Abstract
Glioblastoma (GBM) is a grade IV glioma that is the most malignant brain tumor type. Currently, there are no effective and sufficient therapeutic strategies for its treatment because its pathological mechanism is not fully characterized. With the fast development of the Next Generation Sequencing (NGS) technology, more than 170 kinds of covalent ribonucleic acid (RNA) modifications are found to be extensively present in almost all living organisms and all kinds of RNAs, including ribosomal RNAs (rRNAs), transfer RNAs (tRNAs) and messenger RNAs (mRNAs). RNA modifications are also emerging as important modulators in the regulation of biological processes and pathological progression, and study of the epi-transcriptome has been a new area for researchers to explore their connections with the initiation and progression of cancers. Recently, RNA modifications, especially m6A, and their RNA-modifying proteins (RMPs) such as methyltransferase like 3 (METTL3) and α-ketoglutarate-dependent dioxygenase alkB homolog 5 (ALKBH5), have also emerged as important epigenetic mechanisms for the aggressiveness and malignancy of GBM, especially the pluripotency of glioma stem-like cells (GSCs). Although the current study is just the tip of an iceberg, these new evidences will provide new insights for possible GBM treatments. In this review, we summarize the recent studies about RNA modifications, such as N6-methyladenosine (m6A), N6,2′O-dimethyladenosine (m6Am), 5-methylcytosine (m5C), N1-methyladenosine (m1A), inosine (I) and pseudouridine (ψ) as well as the corresponding RMPs including the writers, erasers and readers that participate in the tumorigenesis and development of GBM, so as to provide some clues for GBM treatment.
Keywords: m6A, RNA modification, glioblastoma, epi-transcriptome, RNA processing
1. Introduction
Glioblastoma (GBM), a World Health Organization (WHO) grade IV glioma, is the most prevalent, malignant and lethal intrinsic tumor in the central nervous system [1]. It accounts for about 50% of all brain neoplasms and has a median survival time of approximately 14.6 months [2]. The annual worldwide occurrence of this disease is about five cases per 100,000 people [3]. Currently, the origin of GBM is mainly the poorly differentiated glial cell, which is characterized with nuclear atypia, cellular polymorphism and high mitotic activity [4]. Since GBM is very aggressive and invasive, it is difficult to remove all the neoplastic lesions from the brain by using surgical resection. Subsequently, the patients typically receive 30-week radiotherapy and an adjuvant chemotherapeutic alkylating drug, temozolomide (TMZ) [3]. However, since GBM is highly heterogeneous, the effect is limited and no remarkable progression has been achieved yet. Therefore, it is urgent to develop new therapeutic strategies to treat GBM; however, the natural characteristics of GBM remain not fully understood.
To know how GBM initiates and develops, the molecular characteristics of GBM should be accurately determined. Actually, many genetic, epigenetic, metabolic and immunologic profiles in GBM have been described in recent years [5,6,7,8,9]. These landscapes of GBM have greatly expanded our knowledge about GBM, thereby providing clues for the treatment of this disease. Particularly, covalent modifications in DNA and histones, also known as that main concept of epigenetics, play important roles in regulating cell fate determination, cell proliferation, cellular metabolism and even many pathological processes [10,11,12,13,14]. Recently, the RNA world has become a new area to be explored for cancer therapy. Both coding and non-coding mRNAs have many diverse functions in addition to their traditional functions. Studies on RNA processing, editing, splicing, polyadenylation, post-transcription of RNA and RNA-binding proteins are developing very fast [15,16,17,18], leading to a new area called the epi-transcriptome. Covalent modifications in RNA and their connections with cancers are also an area of concentrated activity in cancer biology research [19].
Recently, RNA modifications, as known as RNA epigenetics, were found to be present in almost all the cellular RNAs in archaeobacteria, bacteria, prototists, plants, fungi, animals and viruses. Even in mitochondria, there are also some chemical modifications in mitochondrial DNA-encoded RNAs [20]. More than 170 kinds of covalent modifications have been found in RNAs at present [21,22], however, the functional implications of most of them remain unravelled. These kinds of biological modifications exercise important regulation functions during both normal and pathological bioprocesses and tissue development. Importantly, RNA modifications, especially N6-methyladenosine (m6A) modification, are shown to be essential for tumor development [22,23,24,25].
In this review, we summarize the most recent advances in RNA modifications, especially the functions of m6A writers, erasers and readers, and their molecular mechanisms of action in GBM, so as to provide some clues for the development of new strategies for GBM treatment.
2. RNA Modifications
RNA modifications refer to dynamic, reversible covalent alterations present in the oligonucleotides of RNAs, including mRNAs, tRNAs, rRNAs and spliceosomal RNAs (sRNAs). These changes are commonly present in the transcriptome. Since 1957, when pseudouridine (ψ, also known as the fifth RNA nucleotide) was discovered by Davis and Allen [26], at least 170 kinds of RNA modifications have been identified [26]. Actually, m7G (7-methylguanosine, also known as 5′ cap) and 3′ poly(A) tail are well-known modifications in mRNA [27]. In 2012, the NGS method was developed, and since then, many widespread and conserved internal RNA modifications in RNAs were identified [22].
In eukaryotic RNA, the most commonly found modifications are N6-methyladenosine (m6A), N6, 2′O-dimethyladenosine (m6Am), 5-methylcytosine (m5C), 5-hydroxymethylcytosine (hm5C), N1-methyladenosine (m1A), inosine (I), N4-acetylcytidine (ac4C), ψ, uridylation (U-tail), 2-methylthio-N6-(cis-hydroxyisopentenyl)adenosine (ms2i6A), 3-methylcytosine (m3C), m7G, 7-methylguanosine cap (m7Gpp(pN)), queuosine (Q), wybutosine and derivatives (yW), 5-methyluridine (m5U), 5-carbamoylmethyluridine (ncm5U), 5-methoxycarbonylmethyluridine (mcm5U), 5-methoxy-carbonylmethyl-2-thiouridine (mcm5s2U), dihydrouridine (D), 2′-O-methylnucleotide (Nm), N6-isopentenyladenosine (i6A), and 5′-phosphate monomethylation (m(pN)) [28,29,30]. Most of these modifications have been found in mRNAs. Moreover, eukaryotic tRNAs also contain many modifications (13 modifications per molecule on average) to maintain their chemical diversity, thereby contributing to decoding fidelity, folding efficiency, cellular stability and localization [31]. rRNAs also contain abundant modifications (>200 sites per molecule) including 2′-O-methyls, pseudouridines, and base methylations in the peptidyl transferase center, the A, P and E sites of tRNA- and mRNA binding [32], thereby maintaining their functions. In addition, sRNAs also contain moderate amounts of modifications (>50 sites per molecule), some of which play important roles in the RNA splicing actions [33].
Each modification has its writers, the enzymes that deposit RNA chemical marks; erasers, the enzymes that remove these marks; and readers, the proteins that selectively recognize and bind to these marks (Figure 1). Collectively, the writers, erasers and readers also refer to as RNA-modifying proteins (RMPs). These modifications are not only needed for normal function but also play essential roles in pathological processes, such as tumorigenesis.
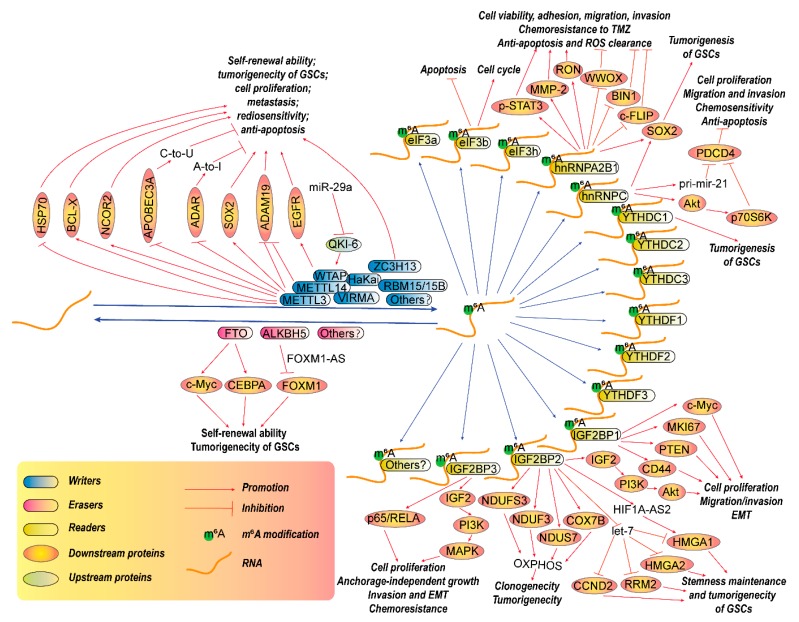
Figure 1.
Functions of m6A modification and its writers, erasers and readers in GBM. Abbreviations: RNA-modifying proteins (RMPs): ALKBH5, α-Ketoglutarate-dependent dioxygenase alkB homolog 5; eIF3a/b/h, eukaryotic translation initiation factor 3a/b/h; FTO, Fat mass and obesity-associated protein; Hakai, Cbl proto-oncogene like 1, CBLL1; hnRNPA2B1, heterogeneous nuclear ribonucleoprotein A2/B1; hnRNPC, heterogeneous nuclear ribonucleoprotein C1/C2; IGF2BP1/2/3, insulin-like growth factor 2 mRNA-binding protein 1/2/3; METTLE3/14, methyltransferase like 3/14; RBM15/15B, RNA-binding motif protein 15/15B; VIRMA, Virilizer like m6A methyltransferase associated protein; WTAP, Wilms’ tumor 1-associated protein; YTHDF1/2/3, YTH N6-methyladenosine RNA binding protein 1/2/3; YTHDC1/2/3, YTH domain-containing 1/2/3; ZC3H13, zinc finger CCCH domain-containing protein 13. Others: A-to-I, adenosine-to-inosine; ADAM19, a disintegrin and metallopeptidase domain 19; ADAR1,adenosine deaminase RNA specific 1; APOBEC3A, apolipoprotein B mRNA editing enzyme catalytic subunit 3A; BCL-X, Bcl-2-like protein 1, BCL2L1; BIN1, bridging integrator 1; c-FLIP, cellular FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein; c-Myc, avian myelocytomatosis viral oncogene homolog; C-to-U, cytidine to uridine; CCND2, cyclin D2; CEBPA, CCAAT enhancer binding protein alpha; COX7B, mitochondrial cytochrome c oxidase subunit 7B; EGFR, epidermal growth factor receptor; EMT, epithelial-to-mesenchymal transition; FOXM1, forkhead box M1; FOXM1-AS, forkhead box M1-antisense RNA; GSCs, glioma stem-like cells; HIF1A-AS2, hypoxia-inducible factor 1 alpha-antisense RNA 2; HMGA1/2, high mobility group AT-hook 1/2; HSP70, heat shock protein 70; IGF2, insulin-like growth factor 2; m6A, N6-methyladenosine; MAPK, mitogen-activated protein kinase; MKI67, marker of proliferation Ki-67; MMP-2, matrix metallopeptidase 2; NCOR2, nuclear receptor corepressor 2; NDUF3,NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3; NDUFS3, mitochondrial NADH dehydrogenase [ubiquinone] iron-sulfur protein 3; NDUS7, NADH dehydrogenase iron-sulfur protein 7; OXPHOS, oxidative phosphorylation; PI3K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homolog; QKI-6, Quaking gene isoform 6; RON, macrophage stimulating 1 receptor, MST1R; ROS, reactive oxygen species; RRM2, ribonucleotide reductase regulatory subunit M2; SOX2, SRY-box transcription factor 2; STAT3, signal transducer and activator of transcription 3; TMZ, temozolomide; WWOX, WW domain containing oxidoreductase.
3. RNA Modifications in GBM
With the developing of sequencing technology in RNA modifications, novel markers regarding RNA modifications in cancers are also emerging [34]. GBM is the most malignant tumors in the brain that has no effect therapy. Therefore, it is urgent to develop new strategies to treat this disease. Since m6A is the best-known RNA mark, it is highly connected with GBM progression and aggressiveness and some regulators may be potential drug targets. Recently, other kinds of RNA marks, including m6Am, m1A, m5C, hm5C, I and ψ, as well as their modulators are also emerging to be correlated with GBM progression (Table 1).
Table 1.
Functions of RNA-modifying proteins (RMPs) in GBM.
| Gene Symbol | Gene Description | Role in RNA Modification | Expression | Biological Function | Mechanism | Reference |
|---|---|---|---|---|---|---|
| METTL3 | Methyltransferase-like 3 | m6A writer | Elevated or decreased (controversial) | Maintains or prevents stemness of GSSs; transforms or reverse malignancy; enhances radiosensitivity; reduces migration and induces apoptosis (controversial) | Installs m6A marks in GSCs-specifically-expressed genes, such as SOX2; modulates ADAR1 and APOBEC3A to induce A-to I and C-to-U editing; upregulates BCL-X and NCOR2 to affect the levels of SRSFs; prevents YTHDC1-dependent NMD; introduces m6A modifications in critical factors, such as ADAM19; downregulates HSP70 expression | [56,57,58,59,60] |
| METTL14 | Methyltransferase-like 14 | m6A writer | Elevated or Decreased (controversial) | Prevents growth, self-renewal, and tumorigenesis of GSCs | Introduces m6A modifications in critical factors, such as ADAM19 | [58,59] |
| WTAP | Wilms’ tumor 1-associated protein | m6A writer | Elevated | Stimulates tumorigecity, migration and invasion and inhibits apoptosis | Increases EGFR expression; | [58,61,62,63] |
| VIRMA/KIAA1429 | Virilizer like m6A methyltransferase associated protein | m6A writer | Decreased | - | - | [58] |
| ZC3H13/KIAA0853/Flacc | Zinc finger CCCH domain-containing protein 13 | m6A writer | Decreased | Inhibits malignant tansformation and reduces chemosensitivity | - | [58] |
| RBM15 | RNA binding motif protein 15 | m6A writer | Elevated | - | - | [58] |
| RBM15B | RNA binding motif protein 15B | m6A writer | Elevated | - | - | [58] |
| FTO | Fat mass and obesity-associated protein | m6A and m6Am eraser | Decreased or elevated (controversial) | Promotes self-renewal ability and tumorigenecity of GSCs | Maintains the stability of c-Myc and CEBPA transcripts | [58,59,64] |
| ALKBH5 | α-Ketoglutarate-dependent dioxygenase alkB homolog 5 | m6A eraser | Elevated | Inhibits cell proliferation of GSCs | Demethylates FOXM1 nascent transcripts to enhance its expression by the aid of lncRNA FOXM1-AS | [58,65] |
| YTHDF1 | YTH N6-methyladenosine RNA binding protein 1 | m6A reader | Elevated | - | - | [58] |
| YTHDF2 | YTH N6-methyladenosine RNA binding protein 2 | m6A reader | Elevated | - | - | [58] |
| YTHDF3 | YTH N6-methyladenosine RNA binding protein 3 | m6A reader | Elevated | - | - | [58] |
| YTHDC1 | YTH domain-containing 1 | m6A reader | - | Maintains stemness of GSCs | - | [57] |
| YTHDC2 | YTH domain-containing 2 | m6A reader | Elevated in LGGs; decreased in GBM | - | - | [58] |
| IGF2BP1/IMP1 | Insulin like growth factor 2 mRNA binding protein 1 | m6A reader | Elevated | Promotes cell proliferation and invasion | Stabilizes the mRNA transcripts of its target genes, including c-Myc, MKI67, PTEN and CD44 | [66,67]. |
| IGF2BP2/IMP2 | Insulin like growth factor 2 mRNA binding protein 2 | m6A reader | Elevated | Promotes cell proliferation, migration, invasion and EMT; sensitizes GBM to TMZ treatment; maintains stemness of GSCs; promotes OXPHOS; promotes hypoxia-dependent molecular reprogramming | Activates IGF2-mediated PI3K/AKT signaling; binds to let-7 MREs and prevents LIN28-independent let-7 target gene silencing, including HMGA1, HMGA2, CCND2 and RRM2; binds to several mRNAs such as COX7B, NDUS7, NDUF3 and NDUFS3; interacts with HIF1A-AS2 to maintain HMGA1 expression | [68,69,70,71] |
| IGF2BP3/IMP3 | Insulin like growth factor 2 mRNA binding protein 3 | m6A reader | Elevated | Promotes cell proliferation, anchorage-independent growth, invasion, EMT and chemoresistance | Activates PI3K/MAPK via promoting IGF2 translation; enhances translation of p65 (RELA) | |
| eIF3b | Eukaryotic translation initiation factor 3 subunit b | m6A reader | Dcreased | Inhibits cell proliferation and induces G0/G1 cell cycle arrest and apoptosis | - | [72] |
| eIF3h | Eukaryotic translation initiation factor 3 subunit h | m6A reader | Dublicated | - | - | [73] |
| hnRNPA2B1 | Heterogeneous nuclear ribonucleoprotein A2/B1 | m6A reader | Elevated | Promotes GBM cell viability, adhesion, migration, invasion, chemoresistance to TMZ and tumorigenecity as well as protects cells from apoptosis and reactive oxygen species (ROS) generation | Interacts with SOX2; downregulates tumor suppressors such as c-FLIP, BIN1 and WWOX, and upregulates phospho-STAT3, MMP-2, and the proto-oncogene RON | [58,74,75] |
| hnRNPC | Heterogeneous nuclear ribonucleoprotein C | m6A reader | Elevated in LGGs but decreased in GBM | Promotes migratory and invasive activities, increases cell proliferation and protects GBM cells from etoposide-induced apoptosis | Interacts with SOX2; binds directly to pri-mir-21 and promotes the processing of miR-21 that targets PDCD4, supporting Akt and p70S6K activation, | [58,76] |
| METTL4 | Methyltransferase-like 4 | m6Am writer | Missensely mutated | - | - | [77] |
| DCP2 | Decapping mRNA 2 | m6Am reader | Participates in radiosensitivity and DNA damage response | - | [78] | |
| TRMT6/TRM6 | tRNA methyltransferase 6 | m1A writer | Elevated | - | - | [79] |
| TRMT61A/TRM61A | tRNA methyltransferase 61A | m1A writer | Elevated | - | - | [79] |
| ALKBH1 | α-Ketoglutarate-dependent dioxygenase alkB homolog 1 | m1A eraser | Elevated | Promotes cell proliferation and tumorigenecity | Induces DNA demethylation on hypoxia response genes, | [80] |
| NSUN5 | NOP2/Sun RNA methyltransferase 5 | m5C writer | Decreased | Leads to an adaptive translational program for survival under cellular stress and renders these gliomas sensitive to bioactivatable substrates of the stress-related enzyme NQO1 | Drives an overall depletion of protein synthesis | [81,82] |
| LYREF | Aly/REF export factor | m5C reader | Elevated | - | - | [83,84] |
| TET1 | Tet methylcytosine dioxygenase 1 | hm5C writer | Elevated | - | - | [85] |
| TET2 | Tet methylcytosine dioxygenase 2 | hm5C writer | Decreased | - | - | [86] |
| TET3 | Tet methylcytosine dioxygenase 3 | hm5C writer | Decreased | - | - | [87] |
| ADAR1/ADAR | Adenosine deaminase RNA specific 1 | I writer | Decreased | Inhibits cell proliferation | Downregulates A-to-I RNA editing | [88] |
| ADAR2/ADARB1 | Adenosine deaminase RNA specific 2 | I writer | Decreased | Inhibits cell proliferation | Downregulates A-to-I RNA editing in the pre-mRNA of a phosphatase CDC14B, thereby downregulates Skp2/p21/p27 proteins | [88,89,90] |
| ADAR3/ADARB2 | Adenosine deaminase RNA specific 3 | I writer | Decreased | - | - | [88] |
| DKC1 | Dyskerin pseudouridine synthase 1 | ψ writer | Elevated | Promotes cell growth by stimulating cell cycle progression and activates migration | Upregulates N-cadherin, HIF1A, and MMP-2 expression | [91] |
Modification abbreviations: A-to-I, adenosine-to-inosine; C-to-U, cytidine to uridine, m1A, N1-methyladenosine; m5C, 5-methylcytosine; m6A, N6-methyladenosine; m6Am, N6, 2′O-dimethyladenosine; hm5C, 5-hydroxymethylcytosine; I, inosine; ψ, pseudouridine. Others: ADAM19, a disintegrin and metallopeptidase domain 19; ADAR1,adenosine deaminase RNA specific 1; APOBEC3A, apolipoprotein B mRNA editing enzyme catalytic subunit 3A; BCL-X, Bcl-2-like protein 1, BCL2L1; BIN1, bridging integrator 1; c-FLIP, cellular FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein; c-Myc, avian myelocytomatosis viral oncogene homolog; CCND2, cyclin D2; CDC14B, cell division cycle 14B; CEBPA, CCAAT enhancer binding protein alpha; COX7B, mitochondrial cytochrome c oxidase subunit 7B; EGFR, epidermal growth factor receptor; EMT, epithelial-to-mesenchymal transition; FOXM1, forkhead box M1; FOXM1-AS, forkhead box M1-antisense RNA; GBM, glioblastoma; GSCs, glioma stem-like cells; HIF1A, hypoxia-inducible factor 1 alpha; HIF1A-AS2, hypoxia-inducible factor 1 alpha-antisense RNA 2; HMGA1/2, high mobility group AT-hook 1/2; HSP70, heat shock protein 70; IGF2, insulin-like growth factor 2; LGG, low-grade glioma; MAPK, mitogen-activated protein kinase; MKI67, marker of proliferation Ki-67; MMP-2, matrix metallopeptidase 2; MRE, miRNA recognition element; NCOR2, nuclear receptor corepressor 2; NDUF3,NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3; NDUFS3, mitochondrial NADH dehydrogenase [ubiquinone] iron-sulfur protein 3; NDUS7, NADH dehydrogenase iron-sulfur protein 7; NMD, nonsense-mediated mRNA decay; NQO1, Quaking gene isoform 6; OXPHOS, oxidative phosphorylation; p70S6K, p70 S6 kinase; PDCD4, programmed cell death 4; PI3K, phosphatidylinositol 3-kinase; PTEN, phosphatase and tensin homolog; RON, macrophage stimulating 1 receptor, MST1R; RRM2, ribonucleotide reductase regulatory subunit M2; Skp2, S-phase kinase associated protein 2; SOX2, SRY-box transcription factor 2; STAT3, signal transducer and activator of transcription 3; TMZ, temozolomide; WWOX, WW domain containing oxidoreductase.
3.1. RNA m6A Modification in GBM
RNA m6A modification is the most prevalent and abundant modifications that occur in the mRNAs, rRNAs and small nuclear RNAs (snRNAs) [35]. m6A modification of mRNA usually occurs in nuclear speckles where the methyltransferases and demethylases are concentrated [29] and are enriched in single nucleotide polymorphisms (SNPs) [36]. Generally, the enzymatic core METTLE3 and methyltransferase like 14 (METTL14), as well as Wilms’ tumor 1-associated protein (WTAP), Virilizer like m6A methyltransferase associated protein (VIRMA/KIAA1429), RNA-binding motif protein 15 (RBM15), RNA-binding motif protein 15B (RBM15B/OTT3), zinc finger CCCH domain-containing protein 13 (ZC3H13, as known as Flacc and KIAA0853) and Hakai (also known as Cbl proto-oncogene like 1, CBLL1) constitute of a methyltransferase complex, also known as m6A-METTL associated complex (MACOM), that can mediates the m6A methylation. Meanwhile, two demethylases, Fat mass and obesity-associated protein (FTO) and ALKBH5, catalyze the m6A demethylation [37,38,39,40,41,42].
Several m6A-binding proteins such as YTH N6-methyladenosine RNA binding protein 1/2/3 (YTHDF1/2/3) and YTH domain-containing 1/2/3 (YTHDC1/2/3), which have the YTH domain, are readers that mediate the degradation of m6A-labelled RNAs [43,44]. Other factors, such as insulin-like growth factor 2 mRNA-binding protein 1/2/3 (IGF2BP1/2/3, also known as IMP1/2/3), eukaryotic translation initiation factor 3a/b/h (eIF3a/b/h), heterogeneous nuclear ribonucleoprotein A2/B1 (hnRNPA2B1) and heterogeneous nuclear ribonucleoprotein C1/C2 (hnRNPC) were recently shown to be able to read the m6A marks, too [22]. These m6A reader proteins can recognize these marks and also can play key roles in the RNA stabilization of the writers [45]. m6A marks recognized by different readers with different sub-locations may exert different functions: the readers in the nucleus, such as hnRNPC, hnRNPA2B1 and YTHDC1, are responsible for RNA structure switching, alternative splicing, microRNA maturation, RNA stability, RNA export and X chromosome inactivation; while readers in the cytoplasm, such as YTHDF1/2/3 and YTHDC2, are mainly responsible for mRNA translation and decay [46].
As RNA m6A affects RNA stability, processing, splicing, translation, and the epigenetic function of some non-coding RNAs (ncRNAs) [47,48], it plays essential role in cell fate determination, embryonic stem cell maintenance and specification, T-cell homeostasis, neuronal functions, sex determination as well as pathogenesis [49,50,51,52,53,54]. In tumors, m6A also emerges as an important modulator. For instance, METTL3 drives tumorigenicity and metastasis through suppressing suppressor of cytokine signaling 2 (SOCS2) expression in hepatocellular carcinoma [55].
Actually, m6A methylomes of brain tissues are highly specific [35], but the status of m6A may be different in GBM. A bioinformatic analysis in The Chinese Glioma Genome Atlas (CGGA) microarray and RNA sequencing databases [58] show that m6A writers, including METTL3, METTL14, WTAP, RBM15 and RBM15B, as well as m6A readers including YTHDC2, YTHDF1, YTHDF2, YTHDF3, hnRNPA2B1 and hnRNPC are elevated, while m6A writer ZC3H13 and m6A eraser FTO are decreased in gliomas. In GBMs, m6A writers WTAP and RBM15, m6A eraser ALKBH5, and m6A reader YTHDF2 are significantly elevated, while m6A writers METTL3, VIRMA and ZC3H13, m6A eraser FTO, m6A readers YTHDC2 and hnRNPC are significantly decreased, compared with low-grade gliomas (LGGs). In addition, four lncRNAs including MIR9-3HG, LINC00900, MIR155HG, and LINC00515 seem to be related with m6A. Another bioinformatic analysis [92] revealed that the expressions of WTAP, RBM15, YTHDF and ALBKH5 are positively, while FTO expression is negatively associated with WHO grades of glioma. Additionally, expression levels of ALKBH5, YTHDF2, RBM15, METTL3, METTL14, FTO and YTHDC1 in LGGs with or without mutant isocitrate dehydrogenase (IDH) are significantly different. Moreover, FTO, YTHDC1 and METTL3 are also differentially expressed between GBM with and without IDH mutation. Importantly, the expressions of gliomas malignant clinicopathological features-related m6A RNA methylation regulators are tightly correlated with pro-oncogenic genes in gliomas. These evidences imply that m6A modifications are tightly correlated with GBM progression. According to present studies in GBM, m6A modification seems to play pivotal roles since both m6A writers and erasers contribute to the tumorigenesis of GBM (Figure 1), especially glioma stem-like cells (GSCs).
3.1.1. RNA m6A Writers in GBM
The expression of METTL3, the most important m6A writer, is significantly elevated in GSCs, while it is attenuated during GSCs differentiation [56]. Elevated expression of METTL3 are associated with the clinical aggressiveness of malignant gliomas [57]. An integrated analysis of m6A-RNA immunoprecipitation (RIP) and total RNA-Sequencing (RNA-seq) of METTL3-silenced GSCs revealed that METTL3 is the most important writer responsible for m6A modification in GSCs [45]. Moreover, METTL3 silencing also reverses RasV12-mediated malignant transformation of mouse immortalized astrocytes and suppresses GBM tumor growth in vitro and in vivo [56,57]. Furthermore, METTL3 silencing in GSCs enhances their sensitivity to γ-irradiation and reduces DNA repair [56]. Mechanically, METTL3 seems to be responsible for the m6A modification in some GSCs-specifically-expressed genes [45]. Particularly, METTL3 mediates m6A modification of mRNA of SRY-box transcription factor 2 (SOX2), a pluripotent gene, at three METTL3/m6A sites present in its 3′ untranslated region (UTR), leading to the recruitment of human antigen R (HuR) to SOX2 mRNA to enhance its stability, thereby resulting in GSC maintenance and dedifferentiation [56].
In addition to directly affecting the pluripotent genes, an m6A-specific methylated RNA immunoprecipitation with NGS (MeRIP-Seq or m6A-Seq) analysis also shows that m6A modification peaks seem to be enriched at transcripts responsible for metabolic pathways in GSCs, compared with neural progenitor cells [57]. METTL3 also causes alterations such as decrease of adenosine-to-inosine (A-to-I) and cytidine to uridine (C-to-U) RNA editing events in GSCs by downregulating adenosine deaminase RNA specific 1 (ADAR1) and apolipoprotein B mRNA editing enzyme catalytic subunit 3A (APOBEC3A), which are RNA editing enzymes [45]. Furthermore, gene ontology analysis reveals that the direct targets of METTL3 seems to be enriched in some major oncogenic pathways including NOTCH signaling, vascular endothelial growth factor (VEGF) signaling, angiogenesis, glycolysis and Hedgehog signaling pathways, whereas the indirect targets are enriched in the RAS, mitogen-activated protein kinase (MAPK), G protein-coupled receptors (GPCRs), cadherin signaling pathways and cell cycle [45]. Additionally, in GSCs, METTL3-dependent m6A modification also affects the levels of serine- and arginine-rich splicing factors (SRSFs) through upregulating BCL-X (Bcl-2-like protein 1, BCL2L1) or nuclear receptor corepressor 2 (NCOR2) isoforms, which are important to prevent YTHDC1-dependent nonsense-mediated mRNA decay (NMD) [57]. lncRNAs with METTL3-dependent m6A marks is also highly expressed in GSCs, compared with that of protein-coding genes [45]. m6A marks within 3′UTRs seem to hinder the miRNA binding in GSCs [45].
Nevertheless, an opposite study shows that METTL3 and METTL14 may be a tumor suppressor in GBM [59]. Their results show that deficiency of METTL3 or METTL14 significantly enhances the growth, self-renewal, and tumorigenesis of GBM stem cells. Subsequently, m6A sequencing and molecular experiments implies that METTL3 or METTL14 knockdown induced alterations in mRNA m6A enrichment and the mRNA expression of critical factors, such as a disintegrin and metallopeptidase domain 19 (ADAM19), that participate in modulation of GBM stem cells. In contrast, overexpression of METTL3 renders GBM stem cells growth and self-renewal. Another study in established GBM cell line U251 cells also shows the similar results: METTL3 overexpression reduces migration and proliferation ability and induces cellular apoptosis possible by downregulating heat shock protein 70 (HSP70) expression [60]. The controversial conclusion may be due to the samples they used. These results mean that function of m6A modifications regulated by METTL3 in GBM should indeed be further confirmed.
WTAP is also an important part of the m6A methyltransferase complex. It is also highly expressed in glioma, closely correlated with pathological grade and negatively correlated with postoperative survival in glioma patients [61,62]. WTAP stimulates tumorigecity, migration and invasion of GBM cells possibly through enhancing the activity of epidermal growth factor receptor (EGFR), an important oncogenic factors in GBM [61]. In GSCs, WTAP also plays an important role as a downstream target of miR-29a/Quaking gene isoform 6 (QKI-6) axis-mediated inhibition of cell proliferation, migration and invasion, as well as promotion of apoptosis in GSCs [63].
ZC3H13, as a zinc-finger protein, is also an essential regulator in the MACOM complex, which can anchor WTAP, Virilizer, and Hakai in the nucleus [93]. A recent study shows that ZC3H13 mutation combined with retinoblastoma 1 (RB1) mutation can recapture human GBM in a mouse model. Moreover, ZC3H13 mutation also alters the gene expression profiles of RB1 mutants, rendering GBM tumors more resistant to TMZ [94]. These evidences connote that ZC3H13 may be an anti-cancer factor.
Collectively, m6A writers are important for GBM progression. Most of them are upregulated in GBM, and show oncogenic roles by regulating specific signaling pathways, especially factors that contribute to stemness maintenance. However, some opposite results show that the expression alterations of some writers in GBM are downregulated and they may pose anti-cancer properties, indicating that much more work should be done to confirm their expressions and functions in GBM.
3.1.2. RNA m6A Erasers in GBM
Like the writers, m6A erasers also play essential roles in GBM. Inhibition of FTO, an m6A eraser, also impedes the self-renewal ability and tumorigenecity of GBM stem cells both in vitro and in mice models [59]. Other studies also show that FTO functions as an oncogenic role via maintaining the stability of transcripts of avian myelocytomatosis viral oncogene homolog (c-Myc) and CCAAT enhancer binding protein alpha (CEBPA) in glioma, especially IDH1/2 mutant glioma [64]. Besides, inhibition of FTO by its specific inhibitor, MA2, significantly renders the tumorigenecity in GSC-grafted mice [59]. These evidences suggest that the homostasis of m6A modification in cells is important and FTO may be a promising drug target of GBM.
Another m6A demethylase, ALKBH5, is also highly expressed in patient-derived GSCs. Silencing ALKBH5 inhibits cell proliferation of GSCs. An integrated transcriptome and m6A-seq analysis identified several ALKBH5 target genes, including forkhead box M1 (FOXM1), a transcription factor that is critical for GSCs. ALKBH5 demethylates FOXM1 nascent transcripts, thereby enhancing its expression. A lncRNA forkhead box M1-antisense RNA (FOXM1-AS), also enhances the interaction of ALKBH5 with FOXM1 nascent transcripts [65]. These results show that m6A erasers may also play pro-oncogenic roles in GBM and may be potential drug targets for GBM treatment. If both m6A writers and erasers are confirmed to be upregulated in GBM, it is probable that the modulatory capacity of m6A in GBM is enhanced, indicating that GBM may be more flexible to response to complex extracellular or intracellular stresses.
3.1.3. RNA m6A Readers in GBM
The fate of m6A-modified RNA is determined by m6A readers, so they may be more important factors for cancer treatment, because when functions of m6A writers and erasers are settled, the functions of readers depend on what kinds of readers to carry out the effects of m6A marks. Importantly, the aberrant expression of m6A readers may contribute to GBM tumorigenesis independently of m6A alterations to some extent. This means that even in the condition that m6A is not changed, the effect of m6A may be altered by the abnormal expression of m6A readers. Based on current study, several families that exert as m6A readers are tightly related to the tumorigenesis of GBM.
YTHDF and YTHDC Families
YTHDF and YTHDC families are the most important m6A readers that contain an YTH domain that can bind to RNAs. They exhibit different functions: YTHDC1 mediates mRNA splicing; YTHDF2, YTHDF3, and YTHDC2 mediate mRNA decay; YTHDF1, YTHDF3, and YTHDC2 mediate translation; and YTHDC2 mediates RNA structures [22]. YTHDC1 reads the m6A marks dependent on its tryptophan 377 (W377) or W428 sites [95]. Recently, these readers were shown to also participate in tumorigenesis of GBM. For instance, YTHDC1 deficiency reduces sphere number substantially in METTL3 overexpressed U87 cells but not in control cells. Overexpressing W377A/W428A mutant YTHDC1 cannot promotes the sphere formation capacity of U87 cells, suggesting that YTHDC1 promotes GBM phenotype dependently on its m6A binding activity [57].
IGF2BP Family
Another m6A reader protein family, the IGF2BP family, including IGF2BP1/2/3, inhibits the degradation of m6A-modified transcripts and facilitates their translation [96]. For a long time, these factors are recognized as important regulators in GBM progression, even though there are no direct connections between their functions in m6A readers and oncogenic roles in GBM. For example, IGF2BP1 promotes cell proliferation and invasion in GBM cells through stabilizing the mRNA transcripts of its target genes, including c-Myc, marker of proliferation Ki-67 (MKI67), phosphatase and tensin homolog (PTEN) and CD44 [66,67]. However, whether this phenomenon depens on its activity in m6A reading remains unknown.
IGF2BP2 is also upregulated in GBM tissues. It supports GBM cell proliferation, migration, invasion and epithelial-to-mesenchymal transition (EMT) via regulating the activity of insulin-like growth factor 2 (IGF2), which further activates phosphatidylinositol 3-kinase (PI3K)/Akt signaling. Additionally, inhibition of IGF2BP2 sensitizes GBM to TMZ treatment [68]. Another study shows that IGF2BP2 binds to let-7 miRNA recognition elements (MREs) and prevents LIN28-independent let-7 target gene silencing, including high mobility group AT-hook 1 (HMGA1), HMGA2, cyclin D2 (CCND2) and ribonucleotide reductase regulatory subunit M2 (RRM2), thereby ensuring maintenance of stemness in GSCs [69]. In this process, IGF2BP2 is responsible for miRNA maturation. IGF2BP2 also interacts with lncRNAs such as hypoxia-inducible factor 1 α-antisense RNA 2 (HIF1A-AS2), which functions as a subtype-specific hypoxia-inducible lncRNA, thereby maintaining the expression of their target gene, HMGA1, resulting in promoted GSC growth, self-renewal, and hypoxia-dependent molecular reprogramming [71]. In addition, IGF2BP2 also interacts with mRNAs and even proteins. For example, it can promote oxidative phosphorylation (OXPHOS) in primary GBM sphere cultures (gliomaspheres, GSCs) via binding several mRNAs such as mitochondrial cytochrome c oxidase subunit 7B (COX7B), NADH dehydrogenase iron-sulfur protein 7 (NDUS7), NADH dehydrogenase [ubiquinone] 1α subcomplex assembly factor 3 (NDUF3) and mitochondrial NADH dehydrogenase [ubiquinone] iron-sulfur protein 3 (NDUFS3), that encode mitochondrial respiratory chain complex subunits. Additionally, it also interacts with complex I (NADH: ubiquinone oxidoreductase) proteins such as NDUFS3, thereby promoting clonogenicity in vitro and tumorigenicity in vivo [70].
IGF2BP3 mRNA and protein levels are also upregulated in GBMs but not in lower grade astrocytomas [97]. Particularly, IGF2BP3 is one of the genes overexpressed in pilomyxoid astrocytomas (PMAs) compared with a less aggressive pilocytic astrocytomas (PAs), according to a gene expression microarray analysis of 9 PMAs and 13 PAs from infra- and supratentorial sites [98]. A microarray analysis in glioma cells shows that IGF2BP3 mediates regulation of direct targets at transcriptome associated with cell cycle process, whereas direct targets at the translatome participating in apoptosis related pathways [99]. IGF2BP3 also promotes cell proliferation, migration and invasion by inducing EMT via downregulating the expression of E-cadherin and upregulating the expressions of N-cadherin, vimentin, snail, slug and MMP-9 in GBM [100]. In addition, IGF2BP3 promotes cell proliferation, anchorage-independent growth, invasion, and chemoresistance via activating PI3K/MAPK pathways by binding to the 5′-UTR of IGF-2 mRNA to activate its translation [97]. IGF2BP3 also stimulates glioma cell migration by enhancing the translation of p65 (RELA), a subunit of nuclear factor kappa-B (NF-κB) heterodimer, which can also in turn transcriptionally activates IGF2BP3 to form a feedback loop [101].
eIF3 Family
eIF3a/b/h, also function as m6A readers, and can directly, physically and functionally interact with METTL3, thereby enhancing translation, and the formation of densely packed polyribosomes through recognizing m6A modification at the 5′ UTR of mRNA [102,103,104]. A study of cancer predisposition syndromes (CPS) shows that eIF3h is duplicated in a pediatric patient with infantile desmoplastic astrocytoma and low-grade glioma [73]. eIF3b silencing significantly inhibits cell proliferation of U87 cells via inducing G0/G1-phase arrest and apoptosis [72]. These evidences indicate that eIF3 family members may play specific roles in GBM.
hnRNPC and hnRNPA2B1
During nuclear RNA processing period, hnRNPC and hnRNPA2B1 are other types of m6A readers that can bind to unfolded RNA through an m6A structural switch mechanism, since the m6A marked RNA cannot efficiently form secondary structures because the base pairing of m6A-U is weaker of than that of A-U [105,106]. A study on the SOX2 protein-protein interactome reveals that hnRNPC and hnRNPA2B1 can interact with SOX2 in GBM [107], suggesting that they may play key roles in maintaining the stemness of GSCs.
Actually, hnRNPC is one of the important physiological and cancer-related regulators (including GBM) of 3′ UTR processing and miRNA maturation [108]. hnRNPC has a higher abundance in higher grade GBM. Mechanically, hnRNPC directly binds to primary mir-21 (pri-mir-21) and promotes the processing of miR-21 that targets programmed cell death 4 (PDCD4), an important regulator of apoptosis and cellular survival. PDCD4 subsequently supports Akt and p70 S6 kinase (p70S6K) activation, and then promotes migratory and invasive activities, increases cell proliferation and protects GBM cells from etoposide-induced apoptosis [76].
hnRNPA2B1 is also an important modulator of pre-mRNA processing, mRNA metabolism and transportation in cells. hnRNPA2B1 protein is highly expressed in glioma tissues, correlated with glioma grades and related to poor prognosis [75]. Mechanically, hnRNPA2B1 promotes GBM cell viability, adhesion, migration, invasion, chemoresistance to TMZ and tumorigenecity as well as protects cells from apoptosis and reactive oxygen species (ROS) generation possibly via downregulating tumor suppressors such as cellular FLICE (FADD-like IL-1β-converting enzyme)-inhibitory protein (c-FLIP), bridging integrator 1 (BIN1) and WW domain containing oxidoreductase (WWOX), and upregulating phospho-signal transducer and activator of transcription 3 (p-STAT3), matrix metallopeptidase 2 (MMP-2), and the proto-oncogene RON (also known as macrophage stimulating 1 receptor, MST1R) [74,75]. These results indicate that hnRNPC and hnRNPA2B1 are possible m6A readers that contribute to GBM pathogenesis.
Collectively, from the current study, YTHDC1 is the only confirmed m6A reader that exerts its function in GBM dependently on its biological function in m6A RNA modification. Whether other readers’ functions are m6A-dependent remains unknown. However, most of their functions are dependent on their RNA binding capacity, implying that m6A in RNAs may be involved in their binding and their functions in GBM. This hypothesis needs to be further confirmed. As an important mark, m6A may have more readers than we can imagine. These readers are like messengers of m6A marks but beyond that, because they not only deliver message of m6A modified mRNA, but also determine their fate.
3.2. RNA m6Am Modification in GBM
N6,2′-O-dimethyladenosine (m6Am) is a terminal modification at mRNA cap (usually occurs in the first and sometimes the second nucleotide after the N7-methylguanosine (m7G) cap) in higher eukaryotic mRNAs and ncRNAs, expecially snRNAs, as well as viral RNAs [109]. m6Am plays essential roles in RNA splicing [110], snRNA biogenesis [111], mRNA stability [112] and cap-dependent translation [113]. In human beings, phosphorylated CTD interacting factor 1 (PCIF1, also known as cap-specific adenosine methyltransferase, CAPAM) and methyltransferase-like 4 (METTL4) are the m6Am writers, the m6A eraser FTO also functions as an m6Am eraser, and mRNA-decamping enzyme 2 (DCP2) may be one of the m6Am readers [109,110,111,114,115].
Recent evidence shows that FTO-mediated m6Am demethylation is an important regulatory mechanism in adjusting the stem-like properties of colorectal cancer cells [116], and many clues also imply that m6Am also emerges as an essential regulator in GBM (Figure 2, m6Am part). For instance, an integrated proteogenomic analysis in 17 GBM patient samples reveals that METTL4 is one of the top-ranked missense mutation genes [77]. DCP2 is upregulated in miR-338-5p overexpressed GBM cells and may participate in radiosensitivity and DNA damage response induced by miR-338-5p overexpression in GBM cells [78]. DCP2 is also one of top 20 hub genes of the optimal competing endogenous RNA (ceRNA) network in GBM and other cancers [117]. These results imply that m6Am probably participates in the development of GBM.
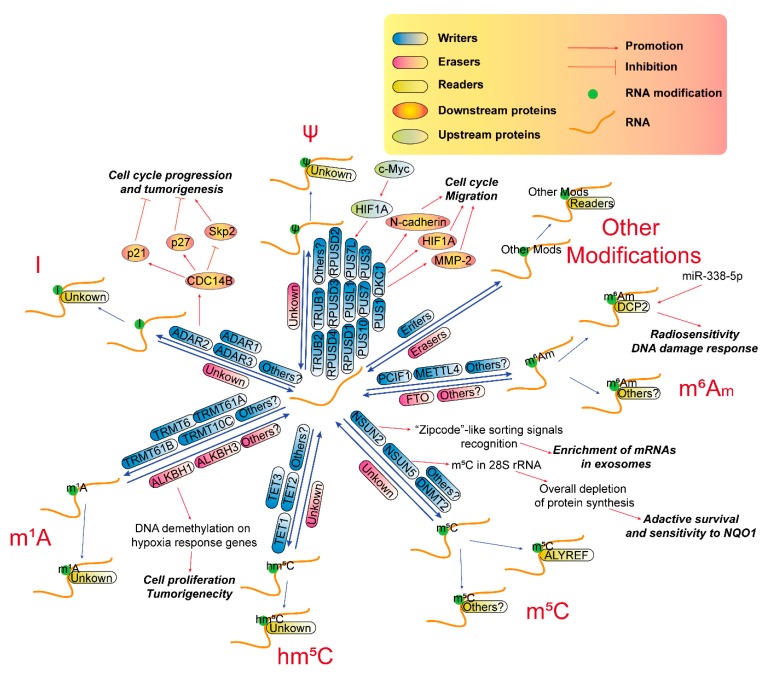
Figure 2.
Emerging functions of RNA modifications (except m6A) and their writers, erasers and readers in GBM. Abbreviations: Modifications: A-to-I, adenosine-to-inosine; m1A, N1-methyladenosine; m5C, 5-methylcytosine; m6Am, N6, 2′O-dimethyladenosine; hm5C, 5-hydroxymethylcytosine; I, inosine; ψ, pseudouridine. RNA-modifying proteins (RMPs): ADAR1/2/3, adenosine deaminase that acts on RNA 1/2/3; ALYREF, ALY/REF export factor; DCP2, mRNA- decapping enzyme 2; DKC1, dyskerin pseudouridine synthase 1; DNMT2, DNA (cytosine-5-)-methyltransferase 2; METTLE4, methyltransferase like 4; NSUN2/5, NOP2/Sun RNA methyltransferase 2/5; PCIF1, phosphorylated CTD interacting factor 1; PUS1/3/7/10, pseudouridine synthase 1/3/7/10; PUSL1, pseudouridine synthase like 1; PUS7L, pseudouridine synthase 7 like; RPUSD1/2/3/4, RNA pseudouridylate synthase domain-containing 1/2/3/4; TET1/2/3, ten-eleven translocation (TET) family; TRMT6/61A, tRNA methyltransferase 6/61A; TRMT61B, tRNA methyltransferase 61 homolog B; TRMT10C, tRNA methyltransferase 10C, mitochondrial RNase P subunit; TRUB1/2, TruB pseudouridine synthase family member 1/2. Others: c-Myc, avian myelocytomatosis viral oncogene homolog; CDC14B, cell division cycle 14B; NQO1, Quaking gene isoform 6; HIF1A, hypoxia-inducible factor 1 alpha; Mods, modifications; p70S6K, p70 S6 kinase;Skp2, S-phase kinase associated protein 2; PDCD4, programmed cell death 4; Skp2, S-phase kinase associated protein 2.
3.3. RNA m5C Modification in GBM
5-Methylcytosine (m5C) in DNA has been well described, however, it is not well understood in RNAs. Actually, m5C is not only confined within tRNAs and rRNAs, but also present in mRNA and ncRNAs [118]. RNA m5C usually responsible for stabilization of tRNA molecules via affecting Mg2+ binding, regulating mRNA translation, preventing recognition of endogenous RNAs by the innate immune system, and modulating RNA-dependent inheritance of certain phenotypes [119]. In human beings, NOP2/Sun RNA methyltransferase 2 (NSUN2, Misu), NSUN5 and DNA (cytosine-5-)-methyltransferase 2 (DNMT2) are confirmed as m5C writers, while ALY/REF export factor (ALYREF) is an m5C reader [119].
As a target of proto-oncogene c-Myc [120], NSUN2 has been shown to play pivotal roles in many neoplasms, such as breast cancer [121], colorectal cancer [122], ovarian cancer [123], head and neck squamous carcinoma [124] as well as esophageal squamous cell carcinoma [125]. In GBM, there is no direct evidences about the biological function of NSUN2. However, “zipcode”-like sorting signals in RNA that mediate the enrichment of mRNAs in exosomes of GBM cells [126] may probably be recognized by cytosolic Y-box binding protein 1 (YB-1) and NSUN2 [127]. NSUN5, another RNA methyltransferase that is responsible for m5C in the C3782 position of human 28S rRNA, undergoes epigenetic loss in gliomas, which drives an overall depletion of protein synthesis, resulting in an adaptive translational program for survival under cellular stress and renders gliomas sensitive to bio-activatable substrates of the stress-related enzyme NAD(P)H quinone dehydrogenase 1 (NQO1) [81,82]. The m5C reader, ALYREF, may be an overexpressed antigen in GBM, according to a study with a nanobody-based anti-proteome approach [83] and is upregulated in recurrent GBM, according to a transcriptome analysis [84], implying it may be a promising drug target for GBM. These evidences show m5C and its RMPs may also play important roles during the tumorigenesis of GBM (Figure 2, m5C part), indicating they are promising pharmaceutical targets for GBM treatment.
3.4. RNA hm5C Modification in GBM
In mammals, both DNA and RNA m5C are subject to oxidative processing by the ten-eleven translocation (TET) family (TET1/2/3), forming 5-hydroxymethylcytidine (hm5C) and 5-formyl-cytidine (f5C), which are considered as mediating substances during the demethylation pathway [128]. However, a recent study shows that m5C oxidation is a conserved process that could have critical regulatory functions inside cells [129]. For example, hm5C is distributed at lncRNA loci, involved in long-range chromatin interactions and positively correlated with lncRNA transcription [130]. TET1 is elevated in GBM [85], while TET2 expression is downregulated in glioma [86], and TET3 is epigenetically repressed in glioma [87]. 5-hydroxymethylcytidine in DNA has been recognized as important markers in GBM [131,132,133], however, the direct evidence of hm5C modification in RNA has never been explored in GBM (Figure 2, hm5C part).
3.5. RNA m1A Modification in GBM
N1-methyladenosine (m1A) is a reversible modification in tRNA, mRNA and lncRNAs, even in mitochondrial transcripts, and is critical for tRNA stability, and translation of mitochondrial DNA-encoded proteins [134]. In nucleus, m1A modifications in pre-tRNA and pre-mRNA are catalyzed by tRNA methyltransferase 6/61A (TRMT6-TRMT61A) catalytic complex and are erased by ALKBH1 and ALKBH3, respectively [135,136]. Whereas in mitochondrion, m1A methylation in mt-tRNA and mt-mRNA are installed by tRNA methyltransferase 61 homolog B (TRMT61B) and tRNA methyltransferase 10C, mitochondrial RNase P subunit (TRMT10C) [137]. Recently, some RMPs of m1A, both writers and erasers, seem to involve in the progression of GBM (Figure 2, m1A part). TRMT6 and TRMT61A expression is significantly upregulated in highly aggressive GBM compared with grade II/III gliomas [79]. TRMT61A is a target of HIF1A and is dowregulated after c-Myc inhibition in GBM under hypoxia [138]. The eraser ALKBH1 also seems to be an important regulator in GBM, since targeting ALKBH1 in patient-derived GBM models induces cell proliferation inhibition and extends the survival of tumor-bearing mice. However, this effect may be caused by ALKBH1-induced DNA demethylation on hypoxia response genes, while not ALKBH1-controlled tRNA stability [80].
3.6. RNA A-To-I Modification in GBM
A-to-I RNA editing is abundant in the human transcriptome and plays essential roles in RNA processing, such as post-transcriptionally altering codons, introducing or removing splice sites, and affecting the base pairing of the RNA molecule with itself or with other RNAs [139]. All 3 editing mediating enzymes, ADAR1 (ADAR), ADAR2 (ADARB1), and ADAR3 (ADARB2), is downregulated in brain tumors and ADARB2 is also negatively correlated with GBM grades. ADAR1 and ADAR2 overexpression in GBM cell line inhibit cell proliferation, suggesting that reduced A-to-I editing contributes to the pathogenesis of this disease [88]. In addition, abnormal expression of an ADAR2 alternative splicing variant also downregulates A-to-I RNA editing in glioma [89], because ADAR2 can prevent GBM tumor growth via modulating an important cell cycle pathway involving S-phase kinase associated protein 2 (Skp2), p21 and p27 proteins by introducing an A-to-I editing in the pre-mRNA of a phosphatase cell division cycle 14B (CDC14B) [90]. Additionally, A-to-I editing in miR-376a-3p is attenuated in GBM, thus promoting invasiveness of this disease [140]. These results show that A-to-I editing is essential for GBM progression (Figure 2, I part), and RMPs of A-to-I modification may be potential targets for GBM treatment.
3.7. RNA ψ Modification in GBM
Pseudouridine (ψ), the most abundant RNA modification, is occurs in almost all kinds of RNAs, including tRNA, mRNA, snRNA, nucleolar RNA (snoRNA), and rRNA [141,142]. Ψ is important for rRNA folding, translational fidelity and rate, stress response, tRNA codon–anticodon base-pairing, snRNP biogenesis and pre-mRNA splicing [143]. There are 13 pseudouridine synthases (PUSs, writers) including PUS1, PUS3, PUS7, PUS10, pseudouridine synthase like 1 (PUSL1), pseudouridine synthase 7 like (PUS7L), TruB pseudouridine synthase family member 1 (TRUB1), TRUB2, RNA pseudouridylate synthase domain-containing 1 (RPUSD1), RPUSD2, RPUSD3, RPUSD4 and dyskerin pseudouridine synthase 1 (DKC1) [144,145]. Recently, emerging evidence also shows that ψ may play pivotal roles during gliomagenesis. One of the ψ writers, DKC1, is significantly upregulated in glioma and correlates with the WHO stages of tumors. DKC1 promotes glioma cell growth by stimulating cell cycle progression and activates migration via upregulating N-cadherin, hypoxia inducible factor 1 subunit α (HIF1A), and MMP-2 expression [91]. PUS7L, as a target of HIF1A, is also downregulated after c-Myc inhibition in GBM under hypoxia [138]. These emerging evidences provide us the importance of pseudouridine modification in RNAs during the pathogenesis of GBM (Figure 2, ψ part).
3.8. Other RNA Modifications in GBM
Although most of the other RNA modifications are not well described, there are also some studies regarding the effect of these RNA modifications on GBM progression. For instance, ADAR2 and ADAR3 binds to the pre-mRNA of glutamate receptor subunit B, glutamate receptor ionotropic AMPA (GRIA2), contributing to a RNA editing at the Q/R site by modifying a codon replacing the glutamine (Q) with arginine (R). ADAR2/3 deficiency in GBM leads to increased unedited GRIA2 subunits, thereby leads to a calcium-permeable glutamate receptor, which can promote cell migration and tumor invasion [146]. Cleavage factor Im 25 (CFIm25) restricts mRNA proximal poly(A) site usage via alternative polyadenylation (APA). CFIm25 knockdown leads to at least 1,450 genes with shortened 3′UTRs and marked increases in the expression of several oncogenes in GBM, including cyclin D1 (CCND1), glutaminase (GLS) and methyl-CpG-binding protein 2 (MECP2), thereby enhancing their tumorigenesis [147]. Mechanically, shortened 3′UTR truncation of oncogenic transcripts relieves intrinsic microRNA- and AU-rich-element-mediated repression. These new findings unravel a wide horizon for us to explore the implements of RNA modifications in cancer treatment. With the developing technology, much more modifications in RNA and their RMPs, as well as their connections with human diseases, especially cancers, will be disclosed.
4. Potential Clinical Implications of RNA Modifications in GBM
The descriptions above indicate that RNA modifications are important for tumorigenesis and aggressiveness in GBM. Chemical modifications in both DNA and proteins have been well studied in cancer biology, and many chemical modifications such as DNA methylation, histone modification and EGFR post-translational modifications, as well as some of their modification enzymes have been shown to have clinical significance in both the diagnosis and therapy [148,149,150,151,152,153,154,155]. Importantly, these chemical modifications are also important for GBM diagnosis and therapy [8,156,157,158]. Actually, as kinds of chemical modifications, RNA modifications and their RMPs also give promise to both the diagnosis and therapeutics of GBM in the future.
4.1. Potential Diagnostic Implications of RNA Modifications in GBM
Epigenetic alterations are considered as promising markers for GBM diagnosis [159,160,161,162], although this study is just the first step for their possible applications in GBM diagnosis. Besides, some epigenetic status really account for the outcome of GBM. For example, the epigenetic silencing of the O6-methylguanine–DNA methyltransferase (MGMT) significantly affects the TMZ treatment of GBM [163]. Therefore, MGMT promoter methylation status is an important biomarker to predict survival and TMZ response in patients with GBM [164].
As a kind of chemical modifications in RNA, the alterations of their regulations may also predict the prognosis or be used for diagnosis of GBM. Actually, m6A in in peripheral blood RNA has shown to be a predictive biomarker for gastric cancer [165]. Methylation levels in miRNAs are also potential diagnostic biomarkers for early-stage gastrointestinal cancers [166]. The m6A erasers ALKBH5 and FTO are also shown to be prognostic biomarkers in patients with clear cell renal carcinoma [167]. From Table 1, some RNA modification enzymes are also tightly related to the grades and may predict the prognosis of GBM. However, the specificity of their expression should be further confirmed. Besides, some RNA modifications in peripheral blood RNA may also be promising methods for GBM diagnosis. In the future, it can be suspected that RNA modification marks as well as their RMPs’ expression may be important biomarkers for the diagnosis of GBM.
4.2. Potential Therapeutical Implications of RNA Modifications in GBM
Covalent modifications in both DNA, histones and other proteins have shown promising probability for cancer treatment. Actually, some epigenetic drugs such as azacitidine, 5-aza-2′-deoxycytidine, suberoylanilide hydroxamic acid (SAHA), romidepsin, belinostat, panobinostat, has been approved by the United States Food and Drug Administration (FDA) and chidamide has been approved by China FDA to treat some T-cell lymphoma or multiple myeloma, and many other epigenetic drugs on cancers are ongoing clinical trials [10]. Besides, drugs that target modulators of protein post-translational modifications, such as E3 ubiquitin ligases including mouse double minute 2 homolog (MDM2), sphase kinaseassociated protein 2 (SKP2), speckle type BTB/POZ protein (SPOP), cellular inhibitor of apoptosis 2 (cIAP) and anaphase-promoting complex/cyclosome (APC/C), are also been evaluated in clinical and preclinical settings [168]. Importantly, some epigenetic drugs such as vorinostat, panobinostat, romidepsin and valproic acid alone or in combination with other chemotherapeutic methods such as TMZ treatment or radiotherapy are studying in phase I/II clinical trials [169].
As kinds of covalent modifications in RNA, their potentials in cancers like malignant GBM are being explored. Since more than 170 kinds of modifications, it is a wider world for us to explore. Specific antagonist against writers, erasers and readers of RNA modifications needs to be screened, designed and identified, as well as their biological functions and clinical applications in cancer treatment also need to be extensively studied. In fact, several pharmaceutical companies are planning to develop small-molecule inhibitors of the METTL3–METTL14 complex and ADAR [170]. Several compounds with piperidine and piperazine rings have exceptionally high docking efficiencies to METTL3-METTL14-WTAP complex, thereby activating RNA methylation [171]. Recently, a small molecule inhibitor of METTL3 is also shown to curb the development of acute myeloid leukemia (AML) in vivo [172,173]. We believe that effective and efficient therapies regarding RNA modifications will soon be developed to treat malignant neoplasms such as GBM.
5. Conclusions and Perspectives
From the collective evidence, we can conclude that RNA modifications play increasingly important roles in the tumorigenesis and development of GBM. Some of the writers, erasers and even readers of RNA modifications are potential biomarkers for the diagnosis and drug targets for treatment of GBM [30]. Despite the fact that the present study of epi-transcriptome is just a tip of the iceberg, its fast-development will comprehensively expand our sights into cancers.
Nevertheless, most of these studies show that no direct connections of their pro-oncogenic or anti-oncogenic roles are dependent on their capacities of RNA modifications. That is to say that the functions of these modulators in GBM may be not dependent on its function in RNA modifications. Therefore, the RNA modifications and their detailed targets regulated by RMPs should be comprehensively explored in the future. To explore these, much fundamental work should be done before a better understanding of RNA modifications in cancers.
The first task is to identify the physiological and biological function of more than 170 kinds of RNA modifications and their RMPs. However, it is a very challenging and difficult work to decode the complex layer of the epitranscriptome. Although some modifications such as m6A can be detected by fast-developing antibody-based NGS through a mechanism of capturing and sequencing of immunoprecipitated RNA targets [35,174,175], however, this technology has some limitations such as low detection accurancy for low-abundance modifications, false-positive mapping results, not-specific capturing antibody, RNA samples contaminations, and Dimroth rearrangement that converse m1A to m6A [176]. Therefore, it is urgent to develop a novel accurate detecting system that are not dependent on immunoprecipitation.
After solving these problems, exploring the connections of RNA modifications with GBM pathogenesis, prognosis and diagnosis, digging the mechanisms underlying their functions, designing of specific small molecules target RNA modifications, and detecting these targeted drugs penetrating the blood-brain barrier for GBM treatment can be possible. The results of basic research about RNA modifications in GBM will also soon be tested in clinical trials and be applied in the clinic.
Author Contributions
Z.D. reviewed the literature, designed the tables and figures, and wrote the manuscript. H.C. reviewed and revised the text. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Nos. 81902664, 81872071 and 81672502), the National Key Research and Development Program of China (Nos. 2016YFC1302204 and 2017YFC1308600), and the Natural Science Foundation of Chongqing (No. cstc2019jcyj-zdxmX0033).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ostrom Q.T., Gittleman H., de Blank P.M., Finlay J.L., Gurney J.G., McKean-Cowdin R., Stearns D.S., Wolff J.E., Liu M., Wolinsky Y., et al. American brain tumor association adolescent and young adult primary brain and central nervous system tumors diagnosed in the United States in 2008–2012. Neuro Oncol. 2016;18(Suppl. 1):i1–i50. doi: 10.1093/neuonc/nov297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jemal A., Siegel R., Xu J., Ward E. Cancer statistics, 2010. CA Cancer J. Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 3.Hottinger A.F., Stupp R., Homicsko K. Standards of care and novel approaches in the management of glioblastoma multiforme. Chin. J. Cancer. 2014;33:32–39. doi: 10.5732/cjc.013.10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alexander B.M., Cloughesy T.F. Adult glioblastoma. J. Clin. Oncol. 2017;35:2402–2409. doi: 10.1200/JCO.2017.73.0119. [DOI] [PubMed] [Google Scholar]
- 5.Indraccolo S., Lombardi G., Fassan M., Pasqualini L., Giunco S., Marcato R., Gasparini A., Candiotto C., Nalio S., Fiduccia P., et al. Genetic, epigenetic, and immunologic profiling of MMR-deficient relapsed glioblastoma. J. Clin. Cancer Res. 2019;25:1828–1837. doi: 10.1158/1078-0432.CCR-18-1892. [DOI] [PubMed] [Google Scholar]
- 6.Neftel C., Laffy J., Filbin M.G., Hara T., Shore M.E., Rahme G.J., Richman A.R., Silverbush D., Shaw M.L., Hebert C.M., et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell. 2019;178:835–849. doi: 10.1016/j.cell.2019.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dong Z., Cui H. Epigenetic modulation of metabolism in glioblastoma. Semin. Cancer Biol. 2019;57:45–51. doi: 10.1016/j.semcancer.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Romani M., Pistillo M.P., Banelli B. Epigenetic targeting of glioblastoma. Front. Oncol. 2018;8:448. doi: 10.3389/fonc.2018.00448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang B., Zhang H., Gu L., Ye B., Jian Z., Stary C., Xiong X. Advances in immunotherapy for glioblastoma multiforme. J. Immunol. Res. 2017;2017:3597613. doi: 10.1155/2017/3597613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng Y., He C., Wang M., Ma X., Mo F., Yang S., Han J., Wei X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal. Transduct. Target. Ther. 2019;4:62. doi: 10.1038/s41392-019-0095-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cavalli G., Heard E. Advances in epigenetics link genetics to the environment and disease. Nature. 2019;571:489–499. doi: 10.1038/s41586-019-1411-0. [DOI] [PubMed] [Google Scholar]
- 12.Zhu S., Dong Z., Ke X., Hou J., Zhao E., Zhang K., Wang F., Yang L., Xiang Z., Cui H. The roles of sirtuins family in cell metabolism during tumor development. Semin. Cancer Biol. 2019;57:59–71. doi: 10.1016/j.semcancer.2018.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Berdasco M., Esteller M. Clinical epigenetics: Seizing opportunities for translation. Nat. Rev. Genet. 2019;20:109–127. doi: 10.1038/s41576-018-0074-2. [DOI] [PubMed] [Google Scholar]
- 14.Yang L., Lei Q., Li L., Yang J., Dong Z., Cui H. Silencing or inhibition of H3K79 methyltransferase DOT1L induces cell cycle arrest by epigenetically modulating c-Myc expression in colorectal cancer. Clin. Epigenet. 2019;11:199. doi: 10.1186/s13148-019-0778-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hsiao Y.-H.E., Bahn J.H., Yang Y., Lin X., Tran S., Yang E.-W., Quinones-Valdez G., Xiao X. RNA editing in nascent RNA affects pre-mRNA splicing. Genome Res. 2018;28:812–823. doi: 10.1101/gr.231209.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanchez-Diaz P., Penalva L. Post-Transcription meets post-genomic: The saga of RNA binding proteins in a new era. RNA Biol. 2006;3:101–109. doi: 10.4161/rna.3.3.3373. [DOI] [PubMed] [Google Scholar]
- 17.Bandziulis R., Swanson M., Dreyfuss G. RNA-Binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- 18.Hentze M.W., Castello A., Schwarzl T., Preiss T. A brave new world of RNA-binding proteins. Nat. Rev. Mol. Cell Biol. 2018;19:327–341. doi: 10.1038/nrm.2017.130. [DOI] [PubMed] [Google Scholar]
- 19.Sullenger B.A., Nair S. From the RNA world to the clinic. Science. 2016;352:1417–1420. doi: 10.1126/science.aad8709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong Z., Pu L., Cui H. Mitoepigenetics and its emerging roles in cancer. Front. Cell Dev. Biol. 2020;8:4. doi: 10.3389/fcell.2020.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boccaletto P., Machnicka M.A., Purta E., Piątkowski P., Bagiński B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A. MODOMICS: A database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018;46:D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang H., Weng H., Deng X., Chen J. RNA modifications in cancer: Functions, mechanisms, and therapeutic implications. Ann. Rev. Cancer. 2019;4:221–240. doi: 10.1146/annurev-cancerbio-030419-033357. [DOI] [Google Scholar]
- 23.He L., Li J., Wang X., Ying Y., Xie H., Yan H., Zheng X., Xie L. The dual role of N6-methyladenosine modification of RNAs is involved in human cancers. J. Cell. Mol. Med. 2018;22:4630–4639. doi: 10.1111/jcmm.13804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thapar R., Bacolla A., Oyeniran C., Brickner J.R., Chinnam N.B., Mosammaparast N., Tainer J. RNA modifications: Reversal mechanisms and cancer. Biochemistry. 2019;58:312–329. doi: 10.1021/acs.biochem.8b00949. [DOI] [PubMed] [Google Scholar]
- 25.Close P., Bose D., Chariot A., Leidel S.A. Cancer and Noncoding RNAs. Elsevier; Amsterdam, The Netherlands: 2018. Dynamic regulation of tRNA modifications in cancer; pp. 163–186. [Google Scholar]
- 26.Davis F.F., Allen F. Ribonucleic acids from yeast which contain a fifth nucleotide. J. Biol. Chem. 1957;227:907–915. [PubMed] [Google Scholar]
- 27.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilbert W.V., Bell T.A., Schaening C. Messenger RNA modifications: Form, distribution, and function. Science. 2016;352:1408–1412. doi: 10.1126/science.aad8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee M., Kim B., Kim V.N. Emerging roles of RNA modification: m6A and U-Tail. Cell. 2014;158:980–987. doi: 10.1016/j.cell.2014.08.005. [DOI] [PubMed] [Google Scholar]
- 30.Esteve-Puig R., Bueno-Costa A., Esteller M. Writers, readers and erasers of RNA modifications in cancer. Cancer Lett. 2020;474:127–137. doi: 10.1016/j.canlet.2020.01.021. [DOI] [PubMed] [Google Scholar]
- 31.Jackman J.E., Alfonzo J. Transfer RNA modifications: nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA. 2013;4:35–48. doi: 10.1002/wrna.1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Decatur W.A., Fournier M. rRNA modifications and ribosome function. Trends Biochem. Sci. 2002;27:344–351. doi: 10.1016/S0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 33.Ganot P., Jády B.E., Bortolin M.-L., Darzacq X., Kiss T. Nucleolar factors direct the 2′-O-ribose methylation and pseudouridylation of U6 spliceosomal RNA. Mol. Cell. Biol. 1999;19:6906–6917. doi: 10.1128/MCB.19.10.6906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Delaunay S., Frye M. RNA modifications regulating cell fate in cancer. Nat. Cell Biol. 2019;21:552–559. doi: 10.1038/s41556-019-0319-0. [DOI] [PubMed] [Google Scholar]
- 35.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 36.Liu J., Li K., Cai J., Zhang M., Zhang X., Xiong X., Meng H., Xu X., Huang Z., Peng J., et al. Landscape and Regulation of m6A and m6Am methylome across human and mouse tissues. Mol. Cell. 2020;77:426–440. doi: 10.1016/j.molcel.2019.09.032. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., et al. A METTL3–METTL14 complex mediates mammalian nuclear RNA N 6-adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.-G., et al. N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Balacco D.L., Soller M. The m6A writer: Rise of a machine for growing tasks. Biochemistry. 2018;58:363–378. doi: 10.1021/acs.biochem.8b01166. [DOI] [PubMed] [Google Scholar]
- 42.Lence T., Paolantoni C., Worpenberg L., Roignant J.-Y. Mechanistic insights into m6A RNA enzymes. Biochim. Biophys. Acta Gene Regul. Mech. 2019;1862:222–229. doi: 10.1016/j.bbagrm.2018.10.014. [DOI] [PubMed] [Google Scholar]
- 43.Stoilov P., Rafalska I., Stamm S. YTH: A new domain in nuclear proteins. Trends Biochem. Sci. 2002;27:495–497. doi: 10.1016/S0968-0004(02)02189-8. [DOI] [PubMed] [Google Scholar]
- 44.Luo S., Tong L. Molecular basis for the recognition of methylated adenines in RNA by the eukaryotic YTH domain. Proc. Natl. Acad. Sci. USA. 2014;111:13834–13839. doi: 10.1073/pnas.1412742111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Visvanathan A., Patil V., Abdulla S., Hoheisel D.J., Somasundaram K. N6-Methyladenosine landscape of glioma stem-like cells: METTL3 is essential for the expression of actively transcribed genes and sustenance of the oncogenic signaling. Genes. 2019;10:141. doi: 10.3390/genes10020141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y., Hsu P.J., Chen Y.-S., Yang Y.-G. Dynamic transcriptomic m6A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 2018;28:616–624. doi: 10.1038/s41422-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meyer K.D., Jaffrey S.R. Rethinking m6A readers, writers, and erasers. Ann. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yu J., Chen M., Huang H., Zhu J., Song H., Zhu J., Park J., Ji S.-J. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2017;46:1412–1423. doi: 10.1093/nar/gkx1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y. m6A mRNA methylation facilitates resolution of naïve pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 50.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao W., Adhikari S., Dahal U., Chen Y.-S., Hao Y.-J., Sun B.-F., Sun H.-Y., Li A., Ping X.-L., Lai W.-Y. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 52.Wang X., Lu Z., Gomez A., Hon G.C., Yue Y., Han D., Fu Y., Parisien M., Dai Q., Jia G. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N6-methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen M., Wei L., Law C.T., Tsang F.H.C., Shen J., Cheng C.L.H., Tsang L.H., Ho D.W.H., Chiu D.K.C., Lee J. RNA N6-methyladenosine methyltransferase-like 3 promotes liver cancer progression through YTHDF2-dependent posttranscriptional silencing of SOCS2. Hepatology. 2018;67:2254–2270. doi: 10.1002/hep.29683. [DOI] [PubMed] [Google Scholar]
- 56.Visvanathan A., Patil V., Arora A., Hegde A.S., Arivazhagan A., Santosh V., Somasundaram K. Essential role of METTL3-mediated m6A modification in glioma stem-like cells maintenance and radioresistance. Oncogene. 2018;37:522–533. doi: 10.1038/onc.2017.351. [DOI] [PubMed] [Google Scholar]
- 57.Li F., Yi Y., Miao Y., Long W., Long T., Chen S., Cheng W., Zou C., Zheng Y., Wu X., et al. N6-Methyladenosine modulates nonsense-mediated mRNA decay in human glioblastoma. J. Cancer Res. 2019;79:5785–5798. doi: 10.1158/0008-5472.CAN-18-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang W., Li J., Lin F., Guo J., Zhao J. Identification of N6-methyladenosine-related lncRNAs for patients with primary glioblastoma. Neurosurg. Rev. 2020 doi: 10.1007/s10143-020-01238-x. [DOI] [PubMed] [Google Scholar]
- 59.Cui Q., Shi H., Ye P., Li L., Qu Q., Sun G., Sun G., Lu Z., Huang Y., Yang C.-G., et al. mA RNA methylation regulates the self-renewal and tumorigenesis of glioblastoma stem cells. Cell Rep. 2017;18:2622–2634. doi: 10.1016/j.celrep.2017.02.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li F., Zhang C., Zhang G. m6A RNA methylation controls proliferation of human glioma cells by influencing cell apoptosis. Cytogenet. Genome Res. 2019;159:119–125. doi: 10.1159/000499062. [DOI] [PubMed] [Google Scholar]
- 61.Jin D.I., Lee S.W., Han M.E., Kim H.J., Seo S.A., Hur G.Y., Jung S., Kim B.S., Oh S.O. Expression and roles of W ilms’ tumor 1-associating protein in glioblastoma. Cancer Sci. 2012;103:2102–2109. doi: 10.1111/cas.12022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xi Z., Xue Y., Zheng J., Liu X., Ma J., Liu Y. WTAP expression predicts poor prognosis in malignant glioma patients. J. Mol. Neurosci. 2016;60:131–136. doi: 10.1007/s12031-016-0788-6. [DOI] [PubMed] [Google Scholar]
- 63.Xi Z., Wang P., Xue Y., Shang C., Liu X., Ma J., Li Z., Li Z., Bao M., Liu Y. Overexpression of miR-29a reduces the oncogenic properties of glioblastoma stem cells by downregulating Quaking gene isoform 6. Oncotarget. 2017;8:24949–24963. doi: 10.18632/oncotarget.15327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Su R., Dong L., Li C., Nachtergaele S., Wunderlich M., Qing Y., Deng X., Wang Y., Weng X., Hu C. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2018;172:90–105. doi: 10.1016/j.cell.2017.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bögler O., et al. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang R.J., Li J.W., Bao B.H., Wu H.C., Du Z.H., Su J.L., Zhang M.H., Liang H.Q. MicroRNA-873 (miRNA-873) inhibits glioblastoma tumorigenesis and metastasis by suppressing the expression of IGF2BP1. J. Biol. Chem. 2015;290:8938–8948. doi: 10.1074/jbc.M114.624700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Luo Y., Sun R., Zhang J., Sun T., Liu X., Yang B. miR-506 inhibits the proliferation and invasion by targeting IGF2BP1 in glioblastoma. Am. J. Transl. Res. 2015;7:2007–2014. [PMC free article] [PubMed] [Google Scholar]
- 68.Mu Q., Wang L., Yu F., Gao H., Lei T., Li P., Liu P., Zheng X., Hu X., Chen Y., et al. Imp2 regulates GBM progression by activating IGF2/PI3K/Akt pathway. Cancer Biol. Ther. 2015;16:623–633. doi: 10.1080/15384047.2015.1019185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Degrauwe N., Schlumpf T.B., Janiszewska M., Martin P., Cauderay A., Provero P., Riggi N., Suva M.L., Paro R., Stamenkovic I. The RNA binding protein IMP2 preserves glioblastoma stem cells by preventing let-7 target gene silencing. Cell Rep. 2016;15:1634–1647. doi: 10.1016/j.celrep.2016.04.086. [DOI] [PubMed] [Google Scholar]
- 70.Janiszewska M., Suva M.L., Riggi N., Houtkooper R.H., Auwerx J., Clement-Schatlo V., Radovanovic I., Rheinbay E., Provero P., Stamenkovic I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26:1926–1944. doi: 10.1101/gad.188292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mineo M., Ricklefs F., Rooj A.K., Lyons S.M., Ivanov P., Ansari K.I., Nakano I., Chiocca E.A., Godlewski J., Bronisz A. The long non-coding RNA HIF1A-AS2 facilitates the maintenance of mesenchymal glioblastoma stem-like cells in hypoxic niches. Cell Rep. 2016;15:2500–2509. doi: 10.1016/j.celrep.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liang H., Ding X., Zhou C., Zhang Y., Xu M., Zhang C., Xu L. Knockdown of eukaryotic translation initiation factors 3B (EIF3B) inhibits proliferation and promotes apoptosis in glioblastoma cells. Neurol. Sci. 2012;33:1057–1062. doi: 10.1007/s10072-011-0894-8. [DOI] [PubMed] [Google Scholar]
- 73.Gambale A., Russo R., Andolfo I., Quaglietta L., De Rosa G., Contestabile V., De Martino L., Genesio R., Pignataro P., Giglio S., et al. Germline mutations and new copy number variants among 40 pediatric cancer patients suspected for genetic predisposition. Clin. Genet. 2019;96:359–365. doi: 10.1111/cge.13600. [DOI] [PubMed] [Google Scholar]
- 74.Deng J., Chen S., Wang F., Zhao H., Xie Z., Xu Z., Zhang Q., Liang P., Zhai X., Cheng Y. Effects of hnRNP A2/B1 knockdown on inhibition of glioblastoma cell invasion, growth and survival. Mol. Neurobiol. 2016;53:1132–1144. doi: 10.1007/s12035-014-9080-3. [DOI] [PubMed] [Google Scholar]
- 75.Golan-Gerstl R., Cohen M., Shilo A., Suh S.S., Bakacs A., Coppola L., Karni R. Splicing factor hnRNP A2/B1 regulates tumor suppressor gene splicing and is an oncogenic driver in glioblastoma. Cancer Res. 2011;71:4464–4472. doi: 10.1158/0008-5472.CAN-10-4410. [DOI] [PubMed] [Google Scholar]
- 76.Park Y.M., Hwang S.J., Masuda K., Choi K.M., Jeong M.R., Nam D.H., Gorospe M., Kim H.H. Heterogeneous nuclear ribonucleoprotein C1/C2 controls the metastatic potential of glioblastoma by regulating PDCD4. Mol. Cell. Biol. 2012;32:4237–4244. doi: 10.1128/MCB.00443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Asif S., Fatima R., Krc R., Bennett J., Raza S. Comparative proteogenomic characterization of glioblastoma. CNS Oncol. 2019;8 doi: 10.2217/cns-2019-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Besse A., Sana J., Lakomy R., Kren L., Fadrus P., Smrcka M., Hermanova M., Jancalek R., Reguli S., Lipina R. MiR-338-5p sensitizes glioblastoma cells to radiation through regulation of genes involved in DNA damage response. Tumor Biol. 2016;37:7719–7727. doi: 10.1007/s13277-015-4654-x. [DOI] [PubMed] [Google Scholar]
- 79.Macari F., El-Houfi Y., Boldina G., Xu H., Khoury-Hanna S., Ollier J., Yazdani L., Zheng G., Bieche I., Legrand N., et al. TRM6/61 connects PKCalpha with translational control through tRNAi(Met) stabilization: Impact on tumorigenesis. Oncogene. 2016;35:1785–1796. doi: 10.1038/onc.2015.244. [DOI] [PubMed] [Google Scholar]
- 80.Xie Q., Wu T.P., Gimple R.C., Li Z., Prager B.C., Wu Q., Yu Y., Wang P., Wang Y., Gorkin D.U., et al. N6-methyladenine DNA Modification in Glioblastoma. Cell. 2018;175:1228–1243. doi: 10.1016/j.cell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Janin M., Ortiz-Barahona V., de Moura M.C., Martínez-Cardús A., Llinàs-Arias P., Soler M., Nachmani D., Pelletier J., Schumann U., Calleja-Cervantes M.E., et al. Epigenetic loss of RNA-methyltransferase NSUN5 in glioma targets ribosomes to drive a stress adaptive translational program. Acta Neuropathol. 2019;138:1053–1074. doi: 10.1007/s00401-019-02062-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou J., Vincent K., Findlay S., Choi D., Godbout R., Postovit L.-M., Fu Y. 61 Functional characterization of ribosomal RNA methyltransferase NSUN5 in glioblastoma. Can. J. Neurol. Sci. 2018;45:S10–S11. doi: 10.1017/cjn.2018.289. [DOI] [Google Scholar]
- 83.Jovčevska I., Zupanec N., Urlep Ž., Vranič A., Matos B., Stokin C.L., Muyldermans S., Myers M.P., Buzdin A.A., Petrov I., et al. Differentially expressed proteins in glioblastoma multiforme identified with a nanobody-based anti-proteome approach and confirmed by OncoFinder as possible tumor-class predictive biomarker candidates. Oncotarget. 2017;8:44141–44158. doi: 10.18632/oncotarget.17390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Narsia N., Ramagiri P., Ehrmann J., Kolar Z. Transcriptome analysis reveals distinct gene expression profiles in astrocytoma grades II-IV. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2017;161:261–271. doi: 10.5507/bp.2017.020. [DOI] [PubMed] [Google Scholar]
- 85.Takai H., Masuda K., Sato T., Sakaguchi Y., Suzuki T., Suzuki T., Koyama-Nasu R., Nasu-Nishimura Y., Katou Y., Ogawa H. 5-Hydroxymethylcytosine plays a critical role in glioblastomagenesis by recruiting the CHTOP-methylosome complex. Cell Rep. 2014;9:48–60. doi: 10.1016/j.celrep.2014.08.071. [DOI] [PubMed] [Google Scholar]
- 86.García M.G., Carella A., Urdinguio R.G., Bayón G.F., Lopez V., Tejedor J.R., Sierra M.I., García-Toraño E., Santamarina P., Perez R. Epigenetic dysregulation of TET2 in human glioblastoma. Oncotarget. 2018;9:25922–25934. doi: 10.18632/oncotarget.25406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Carella A., Tejedor J.R., García M.G., Urdinguio R.G., Bayón G.F., Sierra M., López V., García-Toraño E., Santamarina-Ojeda P., Pérez R. Epigenetic downregulation of TET3 reduces genome-wide 5hmC levels and promotes glioblastoma tumorigenesis. Int. J. Cancer. 2020;146:373–387. doi: 10.1002/ijc.32520. [DOI] [PubMed] [Google Scholar]
- 88.Paz N., Levanon E.Y., Amariglio N., Heimberger A.B., Ram Z., Constantini S., Barbash Z.S., Adamsky K., Safran M., Hirschberg A. Altered adenosine-to-inosine RNA editing in human cancer. Genome Res. 2007;17:1586–1595. doi: 10.1101/gr.6493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wei J., Li Z., Du C., Qi B., Zhao X., Wang L., Bi L., Wang G., Zhang X., Su X., et al. Abnormal expression of an ADAR2 alternative splicing variant in gliomas downregulates adenosine-to-inosine RNA editing. Acta Neurochir. 2014;156:1135–1142. doi: 10.1007/s00701-014-2004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Galeano F., Rossetti C., Tomaselli S., Cifaldi L., Lezzerini M., Pezzullo M., Boldrini R., Massimi L., Di Rocco C.M., Locatelli F., et al. ADAR2-Editing activity inhibits glioblastoma growth through the modulation of the CDC14B/Skp2/p21/p27 axis. Oncogene. 2013;32:998–1009. doi: 10.1038/onc.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Miao F.-A., Chu K., Chen H.-R., Zhang M., Shi P.-C., Bai J., You Y.P. Increased DKC1 expression in glioma and its significance in tumor cell proliferation, migration and invasion. Investig. New Drugs. 2019;37:1177–1186. doi: 10.1007/s10637-019-00748-w. [DOI] [PubMed] [Google Scholar]
- 92.Chai R.-C., Wu F., Wang Q.-X., Zhang S., Zhang K.-N., Liu Y.-Q., Zhao Z., Jiang T., Wang Y.-Z., Kang C.-S. m6A RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging. 2019;11:1204–1225. doi: 10.18632/aging.101829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wen J., Lv R., Ma H., Shen H., He C., Wang J., Jiao F., Liu H., Yang P., Tan L., et al. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Chow R.D., Guzman C.D., Wang G., Schmidt F., Youngblood M.W., Ye L., Errami Y., Dong M.B., Martinez M.A., Zhang S., et al. AAV-Mediated direct In Vivo CRISPR screen identifies functional suppressors in glioblastoma. Nat. Neurosci. 2017;20:1329–1341. doi: 10.1038/nn.4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 96.Huang H., Weng H., Sun W., Qin X., Shi H., Wu H., Zhao B.S., Mesquita A., Liu C., Yuan C.L., et al. Recognition of RNA N 6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell Biol. 2018;20:285–295. doi: 10.1038/s41556-018-0045-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Suvasini R., Shruti B., Thota B., Shinde S.V., Friedmann-Morvinski D., Nawaz Z., Prasanna K.V., Thennarasu K., Hegde A.S., Arivazhagan A., et al. Insulin growth factor-2 binding protein 3 (IGF2BP3) is a glioblastoma-specific marker that activates phosphatidylinositol 3-kinase/mitogen-activated protein kinase (PI3K/MAPK) pathways by modulating IGF-2. J. Biol. Chem. 2011;286:25882–25890. doi: 10.1074/jbc.M110.178012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kleinschmidt-DeMasters B.K., Donson A.M., Vogel H., Foreman N.K. Pilomyxoid astrocytoma (PMA) shows significant differences in gene expression vs. pilocytic astrocytoma (PA) and variable tendency toward maturation to PA. Brain Pathol. 2015;25:429–440. doi: 10.1111/bpa.12239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bhargava S., Patil V., Shah R.A., Somasundaram K. IGF2 mRNA binding protein 3 (IMP3) mediated regulation of transcriptome and translatome in glioma cells. Cancer Biol. Ther. 2018;19:42–52. doi: 10.1080/15384047.2017.1323601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu C., Ma H., Qi G., Chen F., Chu J. Insulin-Like growth factor II mRNA-binding protein 3 promotes cell proliferation, migration and invasion in human glioblastoma. OncoTargets Ther. 2019;12:3661–3670. doi: 10.2147/OTT.S200901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bhargava S., Visvanathan A., Patil V., Kumar A., Kesari S., Das S., Hegde A.S., Arivazhagan A., Santosh V., Somasundaram K. IGF2 mRNA binding protein 3 (IMP3) promotes glioma cell migration by enhancing the translation of RELA/p65. Oncotarget. 2017;8:40469–40485. doi: 10.18632/oncotarget.17118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Choe J., Lin S., Zhang W., Liu Q., Wang L., Ramirez-Moya J., Du P., Kim W., Tang S., Sliz P., et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature. 2018;561:556–560. doi: 10.1038/s41586-018-0538-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meyer K.D., Patil D.P., Zhou J., Zinoviev A., Skabkin M.A., Elemento O., Pestova T.V., Qian S.-B., Jaffrey S.R. 5′ UTR m6A promotes cap-independent translation. Cell. 2015;163:999–1010. doi: 10.1016/j.cell.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lin S., Choe J., Du P., Triboulet R., Gregory R. The m6A methyltransferase METTL3 promotes translation in human cancer cells. Mol. Cell. 2016;62:335–345. doi: 10.1016/j.molcel.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S. HNRNPA2B1 is a mediator of m6A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu B., Su S., Patil D.P., Liu H., Gan J., Jaffrey S.R., Ma J. Molecular basis for the specific and multivariant recognitions of RNA substrates by human hnRNP A2/B1. Nat. Commun. 2018;9:420. doi: 10.1038/s41467-017-02770-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Fang X., Yoon J.G., Li L., Tsai Y.S., Zheng S., Hood L., Goodlett D.R., Foltz G., Lin B. Landscape of the SOX2 protein-protein interactome. Proteomics. 2011;11:921–934. doi: 10.1002/pmic.201000419. [DOI] [PubMed] [Google Scholar]
- 108.Gruber A.J., Schmidt R., Ghosh S., Martin G., Gruber A.R., van Nimwegen E., Zavolan M. Discovery of physiological and cancer-related regulators of 3′ UTR processing with KAPAC. Genome Biol. 2018;19:44. doi: 10.1186/s13059-018-1415-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun H., Zhang M., Li K., Bai D., Yi C. Cap-specific, terminal N6-methylation by a mammalian m6Am methyltransferase. Cell Res. 2019;29:80–82. doi: 10.1038/s41422-018-0117-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen H., Gu L., Orellana E.A., Wang Y., Guo J., Liu Q., Wang L., Shen Z., Wu H., Gregory R.I., et al. METTL4 is an snRNA m6Am methyltransferase that regulates RNA splicing. Cell Res. 2020 doi: 10.1038/s41422-019-0270-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mauer J., Sindelar M., Despic V., Guez T., Hawley B.R., Vasseur J.J., Rentmeister A., Gross S.S., Pellizzoni L., Debart F., et al. FTO controls reversible m6Am RNA methylation during snRNA biogenesis. Nat. Chem. Biol. 2019;15:340–347. doi: 10.1038/s41589-019-0231-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.J., Chen Q., et al. Reversible methylation of m6Am in the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sendinc E., Valle-Garcia D., Dhall A., Chen H., Henriques T., Navarrete-Perea J., Sheng W., Gygi S.P., Adelman K., Shi Y. PCIF1 catalyzes m6Am mRNA methylation to regulate gene expression. Mol. Cell. 2019;75:620–630. doi: 10.1016/j.molcel.2019.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Akichika S., Hirano S., Shichino Y., Suzuki T., Nishimasu H., Ishitani R., Sugita A., Hirose Y., Iwasaki S., Nureki O., et al. Cap-Specific terminal N6-methylation of RNA by an RNA polymerase II–associated methyltransferase. Science. 2019;363 doi: 10.1126/science.aav0080. [DOI] [PubMed] [Google Scholar]
- 115.Boulias K., Toczydłowska-Socha D., Hawley B.R., Liberman N., Takashima K., Zaccara S., Guez T., Vasseur J.-J., Debart F., Aravind L., et al. Identification of the m6Am methyltransferase PCIF1 reveals the location and functions of m6Am in the transcriptome. Mol. Cell. 2019;75:631–643. doi: 10.1016/j.molcel.2019.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Relier S., Ripoll J., Guillorit H., Amalric A., Boissière F., Vialaret J., Attina A., Debart F., Choquet A., Macari F., et al. FTO-Mediated cytoplasmic m6Am demethylation adjusts stem-like properties in colorectal cancer cell. BioRxiv. 2020 doi: 10.1101/2020.01.09.899724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chiu Y.-C., Hsiao T.-H., Chen Y., Chuang E. Parameter optimization for constructing competing endogenous RNA regulatory network in glioblastoma multiforme and other cancers. BMC Genom. 2015;16(Suppl. 4):S1. doi: 10.1186/1471-2164-16-S4-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Squires J.E., Patel H.R., Nousch M., Sibbritt T., Humphreys D.T., Parker B.J., Suter C.M., Preiss T. Widespread occurrence of 5-methylcytosine in human coding and non-coding RNA. Nucleic Acids Res. 2012;40:5023–5033. doi: 10.1093/nar/gks144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Motorin Y., Lyko F., Helm M. 5-Methylcytosine in RNA: Detection, enzymatic formation and biological functions. Nucleic Acids Res. 2010;38:1415–1430. doi: 10.1093/nar/gkp1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hussain S., Benavente S.B., Nascimento E., Dragoni I., Kurowski A., Gillich A., Humphreys P., Frye M. The nucleolar RNA methyltransferase Misu (NSun2) is required for mitotic spindle stability. J. Cell Biol. 2009;186:27–40. doi: 10.1083/jcb.200810180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Frye M., Dragoni I., Chin S.-F., Spiteri I., Kurowski A., Provenzano E., Green A., Ellis I.O., Grimmer D., Teschendorff A. Genomic gain of 5p15 leads to over-expression of Misu (NSUN2) in breast cancer. Cancer Lett. 2010;289:71–80. doi: 10.1016/j.canlet.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 122.Okamoto M., Hirata S., Sato S., Koga S., Fujii M., Qi G., Ogawa I., Takata T., Shimamoto F., Tatsuka M., et al. Frequent increased gene copy number and high protein expression of tRNA (cytosine-5-)-methyltransferase (NSUN2) in human cancers. DNA Cell Biol. 2012;31:660–671. doi: 10.1089/dna.2011.1446. [DOI] [PubMed] [Google Scholar]
- 123.Yang J.-C., Risch E., Zhang M., Huang C., Huang H., Lu L. Association of tRNA methyltransferase NSUN2/IGF-II molecular signature with ovarian cancer survival. Future Oncol. 2017;13:1981–1990. doi: 10.2217/fon-2017-0084. [DOI] [PubMed] [Google Scholar]
- 124.Lu L., Zhu G., Zeng H., Xu Q., Holzmann K. High tRNA transferase NSUN2 gene expression is associated with poor prognosis in head and neck squamous carcinoma. Cancer Investig. 2018;36:246–253. doi: 10.1080/07357907.2018.1466896. [DOI] [PubMed] [Google Scholar]
- 125.Li Y., Li J., Luo M., Zhou C., Shi X., Yang W., Lu Z., Chen Z., Sun N., He J. Novel long noncoding RNA NMR promotes tumor progression via NSUN2 and BPTF in esophageal squamous cell carcinoma. Cancer Lett. 2018;430:57–66. doi: 10.1016/j.canlet.2018.05.013. [DOI] [PubMed] [Google Scholar]
- 126.Bolukbasi M.F., Mizrak A., Ozdener G.B., Madlener S., Ströbel T., Erkan E.P., Fan J.-B., Breakefield X.O., Saydam O. miR-1289 and “Zipcode”-like sequence enrich mRNAs in microvesicles. Mol. Ther. Nucleic Acids. 2012;1:e10. doi: 10.1038/mtna.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kossinova O.A., Gopanenko A.V., Tamkovich S.N., Krasheninina O.A., Tupikin A.E., Kiseleva E., Yanshina D.D., Malygin A.A., Ven’yaminova A.G., Kabilov M.R., et al. Cytosolic YB-1 and NSUN2 are the only proteins recognizing specific motifs present in mRNAs enriched in exosomes. Biochim. Biophys. Acta Proteins Proteom. 2017;1865:664–673. doi: 10.1016/j.bbapap.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 128.Fu L., Guerrero C.R., Zhong N., Amato N.J., Liu Y., Liu S., Cai Q., Ji D., Jin S.-G., Niedernhofer L.J., et al. Tet-Mediated formation of 5-hydroxymethylcytosine in RNA. J. Am. Chem. Soc. 2014;136:11582–11585. doi: 10.1021/ja505305z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Huber S.M., van Delft P., Mendil L., Bachman M., Smollett K., Werner F., Miska E.A., Balasubramanian S. Formation and abundance of 5-hydroxymethylcytosine in RNA. Chembiochem. 2015;16:752–755. doi: 10.1002/cbic.201500013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hu H., Shu M., He L., Yu X., Liu X., Lu Y., Chen Y., Miao X., Chen X. Epigenomic landscape of 5-hydroxymethylcytosine reveals its transcriptional regulation of lncRNAs in colorectal cancer. Br. J. Cancer. 2017;116:658–668. doi: 10.1038/bjc.2016.457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Johnson K.C., Houseman E.A., King J.E., Von Herrmann K.M., Fadul C.E., Christensen B.C. 5-Hydroxymethylcytosine localizes to enhancer elements and is associated with survival in glioblastoma patients. Nat. Commun. 2016;7:13177. doi: 10.1038/ncomms13177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Müller T., Gessi M., Waha A., Isselstein L.J., Luxen D., Freihoff D., Freihoff J., Becker A., Simon M., Hammes J., et al. Nuclear exclusion of TET1 is associated with loss of 5-hydroxymethylcytosine in IDH1 wild-type gliomas. Am. J. Pathol. 2012;181:675–683. doi: 10.1016/j.ajpath.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 133.Raiber E.-A., Beraldi D., Cuesta S.M., McInroy G.R., Kingsbury Z., Becq J., James T., Lopes M., Allinson K., Field S., et al. Base resolution maps reveal the importance of 5-hydroxymethylcytosine in a human glioblastoma. NPJ Genom. Med. 2017;2:6. doi: 10.1038/s41525-017-0007-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhang C., Jia G. Reversible RNA Modification N1-methyladenosine (m1A) in mRNA and tRNA. Genom. Proteom. Bioinform. 2018;16:155–161. doi: 10.1016/j.gpb.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ozanick S., Krecic A., Andersland J., Anderson J. The bipartite structure of the tRNA m1A58 methyltransferase from S. cerevisiae is conserved in humans. RNA. 2005;11:1281–1290. doi: 10.1261/rna.5040605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Liu F., Clark W., Luo G., Wang X., Fu Y., Wei J., Wang X., Hao Z., Dai Q., Zheng G. ALKBH1-Mediated tRNA demethylation regulates translation. Cell. 2016;167:816–828. doi: 10.1016/j.cell.2016.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Vilardo E., Nachbagauer C., Buzet A., Taschner A., Holzmann J., Rossmanith W. A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase—Extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 2012;40:11583–11593. doi: 10.1093/nar/gks910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mongiardi M.P., Savino M., Falchetti M.L., Illi B., Bozzo F., Valle C., Helmer-Citterich M., Ferrè F., Nasi S., Levi A. c-MYC inhibition impairs hypoxia response in glioblastoma multiforme. Oncotarget. 2016;7:33257–33271. doi: 10.18632/oncotarget.8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Slotkin W., Nishikura K. Adenosine-to-inosine RNA editing and human disease. Genome Med. 2013;5:105. doi: 10.1186/gm508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Choudhury Y., Tay F.C., Lam D.H., Sandanaraj E., Tang C., Ang B.-T., Wang S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J. Clin. Investig. 2012;122:4059–4076. doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ge J., Yu Y.-T. RNA pseudouridylation: New insights into an old modification. Trends Biochem. Sci. 2013;38:210–218. doi: 10.1016/j.tibs.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Cohn W. 5-Ribosyl uracil, a carbon-carbon ribofuranosyl nucleoside in ribonucleic acids. Biochim. Biophys. Acta. 1959;32:569–571. doi: 10.1016/0006-3002(59)90644-4. [DOI] [PubMed] [Google Scholar]
- 143.Li X., Ma S., Yi C. Pseudouridine: The fifth RNA nucleotide with renewed interests. Curr. Opin. Chem. Biol. 2016;33:108–116. doi: 10.1016/j.cbpa.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 144.Hamma T., Ferré-D’Amaré A. Pseudouridine synthases. Chem. Biol. 2006;13:1125–1135. doi: 10.1016/j.chembiol.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 145.Rintala-Dempsey A.C., Kothe U. Eukaryotic stand-alone pseudouridine synthases-RNA modifying enzymes and emerging regulators of gene expression? RNA Biol. 2017;14:1185–1196. doi: 10.1080/15476286.2016.1276150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Oakes E., Anderson A., Cohen-Gadol A., Hundley H. Adenosine deaminase that acts on RNA 3 (ADAR3) binding to glutamate receptor subunit B pre-mRNA inhibits RNA editing in glioblastoma. J. Biol. Chem. 2017;292:4326–4335. doi: 10.1074/jbc.M117.779868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Masamha C.P., Xia Z., Yang J., Albrecht T.R., Li M., Shyu A.-B., Li W., Wagner E.J. CFIm25 links alternative polyadenylation to glioblastoma tumour suppression. Nature. 2014;510:412–416. doi: 10.1038/nature13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Hao X., Luo H., Krawczyk M., Wei W., Wang W., Wang J., Flagg K., Hou J., Zhang H., Yi S., et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc. Nat. Acad. Sci. USA. 2017;114:7414–7419. doi: 10.1073/pnas.1703577114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fardi M., Solali S., Farshdousti Hagh M. Epigenetic mechanisms as a new approach in cancer treatment: An updated review. Genes Dis. 2018;5:304–311. doi: 10.1016/j.gendis.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hatzimichael E., Lagos K., Sim V.R., Briasoulis E., Crook T. Epigenetics in diagnosis, prognostic assessment and treatment of cancer: An update. EXCLI J. 2014;13:954–976. [PMC free article] [PubMed] [Google Scholar]
- 151.Kinehara M., Yamamoto Y., Shiroma Y., Ikuo M., Shimamoto A., Tahara H. DNA and histone modifications in cancer diagnosis. In: Kaneda A., Tsukada Y.-I., editors. DNA and Histone Methylation as Cancer Targets. Springer International Publishing; Cham, Switzerland: 2017. pp. 533–584. [Google Scholar]
- 152.Takeshima H., Yamada H., Ushijima T. Chapter 5—Cancer epigenetics: Aberrant DNA methylation in cancer diagnosis and treatment. In: Dammacco F., Silvestris F., editors. Oncogenomics. Academic Press; Cambridge, MA, USA: 2019. pp. 65–76. [Google Scholar]
- 153.Delpu Y., Cordelier P., Cho W.C., Torrisani J. DNA methylation and cancer diagnosis. Int. J. Mol. Sci. 2013;14:15029–15058. doi: 10.3390/ijms140715029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kurdistani S.K. Histone modifications as markers of cancer prognosis: A cellular view. Br. J. Cancer. 2007;97:1–5. doi: 10.1038/sj.bjc.6603844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sigismund S., Avanzato D., Lanzetti L. Emerging functions of the EGFR in cancer. Mol. Oncol. 2018;12:3–20. doi: 10.1002/1878-0261.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kim Y.Z. Altered histone modifications in gliomas. Brain Tumor Res. Treat. 2014;2:7–21. doi: 10.14791/btrt.2014.2.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Petushkova N.A., Zgoda V.G., Pyatnitskiy M.A., Larina O.V., Teryaeva N.B., Potapov A.A., Lisitsa A.V. Post-translational modifications of FDA-approved plasma biomarkers in glioblastoma samples. PLoS ONE. 2017;12:e0177427. doi: 10.1371/journal.pone.0177427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Carén H., Pollard S.M., Beck S. The good, the bad and the ugly: Epigenetic mechanisms in glioblastoma. Mol. Asp. Med. 2013;34:849–862. doi: 10.1016/j.mam.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Klughammer J., Kiesel B., Roetzer T., Fortelny N., Nemc A., Nenning K.-H., Furtner J., Sheffield N.C., Datlinger P., Peter N., et al. The DNA methylation landscape of glioblastoma disease progression shows extensive heterogeneity in time and space. Nat. Med. 2018;24:1611–1624. doi: 10.1038/s41591-018-0156-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hyman G., Manglik V., Rousch J.M., Verma M., Kinkebiel D., Banerjee H.N. Epigenetic approaches in glioblastoma multiforme and their implication in screening and diagnosis. Methods Mol. Biol. 2015;1238:511–521. doi: 10.1007/978-1-4939-1804-1_26. [DOI] [PubMed] [Google Scholar]
- 161.Etcheverry A., Aubry M., de Tayrac M., Vauleon E., Boniface R., Guenot F., Saikali S., Hamlat A., Riffaud L., Menei P., et al. DNA methylation in glioblastoma: Impact on gene expression and clinical outcome. BMC Genom. 2010;11:701. doi: 10.1186/1471-2164-11-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Jia D., Lin W., Tang H., Cheng Y., Xu K., He Y., Geng W., Dai Q. Integrative analysis of DNA methylation and gene expression to identify key epigenetic genes in glioblastoma. Aging. 2019;11:5579–5592. doi: 10.18632/aging.102139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hegi M.E., Diserens A.-C., Gorlia T., Hamou M.-F., de Tribolet N., Weller M., Kros J.M., Hainfellner J.A., Mason W., Mariani L., et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 164.Brandes A.A., Franceschi E., Paccapelo A., Tallini G., De Biase D., Ghimenton C., Danieli D., Zunarelli E., Lanza G., Silini E.M., et al. Role of MGMT Methylation status at time of diagnosis and recurrence for patients with glioblastoma: Clinical implications. Oncologist. 2017;22:432–437. doi: 10.1634/theoncologist.2016-0254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Ge L., Zhang N., Chen Z., Song J., Wu Y., Li Z., Chen F., Wu J., Li D., Li J., et al. Level of N6-methyladenosine in peripheral blood RNA: A novel predictive biomarker for gastric cancer. Clin. Chem. 2020;66:342–351. doi: 10.1093/clinchem/hvz004. [DOI] [PubMed] [Google Scholar]
- 166.Konno M., Koseki J., Asai A., Yamagata A., Shimamura T., Motooka D., Okuzaki D., Kawamoto K., Mizushima T., Eguchi H., et al. Distinct methylation levels of mature microRNAs in gastrointestinal cancers. Nat. Commun. 2019;10:3888. doi: 10.1038/s41467-019-11826-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Strick A., von Hagen F., Gundert L., Klümper N., Tolkach Y., Schmidt D., Kristiansen G., Toma M., Ritter M., Ellinger J., et al. The N6-methyladenosine (m6A) erasers alkylation repair homologue 5 (ALKBH5) and fat mass and obesity-associated protein (FTO) are prognostic biomarkers in patients with clear cell renal carcinoma. BJU Int. 2020 doi: 10.1111/bju.15019. [DOI] [PubMed] [Google Scholar]
- 168.Senft D., Qi J., Ronai Z.E.A. Ubiquitin ligases in oncogenic transformation and cancer therapy. Nat. Rev. Cancer. 2018;18:69–88. doi: 10.1038/nrc.2017.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zang L., Kondengaden S.M., Che F., Wang L., Heng X. Potential Epigenetic-based therapeutic targets for glioma. Front. Mol. Neurosci. 2018;11:408. doi: 10.3389/fnmol.2018.00408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cully M. Chemical inhibitors make their RNA epigenetic mark. Nature reviews. Drug Discov. 2019;18:892–894. doi: 10.1038/d41573-019-00179-5. [DOI] [PubMed] [Google Scholar]
- 171.Selberg S., Blokhina D., Aatonen M., Koivisto P., Siltanen A., Mervaala E., Kankuri E., Karelson M. Discovery of small molecules that activate RNA methylation through cooperative binding to the METTL3-14-WTAP complex active site. Cell Rep. 2019;26:3762–3771.e3765. doi: 10.1016/j.celrep.2019.02.100. [DOI] [PubMed] [Google Scholar]
- 172.Tzelepis K., De Braekeleer E., Yankova E., Rak J., Aspris D., Domingues A.F., Fosbeary R., Hendrick A., Leggate D., Ofir-Rosenfeld Y., et al. Pharmacological inhibition of the RNA m6A writer METTL3 as a novel therapeutic strategy for acute myeloid leukemia. Blood. 2019;134:403. doi: 10.1182/blood-2019-127962. [DOI] [Google Scholar]
- 173.Albertella M., Blackaby W., Fosbeary R., Hendrick A., Leggate D., Ofir-Rosenfeld Y., Sapetschnig A., Tzelepis K., Yankova E., Kouzarides T., et al. Abstract B126: A small molecule inhibitor of the RNA m6A writer METTL3 inhibits the development of acute myeloid leukemia (AML) In Vivo. J. Mol. Cancer Ther. 2019;18:B126. [Google Scholar]
- 174.Dominissini D., Moshitch-Moshkovitz S., Salmon-Divon M., Amariglio N., Rechavi G. Transcriptome-Wide mapping of N6-methyladenosine by m6A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc. 2013;8:176–189. doi: 10.1038/nprot.2012.148. [DOI] [PubMed] [Google Scholar]
- 175.Linder B., Grozhik A.V., Olarerin-George A.O., Meydan C., Mason C.E., Jaffrey S.R. Single-Nucleotide-Resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods. 2015;12:767–772. doi: 10.1038/nmeth.3453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Hartstock K., Rentmeister A. Mapping N6-methyladenosine (m6A) in RNA: Established methods, remaining challenges, and emerging approaches. Chem. Eur. J. 2019;25:3455–3464. doi: 10.1002/chem.201804043. [DOI] [PubMed] [Google Scholar]