Abstract
Disruption of estrogen receptor beta (ESR2) dysregulates granulosa cell genes essential for follicle maturation and ovulation. The datasets presented in this article depict gonadotropin-induced genes, which are differentially expressed in Esr2-null rat granulosa cells. Synchronized follicle development was initiated in four-week-old wildtype and Esr2-null female rats by administration of PMSG. Forty-eight hours after PMSG injection, further maturation of ovarian follicles was induced by hCG treatment. Granulosa cells were collected from the ovaries before gonadotropin administration, 48 h after PMSG treatment, and 4 h after hCG injection to the PMSG-treated rats. Total RNA was purified from granulosa cells and whole transcriptome was assessed by RNA-sequencing on an Illumina HiSeq X platform. RNA-seq data of wildtype and Esr2-null granulosa cells were analyzed and differentially expressed genes were identified by CLC Genomics Workbench. Gonadotropin-induced genes were identified by comparing the transcriptome data of PMSG- or hCG-induced wildtype granulosa cells with those without gonadotropin treatment. Furthermore, differentially expressed genes in Esr2-null granulosa cells were determined by comparing the transcriptome data with that of wildtype granulosa cells. These datasets can be used to recognize the gonadotropin-induced genes in granulosa cells that are Esr2-regulated and important for ovarian follicle maturation.
Keywords: Rat models, ESR2, Gonadotropins, Granulosa cells, RNA-sequencing, Transcriptome-analyses
Specifications table
| Subject | Biology |
| Specific subject area | Reproductive biology |
| Type of data | Table |
| Figure | |
| How data were acquired | Sequencing of RNA from rat granulosa cells |
| Data format | Raw (FASTQ) and analysed (Excel tables) |
| Parameters for data collection | RNA sequencing was performed on granulosa cells collected from the ovaries of gonadotropin treated wildtype and Esr2-null mutant rats. |
| Description of data collection | Four-week-old wildtype and Esr2-null Holtzman rats were treated with 30IU of PMSG. Forty-eight hours after PMSG administration, rats were injected with 30IU of hCG. Granulosa cells were collected from the rat ovaries before and after gonadotropin treatment, RNAs were purified and analysed by mRNA-sequencing. Gonadotropin-induced genes and the differentially expressed genes in Esr2-null granulosa cells were identified by analyses of the transcriptome data using CLC Genomics Workbench. |
| Data source location | University of Kansas Medical Center, Kansas City, KS 66160, USA |
| Data accessibility | Repository name: SRA |
| Data identification number: PRJNA551764 and PRJNA551766 | |
| Direct URL to data: https://www.ncbi.nlm.nih.gov/sra/ PRJNA551764 and https://www.ncbi.nlm.nih.gov/sra/ PRJNA551766 |
Value of the data
-
•
These datasets provide the transcriptome analyses of rat granulosa cells during gonadotropin induced follicle maturation.
-
•
These datasets also represent the differentially expressed gonadotropin-induced genes in Esr2-null rat granulosa cells.
-
•
These data can be used to identify the ESR2-regulated genes in granulosa cells that are involved in follicle maturation and ovulation.
-
•
These data will help understand the molecular mechanisms involved in gonadotropin- induced changes in the granulosa cell transcriptome, and the role of ESR2-signaling during gonadotropin induced ovarian follicle maturation.
-
•
These data are useful to the researchers who study the biology of folliculogenesis.
1. Data
Transcriptome data were generated by RNA-sequencing of wildtype and Esr2-null granulosa cells during different stages of follicle development. The raw data are deposited to NCBI SRA under PRJNA551764 and PRJNA551766 and the analyzed data are presented in this article in Excel format. Analysed data are presented in the following tables:
Supplementary Table 1. Differentially expressed genes in wildtype rat granulosa cells after PMSG treatment.
Transcriptome analyses of PMSG-induced changes in gene expression in rat granulosa cells compared to basal levels before gonadotropin treatment. Of the 1720 differentially expressed genes, 591 were ≥ 2 fold upregulated, whereas the 1129 were ≤ 2 fold downregulated.
Supplementary Table 2. Differentially expressed genes in wildtype rat granulosa cells after hCG treatment.
Transcriptome analyses of hCG-induced changes in gene expression in rat granulosa cells compared to basal levels before gonadotropin treatment. Of the 2804 differentially expressed genes, 1269 were ≥ 2 fold upregulated, whereas the 1535 were ≤ 2 fold downregulated.
Supplementary Table 3. Differentially expressed genes in wildtype rat granulosa cells after hCG treatment following PMSG administration.
Transcriptome analyses of hCG-induced changes in gene expression in rat granulosa cells compared to the levels after PMSG treatment. Of the 3021 differentially expressed genes, 1520 were ≥ 2 fold upregulated, whereas the 1501 were ≤ 2 fold downregulated.
Supplementary Table 4. Differentially expressed genes in Esr2-null granulosa cells before gonadotropin treatment.
Transcriptome data of Esr2-null granulosa cells were compared to that of wildtype granulosa cells isolated from four-week-old rats before gonadotropin treatment. Of the 635 differentially expressed genes, 278 were ≥ 2 fold upregulated, whereas the 357 were ≤ 2 fold downregulated.
Supplementary Table 5. Differentially expressed genes in Esr2-null granulosa cells 48 h after PMSG treatment.
Transcriptome data of Esr2-null granulosa cells were compared to that of wildtype granulosa cells isolated from four-week-old rats 48 h after PMSG administration. Of the 1157 differentially expressed genes, 708 were ≥ 2 fold upregulated, whereas 449 were ≤ 2 fold downregulated.
Supplementary Table 6. Differentially expressed genes in Esr2-null granulosa cells 4 h after hCG treatment.
Transcriptome data of Esr2-null granulosa cells were compared to that of wildtype granulosa cells isolated from four-week-old rats 4 h after hCG treatment following 48 h of PMSG administration. Of the 1369 Differentially expressed genes, 865 were ≥ 2 fold upregulated, whereas rest 504 were ≤ 2 fold downregulated.
2. Experimental design, materials and methods
2.1. Experimental animals
Four-week-old wildtype and age matched Esr2-null female Holtzman Sprague-Dawley (HSD) rats were used to generate these datasets. All procedures were performed in accordance with the protocols by University of Kansas Medical Center Animal Care and Use Committee.
2.2. Gonadotropin treatment
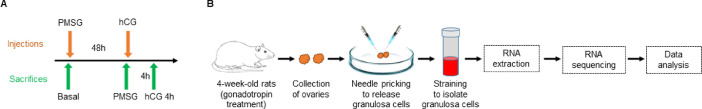
Synchronized follicular growth was initiated by administration of exogenous gonadotropins to four-week-old wildtype and Esr2-null female rats [1,3,4]. Initially, 30 IU of pregnant mare's serum gonadotropin (PMSG, Lee Bioscience, MO) was injected intraperitoneally. 48 h after the PMSG administration, 30 IU of human chorionic gonadotropin (hCG, Lee Bioscience, MO) was injected to the rats (Fig. 1).
Fig. 1.
Schematic presentation of the experimental design. (A) Four-week-old wildtype or Esr2-null female rats were injected intraperitoneally with 30IU of PMSG, and 48 h after the PMSG injection, with 30IU of hCG. (B) Rats were sacrificed at the indicated time points before and after PMSG or hCG injections (A), and the ovaries were collected for granulosa cell isolation. Granulosa cells were released from the ovarian follicles by needle puncture, filtered with a strainer and pelleted out by centrifugation. Then, total RNA was purified from the granulosa cells and used for mRNA sequencing.
2.3. Sample collection and processing
Esr2-null and age-matched wildtype female rats were sacrificed, and ovaries were collected prior to gonadotropin administration (Basal group), 48 h after PMSG injection (PMSG group), and 4 h after hCG injection to PMSG-treated rats (hCG 4 h group) (Fig. 1A). Granulosa cells were collected from the ovaries by needle puncture under microscopic examination followed by filtration with a 40 µM cell strainer (Fig. 1B) [1,3]. Total RNA was extracted from the granulosa cells using TRI Reagent (Sigma-Aldrich, St. Louis, MO) following the manufacturers instruction. RNA quality was assessed by a Bioanalyzer and samples with a RIN value over 9 were selected for mRNA-seq library preparation. Approximately 500 ng of total RNA was used for the RNA-seq library preparation using a TruSeq Standard mRNA kit (Illumina, San Diego, CA) following the manufacturer's instruction [2]. The cDNA libraries were initially evaluated for quality and then sequenced on an Illumina HiSeq X sequencer (Novogene Corporation, Sacramento, CA).
2.4. RNA-seq data analyses
RNA sequencing data were demultiplexed, trimmed, aligned and analyzed using CLC Genomics Workbench 12.2 (Qiagen Bioinformatics, Germantown, MD). Trimming was performed to remove low quality reads and good quality reads were aligned with Rattus norvegicus genome (Rnor_6.0) using default parameters (a) maximum number of allowable mismatches was 2 (b) minimum length and similarity fraction was set at 0.8; and (c) minimum number of hits per read was 10. Expression values were measured in TPM (transcripts per million). The threshold p-value was determined according to the false discovery rate (FDR). Differentially expressed genes were determined if the absolute fold change in expression was 2 with an FDR p-value of ≤ 0.05.
2.5. Statistical analysis
Each RNA-seq library was prepared using pooled RNA samples form three individual wildtype or Esr2-null rats. Each group of RNA sequencing consisted of three libraries. Differentially expressed genes were identified by CLC Genomics Workbench as described previously [2].
Acknowledgments
Generation of these datasets was partially supported by funding from the Centers of Biomedical Research Excellence (COBRE) award from the National Institute of General Medical Sciences (P30 GM122731) from the National Institutes of Health.
Conflict of interest
None.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.dib.2020.105405.
Appendix. Supplementary materials
References
- 1.V.P. Chakravarthi, V. Khristi, S. Ghosh, S. Yerrathota, E. Dai, K.F. Roby, M.W. Wolfe, M.A.K. Rumi, ESR2 is Essential for gonadotropin-induced Kiss1 expression in granulosa cells. Endocrinology159 (2018) 3860–3873. doi: 10.1210/en.2018-00608. [DOI] [PMC free article] [PubMed]
- 2.V. Khristi, V.P. Chakravarthi, P. Singh, S. Ghosh, A. Pramanik, A. Ratri, S. Borosha, K.F. Roby, M.W. Wolfe, M.A.K. Rumi, Differentially regulated genes in Esr2-mutant rat granulosa cells. Data Brief19 (2018) 1008–1011. doi: 10.1016/j.dib.2018.05.098. [DOI] [PMC free article] [PubMed]
- 3.V. Khristi, V.P. Chakravarthi, P. Singh, S. Ghosh, A. Pramanik, A. Ratri, S. Borosha, K.F. Roby, M.W. Wolfe, M.A.K. Rumi, ESR2 regulates granulosa cell genes essential for follicle maturation and ovulation. Mol. Cell. Endocrinol.474 (2018) 214–226. doi: 10.1016/j.mce.2018.03.012. [DOI] [PubMed]
- 4.Rumi M.A.K., Singh P., Roby K.F., Zhao X., Iqbal K., Ratri A., Lei T., Cui W., Borosha S., Dhakal P., Kubota K., Chakraborty D., Vivian J.L., Wolfe M.W., Soares M.J. Defining the role of estrogen receptor beta in the regulation of female fertility. Endocrinology. 2017;158:2330–2343. doi: 10.1210/en.2016-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.