from Hitachi Healthcare, Department Activity: 492100035 Fund: 12300030 to Dr. Kagiyama.
Footnotes
This study was the winner of the Arthur E. Weymann Young Investigator Award at the annual meeting of the American Society of Echocardiography in June 2019.
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102726.
Appendix. Supplementary materials
References
- 1.Benjamin E.J., Virani S.S., Callaway C.W., Chamberlain A.M., Chang A.R., Cheng S. Heart disease and stroke statistics-2018 update: a report from the American Heart Association. Circulation. 2018;137(12):e67–e492. doi: 10.1161/CIR.0000000000000558. [DOI] [PubMed] [Google Scholar]
- 2.Jugdutt B.I. Ventricular remodeling after infarction and the extracellular collagen matrix: when is enough enough? Circulation. 2003;108(11):1395–1403. doi: 10.1161/01.CIR.0000085658.98621.49. [DOI] [PubMed] [Google Scholar]
- 3.Konstam M.A., Kramer D.G., Patel A.R., Maron M.S., Udelson J.E. Left ventricular remodeling in heart failure: current concepts in clinical significance and assessment. JACC Cardiovasc Imaging. 2011;4(1):98–108. doi: 10.1016/j.jcmg.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 4.aus dem Siepen F., Buss S.J., Messroghli D., Andre F., Lossnitzer D., Seitz S. T1 mapping in dilated cardiomyopathy with cardiac magnetic resonance: quantification of diffuse myocardial fibrosis and comparison with endomyocardial biopsy. Eur Heart J Cardiovasc Imaging. 2015;16(2):210–216. doi: 10.1093/ehjci/jeu183. [DOI] [PubMed] [Google Scholar]
- 5.Pashakhanloo F., Herzka D.A., Mori S., Zviman M., Halperin H., Gai N. Submillimeter diffusion tensor imaging and late gadolinium enhancement cardiovascular magnetic resonance of chronic myocardial infarction. J Cardiovasc Magn Reson. 2017;19(1):9. doi: 10.1186/s12968-016-0317-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnson M.R., McPherson D.D., Fleagle S.R., Hunt M.M., Hiratzka L.F., Kerber R.E. Videodensitometric analysis of human coronary stenoses: validation in vivo by intraoperative high-frequency epicardial echocardiography. Circulation. 1988;77(2):328–336. doi: 10.1161/01.cir.77.2.328. [DOI] [PubMed] [Google Scholar]
- 7.Milunski M.R., Mohr G.A., Perez J.E., Vered Z., Wear K.A., Gessler C.J. Ultrasonic tissue characterization with integrated backscatter. Acute myocardial ischemia, reperfusion, and stunned myocardium in patients. Circulation. 1989;80(3):491–503. doi: 10.1161/01.cir.80.3.491. [DOI] [PubMed] [Google Scholar]
- 8.Aerts H.J., Velazquez E.R., Leijenaar R.T., Parmar C., Grossmann P., Carvalho S. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castellano G., Bonilha L., Li L.M., Cendes F. Texture analysis of medical images. Clin Radiol. 2004;59(12):1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Gillies R.J., Kinahan P.E., Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller H., Michoux N., Bandon D., Geissbuhler A. A review of content-based image retrieval systems in medical applications-clinical benefits and future directions. Int J Med Inform. 2004;73(1):1–23. doi: 10.1016/j.ijmedinf.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 12.Hoit B.D. Strain and strain rate echocardiography and coronary artery disease. Circ Cardiovasc Imaging. 2011;4(2):179–190. doi: 10.1161/CIRCIMAGING.110.959817. [DOI] [PubMed] [Google Scholar]
- 13.Muraru D., Niero A., Rodriguez-Zanella H., Cherata D., Badano L. Three-dimensional speckle-tracking echocardiography: benefits and limitations of integrating myocardial mechanics with three-dimensional imaging. Cardiovasc Diagn Ther. 2018;8(1):101–117. doi: 10.21037/cdt.2017.06.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nioche C., Orlhac F., Boughdad S., Reuze S., Goya-Outi J., Robert C. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018;78(16):4786–4789. doi: 10.1158/0008-5472.CAN-18-0125. [DOI] [PubMed] [Google Scholar]
- 15.Sun C., Wee W.G. Neighboring gray level dependence matrix for texture classification. Comput Vis Graphics Image Process. 1983;23(3):341–352. [Google Scholar]
- 16.Simpson J., Jhund P.S., Silva Cardoso J., Martinez F., Mosterd A., Ramires F. Comparing LCZ696 with enalapril according to baseline risk using the MAGGIC and EMPHASIS-HF risk scores: an analysis of mortality and morbidity in PARADIGM-HF. J Am Coll Cardiol. 2015;66(19):2059–2071. doi: 10.1016/j.jacc.2015.08.878. [DOI] [PubMed] [Google Scholar]
- 17.Lang R.M., Badano L.P., Mor-Avi V., Afilalo J., Armstrong A., Ernande L. Recommendations for cardiac chamber quantification by echocardiography in adults: an update from the American Society of Echocardiography and the European Association of Cardiovascular Imaging. J Am Soc Echocardiogr. 2015;28(1):1–39. doi: 10.1016/j.echo.2014.10.003. e14. [DOI] [PubMed] [Google Scholar]
- 18.Lum P.Y., Singh G., Lehman A., Ishkanov T., Vejdemo-Johansson M., Alagappan M. Extracting insights from the shape of complex data using topology. Sci Rep. 2013;3:1236. doi: 10.1038/srep01236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haralick R.M., Shanmugam K., Dinstein I. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;SMC-3(6):610–621. [Google Scholar]
- 20.Guo W., Banerjee A.G. 2016 IEEE international symposium on assembly and manufacturing (ISAM) 2016. Toward automated prediction of manufacturing productivity based on feature selection using topological data analysis; pp. 21–22. Aug. 2016. [Google Scholar]
- 21.Kellman P., Arai A.E., McVeigh E.R., Aletras A.H. Phase-sensitive inversion recovery for detecting myocardial infarction using gadolinium-delayed hyperenhancement. Magn Reson Med. 2002;47(2):372–383. doi: 10.1002/mrm.10051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu W., Fang W. 2008 ISECS international colloquium on computing, communication, control, and management. 2008. Adaptive spam filtering based on fingerprint vectors; pp. 3–4. Aug. 2008. [Google Scholar]
- 23.Casaclang-Verzosa G., Shrestha S., Khalil M.J., Cho J.S., Tokodi M., Balla S. Network tomography for understanding phenotypic presentations in aortic stenosis. JACC Cardiovasc Imaging. 2019;12(2):236–248. doi: 10.1016/j.jcmg.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 24.Hinks T., Zhou X., Staples K., Dimitrov B., Manta A., Petrossian T. Multidimensional endotypes of asthma: topological data analysis of cross-sectional clinical, pathological, and immunological data. Lancet. 2015;385(Suppl 1):S42. doi: 10.1016/S0140-6736(15)60357-9. [DOI] [PubMed] [Google Scholar]
- 25.Diddams S.A., Hollberg L., Mbele V. Molecular fingerprinting with the resolved modes of a femtosecond laser frequency comb. Nature. 2007;445(7128):627–630. doi: 10.1038/nature05524. [DOI] [PubMed] [Google Scholar]
- 26.Johannes M., Brase J.C., Frohlich H., Gade S., Gehrmann M., Falth M. Integration of pathway knowledge into a reweighted recursive feature elimination approach for risk stratification of cancer patients. Bioinformatics. 2010;26(17):2136–2144. doi: 10.1093/bioinformatics/btq345. [DOI] [PubMed] [Google Scholar]
- 27.Shahriari B., Swersky K., Wang Z., Adams R.P., Freitas N.d. Taking the human out of the loop: a review of bayesian optimization. Proc IEEE. 2016;104(1):148–175. [Google Scholar]
- 28.Hutter F., Hoos H.H., Leyton-Brown K. Learning and intelligent optimization. Springer Berlin Heidelberg; Berlin, Heidelberg: 2011. Sequential model-based optimization for general algorithm configuration. 2011// [Google Scholar]
- 29.Yancy C.W., Jessup M., Bozkurt B., Butler J., Casey D.E., Jr., Drazner M.H. 2013 ACCF/AHA guideline for the management of heart failure: a report of the American College of Cardiology Foundation/American Heart Association Task Force on practice guidelines. J Am Coll Cardiol. 2013;62(16):5. doi: 10.1016/j.jacc.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Cueto-Garcia L., Reeder G.S., Kyle R.A., Wood D.L., Seward J.B., Naessens J. Echocardiographic findings in systemic amyloidosis: spectrum of cardiac involvement and relation to survival. J. Am. Coll. Cardiol. 1985;6(4):737–743. doi: 10.1016/s0735-1097(85)80475-7. [DOI] [PubMed] [Google Scholar]
- 31.Chandrasekaran K., Aylward P.E., Fleagle S.R., Burns T.L., Seward J.B., Tajik A.J. Feasibility of identifying amyloid and hypertrophic cardiomyopathy with the use of computerized quantitative texture analysis of clinical echocardiographic data. J. Am. Coll. Cardiol. 1989;13(4):832–840. doi: 10.1016/0735-1097(89)90225-8. [DOI] [PubMed] [Google Scholar]
- 32.Mor-Avi V., Lang R.M., Badano L.P., Belohlavek M., Cardim N.M., Derumeaux G. Current and evolving echocardiographic techniques for the quantitative evaluation of cardiac mechanics: ASE/EAE consensus statement on methodology and indications endorsed by the Japanese Society of Echocardiography. J Am Soc Echocardiogr. 2011;24(3):277–313. doi: 10.1016/j.echo.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 33.Parekh V.S., Jacobs M.A. Deep learning and radiomics in precision medicine. Expert Rev Precis Med Drug Dev. 2019;4(2):59–72. doi: 10.1080/23808993.2019.1585805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu X., Li Y., Chen X., Huang Y., He L., Zhao K. Deep learning features improve the performance of a radiomics signature for predicting KRAS status in patients with colorectal cancer. Acad Radiol. 2020 doi: 10.1016/j.acra.2019.12.007. doi: 10.1016/j.acra.2019.12.007. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Wei L., Osman S., Hatt M., El Naqa I. Machine learning for radiomics-based multimodality and multiparametric modeling. Q J Nucl Med Mol Imaging. 2019;63(4):323–338. doi: 10.23736/S1824-4785.19.03213-8. [DOI] [PubMed] [Google Scholar]
- 36.Oikonomou E.K., Williams M.C., Kotanidis C.P., Desai M.Y., Marwan M., Antonopoulos A.S. A novel machine learning-derived radiotranscriptomic signature of perivascular fat improves cardiac risk prediction using coronary CT angiography. Eur Heart J. 2019;40(43):3529–3543. doi: 10.1093/eurheartj/ehz592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neisius U., El-Rewaidy H., Kucukseymen S., Tsao C.W., Mancio J., Nakamori S. Texture signatures of native myocardial T1 as novel imaging markers for identification of hypertrophic cardiomyopathy patients without scar. J Magn Reson Imaging. 2020 doi: 10.1002/jmri.27048. doi: 10.1002/jmri.27048. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y., Hamilton J., Rajagopalan S., Seiberlich N. Cardiac magnetic resonance fingerprinting: technical overview and initial results. JACC Cardiovasc Imaging. 2018;11(12):1837–1853. doi: 10.1016/j.jcmg.2018.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.