Abstract
Basal or partial resistance has been considered race-non-specific and broad-spectrum. Therefore, the identification of genes or quantitative trait loci (QTLs) conferring basal resistance and germplasm containing them is of significance in breeding crops with durable resistance. In this study, we performed a bulked segregant analysis coupled with whole-genome sequencing (BSA-seq) to identify QTLs controlling basal resistance to blast disease in an F2 population derived from two rice varieties, 02428 and LiXinGeng (LXG), which differ significantly in basal resistance to rice blast. Four candidate QTLs, qBBR-4, qBBR-7, qBBR-8, and qBBR-11, were mapped on chromosomes 4, 7, 8, and 11, respectively. Allelic and genotypic association analyses identified a novel haplotype of the durable blast resistance gene pi21 carrying double deletions of 30 bp and 33 bp in 02428 (pi21-2428) as a candidate gene of qBBR-4. We further assessed haplotypes of Pi21 in 325 rice accessions, and identified 11 haplotypes among the accessions, of which eight were novel types. While the resistant pi21 gene was found only in japonica before, three Chinese indica varieties, ShuHui881, Yong4, and ZhengDa4Hao, were detected carrying the resistant pi21-2428 allele. The pi21-2428 allele and pi21-2428-containing rice germplasm, thus, provide valuable resources for breeding rice varieties, especially indica rice varieties, with durable resistance to blast disease. Our results also lay the foundation for further identification and functional characterization of the other three QTLs to better understand the molecular mechanisms underlying rice basal resistance to blast disease.
Keywords: rice, blast disease, partial resistance, pi21, haplotype
1. Introduction
Rice is one of the most important staple crops for more than half of the population in the world [1]. Rice blast, caused by the fungus Magnaporthe oryzae, is one of the most devastating diseases of rice, causing yield losses of 10%–30% annually [2]. The development and use of resistant varieties appears to be the most economical and environmentally sustainable way to control rice blast [3]. Identification of genes or genetic loci conferring resistance to blast disease could accelerate breeding programs for resistant rice varieties.
Genetically, disease resistance in plants can be categorized into two types, qualitative and quantitative [4,5]. Qualitative resistance is mainly mediated by a single resistance gene (R gene), which confers complete but race-specific resistance through the recognition of pathogen effectors [6,7]. R gene-mediated resistance has been widely deployed in crop breeding programs. However, R gene-mediated resistance is often not durable, as most pathogens are able to rapidly evolve new virulent races lacking the corresponding avirulence effectors to evade recognition by the cognate R protein. Quantitative resistance is mediated by multiple genes or quantitative trait loci (QTLs), providing partial or basal resistance associated with delayed and reduced development of disease lesions [8,9]. Although quantitative resistance has only partial effects, it has been considered race-non-specific and broad-spectrum, and is therefore of particular interest for breeding crops with durable resistance [4,10].
To date, more than 100 rice blast R genes have been identified and at least 28 R genes have been cloned [11,12]. All the cloned major R genes encode nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins, with the exception of Pid2, encoding a B-lectin kinase [13], and Ptr, encoding an Armadillo repeat protein [14]. Several hundred QTLs associated with blast resistance have been identified [15]. However, only a limited number of QTLs for blast resistance have been cloned [16,17,18,19,20]. The cloned QTLs encode proteins that are diverse in their structure and function: pi21 encodes a proline-rich protein with loss-of-function deletions [16]; Pb1 encodes an atypical coiled-coil (CC)-NBS-LRR protein [17]; Pi35 and Pi63 encode NBS-LRR proteins [18,19]; whereas bsr1-d1 encodes a C2H2-type transcription factor with a single nucleotide change in the promoter [20]. These findings indicate that quantitative resistances are controlled by diverse molecular mechanisms.
With the rapid development of next-generation sequencing, approaches based on bulked segregant analysis coupled with whole-genome sequencing (BSA-Seq) have been developed for the mapping of agronomically important loci in rice [21,22,23], including major genes or QTLs responsible for blast resistance [22,24,25]. In the present study, we apply BSA-seq to rapidly map four QTLs, qBBR-4, qBBR-7, qBBR-8, and qBBR-11, responsible for basal resistance to blast disease, and identify a novel haplotype of the durable blast resistance gene pi21 as a candidate gene of qBBR-4 on chromosome 4 in a japonica variety 02428 (pi21-2428). While the resistant pi21 gene was found only in japonica before [16], we identify three Chinese indica varieties carrying the resistant pi21-2428 allele in 325 accessions. Therefore, the novel pi21-2428 allele and the pi21-2428-containing rice varieties identified in the present study provide valuable resources for breeding rice varieties, especially indica rice, which are durably resistant to blast disease. Our results also lay the foundation for further identification and functional characterization of the other three QTLs for a better understanding of rice basal resistance to blast disease.
2. Results
2.1. Evaluation of 02428 and LXG in Basal Resistance to Rice Blast Disease
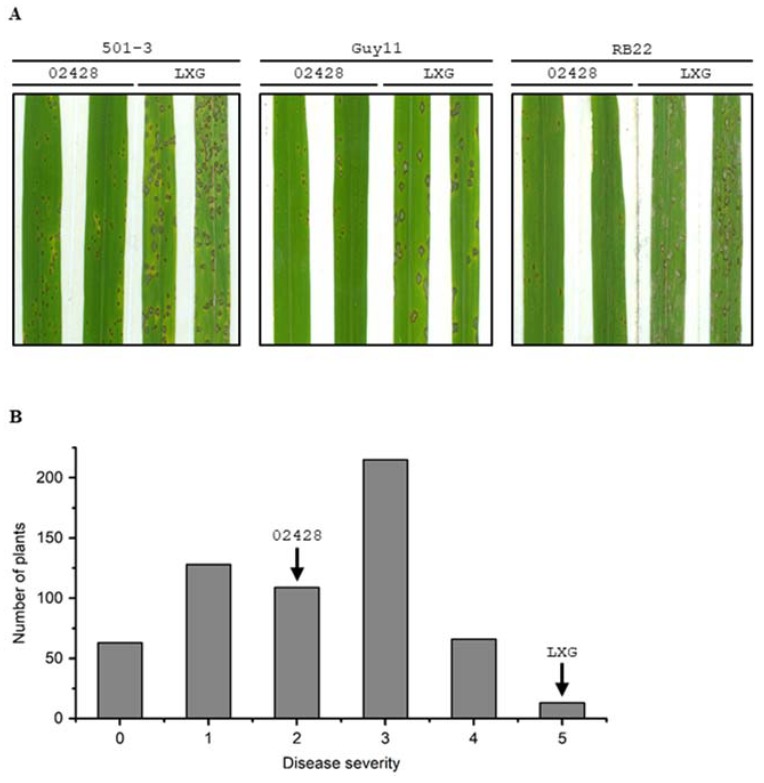
In our earlier evaluations, the rice variety 02428 was observed to possess high basal resistance to the rice blast fungus M. oryzae under natural nursery conditions (data not shown). We further performed artificial inoculations on 02428 seedlings using three virulent isolates of M. oryzae in this study. The results showed that 02428 was moderately susceptible to isolates 501-3 and Guy11, and was moderately resistant to isolate RB22 (Figure 1A). Most lesions on leaves of 02428 were limited in size. In contrast, LiXinGeng (LXG) was highly susceptible to all three isolates. These results suggest that 02428 possesses high basal resistance, preventing blast disease development.
Figure 1.
Resistance reaction of rice varieties 02428, LiXinGeng (LXG) and their F2 population to rice blast disease. (A) Phenotypes of 02428 and LXG inoculated with M. oryzae isolates 501-3, Guy11, and RB22. (B) The frequency distribution of disease severity in the F2 population of 02428 × LXG inoculated with M. oryzae isolate RB22. Disease severity was assessed following a 0–5 scale (0–1: resistant, 2: moderately resistant, 3: moderately susceptible, 4–5: severely susceptible).
An F2 population of 02428 × LXG with 626 individuals was inoculated with RB22. The results show that the frequency distribution of disease severity in the F2 population of 02428 × LXG exhibited continuous variation (Figure 1B), indicating that the resistance to RB22 is likely controlled by multiple genes.
2.2. SNP and Short InDel Polymorphism Profiling
Whole-genome sequencing of extremely resistant (ER) and extremely susceptible (ES) pools derived from the F2 population of 02428 × LXG and the two parental lines 02428 and LXG generated about 90.7 to 158.9 million reads for each sub-pool or parental line (Supplementary Table S1). After filtering, a total of 469,512 bi-allelic single-nucleotide polymorphisms (SNPs), and a total of 65,766 bi-allelic short insertions and deletions (InDels) were identified (Table 1). The average densities of SNP and short InDel markers were about 1.26 SNP/kb (average every 795 bp exists a SNP) and 0.18 InDel/kb (average every 5,675 kb exists a short InDel) (Table 1), respectively. The polymorphic markers were sufficiently distributed across the whole genome, except for one region of about 6.5 Mb on chromosome 3 containing relatively fewer markers (Supplementary Figure S1).
Table 1.
Chromosome-wise distribution of the identified single-nucleotide polymorphisms (SNPs) and short InDels.
| Chr. | Length | SNP | Short InDel | ||
|---|---|---|---|---|---|
| Number | Density (per kb) | Number | Density (per kb) | ||
| Chr1 | 43,270,923 | 48,669 | 1.12 | 7735 | 0.18 |
| Chr2 | 35,937,250 | 67,302 | 1.87 | 9069 | 0.25 |
| Chr3 | 36,413,819 | 20,399 | 0.56 | 3336 | 0.09 |
| Chr4 | 35,502,694 | 42,156 | 1.19 | 5558 | 0.16 |
| Chr5 | 29,958,434 | 67,386 | 2.25 | 9189 | 0.31 |
| Chr6 | 31,248,787 | 34,848 | 1.12 | 5309 | 0.17 |
| Chr7 | 29,697,621 | 25,681 | 0.86 | 4040 | 0.14 |
| Chr8 | 28,443,022 | 56,474 | 1.99 | 7152 | 0.25 |
| Chr9 | 23,012,720 | 23,331 | 1.01 | 2965 | 0.13 |
| Chr10 | 23,207,287 | 36,435 | 1.57 | 4062 | 0.18 |
| Chr11 | 29,021,106 | 31,309 | 1.08 | 4680 | 0.16 |
| Chr12 | 27,531,856 | 15,522 | 0.56 | 2671 | 0.10 |
| Total | 373,245,519 | 469,512 | 1.26 | 65,766 | 0.18 |
2.3. QTL Mapping and Heritability Estimation
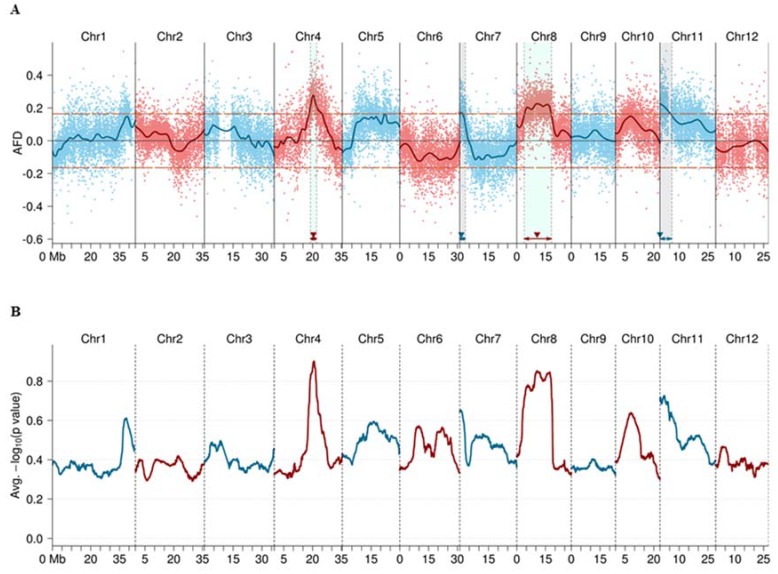
Calculation results of the average allele frequency (AF) value for each marker showed that most of the identified SNP and short InDel markers had an expected AF value of around 0.5 in the four sub-pools ER-1, ER-2, ES-1, and ES-2 (Supplementary Figure S2), indicating no severe segregation distortion of the markers as a whole. Allele frequency difference (AFD) value between ER (ER-1 + ER-2) and ES (ES-1 + ES-2) pools was calculated, and four positive AFD peaks were detected in the fitted curve and exceeded the threshold (0.165 at the overall significance level of p < 0.05) (Figure 2A). Subsequently, unpaired t-tests were performed for the two replicated sub-pools for ER and ES. The p-values of each marker were estimated, and the peaks of negative logarithmic p value (NLP) (Figure 2B) were consistent with the peaks in AFD curve. These results suggest that there were four candidate QTLs located in these regions. The confidence intervals of the four QTLs located on chromosome 4, 7, 8, and 11 were estimated (Figure 2A and Figure 3, Table 2), and the four QTLs were named qBBR-4, qBBR-7, qBBR-8, and qBBR-11, respectively.
Figure 2.
BSA-seq-based identification of four candidate quantitative trait loci (QTLs) conferring basal resistance to rice blast disease. (A) Allele frequency difference (AFD) graph from BSA-seq analysis. The horizontal orange dashed lines indicate the threshold (±0.165) at the overall significance level of p < 0.05. QTL positions estimated are indicated by filled triangles. AFD was obtained by subtraction of allele frequency (AF) of the extremely susceptible (ES) pool from that of the extremely resistant (ER) pool. (B) T-test verification for the two replicated sub-pools for the ER pool and the ES pool. The average negative logarithmic p-values of the markers were smoothed by sliding window (size = 3000 kb and step = 10 kb) across each chromosome. The peaks of the p-value graph were consistent with those in the AFD graph.
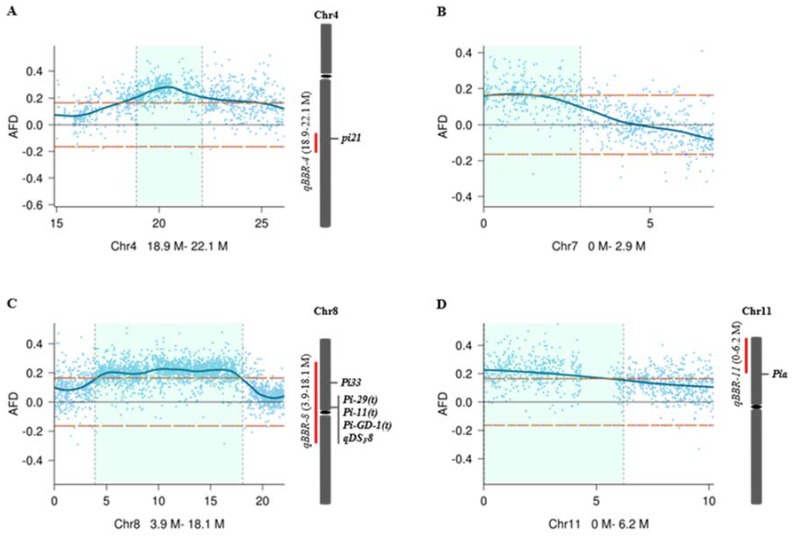
Figure 3.
Estimates of confidence intervals of the four QTLs, qBBR-4 (A), qBBR-7 (B), qBBR-8 (C), and qBBR-11 (D). The horizontal orange dashed lines indicate the threshold (0.165) at the overall significance level of p < 0.05. The light green areas indicate 95% confidence intervals of the QTLs. Previously reported R genes or QTLs within or closely near the confidence intervals of qBBR-4, qBBR-8, and qBBR-11 are indicated on the right. AFD: allele frequency difference.
Table 2.
Estimates of position and heritability of the four identified QTLs.
| QTL | Chr. | AFD value a | Pos. (Mb) b | Interval (Mp) c | Max. NLP d | (%) e | (%) f |
|---|---|---|---|---|---|---|---|
| qBBR-4 | 4 | 0.278 | 20.46 | 18.90–22.10 | 0.90 | 2.09 | 7.82 |
| qBBR-7 | 7 | 0.172 | 0.86 | 0–2.91 | 0.65 | 0.89 | 0.26 |
| qBBR-8 | 8 | 0.227 | 10.63 | 3.89–18.09 | 0.85 | 1.56 | 0.01 |
| qBBR-11 | 11 | 0.226 | 0.02 | 0–6.21 | 0.73 | 1.54 | 3.50 |
a Maximum value of the peak of the AFD curve; b Chromosome position of the peak of the AFD curve; c Estimated based on the 95% confidence; d The most significant p-value of the peak, which was converted to negative logarithmic p (NLP) value; e Heritability attributed to additive effect of the QTL; f Heritability attributed to dominance effect of the QTL.
Heritability estimation showed that the additive heritability and dominance heritability of each QTL varied by 0.89%−2.09% and 0.01%−7.82%, respectively (Table 2). Among the four QTLs, qBBR-4 had the largest effect, with the biggest additive heritability at about 2.09% (Table 2), suggesting that qBBR-4 is a major QTL involved in basal resistance to blast disease.
2.4. Identification of a New Haplotype of pi21 as a Candidate Gene of qBBR-4
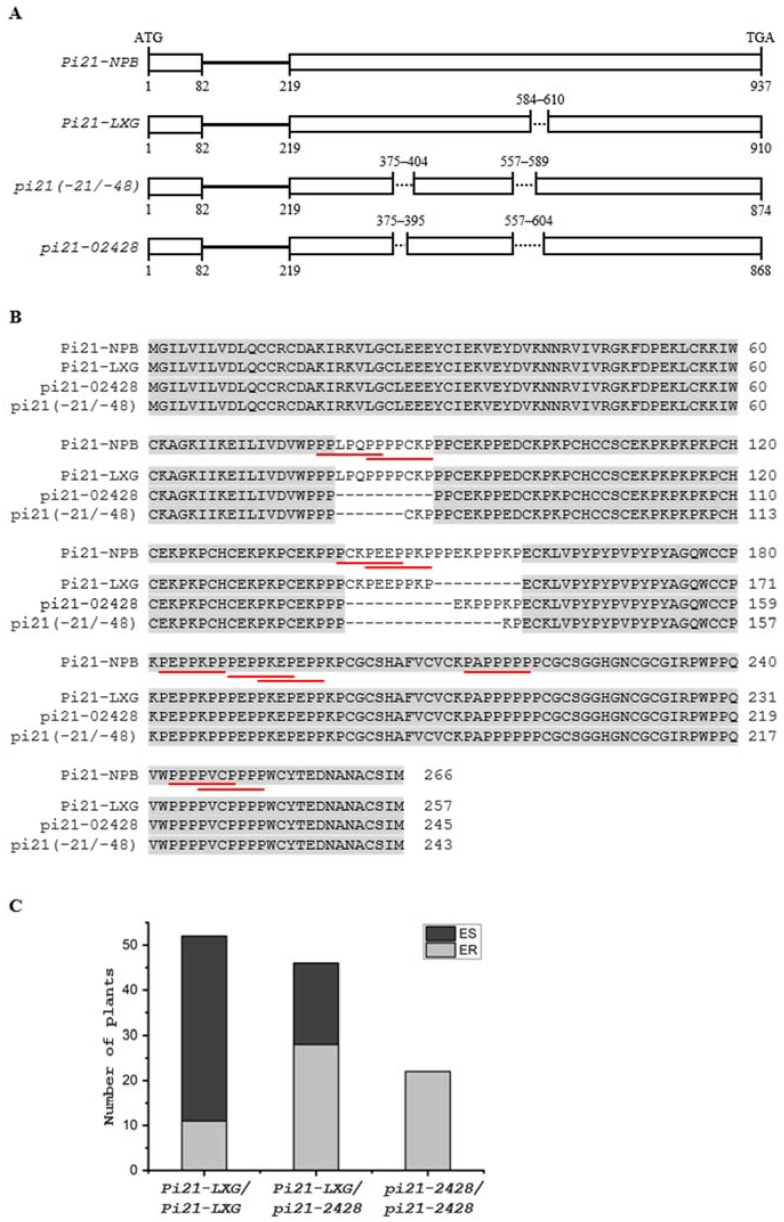
To further refine candidate genes involved in basal resistance to blast disease, we searched previously reported R genes or QTLs within the confidence intervals of the four QTLs (Figure 3). While no previously reported R genes or QTLs were identified within or near the confidence interval of qBBR-7 (Figure 3B), a previously identified Pia [26] gene was located near the confidence interval of qBBR-11 (Figure 3D), and there were several R genes or QTL, including Pi-11(t) [27], Pi-29(t) [28], Pi33 [29], Pi-GD-1(t) [30], and qDSF8 [3], in the qBBR-8 region (Figure 3C). Interestingly, qBBR-4 was observed to be co-localized with a cloned recessive durable blast disease resistance QTL pi21 (Figure 3A). Pi21 has been found to have at least 12 variants (haplotypes A to L) based on InDel polymorphisms in the proline-rich region. While haplotype L containing double deletions of 21 bp and 48 bp (pi21(-21/-48)) resulting in deletions of the core motif “PxxPxxP” in the proline-rich region (Figure 4A) was identified, conferring durable blast disease resistance due to loss of function of Pi21, the other haplotypes carrying one of the two deletions or two smaller deletions did not confer high basal resistance to blast disease [16].
Figure 4.
Identification of a novel haplotype of pi21 as a candidate gene of qBBR-4. (A) Schematic diagram of the genomic coding region of the Pi21 alleles in Nipponbare (Pi21-NPB), LXG (Pi21-LXG), and 02428 (pi21-2428), and the resistant allele of haplotype L containing double deletions of 21 bp and 48 bp (pi21(-21/-48)) [16]. Open boxes represent exons, lines represent introns, and dotted lines represent deletions. Numbers above the diagrams indicate positions of deletions corresponding to the Pi21-NPB allele, and numbers below the diagrams represent the start and end nucleotide positions of exons of each allele. (B) Alignment of the deduced amino acid sequences of Pi21-NPB, Pi21-LXG, pi21-2428, and pi21(-21/-48). Putative proline-rich motifs (PxxPxxP) for protein–protein interaction are underlined in red. (C) Genotype-phenotype correlation analysis of 61 ER individuals and 59 ES individuals. All the homozygous pi21-2428 individuals belonged to the ER bulk.
Sequence analysis of the Pi21 alleles in LXG and 02428 showed that while the allele in LXG (Pi21-LXG) had a 27 bp deletion, the pi21-2428 allele had double deletions of 30 bp and 33 bp, resulting in deletions of 10 aa and 11 aa of the core “PxxPxxP” motif, the same as in the pi21(-21/-48) allele (Figure 4A,B). Both Pi21-LXG and pi21-2428 were different from the 12 identified haplotypes. Genotypic analysis of 61 individual ER and 59 individual ES plants showed that all pi21-2428 homozygous seedlings showed resistance to blast disease. In contrast, about 79% (41 out of 52) of the Pi21-LXG homozygous, and 39% (18 out of 46) of Pi21-LXG/pi21-2428 heterozygous seedlings showed susceptible to blast disease (Figure 4C). These results support pi21-2428 as a candidate gene of qBBR-4, and suggest that while the 27 bp deletion did not affect Pi21-LXG function, double deletions of the 30 bp and 33 bp sequences could cause a defect in pi21-2428 function, leading to high basal resistance to blast disease.
2.5. Assessment of the pi21-2428 Haplotype in 325 Rice Accessions
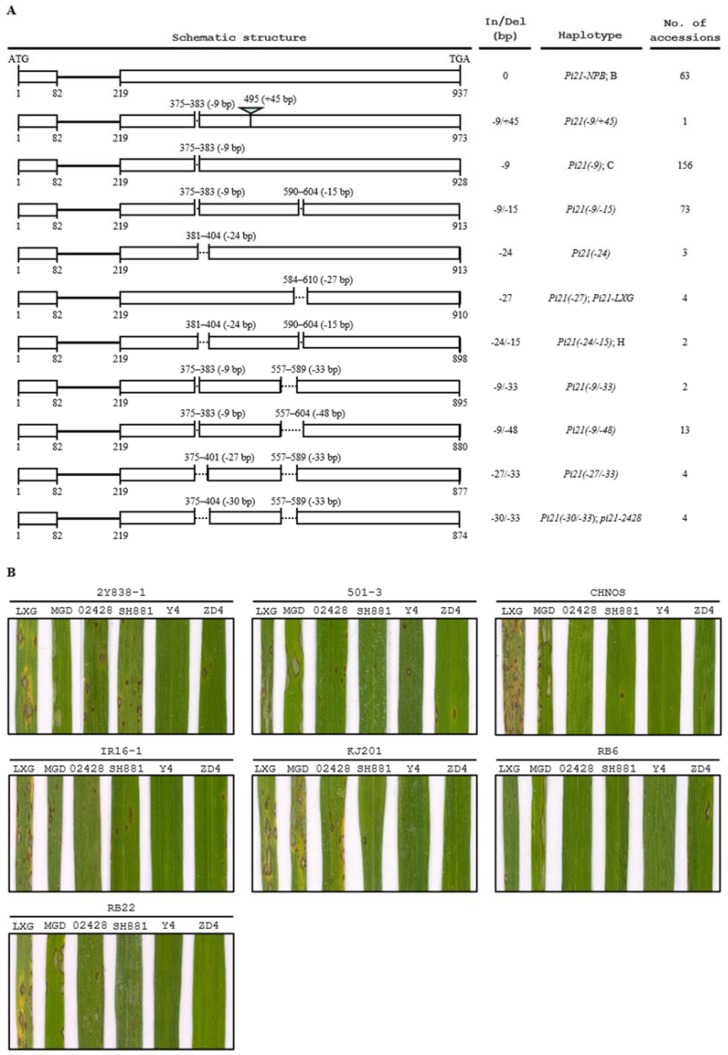
Multiple sequence alignment of the Pi21 alleles in 325 rice accessions revealed a total of 11 haplotypes among the tested accessions (Figure 5A, Supplementary Table S2). However, none of the accessions carried the resistant pi21(-21/-48) allele. The haplotypes were named based on insertion/deletion patterns. Among the 11 haplotypes, Pi21-NPB (the Pi21 allele in Nipponbare), Pi21(-9), and Pi21(-24/-15) were identical to the previously identified haplotypes B, C, and H, respectively (Figure 5A, Supplementary Table S2) [16], and the rest eight were novel types.
Figure 5.
Assessment of haplotypes of Pi21 in 325 rice accessions and blast inoculation of the pi21-2428-containing varieties. (A) A total of 11 haplotypes were identified among 325 rice accessions. The haplotypes were named based on insertion/deletion patterns. Pi21-NPB, Pi21(-9), and Pi21(-24/-15) were identical to the previously identified haplotypes B, C, and H, respectively [16]. Open boxes represent exons, lines represent introns, and dotted lines represent deletions. Numbers above the diagrams indicate positions of deletions corresponding to the Pi21-NPB allele, and numbers below the diagrams represent the start and end nucleotide positions of exons of each allele. (B) Phenotypes of the pi21-2428-containing varieties 02428, ShuHui881 (SH881), Yong4 (Y4), and ZhengDa4Hao (ZD4) inoculated with M. oryzae isolates 2Y838-1, 501-3, CHNOS, IR16-1, KJ201, RB6, and RB22, respectively. Rice varieties LXG and MengGuDao (MGD) were used as highly susceptible controls.
Among the 325 rice accessions, in addition to 02428, three Chinese indica varieties ShuHui881, Yong4, and ZhengDa4Hao were detected carrying the resistant pi21-2428 allele. The pi21-2428-containing varieties were inoculated with seven M. oryzae isolates. As shown in Figure 5B, the two susceptible varieties LXG and MengGuDao (MGD) were highly susceptible to most of the M. oryzae isolates. The four pi21-2428-containing varieties showed complete resistance, moderate resistance, or moderate susceptibility to the isolates. For example, the indica variety ZhengDa4Hao was highly resistant to isolates KJ201 and RB22, and moderately resistant to isolates CHNOS and IR16-1, suggesting that there might be some R genes conferring the high resistance in ZhengDa4Hao against the four isolates. ZhengDa4Hao also showed susceptibility to isolates 2Y838-1, 501-3, and RB6, but the lesions on leaves were limited in size and much less than those on leaves of LXG or MGD. Overall, none of the pi21-2428-containing varieties showed highly susceptible to the seven M. oryzae isolates, suggesting that the four varieties possessed high basal resistance with significantly delayed and reduced development of disease lesions.
3. Discussion
When compared with R gene-mediated, race-specific resistance, basal resistance has been presumed to be more durable [4,10]. Therefore, the identification of genes or QTLs conferring basal resistance is of significance in breeding crops with long-lasting resistance to plant diseases. In the present study, we employed BSA-Seq to identify four candidate QTLs qBBR-4, qBBR-7, qBBR-8, and qBBR-11, conferring basal resistance to rice blast disease on rice chromosomes 4, 7, 8, and 11, respectively. Building on advances in next-generation sequencing, the BSA-Seq method took advantage of pooled sequencing, which does not require the laborious process of genotyping of each individual from a large mapping population, allowing for rapid identification of candidate genes or QTLs controlling important agronomic traits [24,31].
Among the four QTLs detected in this study, qBBR-4 had the largest additive effect (Table 2). The qBBR-4 locus is located in a region of chromosome 4 with a known recessive blast disease resistant QTL pi21, identified in rice variety Owarihatamochi [16]. Pi21 encodes a proline-rich protein, consisting of a putative heavy metal-binding domain and protein-protein interaction motifs. While the dominant Pi21 appears to slow the plant’s defense responses, loss-of-function of Pi21 (double deletions of 21 bp and 48 bp in the proline-rich region; haplotype L) confers durable resistance to blast disease in rice [16]. Sequence analysis revealed that the pi21 allele in 02428, the parental line with high basal resistance, had double deletions of 30 bp and 33 bp, resulting in deletions of 10 aa and 11 aa in the proline-rich region, which house the core motif “PxxPxxP” (Figure 4A,B) for protein–protein interaction in multicellular organisms [16,32,33]. We further performed genotype-phenotype correlation analysis of 61 ER individuals and 59 ES individuals, and the results showed that all the homozygous pi21-2428 individuals belonged to the ER bulk (Figure 4C). Taken together, these results suggest that double deletions of the 30 bp and 33 bp sequences of Pi21 led to high basal resistance to blast disease, and that pi21-2428 was the candidate gene of qBBR-4.
Previously, sequence analysis of the Pi21 locus revealed 12 haplotypes (A to L) among 80 Asian cultivated rice varieties. Eleven of the haplotypes (A to K) carried an insertion or smaller deletions compared with the resistant pi21(-21/-48) allele, and did not confer resistance to blast disease [16]. In the present study, we detected a total of 11 haplotypes of Pi21 among 325 rice accessions (Figure 5A, Supplementary Table S2), of which three were identical to the previously identified haplotypes B, C, and H [16], and eight were novel. Interestingly, the DNA variations of all the 20 detected haplotypes identified by Fukuoka et al. [16] and in this study were found to result in amino acid insertion/deletions, but not to cause premature termination of the predicted Pi21 product, implying that the Pi21 or pi21 alleles maintain certain functions important for rice [34]. Besides 02428, we identified three more varieties, ShuHui881, Yong4, and ZhengDa4Hao, possessing the resistant pi21-2428 allele. Inoculation testing with seven M. oryzae isolates indicated that, while the susceptible varieties LXG and MGD were highly susceptible to most of the M. oryzae isolates, the four pi21-2428-containing varieties showed complete resistance moderate resistance or moderate susceptibility to the M. oryzae isolates (Figure 5B). The results suggest that there should be some R genes other than pi21-2428 conferring the complete or high resistance in the four pi21-2428-containing varieties. On the other hand, the moderately susceptible reactions with limited lesion size and number, to virulent M. oryzae isolates indicated that ShuHui881, Yong4, ZhengDa4Hao, as well as 02428, possessed high basal resistance to blast disease (Figure 5B). It is worth noting that while the resistant pi21(-21/-48) allele was found only in japonica rice [16], the three pi21-2428-containing varieties ShuHui881, Yong4, and ZhengDa4Hao were indica rice (Supplementary Table S2). Therefore, the varieties identified in the present study provide valuable resources for breeding rice varieties, especially indica varieties, with durable resistance to blast disease.
Transgressive segregation was observed in the F2 population of 02428 × LXG, where some F2 segregants showed more resistance than both parents (Figure 1B). This phenomenon implies that favorable alleles from both resistant and susceptible parents could be combined in the progeny, leading to a higher resistance than in the parents. In the present study, in addition to pi21-2428, we detected three other QTLs, qBBR-7, qBBR-8, and qBBR-11, on chromosomes 7, 8, and 11, respectively. Further identification and functional characterization of the three QTLs should be helpful to better understand the mechanisms underlying rice basal resistance to blast disease. Furthermore, the finding of transgressive segregation for blast resistance in the F2 population of 02428 × LXG indicates that pyramiding more basal resistance genes or QTL alleles with pi21-2428 would be an effective approach to enhance durable resistance to rice blast disease [10,35].
4. Materials and Methods
4.1. Plant Materials and Blast Isolates
Rice cvs. 02428 and LXG were used to construct an F2 population to evaluate the segregation of blast disease reaction and for QTL mapping. Besides 02428 and LXG, a collection of 323 rice accessions consisting mainly of Chinese rice varieties or breeding materials were used for haplotype assessment in this study (Supplementary Table S2). The M. oryzae isolates CHNOS, Guy11, and KJ201 were kindly provided by Dr. Guo-Liang Wang (Department of Plant Pathology, Ohio State University, Ohio, USA), and isolates 2Y838-1, 501-3, IR16-1, RB6, and RB22 were collected from Fujian province, China.
4.2. Rice Blast Inoculations
Rice blast inoculations were carried out following a previously described spraying method [36]. Rice seedlings were grown in a greenhouse for about 12–14 days and were inoculated with M. oryzae spores at a concentration of 5 × 105 spores mL−1. After inoculation, the seedlings were grown at 25 °C under high humidity for 4–5 days. Blast disease reactions were scored following a 0–5 scale (0–1: resistant, 2: moderately resistant, 3: moderately susceptible, 4–5: severely susceptible) [37].
4.3. Bulking, DNA Extraction, and Whole-Genome Resequencing
To understand the genetic basis of basal resistance to rice blast disease in 02428, a total of 626 F2 plants derived from a cross between 02428 and LXG were inoculated by RB22, and were investigated for the segregation of disease reaction. For bulked segregant analysis, about 10,000 individuals of the F2 population of 02428 × LXG were inoculated with the M. oryzae isolate RB22. About 126 highly resistant and 120 highly susceptible F2 individuals were screened to generate the ER and ES bulks, respectively. Both the ER and ES bulks were divided into two replicates, ER-1 (56 individuals) and ER-2 (70 individuals) for the ER bulk, and ES-1 (60 individuals) and ES-2 (60 individuals) for the ES bulk. The genomic DNAs of the four bulked samples were extracted and were mixed with equal amounts. DNA samples of the two parents, 02428 and LXG, and the four pools were subjected to whole-genome resequencing using the Illumina HiSeq X Ten platform, followed by standard paired-end 150 bp sequencing library construction protocols.
4.4. Analysis of Reads and Variants
The raw reads were cleaned and trimmed using BBDuk program of BBTools (http://jgi.doe.gov/data-and-tools/bbtools/). The paired reads were mapped to the IRGSP-1.0 reference rice genome (http://rapdb.dna.affrc.go.jp) by using Burrows-Wheeler Aligner based on the Maximal Exact Matches algorithm (BWA MEM) and the alignments were processed by SAMTools [38,39,40]. Freebayes was used to call SNPs and InDels, with default parameters [41]. To obtain reliable polymorphic markers, variant filtering was performed by custom perl scripts: firstly, SNPs or short InDels exhibiting polymorphism between the two parents were screened; secondly, to further avoid severe segregation distortion, SNPs or short InDels with AF values from 0.3 to 0.7 were retained. These markers were annotated by snpEff [42].
4.5. QTL Analysis
The marker set was employed to map QTLs. The replicated sub-pools ER-1, ER-2, and ES-1, ES-2 were firstly incorporated into one ER pool and one ES pool, respectively. AFD value between the ER and ES pools was calculated and then smoothed by block regression, following the Block Regression Mapping methodology [43]. The block size used for the regression was set to be 20 kb. The AFD curve threshold at the overall significance level of 0.05 was estimated under the assumption of theoretical allele frequency (= 0.5) in the F2 population. For each significant AFD peak (candidate QTL), the 95% confidence interval was estimated. In addition, unpaired t-tests based on the two biological replicates of the ER and ES pools were performed to validate the candidate QTLs following the X-QTL-seq method [44]. According to the peak AFD value, the heritability of each QTL was estimated using the method of Pooled QTL Heritability Estimator (PQHE) [45].
4.6. Detection of Haplotypes of Pi21
Genomic DNAs of the rice accessions (Supplementary Table S2) were subjected to genotyping for haplotypes of Pi21. PCR products were amplified with primers P21-F (5′-CAAGGCTAATCAGCAGTGT-3′) and P21-R (5′-TTGGCGTTGTCCTCGGTGT-3′). DNA sequences of PCR products were aligned using Clustal W [46].
Acknowledgments
This work was supported by grant from National Natural Science Foundation of China (U1405212), grants from Fujian Provincial Science and Technology Program (2017R1019-1, 2018R1019-9), and open project of State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops (SKL018002).
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/6/2162/s1.
Author Contributions
W.T., Z.W. and S.C. conceived and designed the experiments. T.L., S.Z. and S.C. constructed the F2 population. T.L., Z.-Q.C., Z.-J.C., D.T., X.C. and S.C. conducted rice blast inoculation. W.C., L.H. and W.T. analyzed the sequencing data and performed QTL mapping. T.L., M.Q. and Y.G. conducted haplotype analysis. W.T. and S.C. wrote and revised the manuscript. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Khush G.S. What it will take to feed 5.0 billion rice consumers in 2030. Plant Mol. Biol. 2005;59:1–6. doi: 10.1007/s11103-005-2159-5. [DOI] [PubMed] [Google Scholar]
- 2.Talbot N.J. On the trail of a cereal killer: Exploring the biology of Magnaporthe grisea. Annu. Rev. Microbiol. 2003;57:177–202. doi: 10.1146/annurev.micro.57.030502.090957. [DOI] [PubMed] [Google Scholar]
- 3.Sun P., Liu J., Wang Y., Jiang N., Wang S., Dai Y., Gao J., Li Z., Pan S., Wang D., et al. Molecular mapping of the blast resistance gene Pi49 in the durably resistant rice cultivar Mowanggu. Euphytica. 2013;192:45–54. doi: 10.1007/s10681-012-0829-3. [DOI] [Google Scholar]
- 4.Poland J.A., Balint-Kurti P.J., Wisser R.J., Pratt R.C., Nelson R.J. Shades of gray: The world of quantitative disease resistance. Trends Plant Sci. 2009;14:21–29. doi: 10.1016/j.tplants.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 5.Kou Y., Wang S. Broad-spectrum and durability: Understanding of quantitative disease resistance. Curr. Opin. Plant Biol. 2010;13:181–185. doi: 10.1016/j.pbi.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 6.Chisholm S.T., Coaker G., Day B., Staskawicz B.J. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124:803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Jones J.D., Dangl J.L. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 8.Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 9.Aghnoum R., Marcel T.C., Johrde A., Pecchioni N., Schweizer P., Niks R.E. Basal host resistance of barley to powdery mildew: Connecting quantitative trait Loci and candidate genes. Mol. Plant-Microbe Interact. 2010;23:91–102. doi: 10.1094/MPMI-23-1-0091. [DOI] [PubMed] [Google Scholar]
- 10.Yasuda N., Mitsunaga T., Hayashi K., Koizumi S., Fujita Y. Effects of pyramiding quantitative resistance genes Pi21, Pi34, and Pi35 on rice leaf blast disease. Plant Dis. 2015;99:904–909. doi: 10.1094/PDIS-02-14-0214-RE. [DOI] [PubMed] [Google Scholar]
- 11.Tian D., Yang L., Chen Z., Chen Z., Wang F., Zhou Y., Lou Y., Yang L., Chen S. Proteomic analysis of the defense response to Magnaporthe oryzae in rice harboring the blast resistance gene Piz-t. Rice. 2018;11:47. doi: 10.1186/s12284-018-0240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C., Wang D., Peng S., Chen Y., Su P., Chen J., Zheng L., Tan X., Liu J., Xiao Y., et al. Genome-wide association mapping of resistance against rice blast strains in South China and identification of a new Pik allele. Rice. 2019;12:47. doi: 10.1186/s12284-019-0309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X., Shang J., Chen D., Lei C., Zou Y., Zhai W., Liu G., Xu J., Ling Z., Cao G., et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006;46:794–804. doi: 10.1111/j.1365-313X.2006.02739.x. [DOI] [PubMed] [Google Scholar]
- 14.Zhao H., Wang X., Jia Y., Minkenberg B., Wheatley M., Fan J., Jia M.H., Famoso A., Edwards J.D., Wamishe Y., et al. The rice blast resistance gene Ptr encodes an atypical protein required for broad-spectrum disease resistance. Nat. Commun. 2018;9:2039. doi: 10.1038/s41467-018-04369-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu D., Kang H., Li Z., Liu M., Zhu X., Wang Y., Wang D., Wang Z., Liu W., Wang G.L. A genome-wide association study of field resistance to Magnaporthe oryzae in rice. Rice. 2016;9:44. doi: 10.1186/s12284-016-0116-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuoka S., Saka N., Koga H., Ono K., Shimizu T., Ebana K., Hayashi N., Takahashi A., Hirochika H., Okuno K., et al. Loss of function of a proline-containing protein confers durable disease resistance in rice. Science. 2009;325:998–1001. doi: 10.1126/science.1175550. [DOI] [PubMed] [Google Scholar]
- 17.Hayashi N., Inoue H., Kato T., Funao T., Shirota M., Shimizu T., Kanamori H., Yamane H., Hayano-Saito Y., Matsumoto T., et al. Durable panicle blast-resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication. Plant J. 2010;64:498–510. doi: 10.1111/j.1365-313X.2010.04348.x. [DOI] [PubMed] [Google Scholar]
- 18.Fukuoka S., Yamamoto S.I., Mizobuchi R., Yamanouchi U., Ono K., Kitazawa N., Yasuda N., Fujita Y., Nguyen T.T.T., Koizumi S., et al. Multiple functional polymorphisms in a single disease resistance gene in rice enhance durable resistance to blast. Sci. Rep. 2014;4:4550. doi: 10.1038/srep04550. [DOI] [Google Scholar]
- 19.Xu X., Hayashi N., Wang C.T., Fukuoka S., Kawasaki S., Takatsuji H., Jiang C.J. Rice blast resistance gene Pikahei-1(t), a member of a resistance gene cluster on chromosome 4, encodes a nucleotide-binding site and leucine-rich repeat protein. Mol. Breed. 2014;34:691–700. doi: 10.1007/s11032-014-0067-6. [DOI] [Google Scholar]
- 20.Li W., Zhu Z., Chern M., Yin J., Yang C., Ran L., Cheng M., He M., Wang K., Wang J., et al. A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell. 2017;170:114–126. doi: 10.1016/j.cell.2017.06.008. [DOI] [PubMed] [Google Scholar]
- 21.Abe A., Kosugi S., Yoshida K., Natsume S., Takagi H., Kanzaki H., Matsumura H., Yoshida K., Mitsuoka C., Tamiru M., et al. Genome sequencing reveals agronomically important loci in rice using MutMap. Nat. Biotechnol. 2012;30:174–178. doi: 10.1038/nbt.2095. [DOI] [PubMed] [Google Scholar]
- 22.Takagi H., Abe A., Yoshida K., Kosugi S., Natsume S., Mitsuoka C., Uemura A., Utsushi H., Tamiru M., Takuno S., et al. QTL-seq: Rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J. 2013;74:174–183. doi: 10.1111/tpj.12105. [DOI] [PubMed] [Google Scholar]
- 23.Zhang H., Wang X., Pan Q., Li P., Liu Y., Lu X., Zhong W., Li M., Han L., Li J., et al. QTG-Seq accelerates QTL fine mapping through QTL partitioning and whole-genome sequencing of bulked segregant samples. Mol. Plant. 2019;12:426–437. doi: 10.1016/j.molp.2018.12.018. [DOI] [PubMed] [Google Scholar]
- 24.Zheng W., Wang Y., Wang L., Ma Z., Zhao J., Wang P., Zhang L., Liu Z., Lu X. Genetic mapping and molecular marker development for Pi65 (t), a novel broad-spectrum resistance gene to rice blast using next-generation sequencing. Theor. Appl. Genet. 2016;129:1035–1044. doi: 10.1007/s00122-016-2681-7. [DOI] [PubMed] [Google Scholar]
- 25.Wu S., Qiu J., Gao Q. QTL-BSA: A bulked segregant analysis and visualization pipeline for QTL-seq. Interdiscip. Sci. Comput. Life Sci. 2019 doi: 10.1007/s12539-019-00344-9. [DOI] [PubMed] [Google Scholar]
- 26.Okuyama Y., Kanzaki H., Abe A., Yoshida K., Tamiru M., Saitoh H., Fujibe T., Matsumura H., Shenton M., Galam D.C., et al. A multifaceted genomics approach allows the isolation of the rice Pia-blast resistance gene consisting of two adjacent NBS-LRR protein genes. Plant J. 2011;66:467–479. doi: 10.1111/j.1365-313X.2011.04502.x. [DOI] [PubMed] [Google Scholar]
- 27.Zhu L., Chen Y., Xu Y., Xu J., Cai H., Ling Z. Construction of a molecular map of rice and gene mapping using a double-haploid population of a cross between indica and japonica varieties. Rice Genet. Newsl. 1993;10:132–134. [Google Scholar]
- 28.Sallaud C., Lorieux M., Roumen E., Tharreau D., Berruyer R., Svestasrani P., Garsmeur O., Ghesquiere A., Notteghem J.L. Identification of five new blast resistance genes in the highly blast-resistant rice variety IR64 using a QTL mapping strategy. Appl. Genet. 2003;106:794–803. doi: 10.1007/s00122-002-1088-9. [DOI] [PubMed] [Google Scholar]
- 29.Berruyer R., Adreit H., Milazzo J., Gaillard S., Berger A., Dioh W., Lebrun M.H., Tharreau D. Identification and fine mapping of Pi33, the rice resistance gene corresponding to the Magnaporthe grisea avirulence gene ACE1. Appl. Genet. 2003;107:1139–1147. doi: 10.1007/s00122-003-1349-2. [DOI] [PubMed] [Google Scholar]
- 30.Liu B., Zhang S., Zhu X., Yang Q., Wu S., Mei M., Mauleon R., Leach J., Mew T., Leung H. Candidate defense genes as predictors of quantitative blast resistance in rice. Mol. Plant-Microbe Interact. 2004;17:1146–1152. doi: 10.1094/MPMI.2004.17.10.1146. [DOI] [PubMed] [Google Scholar]
- 31.Gao J., Dai G., Zhou W., Liang H., Huang J., Qing D., Chen W., Wu H., Yang X., Li D., et al. Mapping and identifying a candidate gene Plr4, a recessive gene regulating purple leaf in rice, by using bulked segregant and transcriptome analysis with next-generation sequencing. Int. J. Mol. Sci. 2019;20:4335. doi: 10.3390/ijms20184335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawson T. Protein modules and signalling networks. Nature. 1995;373:573–580. doi: 10.1038/373573a0. [DOI] [PubMed] [Google Scholar]
- 33.Ball L.J., Kuhne R., Schneider-Mergener J., Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem. Int. Ed. Engl. 2005;44:2852–2869. doi: 10.1002/anie.200400618. [DOI] [PubMed] [Google Scholar]
- 34.Fukuoka S., Okuno K. Strategies for breeding durable resistance to rice blast using pi21. Crop Breed. Genet. Genom. 2019;1:e190013. [Google Scholar]
- 35.Fukuoka S., Saka N., Mizukami Y., Koga H., Yamanouchi U., Yoshioka Y., Hayashi N., Ebana K., Mizobuchi R., Yano M. Gene pyramiding enhances durable blast disease resistance in rice. Sci. Rep. 2015;5:7773. doi: 10.1038/srep07773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tian D., Chen Z., Chen Z., Zhou Y., Wang Z., Wang F., Chen S. Allele-specific marker-based assessment revealed that the rice blast resistance genes Pi2 and Pi9 have not been widely deployed in Chinese indica rice cultivars. Rice. 2016;9:19. doi: 10.1186/s12284-016-0091-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackill D.J., Bonman J.M. Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology. 1992;82:746–749. doi: 10.1094/Phyto-82-746. [DOI] [Google Scholar]
- 38.Kawahara Y., de la Bastide M., Hamilton J.P., Kanamori H., McCombie W.R., Ouyang S., Schwartz D.C., Tanaka T., Wu J., Zhou S., et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H., Durbin R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrison E., Marth G. Haplotype-based variant detection from short-read sequencing. arXiv. 20121207.3907 [Google Scholar]
- 42.Cingolani P., Platts A., Wang L., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang L., Tang W., Bu S., Wu W. BRM: A statistical method for QTL mapping based on bulked segregant analysis by deep sequencing. Bioinformatics. 2019 doi: 10.1093/bioinformatics/btz861. [DOI] [PubMed] [Google Scholar]
- 44.Ehrenreich I.M., Torabi N., Jia Y., Kent J., Martis S., Shapiro J.A., Gresham D., Caudy A.A., Kruglyak L. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature. 2010;464:1039–1042. doi: 10.1038/nature08923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang W., Huang L., Bu S., Zhang X., Wu W. Estimation of QTL heritability based on pooled sequencing data. Bioinformatics. 2018;34:978–984. doi: 10.1093/bioinformatics/btx703. [DOI] [PubMed] [Google Scholar]
- 46.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.