Abstract
Changes in plant architecture, such as leaf size, leaf shape, leaf angle, plant height, and floral organs, have been major factors in improving the yield of cereal crops. Moreover, changes in grain size and weight can also increase yield. Therefore, screens for additional factors affecting plant architecture and grain morphology may enable additional improvements in yield. Among the basic Helix-Loop-Helix (bHLH) transcription factors in rice (Oryza sativa), we found an enhancer-trap T-DNA insertion mutant of OsbHLH079 (termed osbhlh079-D). The osbhlh079-D mutant showed a wide leaf angle phenotype and produced long grains, similar to the phenotypes of mutants with increased brassinosteroid (BR) levels or enhanced BR signaling. Reverse transcription-quantitative PCR analysis showed that BR signaling-associated genes are largely upregulated in osbhlh079-D, but BR biosynthesis-associated genes are not upregulated, compared with its parental japonica cultivar ‘Dongjin’. Consistent with this, osbhlh079-D was hypersensitive to BR treatment. Scanning electron microscopy revealed that the expansion of cell size in the adaxial side of the lamina joint was responsible for the increase in leaf angle in osbhlh079-D. The expression of cell-elongation-associated genes encoding expansins and xyloglucan endotransglycosylases/hydrolases increased in the lamina joints of leaves in osbhlh079-D. The regulatory function of OsbHLH079 was further confirmed by analyzing 35S::OsbHLH079 overexpression and 35S::RNAi-OsbHLH079 gene silencing lines. The 35S::OsbHLH079 plants showed similar phenotypes to osbhlh079-D, and the 35S::RNAi-OsbHLH079 plants displayed opposite phenotypes to osbhlh079-D. Taking these observations together, we propose that OsbHLH079 functions as a positive regulator of BR signaling in rice.
Keywords: bHLH transcription factor, lamina joint, leaf angle, long grain, brassinosteroid signaling
1. Introduction
In cereal crops, leaf angle (defined as the angle between the leaf blade and the leaf sheath) is a key factor determining plant architecture, which also includes plant height, tiller number, and panicle morphology [1,2]. In cereal crops including rice (Oryza sativa), plant architecture has been an important agronomic trait for increasing crop yield. In particular, leaf angle is closely associated with photosynthetic capacity [3]. Plants with erect leaves capture more sunlight for photosynthesis and are amenable to much denser planting in populations with a high leaf area index for increasing total grain yield. The lamina joint, which connects the leaf blade and leaf sheath, is central in controlling leaf angle [4], as the degree of leaf inclination largely depends on cell proliferation or cell expansion as well as the cell wall composition at the lamina joint.
Brassinosteroid (BR) phytohormones affect lamina joint morphology and increase leaf angle in rice [5]. BRs are a group of steroid phytohormones that are widely distributed in plants; more than 69 types of BRs have been isolated from diverse plants [6]. BRs play pivotal roles in cell expansion, cell division, vascular bundle differentiation, male fertility, senescence, seed germination, grain filling, photomorphogenesis, flowering time, root growth, and abiotic/biotic stress responses [7,8,9,10,11,12,13]. In rice, BR functions in the regulation of grain size, leaf angle, and yield potential. For instance, several mutants with low BR contents or weak BR signaling, such as dwarf2 (d2), d11, and d61, exhibit dwarfism and produce short grains and erect leaves [14,15,16]. Additionally, many genes have a role in controlling leaf angle, such as TILLER ANGLE1 (Ta1), EBISU DWARF (D2), INCREASED LAMINA INCLINATION1 (ILI1), LEAF INCLINATION2 (LC2), INCREASED LEAF ANGLE1 (ILA1), and SLENDER GRAIN (SLG) [1,17,18,19,20,21]. Moreover, loss-of-function mutants of BR-related genes, including OsDWARF4 and OsBRI1, show improved grain yield due to their ability to be planted at a higher density and their enhanced photosynthetic rate [1,22]. Therefore, understanding the effects of BR on rice architecture has important implications for improving yield.
BR signal transduction has been intensively studied in Arabidopsis thaliana [23]. Under normal BR levels, BR interacts with BRASSINOSTEROID INSENSITIVE1 (BRI1) and BRASSINOSTEROID ASSOCIATED RECEPTOR KINASE1 (BAK1), forming a BRI–BR–BAK1 complex [24,25]. This complex inhibits the activity of BRASSINOSTEROID INSENSITIVE2 (BIN2) and activates PHOSPHATASE 2A (PP2A) for the activation of BRASSINAZOLE RESISTANT1 (BZR1). The activated BZR1 is translocated into the nucleus and regulates its downstream genes at the transcriptional level [23,26,27,28].
In rice, the BR signaling pathway remains largely unknown, since only a few components have been reported [23]. BR interacts with OsBRI1 and is involved in the formation of the OsBRI1–OsBAK1 complex [29,30], which inactivates OsBIN2 by an unknown pathway [23]. OsBIN2 phosphorylates OsBZR1, LEAF AND TILLER ANGLE INCREASED CONTROLLER (LIC), and DWARF AND LOW TILLERING (DLT) and inhibits their activities. OsBZR1 upregulates ILI1 and downregulates LIC and DLT, thus transmitting the BR signal to their downstream genes, which affect plant growth and development [23].
BR mainly affects cell elongation and cell division; moreover, cell number and cell size largely determine organ size during organogenesis [31,32]. Grain size (GS), another key trait determining yield, is mainly determined by grain length (GL), grain width (GW), and grain thickness, all of which are closely related to cell elongation or cell division. Various genes and quantitative trait loci (QTLs) in rice, such as GS3, GS5, GW2, GW5, GW8, GW6a, qGL3, THOUSAND-GRAIN WEIGHT6 (TGW6), and BIG GRAIN1 (BG1), affect grain size by regulating cell number [33,34,35,36,37,38,39,40,41]. In addition, GS2/GL2, GL7, and POSITIVE REGULATOR OF GRAIN LENGTH1 (PGL1) regulate grain size by influencing cell size in rice [40,42,43].
The basic helix–loop–helix (bHLH) domain transcription factors act in various biological processes in animals and plants [44]. In flowering plants, 162 bHLH proteins have been identified in Arabidopsis thaliana and 167 in rice [45]. These proteins are divided into two groups: typical bHLH proteins harboring both motifs (basic and HLH motif) bind to DNA through the basic region, whereas atypical, non-DNA-binding bHLH proteins lacking the basic region require other bHLH proteins to bind to DNA as protein dimers [46]. For example, rice ILI1 is an atypical bHLH protein that interacts with the typical bHLH protein OsIBH1 and represses OsIBH1 function [47]. This antagonistic regulation controls cell length in the lamina joint. Several bHLH transcription factors, such as BRASSINOSTEROID UPREGULATED1 (BU1), O. sativa BU1-LIKE1 (OsBUL1), and OsbHLH107, are involved in controlling leaf angle or grain size in rice [47,48,49,50].
In this study, we show that OsbHLH079 acts as a key regulator in determining leaf angle and grain length. OsbHLH079-overexpressing lines exhibited exaggerated leaf inclination, with longer cells on the adaxial surface of lamina joint. In addition, OsbHLH079 is involved in modulating grain shape because the OsbHLH079-overexpressing mutant produced long grains. Several molecular genetic approaches showed that the function of OsbHLH079 is closely associated with the BR signaling pathway. This study provides new insight into the roles of OsbHLH079 in determining leaf angle and grain shape.
2. Results and Discussion
2.1. OsbHLH079 Increases Leaf Angle in Rice
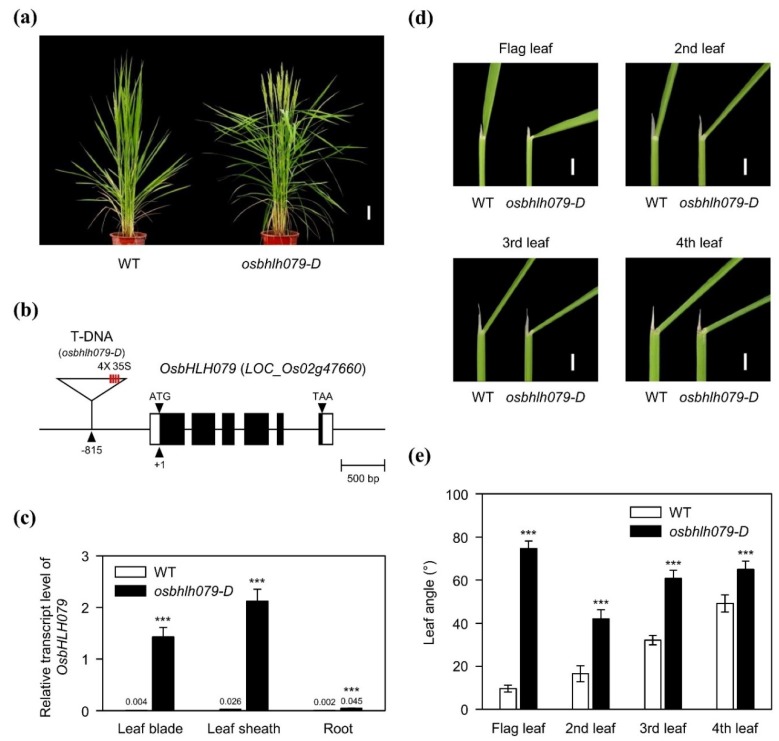
To identify new components that regulate plant architecture, we screened a collection of T-DNA insertion lines in rice in the Rice Functional Genomic Express Database [51]. We isolated a new mutant with increased leaf angle phenotype (Figure 1a), and found that an enhancer-trap line, PFG_3A-01275, which is derived from the Korean japonica rice cultivar ‘Dongjin (hereafter wild type, WT)’ harbors a T-DNA containing four tandem repeats of the CaMV 35S promoter in the promoter of OsbHLH079 (LOC_Os02g47660) (Figure 1b). To check whether the T-DNA insertion alters the expression of OsbHLH079, we compared OsbHLH079 transcript levels in various organs between WT and the enhancer-trap T-DNA insertion line. RT-qPCR analysis revealed that the transcript levels of OsbHLH079 in the T-DNA line were much higher in the leaf blade, leaf sheath, and root, compared with WT, although the degrees of overexpression varied among tissues (Figure 1c). Thus, the gain-of-function mutant was termed osbhlh079-D.
Figure 1.
Phenotypic characterization of the osbhlh079-D mutant in rice. (a) Phenotypes of wild-type (WT) and osbhlh079-D at heading stage in plants grown under natural long day (NLD) conditions in the paddy field. Scale bar = 10 cm. (b) Schematic diagram illustrating the position of the T-DNA insertion in OsbHLH079 (LOC_Os02g47660). Open boxes and filled boxes represent the untranslated region and coding sequence of OsbHLH079, respectively. (c) Comparison of the OsbHLH079 transcript levels between 3-week-old plants of WT and osbhlh079-D grown under natural sunlight in the greenhouse. The transcript level of OsbHLH079 was measured by RT-qPCR and normalized to UBQ5. Means and standard deviations were obtained from five biological replicates. (d) The leaf angle phenotypes of WT and osbhlh079-D at heading stage grown under NLD conditions in the paddy field. Scale bar = 1 cm. (e) Statistical analysis of leaf angles between WT and osbhlh079-D at heading stage grown under NLD conditions in the paddy field. Means and standard deviations were obtained from ten biological replicates. Significant differences between means were analyzed using Student’s t-test (*** p < 0.001). These experiments were repeated twice with similar results.
Next, to characterize the leaf angle phenotype of osbhlh079-D in more detail, we compared the leaf angles of the top four leaves between WT and osbhlh079-D in field-grown plants at heading stage. The leaf angles of all four top leaves in osbhlh079-D were significantly enlarged compared to those in WT, especially those of flag leaves (Figure 1d,e). These results indicate that the overexpression of OsbHLH079 is closely associated with increase in leaf angle in rice.
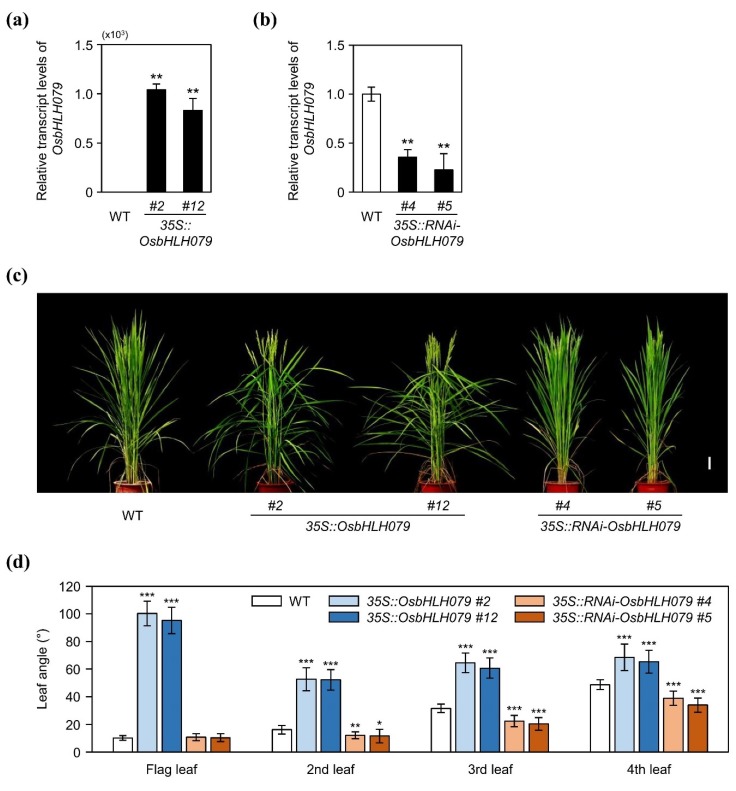
To verify if the overexpression of OsbHLH079 leads to an increase in leaf angle, we generated two independent transgenic rice lines overexpressing the full-length coding sequence of OsbHLH079 (35S::OsbHLH079 #2 and #12) as well as two individual RNAi-mediated knockdown lines of OsbHLH079 (35S::RNAi-OsbHLH079 #4 and #5). First, we checked whether the expression of OsbHLH079 is altered in the 35S::OsbHLH079 and 35S::RNAi-OsbHLH079 lines. RT-qPCR analysis revealed that the transcript levels of OsbHLH079 were upregulated in two 35S::OsbHLH079 lines (Figure 2a) and downregulated in two 35S::RNAi-OsbHLH079 lines (Figure 2b). Next, we compared the leaf angles of top four leaves among WT, 35S::OsbHLH079, and 35S::RNAi-OsbHLH079 at heading stage grown under NLD conditions in the paddy field. Indeed, all the leaf angles of 35S::OsbHLH079 were much larger than WT, especially for the flag leaf, as is the case for osbhlh079-D. By contrast, all the leaf angles of 35S::RNAi-OsbHLH079 were significantly smaller, except for the flag leaf angle (Figure 2c,d). Collectively, these results suggested that OsbHLH079 increases leaf angle during leaf blade growth.
Figure 2.
The leaf angles of WT, 35S::OsbHLH079, and 35S::RNAi-OsbHLH079. (a) Relative transcript levels of OsbHLH079 in WT, 35S::OsbHLH079 #2, and 35S::OsbHLH079 #12. (b) Relative transcript levels of OsbHLH079 in WT, 35S::RNAi-OsbHLH079 #4, and 35S::RNAi-OsbHLH079 #5. (a,b) Total RNA was extracted from the 2-cm lamina joint tissues between the leaf blade and leaf sheath of WT, 35S::OsbHLH079 #2, 35S::OsbHLH079 #12, 35S::RNAi-OsbHLH079 #4, and 35S::RNAi-OsbHLH079 #5 at heading stage in plants grown under NLD conditions in the paddy field. Relative expression levels of OsbHLH079 were determined by RT-qPCR analysis and normalized to UBQ5. Means and standard deviations were obtained from five biological replicates. Differences between means were compared using Student’s t-test (** p < 0.01). (c) Plant phenotypes of WT, 35S::OsbHLH079 #2, 35S::OsbHLH079 #12, 35S::RNAi-OsbHLH079 #4, and 35S::RNAi-OsbHLH079 #5 at heading stage in plants grown under NLD conditions in the paddy field. Scale bar = 10 cm. (d) Statistical analysis of leaf angles among WT, 35S::OsbHLH079 #2, 35S::OsbHLH079 #12, 35S::RNAi-OsbHLH079 #4, and 35S::RNAi-OsbHLH079 #5 at heading stage in plants grown under NLD conditions in the paddy field. Means and standard deviations were obtained from ten biological replicates. Significant differences between means were analyzed using Student’s t-test (* p < 0.05, ** p < 0.01, *** p < 0.001). These experiments were repeated twice with similar results.
2.2. OsbHLH079 Increases Grain Length in Rice
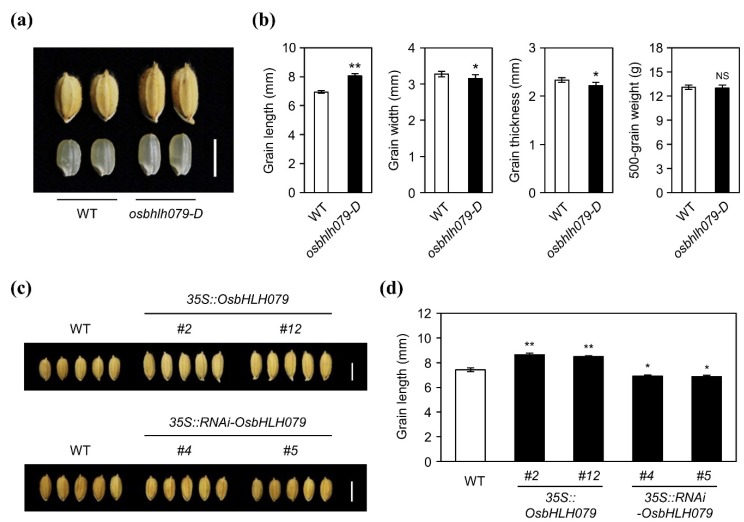
In addition to their increased leaf angle, osbhlh079-D plants produced long grains (Figure 3a). The grain length of osbhlh079-D was longer than WT, while the grain width and grain thickness of osbhlh079-D were smaller, resulting in no significant difference in 500-grain weight between WT and osbhlh079-D (Figure 3b). To confirm if the long grain phenotype of osbhlh079-D is caused by the overexpression of OsbHLH079, we compared the grain length among WT, 35S::OsbHLH079, and 35S::RNAi-OsbHLH079. The grain lengths of two independent 35S::OsbHLH079 lines were much longer than WT, whereas the grain lengths of two independent 35S::RNAi-OsbHLH079 lines were significantly shorter than WT (Figure 3c,d). Collectively, these results suggested that OsbHLH079 is also involved in the regulation of grain length in rice.
Figure 3.
The grain phenotypes of WT, osbhlh079-D, 35S::OsbHLH079, and 35S::RNAi-OsbHLH079. (a) Unhulled and hulled grain phenotypes of the osbhlh079-D mutant compared to those of WT. Scale bar = 0.5 cm. (b) Comparison of grain length, grain width, grain thickness, and 500-grain weight between WT and the osbhlh079-D mutant. Means and standard deviations were obtained from twenty biological replicates. Asterisks indicate statistically significant differences (* p < 0.05, ** p < 0.01, Student’s t-test) compared to WT. NS, not significant. (c) The unhulled grain phenotype of 35S::OsbHLH079 (upper panel), and 35S::RNAi-OsbHLH079 (lower panel) compared to that of WT. Scale bar = 0.5 cm. (d) Statistical analysis of grain lengths among the WT, 35S::OsbHLH079 #2, 35S::OsbHLH079 #12, 35S::RNAi-OsbHLH079 #4, and 35S::RNAi-OsbHLH079 #5. Means and standard deviations were obtained from twenty biological replicates. Significant differences between means were analyzed using Student’s t-test (* p < 0.05, ** p < 0.01). These experiments were repeated twice with similar results.
2.3. OsbHLH079 is a Transcription Factor of the Basic Helix-Loop-Helix (bHLH) Family in Rice
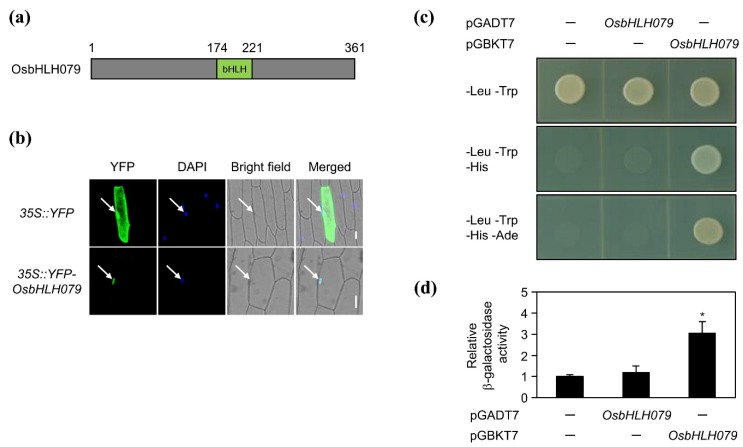
The domains of OsbHLH079 were analyzed using the NCBI-BLASTP program [52]. OsbHLH079 has a conserved basic helix–loop–helix (bHLH) domain from the 174th to 221th amino acids (Figure 4a). Moreover, the bHLH domain was found to be a putative G-box binding type, which directly binds to the G-box motif in the rice genome, in a previous genome-wide analysis [45]. These data suggested that OsbHLH079 is a bHLH-type G-box binding transcription factor. To determine if OsbHLH079 acts as a transcription factor, we first examined its subcellular localization in onion epidermal cells. The 35S::YFP (control) and 35S::YFP-OsbHLH079 constructs were introduced into the onion epidermal cells by particle bombardment, and, at 18 h after particle bombardment, onion nuclei were stained with DAPI to detect the nucleus. Confocal laser scanning microscopy showed that YFP-OsbHLH079 fusion proteins exclusively localized in the DAPI-stained nuclei, while YFP proteins were detected throughout the cells (Figure 4b). Next, we performed a transactivation activity assay for OsbHLH079 in yeast. The full-length cDNA of OsbHLH079 was fused with the yeast GAL4 activation domain in the pGADT7 vector, or with the yeast GAL4 DNA-binding domain in the pGBKT7 vector. Then, the yeast strain AH109, harboring the HIS3, ADE2, and LacZ reporter genes, was co-transformed with a pair of plasmids and plated on each selective medium, as shown in Figure 4c. Only the yeast expressing GAL4BD-OsbHLH079 grew on the selective medium lacking histidine and adenine (Figure 4c). Furthermore, in the β-galactosidase liquid assay, LacZ activity was highly upregulated in the yeast expressing GAL4BD-OsbHLH079 compared to that in the negative control (Figure 4d), indicating that OsbHLH079 has transactivation activity. Taking these observations together, it can be concluded that OsbHLH079 functions as a transcription factor of the basic helix–loop–helix (bHLH) family in rice.
Figure 4.
OsbHLH079 as a putative transcription factor. (a) Domain analysis of the 361-amino-acid-long OsbHLH079 protein. The green box indicates a basic helix–loop–helix domain. (b) Subcellular localization of OsbHLH079 in onion epidermal cells. The 35S::YFP and 35S::YFP-OsbHLH079 constructs were introduced into onion epidermal cells and the cells were analyzed by confocal laser scanning microscopy at 18 h after particle bombardment. Onion nuclei were stained with DAPI. Arrows indicate the nucleus. Scale bar = 50 μm. DAPI, 4′,6-diamidino-2-phenylindole. (c,d) Transactivation activity assay of OsbHLH079. The full-length cDNA of OsbHLH079 was fused with the yeast GAL4 activation domain in the pGADT7 vector, or with the yeast GAL4 DNA-binding domain in the pGBKT7 vector, and the fusion proteins were expressed in the yeast strain AH109. (c) Transformed yeasts were grown on the Leu– Trp–, Leu– Trp– His–, and Leu– Trp– His– Ade– agar media for yeast cell survival assay. (d) LacZ activity was obtained using the β-galactosidase liquid assay. The relative β-galactosidase activity was obtained by normalizing to the activity level of the negative control. Means and standard deviations were obtained from three biological samples. Significant differences between means were analyzed using Student’s t-test (* p < 0.05). These experiments were repeated twice with similar results. -, empty vector.
2.4. OsbHLH079 Enlarges Cell Size in the Adaxial Side of Leaf Lamina Joints by Upregulating Cell Expansion-Related Genes
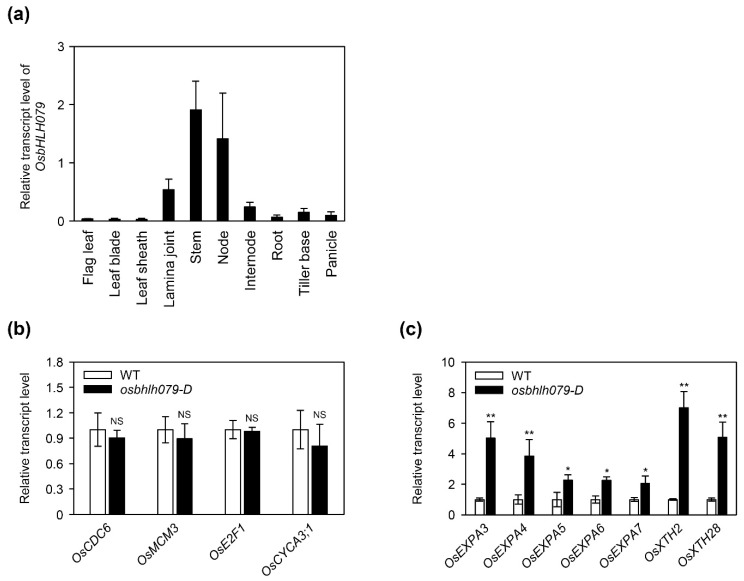
In general, the tissue-specific expression of genes is closely associated with their biological functions. Therefore, we first checked the spatial expression patterns of OsbHLH079 in field-grown (NLD conditions) WT at heading stage. This revealed that OsbHLH079 is mainly expressed in the stem, node, internode, and lamina joint (Figure 5a). Previous studies showed that several genes controlling leaf angle are highly expressed in the lamina joint, as is the case of OsbHLH079, and the degree of leaf inclination is mainly regulated by cell proliferation and/or cell expansion in the lamina joint, especially in the adaxial side of the lamina joint [5,20,21,41,53,54,55,56,57]. Therefore, we speculated that the expression levels of cell proliferation- or expansion-related genes in lamina joint would be altered in osbhlh079-D, and thus compared the transcript levels of those genes in the lamina joint between WT and osbhlh079-D by RT-qPCR analysis. The expression levels of cell proliferation-related genes, including OsCDC6, OsMCM3, OsE2F1, and OsCYCA3;1 [58], in the lamina joint were not significantly different between WT and osbhlh079-D (Figure 5b). However, the transcript levels of cell expansion-related genes, such as OsEXPAs and OsXTHs [59,60], were highly upregulated in the lamina joint of osbhlh079-D compared to WT (Figure 5c). Therefore, we hypothesized that the increased leaf angle of osbhlh079-D might be caused by expansion of cell size, mainly in the adaxial side of lamina joints.
Figure 5.
Expression of cell cycle- and cell elongation-related genes in osbhlh079-D. (a) Spatial expression patterns of OsbHLH079 in WT at the heading stage grown under NLD conditions in the paddy field. The transcript level of OsbHLH079 was determined by RT-qPCR analysis and normalized to UBQ5. Means and standard deviations were obtained from three biological replicates. (b) Expression patterns of cell cycle-related genes in the osbhlh079-D mutant compared to those in WT. (b) Altered expressions of cell elongation-related genes in the osbhlh079-D mutant compared to those in WT. (b,c) Total RNA was extracted from the 2-cm lamina joint segments between leaf blade and leaf sheath of 2-week-old WT and osbhlh079-D grown under long day (LD) conditions (14.5 h light, 30 °C/9.5 h dark, 24 °C) with 60% relative humidity in a growth chamber. The transcript level of each gene was determined by RT-qPCR analysis and normalized to that of UBQ5. Means and standard deviations were obtained from three biological replicates Significant differences between means were analyzed using Student’s t-test (* p < 0.05, ** p < 0.01). These experiments were repeated twice with similar results. NS, not significant.
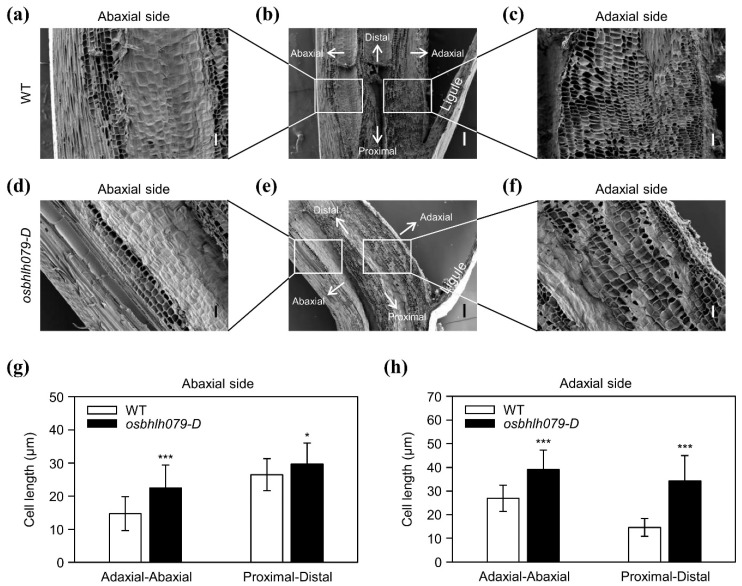
Table 079. D, we observed longitudinal sections of flag–leaf lamina joints in WT and osbhlh079-D by scanning electron microscopy. The cell length on the adaxial side in osbhlh079-D was much larger than WT along the adaxial–abaxial and proximal–distal axes; the abaxial cell size in osbhlh079-D was also slightly increased in both axes (Figure 6a–h). These results suggested that OsbHLH079 increases leaf angle by expanding the cell size on the adaxial side of the lamina joint through the upregulation of OsEXPA and OsXTH genes (Figure 5c).
Figure 6.
Scanning electron microscopy of the lamina joints of leaves in WT and osbhlh079-D. (a–f) Longitudinal sections of the lamina joint of flag leaf in WT (a–c) or osbhlh079-D (d–f) at heading stage grown under NLD conditions in the paddy field. (a,d) Close-up of abaxial regions denoted by rectangles (left side) in (b,e), respectively. (c,f) Close-up of adaxial regions denoted by rectangles (right side) in (b,e), respectively. Scale bar = 200 μm in (b,e). Scale bar = 50 μm in (a,c,d,f). (g) Statistical analysis of cell lengths in (a,d). (h) Statistical analysis of cell lengths in (c,f). (g,h) Cell lengths along the adaxial–abaxial axis and proximal–distal axis were measured on the abaxial and adaxial sides of lamina joints. Means and standard deviations were obtained from thirty cells. Significant differences between means were analyzed using Student’s t-test (* p < 0.05, *** p < 0.001). These experiments were repeated twice with similar results.
2.5. OsbHLH079 Regulates the Expression of BR Signaling-Related Genes
The wide leaf angle phenotype of osbhlh079-D resembles that of mutants with elevated BR accumulation or enhanced BR signaling [11,14,15,61,62,63]. Moreover, the transcript levels of several XTHs and expansin genes, which are upregulated in osbhlh079-D (Figure 5c), are significantly increased by BR treatment in Arabidopsis thaliana, rice, soybean (Glycine max), maize (Zea mays), and wheat (Triticum aestivum) [59,64,65,66,67,68,69,70]. Therefore, we speculated that the increased leaf angle of osbhlh079-D is caused by either elevated endogenous BR accumulation or enhanced BR signaling.
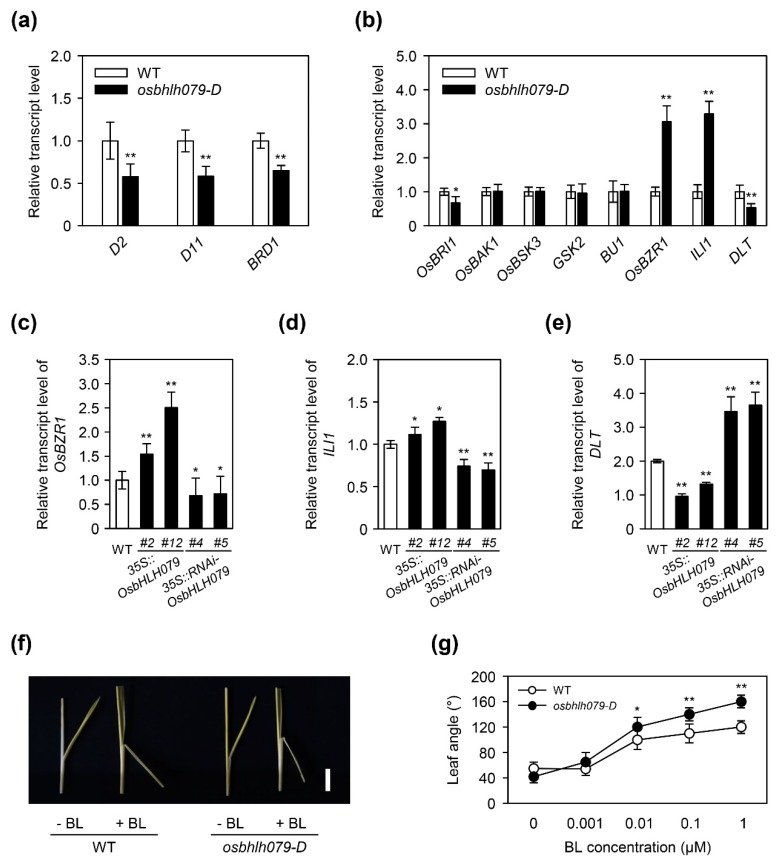
To investigate whether the expression of BR biosynthesis- or BR signaling-related genes is altered in osbhlh079-D, we compared their transcript levels in the lamina joints of leaf blades between WT and osbhlh079-D. In the lamina joints, the expression of BR biosynthesis-related genes, such as D2, D11, and BRD1 [15,16,61], was significantly downregulated in osbhlh079-D compared to that of WT (Figure 7a). In addition, the transcript level of OsBRI1, the BR receptor, was also significantly downregulated compared to that of WT (Figure 7b), indicating a negative feedback regulation by enhanced BR signaling [14,63,71]. Among the BR signaling-related genes, including OsBAK1, OsBSK3, GSK2, BU1, OsBZR1, ILI1, and DLT [14,30,47,48,72,73,74,75], the expression of OsBZR1, and its downstream genes, such as ILI1, and DLT, was significantly altered in the lamina joint of osbhlh079-D compared to WT (Figure 7b). For example, the expression of genes encoding positive regulators of the BR signaling pathway, such as OsBZR1, and ILI1, was highly upregulated, but the transcript level of DLT, which also encodes a positive regulator of BR signaling pathway but is repressed directly by OsBZR1, was significantly downregulated in osbhlh079-D (Figure 7b). To confirm whether the expression levels of OsBZR1, ILI1, and DLT are altered by the ectopic or knockdown expression of OsbHLH079, we compared the expression levels of OsBZR1, ILI1, and DLT in the lamina joint among WT, 35S::OsbHLH079, and 35S::RNAi-OsbHLH079. The transcript levels of OsBZR1, and ILI1 were highly upregulated, while DLT expression was significantly downregulated in the lamina joint of 35S::OsbHLH079 lines, as in osbhlh079-D (Figure 7c–e). By contrast, the expression levels of OsBZR1, and ILI1 were significantly decreased, while the transcript level of DLT was highly increased in the lamina joint of 35S::RNAi-OsbHLH079 lines (Figure 7c–e). These results indicated that the increased activity of OsbHLH079 enhances the BR signaling pathway by altering the expression of OsBZR1 and its downstream genes, such as ILI1, and DLT.
Figure 7.
OsbHLH079 acts as a positive regulator of the brassinosteroid signaling pathway. (a) Expression patterns of brassinosteroid (BR) biosynthesis-related genes in the osbhlh079-D mutant compared to those in WT. (b) Altered expressions of BR signaling-related genes in the osbhlh079-D mutant compared to those in WT. (c–e) Altered expressions of OsBZR1 (c), ILI1 (D), and DLT (E) in 35S::OsbHLH079 and 35S::RNAi-OsbHLH079 compared to those in WT. (a–e) Total RNA was extracted from the 2-cm lamina joints between leaf blade and leaf sheath of 4-week-old plants of WT, osbhlh079-D, 35S::OsbHLH079, and 35S::RNAi-OsbHLH079 grown under LD conditions (14.5 h light, 30 °C/9.5 h dark, 24 °C) with 60% relative humidity in a growth chamber. The transcript level of each gene was determined by RT-qPCR analysis and normalized to UBQ5. Means and standard deviations were obtained from three biological replicates. Asterisks indicate statistically significant differences (* p < 0.05, ** p < 0.01, Student’s t-test) compared to WT. (f) BR-induced lamina joint inclination in WT and the osbhlh079-D mutant. The 2-cm lamina joint segments of 10-day-old seedlings of WT and osbhlh079-D grown at 30 °C in darkness were treated with 1 µM BL for 48 h in darkness. Scale bar = 0.5 cm. BL, 24-epibrassinolide. (g) Dose-dependent responses of the lamina joint of WT and osbhlh079-D to various concentrations of BL. Means and standard deviations were obtained from more than ten biological replicates. Significant differences between means were analyzed using Student’s t-test (* p < 0.05, ** p < 0.01). These experiments were repeated twice with similar results. BL, 24-epibrassinolide.
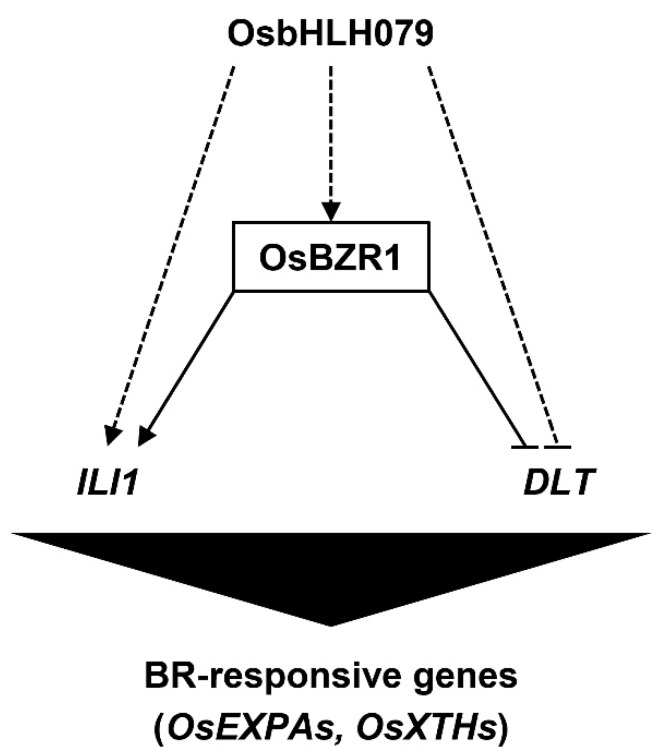
To verify whether the response to BR treatment is enhanced by the overexpression of OsbHLH079, we carried out a BR-induced lamina joint inclination assay. For this assay, 2-cm lamina joint segments were detached from 10-day-old seedlings of WT and osbhlh079-D grown in darkness and treated with 1 µM BL for 48 h in darkness. Then, we compared the extent of lamina inclination of osbhlh079-D with WT. As shown in Figure 7f, osbhlh079-D was more sensitive to BR (24-epibrassinolide) treatment. Moreover, the difference in the extents of lamina inclination between WT and osbhlh079-D increased as the BR concentration increased (Figure 7b). These data indicated that BR signaling is enhanced in osbhlh079-D. Therefore, we concluded that OsbHLH079 enhances the BR signaling pathway, which leads to the expansion of cell size in the adaxial side of lamina joints via upregulation of OsEXPAs and OsXTHs, resulting in an increase in leaf angle in rice (Figure 8).
Figure 8.
A proposed model of the OsbHLH079-mediated regulatory network in the BR signaling pathway. OsbHLH079 enhances brassinosteroid signaling by upregulating genes encoding positive regulators of the BR signaling pathway, such as OsBZR1, and ILI1, and downregulating DLT, which also encodes a positive regulator of BR signaling and downregulated directly by OsBZR1. Then, altered expressions of BR-responsive genes such as OsEXPA, and OsXTH produce changes in leaf angle and grain length. Arrows and bars indicate positive and negative regulation, respectively. Solid and dashed lines indicate direct regulation and possible feed-forward regulation, respectively.
2.6. OsbHLH079 Might Indirectly Regulate Expressions of OsBZR1, and ILI1
As OsBZR1 directly regulates the expression of ILI1, and DLT [23], it is possible that (1) OsbHLH079 modulates the expression and/or activity of OsBZR1, (2) OsbHLH079 directly regulates other genes because OsBZR1 has a limited function, in which the effect of overexpressed OsBZR1 occurs only when the binding site of negative regulators, such as 14-3-3 or GSK2, is mutated in plants [23,63,76], or (3) OsbHLH079 directly regulates downstream genes of OsBZR1, such as ILI1, and DLT, and expression of OsBZR1 is increased by a feedback regulatory loop.
To examine the possible roles of OsbHLH079 in the transcription of OsBZR1, ILI1, and DLT in rice, we investigated the promoter sequences of OsBZR1, ILI1, and DLT (−2000 to −1 from the ATG). It revealed that OsBZR1 or ILI1 did not contain the G-box sequence (CACGTG; a putative binding site of bHLH-type transcription factors), but only one CACGTG sequence in the promoter region (-989 to −984 from ATG) of DLT. These findings suggest that OsbHLH079 regulates OsBZR1 and ILI1 indirectly, although we cannot exclude the possibility that OsbHLH079 binds to the promoter regions of OsBZR1 and ILI1.
2.7. OsbHLH079 Might Directly Regulate the Cell Expansion-Associated Genes
PHYTOCHROME INTERACTING FACTOR LIKE1 (OsPIL1) functions as a key regulator of internode elongation [77]. OsPIL1-overexpressing rice plants (Ubi::OsPIL1) formed elongated internodes via larger cells through direct regulation of its downstream genes, such as OsEXPA4 and 1-ACC OXIDASE, via binding to the G-box element. Both OsPIL1 and OsbHLH079 are bHLH-type transcription factors. Thus, it can be speculated that OsbHLH079 directly binds to the promoter regions of cell expansion-related genes, such as OsEXPAs and OsXTHs. The expression of cell expansion-related genes was increased to a much greater extent in the osbhlh079-D mutant compared to the increased expression of BR signaling-associated genes (OsBZR1, and ILI1) (Figure 5c, Figure 7b). The increased expression of BR-related genes in osbhlh079-D and OsbHLH079-overexpressing plants may have caused a significant change in the expression of the downstream genes, but we could not exclude the possibility that OsbHLH079 directly regulates the cell expansion-associated genes.
2.8. OsbHLH079 Increases Grain Length by Altering TGW6 Expression
In rice, several genes and QTLs, including GS3, GS5, GW2, GW5, GW8, TGW6, GW6a, qGL3, and BG1, affect grain size by regulating cell number, and GS2/GL2, GL7, and PGL1 affect grain size by influencing cell size [33,34,35,36,37,38,39,40,41,42,43]. GW2 is a RING-type E3 ubiquitin ligase and acts as a negative regulator of cell division [33]. GW2-overexpressing transgenic rice showed a reduced grain-width phenotype. Increased expression of GW2 could be one of the reasons for the slender-grain phenotype of the osbhlh079-D mutant (Figure S1). TGW6 encodes an IAA-glucose hydrolase, and loss of function of TGW6 results in enhanced grain weight, resulting in increased grain yield [38]. RNAi-TGW6 rice plants showed an increased-grain-length phenotype, which could be one of the reasons for the long-grain phenotype of the osbhlh079-D mutant (Figure S1). It is possible that the regulatory function of OsbHLH079 is associated with not only the BR signaling pathway, but also the auxin-related pathway, which is closely involved in the control of grain size and grain yield [38].
2.9. OsbHLH079 is an Ortholog of Arabidopsis CRYPTOCHROME-INTERACTING bHLH 1
In the genome-wide analysis, 167 bHLH genes in rice and 162 bHLH genes in Arabidopsis were identified and analyzed by amino acid sequence-based alignments. As a result, they were divided into 25 subfamilies [45]. OsbHLH079 belongs to the C group with 24 genes in rice and 23 genes in Arabidopsis. The C group contains all the Arabidopsis CRYPTOCHROME-INTERACTING bHLH (AtCIB) genes (AtCIB1, AtCIB2, AtCIB3, AtCIB4, and AtCIB5), and OsbHLH079 was annotated to be an AtCIB1-like gene [45]. In Arabidopsis, CIB1 interacts with cryptochrome 2 (CRY2), and this complex affects various developmental processes, such as hypocotyl elongation, flowering time, stomata opening, hypocotyl bending, programmed cell death, plastid development, and silique elongation [35,78,79,80,81,82,83,84,85]. In soybean, GmCIB1 binds to the E-box (CANNTG) motif and acts as a transcriptional activator to regulate leaf senescence-associated genes [86]. In rice, OsbHLH079 might be an ortholog of CIB1, and the bHLH domain was a highly conserved among amino acid sequences of AtCIB1, GmCIB1, and OsbHLH079 (Figure S2). Additionally, the osbhlh079-D mutant flowered earlier than WT under long-day and short-day conditions (Figure S3), which provides more insight into additional regulatory functions of OsbHLH079 in rice growth and development, similar to AtCIB1.
3. Materials and Methods
3.1. Plant Materials and Growth Conditions
The enhancer-trap T-DNA insertion mutant of OsbHLH079 (LOC_Os02g47660; PFG_3A-01275; designated as osbhlh079-D) in rice was isolated from the Korean japonica cultivar ‘Dongjin’, and obtained from the Rice Functional Genomic Express Database [51]. For phenotypic characterization, rice plants were grown under natural long day (NLD) conditions in the paddy field (37°N latitude, Suwon, Republic of Korea). Rice was also grown in growth chambers under long-day (14.5 h light, 30 °C/9.5 h dark, 24 °C) conditions or short-day (10 h light, 30 °C/14 h dark, 24 °C) conditions with 60% relative humidity. The light sources used in the artificial growth chamber were light-emitting diodes (LEDs), and the average photon flux density was around 500 µmol m−2 s−1.
3.2. Vector Construction and Rice Transformation
To generate the 35S::OsbHLH079, and 35S::RNAi-OsbHLH079 transgenic rice plants, the full-length cDNA of OsbHLH079, and the partial cDNA fragment of OsbHLH079 were amplified from the first-strand cDNA obtained from leaves of WT by reverse-transcription polymerase chain reaction (RT-PCR) using gene-specific primers (Table S1), and subcloned into pCR8/GW/TOPO (Invitrogen, USA). After confirming the sequences, the full-length cDNA of OsbHLH079, and the partial cDNA fragment of OsbHLH079 were transferred into the pMDC32 Gateway binary vector [87], and the pANDA vector [88], respectively, by LR reaction using Gateway LR Clonase II Enzyme Mix (Invitrogen, USA). The resulting constructs, 35S::OsbHLH079, and 35S::RNAi-OsbHLH079, were transformed into Agrobacterium tumefaciens strain LBA4404, and then introduced into calli generated from mature embryos of WT through Agrobacterium-mediated transformation, respectively [89].
3.3. RNA Extraction and Reverse Transcription-Quantitative PCR (RT-qPCR) Analysis
Total RNA was extracted from 2-cm lamina joint segments or other tissues using the MG Total RNA Extraction Kit (Macrogen, Seoul, Republic of Korea) according to the manufacturer’s instructions. First-strand cDNAs were synthesized from 2 µg of total RNA using oligo(dT)15 primers and M-MLV reverse transcriptase (Promega, USA). The relative transcript levels of each gene were measured by quantitative PCR (qPCR) using gene-specific primers, and rice Ubiquitin5 (UBQ5) was used as an internal control (Table S1) [90]. GoTaq qPCR Master Mix (Promega, USA) was used in a 20 µl total reaction volume, and quantitative PCR was performed using a LightCycler 480 (Roche, Switzerland). qPCR conditions were 95 °C for 2 min, and then 45 cycles of 95 °C for 10 s and 60 °C for 1 min.
3.4. Subcellular Localization of OsbHLH079
To investigate the subcellular localization of OsbHLH079, the 35S::YFP-OsbHLH079 construct was prepared. The full-length coding sequence of OsbHLH079 was amplified with gene-specific primers (Table S1), and fused with YFP in the pEarleyGate 104 (pEG104) vector through LR reactions using Gateway LR Clonase II Enzyme Mix (Invitrogen, USA). The resultant construct, 35S::YFP-OsbHLH079, and the 35S::YFP construct were introduced into onion epidermal cells using a DNA particle delivery system (Biolistic PDS-1000/He; Bio-Rad, Hercules, CA, USA), respectively. The transformed onion epidermal cells were incubated on Murashige and Skoog phytoagar medium (pH 5.7) under dark conditions at 25 °C for 18 h, and then onion nuclei were stained with 300 nM 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen, USA) in phosphate-buffered saline for 5 min. YFP and DAPI fluorescence were observed using a confocal laser scanning microscope (SP8X, Leica, Germany) with excitation wavelengths of 458 and 405 nm and emission wavelengths of 514 and 488 nm for YFP and DAPI, respectively.
3.5. Transactivation Activity Assay
Transactivation activity assay was performed as previously described with some modifications [91]. The full-length coding sequence of OsbHLH079 was amplified by PCR and fused with the yeast GAL4 activation domain in the pGADT7 vector (Biosciences Clontech, Palo Alto, CA, USA), or with the yeast GAL4 DNA binding domain in the pGBKT7 vector (Biosciences Clontech, Palo Alto, CA, USA), respectively. Then, the yeast strain AH109 was co-transformed with a pair of plasmids, and plated on each medium, as shown in Figure 4c. The yeast β-galactosidase liquid assay was carried out according to the Yeast Protocols Handbook (Clontech) using chlorophenol red-β-D-galactopyranoside (CPRG, Roche Biochemical) as the substrate.
3.6. Scanning Electron Microscopy
Scanning electron microscopy was conducted as previously described with some modifications [92]. The lamina joints of flag leaves of WT and osbhlh079-D at heading stage grown under NLD conditions in the paddy field were excised and sectioned longitudinally as previously described [56]. The samples were fixed with modified Karnovsky’s fixative (2% paraformaldehyde, 2% glutaraldehyde, and 50 mM sodium cacodylate buffer, pH 7.2) at 4 °C for 24 h, and washed with 50 mM sodium cacodylate buffer (pH 7.2) three times at 4 °C for 10 min each. Next, the samples were post-fixed at 4 °C for 2 h with 1% osmium tetroxide in 50 mM sodium cacodylate buffer (pH 7.2), and washed twice with distilled water at room temperature, followed by dehydration with a gradient series of ethanol. After dehydration, the samples were processed as follows: dried in liquid CO2 using a critical point dryer (EM CPD300, Leica, Germany), and coated with platinum using a sputter coater (EM ACE200, Leica, Austria). The processed samples were observed by scanning electron microscope (AURIGA, Carl Zeiss, Germany).
3.7. BR-Induced Lamina Joint Inclination Assay
The BR-induced lamina joint inclination assay was performed as previously described with some modifications [93]. Sterilized seeds of WT and the osbhlh079-D mutant were grown on Murashige and Skoog (MS) medium in an artificial growth chamber at 30 °C under dark conditions for 10 days. Then, the 2-cm lamina joint segments of WT and osbhlh079-D were excised, and incubated on distilled water containing various concentrations of 24-epibrassinolide (BL), an active form of brassinosteroid (Sigma), at 30 °C in darkness for 48 h. The angle between lamina and sheath was measured using ImageJ software [94].
3.8. Gene Information
Sequence data from this article can be found in the National Center for Biotechnology Information (NCBI): OsbHLH079, Os02g0705500; UBQ5, Os01g0328400; OsCDC6, Os01g0856000; OsMCM3, Os05g0476200; OsE2F1, Os02g0537500; OsCYCA3;1, Os03g0607600; OsEXPA3, Os05g0276500; OsEXPA4, Os05g0477600; OsEXPA5, Os02g0744200; OsEXPA6, Os03g0336400; OsEXPA7, Os03g0822000; OsXTH2, Os11g0539200; OsXTH28, Os03g0239000; D2, Os01g0197100; D11, Os04g0469800; BRD1, Os03g0602300; OsBRI1, Os01g0718300; OsBAK1, Os08g0174700; OsBSK3, Os04g0684200; GSK2, Os05g0207500; BU1, Os06g0226500; OsBZR1, Os07g0580500; ILI1, Os04g0641700; DLT, Os06g0127800; GS2, Os02g0701300; GS3, Os03g0407400; GS5, Os05g0158500; GS6, Os06g0127800; GW2, Os02g0244100; GW6a, Os06g0650300; GLW7, Os07g0505200; GW8, Os08g0531600; TGW6, Os06g0623700; GL7, Os07g0603300; PGL1, Os03g0171300; PGL2, Os02g0747900; qGL3, Os03g0646900; RGA1, Os05g0333200; RGB1, Os03g0669200; SRS5, Os11g0247300; TH1, Os02g0811000; BG1, Os03g0175800; DEP1, Os09g0441900; OsMAPK6, Os06g0154500.
4. Conclusions
We found that OsbHLH079 increases leaf angle and grain length in rice. Rice overexpressing OsbHLH079 (osbhlh079-D and 35S::OsbHLH079) showed wider leaf angle and longer grain length, and RNA-mediated knockdown lines of OsbHLH079 (35S::RNAi-OsbHLH079) exhibited narrower leaf angle and shorter grain length compared to those of WT. Our data also revealed that OsbHLH079 enhances BR signaling by modulating the expression levels of BR signaling-related genes (OsBZR1, ILI1, and DLT) which lead to increases in leaf angle and grain length. This study opens up the possibility to improve grain yield per unit area in rice by controlling plant architecture and seed shape.
Abbreviations
| bHLH | basic Helix-Loop-Helix |
| BL | 24-epibrassinolide |
| BR | Brassinosteroid |
| DAPI | 4′,6-diamidino-2-phenylindole |
| GL | Grain length |
| GS | Grain size |
| GW | Grain width |
| LD | Long day |
| NLD | Natural long day |
| QTL | Quantitative trait loci |
| SD | Short day |
| WT | Wild-type |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/6/2090/s1.
Author Contributions
B.-D.L. and N.-C.P. designed and supervised the research. H.S., S.-H.K., and J.-H.L. performed the experiments and analyzed data. S.-H.K. and S.-J.L. performed the revision experiments. G.A. developed osbhlh079-D mutant and provided critical advice. H.S., S.-H.K., B.-D.L., and N.-C.P. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by grants from the Next-Generation BioGreen21 Program (PJ013656 to B.-D.L.), Rural Development Administration, and the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (NRF-2017R1A2B3003310 to N.-C.P.), Republic of Korea.
Conflicts of Interest
The authors have no potential conflicts of interest.
References
- 1.Sakamoto T., Morinaka Y., Ohnishi T., Sunohara H., Fujioka S., Ueguchi-Tanaka M., Mizutani M., Sakata K., Takatsuto S., Yoshida S., et al. Erect leaves caused by brassinosteroid deficiency increase biomass production and grain yield in rice. Nat. Biotechnol. 2006;24:105–109. doi: 10.1038/nbt1173. [DOI] [PubMed] [Google Scholar]
- 2.Yoon J., Cho L.H., Lee S., Pasriga R., Tun W., Yang J., Yoon H., Jeong H.J., Jeon J.S., An G. Chromatin interacting factor OsVIL2 is required for outgrowth of axillary buds in rice. Mol. Cells. 2019;42:858–868. doi: 10.14348/molcells.2019.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinclair T.R., Sheehy J.E. Erect leaves and photosynthesis in rice. Science. 1999;283:1455. doi: 10.1126/science.283.5407.1455c. [DOI] [Google Scholar]
- 4.Nakamura A., Fujioka S., Takatsuto S., Tsujimoto M., Kitano H., Yoshida S., Asami T., Nakano T. Involvement of C-22-hydroxylated brassinosteroids in auxin-induced lamina joint bending in rice. Plant Cell Physiol. 2009;50:1627–1635. doi: 10.1093/pcp/pcp106. [DOI] [PubMed] [Google Scholar]
- 5.Wada K., Marumo S., Ikekawa N., Morisaki M., Mori K. Brassinolide and homobrassinolide promotion of lamina inclination of rice seedling. Plant Cell Physiol. 1981;22:323–325. [Google Scholar]
- 6.Bajguz A. Brassinosteroids—Occurrence and chemical structures in plants. In: Hayat S., Ahmad A., editors. Brassinosteroids: A Class of Plant Hormone. Springer; Dordrecht, The Netherlands: 2011. [Google Scholar]
- 7.Szekeres M., Nemeth K., Koncz-Kalman Z., Mathur J., Kauschmann A., Altmann T., Redei G.P., Nagy F., Schell J., Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/S0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 8.Clouse S.D., Sasse J.M. Brassinosteroids: Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 9.Fujioka S., Yokota T. Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 2003;54:137–164. doi: 10.1146/annurev.arplant.54.031902.134921. [DOI] [PubMed] [Google Scholar]
- 10.Nakashita H., Yasuda M., Nitta T., Asami T., Fujioka S., Arai Y., Sekimata K., Takatsuto S., Yamaguchi I., Yoshida S. Brassinosteroid functions in a broad range of disease resistance in tobacco and rice. Plant J. 2003;33:887–898. doi: 10.1046/j.1365-313X.2003.01675.x. [DOI] [PubMed] [Google Scholar]
- 11.Wu C.Y., Trieu A., Radhakrishnan P., Kwok S.F., Harris S., Zhang K., Wang J., Wan J., Zhai H., Takatsuto S., et al. Brassinosteroids regulate grain filling in rice. Plant Cell. 2008;20:2130–2145. doi: 10.1105/tpc.107.055087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gudesblat G.E., Russinova E. Plants grow on brassinosteroids. Curr. Opin. Plant Biol. 2011;14:530–537. doi: 10.1016/j.pbi.2011.05.004. [DOI] [PubMed] [Google Scholar]
- 13.Kim S., Moon J., Roh J., Kim S.K. Castasterone can be biosynthesized from 28-homodolichosterone in Arabidopsis thaliana. J. Plant Biol. 2018;61:330–335. doi: 10.1007/s12374-018-0194-4. [DOI] [Google Scholar]
- 14.Yamamuro C., Ihara Y., Wu X., Noguchi T., Fujioka S., Takatsuto S., Ashikari M., Kitano H., Matsuoka M. Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell. 2000;12:1591–1606. doi: 10.1105/tpc.12.9.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong Z., Ueguchi-Tanaka M., Umemura K., Uozu S., Fujioka S., Takatsuto S., Yoshida S., Ashikari M., Kitano H., Matsuoka M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell. 2003;15:2900–2910. doi: 10.1105/tpc.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanabe S., Ashikari M., Fujioka S., Takatsuto S., Yoshida S., Yano M., Yoshimura A., Kitano H., Matsuoka M., Fujisawa Y., et al. A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell. 2005;17:776–790. doi: 10.1105/tpc.104.024950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z.K., Paterson A.H., Pinson S.R.M., Khush G.S. A major gene, Ta1 and QTLs affecting tiller and leaf anlgles in rice. Rice Genet. Newsl. 1998;15:154–156. [Google Scholar]
- 18.Li Z., Paterson A.H., Pinson S.R.M., Stansel J.W. RFLP facilitated analysis of tiller and leaf angles in rice. Euphytica. 1999;109:79–84. doi: 10.1023/A:1003533001014. [DOI] [Google Scholar]
- 19.Zhao S.Q., Hu J., Guo L.B., Qian Q., Xue H.W. Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 2010;20:935–947. doi: 10.1038/cr.2010.109. [DOI] [PubMed] [Google Scholar]
- 20.Ning J., Zhang B., Wang N., Zhou Y., Xiong L. Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice. Plant Cell. 2011;23:4334–4347. doi: 10.1105/tpc.111.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feng Z., Wu C., Wang C., Roh J., Zhang L., Chen J., Zhang S., Zhang H., Yang C., Hu J., et al. SLG controls grain size and leaf angle by modulating brassinosteroid homeostasis in rice. J. Exp. Bot. 2016;67:4241–4253. doi: 10.1093/jxb/erw204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morinaka Y., Sakamoto T., Inukai Y., Agetsuma M., Kitano H., Ashikari M., Matsuoka M. Morphological alteration caused by brassinosteroid insensitivity increases the biomass and grain production of rice. Plant Physiol. 2006;141:924–931. doi: 10.1104/pp.106.077081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C., Bai M.Y., Chong K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014;33:683–696. doi: 10.1007/s00299-014-1578-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li J., Nam K.H. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 25.Kinoshita T., Cano-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 26.He J.X., Gendron J.M., Sun Y., Gampala S.S., Gendron N., Sun C.Q., Wang Z.Y. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yin Y., Vafeados D., Tao Y., Yoshida S., Asami T., Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 28.Sun Y., Fan X.Y., Cao D.M., Tang W., He K., Zhu J.Y., He J.X., Bai M.Y., Zhu S., Oh E., et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell. 2010;19:765–777. doi: 10.1016/j.devcel.2010.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Xu Y.Y., Li J., Powell R.A., Xu Z.H., Chong K. Transgenic rice plants ectopically expressing AtBAK1 are semi-dwarfed and hypersensitive to 24-epibrassinolide. J. Plant Physiol. 2007;164:655–664. doi: 10.1016/j.jplph.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Li D., Wang L., Wang M., Xu Y.Y., Luo W., Liu Y.J., Xu Z.H., Li J., Chong K. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield. Plant Biotechnol. J. 2009;7:791–806. doi: 10.1111/j.1467-7652.2009.00444.x. [DOI] [PubMed] [Google Scholar]
- 31.Potter C.J., Xu T. Mechanisms of size control. Curr. Opin. Genet. Dev. 2001;11:279–286. doi: 10.1016/S0959-437X(00)00191-X. [DOI] [PubMed] [Google Scholar]
- 32.Sugimoto-Shirasu K., Roberts K. “Big it up”: Endoreduplication and cell-size control in plants. Curr. Opin. Plant. Biol. 2003;6:544–553. doi: 10.1016/j.pbi.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 33.Song X.J., Huang W., Shi M., Zhu M.Z., Lin H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 2007;39:623–630. doi: 10.1038/ng2014. [DOI] [PubMed] [Google Scholar]
- 34.Weng J., Gu S., Wan X., Gao H., Guo T., Su N., Lei C., Zhang X., Cheng Z., Guo X., et al. Isolation and initial characterization of GW5, a major QTL associated with rice grain width and weight. Cell Res. 2008;18:1199–1209. doi: 10.1038/cr.2008.307. [DOI] [PubMed] [Google Scholar]
- 35.Mao J., Zhang Y.C., Sang Y., Li Q.H., Yang H.Q. A role for Arabidopsis cryptochromes and COP1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA. 2005;102:12270–12275. doi: 10.1073/pnas.0501011102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y., Fan C., Xing Y., Jiang Y., Luo L., Sun L., Shao D., Xu C., Li X., Xiao J., et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011;43:1266–1269. doi: 10.1038/ng.977. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X., Wang J., Huang J., Lan H., Wang C., Yin C., Wu Y., Tang H., Qian Q., Li J., et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA. 2012;109:21534–21539. doi: 10.1073/pnas.1219776110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ishimaru K., Hirotsu N., Madoka Y., Murakami N., Hara N., Onodera H., Kashiwagi T., Ujiie K., Shimizu B., Onishi A., et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013;45:707–711. doi: 10.1038/ng.2612. [DOI] [PubMed] [Google Scholar]
- 39.Liu L., Tong H., Xiao Y., Che R., Xu F., Hu B., Liang C., Chu J., Li J., Chu C. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. USA. 2015;112:11102–11107. doi: 10.1073/pnas.1512748112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y., Xiong G., Hu J., Jiang L., Yu H., Xu J., Fang Y., Zeng L., Xu E., Xu J., et al. Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 2015;47:944–948. doi: 10.1038/ng.3346. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S., Wang S., Xu Y., Yu C., Shen C., Qian Q., Geisler M., de Jiang A., Qi Y. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015;38:638–654. doi: 10.1111/pce.12397. [DOI] [PubMed] [Google Scholar]
- 42.Heang D., Sassa H. Antagonistic actions of HLH/bHLH proteins are involved in grain length and weight in rice. PLoS ONE. 2012;7:e31325. doi: 10.1371/journal.pone.0031325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duan P., Ni S., Wang J., Zhang B., Xu R., Wang Y., Chen H., Zhu X., Li Y. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nat. Plants. 2015;2:15203. doi: 10.1038/nplants.2015.203. [DOI] [PubMed] [Google Scholar]
- 44.Ledent V., Vervoort M. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res. 2001;11:754–770. doi: 10.1101/gr.177001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., Duan X., Jiang H., Sun Y., Tang Y., Yuan Z., Guo J., Liang W., Chen L., Yin J., et al. Genome-wide analysis of basic/helix-loop-helix transcription factor family in rice and Arabidopsis. Plant Physiol. 2006;141:1167–1184. doi: 10.1104/pp.106.080580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Massari M.E., Murre C. Helix-loop-helix proteins: Regulators of transcription in eucaryotic organisms. Mol. Cell Biol. 2000;20:429–440. doi: 10.1128/MCB.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L.Y., Bai M.Y., Wu J., Zhu J.Y., Wang H., Zhang Z., Wang W., Sun Y., Zhao J., Sun X., et al. Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell. 2009;21:3767–3780. doi: 10.1105/tpc.109.070441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka A., Nakagawa H., Tomita C., Shimatani Z., Ohtake M., Nomura T., Jiang C.J., Dubouzet J.G., Kikuchi S., Sekimoto H., et al. BRASSINOSTEROID UPREGULATED1, encoding a helix-loop-helix protein, is a novel gene involved in brassinosteroid signaling and controls bending of the lamina joint in rice. Plant Physiol. 2009;151:669–680. doi: 10.1104/pp.109.140806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jang S., An G., Li H.Y. Rice leaf angle and grain size are affected by the OsBUL1 transcriptional activator complex. Plant Physiol. 2017;173:688–702. doi: 10.1104/pp.16.01653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang X., Ren Y., Cai Y., Niu M., Feng Z., Jing R., Mou C., Liu X., Xiao L., Zhang X., et al. Overexpression of OsbHLH107, a member of the basic helix-loop-helix transcription factor family, enhances grain size in rice (Oryza sativa L.) Rice. 2018;11:41. doi: 10.1186/s12284-018-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rice Functional Genomic Express Database. [(accessed on 21 July 2017)]; Available online: http://signal.salk.edu/cgi-bin/RiceGE.
- 52.NCBI-BLASTP Program. [(accessed on 20 June 2018)]; Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastp&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome.
- 53.Cao H., Chen S. Brassinosteroid-induced rice lamina joint inclination and its relation to indole-3-acetic acid and ethylene. Plant Growth Regul. 1995;16:189–196. doi: 10.1007/BF00029540. [DOI] [Google Scholar]
- 54.Zhao S.Q., Xiang J.J., Xue H.W. Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol. Plant. 2013;6:174–187. doi: 10.1093/mp/sss064. [DOI] [PubMed] [Google Scholar]
- 55.Sun S., Chen D., Li X., Qiao S., Shi C., Li C., Shen H., Wang X. Brassinosteroid signaling regulates leaf erectness in Oryza sativa via the control of a specific U-type cyclin and cell proliferation. Dev. Cell. 2015;34:220–228. doi: 10.1016/j.devcel.2015.05.019. [DOI] [PubMed] [Google Scholar]
- 56.Zhou L.J., Xiao L.T., Xue H.W. Dynamic cytology and transcriptional regulation of rice lamina joint development. Plant Physiol. 2017;174:1728–1746. doi: 10.1104/pp.17.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tang Y., Liu H., Guo S., Wang B., Li Z., Chong K., Xu Y. OsmiR396d affects gibberellin and brassinosteroid signaling to regulate plant architecture in rice. Plant Physiol. 2018;17:946–959. doi: 10.1104/pp.17.00964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W., Li G., Zhao J., Chu H., Lin W., Zhang D., Wang Z., Liang W. DWARF TILLER1, a WUSCHEL-related homeobox transcription factor, is required for tiller growth in rice. PLoS Genet. 2014;10:e1004154. doi: 10.1371/journal.pgen.1004154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokoyama R., Rose J.K., Nishitani K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 2004;134:1088–1099. doi: 10.1104/pp.103.035261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Choi D., Cho H.T., Lee Y. Expansins: Expanding importance in plant growth and development. Physiol. Plant. 2006;126:511–518. doi: 10.1111/j.1399-3054.2006.00612.x. [DOI] [Google Scholar]
- 61.Hong Z., Ueguchi-Tanaka M., Shimizu-Sato S., Inukai Y., Fujioka S., Shimada Y., Takatsuto S., Agetsuma M., Yoshida S., Watanabe Y., et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002;32:495–508. doi: 10.1046/j.1365-313X.2002.01438.x. [DOI] [PubMed] [Google Scholar]
- 62.Sakamoto T., Matsuoka M. Characterization of CONSTITUTIVE PHOTOMORPHOGENESIS AND DWARFISM homologs in rice (Oryza sativa L.) J. Plant Growth Regul. 2006;25:245–251. doi: 10.1007/s00344-006-0041-6. [DOI] [Google Scholar]
- 63.Bai M.Y., Zhang L.Y., Gampala S.S., Zhu S.W., Song W.Y., Chong K., Wang Z.Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. USA. 2007;104:13839–13844. doi: 10.1073/pnas.0706386104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zurek D.M., Clouse S.D. Molecular cloning and characterization of a brassinosteroid-regulated gene from elongating soybean (Glycine max L.) epicotyls. Plant Physiol. 1994;104:161–170. doi: 10.1104/pp.104.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Uozu S., Tanaka-Ueguchi M., Kitano H., Hattori K., Matsuoka M. Characterization of XET-related genes of rice. Plant Physiol. 2000;122:853–860. doi: 10.1104/pp.122.3.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu Y., Liu D., Zhang H., Gao H., Guo X., Wang D., Zhang X., Zhang A. The α-and β-expansin and xyloglucan endotransglucosylase/hydrolase gene families of wheat: Molecular cloning, gene expression, and EST data mining. Genomics. 2007;90:516–529. doi: 10.1016/j.ygeno.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 67.Genovesi V., Fornalé S., Fry S.C., Ruel K., Ferrer P., Encina A., Sonbol F.M., Bosch J., Puigdomenech P., Rigau J. ZmXTH1, a new xyloglucan endotransglucosylase/hydrolase in maize, affects cell wall structure and composition in Arabidopsis thaliana. J. Exp. Bot. 2008;59:875–889. doi: 10.1093/jxb/ern013. [DOI] [PubMed] [Google Scholar]
- 68.Kozuka T., Kobayashi J., Horiguchi G., Demura T., Sakakibara H., Tsukaya H., Nagatani A. Involvement of auxin and brassinosteroid in the regulation of petiole elongation under the shade. Plant Physiol. 2010;153:1608–1618. doi: 10.1104/pp.110.156802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abuqamar S., Ajeb S., Sham A., Enan M.R., Iratni R. A mutation in the expansin-like A2 gene enhances resistance to necrotrophic fungi and hypersensitivity to abiotic stress in Arabidopsis thaliana. Mol. Plant Pathol. 2013;14:813–827. doi: 10.1111/mpp.12049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rao X., Dixon R.A. Brassinosteroid mediated cell wall remodeling in grasses under abiotic stress. Front. Plant Sci. 2017;8:806. doi: 10.3389/fpls.2017.00806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tong H., Liu L., Jin Y., Du L., Yin Y., Qian Q., Zhu L., Chu C. DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-Like Kinase to medate brassinosteroid responses in rice. Plant Cell. 2012;24:2562–2577. doi: 10.1105/tpc.112.097394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tong H., Jin Y., Liu W., Li F., Fang J., Yin Y., Qian Q., Zhu L., Chu C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009;58:803–816. doi: 10.1111/j.1365-313X.2009.03825.x. [DOI] [PubMed] [Google Scholar]
- 73.Bai M.Y., Shang J.X., Oh E., Fan M., Bai Y., Zentella R., Sun T.P., Wang Z.Y. Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat. Cell Biol. 2012;14:810–817. doi: 10.1038/ncb2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu J.Y., Sae-Seaw J., Wang Z.Y. Brassinosteroid signalling. Development. 2013;140:1615–1620. doi: 10.1242/dev.060590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang B., Wang X., Zhao Z., Wang R., Huang X., Zhu Y., Yuan L., Wang Y., Wang Y., Xu X., et al. OsBRI1 activates BR signaling by preventing binding between the TPR and kinase domains of OsBSK3 via phosphorylation. Plant Physiol. 2016;170:1149–1161. doi: 10.1104/pp.15.01668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang Z.Y., Nakano T., Gendron J., He J., Chen M., Vafeados D., Yang Y., Fujika S., Yoshida S., Asami T., et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev. Cell. 2002;2:505–513. doi: 10.1016/S1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 77.Todaka D., Nakashima K., Maruyama K., Kidokoro S., Osakabe Y., Ito Y., Matsukura S., Fujita Y., Yoshiwara K., Ohme-Takagi M., et al. Rice phytochrome-interacting factor-like protein OsPIL1 functions as a key regulator of internode elongation and induces a morphological response to drought stress. Proc. Natl. Acad. Sci. USA. 2012;109:15947–15952. doi: 10.1073/pnas.1207324109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.El-Din El-Assal S., Alonso-Blanco C., Peeters A.J., Raz V., Koornneef M. A QTL for flowering time in Arabidopsis reveals a novel allele of CRY2. Nat. Genet. 2001;29:435–440. doi: 10.1038/ng767. [DOI] [PubMed] [Google Scholar]
- 79.Ahmad M., Grancher N., Heil M., Black R.C., Giovani B., Galland P., Lardemer D. Action spectrum for cryptochrome-dependent hypocotyl growth inhibition in Arabidopsis. Plant Physiol. 2002;129:774–785. doi: 10.1104/pp.010969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ohgishi M., Saji K., Okada K., Sakai T. Functional analysis of each blue light receptor, cry1, cry2, phot1, and phot2, by using combinatorial multiple mutants in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2004;101:2223–2228. doi: 10.1073/pnas.0305984101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Usami T., Mochizuki N., Kondo M., Nishimura M., Nagatani A. Cryptochromes and phytochromes synergistically regulate Arabidopsis root greening under blue light. Plant Cell Physiol. 2004;45:1798–1808. doi: 10.1093/pcp/pch205. [DOI] [PubMed] [Google Scholar]
- 82.Danon A., Coll N.S., Apel K. Cryptochrome-1-dependent execution of programmed cell death induced by singlet oxygen in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 2006;103:17036–17041. doi: 10.1073/pnas.0608139103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu H., Yu X., Li K., Klejnot J., Yang H., Lisiero D., Lin C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science. 2008;322:1535–1539. doi: 10.1126/science.1163927. [DOI] [PubMed] [Google Scholar]
- 84.Kennedy M.J., Hughes R.M., Peteya L.A., Schwartz J.W., Ehlers M.D., Tucker C.L. Rapid blue-light-mediated induction of protein interactions in living cells. Nat. Methods. 2010;7:973–975. doi: 10.1038/nmeth.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Idevall-Hagren O., Dickson E.J., Hille B., Toomre D.K., De Camilli P. Optogenetic control of phosphoinositide metabolism. Proc. Natl. Acad. Sci. USA. 2012;109:E2316–E2323. doi: 10.1073/pnas.1211305109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meng Y., Li H., Wang Q., Liu B., Lin C. Blue light-dependent interaction between cryptochrome2 and CIB1 regulates transcription and leaf senescence in soybean. Plant Cell. 2013;25:4405–4420. doi: 10.1105/tpc.113.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Curtis M.D., Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miki D., Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- 89.Jeon J.S., Lee S., Jung K.H., Jun S.H., Jeong D.H., Lee J., Kim C., Jang S., Yang K., Nam J., et al. T-DNA insertional mutagenesis for functional genomics in rice. Plant J. 2000;22:561–570. doi: 10.1046/j.1365-313x.2000.00767.x. [DOI] [PubMed] [Google Scholar]
- 90.Jain M., Nijhawan A., Tyagi A.K., Khurana J.P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem. Biophys. Res. Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- 91.Wu T., Zhang M., Zhang H., Huang K., Chen M., Chen C., Yang X., Li Z., Chen H., Ma Z., et al. Identification and characterization of EDT1 conferring drought tolerance in rice. J. Plant Biol. 2019;62:39–47. doi: 10.1007/s12374-018-0203-7. [DOI] [Google Scholar]
- 92.Lee D.W., Lee S.K., Rahman M.M., Kim Y.J., Zhang D., Jeon J.S. The role of rice vacuolar invertase2 in seed size control. Mol. Cells. 2019;42:711–720. doi: 10.14348/molcells.2019.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang C., Xu Y., Guo S., Zhu J., Huan Q., Liu H., Wang L., Luo G., Wang X., Chong K. Dynamics of brassinosteroid response modulated by negative regulator LIC in rice. PLoS Genet. 2012;8:e1002686. doi: 10.1371/journal.pgen.1002686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.ImageJ Software. [(accessed on 18 May 2019)]; Available online: https://imagej.nih.gov/ij/
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.