Abstract
Background
This study was designed to investigate the role of long non-coding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) in the proliferation as well as apoptosis of human umbilical vein endothelial cells (HUVECs), to offer a basis for therapy of hypertension.
Material/Methods
The lncRNA MALAT1 expression, hsa-miR-124-3p, hsa-miR-135a-5p, hsa-miR-135b-5p, and hsa-miR-455-5p in plasma were measured from 230 patients with hypertension and 230 non-hypertensive controls. The mechanism for lncRNA MALAT1 modulating the proliferation and apoptosis of HUVECs was explored by cell transfection, Cell Counting Kit-8 (CCK-8), quantitative real-time polymerase chain reaction (qRT-PCR), western blot, and dual-luciferase reporter assays.
Results
The expression of hsa-miR-124-3p and hsa-miR-135a-5p was reduced and the expression of lncRNA MALAT1 was increased in the plasma of hypertensive patients. Moreover, the plasma levels of hsa-miR-124-3p and hsa-miR-135a-5p of hypertensive patients were negatively correlated with lncRNA MALAT1 (r=−0.64, −0.72; P<0.01, P<0.01, respectively). The level of nuclear receptor subfamily 3, group C, member 2 (NR3C2) protein was negatively correlated with hsa-miR-124-3p and hsa-miR-135a-5p (r=−0.74, −0.84; P<0.01, P<0.01, respectively). The proliferation of HUVECs was inhibited after the inhibition of MALAT. Additionally, after knocking down MALAT, the levels of hsa-miR-124-3p and hsa-miR-135a-5p in HUVECs were markedly increased, while the expression level of NR3C2 protein was decreased. The apoptotic rate of HUVECs after the transfection of MALAT1 small interfering RNA (si-MALAT1) (3.64±0.21%) was significantly reduced compared to that of transfected si-MALAT1 no template control (NC) (3.76±0.19%) and the control group (10.51±1.24%).
Conclusions
LncRNA MALAT1 regulates proliferation and apoptosis of HUVECs through the hsa-miR-124-3p/NR3C2 and/or hsa-miR-135a-5p/NR3C2 axis.
MeSH Keywords: MicroRNAs; RNA, Long Noncoding; White Coat Hypertension
Background
Essential hypertension is a chronic disease and a key risk factors for cardiovascular and cerebrovascular diseases. The common complications of hypertension are stroke, chronic kidney disease, heart failure, and myocardial infarction. The disability and mortality rates of hypertension are relatively high, and this disease severely strains medical and social resources, causing a burden on the patients’ families and on society [1–3]. Recently, with the aging of the population in China, and improvement in living standards as well as an accelerated pace of life, the incidence of essential hypertension has increased. Moreover, the proportion of younger patients has increased significantly. Researchers are now strongly focusing on the prevention and treatment of essential hypertension.
The pathophysiology of the development of hypertension includes vascular endothelial cell injury, smooth muscle cell proliferation, and vascular remodeling [4,5]. Previous studies found that vascular endothelial cell injury contributes to the occurrence and progression of hypertension; and hypertension-induced vascular endothelial injury further promotes the development of hypertension [6]. It has been reported that hypertension-induced dysfunction of vascular endothelial cells disrupts the secretion of nitric oxide and endothelin, which induces vasoconstriction, thereby increasing circulation resistance and promoting the development of hypertension [7,8]. Additionally, in hypertension, the apoptotic rate of microvascular endothelial cells is significantly increased, resulting in a decrease in parallel blood pathways, an increase in peripheral circulation resistance, and an increase in blood pressure [9–11]. Although accumulating evidence shows that hypertension-induced vascular endothelial cell injury participates in the progression of hypertension, its underlying molecular mechanism has not yet been elucidated and requires further studies.
Long non-coding RNAs (lncRNAs) belong to a family of non-coding transcripts (above 200 nucleotides in length), which play an important role in gene expression at various levels, including pre-transcriptional, transcriptional, and post-transcriptional processes [12,13]. Reportedly, lncRNAs are involved in the cardiovascular system. For example, the expression of lncRNAs in endothelial cells can regulate blood vessel growth and functions [14]. During the progression of hypertension, abnormal vascular tone is usually found, and vascular tone is regulated by vascular smooth muscle-mediated vasoconstriction and endothelial cells. We speculate that lncRNAs may regulate vascular tone by affecting endothelial and vascular smooth muscle cellular functions.
Metastasis associated lung adenocarcinoma transcript 1 (MALAT1) is an important lncRNA. It is 6.7 kb in length and is ubiquitously expressed in human and murine cells [15]. It is also involved in tumor proliferation and apoptosis [16]. However, it is unclear whether the proliferation and apoptosis of human umbilical vein endothelial cells (HUVECs) are impacted by MALAT1. According to a previous study using HUVECs, lncRNA MALAT1 increased oxidized low-density lipoprotein (ox-LDL) induced autophagy via the phosphatidylinositol 3-kinase/(protein kinase B) (PI3K/AKT) signaling pathway [17].
Hsa-miR-124-3p is a brain-specific microRNAs (miRNAs) [18]; it has been suggested that hsa-miR-124-3p may play an important role in neuronal differentiation. The functions of nuclear receptor subfamily 3, group C, member 2 (NR3C2) in the brain are very important. Previous studies have found that hsa-miR-124-3p can bind to the 3 ‘untranslated region (UTR) of NR3C1 to regulate the expression of NR3C1 [19]. In addition, another study [20] found that hsa-miR-124-3p and hsa-miR-135a-5p were potential regulators of NR3C2 expression. We found that the binding sites of hsa-miR-124-3p and hsa-miR-135a-5p also existed on NR3C2 3′UTR by bioinformatics screening. LncRNA, miRNA, and other non-coding RNAs (ncRNAs) play an important role in hypertension, and ncRNAs have become widely recognized as diagnostic markers for hypertension [21]. Therefore, in this study we aimed to elucidate whether lncRNA MALAT1 regulates the proliferation and apoptosis of HUVECs through the hsa-miR-124-3p and hsa-miR-135a-5p/NR3C2 axis, which could provide a theoretical target for the treatment of hypertension.
Material and Methods
Study participants
A total of 230 patients with essential hypertension who were admitted to the Hangzhou Hospital of Traditional Chinese Medicine from June 2016 and February 2019 were randomly selected for this study (153 males and 77 females, age range was 40 to 85 years, mean age: 58.03±8.65 years). The diagnostic criteria for essential hypertension were as follows: sitting systolic blood pressure (SBP) ≥140 mmHg and/or diastolic blood pressure (DBP) ≥90 mmHg [22]. Patients were excluded if they had secondary hypertension, coronary heart disease, valvular heart disease, diabetes, cancer, or immune system diseases. A total of 230 non-hypertensive study participants were recruited as the control group, with age and gender matched to hypertensive patients. The control group age ranged from 39 to 82 years (mean age: 57.56±8.06 years). The SBP of the control group was <140 mmHg, and the DBP was <90 mmHg. The exclusion criteria for the control group were as follows: diabetes, heart, kidney, and other organ damage, coronary heart disease, cancer, and immune system disorders. This study was approved by the Medical Ethics Committee of the Hangzhou Hospital of Traditional Chinese Medicine. All participants signed the informed consent form.
Cell culture
HUVECs and human embryonic kidney 293 cells (HEK293) were bought from the American Type Culture Collection and cultured with RPMI-1640 medium (Gibco-BRL, USA) containing 10% fetal bovine serum, and cultured in an incubator with 5% CO2 at 37°C.
Cell transfection
MALAT1 interfering RNA (si-MALAT1) (5′-CAC AGG GAA AGC GAG TGG TTG GTA A-3′ and 5′-TTA CCA ACC ACT CGC TTT CCC TGT G-3′), and si-MALAT1 negative control (NC) (5′-GGC CUA AAG UAG CUA UTT-3′ and 5′-AUA GCU ACU UUA GGC CTT-3′) were purchased from Shanghai GenePharma Company (Shanghai, China) [23]. To study the effect of lncRNA MALAT1 on cell proliferation in HUVECs, we diluted si-MALAT1 or si-MALAT1 NC to a working concentration of 20 nM. The plasmid containing si-MALAT1 or si-MALAT1 NC was transfected by Lipofectamine 2000 (Invitrogen, USA), a blank template was transfected as a control (Blank). 48 hours after transfection, the cell cultures were harvested after culture in an incubator at 37°C with 5% CO2.
Detecting the proliferation of HUVECs using Cell Counting Kit-8 (CCK-8) assay
After the transfected HUVECs were cultured for the designated period of time, the cells were digested, isolated, and counted. The cell density was adjusted to 1 × 105/mL and then seeded into a 96-well plate at 100 μL per well (around 1×104 cells), cultured in an incubator with 5% CO2 at 37°C. The cells were collected at 0, 24, and 72 hours, and cultured for 4 hours. Cell viability was detected by Cell Counting Kit-8 (CCK-8) assay and analyzed with a Multiscan MK3 microplate reader and the absorbance at 490 nm (A490) was detected.
Luciferase reporter gene assay
Amplification of cDNA fragments of MALAT1 wild type (WT) and mutant (MUT) containing binding sites of hsa-miR-124-3p and hsa-miR-135a-5p, as well as WT and MUT of NR3C2 3′UTR were cloned into a pmirGLO vector (Promega, USA). HEK293 cells were seeded into a 48-well plate, 24-hours prior to the transfection to detect the luciferase activity. The recombinant vector pmirGLO-KLF4 (10 ng) and hsa-miR-124-3p mimics, hsa-miR-124-3p NC, hsa-miR-135a-5p mimics, and hsa-miR-135a-5p NC (50 nM) were separately transfected by Lipofectamine 2000 according to the manufacturer’s instructions. The empty vector was transfected in a separated group as a control. After 24 hours of culture, the luciferase intensity was detected by the Promega dual-luciferase assay system.
To further verify that NR3C2 is a target binding gene of hsa-miR-124-3p and hsa-miR-135a-5p, we transfected WT NR3C2, which contains binding sites of hsa-miR-124-3p, WT+hsa-miR-124-3p NC, WT+hsa-miR-124-3p mimic, hsa-miR-124-3p NC, MUT+hsa-miR-124-3p NC, and MUT+hsa-miR-124-3p mimic, as well as WT NR3C2, which contains binding sites of hsa-miR-135a-5p, WT+hsa-miR-135a-5p NC, WT+hsa-miR-135a-5p mimic, hsa-miR-135a-5p NC, MUT+hsa-miR-135a-5p NC, and MUT+hsa-miR-135a-5p mimic, respectively. The luciferase intensities were measured after 24 hours of culture by the Promega dual-luciferase assay system.
Detection of lncRNA MALAT1 and miRNAs by quantitative real-time PCR (qRT-PCR)
Total RNA was extracted from the plasma of patients. HUVECs were cultured with TRIzol LS reagent (Invitrogen, USA). Then the cDNA was produced by the M-MLV reverse transcriptase (Clontech, USA). Quantitative real-time PCR (qRT-PCR) was performed on an ABI 7500 PCR system (Applied Biosystems) using SYBRGreen MasterMix (Bio-Rad, USA). And β-actin was selected as a control to detect relative expression of lncRNA MALAT1, hsa-miR-124-3p, hsa-miR-135a-5p, hsa-miR-135b-5p, and hsa-miR-455-5p in plasma and relative expression levels of hsa-miR-124-3p and hsa-miR-135a-5p in cells. The primers information is shown in Table 1.
Table 1.
Primer sequences for qRT-PCR.
| miRNA | Primer sequence (5′ to 3′) |
|---|---|
| hsa-miR-124-3p | Forward primer: CGTGTTCACAGCGGACCTTG; Reverse primer: GGCGCCTCTCTTGGCATTC |
| hsa-miR-135a-5p | Forward primer: GGCCTCGCTGTTCTCTATGG; Reverse primer: GCCACGGCTCCAATCCCTAT |
| hsa-miR-135b-5p | Forward primer: TGGCCTATGGCTTTTCATTCCT; Reverse primer: AGCTCGCCCCTCACTGTA |
| hsa-miR-455-5p | Forward primer: GCCTTTGGACTACATCGTGGA; Reverse primer: ACATAGGCCTTGAGGCAAGT |
| lncRNA MALAT1 | Forward primer: ATGCGAGTTGTTCTCCGTCT; Reverse primer: TATCTGCGGTTTCCTCAAGC |
| β-actin | Forward primer: CATCGTCCACCGCAAATGCTTC; Reverse primer: AACCGACTGCTGTCACCTTCAC |
Detection of protein expression by western blot
In HUVECs with si-MALAT1, si-MALAT1 NC, or blank control transfection, total protein was extracted. Then a BCA protein assay kit (Thermo Fisher Scientific, Germany) was used to quantify the concentration of the extracted total protein. The lysate was separated by a 12% SDS-PAGE gel and the separated protein was then transferred to a nitrocellulose membrane. We used 40 μg of the protein sample for the electrophoresis. Western blot analysis was performed using NR3C2 antibody (Rabbit Polyclonal, catalog: 21854-1-AP, Proteintech) and horse radish peroxidase (HRP)-conjugated secondary antibody. The β-actin protein level was measured as an internal control.
Detection of HUVEC apoptosis by Annexin V FITC/PI double staining flow cytometry
After collecting the cells, the concentration of each group was calibrated to 1×106/mL and was rinsed with cold phosphate buffer saline and centrifuged. Annexin V FITC (5 μL) and PI (5 μL) were added to 500 μL cell suspension. The mixture was then incubated at 4°C for 30 minutes in the dark and analyzed using a NovoCyte™ 2060 flow cytometer (ACEA Biosciences, USA).
Statistical analyses
The statistical analyses in this study were performed using SPSS v22.0. The continuous variables in this study were showed as mean±standard deviation (SD). One-way ANOVA or t-test was performed to compare the differences between groups. The categorical variables were showed as n%, and chi-squared test was performed to analyze the differences between groups. Spearman correlation coefficient was used to evaluate the correlation between lncRNA MALAT1 and hsa-miR-124-3p, hsa-miR-135a-5p levels, as well as the levels of hsa-miR-124-3p, hsa-miR-135a-5p and NR3C2 protein. A statistical significance was considered when P<0.05.
Results
General characteristics of subjects
A total of 230 hypertensive patients and 230 healthy non-hypertensive subjects were selected for the study. No significant difference in age, gender, and body mass index (BMI) was observed between hypertensive patients and healthy subjects (P>0.05), as shown in Table 2. Among patients with hypertension, the proportion of patients with smoking, drinking, and family history of hypertension were significant increased compared to that of control study participants (P<0.05). Moreover, DBP and SBP of hypertensive patients were also significantly enhanced compared to that in the control participants (P<0.05).
Table 2.
Comparison of general characteristics between hypertensive patients and control subjects.
| Case (n=230) | Control (n 230) | p | |
|---|---|---|---|
| Age (years, mean±SD) | 58.03±8.65 | 57.56±8.06 | 0.55 |
| Gender | 0.33 | ||
| Men | 153 (66.52%) | 143 (62.17%) | |
| Women | 77 (33.48%) | 87 (37.83%) | |
| BMI (kg/m2, mean±SD) | 25.57±2.74 | 25.09±2.79 | 0.06 |
| Smoking | <0.01 | ||
| Ever | 120 (52.17%) | 73 (31.74%) | |
| Never | 110 (47.83%) | 157 (68.26%) | |
| Drinking | 0.03 | ||
| Ever | 102 (44.35%) | 79 (34.35%) | |
| Never | 128 (55.65%) | 151 (65.65%) | |
| DBP (mmHg, mean±SD) | 93.43±8.84 | 83.06±9.28 | <0.01 |
| SBP (mmHg, mean±SD) | 145.00±14.15 | 136.33±15.89 | <0.01 |
| Family history | <0.01 | ||
| Yes | 100 (43.48%) | 25 (10.87%) | |
| No | 130 (56.52%) | 205 (89.13%) |
BMI – body mass index; DBP – diastolic blood pressure; SBP – systolic blood pressure.
Inhibited expression of hsa-miR-124-3p, hsa-miR-135a-5p, and upregulation of lncRNA MALAT1 in hypertensive patients
We performed qRT-PCR to detect the plasma levels of hsa-miR-124-3p, hsa-miR-135a-5p, hsa-miR-135b-5p, hsa-miR-455-5p, and lncRNA MALAT1 in all participants. It was revealed that there was a significant decrease in the plasma level of hsa-miR-124-3p and hsa-miR-135a-5p in hypertensive patients compared to controls. While we also detected a significant increase in the level of lncRNA MALAT1 in patients with hypertension compared to controls (P<0.05). Furthermore, there were no considerable differences in the plasma levels of hsa-miR-135b-5p and hsa-miR-455-5p among hypertensive patients and healthy study participants (P>0.05; Figure 1).
Figure 1.
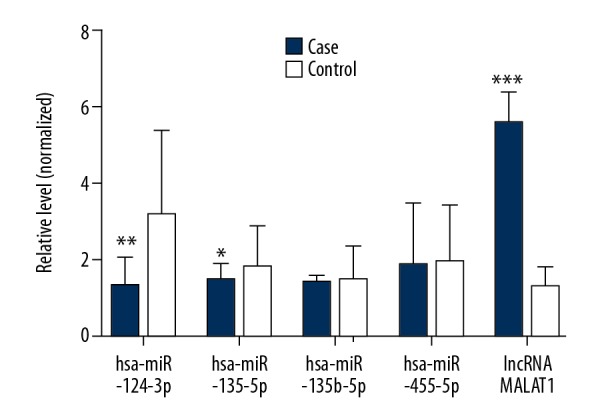
Comparison of the plasma levels of hsa-miR-124-3p, hsa-miR-135a-5p, hsa-miR-135b-5p, hsa-miR-455-5p, and lncRNA MALAT1 between hypertensive patients and controls. * P<0.05, ** P<0.01, *** P<0.001. lncRNA – long noncoding RNA; MALAT1 – metastasis associated lung adenocarcinoma transcript 1.
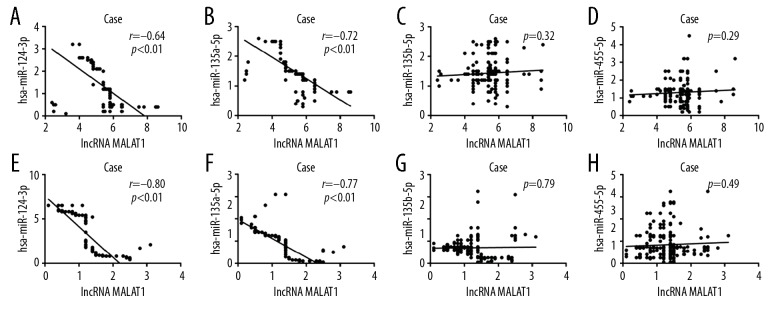
A negative correlation of lncRNA MALAT1 with hsa-miR-124-3p and hsa-miR-135a-5p
We also found that there was a negative correlation of lncRNA MALAT1 with hsa-miR-124-3p and hsa-miR-135a-5p in both hypertensive patients and healthy controls (r=−0.64, −0.72, −0.80, −0.77; P<0.01, P<0.01, P<0.01, P<0.01, respectively; Figure 2A, 2B, 2E, 2F). However, lncRNA MALAT1 levels were not significantly correlated with hsa-miR-135b-5p and hsa-miR-455-5p levels (P>0.05; Figure 2C, 2D).
Figure 2.
Correlation of lncRNA MALAT1 with the levels of miRNAs in the plasma. (A–D) Correlation observed between the levels of hsa-miR-124-3p, hsa-miR-135a-5p, hsa-miR-135b-5p, hsa-miR-455-5p, and lncRNA MALAT1 in hypertensive patients. (E–H) Correlation observed between the levels of hsa-miR-124-3p, hsa-miR-135a-5p, hsa-miR-135b-5p, hsa-miR-455-5p, and lncRNA MALAT1 in the control group. lncRNA – long noncoding RNA; MALAT1 – metastasis associated lung adenocarcinoma transcript 1; miRNAs – microRNAs.
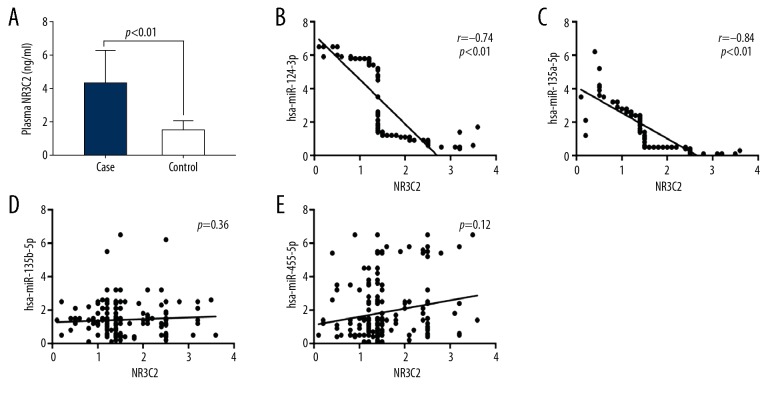
A negative correlation of NR3C2 protein with hsa-miR-124-3p and hsa-miR-135a-5p
After analyzing the plasma levels of NR3C2 protein, we revealed an upregulation of NR3C2 protein in the plasma of hypertensive patients compared to that of controls (P<0.01; Figure 3A). We further analyzed the correlation between plasma miRNA and NR3C2 protein in hypertensive patients, and found a negative correlation of NR3C2 protein with hsa-miR-124-3p and hsa-miR-135a-5p (r=−0.74, −0.84; P<0.01, P<0.01, respectively; Figure 3B, 3C). However, there was no significant correlations between hsa-miR-135b-5p, hsa-miR-455-5p levels, and NR3C2 protein expression level (P>0.05; Figure 3D, 3E).
Figure 3.
Correlation between NR3C2 protein expression levels and hsa-miR-124-3p, hsa-miR-135a-5p. (A) The plasma NR3C2 protein was significantly increased in hypertensive patients. (B) A negative correlation of the plasma NR3C2 protein expression level with hsa-miR-124-3p level in patients with hypertension. (C) A negative correlation of the plasma NR3C2 protein expression level with hsa-miR-135a-5p in hypertensive patients. (D) The plasma NR3C2 protein expression was not associated with hsa-miR-135b-5p in hypertensive patients. (E) The plasma NR3C2 protein expression level was not associated with hsa-miR-455-5p in hypertensive patients. NR3C2 – nuclear receptor subfamily 3, group C, member 2.
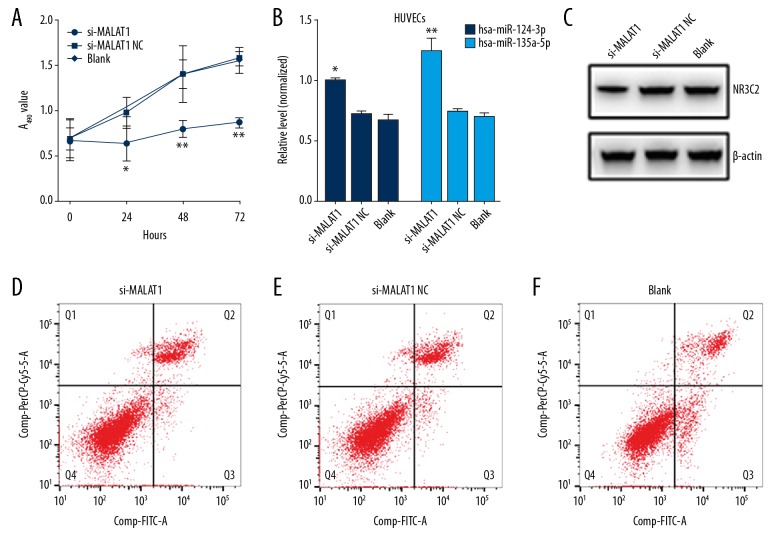
Regulation of HUVEC proliferation and apoptosis by lncRNA MALAT1 via the hsa-miR-124-3p/NR3C2 and/or hsa-miR-135a-5p/NR3C2 axis
Through cellular transfection experiments, we found that cell proliferation was significantly inhibited in HUVECs after the inhibition of MALAT1 (Figure 4A). After knocking down MALAT1, the expressions of hsa-miR-124-3p and hsa-miR-135a-5p were significantly elevated in HUVECs (Figure 4B), and the NR3C2 protein level was decreased (Figure 4C). The apoptotic rate of HUVECs after transfection of si-MALAT1 was significantly lower than that of transfected si-MALAT1 NC and blank control group (si-MALAT1: 3.64±0.21% vs. si-MALAT1 NC: 3.76±0.19%, blank control: 10.51±1.24%; Figure 4D).
Figure 4.
Results of cell transfection experiments. (A) Detecting the proliferation of cells after transfection by CCK-8. (B) Detection of hsa-miR-124-3p and hsa-miR-135a-5p levels by qRT-PCR, compared to the blank group, * P<0.05, ** P<0.01. (C) Detecting the levels of NR3C2 protein in HUVECs after transfection and confirmation by western blot. (D–F) The apoptosis rate of HUVECs after transfection of si-MALAT1 (D), si-MALAT1 NC (E) and blank control (F). CCK-8 – Cell Counting Kit-8; qRT-PCR – quantitative real-time polymerase chain reaction; HUVECs – human umbilical vein endothelial cells; MALAT1 – metastasis associated lung adenocarcinoma transcript 1; NR3C2 – nuclear receptor subfamily 3, group C, member 2.
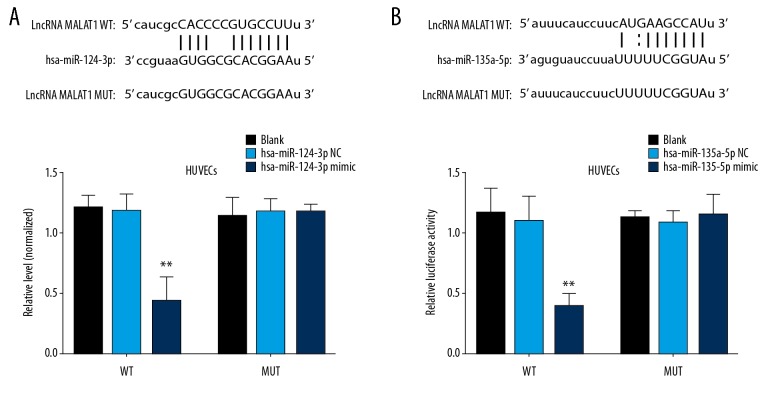
LncRNA MALAT1 was served as a molecular sponge for hsa-miR-124-3p and hsa-miR-135a-5p
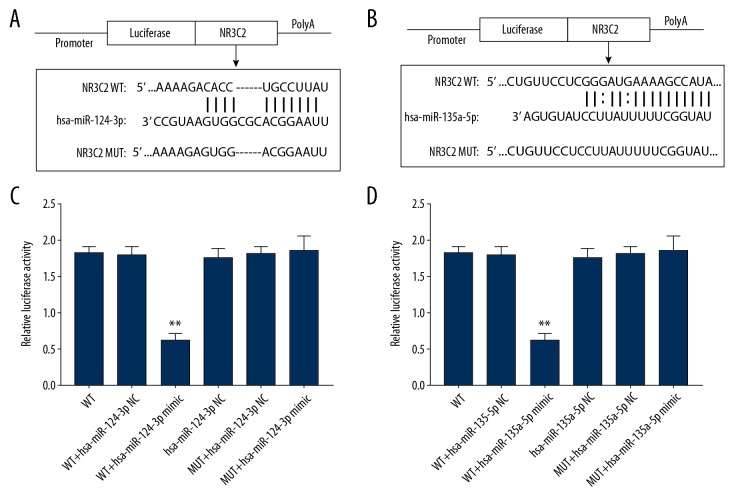
By using an online prediction tool, starbase (http://starbase.sysu.edu.cn/agoClipRNA.php?source=lncRNA), for predicting lncRNA-miRNA interaction, we found that there are binding sites of both hsa-miR-124-3p and hsa-miR-135a-5p in the lncRNA MALAT1 (Figure 5A, 5B). To further identify whether hsa-miR-124-3p and hsa-miR-135a-5p bind to the predicted sites of MALAT1, we constructed WT and MUT MALAT1 luciferase reporter vectors, respectively. After the co-transfection of the hsa-miR-124-3p binding site WT and hsa-miR-124-3p mimic or hsa-miR-135a-5p binding site WT and hsa-miR-135a-5p mimic, but not the hsa-miR-124-3p binding site MUT and the hsa-miR-135a-5p binding site MUT, the luciferase activity in HUVECs was significantly reduced (Figure 5C, 5D).
Figure 5.
Results of the dual-luciferase reporter gene assay. (A) The hsa-miR-124-3p interacted with lncRNA MALAT1 at the predicted site in HUVECs. (B) The hsa-miR-135-5p interacted with lncRNA MALAT1 at the predicted site in HUVECs. ** P<0.01, compared to the blank control group. lncRNA – long noncoding RNA; MALAT1 – metastasis associated lung adenocarcinoma transcript 1; HUVECs – human umbilical vein endothelial cells.
NR3C2 was acted as a target gene of hsa-miR-124-3p and hsa-miR-135a-5p
To further demonstrate whether hsa-miR-124-3p and hsa-miR-135a-5p could target the 3′UTR of NR3C2, we conducted a dual-luciferase reporter gene assay. A fragment of the 3′ UTR region of NR3C2 containing about±50 bp centered at the predicted target site was cloned into the downstream region of the luciferase reporter gene in a reporter vector – pmirGLO – to obtain a WT reporter vector (Figure 6A, 6B). In addition, an MT reporter vector was constructed, and the sequence of the reporter vector was confirmed by sequencing. Co-transfection of hsa-miR-124-3p mimic and a reporter vector containing the 3-UTR of NR3C2, hsa-miR-135a-5p mimic containing the 3′UTR of NR3C2 reporter vector into HEK 293T cells was performed, and the effects of hsa-miR-124-3p and hsa-miR-135a-5p on the luciferase activity of NR3C2 were detected using the dual fluorescence reporter system. We revealed that after the upregulation of hsa-miR-124-3p and hsa-miR-135a-5p and the luciferase activity of the corresponding WT reporter vector was reduced, while the MT reporter vector had no significant changes on the luciferase activity (Figure 6C, 6D).
Figure 6.
Outcomes of dual luciferase reporter gene assay. (A, C) NR3C2 is the target gene of hsa-miR-124-3p, ** P<0.01, compared to the WT. (B, D) NR3C2 is the target gene of hsa-miR-135a-5p, ** P<0.01, compared to the WT. NR3C2 – nuclear receptor subfamily 3, group C, member 2; WT – wild type.
Discussion
In the past years, China’s economy has experienced rapid growth, and along with the changes in lifestyle and diet structure, cardiovascular diseases have become the leading cause of death in China. Hypertension is a “cardiovascular syndrome” characterized by a sustained increase in arterial blood pressure and has been recognized as an essential risk factor for cardiovascular diseases and other vascular diseases such as heart failure, coronary heart disease, peripheral vascular disease, stroke, and chronic kidney disease [24,25]. Demonstrated previous study has demonstrated that even the slightest increase in blood pressure (>115/70 mmHg), can increase the risk of cardiovascular disease [26]. Thus, early prevention and treatment of hypertension are of great significance for improving the prognosis of such patients. Endothelial dysfunction is considered an early pathophysiological feature of hypertension. In hypertensive states, high pressure in the vascular cavity results in the activation of endothelial cells, release of inflammation and procoagulant factors, and adhesion of neutrophils and platelets. Endothelial cells not only have a substantial influence on vascular remodeling via abnormal proliferation and apoptosis, but also on the pathological migration of vascular endothelial cells that leads to changes in vascular structure, thereby initiating and maintaining hypertension and vascular remodeling [27]. Moreover, endothelial cell dysfunction is more closely related to organ damage and clinical diagnosis of patients with hypertension. Hence, it is critical to investigate the development of hypertensive vascular endothelial injury for the prevention as well as treatment of hypertension. To date, its underlying molecular mechanism has not been completely elucidated and requires to be further studied.
The role of lncRNA in cardiovascular diseases has become an important topic of research. The expression of lncRNA is elevated not only in many types of tumor cells but also is in endothelial cells [28–30]. Although MALAT1 is involved in diverse pathways, the ceRNA hypothesis offers a new direction for the control of gene expression [31]. Previous studies showed that miRNAs are negatively correlated with MALAT1 [30,32], and the binding of miRNAs to MALAT1 reduces the level of miRNAs, accompanied by an increased expression of miRNA targeted genes [33]. Zhu et al. [34] found that MALAT1 can inhibit H/R-stimulated HUVEC damage by targeting the miR-320a/RAC1 axis. Li et al. [35] showed that lncRNA MALAT1 contributed essentially to hypertension. The current study revealed that MALAT1 serves as a miRNAs molecular sponge. Here, a negative association of MALAT1 levels with the plasma levels of hsa-miR-124-3p and hsa-miR-135a-5p was found in hypertensive patients. Meanwhile, dual-luciferase reporter gene experiments showed that MALAT1 can also target hsa-miR-124-3p and hsa-miR-135a-5p. It was shown that MALAT1 acts as an endogenous modulator via directly targeting miR-124 [36].
In the cell transfection experiments, we found that the proliferation of HUVECs was markedly reduced, and the apoptosis level of HUVECs was significantly increased after the downregulation of MALAT1. After the knock-down of MALAT1, the levels of hsa-miR-124-3p and hsa-miR-135a-5p in HUVECs were elevated, and the expression level of NR3C2 protein was significantly decreased, suggesting that MALAT1 can downregulate the expression level of hsa-miR-124-3p and hsa-miR-135a-5p. Meanwhile, MALAT1 also upregulates the expression level of NR3C2 protein.
The nuclear receptor subfamily 3, group C, member 2 (NR3C2) gene is located on chromosome 4q31.23 and plays an important role in the progress of hypertension [20,37]. In the present study, using the Targetscan tool, we revealed that NR3C2 was a target gene of hsa-miR-124-3p and hsa-miR-135a-5p. This was also confirmed using dual-luciferase reporter assays. Moreover, it was showed that hsa-miR-124-3p and hsa-miR-135a-5p reduce NR3C2 expression and regulate the renin-angiotensin-aldosterone system [20].
However, some limitations restricted the objectives of this study. First, this study needs to be further validated in animal models. Second, we could not exclude the possibility that other miRNAs also contributed to NR3C2 expression, since their roles were blocked by endogenous miRNAs.
Conclusions
Bioinformatics screening revealed that a variety of miRNAs may serve as molecular sponges of MALAT1; therefore, we selected the 2 miRNAs, hsa-miR-124-3p and hsa-miR-135a-5p. We further demonstrated that the 3′UTR of NR3C2 was located at the binding sites of hsa-miR-124-3p and hsa-miR-135a-5p. We showed that lncRNA MALAT1 might control the proliferation and apoptosis of HUVECs via hsa-miR-124-3p/NR3C2 and/or hsa-miR-135a-5p/NR3C2 axis. However, further studies in animal models are necessary to further validate whether the regulation of lncRNA MALAT1 through the hsa-miR-124-3p/NR3C2 and/or hsa-miR-135a-5p/NR3C2 axis is relevant to the development of hypertension.
Footnotes
Conflicts of interest
None.
Source of support: This study was funded by the Natural Science Foundation of Zhejiang Province (LY18H270008)
References
- 1.Judd E, Calhoun DA. Apparent and true resistant hypertension: Definition, prevalence and outcomes. J Hum Hypertens. 2014;28(8):463–68. doi: 10.1038/jhh.2013.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ferini-Strambi L, Walters AS, Sica D. The relationship among restless legs syndrome (Willis-Ekbom Disease), hypertension, cardiovascular disease, and cerebrovascular disease. J Neurol. 2014;261(6):1051–68. doi: 10.1007/s00415-013-7065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riise HKR, Sulo G, Tell GS, et al. Association between gestational hypertension and risk of cardiovascular disease among 617 589 Norwegian women. J Am Heart Assoc. 2018;7(10) doi: 10.1161/JAHA.117.008337. pii: e008337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caillon A, Mian MOR, Fraulob-Aquino JC, et al. Gamma delta T cells mediate angiotensin II-induced hypertension and vascular injury. Circulation. 2017;135(22):2155–62. doi: 10.1161/CIRCULATIONAHA.116.027058. [DOI] [PubMed] [Google Scholar]
- 5.Pandey AK, Singhi EK, Arroyo JP, et al. Mechanisms of VEGF (vascular endothelial growth factor) inhibitor-associated hypertension and vascular disease. Hypertension. 2018;71(2):E1–8. doi: 10.1161/HYPERTENSIONAHA.117.10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang XW, Yang YC, Yang DP, et al. Tetrandrine prevents monocrotaline-induced pulmonary arterial hypertension in rats through regulation of the protein expression of inducible nitric oxide synthase and cyclic guanosine monophosphate-dependent protein kinase type 1. J Vasc Surg. 2016;64(5):1468–77. doi: 10.1016/j.jvs.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Vaisman BL, Andrews KL, Khong SML, et al. Selective endothelial overexpression of arginase ii induces endothelial dysfunction and hypertension and enhances atherosclerosis in mice. PLoS One. 2012;7(7):e39487. doi: 10.1371/journal.pone.0039487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huby AC, Otvos L, de Chantemele EJB. Leptin induces hypertension and endothelial dysfunction via aldosterone-dependent mechanisms in obese female mice. Hypertension. 2016;67(5):1020–28. doi: 10.1161/HYPERTENSIONAHA.115.06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kriegel AJ, Baker MA, Liu Y, et al. Endogenous microRNAs in human microvascular endothelial cells regulate mRNAs encoded by hypertension-related genes. Hypertension. 2015;66(4):793–99. doi: 10.1161/HYPERTENSIONAHA.115.05645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao Z, Hu J, Gao X, et al. Hyperglycemia via activation of thromboxane A2 receptor impairs the integrity and function of blood-brain barrier in microvascular endothelial cells. Oncotarget. 2017;8(18):30030–38. doi: 10.18632/oncotarget.16273. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 11.Dong Q, Xing W, Su F, et al. Tetrahydroxystilbene glycoside improves microvascular endothelial dysfunction and ameliorates obesity-associated hypertension in obese ZDF rats via inhibition of endothelial autophagy. Cell Physiol Biochem. 2017;43(1):293–307. doi: 10.1159/000480410. [DOI] [PubMed] [Google Scholar]
- 12.Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116(4):751–62. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 13.Khorkova O, Hsiao J, Wahlestedt C. Basic biology and therapeutic implications of lncRNA. Adv Drug Deliv Rev. 2015;87:15–24. doi: 10.1016/j.addr.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Michalik KM, You X, Manavski Y, et al. Long noncoding RNA MALAT1 regulates endothelial cell function and vessel growth. Circ Res. 2014;114(9):1389–97. doi: 10.1161/CIRCRESAHA.114.303265. [DOI] [PubMed] [Google Scholar]
- 15.Zhang X, Hamblin MH, Yin KJ. The long noncoding RNA MALAT1: Its physiological and pathophysiological functions. RNA Biol. 2017;14(12):1705–14. doi: 10.1080/15476286.2017.1358347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu S, Song L, Zeng S, Zhang L. MALAT1-miR-124-RBG2 axis is involved in growth and invasion of HR-HPV-positive cervical cancer cells. Tumour Biol. 2016;37(1):633–40. doi: 10.1007/s13277-015-3732-4. [DOI] [PubMed] [Google Scholar]
- 17.Li S, Pan X, Yang S, et al. LncRNA MALAT1 promotes oxidized low-density lipoprotein-induced autophagy in HUVECs by inhibiting the PI3K/AKT pathway. J Cell Biochem. 2019;120(3):4092–101. doi: 10.1002/jcb.27694. [DOI] [PubMed] [Google Scholar]
- 18.Sempere LF, Freemantle S, Pitha-Rowe I, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5(3):R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vreugdenhil E, Verissimo CS, Mariman R, et al. MicroRNA 18 and 124a downregulate the glucocorticoid receptor: Implications for glucocorticoid responsiveness in the brain. Endocrinology. 2009;150(5):2220–28. doi: 10.1210/en.2008-1335. [DOI] [PubMed] [Google Scholar]
- 20.Sober S, Laan M, Annilo T. MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochem Biophys Res Commun. 2010;391(1):727–32. doi: 10.1016/j.bbrc.2009.11.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu G, Jose PA, Zeng C. Noncoding RNAs in the regulatory network of hypertension. Hypertension. 2018;72(5):1047–59. doi: 10.1161/HYPERTENSIONAHA.118.11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reboussin DM, Allen NB, Griswold ME, et al. Systematic review for the 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA guideline for the prevention, detection, evaluation, and management of high blood pressure in adults: A report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Circulation. 2018;138(17):e595–616. doi: 10.1161/CIR.0000000000000601. [DOI] [PubMed] [Google Scholar]
- 23.Sun JY, Zhao ZW, Li WM, et al. Knockdown of MALAT1 expression inhibits HUVEC proliferation by upregulation of miR-320a and downregulation of FOXM1 expression. Oncotarget. 2017;8(37):61499–509. doi: 10.18632/oncotarget.18507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kokubo Y, Matsumoto C. Hypertension is a risk factor for several types of heart disease: Review of prospective studies. Adv Exp Med Biol. 2017;956:419–26. doi: 10.1007/5584_2016_99. [DOI] [PubMed] [Google Scholar]
- 25.Gorgui J, Gorshkov M, Khan N, Daskalopoulou SS. Hypertension as a risk factor for ischemic stroke in women. Can J Cardiol. 2014;30(7):774–82. doi: 10.1016/j.cjca.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 26.Franklin SS, Wong ND, Kannel WB. Age-specific relevance of usual blood pressure to vascular mortality. Lancet. 2003;361(9366):1389. doi: 10.1016/S0140-6736(03)13059-0. author reply 1391–92. [DOI] [PubMed] [Google Scholar]
- 27.Kumar A, Hoover JL, Simmons CA, et al. Remodeling and neointimal formation in the carotid artery of normal and P-selectin-deficient mice. Circulation. 1997;96(12):4333–42. doi: 10.1161/01.cir.96.12.4333. [DOI] [PubMed] [Google Scholar]
- 28.Yin D, Fu C, Sun D. Silence of lncRNA UCA1 represses the growth and tube formation of human microvascular endothelial cells through miR-195. Cell Physiol Biochem. 2018;49(4):1499–511. doi: 10.1159/000493454. [DOI] [PubMed] [Google Scholar]
- 29.Hirata H, Hinoda Y, Shahryari V, et al. Long noncoding RNA MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and interacts with miR-205. Cancer Res. 2015;75(7):1322–31. doi: 10.1158/0008-5472.CAN-14-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Li M, Wang Z, et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J Biol Chem. 2015;290(7):3925–35. doi: 10.1074/jbc.M114.596866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puthanveetil P, Chen S, Feng B, et al. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med. 2015;19(6):1418–25. doi: 10.1111/jcmm.12576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leucci E, Patella F, Waage J, et al. microRNA-9 targets the long non-coding RNA MALAT1 for degradation in the nucleus. Sci Rep. 2013;3:2535. doi: 10.1038/srep02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon JH, Abdelmohsen K, Srikantan S, et al. LncRNA-p21 suppresses target mRNA translation. Mol Cell. 2012;47(4):648–55. doi: 10.1016/j.molcel.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu R, Hu X, Xu W, et al. LncRNA MALAT1 inhibits hypoxia/reoxygenation-induced human umbilical vein endothelial cell injury via targeting the microRNA-320a/RAC1 axis. Biol Chem. 2019 doi: 10.1515/hsz-2019-0316. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Li D, Zhang C, Li J, et al. Long non-coding RNA MALAT1 promotes cardiac remodeling in hypertensive rats by inhibiting the transcription of MyoD. Aging. 2019;11(20):8792–809. doi: 10.18632/aging.102265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng T, Shao F, Wu Q, et al. miR-124 downregulation leads to breast cancer progression via lncRNA-MALAT1 regulation and CDK4/E2F1 signal activation. Oncotarget. 2016;7(13):16205–16. doi: 10.18632/oncotarget.7578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo JQ, Wang LY, He FZ, et al. Effect of NR3C2 genetic polymorphisms on the blood pressure response to enalapril treatment. Pharmacogenomics. 2014;15(2):201–8. doi: 10.2217/pgs.13.173. [DOI] [PubMed] [Google Scholar]